Abstract
Osteoclasts are multinuclear cells that resorb bone. Osteoclast differentiation is regulated by multiple transcription factors which may be acting in a single or multiple factor complex to regulate gene expression. Myocyte enhancer factor 2 (MEF2) is a family of transcription factors whose role during osteoclast differentiation has not been well characterized. Because MEF2A and MEF2D are the family members most highly expressed during osteoclast differentiation, we created conditional knockout mice models for MEF2A and/or MEF2D. In vitro cultures of A- and D-KO osteoclasts were smaller and less numerous than wild type cultures, while AD-KO osteoclasts were almost completely devoid of TRAP positive mononuclear cells. Female A-KO mice are osteopetrotic while male A- and D-KO mice of either sex had no significant in vivo skeletal phenotype, suggesting a sex-specific regulation of osteoclasts by MEF2A. Lastly, in vivo male AD-KO mice are osteopenic, indicating while MEF2 is required for M-CSF and RANKL-stimulated osteoclastogenesis in vitro, osteoclasts can form in the absence of MEF2 in vivo via an RANKL-alternative pathway.
Keywords: Transcription factors, osteoclasts, MEF2, sex specific, HDACs, estrogen
1. Introduction
Bone is an ever-changing organ that is remodeled through tight coupling of bone resorption followed by formation of new bone [1–3]. These processes are performed by bone resorbing osteoclasts and bone forming osteoblasts. Since osteoclasts are a perpetrator of many skeletal diseases, understanding the mechanisms that regulate their activity during the bone remodeling process is necessary [4, 5]. The dynamic and responsive nature of bone during the remodeling process requires temporal changes in gene expression within the osteoclast lineage [6]. Combinations of transcription factors and co-factors binding to DNA sequences plays an important role in chromatin remodeling and cellular signaling events in regulating osteoclasts, thus impacting bone remodeling. Understanding how these molecular switches work has become an important mechanism to consider in regard to developing targeted bone therapies.
Previous work from various labs identified histone deacetylase 7 (HDAC7) as a negative regulator of osteoclast differentiation [7–9]. Deletion of HDAC7 in the osteoclast lineage enhanced osteoclast differentiation and activity, which led to decreased bone mass in mice at three and six months of age [7, 9]. While continuing our investigation into HDAC7’s inhibitory role in osteoclast development, we discovered that overexpression of a short N-terminal fragment of HDAC7 could repress differentiation to a similar degree as full-length HDAC7. This N-terminal fragment contained a motif known to bind to and repress myocyte enhancer factor 2 (MEF2) transcription factors [10]. HDACs often associate with and inhibit the activity of MEF2 in other cell types in order to block differentiation [11–13]. MEF2A and MEF2D promote terminal differentiation of monocytes into macrophages [14]. MEF2 has only been superficially characterized in osteoclasts [15]. For these reasons we investigated the importance of MEF2 in osteoclast differentiation using a conditional mouse model.
Our work presented here demonstrates MEF2A and MEF2D have essential roles in promoting osteoclast development. Primary bone marrow macrophages (BMMs) from Mef2afl/fl; c-Fms-Cre+/Tg (A-KO) and Mef2dfl/fl; c-Fms-Cre+/Tg (D-KO) mice were differentiated into osteoclasts in vitro. A-KO and D-KO osteoclasts showed reductions in osteoclast size, number, and activity, which were also accompanied by lowered levels of NFATc1. Three-month old male A-KO and D-KO mice presented no overt skeletal phenotype compared to wild-type (WT) mice by micro-CT, mechanical testing of bones, and histology, while A-KO female mice presented the expected osteopetrotic phenotype. Due to potential redundancy among the MEF2 family and the commonality of MEF2 heterodimers [14, 15], we bred Mef2afl/fl; Mef2dfl/fl; c-Fms-Cre+/Tg (AD-KO) mice. Primary BMMs from AD-KO mice could not develop into multinuclear osteoclasts and showed dramatic reductions in osteoclast markers. Surprisingly, three-month old AD-KO male mice had reduced BV/TV as well as cortical thickness. Our results indicate that deletion of both MEF2A and MEF2D in osteoclasts produces distinct phenotypes in vitro versus in vivo, and a RANKL independent mechanism of osteoclast differentiation in vivo.
2. Material and Methods
2.1. Ethics
The use and care of the mice was reviewed and approved by the University of Minnesota Institutional Animal Care and Use Committee, IACUC protocol number 180636053A. Euthanasia was performed by CO2 inhalation.
2.2. Mice
Mef2afl/fl; Mef2dfl/fl mice in a C57Bl/6 background were obtained from Dr. Michael Greenberg (Harvard University, [16, 17]. Mef2afl/fl; Mef2dfl/fl mice were bred with cFms-Cre+/Tg mice (Jackson Laboratory) in a FVB/NJ background, and their progeny were crossed to separate the Mef2afl/fl and Mef2dfl/fl alleles for individual analysis and to obtain Mef2afl/fl; cFms-Cre+/Tg (A-KO), Mef2dfl/fl; cFms-Cre+/Tg (D-KO), Mef2afl/fl (A-WT), or Mef2dfl/fl (D-WT) mice for continued breeding and experiments. MEF2A and MEF2D protein depletion was verified by Western blot. Mef2afl/fl; Mef2dfl/fl (AD-WT) and Mef2afl/fl; Mef2dfl/fl; cFms-Cre+/Tg (AD-KO) mice were obtained from backcrossing progeny from Mef2dfl/fl; cFms-Cre+/Tg and Mef2afl/fl; cFms-Cre+/+ mice.
2.3. In vitro analysis
2.3.1. Primary Osteoclast Cell Culture
BMMs were harvested from femora and tibiae of both male and female mice and differentiated into osteoclasts as done previously with minor modifications [7]. Beginning two days after re-plating BMMs (referred to as day 0), cells were fed 1% CMG 14–12 supernatant (Dr. Sunao Takeshita, Nagoya City University, Nagoya, Japan) containing M-CSF and 10 ng/ml RANKL (R&D Systems) every 48 hours (on days 0, 2, 4, and so on) until the desired experimental end point. Fusion normally occurs on day 3.
2.3.2. Culturing of RAW 264.7 cells
RAW 264,.7 cells were plated at 100,000 cells in a well of a 6 well plate in 1ml of RAW media and 1ml of 2X RANKL media per well. 2X RANKL media is 80 ng/ml of RANKL in RAW media. Cells were incubated at 37°C for 3 days. On the third day of culture, 1 ml of media is removed from each well and 1 ml of 2X RANKL media was added to each well. Cells were incubated at 37°C for an additional 2 days.
2.3.3. Demineralization assay
BMMs were plated on 24-well Corning Osteoassay surface plates at a density of 100,000 cells per well and fed as described above. At day 4–6 cells were washed away with 10% bleach as per manufacturer’s recommendations. Wells were imaged by light microscopy, and demineralization measurements were determined using NIH ImageJ.
