Abstract
A number of treatments have been developed for HER1, 2 and 3-driven non-small cell lung cancer (NSCLC), of which the most successful have been the epidermal growth factor receptor-tyrosine kinase inhibitors in HER1-mutant tumours resulting in highly improved progression-free survival. Human epidermal growth factor (HER)2 and 3-driven tumours represent the minority of NSCLC, and effective therapies in these patients still represent an unmet medical need. The encouraging results seen with anti-HER2 and anti-HER3 monoclonal antibodies need to be validated in larger studies, even if the greatest obstacle is represented by the exiguous number of patients bearing deregulated HER2/3 system and abnormalities of signal transduction pathway. Considering NSCLC tumour heterogeneity, which affects response and resistance to treatment, combined multiparametric approaches, such as liquid biopsy together with radiomics, may provide a better understanding of the tumour dynamics and clonal selection during the treatments.
Keywords: NSCLC, erbB, TKIs, liquid biopsy, radiomics
The biological role of the erbB family of proteins in non-small cell lung cancer
ErbB1/HER1-driven non-small cell lung cancer
The epidermal growth factor receptor 1 (HER1, EGFR) belongs to the HER/ErbB family of membrane-bound proteins (ErbB1-4) and plays a pivotal role in cell proliferation and survival. These receptors are characterised by four different domains with diverse functions: the extracellular domain is involved in ligand biding, the transmembrane domain serves for receptor anchorage, the cytoplasmic domain contains the tyrosine kinase activity and the carboxy terminal and is involved in signal transduction.1 HER1 is associated with a complex signalling network, activated both in a ligand-dependent or independent manner.2 Ligand binding induces receptor homodimerisation (HER1/HER1) or heterodimerisation (HER1/HER2) that, in turn, promotes autophosphorylation at the tyrosine kinase domain. HER1 recognises different ligands, including EGF and transforming growth factor-α. EGF family ligands are present as preproteins in a membrane-docked form and membrane-anchorage metalloproteases were found to be involved in their cleavage. The release of ligands and the consequent EGFR dimerisation can trigger multiple signalling transduction pathways, including Rat Sarcoma (RAS), Rapidly Accelerated Fibrosarcoma (RAF), Mitogen Activated Protein Kinases (MAPKs), phosphatidylinositol-3 kinase (PI3K/Akt), signal transducer and activator of transcription (STAT) and mammalian target of rapamycin (mTOR).3
Ligand-independent (constitutive) activation of ErbB1/HER1 due to receptor aberration was found in multiple cancer types, particularly in lung cancer.2 For example, EGFRvIII is generated by an in-frame deletion of the extracellular domain and this mutant receptor neither requires a ligand to be activated nor forms dimers. Furthermore, EGFRvIII was only found in cancer cells and its constitutive state was associated with an invasive phenotype.4 Somatic HER1 mutations at the tyrosine-kinase domain make non-small cell lung cancer (NSCLC) sensitive to selective EGFR tyrosine kinase inhibitors (TKIs).5 Class I mutations are in-frame deletions encoded by exon 19; class II are single-nucleotide polymorphisms in exons 18–21, and class III mutations are in-frame duplications and/or insertions in exon 20.
EGFR activation is also involved in the immune system regulation. Stimulation of EGFR or its constitutive activation can trigger the MAPK cascade and upregulate Programmed Death-Ligand 1 (PD-L1), a checkpoint protein that plays a key role as a negative regulator of the antitumor immune response.6 Furthermore, constitutive EGFR activation also plays a role in tumour angiogenesis through the upregulation of hypoxia inducible factor-1α (HIF-1α) and vascular endothelial growth factor (VEGF).7
ErbB2/HER2-driven NSCLC
HER2 is a member of the HER/erbB family and it is well characterised in breast cancer, in which HER2 overexpression is associated with sensitivity to anti-HER2 drugs.8 HER2 does not have a specific ligand and can transmit signals downstream through the formation of homodimers and heterodimers with HER1 and HER3. Furthermore, HER2 is less incline to internalisation and degradation than other EGFR members and can remain activated for a long time on the cell membrane.9 It is worth noting that increased signalling of HER2/HER1 heterodimer compared with HER1/HER1 homodimer may account for the high sensitivity of EGFR-activating NSCLC with HER2 overexpression to selective EGFR inhibitors.10 Somatic ErbB2 mutations were found in 1%–4% of patients with lung adenocarcinoma, particularly in exon 18–21, and were associated with worse prognosis.11 12 HER2 YVMA consists in a 12 bp duplication/insertion of the amino acid sequence YVMA in exon 20 at codon 776 that change receptor conformation leading to an increased tyrosine kinase activity, compared with the wild-type.10 13 NSCLCs harbouring this type of mutation were found to be less sensitive to osimertinib than those having HER1 amplification.14
ErbB3/HER3-driven NSCLC
HER3 is characterised by a reduced kinase activity, compared with other EGFR members. HER3 binding to heregulin led to the formation of HER2 heterodimers able to activate signalling pathways (ie, PI3K/AKT) involved in tumour progression15 and resistance to fist-generation EGFR TKIs in NSCLC.16 Somatic HER3 mutations were found in multiple cancer types, including NSCLC; among them, HER3 V855A is a novel mutation homologous to EGFR L858R found in a 14-year-old patient with NSCLC.17 HER3 V855A can enhance heregulin-induced transactivation of HER2, compared with HER3 wild type, thus increasing the transforming potential of cancer cells. Co-transfection experiments demonstrated that HER3 V855A were sensitive to afatinib and pertuzumab,18 suggesting a possible targeting strategy in patients harbouring this type of mutation. Notably, hepatocyte growth factor receptor (HGFR or MET) amplification was found to induce resistance to TKIs in NSCLC by driving HER3-dependent activation of PI3K pathway.19
erbB receptor activation as a target
Anti-HER1 TKIs
EGFR is mutated in 10%–20% of lung adenocarcinomas in Caucasian patients, being more common in never smokers, younger, female and Asian patients; whereas EGFR mutations are uncommon in other lung cancer histotypes.20 21 The most common EGFR mutations are inframe deletions of exon 19 (ex19del; 45%) and the missense mutation L858R of exon 21 (40%–45%).5 These mutations are predictive factors of response to treatment with EGFR TKIs. Indeed, gefitinib and erlotinib demonstrated only a mild activity in unselected NSCLC populations.22 23 On the contrary, gefitinib and erlotinib were effective treatments in EGFR mutant NSCLC.24–27 Different randomised clinical trials evaluated the role of gefitinib and erlotinib comparing with standard chemotherapy for the first line of metastatic NSCLC, harbouring common EGFR mutations. Gefitinib and erlotinib demonstrated a better progression-free survival (PFS) compared with chemotherapy,24 27 however, none of these trials showed an improvement in overall survival (OS), due to the high rate of crossover between treatments. Since TKIs were less toxic than chemotherapy, erlotinib and gefitinib become the standard of care for the first line treatment of advanced NSCLC with EGFR mutations.28–30
Second-generation TKIs include afatinib and dacomitinib, and are irreversible EGFR inhibitors. Afatinib prolonged PFS compared with chemotherapy in LuxLung3 and LuxLung6, and in the LuxLung7 demonstrated no significant differences compared with gefitinib/erlotinib.31–34 However, any difference in OS was not observed in both trials. The combined data of these trials suggested a better OS in patients with ex19del mutation treated with afatinib compared with chemotherapy; whereas, differences in OS were not significant in patients with L858R mutation. Dacomitinib was compared with gefitinib obtaining a longer PFS and OS. Second generation TKIs, afatinib and dacomitinib, were more toxic with higher grade of diarrhoea, paronychia, acneiform dermatitis and stomatitis.35
Osimertinib is a third-generation TKI, specifically designed to interact with mutated EGFR and to spare wild type receptors, and is also effective in patients harbouring the T790M EGFR mutations. In patients who progress to first line TKIs that developed T790M mutation as mechanism of acquired resistance, osimertinib obtained a better overall response rate (ORR) compared with chemotherapy (71% vs 21%) and prolonged PFS (median 10.1 vs 4.4 months).36 Osimertinib was compared with first generation TKIs,37 obtaining improvement in ORR (80% vs 76%), a better PFS (18.9 vs 10.2 months) and a better OS (38.6 vs 34.5 months),37 becoming the new standard of treatment for these patients.
