Abstract
Background
The immunological microenvironment of primary high-grade serous carcinomas (HGSCs) has a major impact on disease outcome. Conversely, little is known on the microenvironment of metastatic HGSCs and its potential influence on patient survival. Here, we explore the clinical relevance of the immunological configuration of HGSC metastases.
Methods
RNA sequencing was employed on 24 paired primary tumor microenvironment (P-TME) and metastatic tumor microenvironment (M-TME) chemotherapy-naive HGSC samples. Immunohistochemistry was used to evaluate infiltration by CD8+ T cells, CD20+ B cells, DC-LAMP+ (lysosomal-associated membrane protein 3) dendritic cells (DCs), NKp46+ (natural killer) cells and CD68+CD163+ M2-like tumor-associated macrophages (TAMs), abundance of PD-1+ (programmed cell death 1), LAG-3+ (lymphocyte-activating gene 3) cells, and PD-L1 (programmed death ligand 1) expression in 80 samples. Flow cytometry was used for functional assessments on freshly resected HGSC samples.
Results
1468 genes were differentially expressed in the P-TME versus M-TME of HGSCs, the latter displaying signatures of extracellular matrix remodeling and immune infiltration. M-TME infiltration by immune effector cells had little impact on patient survival. Accordingly, M-TME-infiltrating T cells were functionally impaired, but not upon checkpoint activation. Conversely, cytokine signaling in favor of M2-like TAMs activity appeared to underlie inhibited immunity in the M-TME and poor disease outcome.
Conclusions
Immunosuppressive M2-like TAM infiltrating metastatic sites limit clinically relevant immune responses against HGSCs.
Keywords: tumor microenvironment, tumor biomarkers, macrophages
Introduction
Tumorigenesis is a complex and dynamic process, involving different steps: malignant transformation, local progression and metastatic dissemination.1 It is now clear that all these stages are influenced by a multitude of interactions between neoplastic cells and their environment (including epithelial, endothelial, stromal and immune cells, as well as the extracellular matrix), overall determining whether a newly formed lesion will be eradicated by the immune system, progress uncontrollably or be preserved temporarily under control as a dormant micrometastasis.2 3 Of note, the composition, localization and functional orientation of the tumor microenvironment (TME) exhibit considerable degrees of heterogeneity, not only across patients, cancer types and disease stages but also across distinct lesions of the same tumor.4–8 High-grade serous carcinoma (HGSC) represents one of the most lethal peritoneal cancers.9 The poor prognosis of patients with HGSC is mainly linked to early dissemination into the peritoneal cavity and establishment of therapy-resistant metastases.10 HGSCs display indeed a characteristic intraperitoneal metastatic spread, with a clear preference for the omentum.10 The omental microenvironment, which is surrounded by a single layer of mesothelial cells, is primarily composed of adipocytes embedded in a collagen matrix, with interspersed fibroblasts, endothelial cells, and resident immune cells.11 12 During the metastatic process, these stromal cells are recruited and reprogrammed by their malignant counterparts to support cancer cell survival and proliferation.10 Accumulating data indicate that both the cellular and non-cellular components of the primary TME (P-TME) are dynamic, evolve with disease progression, and convey robust prognostic and predictive value.13–15 However, the HGSC TME at metastatic sites (M-TME) is poorly defined. Similarly, it is unclear whether the immunological characteristics of the M-TME have prognostic or predictive value in patients with HGSC.
Here, we compared the transcriptional profile of 24 paired P-TME and M-TME of patients with HGSC, with the aim to identify molecular signatures that can predict TME composition and impact on clinical outcome. Functional analyses of differentially expressed genes (DEGs) revealed a strong association with extracellular matrix organization and immune responses. By combining biomolecular, cytofluorometric, and immunohistochemical studies, we demonstrate that the M-TME contain a higher density of tumor-infiltrating lymphocytes (TILs) as compared with the P-TME. However, TIL density in the M-TME is not associated with improved clinical outcome, largely due to the functional impairment imposed by robust immunosuppression at metastatic sites by tumor-associated macrophages (TAMs). Although the dichotomic distinction between antitumor (M1) and protumor (M2) TAMs does not properly reflect the functional complexity of this cellular compartment,16 17 the M-TME of patients with HGSC was indeed enriched with immunosuppressive CD68+CD163+ TAMs that were herein refer to as M2-like TAMs.
Materials and methods
Patients
Study group 1
A retrospective series of paired 80 primary tumors and 80 peritoneal metastatic formalin-fixed paraffin-embedded samples were obtained from patients with HGSC who underwent primary surgery in the absence of neoadjuvant chemotherapy between 2008 and 2014 at the University Hospital Hradec Kralove (Czech Republic) (study group 1). Baseline characteristics of these patients are summarized in table 1. Pathology staging was performed according to the eighth tumor, node, metastasis (TNM) classification (2017), and histological types were determined according to the current WHO classification.18 19 Data on long-term clinical outcome were obtained retrospectively by interrogation of municipality registers or families. An additional series of paired 15 primary tumor samples and 15 peritoneal metastases from patients with HGSC was prospectively collected at the University Hospital Motol (Prague, Czech Republic) and the University Hospital Kralovske Vinohrady (Prague, Czech Republic) (study group 2). Written informed consent was obtained from patients before inclusion in the study. The protocol was approved by the local ethical committee. The experimental design of study is summarized in online supplementary figure 1.
Table 1.
Main clinical and biological characteristic of study group 1
| Variable | Overall cohort (n=80) |
| Age (years) | |
| Median±SD | 62±10 |
| Range | 49–83 |
| pTNM stage | |
| Stage III | 70 (87.5%) |
| Stage IV | 10 (12.5%) |
| Debulking | |
| R0 | 28 (35%) |
| R1 | 8 (10%) |
| R2 | 44 (55%) |
| Vital status of patients | |
| Alive | 27 (33.7%) |
| Dead | 53 (66.3%) |
pTNM, pathological tumor, node, metastasis.
jitc-2020-000979supp001.pdf (1.3MB, pdf)
Immunohistochemistry
Immunostaining with antibodies specific for programmed cell death 1 (PDCD1, best known as PD-1), CD274 (best known as programmed death ligand 1 (PD-L1)), lymphocyte-activating gene 3 (LAG-3), lysosomal-associated membrane protein 3 (LAMP3; best known as DC-LAMP), CD8, CD20 and NKp46 was performed according to conventional protocols as published previously (online supplementary table 1).13 20 21 Briefly, tumor specimens from study group 1 were fixed in neutral buffered 10% formalin solution and embedded in paraffin as per standard procedures. Tissue sections were deparaffinized, followed by antigen retrieval with Target Retrieval Solution (Leica) in Tris EDTA at pH 6.0 (for PD-L1), pH 8.0 (for CD8, CD20, DC-LAMP and NKp46), or pH 9.0 (for LAG-3 and PD-1) in a heated water bath (98°C, 30 min). Endogenous peroxidase and alkaline phosphatase were blocked with 3% H2O2 and levamisole, respectively, for 15 min. Thereafter, sections were treated with protein block (DAKO) for 15 min and incubated with primary antibodies, followed by the revelation of enzymatic activity. Sections were counterstained with hematoxylin (DAKO) for 30 s. Images of whole tumor sections were acquired using a Leica Aperio AT2 scanner (Leica).