2.3.4. Tartrate-resistant acid phosphatase (TRAP) and DAPI staining
Cells were cultured to the desired time point and fixed in 4% paraformaldehyde for 20 minutes at 4°C and then washed in PBS. Nuclei were stained using DAPI for five minutes at room temperature. The DAPI stain was then replaced with PBS. TRAP activity was stained using Naphthol AS-MX phosphate and Fast Violet LB salt. Cells were imaged using cellSens software (Olympus) and analyzed using NIH ImageJ.
2.3.5. RNA extraction and analysis
RNA was isolated from cells plated in triplicate using Trizol reagent (Ambion, Life Technologies) and quantified using UV spectroscopy. 1 μg of RNA was used to synthesize cDNA with the iScript cDNA synthesis kit (Bio-Rad) as per the manufacturer’s protocol. Quantitative real-time PCR (RT-qPCR) was performed using the MyiQ Single Color Real-Time PCR Detection System (Bio-Rad). Each 20 μl reaction contained 1 ul of cDNA, 10 μl of iTaq Universal SYBR Green Supermix, and 25 μM forward and reverse primers. The PCR conditions were as follows: 95°C for 3 minutes, and the 40 cycles of 94°C for 15 seconds, 58°C for 30 seconds and 72°C for 30 seconds, followed by melting curve analysis (95°C for 5 sec, 65°C for 5 sec and then 65°C to 95°C with 0.5°C increase every 5 seconds). Experimental genes were normalized to Hprt. All measurements were performed in triplicate and analyzed using the ΔΔCT method. A list of primers is included in Table 1.
Table 1.
qPCR primer sequences.
| Primer Name | 5'-3' Sequence |
|---|---|
| Acp5 qPCR F | CGT CTC TGC ACA GAT TGC A |
| Acp5 qPCR R | GAG TTG CCA CAC AGC ATC AC |
| Atp6v0d2 qPCR F | TCA GAT CTC TTC AAG GCT GTG CTG |
| Atp6v0d2 qPCR R | GTG CCA AAT GAG TTC AGA GTG ATG |
| Ctsk qPCR F | AGG GAA GCA AGC ACT GGA TA |
| Ctsk qPCR R | GCT GGC TGG AAT CAC ATC TT |
| Dcstamp qPCR F | CAG ACT CCC AAA TGC TGG AT |
| Dcstamp qPCR R | CTT GTG GAG GAA CCT AAG CG |
| Hprt qPCR F | GAG GAG TCC TGT TGA TGT TGC CAG |
| Hprt qPCR R | GGC TGG CCT ATA GGC TCA TAG TGC |
| Nfatc1 qPCR F | TCA TCC TGT CCA ACA CCA AA |
| Nfatc1 qPCR R | TCA CCC TGG TGT TCT TCC TC |
| Mef2a qPCR F | GTG TAC TCA GCA ATG CCG AC |
| Mef2a qPCR R | AAC CCT GAG ATA ACT GCC CTC |
| Mef2b qPCR F | CAT CAG GCT CCA CAG ATT GC |
| Mef2b qPCR R | GAC TTGATG CTG ACC GGA GG |
| Mef2c qPCR F | AAG AAA CAC GGG GAC TAT GG |
| Mef2c qPCR R | ACA GCT TGT TGG TGC TGT TG |
| Mef2d qPCR F | GAG AAG ATG GGG AGG AAA AAG |
| Mef2d qPCR R | GCC TTC TTC ATC AGT CCA AAC |
2.3.6. Immunoblotting
Protein cell lysates were harvested from primary osteoclasts in modified RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% IGEPAL, 0.25% sodium deoxycholate, 1 mM EDTA) supplemented with Halt Protease & Phosphatase Inhibitor Cocktail (Thermo Scientific). Lysates were cleared by centrifugation. Proteins were resolved by SDS-PAGE, transferred to PVDF membrane (Millipore). Blots were blocked in TBS/0.1% Tween-20 (TBST) plus 5% nonfat dry milk then incubated at 4°C overnight with primary antibodies diluted in TBST plus 5% bovine serum albumin. Primary antibodies used are included in Table 2. The next day, blots were washed with TBST, and then they were incubated for 1 hour at room temperature with horseradish-peroxidase conjugated secondary antibodies diluted in TBST plus 5% nonfat dry milk. Secondary antibodies used were from G.E. Health Systems: Amersham ECL anti-mouse (NA-931) and anti-rabbit (NA-934) at 1:6000, or Santa Cruz: anti-goat (SC-2020) at 1:6,000 dilution. Blots were then washed in TBST again before antibody binding was detected using a western blotting detection kit (Western Bright Quantum, Advansta) and the ChemiDoc Imaging System (Bio-Rad). Histone 3 (H3) or actin was used as a loading control for all blots.
Table 2.
Antibodies used for immunoblotting.
| Antibody Target | Host Species | Vendor | Catalog Number | Lot Number |
|---|---|---|---|---|
| c-Fos | Rabbit | CST | 2250S | 9 |
| c-Jun | Rabbit | CST | 9165S | 11 |
| C/EBPα | Rabbit | CST | 8178S | 3 |
| CREB | Rabbit | CST | 9197S | 16 |
| H3 | Rabbit | CST | 2650S | 3 |
| MEF2A | Rabbit | Abcam | 227120 | GR3210298–1 |
| MEF2D | Rabbit | Abcam | 32845 | GR3176036–1 |
| MITF | Rabbit | Sigma | HPA003259 | B57783 |
| NFATc1 | Mouse | Santa Cruz | 7294 | H0508 |
| NFATc2 | Rabbit | CST | 5861T | 4 |
| HA-Tag | Rabbit | CST | C29F4 | |
| FLAG-Tag | Rabbit | Sigma | A2220 | |
| PU.1 | Rabbit | Santa Cruz | 352 | D1113 |
2.3.7. Co-immunoprecipitations of PU.1 and HDAC7
HEK 293T cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% bovine calf serum, 2% L-glutamine and 0.5% penicillin/streptomycin. Cells were seeded in 60 mm plates (Corning) at 1.3X104 cells/cm2 density and transfected by Lipofectamine Plus Reagent (Invitrogen) to overexpress the indicated HA-tagged PU.1 and/or FLAG-tagged HDAC7constructs. 5 μg of each plasmid were transfected using 12 μL of Lipofectamine and 8 μL of Plus reagents per dish according to the manufacturer’s recommended protocol. Twenty-four hours after transfection, cells were harvested in NP-40 lysis buffer (20 mMTris, pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40 and protease and phosphatase inhibitors). Extracts were incubated with target antibody and Protein A/G beads (Pierce) overnight at 4°C. The following day, immunoprecipitates were precipitated and washed three times in lysis buffer. Bound proteins were resuspended in sample buffer and resolved by SDS-PAGE.