To improve efficacy, EGFR TKIs have been combined with anti-angiogenic agents and chemotherapy. The combination of bevacizumab plus erlotinib improved PFS compared with erlotinib in a preplanned interim analysis.38 Similarly, the combination of ramucirumab with erlotinib improved PFS compared with erlotinib.39 In both reports, data were still not mature for OS analysis.
Several attempts were aimed at overcame the emergence of resistance by combining chemotherapy with an EGFR TKI. Based on that strategy, in the NEJ009 phase III trial, the combination of carboplatin and pemetrexed with gefitinib prolonged PFS and OS compared with gefitinib monotherapy, in EGFR mutant NSCLC patients.40 Noronha et al performed a phase III randomised trial adding pemetrexed and carboplatin chemotherapy to geftinib as first-line treatment, demonstrating that the association significantly prolonged PFS (16 vs 8 months, combination vs gefitinib alone, respectively; p<0.001) and OS (not reached vs 17 months, combination vs gefitinib alone, respectively; p<0.001), despite an increased toxicity (51% vs 25%, combination vs gefitinib alone, respectively; p<0.001).41Anti PD-1 and PD-L1 antibodies (Abs) have a mild activity in EGFR mutated tumours with 4% ORR compared with 23% in the wild type population.42 However, there is a biological rationale for the combination of TKIs with immunotherapy and, in the IMPower150 trial, the combination of atezolizumab with carboplatin, paclitaxel and bevacizumab seemed to improve PFS and OS in the small subgroup of patients with EGFR mutations. The combination of osimertinib or gefitinib with durvalumab was too toxic for clinical development because of a high rate of pneumonitis, whereas preliminary data suggest that the combination of erlotinib with atezolizumab or pembrolizumab could be feasible.43 A single-centre experience on the KEYNOTE-001 trial suggested that TKI naïve patients had superior outcome when treated with pembrolizumab. Therefore, a phase II trial evaluated pembrolizumab efficacy in TKI naive patients with EGFR mutant, PD-L1-positive (≥1%), advanced NSCLC. The study was interrupted due to lack of efficacy after 11 of 25 planned patients were treated (ORR 0%; 46% of treatment adverse events).44
Beside ex19del and L858R mutations, less common EGFR mutations can be diagnosed. Only some of them are predictive factors of response to EGFR-TKIs. Objective responses have been obtained using afatinib in tumours with G719X, L861Q and S768I EGFR mutations.45 Considering the remaining EGFR mutations two possibilities can be presumed: (1) the mutation does not transform the proto-oncogene EGFR in an oncogene; (2) TKIs are ineffective to inhibit tyrosine phosphorylation in that specific mutation. This second option seems to be the case of exon 20 insertion. Preliminary data suggest that specific inhibitors, such as poziotinib and TAK788, can be effective in this subgroup of patients. Although poziotinib obtained 58% ORR in the first treated patients, it failed to meet its primary endpoint in the phase II ZENITH20 trial with only 15% ORR. TAK788 obtained an interesting 50% ORR in the first 14 treated patients.46 A selection of the most important trials in EGFR mutant tumors is reported in table 1.
Table 1.
Summary of EGFR TKI trials for the first line treatment of NSCLC patients harbouring EGFR mutations
| Trial | Arms | Patients | ORR | mPFS (months) |
HR (95% CI) | mOS (months) |
HR (95% CI) | Reference |
| EURTAC | Erlotinib | 86 | 58% | 9.7 | 0.37 (0.25 to 0.54) | 19.3 | 1.04 (0.65 to 1.68) | 28 |
| Chemotherapy | 87 | 15% | 5.2 | 19.5 | ||||
| NEJ002 | Gefitinib | 115 | 74% | 10.8 | 0.31 (0.24 to 0.44) | 27.7 | 0.89 (0.63 to 1.24) | 27 29 |
| CBDCA+TXL | 115 | 31% | 5.4 | 26.6 | ||||
| IPASS† | Gefitinib | 132 | 71% | 9.5 | 0.48 (0.26 to 0.64) | 21.6 | 1 (0.76 to 1.33) | 30 |
| CBDCA+TXL | 129 | 47% | 6.3 | 21.9 | ||||
| LuxLung3 | Afatinib | 230 | 56% | 11.1 | 0.58 (0.43 to 0.78) | 28.2 | 0.88 (0.66 to 1.17) | 32 33 |
| CDDP+Pem | 115 | 23% | 6.9 | 28.2 | ||||
| LuxLung6 | Afatinib | 242 | 70% | 11 | 0.28 (0.2 to 0.34) | 23.1 | 0.93 (0.72 to 1.22) | 33 34 |
| CDDP+Gem | 122 | 23% | 5.6 | 23.5 | ||||
| LuxLung7 | Afatinib | 160 | 70% | 11.1 | 0.73 (0.57 to 0.95) | 27.9 | 0.88 (0.66 to 1.12) | 31 |
| Gefitinib | 159 | 56% | 10.9 | 24.5 | ||||
| Archer1050 | Dacomitinib | 227 | 75% | 14.7 | 0.59 (0.47 to 0.74) | 34.1 | 0.76 (0.58 to 0.99) | 35 |
| Gefitinib | 225 | 70% | 9.2 | 26.8 | ||||
| FLAURA | Osimertinib | 279 | 80% | 18.9 | 0.46 (0.37 to 0.57) | 38.6 | 0.8 (0.64 to 1.00) | 37 |
| Gefitinib | 277 | 76% | 10.2 | 34.5 | ||||
| NEJ026 | Erlotinib+Bev | 114 | 72% | 16.9 | 0.61 (0.42 to 0.88) | Not mature |
38 | |
| Erlotinib | 114 | 66% | 13.3 | |||||
| RELAY | Erlotinib+Ram | 224 | 76% | 19.4 | 0.59 (0.46 to 0.76) | Not mature |
39 | |
| Erlotinib | 225 | 75% | 12.4 | |||||
| NEJ009 | Gef +CBDCA+Pem | 172 | 84% | 20.9 | 0.49 (0.39 to 0.62) | 50.9 | 0.72 (0.55 to 0.95) | 40 |
| Gefitinib | 173 | 67% | 11.9 | 38.8 |
EGFR, epidermal growth factor receptor; mPFS, median progression free survival; NSCLC, non-small cell lung cancer; ORR, overall response rate; OS, median overall survival; TKIs, tyrosine kinase inhibitors.