Immunofluorescence
Slides were incubated for 20 min in NH4Cl (50 mmol/L) to reduce autofluorescence and blocked in phosphate-buffered saline (PBS) supplemented with 2.5% bovine serum albumin and 2.5% goat serum for 40 min at room temperature (RT). Subsequently, primary antibodies specific for CD68 and CD163 were applied for 1 hour at RT (online supplementary table 1). Thereafter, slides were incubated with appropriate horseradish peroxidase polymer secondary antibodies for 1 hour at RT, followed by Tyramide Signal Amplification (Thermo Fisher Scientific). Finally, sections were mounted with ProLong Gold antifade reagent containing DAPI (Thermo Fisher Scientific). The specificity of the staining was determined using appropriate isotype controls. Images of whole tumor sections were acquired using a Leica Aperio AT2 scanner (Leica).
Cell quantification
Infiltration of tumor nests by CD8+ T cells, DC-LAMP+ DCs, CD20+ B cells, PD-1+, LAG-3+, NKp46+ cells and CD68+CD163+ cells was quantified in whole tumor sections with Calopix software (Tribvn) as published previously.13 20 Data are reported as absolute number of positive cells/mm2 (for PD-1+, LAG-3+, DC-LAMP+, CD8+, NKp46+, CD68+CD163+ cells) or cell surface/tumor section surface (for CD20+ cells), as previously described.20 21 PD-L1 expression was scored in the intratumoral and stromal compartments as a percentage of tumor area and categorized as 1 (0%), 2 (1%–4%), 3 (5%–9%), and 4 (>10%), as previously described.22 Quantitative assessments by Calopix software were performed by three independent investigators (JF, JR, LK) and reviewed by an expert pathologist (JL, PS).
Flow cytometry
Total live mononuclear cells were isolated from fresh tumor specimens, as previously described.20 Mononuclear cells were stained with multiple panels of fluorescent primary antibodies (online supplementary table 2) or appropriate isotype controls for 20 min at 4°C in the dark, following by washing and acquisition on a Fortessa flow cytometer (BD Bioscience). Flow cytometry data were analyzed with the FlowJo software (TreeStar).
Degranulation and interferon gamma (IFN-γ) production
Mononuclear cells isolated from fresh tumor specimens (study group 2) were incubated with 10 µg/mL anti-PD-1 and 10 µg/mL anti-TIM-3 antibodies for 24 hours at 37°C and 5% CO2. Thereafter, mononuclear cells were stimulated with 50 ng/mL phorbol 12-myristate 13-acetate (PMA)+1 µg/mL ionomycin for 1 hour followed by 3-hour incubation with brefeldin A (BioLegend). Unstimulated cells were used as control. Cells were washed in PBS, stained with anti-CD3 Alexa Fluor 700 (EXBIO), anti-CD4 ECD (Beckman Coulter), and anti-CD8 HV500 (BD Biosciences) monoclonal antibodies, fixed in fixation/permeabilization buffer (eBioscience), further permeabilized with permeabilization buffer (eBioscience) and intracellularly stained with an anti-IFN-γ PE-Cy7 (eBioscience) and anti-GZMB BV421 (BD Bioscience) monoclonal antibody. Flow cytometry was performed on the LSRFortessa analyzer, and data were analyzed with the FlowJo software package (Tree Star) (online supplementary figure 2).
NGS data analysis
Raw FASTQ sequencing files were aligned to human reference genome (build h19) with bowtie2 (version 2.3.2) and tophat2 (version 2.1). Expression levels as raw “counts” were calculated from aligned reads with mapping quality ≥10 using htseq-count (version 0.6.0). Differential gene expression analyses were performed using DESeq2 (version 1.24.0) in R. Heatmaps with hierarchical clustering analysis were assembled for DEGs using the ComplexHeatmap package in R23 based on the Pearson distance and average clustering method. Functional and enrichment analysis of DEGs was performed using the ClusterProfiler24 and the web-based tool REACTOME and METASCAPE. To estimate the relative abundance of immune cell populations, we used “metagene” markers.25
Statistical analysis
Survival analysis was performed using the Survival and Survminer R packages, using both log-rank tests and Cox proportional hazard regressions. For log-rank tests, the prognostic value of continuous variables was assessed using median cut-offs. For Cox proportional hazard regressions, cell densities were log-transformed. In multivariate Cox regressions, variables that were not significantly associated with prognosis in univariate analysis, as well as variables intrinsically correlated to each other, were not included. Cox proportional hazard regression analysis, log-rank analysis, Fisher’s exact tests, Student’s t-tests, the Wilcoxon and Mann-Whitney tests were used to assess statistical significance and p values were reported (considered not significant when >0.05).