2.3.8. HDAC7 Lentiviral Constructs
Green fluorescent protein (GFP) or HDAC7 fragments were cloned into pLex-MCS (Open Biosystems) using the NotI and XhoI sites and replication defective lentiviruses were produced using Trans-lentiviral ORF packaging mix (Dharmacon). BMMs were isolated as described above. Prior to stimulation with RANKL, the cells were incubated with lentivirus expressing either GFP or HDAC7 fragment at 37°C in the presence of CMG14–12 supernatant containing M-CSF. After 24 hours, medium containing the lentivirus was removed, and cells were fed with 1% CMG 14–12 culture supernatant and RANKL (10 ng/ml). On day five, cells were fixed in 4% paraformaldehyde and stained for TRAP as described in the above section.
2.3.9. Measurement of Apoptosis
Caspase-Glo 3/7 (Promega) was used to evaluate steady-state apoptosis in in vitro cultures as per the manufacturer’s protocol with minor changes. Cells were seeded at 50,000 cells per well in 48-well plates in quadruplicate. On the indicated day, room temperature Caspase-Glo 3/7 reagent was added to each well to lyse cells for 30 minutes at room temperature. After lysis, caspase activity for each sample was measured using the GloMax20/20 Luminometer (Promega). Since Caspase-Glo 3/7 activity is highly dependent on the number of cells in each reaction (see manufacturer’s datasheet), the average nuclei number per culture was assessed in additional cells plated and cultured in parallel. Graphed data represents Caspase-Glo3/7 activity normalized to the average number of nuclei in each culture condition.
2.4. In vivo analysis
2.4.1. Sample Harvest
One- and three-month old mice were used for in vivo experiments. Upon euthanasia, whole blood from the heart was drawn and centrifuged to obtain serum, which was immediately frozen until use. Femora and tibiae were removed and de-fleshed. For each mouse the right femur and tibia were immediately stored in PBS and frozen without fixation for micro-CT and mechanical testing. The left tibia was fixed in Z-fix (Anatech LTD) and decalcified in 10% EDTA (pH 7.4) for paraffin-embedded sectioning and histological staining. The left femur was used to harvest BMMs for in vitro culture. Lastly, a piece of the tail was cut to verify the genotype of each mouse.
2.4.2. Micro-CT Analysis
Frozen right femora were equilibrated to room temperature and scanned in PBS with a 1 mm aluminum filter using the XT H 225 micro-computed tomography machine (Nikon Metrology Inc., Brighton, MI, USA) at an isotropic voxel size of 7.4 μm. The scan settings were 120 kV, 61 μA, 720 projections, 2 frames per projection, and an integration time of 708 milliseconds. CT Pro 3D (Nikon metrology, Inc., Brighton, MI, USA) was used to make 3D reconstruction volumes for each scan. VGStudio MAX 3.2 (Volume Graphics GmbH, Heidelberg, Germany) was used to convert 3D reconstruction volumes to bitmap datasets for each scan. Morphometric analysis was completed with the SkyScan CT-Analyser (CTAn, Bruker micro-CT, Belgium) following Bruker’s instructions and reported guidelines for the field [18]. The region of interest for trabecular bone analysis in the distal metaphysis started 0.5 mm proximal to the growth plate and extended 1.5 mm proximally towards the diaphysis. The region for cortical bone analysis was a 0.5 mm region at the mid-diaphysis. Automated contouring was used to determine the region of interest boundaries for both trabecular and cortical bone with manual editing as needed. Global thresholding was used to segment bone from surrounding tissue for both 3D trabecular and 2D cortical analyses. One threshold value was used for all cortical analyses, and a different threshold value was used for all trabecular analyses. CT-Volume (Bruker micro-CT, Belgium) was used to create all 3D models from bitmaps corresponding to cortical and trabecular regions analyzed; however, only a 1 mm region of the most distal trabecular selection was used to create a model. Femora were re-frozen after scanning until mechanical testing was performed.
2.4.3. Staining of paraffin-embedded sections
Decalcified bone sections were de-paraffinized in xylene, rehydrated through an ethanol gradient, and stained for TRAP at 37oC for 1 hour as described above. Sections were then counterstained with methyl-green for 15 seconds, coverslipped using Permount mounting media (Electron Microscopy Sciences) and allowed to rest for 24 hours before imaging.
2.4.4. ELISA Assays
Detection of mouse CTX (IDS), TRAcP5b (IDS) and P1NP (IDS) in serum by ELISA was done following the manufacturer’s protocols. Each sample was read in duplicate and averaged before analysis.
2.4.5. Three-point bending test
The three-point bending tests were applied on the sample for assessing mechanical properties of the mice femur. Before testing, the length of each bone was measured and recorded with an electrical digital Vernier caliper. Then, the femurs were placed on the lower sample supports, which had a 10 mm span length. Pictures were taken for the placed samples to record the position and orientation of the bone. Loads were applied with a 2 mm/min loading rate using an MTS 858 Mini Bionix II testing system (Eden Prairie, MN, USA) until the fracture of the femur occurred. The recorded load-displacement curve was analyzed to determine the values of ultimate load, stiffness and elastic modulus. The ultimate load was determined as the maximum load that the femur was able to resist during the test. The stiffness was determined by calculating the slope of the linear region of the load-displacement curve. The elastic modulus was calculated based on the beam theory applied to bone mechanical properties characterization (Equation 1[19].
| Equation 1. Elastic Modulus equation. |
The minimum moment of inertia was calculated for the bone cross section at the point of fracture with the SkyScan CT-Analyser using micro-CT data from the cortical bone. To identify the cross-section used for calculation of the moment of inertia, the distance from the fracture location to each end of the bone was measured with the electrical digital Vernier caliper right after each femur was fractured.
2.5. Statistical Testing
Data presented in graphs for in vitro experiments represents an average of at least three repeated experiments performed with bone marrow cells from independent mice cultured at different times. N=4 for all in vitro DAPI-stained, TRAP-stained, and demineralization assays. N = 3 for all RT-qPCR experiments and caspase 3/7 activity. In vivo data represents all samples harvested for that specific experiment graphed together. N = 8–14 for all groups in micro-CT and mechanical testing experiments. N = 6–11 per group for ELISA assays and N = 5–9 for histology. No samples were removed as outliers in any experiment. Unpaired T-test with Welch’s correction was used when comparing only two groups. One-way ANOVA with Tukey’s multiple comparisons test was used when comparing three or more groups. All statistical testing was performed in GraphPad Prism 7 or 8.