Anti-HER2 TKI and moAbs
Trastuzumab emtansine (T-DM1), an Ab drug conjugate of trastuzumab and a mytansinoid potent inhibitor of cellular microtubules, has been evaluated in NSCLC patients.
A study evaluated 18 patients with advanced, previously treated HER2-mutant NSCLC. The response rate was remarkably high, 44%, and met the primary endpoint of the study. Subjects with mutations of the HER2 exon 20, consisting of insertions and point mutations in the different domains of HER2, including the kinase, transmembrane and extracellular, responded to the treatment. The selection of patients based on the immunohistochemical evaluation of HER2 expression did not predict response, since responders also had low HER2 score. Median PFS was 5 months and adverse events consisted of infusion reactions (grade 1 or 2), thrombocytopenia, and elevated AST/ALT. No patient discontinued treatment due to toxicity and no toxic deaths were recorded.47 This study demonstrated that T-DM1 is an active treatment in patients with NSCLC harbouring mutations of HER2.47
The EUHER2 retrospective study was conducted in patients with advanced NSCLC harbouring HER2 exon 20 insertion and administered chemotherapy with or without anti-HER2 drugs.48 The largest group of patients received trastuzumab or T-DM1 (n=57/1), while the remaining were treated with the EGFR TKIs neratinib (n=14), afatinib (n=11), or lapatinib (n=5). The ORR was 50.9% and PFS was 4.8 months with trastuzumab or T-DM1 while the group receiving neratinib, lapatinib, and afatinib showed an ORR of 7.4% and PFS of 3.4 months.48 Overall, the study demonstrated good clinical activity of trastuzumab/T-DM1, while TKIs showed only modest effect.
A phase II study evaluated the activity of single agent T-DM1 in patients with advanced, multitreated NSCLC with HER2 overexpression or mutation.49 50 The HER2 status assessed by immunohistochemistry was 3+in 33% subjects; 2+ combined with positive fluorescence in situ hybridisation (20%), while 47% of patients had exon 20 mutation. Only one subject achieved partial response (ORR 6.7%) and PFS was 2 months; thrombocytopenia (40%) and liver toxicity (20%) were the most serious adverse events.50 In conclusion, T-DM1 had a limited efficacy for HER2-positive NSCLC. A clinical trial on HER2-overexpressing NSCLC patients evaluated the clinical effect of T-DM1 in subjects whose tumours scored 2+/3+ at immunohistochemistry.51 No responses were detected in the HER2 2+ cohort, while four patients with HER2 3+ NSCLC showed partial response (ORR, 20%). Response duration ranged from 2.9 to 10.8 months and no differences were observed in PFS between 2+ and 3+ NSCLC.51 The study demonstrated limited activity of T-DM1 in HER2 overexpressing tumours and concluded that HER2 overexpression as a single marker had unsatisfactory predictivity.
The DESTINY-Lung01 study enrolled two cohorts of patients (HER2-overexpressing (IHC3 +or IHC2+) and HER activating mutations) to be treated with trastuzumab–deruxtecan. Trastuzumab deruxtecan is an Ab drug conjugate composed of a humanised anti-HER2 IgG1 moAb with the same aminoacid sequence of trastuzumab, a topoisomerase I inhibitor payload, and a tetrapeptide-based cleavable linker. Trastuzumab deruxtecan has a higher drug-to-Ab ratio, retaining a favourable pharmacokinetic profile. The tetrapeptide-based linker is stable in plasma and selectively cleaved by upregulated cathepsines in tumour cells; the payload easily crosses the cell membrane and has a short half-life, allowing higher cytotoxic effect with minimally systemic exposure. The interim analysis on the HER2 mutant cohort showed an ORR of 61.9%, disease control rate (DCR) 90.5% and an estimated mPFS of 14 months. The safety profile was generally consistent with previously reported studies. Based on the clinical activity demonstrated in the interim analysis and the safety profile, trastuzumab–deruxtecan represent a new treatment option for patients with HER2-mutated NSCLC.52
The administration of pertuzumab to 43 patients with advanced, pretreated NSCLC unselected for HER2 expression was associated with lack of responses and only 20.9% patients had stable disease at 12 weeks.53 The PFS was 6.1 weeks and four patients (9.3%) reported grade 3/4 adverse events but no cardiac toxicity.53 Overall, the administration of pertuzumab yielded modest clinical effect in an unselected population.
Preclinical evidence on neratinib shoedw that the drug has remarkable antitumour activity in mouse xenografts using NSCLC cells overexpressing wild-type HER2 or bearing HER2 mutations.54 The results of the present study strongly suggest that neratinib has potential as promising therapeutic option for the treatment of HER2-altered NSCLC. However, no robust clinical data on HER2-altered NSCLC patients are available.
Anti-HER3 agents in NSCLC
To exploit the concept that HER3 dimerises with other HER family members and is involved in the development of resistance to HER-targeting drugs, the human anti-HER3 monoclonal Ab patritumab was evaluated in combination with erlotinib in 24 patients with previously treated advanced NSCLC.55 PFS was 44 days in nine NSCLC patients with wild-type EGFR and 107.0 days in 13 subjects bearing EGFR-activating mutation. No relationship was found between efficacy and HER3 expression in NSCLC tissues. The most frequent toxicities were gastrointestinal or dermatological, all characterised by a manageable profile.55 The preliminary results of the U3-1402, a HER3-Ab conjugated with the topoisomerase I inhibitor DX-8951 in patients with advanced or metastatic EGFR TKI-resistant, EGFR-mutant NSCLC demonstrated six partial responses in 26 patients evaluable for efficacy; HER3 overexpression was demonstrated in all tumour tissues.56 The dual HER1/HER3 inhibitor duligotuzumab was evaluated in a phase I trial and demonstrated encouraging activity in patients affected by squamous cell carcinomas of head and neck displaying high tumour expression heregulin, the HER3 ligand; three patients with NSCLC had stable disease lasting ≥8 weeks.57 Finally, the humanised anti-HER3 monoclonal Ab lumretuzumab was evaluated in combination with carboplatin and paclitaxel in first-line treatment of 12 patients with squamous NSCLC.58 Mild and manageable adverse events were gastrointestinal, haematological and neurological (central); partial responses were observed in three subjects with high heregulin mRNA tumour levels and lasted from 81 to 207 days.58 Overall, lumretuzumab in combination with carboplatin and paclitaxel showed promising activity in tumours characterised by high HER3 mRNA levels.