Results
Transcriptional characterization of primary versus metastatic HGSC microenvironments
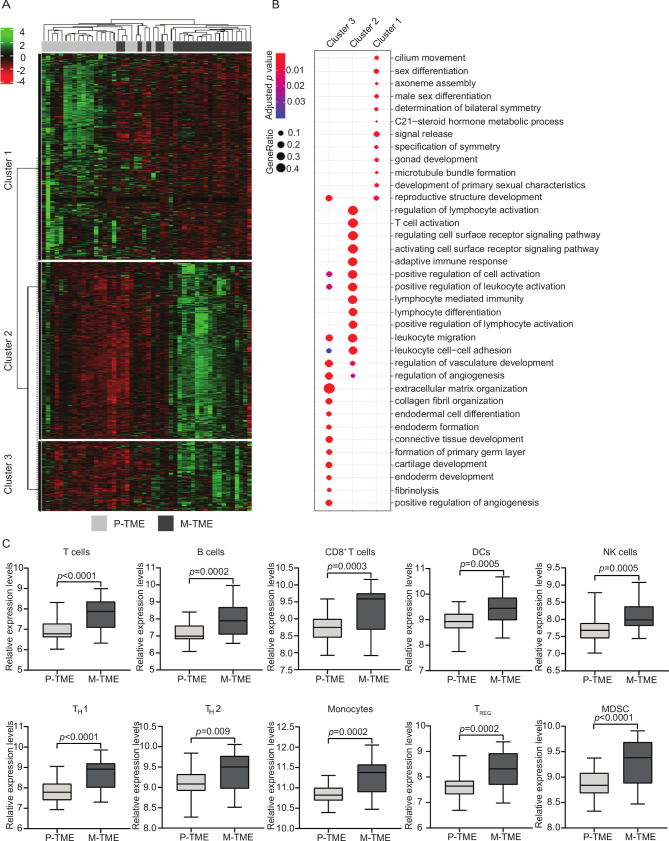
We first set to harness RNA sequencing (RNA-seq) to compare the gene expression profile of 24 paired primary (P-TME) and metastatic (M-TME) tumor samples from patients with HGSC who did not receive neoadjuvant chemotherapy (study group 1) (table 1). We identified a set of 1468 DEGs (online supplementary table 3), associated with three main functional clusters (figure 1A). Enrichment analysis of Gene Ontology (GO) terms from cluster 1, which was over-represented in the P-TME, mainly identified DEGs linked to the female reproductive system (figure 1B). On the contrary, clusters 2 and 3, which were over-represented in the M-TME were enriched for genes linked to extracellular matrix organization, immune response, lymphocyte trafficking, and immune system regulation (figure 1B). These data indicate that while the P-TME is dominated by ovary-associated genes, the M-TME contains an abundant immune infiltrate.
Figure 1.
Transcriptional characterization of primary versus metastatic high-grade serous carcinomas (HGSCs). (A) Hierarchical clustering of 1468 transcripts (fold-change >2, adjusted p-value <0.05) that were significantly changed in paired 24 metastatic tumor microenvironment (M-TME) versus 24 primary tumor microenvironment (P-TME) samples from study group 1, as determined by RNA sequencing. (B) Gene Ontology (biological processes) enrichment analysis of differentially expressed in M-TME versus P-TME. See also online supplementary table 3. (C) Relative expression levels of gene sets associated with T cells, B cells, CD8+ T cells, dendritic cells (DCs), natural killer (NK) cells, TH1 cells, TH2 cells, monocytes, regulatory T (TREG) cells, and myeloid-derived suppressor cells (MDSCs) across paired 24 P-TME and 24 M-TME samples, as determined by metagenes on RNA sequencing data from study group 1. Box plots: lower quartile, median, upper quartile; whiskers, minimum, maximum.
As published previously by us and others,13 20 26 27 the composition, localization, and functional orientation of the immunological P-TME of patients with HGSC convey robust prognostic and predictive information. However, little is known on the potential impact of the immunological M-TME on the clinical outcome of these patients. Driven by these premises, we employed “metagene” markers to estimate the relative abundance of different immune cell populations in the P-TME versus M-TME of study group 1. In line with whole-transcriptome findings (figure 1B), metastatic samples were enriched for gene sets associated with T cells (p<0.0001), B cells (p=0.0002), CD8+ T cells (p=0.0003), dendritic cells (DCs, p=0.0005), natural killer (NK) cells (p=0.0005), TH1 cells (p<0.0001), TH2 cells (p=0.009), monocytes (p=0.0002), as well as regulatory T (TREG) cells (p=0.0002) and myeloid-derived suppressor cells (MDSCs, p<0.0001) (figure 1C). These findings indicate that both immunostimulatory and immunosuppressive cell subsets are increased in the M-TME of HGSCs as compared with their P-TME.
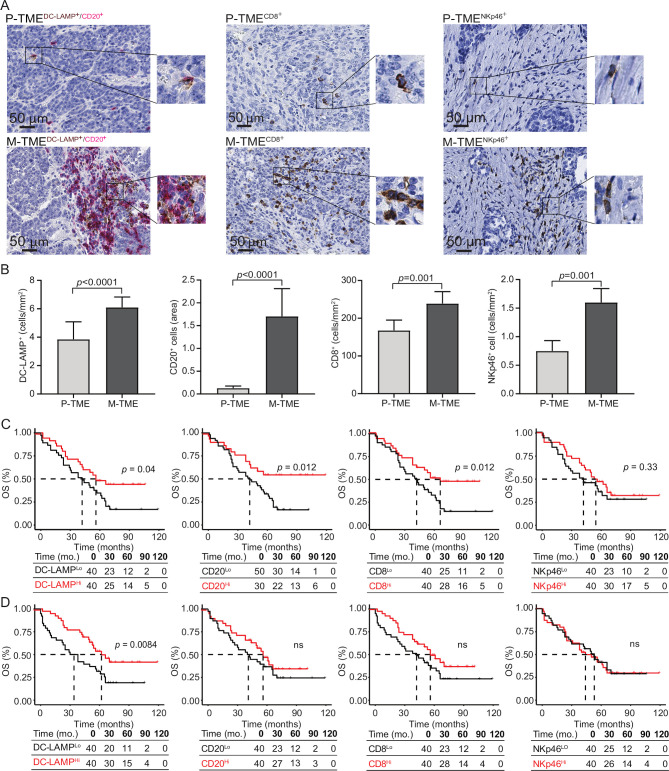
To validate these data with another technological approach and in a larger cohort of patients with HGSC, we next analyzed the composition of the immune infiltrate in paired 80 P-TME and 80 M-TME samples (from study group 1) by immunohistochemistry (figure 2A). Confirming transcriptomic observations, we detected a higher density of DC-LAMP+ mature DCs (p<0.0001), CD20+ B cells (p<0.0001), CD8+ T cells (p=0.001), and NKp46+ NK cells (p=0.001) in the M-TME as compared with the P-TME (figure 2B). To evaluate the prognostic impact of the P-TME and M-TME immune infiltrate, we investigated overall survival (OS) on stratifying the entire patient cohort based on median values of DC-LAMP+ DCs, CD20+ B cells, CD8+ T cells, and NKp46+ NK cells (figure 2C, D). As shown previously by us, robust tumor infiltration by DC-LAMP+ DCs, CD20+ B cells and CD8+ T cells in P-TME was associated with favorable OS in study group 1 (figure 2C). Moreover, patients with an abundant DC-LAMP+ DC infiltrate in the M-TME tissue exhibited significantly longer OS as compared with their DC-LAMPLo counterparts (p=0.0084). However, the density of CD20+ B cells, CD8+ T cells, and NKp46+ NK cells in the M-TME failed to influence OS in this patient cohort (figure 2D). Univariate Cox analyses confirmed the prognostic impact of mature DC infiltration in the M-TME (HR=0.74, 95% CI 0.60 to 0.92; p=0.006) (table 2).