3. Results
3.1. HDAC7 amino acids 1–178 represses osteoclast differentiation
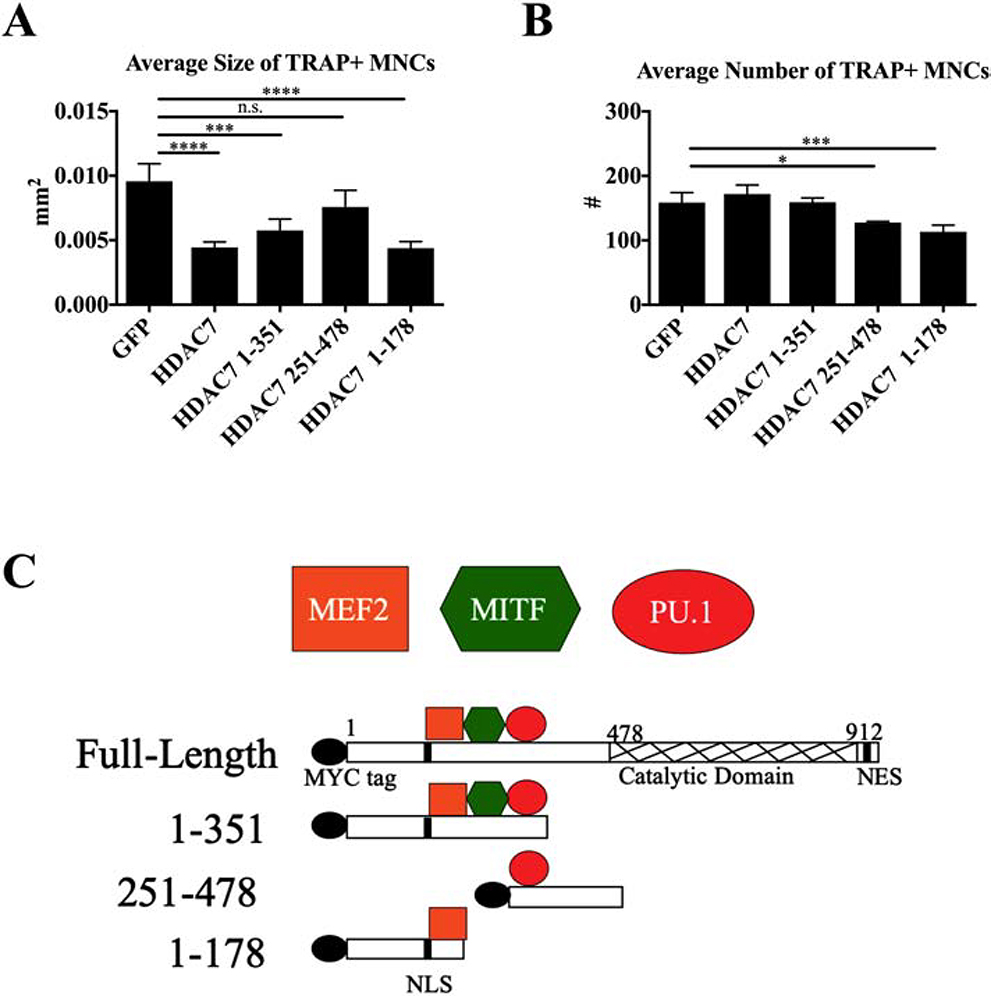
To investigate the mechanism of HDAC7’s repressive function during osteoclast differentiation, we overexpressed full-length HDAC7, three N-terminal HDAC7 fragments missing the deacetylase domain, and a GFP control construct in bone marrow macrophages (BMMs) undergoing osteoclast differentiation. Tartrate-resistant acid phosphatase (TRAP) staining was performed on osteoclasts at day four of differentiation, followed by measurements of the average size and number of TRAP-positive multi-nuclear cells (TRAP+ MNCs). As reported previously, full-length HDAC7 reduced osteoclast size by 50% compared to GFP control cells [8] (Fig. 1A, p≤0.001). Amino acids 1–351 and 1–178 of HDAC7 also reduced osteoclast size to a similar degree as full-length HDAC7, while amino acids 251–478 of HDAC7 did not reduce cell size and number by a significant degree (Fig. 1A). Amino acids 1–351 of HDAC7, but not 1–178, interact with the transcription factors PU.1 (Fig S1) and MITF in vitro [7], which suggests HDAC7’s repressive function in this context is not due to interaction with PU.1 or MITF in osteoclasts. Since efficient repression of osteoclastogenesis was seen with the 1–178 fragment but not the 251–478 fragment, we hypothesized that the 1–178 fragment contained a motif capable of interacting with and repressing a target necessary for osteoclastogenesis. HDAC7’s interaction with the myocyte enhancer factor 2 (MEF2) family has previously been mapped to amino acids 1–121, suggesting that HDAC7 might repress MEF2 activity in osteoclasts [10] (Fig. 1C).
Figure. 1. 1–178 aa of HDAC7 represses osteoclast differentiation.
(A) Average size of TRAP-positive multinuclear cells (TRAP+ MNCs) and (B) average number of TRAP+ MNCs five days after infection with control GFP or HDAC7 lentiviral constructs. (C) HDAC7 fragments showing binding regions for three transcription factors, MEF2, MITF, and PU.1. N = 4 independent experiments. Statistical testing by one-way ANOVA with Tukey’s multiple comparisons test. * P ≤ 0.05, *** P ≤ 0.001, **** P ≤ 0.0001. Significance compared to GFP control only. Bars show mean ± standard deviation (SD). n.s. = not significant.