Mechanisms of resistance to anti-HER1 agents
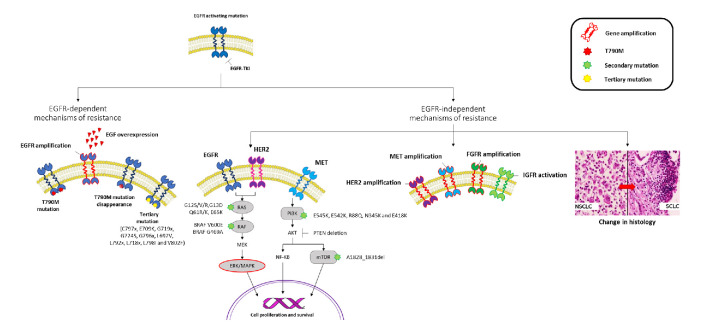
The mechanism underlying the resistance to an EGFR-TKI reflects the TKI potency against the target and its pharmacological characteristics. First-generation TKIs are characterised by reversible binding to both the wild type and the mutant EGFR, while second-generation TKIs are irreversible EGFR inhibitors covalently binding to HER1, HER2 and HER4. Third-generation TKIs are irreversible mutant EGFR inhibitors.59 Since TKIs activity against the EGFR is not equal, it is reasonable that also the relative mechanisms of resistance are not equal as well. In detail, the highest is the potency of the TKI, the highest is the possibility that the acquired resistance may occur through an EGFR independent mechanism. Therefore, mechanisms of resistance to EGFR-TKIs may be divided as EGFR dependent and independent. The EGFR dependent mechanisms include the appearance of EGFR secondary/third mutations and EGFR overexpression; while the EGFR independent mechanisms include HER-2 and MET amplification, appearance of mutations in alternative pathways, and the small cell histologic transformation (figure 1).60
Figure 1.
EGFR dependent and independent mechanisms of resistance to EGFR TKIs. EGFR, epidermal growth factor receptor; HER2, human epidermal growth factor 2; IGFR, insulin-like growth factor receptor; mTOR, mammalian target of rapamycin; NF-KB, nuclear factor kappa-light-chain-enhancer of activated B cells; NSCLC, non-small cell lung cancer; PI3T, phosphatidylinositol-3 kinase; TKIs, tyrosine kinase inhibitors.
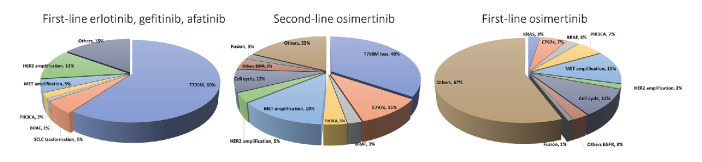
In patients progressed under first/second-generation TKIs, the molecular aberrations include the T790M mutation (~50%),61 MET (5%–15%)62 and HER2 (12%) amplification,63 PIK3CA mutations (5%),64 BRAF (1%)65 and transformation into small-cell histology (3%–14%).64 Moreover, the epithelial to mesenchymal transition has been related to acquired resistance (figure 2).64
Figure 2.
Incidence of mechanisms of resistance to EGFR TKIs based on first, second and third-generation TKIs. EGFR, epidermal growth factor receptor; HER2, human epidermal growth factor 2; TKIs, tyrosine kinase inhibitors.
The T790M mutation plays a major role in resistance to first/second-generation EGFR TKIs (~50%).66 In the case of gefitinib and erlotinib, EGFR reversible binders, it is expected that the main mechanism of resistance is EGFR dependent. The T790M is a gatekeeper mutation affecting the ATP binding pocket of the EGFR kinase domain, able to confer resistance by increasing the affinity for the ATP, so that the inhibitors are outcompeted. In addition, being the methionine larger than threonine, it directly blocks the inhibitor binding to the active site.67
A preclinical study demonstrated that afatinib is the most potent inhibitor of EGFR ex19del or L858R mutant, followed by gefitinib, erlotinib and osimertinib.68 Being afatinib an irreversible EGFR binder, the appearence of the T790M as a mechanism of resistance may be reduced in patients treated with afatinib.69 However, while the mechanisms of resistance to gefitinib/erlotinib are well known and described, few data are available for afatinib.
MET gene amplification is also reported as acquired mechanism of resistance, causing ERBB3 phosphorylation, which activates the PI3K/Akt signal downstream. Therefore, even with the TKI inhibiting ERBB3 phosphorylation by EGFR, the proliferation signal is not inhibited because of the maintenance of the ERBB3 phosphorylation by MET.19 MET amplification has been reported in 5% of patients treated with gefitinib, erlotinib, afatinib as first line. Other mechanisms of resistance, that is, PI3K, BRAF mutations are reported as lower than 3% (figure 2).
The landscape of mechanisms of resistance dramatically changes considering osimertinib. Multiple coexisting EGFR-dependent or independent mechanisms frequently occur when osimertinib is administered as first/second line. The appearance of the EGFR C797S mutation accounts for 6%–10% after osimertinib as first line and 10%–26% as second line.36 C797S acquisition has potential implications for treatment: when C797S and T790M occur on the same allele (cis), no response to EGFR TKIs alone or in combination can be expected, while the C797S in trans with the T790M mutation confers sensitivity to a combination of first/third-generation drugs.70–72 A number of rare point mutations have been identified in circulating tumour DNA (ctDNA). A study on 93 osimertinib-resistant NSCLC patients showed the coexistence of C797S with novel tertiary EGFR C797G in 24% of cases73; and besides C797X, other mutations such as those in the G796, L792, L718, G719 and G724 residue have been showed to sterically interfere with the osimertinib-EGFR interaction.74–78 T790M loss is another common mechanism demonstrated in about 50%–60% of patients at osimertinib progression.79–81 Acquired EGFR mutations (C797S, 14%) was observed in 21% of cases while 49% of patients showed the T790M loss at progression in ctDNA. An association has been demonstrated between the T790M loss and a shorter time to treatment discontinuation (6.1 vs 15.2 months).82 Lastly, other EGFR-dependent mechanisms of resistance to osimertinib include exon 20 insertion (1%), EGFR S768I (<1%),83 and EGFR amplification (4%–35%).79 84
TKIs resistance is also mediated by activation of alternative pathways or histological transformation, together with the afore-mentioned mutations. The ErbB2 overexpression, coexisting with EGFR G796S+MET amplification (1%) and PIK3CA amplifications (1%), was identified in 5% of patients who acquired secondary resistance to osimertinib.85 86 BRAF mutation has been identified as responsible for osimertinib resistance in ~5%–7% of cases65 87 both as first and second line. V600E and G469A mutations seem to coexist with EGFR T790M.65 KRAS and NRAS mutations have also been reported after osimertinib failure.86 MET amplification was observed in nearly 19% of the samples at disease progression, opening-up the possibility for future combinations to overcome resistance.88 Concluding, oncogenic fusions (ie, FGFR3–TACC3, RET–ERC1, NTRK1–TPM3, ESYT2–BRAF) and histological and phenotypic transformation have also been confirmed after progression on second-line osimertinib.89
Multiparametric approach to treatment monitoring
Liquid biopsy
The analysis of circulating nucleic acids through liquid biopsy may be sufficiently sensitive and comprehensive to understand the clonal evolution driving resistance mechanisms in EGFR NSCLC and to identify new genomic targets. Liquid biopsy was first introduced in NSCLC for the research of the T790M in patients with metastatic NSCLC progressing on a first/second-generation TKI. The analysis of ctDNA quickly replaced the molecular analysis of tumour tissue, allowing real-time sampling of multifocal clonal evolution, catching tumour heterogeneity.90 91 Several studies demonstrated that liquid biopsy has good sensitivity and specificity compared with tissue, able to detect single nucleotide polymorphisms, insertions/deletions, amplifications and rearrangements; therefore, its potential in clinical practice increased, becoming an useful tool for the appearance of new mutations, and to monitor tumour dynamics and clonal evolution.92 93 Liquid biopsy is usually performed on plasma samples, however, data on ctDNA testing in other body fluids such cerebrospinal fluid and urine are also available.94 From a technical point of view, there is still a lack of standardisation across platforms. The complexity of Next Generation Sequencing (NGS) workflow, data analysis and costs can be challenging, and on the other side PCR-based tests are more accessible and cheaper and have a shorter turnaround time, and allow a limited number of target to analyse. However, mutual agreement has been demonstrated between NGS and digital PCR in ctDNA on specific mutations with comparable results.95 The chance to find a mutation in liquid biopsy is strictly dependent both on the analytical method and the clinical characteristics of patients. It is well known that intrathoracic lesions, CNS and bone metastasis yield lower amounts of ctDNA.96 Moreover, a proportion of tumours appear not to shed ctDNA into the peripheral blood and seem to have a better prognosis, being correlated with tumour burden.92
Furthermore, ctDNA is a possible tool for detection of minimal residual disease among treated early-stage NSCLC patients,97 and it is possible to follow the amount of mutant DNA during anti-EGFR treatments. The amount of EGFR mutation in plasma decreases during the treatment, and its disappearance is correlated with tumour responses according to RECIST criteria and radiological evaluation.98 99
However, despite several advantages of ctDNA analysis, mechanisms of resistance involving histological transformation (ie, SCLC) can be captured by tissue biopsy only. Recent data showed that ctDNA dynamics in global copy number may predict the histological transformation into SCLC, however, its clinical validation is still needed.12 The major issue for liquid biopsy remains the risk of false negative results. Moreover, being tumour heterogeneity a complex entity to consider to personalised treatment, a multiomics approach may be the best solution for a good predictive biomarker.