Figure 2.
Prognostic impact of immune infiltrate in tumor microenvironment of primary and metastatic tissue of high-grade serous carcinoma (HGSC). (A) Representative images of lysosomal-associated membrane protein (DC-LAMP), CD20, CD8, and natural killer (NK) p46 immunostaining in primary tumor microenvironment (P-TME) and metastatic tumor microenvironment (M-TME) samples from study group 1. Scale bar=50 µm. (B) Density of DC-LAMP+, CD20+, CD8+ cells, and NKp46+ cells in paired 80 P-TME versus 80 M-TME samples from study group 1, as determined by immunostaining. Box plots: lower quartile, median, upper quartile; whiskers, minimum, maximum. (C and D) Overall survival (OS) of 80 patients from study group 1 on stratification based on median density of DC-LAMP+ cells, CD20+ cells, CD8+ cells, and NKp46+ cells in the P-TME (C) and M-TME (D). Survival curves were estimated by the Kaplan-Meier method, and differences between groups were evaluated using log-rank test. Number of patients at risk and p values are reported. ns, not significant.
Table 2.
Univariate Cox proportional hazard analysis
| Overall survival | ||
| Variable | HR (95% Cl) | P value |
| Age | 1.01 (0.98 to 1.04) | 0.51 |
| Stage 4 | 1.53 (0.65 to 3.58) | 0.33 |
| CA-125 | 1.00 (1.00 to 1.00) | 0.036 |
| Debulking | ||
| R1 | 1.00 (0.33 to 2.98) | 0.99 |
| R2 | 2.06 (1.12 to 3.78) | 0.019 |
| P-TME | M-TME | |||
| DC-LAMP | 0.74 (0.58 to 0.95) | 0.018 | 0.74 (0.60 to 0.92) | 0.006 |
| CD8 | 0.87 (0.75 to 0.99) | 0.039 | 0.90 (0.78 to 1.03) | 0.11 |
| CD20 | 0.35 (0.09 to 1.33) | 0.12 | 0.85 (0.64 to 1.13) | 0.26 |
| NKp46 | 0.83 (0.54 to 1.27) | 0.39 | 0.95 (0.72 to 1.25) | 0.70 |
| M2-like TAMs | 0.15 (0.01 to 3.20) | 0.22 | 6.74 (1.58 to 28.68) | 0.009 |
CA-125, cancer antigen 125; DC-LAMP, lysosomal-associated membrane protein; M-TME, metastatic tumor microenvironment; NK, natural killer; P-TME, primary tumor microenvironment; TAMs, tumor-associated macrophages.
Functional impairment of CD8+ T cells from the M-TME of patients with HGSC
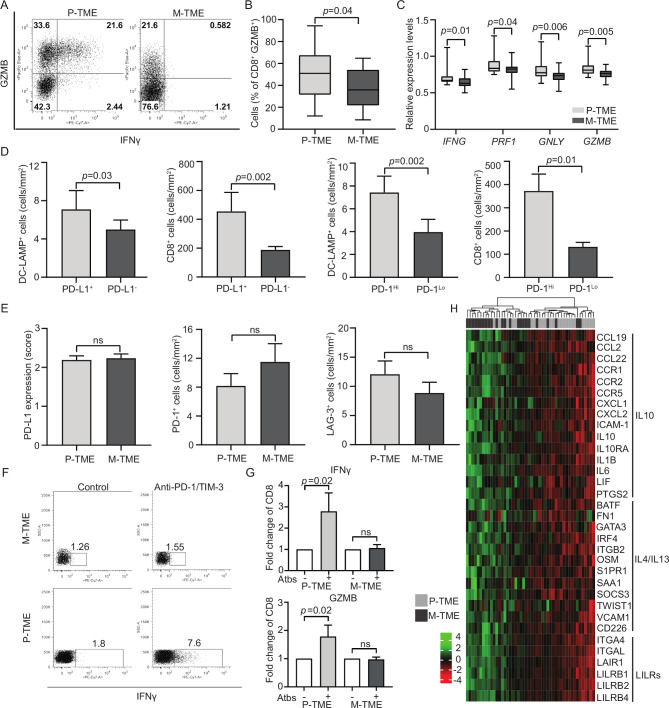
Surprised by the fact that M-TME-infiltrating CD8+ T cells had no impact on clinical outcome, at odds with their P-TME-infiltrating counterparts,13 20 we set to compare the functional capacity of these two CD8+ T-cell populations. To this aim, we focused on freshly resected tumors from a prospective cohort of 15 patients with HGSCs (study group 2). In this setting, non-specific stimulation of tumor-infiltrating mononuclear cells with PMA and ionomycin caused a more pronounced increase in CD8+ cells staining positively for the cytotoxic effector granzyme B (GZMB) when mononuclear cells were isolated from the P-TME versus the M-TME (p=0.04) (figure 3A, B). Consistently, the mRNAs coding for multiple CD8+ T-cell effector molecules including IFNG (p=0.01), perforin 1 (PRF1; p=0.04), granulysin (GNLY; p=0.006) and GZMB (p=0.005) were over-represented in P-TME versus M-TME samples, as determined by RNA-seq in samples from study group 1 (figure 3C). These findings suggest that CD8+ T cells from the M-TME are functionally impaired and exhibit limited effector functions.
Figure 3.
Functional T-cell exhaustion in the metastatic tumor microenvironment (M-TME) of patients with high-grade serous carcinoma (HGSC). (A and B) Percentage of interferon gamma (IFN-γ+) and granzyme B (GZMB)+ cells in the CD3+CD8+ population of the paired 15 primary (P-TME) and 15 metastatic tumor microenvironment (M-TME) microenvironment of patients from study group 2 on non-specific stimulation with phorbol 12-myristate13-acetate (PMA) and ionomycin, as determined by flow cytometry. Gating strategy (A) and quantitative results (B) are reported. Box plots: lower quartile, median, upper quartile; whiskers, minimum, maximum. (C) Expression levels of IFNG, perforin 1 (PRF1), granulysin (GNLY) and GZMB relative to CD3E in 24 P-TME versus 24 M-TME samples from study group 1, as determined by RNA sequencing. P values are reported. (D) Density of CD8+ T cells and lysosomal-associated membrane protein (DC-LAMP+) dendritic cells (DCs) in the M-TME of programmed death ligand 1 (PD-L1)− versus PD-L1+ and programmed cell death 1 (PD-1)Lo versus PD-1Hi patients from study group 1 (n=80), as determined by immunostaining. Box plots: lower quartile, median, upper quartile; whiskers, minimum, maximum. (E) Distribution of PD-L1 levels and density of PD-1+ and lymphocyte-activating gene 3 (LAG-3)+ cells in paired 80 P-TME and 80 M-TME samples from study group 1, as determined by immunostaining. Box plots: lower quartile, median, upper quartile; whiskers, minimum, maximum. (F) Gating strategy of IFN-γ+ CD3+CD8+ T cells in paired 15 P-TME and 15 M-TME samples (study group 2). The percentage of cells in each gate is reported. (G) Fold change of IFN-γ+ and GZMB+ CD8+ T cells isolated from paired 15 P-TME and 15 M-TME samples (study group 2) after non-specific stimulation with PMA and ionomycin in the context of PD-1 and TIM-3 blockage (Atbs) as determined by flow cytometry. P values are indicated. (H) Hierarchical clustering of 33 genes linked to cytokine/chemokine signaling that were differentially expressed in paired 24 M-TME versus 24 P-TME samples from study group 1. P values are reported. IL, interleukin; ns, not significant.