3.2. MEF2 Expression During Osteoclast Differentiation
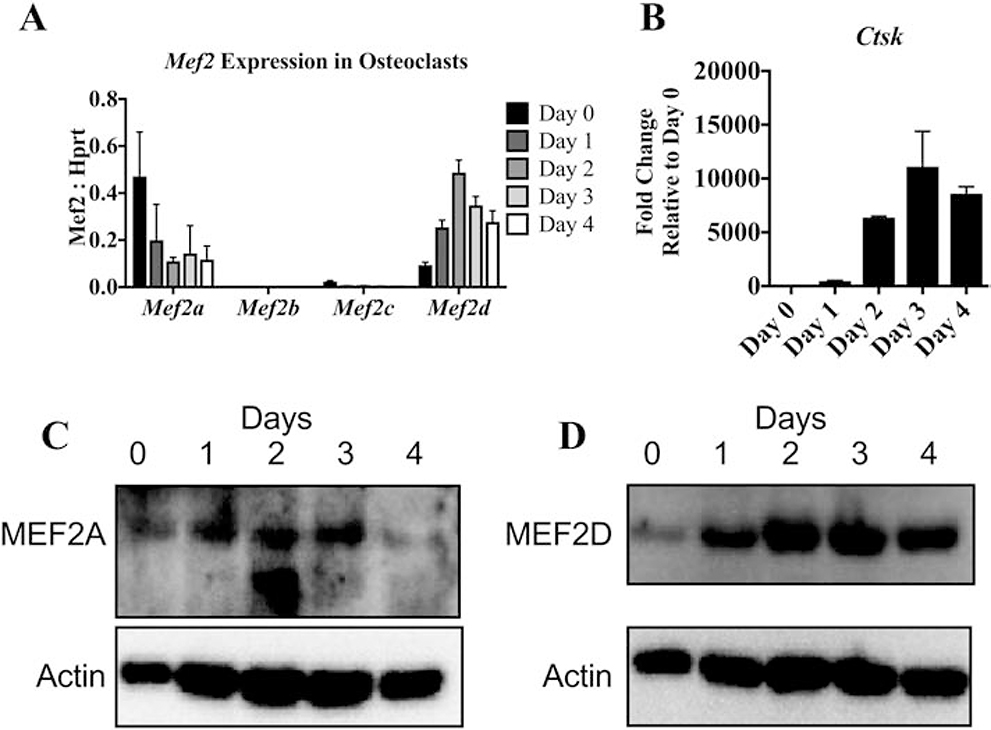
MEF2 is a family of transcription factors that consists of four proteins, MEF2A, -B, -C and -D [11, 12, 20]. As was previously done in the Feng et al. study and to begin to address the significance of MEF2 in osteoclast differentiation, we measured MEF2 expression pattern in RAW 264.7 cells [15]. In RAW 264.7 Mef2c is the mostly highly expressed family member followed by Mef2a and Mef2d. (Fig. S2A) Next, we evaluated expression of Mef2a, Mef2b, Mef2c, and Mef2d by qRT-PCR in BMMs induced to form osteoclasts. Of the four genes, Mef2a showed the highest expression in BMMs (day 0) and decreased during differentiation (Fig. 2A). Mef2d expression increased after RANKL stimulation. Mef2b was not detected, and Mef2c was expressed at very low levels. Evaluation of Ctsk expression was used to confirm that osteoclast differentiation proceeded normally in these samples (Fig. 2B). Protein levels of MEF2A and MEF2D follow similar patterns to the mRNA (Fig. 2C and D). These results indicate MEF2A, MEF2D and MEF2C are more likely than MEF2B to be involved in osteoclast differentiation.
Figure. 2. Expression of MEF2 transcription factors during osteoclast differentiation.
RT-qPCR showing relative expression of Mef2a-d (A) and fold change of Ctsk (B) in wild-type osteoclasts throughout differentiation normalized to Hprt levels. N = 3 independent experiments. Western blots showing MEF2A (C) and MEF2D (D) protein levels throughout differentiation with actin as a loading control.
3.3. Deletion of Mef2a or Mef2d compromises osteoclast development
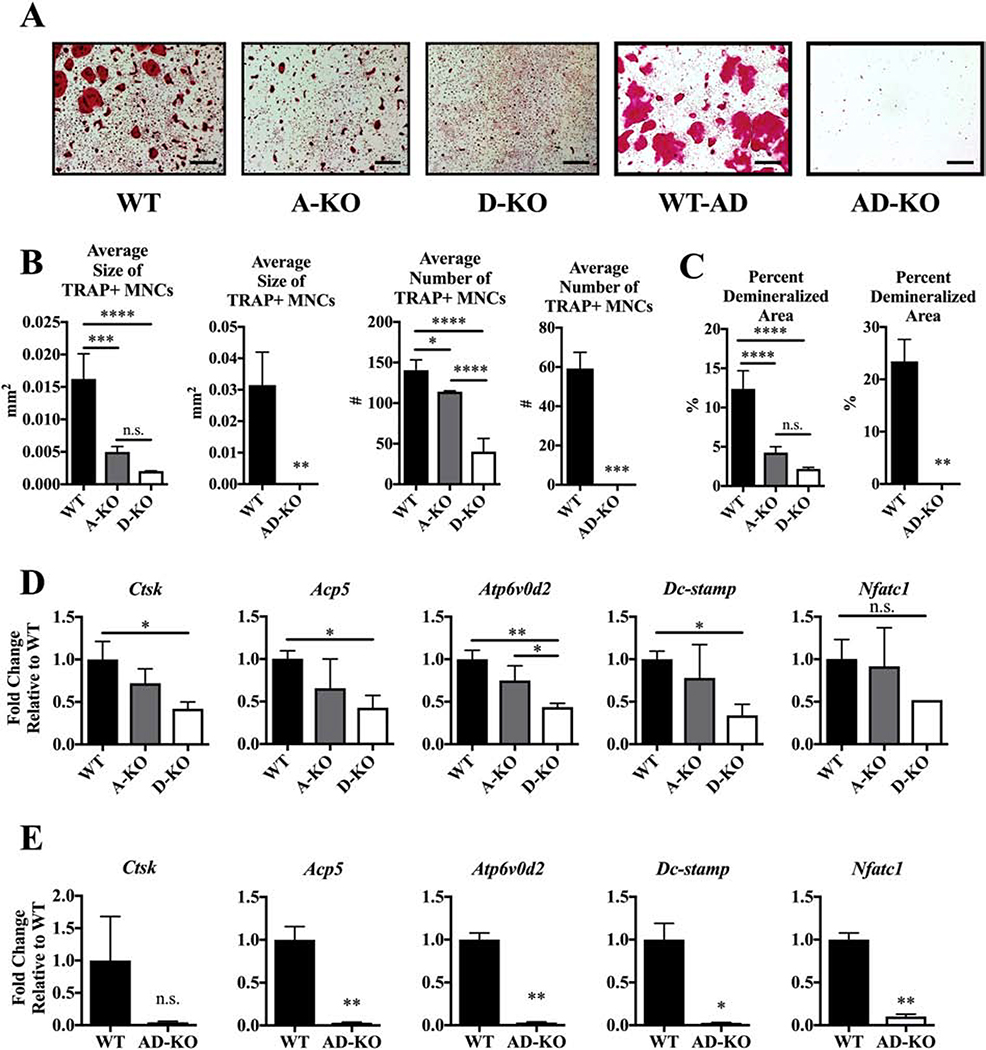
Due to the expression profile of Mef2a and Mef2d, and the lack of studies examining their importance in osteoclast development, we chose to investigate how genetic ablation of Mef2a and Mef2d in osteoclasts would impact differentiation. Mef2a-flox/flox (Mef2afl/fl) and Mef2d-flox/flox (Mef2dfl/fl) mice (obtained from Dr. Michael Greenberg, Harvard University [16, 17], were bred with c-Fms-Cre mice (c-Fms-Cre+/Tg; Jackson Laboratory,[21] to obtain Mef2afl/fl; c-Fms-Cre+/Tg (A-KO) and Mef2dfl/fl; c-Fms-Cre+/Tg (D-KO) mice and compared to Mef2afl/fl or Mef2dfl/fl (WT) mice. Depletion of MEF2A and MEF2D were confirmed by Western blot (Fig. S3A). A-KO osteoclasts were smaller and slightly less in number compared to WT osteoclasts, and D-KO osteoclasts showed a similar but more severe phenotype (Fig. 3A–B). Expectedly, A-KO and D-KO cultures showed significant reductions in the percent area demineralized on calcium phosphate osteoassay plates (Fig. 3C).