Radiogenomics and artificial intelligence
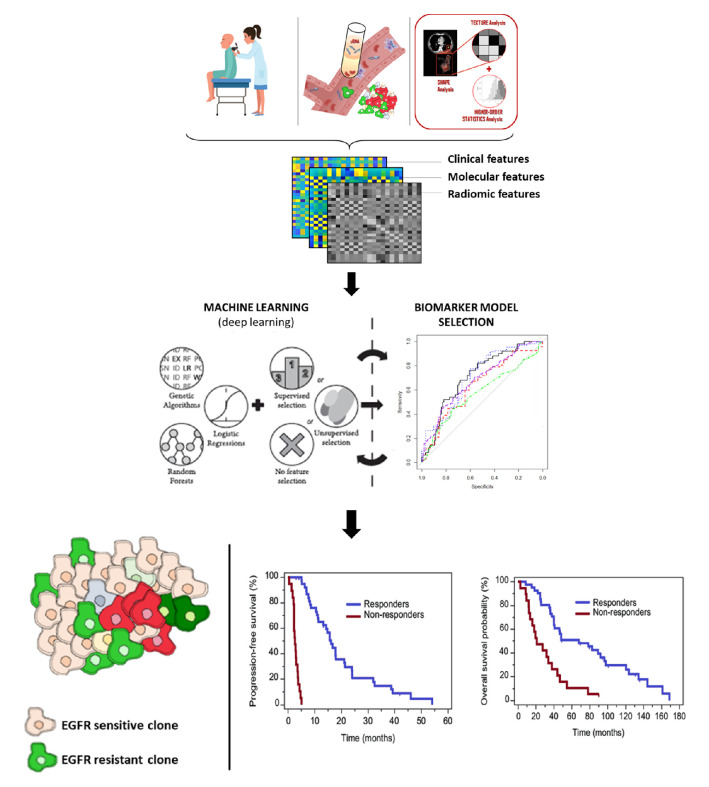
Image processing and machine learning methods can be proposed in support to understand the molecular profile and cancer dynamics during treatment. In this regard, radiomics is emerging as a novel tool for the extraction of qualitive and quantitative features from images, to develop potential non-invasive biomarkers for detection and characterisation of disease.100 Radiomic features provide information about the grey-scale patterns, interpixel relationships, shape and spectral properties within regions of interest on radiological images,101 which are able to reflect the patho-physiological processes and the heterogeneity of tumours, including transcriptomics, metabolomics, proteomics and genomics.100 Radiomics deals with development of deep artificial neural networks, inspired by biological neural networks in our brain to perform accurate segmentations and recognise patterns,102 succeeding an appropriate auto-training of the underlying functions, minimising the difference between ground truth and prediction (ie, deep learning).103 Several data showed that NSCLC texture analysis can classify tumour with EGFR mutations.104 However, the lack of standardised statistical processes for features selection and the heterogeneity of the analysed cohorts allowed several selected radiomic features among different research groups. Deep learning approaches may overcome these limitations by computing the most distinguishable radiomic features through n-fold bootstrap training sets, incorporating them into a model for EGFR status prediction. Most frequently, top ranking features were incorporated in a signature via multinomial logistic regression,104 naive bayesian classifier, K-nearest neighbour, random forest, support vector machine and decision tree.105 Performance models are measured by using area under the curve of receiver operating characteristic curve analysis in the validation cohort, evidencing good capability with acceptable representativeness for predicting EGFR status.
Since radiomic analysis may be performed using routinely diagnostic scans, some limitation may come from the heterogeneous scanning protocols: images are acquired using scanners manufactured by different companies, with a range of image reconstruction algorithms, different slice thicknesses, with and without contrast, using different dosages; despite normalisation, these factors potentially adding noise to the data.106
Radiomics together with liquid biopsy may have great potential, since are both minimally invasive, easy to perform, and can be repeated over time, enabling the extraction of the overall tumour load. This approach, may help decode tumour information regarding type, aggressiveness, progression and response to treatment.107 This approach may provide new diagnostic support, being able to suggest a change in treatment strategy earlier than with conventional methods (figure 3).
Figure 3.
Multiomics approach as predictive biomarker in NSCLC. EGFR, epidermal growth factor receptor; NSCLC, non-small cell lung cancer.
Conclusions
A wide array of treatments have been developed for NSCLC driven by HER1, 2 and 3, the most successful being EGFR-TKI in HER1-mutant tumours. HER2 and 3-driven tumours represent the minority of NSCLC and in these patients effective therapies still represent an unmet medical need. The encouraging results seen with anti-HER2 and with anti-HER3 monoclonal Abs need to be validated in larger studies even if the greatest obstacle is represented by the scarce number of patients bearing deregulated HER2/3 expression/activation and corresponding abnormalities of the signal transduction pathway.
Considering NSCLC tumour heterogeneity, which affects response and resistance to treatment, combined multiparametric approaches, such as liquid biopsy together with radiomics, may provide a better understanding of the tumour dynamics during treatments.
Footnotes
Twitter: @Marzia_Del_Re, @apassaroMD
Contributors: MDR and RD contributed to the conception of the work and to the interpretation of data. All the authors contributed to write the work and revised it critically for important intellectual content. All the authors gave their final approval of the version published, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Commissioned; externally peer reviewed.