Since the expression of coinhibitory receptors is a major driver of functional T-cell exhaustion in multiple malignancies including ovarian cancer,28 we next profiled immune checkpoint status in the P-TME versus M-TME of patients with HGSC. To this aim, paired 80 P-TME and 80 M-TME samples from study group 1 were analyzed by immunohistochemistry for (1) expression of the coinhibitory ligand PD-L1 in the tumor nest and (2) infiltration by immune cells expressing the coinhibitory receptors PD-1 (the receptor for PD-L1) and LAG-3 (online supplementary figure 3). Both PD-L1 expression and PD-1+ cell density in the M-TME positively associated with infiltration by DC-LAMP+ DCs (p=0.03 and p=0.002, respectively). Similar results were obtained for CD8+ T cells (p=0.002 and p=0.01, respectively) (figure 3D). However, PD-L1 levels as well as infiltration by PD-1+ and LAG-3+ cells were heterogeneous across samples and did not differ based on site of assessment (figure 3E), suggesting that the PD-1/PD-L1 axis and LAG-3 systems are not the major determinants of functional exhaustion in this setting.
These findings recapitulate previous results from our group on the P-TME, where functional exhaustion appears to mainly depend on the coinhibitory receptor hepatitis A virus cellular receptor 2 (best known as TIM-3).13 20 We therefore harnessed flow cytometry on samples from study group 2 to investigate whether TIM-3 is also operational in the M-TME (figure 3F). We found that simultaneously blocking PD-1 and TIM-3 improves the ability of CD8+ T cells from the P-TME, but not the M-TME, to respond to non-specific stimulation by synthesizing effector molecules including IFN-γ and GZMB (figure 3G). Moreover, the P-TME contains increased amount of PD-1+TIM-3+cells as compared with the M-TME (online supplementary figure 3B), despite the fact that the overall T-cell density is decreased (figure 2B). Thus, TIM-3 does not stand out as a major target for immunostimulation in the M-TME of patients with HGSC.
We therefore hypothesized that immunosuppression in the M-TME would mostly originate from cytokine signaling. We investigated this possibility in our transcriptomic dataset from study group 1, identifying 33 DEGs linked to cytokine/chemokine signaling that were over-represented in the M-TME over the P-TME (figure 3H). These DEGs encompass major players in cancer-associated immunosuppression by TAMs, including (but not limited to): C-C motif chemokine ligand 22 (CCL22), interleukin 6, interleukin 10 (IL-10), IL-10 receptor subunit alpha, GATA-binding protein 3, and interferon regulatory factor 4 (figure 3H), supporting the possibility that the immunosuppressive circuitries at play in the M-TME stem from cytokine signaling.16
M2-like TAMs dictate clinically relevant immunosupression in the M-TME of HGSC
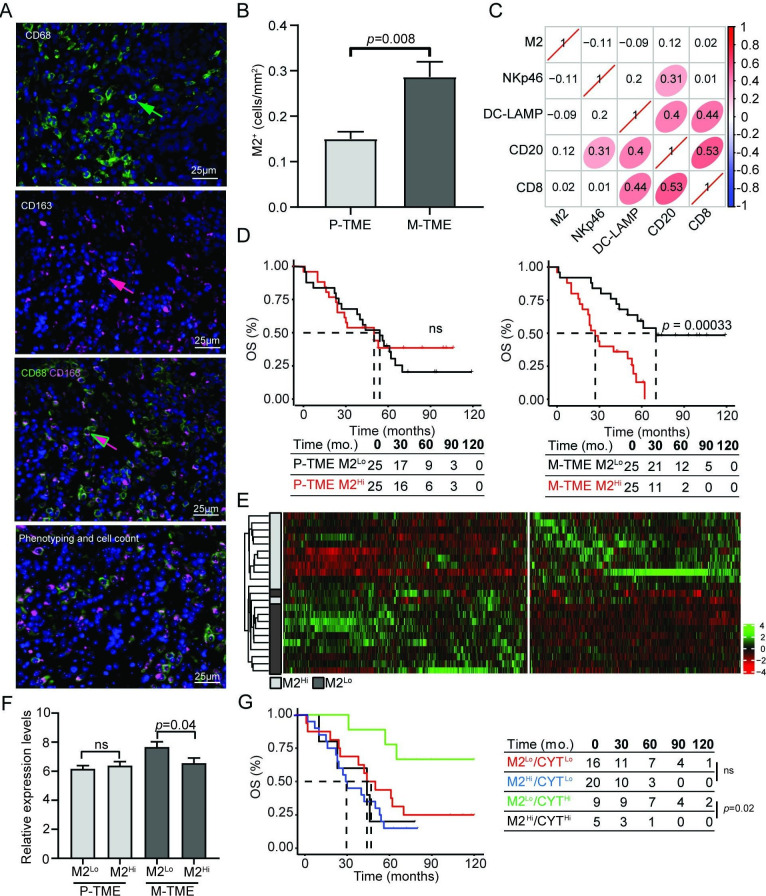
Driven by the potential implication of TAM-dependent cytokine signaling in immunosupression at metastatic sites, we harnessed immunofluorescence microscopy to study tumor infiltration by CD68+CD163+ M2-like TAMs in paired 50 P-TME and 50 M-TME samples from study group 1 (figure 4A). We detected a higher density of M2-like TAMs in the M-TME as compared with the P-TME (p=0.008) (figure 4B), but the abundance of M2-like TAMs did not correlate with the frequency of various immune components of the M-TME including NKp46+, DC-LAMP+, CD20+, and CD8+ cells (figure 4C). To evaluate the prognostic impact of M2-like TAMs in this patient cohort, we stratified patients from study group 1 based on median density of M2-like TAMs in the P-TME and M-TME, followed by retrospective OS analysis. Although, the density of M2-like TAMs in the P-TME failed to influence OS in this patient cohort, patients with a high density of M2 TAMs in the M-TME had poor OS as compared with their M2Lo counterparts (p=0.00033) (figure 4D). Univariate Cox regression confirmed the negative prognostic impact of M-TME infiltration by M2-like TAMs (HR=6.74, 95% CI 1.58 to 28.68, p=0.009; table 2) and the significance of this association was further corroborated by multivariate Cox regression (HR=10.85, 95% CI 2.28 to 51.44, p=0.002; table 3). We next compared the transcriptional profile of 12 patients with M2Hi versus 12 M2Lo M-TMEs (study group 1). We identified 1016 genes that were significantly over-represented in samples from M2Lo patients as compared with their M2Hi TAMs counterparts (figure 4E, online supplementary table 4), and functional studies revealed a strong association between such DEGs and immune system activation and T-cell proliferation (online supplementary figure 4). Moreover, we observed a negative correlation between M2-like TAM infiltration of the M-TME and cytolytic activity (CYT) based on the levels of transcripts encoding the cytolytic effectors granzyme A and PRF1 (figure 4F).29 These findings confirm the negative impact of M2-like TAMs on the immune effector functions of the M-TME in patients with HGSC. On stratifying patients from study group 1 into four subsets based on M2 TAM infiltration and CYT in the M-TME (M2Hi/CYTHi, M2Hi/CYTLo, M2Lo/CYTHi, M2Lo/CYTLo), we found that M2Lo/CYTHi individuals had improved OS as compared with their M2Hi/CYTHi counterparts (p=0.02) (figure 4G). Altogether, these findings document a critical role for M2-like TAMs in the establishment of clinically relevant immunosupression in the M-TME of patients with HGSC.