Figure. 3. MEF2A and D expression are necessary for in vitro osteoclast differentiation.
(A) Representative TRAP-stained images at day 5 after addition of RANKL. Scale bar = 0.5 mm. (B) Average size and number of TRAP+ MNCs and (C) average percent demineralized area at day 4 after RANKL stimulation. Fold change of osteoclast marker genes normalized to WT controls against Hprt (D) WT, A-KO, and D-KO cells, and (E) WT and AD-KO cells. Bone marrow cells for osteoclast cultures were obtained from both male and female mice. Bars show means ± SD. Statistical testing by one-way ANOVA with Tukey’s multiple comparisons test for WT, A-KO and D-KO comparisons. Statistical testing by unpaired T-test with Welch’s correction for WT and AD-KO comparisons. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001 vs. wild type cultures. n.s. = not significant.
Due to the high degree of amino acid sequence homology in the amino terminus between MEF2 family members, we also bred Mef2afl/fl; Mef2dfl/fl (AD-WT) and Mef2afl/fl; Mef2dfl/fl; c-Fms-Cre+/Tg (AD-KO) mice. Expression of MEF2A and MEF2D were strikingly reduced in these mice (Fig. S3B). Osteoclast differentiation assays from primary AD-KO BMMs display a striking absence of multinuclear cells and faint TRAP staining (Fig. 3A–B). Consistent with the TRAP-staining data, AD-KO cells show no activity on calcium-phosphate plates (Fig. 3C, right graphs).
To understand whether reductions in osteoclast differentiation were due to reductions in BMM numbers prior to and during differentiation, we performed DAPI staining to count total nuclei number. DAPI staining showed no difference in the number of nuclei at day 0 among the three cultures but an increased number of nuclei in D-KO cultures relative to both A-KO and WT cultures at day 3 (Fig. S4A), suggesting the reduced differentiation in D-KO cells is caused by a defect in the process of differentiation instead of a loss of viable precursor cells. The same was true for the AD-KO cultures since nuclei number by DAPI staining was consistently and significantly increased in AD-KO compared to AD-WT cultures (Fig. S4B). To further confirm the changes in osteoclast differentiation were not due to apoptosis, we performed a caspase 3/7 assay. We detected no difference in levels of caspase 3/7 activity in the A-, D- or AD-KO cultures compared to wild type at either day 1 or day 3 after RANKL stimulation (Fig S4C–D).
To further characterize changes in osteoclast differentiation after the loss of MEF2A and/or MEF2D, we evaluated expression of vital osteoclast markers by RT-qPCR. While we did not measure a significant change in osteoclast genes in A-KO culures, D-KO and AD-KO osteoclasts presented significant reductions in expression of the markers for resorption and fusion Ctsk, Atp6v0d2, Dc-stamp, and Acp5, as well as the transcription factor Nfatc1 (Fig. 3D and E).
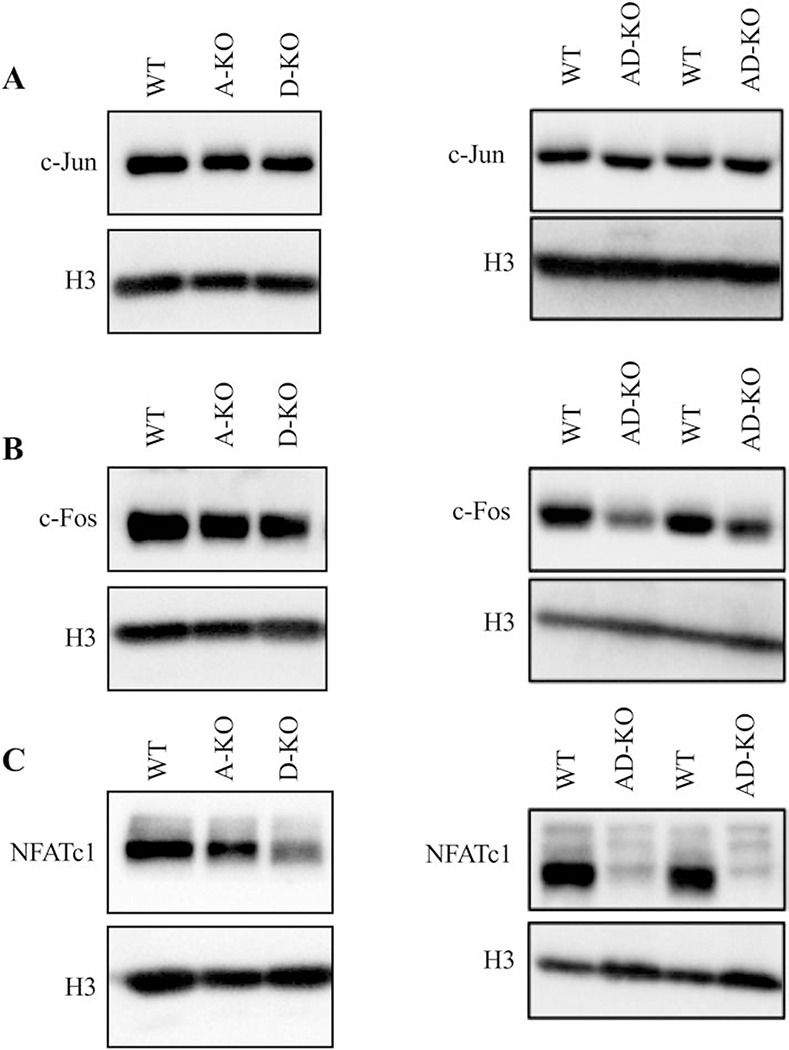
Osteoclast gene expression is regulated by a complex of multiple transcription factors that activate promoters at the appropriate times during differentiation. NFATc1 is termed the “master regulator” and is sufficient for osteoclast differentiation; however, ChIP experiments demonstrated that NFATc1 regulates osteoclast gene expression in a complex with PU.1, MITF and c-Fos [22–24]. Western blots showed D-KO and AD-KO osteoclasts had reduced protein levels of NFATc1 while A-, D- and AD-KO mice had unaffected levels of c-Jun. c-FOS was only affected in AD-KO osteoclasts (Fig. 4). Other transcription factors involved in osteoclast differentiation such as MITF, PU.1, CREB and C/EBPα had similar to increased expression levels in the AD-KO osteoclasts when compared to WT (Fig. S5). Together, these results demonstrate that deletion of Mef2a and Mef2d negatively affects osteoclast differentiation and activity by altering expression of key proteins that regulate osteoclast gene expression.
Figure. 4. AD-KO cells have reduced levels of NFATc1.