References
- 1.da Cunha Santos G, Shepherd FA, Tsao MS. Egfr mutations and lung cancer. Annu Rev Pathol 2011;6:49–69. 10.1146/annurev-pathol-011110-130206 [DOI] [PubMed] [Google Scholar]
- 2.Guo G, Gong K, Wohlfeld B, et al. Ligand-Independent EGFR signaling. Cancer Res 2015;75:3436–41. 10.1158/0008-5472.CAN-15-0989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roskoski R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res 2014;79:34–74. 10.1016/j.phrs.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 4.Okamoto I, Kenyon LC, Emlet DR, et al. Expression of constitutively activated EGFRvIII in non-small cell lung cancer. Cancer Sci 2003;94:50–6. 10.1111/j.1349-7006.2003.tb01351.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169–81. 10.1038/nrc2088 [DOI] [PubMed] [Google Scholar]
- 6.Chen N, Fang W, Zhan J, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-Driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol 2015;10:910–23. 10.1097/JTO.0000000000000500 [DOI] [PubMed] [Google Scholar]
- 7.Ranayhossaini DJ, Lu J, Mabus J, et al. EGF potentiation of VEGF production is cell density dependent in H292 EGFR wild type NSCLC cell line. Int J Mol Sci 2014;15:17686–704. 10.3390/ijms151017686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Melo Gagliato D, Jardim DLF, Marchesi MSP, et al. Mechanisms of resistance and sensitivity to anti-HER2 therapies in HER2+ breast cancer. Oncotarget 2016;7:64431–46. 10.18632/oncotarget.7043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidhanth C, Manasa P, Krishnapriya S, et al. A systematic understanding of signaling by ErbB2 in cancer using phosphoproteomics. Biochem Cell Biol 2018;96:295–305. 10.1139/bcb-2017-0020 [DOI] [PubMed] [Google Scholar]
- 10.Garrido-Castro AC, Felip E. HER2 driven non-small cell lung cancer (NSCLC): potential therapeutic approaches. Transl Lung Cancer Res 2013;2:122–7. 10.3978/j.issn.2218-6751.2013.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cappuzzo F, Bemis L, Varella-Garcia M. HER2 mutation and response to trastuzumab therapy in non-small-cell lung cancer. N Engl J Med 2006;354:2619–21. 10.1056/NEJMc060020 [DOI] [PubMed] [Google Scholar]
- 12.Pillai RN, Behera M, Berry LD, et al. HER2 mutations in lung adenocarcinomas: a report from the lung cancer mutation Consortium. Cancer 2017;123:4099–105. 10.1002/cncr.30869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robichaux JP, Elamin YY, Tan Z, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med 2018;24:638–46. 10.1038/s41591-018-0007-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, Li S, Hai J, et al. Targeting HER2 Aberrations in Non-Small Cell Lung Cancer with Osimertinib. Clin Cancer Res 2018;24:2594–604. 10.1158/1078-0432.CCR-17-1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra R, Patel H, Alanazi S, et al. Her3 signaling and targeted therapy in cancer. Oncol Rev 2018;12:355. 10.4081/oncol.2018.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonesaka K, Iwama E, Hayashi H, et al. Heregulin expression and its clinical implication for patients with EGFR-mutant non-small cell lung cancer treated with EGFR-tyrosine kinase inhibitors. Sci Rep 2019;9:19501. 10.1038/s41598-019-55939-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verlingue L, Hollebecque A, Lacroix L, et al. Human epidermal receptor family inhibitors in patients with ErbB3 mutated cancers: entering the back door. Eur J Cancer 2018;92:1–10. 10.1016/j.ejca.2017.12.020 [DOI] [PubMed] [Google Scholar]
- 18.Umelo I, Noeparast A, Chen G, et al. Identification of a novel HER3 activating mutation homologous to EGFR-L858R in lung cancer. Oncotarget 2016;7:3068–83. 10.18632/oncotarget.6585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039–43. 10.1126/science.1141478 [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsao AS, Tang XM, Sabloff B, et al. Clinicopathologic characteristics of the EGFR gene mutation in non-small cell lung cancer. J Thorac Oncol 2006;1:231–9. 10.1016/S1556-0864(15)31573-2 [DOI] [PubMed] [Google Scholar]
- 22.Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa survival evaluation in lung cancer). Lancet 2005;366:1527–37. 10.1016/S0140-6736(05)67625-8 [DOI] [PubMed] [Google Scholar]
- 23.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005;353:123–32. 10.1056/NEJMoa050753 [DOI] [PubMed] [Google Scholar]
- 24.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239–46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 25.Zhou C, Wu Y-L, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735–42. 10.1016/S1470-2045(11)70184-X [DOI] [PubMed] [Google Scholar]
- 26.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121–8. 10.1016/S1470-2045(09)70364-X [DOI] [PubMed] [Google Scholar]
- 27.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380–8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 28.Wu Y-L, Zhou C, Liam C-K, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol 2015;26:1883–9. 10.1093/annonc/mdv270 [DOI] [PubMed] [Google Scholar]
- 29.Inoue A, Kobayashi K, Maemondo M, et al. Updated overall survival results from a randomized phase III trial comparing gefitinib with carboplatin-paclitaxel for chemo-naïve non-small cell lung cancer with sensitive EGFR gene mutations (NEJ002). Ann Oncol 2013;24:54–9. 10.1093/annonc/mds214 [DOI] [PubMed] [Google Scholar]
- 30.Mok TS, Wu Y-L, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947–57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 31.Park K, Tan E-H, O'Byrne K, et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol 2016;17:577–89. 10.1016/S1470-2045(16)30033-X [DOI] [PubMed] [Google Scholar]
- 32.Sequist LV, Yang JC-H, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327–34. 10.1200/JCO.2012.44.2806 [DOI] [PubMed] [Google Scholar]
- 33.Yang JC-H, Wu Y-L, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141–51. 10.1016/S1470-2045(14)71173-8 [DOI] [PubMed] [Google Scholar]
- 34.Wu Y-L, Zhou C, Hu C-P, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213–22. 10.1016/S1470-2045(13)70604-1 [DOI] [PubMed] [Google Scholar]
- 35.Wu Y-L, Cheng Y, Zhou X, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol 2017;18:1454–66. 10.1016/S1470-2045(17)30608-3 [DOI] [PubMed] [Google Scholar]
- 36.Mok TS, Wu Y-L, Ahn M-J, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive lung cancer. N Engl J Med 2017;376:629–40. 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41–50. 10.1056/NEJMoa1913662 [DOI] [PubMed] [Google Scholar]
- 38.Saito H, Fukuhara T, Furuya N, et al. Erlotinib plus bevacizumab versus erlotinib alone in patients with EGFR-positive advanced non-squamous non-small-cell lung cancer (NEJ026): interim analysis of an open-label, randomised, multicentre, phase 3 trial. Lancet Oncol 2019;20:625–35. 10.1016/S1470-2045(19)30035-X [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa K, Garon EB, Seto T, et al. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (relay): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20:1655–69. 10.1016/S1470-2045(19)30634-5 [DOI] [PubMed] [Google Scholar]