Figure 4.
M2-like tumor-associated macrophages (TAMs) dictate clinically relevant immunosuppression in the metastatic tumor microenvironment (M-TME) of high-grade serous carcinoma (HGSC). (A) Representative images of CD68 and CD163 immunofluorescence. Cells expressing CD68 (green arrow), cell expressing CD163 (red arrow), and cells coexpressing CD68 and CD163 (red/green arrow). For automated counting, Calopix software allows cell segmentation based on DAPI staining of the nucleus and morphometric characteristics. (B) Density of M2-like TAMs determined as CD68+CD163+ cells in the 50 primary tumor microenvironment (P-TME) and 50 M-TME of HGSCs from study group 1. (C) The correlation matrix for M2-like TAMs, natural killer (NK) p46+, lysosomal-associated membrane protein (DC-LAMP)+, CD20+, and CD8+ cells in the tumor nest of patients with HGSC (study group 1). The correlation coefficient is displayed. (D) Overall survival (OS) of 50 patients with HGSC from study group 1 on stratification based on the median density of M2-like TAMs in P-TME and M-TME. Survival curves were estimated by the Kaplan-Meier method, and differences between groups were evaluated using log-rank test. Number of patients at risk and p values are reported. (E) Hierarchical clustering of 1016 transcripts that were significantly changed in 12 M2Hi M-TME patients as compared with their 12 M2Lo M-TME counterparts from study group 1, as determined by RNA sequencing. (F) Relative expression levels of immune cytolytic activity (CYT) index based on transcript levels of granzyme A (GZMA) and perforin (PRF1) in 24 M2Lo and 24 M2Hi P-TME and M-TME samples of HGSC from study group 1, as determined by RNA sequencing. (G) OS of 50 patients with HGSC from study group 1 on stratification based on the median density of M2-like TAMs and immune CYT index. Survival curves were estimated by Kaplan-Meier method, and difference between groups were evaluated suing the log-rank test. Number of patients at risk and p values are reported. ns, not significant.
Table 3.
Multivariate Cox proportional hazard analysis
| Variable | Overall survival | |
| HR (95% Cl) | P value | |
| M2-like TAMs (M-TME) | 10.85 (2.28 to 51.44) | 0.002 |
| DC-LAMP (P-TME) | 0.32 (0.14 to 0.73) | 0.006 |
DC-LAMP, lysosomal-associated membrane protein; M-TME, metastatic tumor microenvironment; P-TME, primary tumor microenvironment; TAMs, tumor-associated macrophages.
Finally, we investigated the potential role of other immunosuppressive networks, including the networks orchestrated by transforming growth factor beta 1 (best known as TGF-β1),28 cancer-associated fibroblasts (CAFs),30 and endothelial cells. Harnessing our transcriptomic dataset from study group 1, we observed a significant increase in DEGs associated with TGF-β and CAF signatures in the M-TME of patients with HGSC as compared with the P-TME (p=0.001, p=0.0002, respectively) (online supplementary figure 5A). Conversely, the transcriptomic signature of endothelial cells did not differ between the M-TME and P-TME. Moreover, we observed a strong correlation between the TGF-β (and CAF) signature and signatures of MDSCs and TREG cells in M-TME (but much less so) in the P-TME of HGSCs (online supplementary figure 5B). However, neither the TGF-β nor the CAF signature in the M-TME influenced OS in our patient cohort based on univariate Cox analysis (online supplementary table 5). Conversely, a signature of M2-like macrophages in the M-TME was associated with an increased risk for death (HR=56.85, 95% CI 1.41 to 2291.5; p=0.032) (online supplementary table 5).
Discussion
Despite recent therapeutic advances, ovarian cancer still represents the most lethal of gynecological malignancies, mainly reflecting presentation at advanced disease stage with large tumor burden and extensive peritoneal involvement.12 31 Both immunological and non-immunological components of the TME have been attributed prognostic and predictive value in patients with HGSC.13 20 26 27 32–36 In most cases, however, attention has been focused on the P-TME, largely reflecting sample availability from diagnostic or surgical procedures.37 38 However, prognosis is largely determined by the establishment of metastatic sites with a complex TME.10 Thus, there is a strong need to improve our understanding of the different components of the M-TME and how they impact the tumor progression and disease outcome.