Western blots showing protein levels of (A) c-Jun, (B) c-Fos, and (C) NFATc1 with H3 as a loading control. Lysates were prepared 2 days after RANKL stimulation.
3.4. Three-month AD-KO male mice are osteopenic
Given the significant in vitro osteoclast phenotype of A-KO, D-KO and AD-KO mice, we collected long bones and serum from three-month old male WT, A-KO, D-KO and AD-KO mice for micro-CT, mechanical testing, static histology, and ELISA assays. Since the A-KO, D-KO and AD-KO mice were maintained as separate lines, each line was compared to its own WT control, designated either A-WT, D-WT, or AD-WT. Analysis of trabeculae within the metaphysis showed no change in trabecular number, trabecular spacing, or the bone volume to total volume ratio but a significant increase in trabecular thickness for D-KO versus D-WT bones (Fig. 5A and B, p≤0.05). A-KO bones showed no difference in micro-CT parameters relative to A-WT bones (Fig. 5A and B). Mechanical testing demonstrated that A- and D-KO bones were similar in all mechanical testing parameters (Fig. S6G).
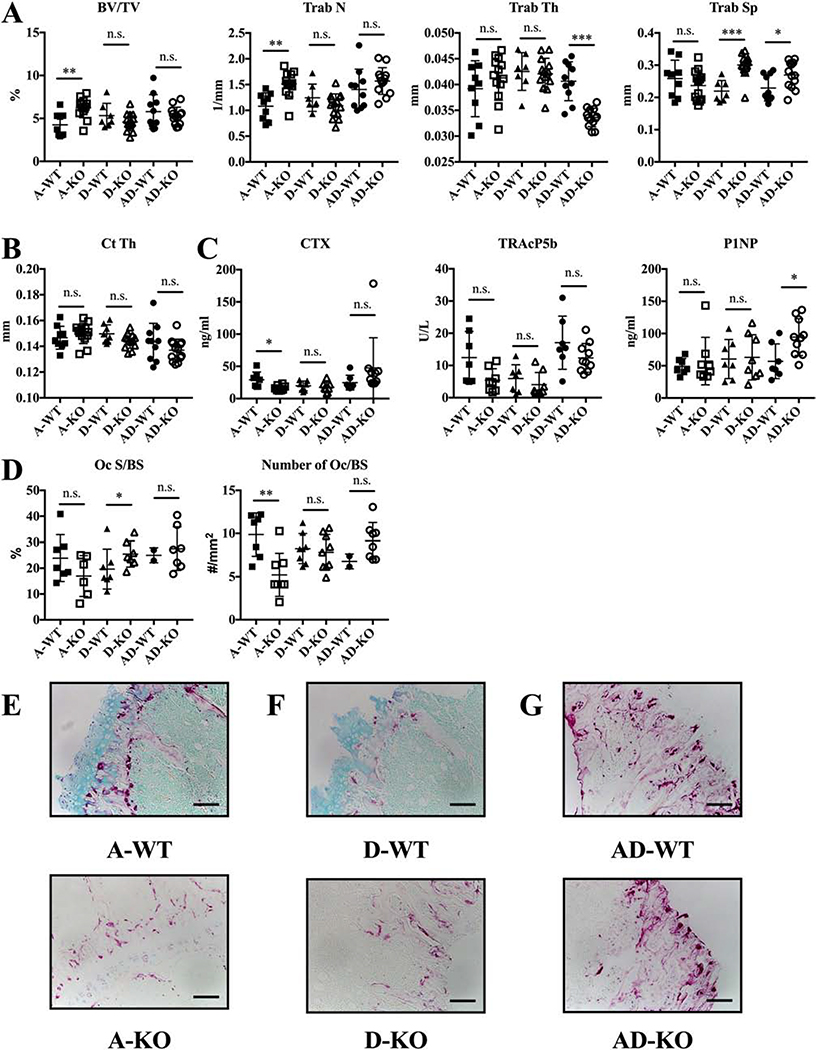
Figure. 5. Male AD-KO mice are osteopenic.
Data shown are for three-month old A-WT, A-KO, D-WT, D-KO, AD-WT, and AD-KO males only. (A) Trabecular and (B) cortical micro-CT measurements of bones. (C) ELISA assays for bone turnover markers. (D) Histological measurements of osteoclast surface area to bone surface and number to bone surface with (E, F, and G) representative images from decalcified, TRAP-stained bones for all. Scale bar = 10 mm. N = 8–12 for all groups in micro-CT and mechanical testing experiments. N = 6–11 per group for ELISA assays and N = 5–9 for histology. Bars show means ± SD. Statistical testing by unpaired T-test with Welch’s correction to compare A-WT with A-KO, D-WT with D-KO, and AD-WT with AD-KO. * P ≤ 0.05, ** P ≤ 0.01, n.s. = not significant.
Intriguingly, AD-KO male mice present with an osteopenic phenotype compared to AD-WT mice, seen as reductions of 24.5% in trabecular bone volume to total volume and 13.2% in cortical thickness in AD-KO versus AD-WT mice (Fig. 5A, p≤0.05 and B, p≤0.01). Thinner trabeculae mostly account for the decreased trabecular bone volume. Despite the osteopenic phenotype, AD-KO bones had no changes in reductions in stiffness and the elastic modulus from mechanical testing (Fig. S6G).
CTX TRAcP5b, P1NP levels were similar in A- and D-WT and A and D-KO mice (Fig. 5C). While CTX and P1NP were not changed in the AD-KO mice, TRAcP5b was significantly decreased (Fig. 5C, p≤0.05). Osteoclast surface and number, measured from TRAP-stained sectioned long bones, indicated no change in A-KO relative to A-WT osteoclast populations in vivo. However, D-KO bones compared to D-WT had significantly reduced osteoclast surface and number, which agrees with the significant increases in trabecular thickness (Fig. 5A, p≤0.05). In AD-KO mice, osteoclast surface area and osteoclast number is not significantly different from AD-WT mice. As can be seen from TRAP-stained bones (Fig. 5D and G), osteoclasts do form in vivo in AD-KO males. In sum, these results indicate the in vitro results seen in A- and D-KO cultures do not extend to three-month old male mice. Additionally, since AD-KO BMMs could not form multinuclear osteoclasts in the presence of RANKL in vitro yet AD-KO mice did have osteoclasts, we hypothesize that AD-KO BMMs require an additional cytokine besides RANKL to undergo differentiation in vitro.
3.5. Three-month female A-KO mice display osteopetrosis in vivo
Since the skeleton in males and females can be regulated differentially depending on hormones and other factors [25], we also investigated whether female A-KO, D-KO and AD-KO mice presented with the same phenotype as male mice at three months of age. Surprisingly, phenotypes for A-KO females were different from their male counterparts. Most noticeable was a significant increase in trabecular number and bone volume to total volume ratio in A-KO versus A-WT bones (Fig. 6A, p≤0.01). In agreement with the osteopetrotic phenotype, A-KO mice had reductions in serum markers for osteoclast activity (Fig. 6C, p≤0.05) and number of osteoclasts per bone surface by histology (Fig. 6D–E, p≤0.01).