- 40.Hosomi Y, Morita S, Sugawara S, et al. Gefitinib alone versus gefitinib plus chemotherapy for non-small-cell lung cancer with mutated epidermal growth factor receptor: NEJ009 study. J Clin Oncol 2020;38:115–23. 10.1200/JCO.19.01488 [DOI] [PubMed] [Google Scholar]
- 41.Noronha V, Patil VM, Joshi A, et al. Gefitinib Versus Gefitinib Plus Pemetrexed and Carboplatin Chemotherapy in EGFR-Mutated Lung Cancer. J Clin Oncol 2020;38:124–36. 10.1200/JCO.19.01154 [DOI] [PubMed] [Google Scholar]
- 42.Gainor JF, Shaw AT, Sequist LV, et al. Egfr mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res 2016;22:4585–93. 10.1158/1078-0432.CCR-15-3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang JC-H, Gadgeel SM, Sequist LV, et al. Pembrolizumab in combination with erlotinib or gefitinib as first-line therapy for advanced NSCLC with sensitizing EGFR mutation. J Thorac Oncol 2019;14:553–9. 10.1016/j.jtho.2018.11.028 [DOI] [PubMed] [Google Scholar]
- 44.Lisberg A, Cummings A, Goldman JW, et al. A phase II study of pembrolizumab in EGFR-mutant, PD-L1+, tyrosine kinase inhibitor naïve patients with advanced NSCLC. J Thorac Oncol 2018;13:1138–45. 10.1016/j.jtho.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masood A, Kancha RK, Subramanian J. Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors in non-small cell lung cancer harboring uncommon EGFR mutations: focus on afatinib. Semin Oncol 2019;46:271–83. 10.1053/j.seminoncol.2019.08.004 [DOI] [PubMed] [Google Scholar]
- 46.Romero D. Poziotinib for uncommon ERBB mutations. Nat Rev Clin Oncol 2018;15:404. 10.1038/s41571-018-0038-7 [DOI] [PubMed] [Google Scholar]
- 47.Li BT, Shen R, Buonocore D, et al. Ado-Trastuzumab emtansine for patients with HER2-Mutant lung cancers: results from a phase II basket trial. J Clin Oncol 2018;36:2532–7. 10.1200/JCO.2018.77.9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazières J, Barlesi F, Filleron T, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: results from the European EUHER2 cohort. Ann Oncol 2016;27:281–6. 10.1093/annonc/mdv573 [DOI] [PubMed] [Google Scholar]
- 49.Hotta K, Aoe K, Kozuki T, et al. A phase II study of trastuzumab emtansine in HER2-positive non-small cell lung cancer. J Thorac Oncol 2018;13:273–9. 10.1016/j.jtho.2017.10.032 [DOI] [PubMed] [Google Scholar]
- 50.Ohashi K, Hotta K, Hirata T, et al. Trastuzumab emtansine in HER2+ recurrent metastatic non-small-cell lung cancer: study protocol. Clin Lung Cancer 2017;18:92–5. 10.1016/j.cllc.2016.06.014 [DOI] [PubMed] [Google Scholar]
- 51.Peters S, Stahel R, Bubendorf L, et al. Trastuzumab emtansine (T-DM1) in patients with previously treated HER2-overexpressing metastatic non-small cell lung cancer: efficacy, safety, and biomarkers. Clin Cancer Res 2019;25:64–72. 10.1158/1078-0432.CCR-18-1590 [DOI] [PubMed] [Google Scholar]
- 52.Smit EF, Nakagawa K, Nagasaka M, et al. Trastuzumab deruxtecan (T-DXd; DS-8201) in patients with HER2-mutated metastatic non-small cell lung cancer (NSCLC): interim results of DESTINY-Lung01. J Clin Oncol 2020;38:9504 10.1200/JCO.2020.38.15_suppl.9504 [DOI] [Google Scholar]
- 53.Herbst RS, Davies AM, Natale RB, et al. Efficacy and safety of single-agent pertuzumab, a human epidermal receptor dimerization inhibitor, in patients with non small cell lung cancer. Clin Cancer Res 2007;13:6175–81. 10.1158/1078-0432.CCR-07-0460 [DOI] [PubMed] [Google Scholar]
- 54.Ogoshi Y, Shien K, Yoshioka T, et al. Anti-tumor effect of neratinib against lung cancer cells harboring HER2 oncogene alterations. Oncol Lett 2019;17:2729–36. 10.3892/ol.2019.9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishio M, Horiike A, Murakami H, et al. Phase I study of the HER3-targeted antibody patritumab (U3-1287) combined with erlotinib in Japanese patients with non-small cell lung cancer. Lung Cancer 2015;88:275–81. 10.1016/j.lungcan.2015.03.010 [DOI] [PubMed] [Google Scholar]
- 56.Yu H, Johnson M, Steuer C, et al. Preliminary phase 1 results of U3-1402 — a novel HER3-Targeted Antibody–Drug Conjugate—in EGFR TKI-Resistant, EGFR-mutant NSCLC. J Thoracic Oncol. [Google Scholar]
- 57.Juric D, Dienstmann R, Cervantes A, et al. Safety and Pharmacokinetics/Pharmacodynamics of the first-in-class dual action HER3/EGFR antibody MEHD7945A in locally advanced or metastatic epithelial tumors. Clin Cancer Res 2015;21:2462–70. 10.1158/1078-0432.CCR-14-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cejalvo J-M, Jacob W, Fleitas Kanonnikoff T, et al. A phase Ib/II study of HER3-targeting lumretuzumab in combination with carboplatin and paclitaxel as first-line treatment in patients with advanced or metastatic squamous non-small cell lung cancer. ESMO Open 2019;4:e000532. 10.1136/esmoopen-2019-000532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Girard N. Optimizing outcomes in EGFR mutation-positive NSCLC: which tyrosine kinase inhibitor and when? Future Oncol 2018;14:1117–32. 10.2217/fon-2017-0636 [DOI] [PubMed] [Google Scholar]
- 60.Lim SM, Syn NL, Cho BC, et al. Acquired resistance to EGFR targeted therapy in non-small cell lung cancer: mechanisms and therapeutic strategies. Cancer Treat Rev 2018;65:1–10. 10.1016/j.ctrv.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 61.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. 10.1371/journal.pmed.0020073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bean J, Brennan C, Shih J-Y, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932–7. 10.1073/pnas.0710370104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov 2012;2:922–33. 10.1158/2159-8290.CD-12-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A 2012;109:E2127–33. 10.1073/pnas.1203530109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen K-SH, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer 2009;10:281–9. 10.3816/CLC.2009.n.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yun C-H, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070–5. 10.1073/pnas.0709662105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cross DAE, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014;4:1046–61. 10.1158/2159-8290.CD-14-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Del Re M, Petrini I, Mazzoni F, et al. Incidence of T790M in patients with NSCLC progressed to gefitinib, erlotinib, and afatinib: a study on circulating cell-free DNA. Clin Lung Cancer 2020;21:232–7. 10.1016/j.cllc.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 70.Wang Z, Yang J-J, Huang J, et al. Lung Adenocarcinoma Harboring EGFR T790M and In Trans C797S Responds to Combination Therapy of First- and Third-Generation EGFR TKIs and Shifts Allelic Configuration at Resistance. J Thorac Oncol 2017;12:1723–7. 10.1016/j.jtho.2017.06.017 [DOI] [PubMed] [Google Scholar]
- 71.Arulananda S, Do H, Musafer A, et al. Combination Osimertinib and gefitinib in C797S and T790M EGFR-mutated non-small cell lung cancer. J Thorac Oncol 2017;12:1728–32. 10.1016/j.jtho.2017.08.006 [DOI] [PubMed] [Google Scholar]
- 72.Niederst MJ, Hu H, Mulvey HE, et al. The allelic context of the C797S mutation acquired upon treatment with third-generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin Cancer Res 2015;21:3924–33. 10.1158/1078-0432.CCR-15-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang Z, Yang N, Ou Q, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor Osimertinib in non-small cell lung cancer patients. Clin Cancer Res 2018;24:3097–107. 10.1158/1078-0432.CCR-17-2310 [DOI] [PubMed] [Google Scholar]