Here, we compared the transcriptomic profile of 24 paired primary and metastatic tumor samples from patients with HGSC who did not receive neoadjuvant chemotherapy. Enrichment analysis identified two gene clusters associated with extracellular matrix organization and immune responses that were over-represented in M-TME versus P-TME samples (figure 1). Using functional and immunohistochemical analyses, we confirmed that the M-TME contains a higher amount of immune cells than the P-TME (figures 1 and 2), but the density of CD8+ T cells, CD20+ B cells, and NKp46+ NK cells in the M-TME largely failed to affect OS, with the only exception of DC-LAMP+ DCs on OS (figure 2). This is at odds with the well-established prognostic role of these cell populations in the P-TME.20 27 35 39
We therefore postulated that immune effectors are functionally impaired in the M-TME, but we could not attribute such an impairment to PD-1, LAG-3, or TIM-3 signaling (figure 3), despite our previous findings on the key role of the latter in the P-TME.13 These findings are in line with the poor clinical activity of immune checkpoint blockers in patients with late-stage HGSC.40 Conversely, we identified several genes involved in cytokine/chemokine signaling by M2-like TAMs as potential drivers of T-cell exhaustion in the M-TME, including IL-10, CCL22, and components of the IL-4 and IL-13 signaling pathways.33 34 Consistent with this, we found that the M-TME contains a higher amount of M2-like TAMs than the P-TME (figure 4). Moreover, patients with HGSC with robust M-TME infiltration by M2-like TAMs exhibited inhibited immune responses at metastatic sites, correlating with poor disease outcome (figure 4). These findings are in line with previous preclinical data showing that M2-like TAMs facilitate the metastatic dissemination of ovarian cancer cells,41 42 as well as with clinical data correlating the abundance of immunosuppressive M2-like TAMs with poor disease outcome in a variety of tumors.43–45 That said, TAMs are a highly plastic population of the immunological TME, and their dichotomic classification in immunostimulatory (M1) versus immunosuppressive (M2) fails to recapitulate such a functional plasticity.16 17 Instead, TAMs are expected to exist along a continuum of phenotypic and functional states that continuously evolve during disease progression. Thus, it will be interesting to determine how the TAM compartment of HGSC evolves over space (at different regions of the tumor) and time (along with progression and response to treatment).
Irrespective of these and other unknowns, our present findings demonstrate that the microenvironment of primary and metastatic HGSC lesions from the same patient differ from each other with respect to immunological competence and prognostic impact, as they delineate for the first time the negative impact of M2-like TAMs on the establishment of clinically relevant anticancer immune responses at the M-TME of patients with HGSCs. We have recently demonstrated that TIM-3 is the main coinhibitory receptor that supports clinically relevant immunosuppression in patients with HGSC,13 and a link between immunosuppressive TAMs expressing galectin 9 (LGALS9) and exhausted PD-1+TIM-3+ CD8+ T cells has been documented in patients with muscle-invasive bladder cancer.46 Thus, it is tempting to speculate that patients with metastatic HGSC may benefit from dual PD-1/TIM-3 blockage, potentially in the context of chemotherapeutic regimens that deplete M2-like TAMs.47 Clinical trials dissecting these possibilities are urgently awaited.
Footnotes
Contributors: Concept and design: MH, PS, JL, LG, RS, JF; development of methodology: MH, LK, KF, IT, JF; acquisition of data: MH, LK, JR, PS, JL, TL, LP, IT, SV, IM, IP, VD, MR, TB, JP, VB, LS, JF; analysis and interpretation of data: MH, LK, KF, JR, LP, IT, JF; writing, review, and/or revision of the manuscript: MH, LK, WHF, CS-F, LG, RS, JF; study supervision: MH, LG, RS, JF.
Funding: This study was sponsored by Sotio, Prague. JL and AR were supported by program PROGRES Q40/11, by the project BBMRI-CZ LM2015089 and by the European Regional Development Fund-Project BBMRI-CZ: Biobank network—a versatile platform for research on the etiopathogenesis of diseases, No: EF16 013/0001674. LG is supported by a Breakthrough Level 2 grant from the US Department of Defense, Breast Cancer Research Program (no: BC180476P1), by the 2019 Laura Ziskin Prize in Translational Research (no: ZP-6177, Principal Investigator (PI): Formenti) from the Stand Up to Cancer, by a Mantle Cell Lymphoma Research Initiative (PI: Chen-Kiang) grant from the Leukemia and Lymphoma Society, by a start-up grant from the Department of Radiation Oncology at Weill Cornell Medicine (New York, USA), by a Rapid Response Grant from the Functional Genomics Initiative (New York, USA), by industrial collaborations with Lytix (Oslo, Norway) and Phosplatin (New York, USA), and by donations from Phosplatin (New York, USA), the Luke Heller TECPR2 Foundation (Boston, USA) and Sotio a.s. (Prague, Czech Republic). KF was supported by Ministry of Health of the Czech Republic, grant no: NV18-08-00385.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: The protocol was approved by the local ethical committee (201 607 S14P).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as supplementary information. Not applicable.
References
- 1.Fridman WH, Zitvogel L, Sautès-Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717–34. 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 3.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321–30. 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 4.Maley CC, Aktipis A, Graham TA, et al. Classifying the evolutionary and ecological features of neoplasms. Nat Rev Cancer 2017;17:605–19. 10.1038/nrc.2017.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell 2011;147:275–92. 10.1016/j.cell.2011.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giraldo NA, Becht E, Pagès F, et al. Orchestration and prognostic significance of immune checkpoints in the microenvironment of primary and metastatic renal cell cancer. Clin Cancer Res 2015;21:3031–40. 10.1158/1078-0432.CCR-14-2926 [DOI] [PubMed] [Google Scholar]
- 7.Remark R, Alifano M, Cremer I, et al. Characteristics and clinical impacts of the immune environments in colorectal and renal cell carcinoma lung metastases: influence of tumor origin. Clin Cancer Res 2013;19:4079–91. 10.1158/1078-0432.CCR-12-3847 [DOI] [PubMed] [Google Scholar]
- 8.Tanchot C, Terme M, Pere H, et al. Tumor-infiltrating regulatory T cells: phenotype, role, mechanism of expansion in situ and clinical significance. Cancer Microenviron 2013;6:147–57. 10.1007/s12307-012-0122-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torre LA, Siegel RL, Ward EM, et al. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev 2016;25:16–27. 10.1158/1055-9965.EPI-15-0578 [DOI] [PubMed] [Google Scholar]
- 10.Lengyel E. Ovarian cancer development and metastasis. Am J Pathol 2010;177:1053–64. 10.2353/ajpath.2010.100105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nieman KM, Kenny HA, Penicka CV, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med 2011;17:1498–503. 10.1038/nm.2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423–37. 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fucikova J, Rakova J, Hensler M, et al. TIM-3 dictates functional orientation of the immune infiltrate in ovarian cancer. Clin Cancer Res 2019;25:4820–31. 10.1158/1078-0432.CCR-18-4175 [DOI] [PubMed] [Google Scholar]
- 14.Kasikova L, Hensler M, Truxova I, et al. Calreticulin exposure correlates with robust adaptive antitumor immunity and favorable prognosis in ovarian carcinoma patients. J Immunother Cancer 2019;7:312. 10.1186/s40425-019-0781-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bantug GR, Galluzzi L, Kroemer G, et al. The spectrum of T cell metabolism in health and disease. Nat Rev Immunol 2018;18:19–34. 10.1038/nri.2017.99 [DOI] [PubMed] [Google Scholar]
- 16.Vitale I, Manic G, Coussens LM, et al. Macrophages and metabolism in the tumor microenvironment. Cell Metab 2019;30:36–50. 10.1016/j.cmet.2019.06.001 [DOI] [PubMed] [Google Scholar]
- 17.Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov 2018;17:887–904. 10.1038/nrd.2018.169 [DOI] [PubMed] [Google Scholar]
- 18.Meinhold-Heerlein I, Fotopoulou C, Harter P, et al. The new WHO classification of ovarian, fallopian tube, and primary peritoneal cancer and its clinical implications. Arch Gynecol Obstet 2016;293:695–700. 10.1007/s00404-016-4035-8 [DOI] [PubMed] [Google Scholar]
- 19.O'Sullivan B, Brierley J, Byrd D, et al. The TNM classification of malignant tumours-towards common understanding and reasonable expectations. Lancet Oncol 2017;18:849–51. 10.1016/S1470-2045(17)30438-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truxova I, Kasikova L, Hensler M, et al. Mature dendritic cells correlate with favorable immune infiltrate and improved prognosis in ovarian carcinoma patients. J Immunother Cancer 2018;6:139. 10.1186/s40425-018-0446-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goc J, Germain C, Vo-Bourgais TKD, et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res 2014;74:705–15. 10.1158/0008-5472.CAN-13-1342 [DOI] [PubMed] [Google Scholar]
- 22.Solomon B, Young RJ, Bressel M, et al. Prognostic Significance of PD-L1 + and CD8 + Immune Cells in HPV + Oropharyngeal Squamous Cell Carcinoma. Cancer Immunol Res 2018;6:295–304. 10.1158/2326-6066.CIR-17-0299 [DOI] [PubMed] [Google Scholar]
- 23.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 24.Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angelova M, Charoentong P, Hackl H, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol 2015;16:64. 10.1186/s13059-015-0620-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milne K, Köbel M, Kalloger SE, et al. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS One 2009;4:e6412. 10.1371/journal.pone.0006412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Conejo-Garcia JR, Katsaros D, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med 2003;348:203–13. 10.1056/NEJMoa020177 [DOI] [PubMed] [Google Scholar]
- 28.Galluzzi L, Chan TA, Kroemer G, et al. The hallmarks of successful anticancer immunotherapy. Sci Transl Med 2018;10. 10.1126/scitranslmed.aat7807. [Epub ahead of print: 19 Sep 2018]. [DOI] [PubMed] [Google Scholar]
- 29.Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitale I, Manic G, Galassi C, et al. Stress responses in stromal cells and tumor homeostasis. Pharmacol Ther 2019;200:55–68. 10.1016/j.pharmthera.2019.04.004 [DOI] [PubMed] [Google Scholar]
- 31.Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284–96. 10.3322/caac.21456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senovilla L, Vacchelli E, Galon J, et al. Trial watch: prognostic and predictive value of the immune infiltrate in cancer. Oncoimmunology 2012;1:1323–43. 10.4161/onci.22009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnett JC, Bean SM, Whitaker RS, et al. Ovarian cancer tumor infiltrating T-regulatory (T(reg)) cells are associated with a metastatic phenotype. Gynecol Oncol 2010;116:556–62. 10.1016/j.ygyno.2009.11.020 [DOI] [PubMed] [Google Scholar]
- 34.Curiel TJ, Coukos G, Zou L, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004;10:942–9. 10.1038/nm1093 [DOI] [PubMed] [Google Scholar]
- 35.Hwang W-T, Adams SF, Tahirovic E, et al. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol Oncol 2012;124:192–8. 10.1016/j.ygyno.2011.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen JS, Sahota RA, Milne K, et al. CD20+ tumor-infiltrating lymphocytes have an atypical CD27- memory phenotype and together with CD8+ T cells promote favorable prognosis in ovarian cancer. Clin Cancer Res 2012;18:3281–92. 10.1158/1078-0432.CCR-12-0234 [DOI] [PubMed] [Google Scholar]
- 37.Bonome T, Levine DA, Shih J, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res 2008;68:5478–86. 10.1158/0008-5472.CAN-07-6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshihara K, Tajima A, Yahata T, et al. Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. PLoS One 2010;5:e9615. 10.1371/journal.pone.0009615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson BH. CD20+ B cells: the other tumor-infiltrating lymphocytes. J Immunol 2010;185:4977–82. 10.4049/jimmunol.1001323 [DOI] [PubMed] [Google Scholar]
- 40.Hamanishi J, Mandai M, Konishi I. Immune checkpoint inhibition in ovarian cancer. Int Immunol 2016;28:339–48. 10.1093/intimm/dxw020 [DOI] [PubMed] [Google Scholar]
- 41.Robinson-Smith TM, Isaacsohn I, Mercer CA, et al. Macrophages mediate inflammation-enhanced metastasis of ovarian tumors in mice. Cancer Res 2007;67:5708–16. 10.1158/0008-5472.CAN-06-4375 [DOI] [PubMed] [Google Scholar]
- 42.Yin M, Li X, Tan S, et al. Tumor-associated macrophages drive spheroid formation during early transcoelomic metastasis of ovarian cancer. J Clin Invest 2016;126:4157–73. 10.1172/JCI87252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimura S, Yang G, Ebara S, et al. Reduced infiltration of tumor-associated macrophages in human prostate cancer: association with cancer progression. Cancer Res 2000;60:5857–61. [PubMed] [Google Scholar]
- 44.Medrek C, Pontén F, Jirström K, et al. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012;12:306. 10.1186/1471-2407-12-306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrera M, Herrera A, Domínguez G, et al. Cancer-associated fibroblast and M2 macrophage markers together predict outcome in colorectal cancer patients. Cancer Sci 2013;104:437–44. 10.1111/cas.12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi Y, Chang Y, Wang Z, et al. Tumor-associated macrophages expressing galectin-9 identify immunoevasive subtype muscle-invasive bladder cancer with poor prognosis but favorable adjuvant chemotherapeutic response. Cancer Immunol Immunother 2019;68:2067–80. 10.1007/s00262-019-02429-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galluzzi L, Humeau J, Buqué A, et al. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat Rev Clin Oncol. In Press 2020;136 10.1038/s41571-020-0413-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-000979supp001.pdf (1.3MB, pdf)