- 74.Ou S-HI, Cui J, Schrock AB, et al. Emergence of novel and dominant acquired EGFR solvent-front mutations at Gly796 (G796S/R) together with C797S/R and L792F/H mutations in one EGFR (L858R/T790M) NSCLC patient who progressed on osimertinib. Lung Cancer 2017;108:228–31. 10.1016/j.lungcan.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 75.Zheng D, Hu M, Bai Y, et al. EGFR G796D mutation mediates resistance to osimertinib. Oncotarget 2017;8:49671–9. 10.18632/oncotarget.17913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fassunke J, Müller F, Keul M, et al. Overcoming EGFRG724S-mediated osimertinib resistance through unique binding characteristics of second-generation EGFR inhibitors. Nat Commun 2018;9:4655. 10.1038/s41467-018-07078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. 10.1038/ncomms11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, He B, Zhou D, et al. Newly emergent acquired EGFR exon 18 G724S mutation after resistance of a T790M specific EGFR inhibitor osimertinib in non-small-cell lung cancer: a case report. Onco Targets Ther 2019;12:51–6. 10.2147/OTT.S188612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fogli S, Polini B, Del Re M, et al. EGFR-TKIs in non-small-cell lung cancer: focus on clinical pharmacology and mechanisms of resistance. Pharmacogenomics 2018;19:727–40. 10.2217/pgs-2018-0038 [DOI] [PubMed] [Google Scholar]
- 80.Dal Maso A, Lorenzi M, Roca E, et al. Clinical features and progression pattern of acquired T790M-positive compared with T790M-negative EGFR mutant non-small-cell lung cancer: catching tumor and clinical heterogeneity over time through liquid biopsy. Clin Lung Cancer 2020;21:e13:1–14. 10.1016/j.cllc.2019.07.009 [DOI] [PubMed] [Google Scholar]
- 81.Matsumoto Y, Sawa K, Fukui M, et al. Predictive impact of low-frequency pretreatment T790M mutation in patients with EGFR-mutated non-small cell lung cancer treated with EGFR tyrosine kinase inhibitors. Lung Cancer 2020;139:80–8. 10.1016/j.lungcan.2019.10.029 [DOI] [PubMed] [Google Scholar]
- 82.Oxnard GR, Hu Y, Mileham KF, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-Positive lung cancer and acquired resistance to Osimertinib. JAMA Oncol 2018;4:1527–34. 10.1001/jamaoncol.2018.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leventakos K, Kipp BR, Rumilla KM, et al. S768I mutation in EGFR in patients with lung cancer. J Thorac Oncol 2016;11:1798–801. 10.1016/j.jtho.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakatani K, Yamaoka T, Ohba M, et al. KRAS and EGFR Amplifications Mediate Resistance to Rociletinib and Osimertinib in Acquired Afatinib-Resistant NSCLC Harboring Exon 19 Deletion/T790M in EGFR. Mol Cancer Ther 2019;18:112–26. 10.1158/1535-7163.MCT-18-0591 [DOI] [PubMed] [Google Scholar]
- 85.Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous mechanisms of primary and acquired resistance to third-generation EGFR inhibitors. Clin Cancer Res 2016;22:4837–47. 10.1158/1078-0432.CCR-15-1915 [DOI] [PubMed] [Google Scholar]
- 86.Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725–37. 10.1038/s41416-019-0573-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paik PK, Arcila ME, Fara M, et al. Clinical Characteristics of Patients With Lung Adenocarcinomas Harboring BRAF Mutations. J Clin Oncol 2011;29:2046–51. 10.1200/JCO.2010.33.1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Le X, Puri S, Negrao MV, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin Cancer Res 2018;24:6195–203. 10.1158/1078-0432.CCR-18-1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.La Monica S, Minari R, Cretella D, et al. Acquired BRAF G469A mutation as a resistance mechanism to first-line Osimertinib treatment in NSCLC cell lines harboring an EGFR exon 19 deletion. Target Oncol 2019;14:619–26. 10.1007/s11523-019-00669-x [DOI] [PubMed] [Google Scholar]
- 90.Murtaza M, Dawson S-J, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015;6:8760. 10.1038/ncomms9760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223–38. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 92.Mok T, Wu Y-L, Lee JS, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015;21:3196–203. 10.1158/1078-0432.CCR-14-2594 [DOI] [PubMed] [Google Scholar]
- 93.Del Re M, Rofi E, Cappelli C, et al. The increase in activating EGFR mutation in plasma is an early biomarker to monitor response to osimertinib: a case report. BMC Cancer 2019;19:410. 10.1186/s12885-019-5604-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reckamp KL, Melnikova VO, Karlovich C, et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol 2016;11:1690–700. 10.1016/j.jtho.2016.05.035 [DOI] [PubMed] [Google Scholar]
- 95.Steendam CMJ, Atmodimedjo P, de Jonge E, et al. Plasma cell-free DNA testing of patients with EGFR mutant Non–Small-Cell lung cancer: droplet digital PCR versus next-generation sequencing compared with tissue-based results. JCO Precis Oncol 2019:1–9. 10.1200/PO.18.00401 [DOI] [PubMed] [Google Scholar]
- 96.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579–86. 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446–51. 10.1038/nature22364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yung TKF, Chan KCA, Mok TSK, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009;15:2076–84. 10.1158/1078-0432.CCR-08-2622 [DOI] [PubMed] [Google Scholar]
- 99.Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 2014;120:3896–901. 10.1002/cncr.28964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Neri E, Del Re M, Paiar F, et al. Radiomics and liquid biopsy in oncology: the holons of systems medicine. Insights Imaging 2018;9:915–24. 10.1007/s13244-018-0657-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563–77. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Parekh VS, Jacobs MA. Deep learning and radiomics in precision medicine. Expert Rev Precis Med Drug Dev 2019;4:59–72. 10.1080/23808993.2019.1585805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang S, Yang DM, Rong R, et al. Artificial intelligence in lung cancer pathology image analysis. Cancers 2019;11. 10.3390/cancers11111673. [Epub ahead of print: 28 Oct 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu Y, Kim J, Balagurunathan Y, et al. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer 2016;17:e446:441–8. 10.1016/j.cllc.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hong D, Xu K, Zhang L, et al. Radiomics signature as a predictive factor for EGFR mutations in advanced lung adenocarcinoma. Front Oncol 2020;10:28. 10.3389/fonc.2020.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, et al. Radiomics of CT features may be Nonreproducible and redundant: influence of CT acquisition parameters. Radiology 2018;288:407–15. 10.1148/radiol.2018172361 [DOI] [PubMed] [Google Scholar]
- 107.Aerts HJWL, Grossmann P, Tan Y, et al. Defining a radiomic response phenotype: a pilot study using targeted therapy in NSCLC. Sci Rep 2016;6:33860. 10.1038/srep33860 [DOI] [PMC free article] [PubMed] [Google Scholar]