Figure. 6. Female MEF2A-null mice are osteopetrotic.
Data shown are for three-month old A-WT, A-KO, D-WT, D-KO, AD-WT, and AD-KO females only. (A) Trabecular and (B) cortical micro-CT measurements of bones. (C) ELISA assays for bone turnover markers. (D) Histological measurements of osteoclast surface area to bone surface and number to bone surface with (E, F, and G) representative images from decalcified, TRAP-stained bones for all. Scale bar = 10 mm. N = 8–14 for all groups in micro-CT experiments. N = 7–8 per group for ELISA assays. N = 7–9 for histology. Statistical testing by unpaired T-test with Welch’s correction to compare A-WT with A-KO, D-WT with D-KO, and AD-WT with AD-KO. Bars show means ± SD. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001. n.s. = not significant.
D-KO female mice presented with non-significant reductions in trabecular bone volume to total volume, trabecular number, and cortical thickness (Fig. 6A and B). Interestingly, there was a large and highly significant increase in trabecular spacing in D-KO versus D-WT female mice (Fig. 6A, p≤0.001). Furthermore, serum markers of bone turnover were unchanged (Fig. 6C), but osteoclast surface area per bone surface was significantly increased in D-KO bones versus D-WT bones (Fig. 6D, p≤0.05). Lastly, elastic modulus was significantly increased in A-KO females while stiffness was significantly decreased in D-KO female mice; however, ultimate load was not significantly changed in either the A- or D-KO mice (Fig. S7, p≤0.05).
Similar to AD-KO males, AD-KO females had significant reductions trabecular thickness and significant increases in trabecular spacing compared to AD-WT females (Fig. 6A, p≤0.001 and p≤0.05). AD-KO females had significantly increased P1NP (Fig. 6C, p≤0.05). Stiffness and elastic modulus but not ultimate load were significantly decreased in the AD-KO females (Fig. S7, p≤0.001).
These results indicate that loss of Mef2a in female mice produces a substantial increase in trabecular bone, potentially as a result of decreased osteoclast numbers in vivo, which would agree with the in vitro model presented in Figure 3. Additionally, phenotypes of male and female mice contradict one another, suggesting sex-specific differences may contribute to altered activity of MEF2A in male versus female mice.
3.6. One-month AD-KO female mice are osteopetrotic
Since AD-KO males and females had reduced trabecular and cortical thickness, we analyzed one-month AD-KO mice to determine whether the reductions in bone were caused early in skeletal development or present only in later stages of skeletal development. AD-KO males exhibit no significant differences with their wild type littermates; however, the female AD-KO mice were osteopetrotic with a significant increase in trabecular number but no significant change in trabecular thickness or separation (Fig. 7A, p≤0.05). Lastly neither males nor females demonstrated a significant change in cortical thickness at this time point. Histology shows female AD-KO bones have slightly smaller osteoclasts than their WT littermates, and female AD-KO bones have fewer osteoclasts than female WT bones (Fig. 7F and H). Tables 3 and 4 demonstrate the results from the 1- and 3-month AD-WT and KO male and female mice. These data suggest that increases in bone volume seen at the three-month time point in female A-KO is similar to one-month female AD-KO mice.
Figure. 7. AD-KO female mice are osteopetrotic at one month.
(A) Trabecular and cortical micro-CT measurements and (B-E) representative models from each group. N = 8–13 for all groups in micro-CT experiments. (F) Osteoclast surface and number and (G and H) representative images from TRAP-stained decalcified bones. Scale bar = 10 mm. N = 5–11 for histology. Statistical testing by unpaired T-test with Welch’s correction to compare male WT to male AD-KO and female WT to female AD-KO mice. Bars show means ± SD. * P ≤ 0.05, *** P ≤ 0.001. n.s. = not significant.
Table 3.
Summary of Male AD-WT and AD-KO Micro-CT Measures
| Male 1-month | Male 3-month | |||
|---|---|---|---|---|
| Micro-CT Measures | AD-WT | AD-KO | AD-WT | AD-KO |
| BV/TV | 13.69 ± 2.49 | 14.95 ± 4.37 | 13.32±3.03 | 10.06± 2.74 |
| Tb. Th | 0.040 ± 0.002 | 0.039 ± 0.002 | 0.0043 ± 0.001 | 0.0046 ± 0.001 |
| Tb. Sp | 0.128 ± 0.035 | 0.115 ± 0.028 | 0.192 ± 0.008 | 0.218 ± 0.009 |
| Tb. N | 3.35 ± 0.493 | 3.76 ± 0.885 | 2.889 ±0.61 | 2.474 ± 0.46 |
| Ct.Th | 0.108 ± 0.007 | 0.099 ± 0.012 | 0.168 ± 0.004 | 0.146 ± 0.004 |
Table 4.
Summary of Female AD-WT and AD-KO Micro-CT Measures
| Female 1-month | Female 3-month | |||
|---|---|---|---|---|
| Micro-CT Measures | AD-WT | AD-KO | ADWT | AD-KO |
| BV/TV | 9.65 ± 2.35 | 12.75 ± 3.37 | 5.791 ± 1.98 | 5.287 ± 1.01 |
| Tb. Th | 0.037± 0.002 | 0.038 ± 0.004 | 0.040 ± 0.003 | 0.0336 ± 0.001 |
| Tb. Sp | 0.121 ± 0.027 | 0.133 ± 0.037 | 0.229 ± 0.040 | 0.269 ± 0.040 |
| Tb. N | 2.58 ± 0.196 | 3.26 ± 0.616 | 1.406 ± 0.392 | 1.569 ± 0.258 |
| Ct.Th | 0.097 ± 0.004 | 0.096 ± 0.007 | 0.143 ± 0.014 | 0.136 ± 0.008 |
To determine if changes in expression of MEF2 family members accounted for the in vivo phenotypes, we measured Mef2a, d and c expression during osteoclast differentiation in A-, D- and AD-KO cultures. We measured a significant increase in Mef2c at day 0 and no significant change in Mef2d expression in our A-KO cultures (Fig 8A). There was a significant decrease in Mef2a expression in our D-KO cultures with no significant changes in Mef2c expression in either our D- or AD-KO cultures (Fig 8B–C). These changes agree with the MEF2A and D expression data presented in this study (Fig S3).
Figure 8. MEF2C expression increases in the absence of MEF2A.
Relative expression of Mef2a, d and c in BMMs from both male and female mice at day 0 (M-CSF stimulation only) or days 1–4 (M-CSF and RANKL stimulation) in (A) A-KO, (B) D-KO or (C) AD-KO cultures.
4. Discussion
Based on our studies with HDAC7, we determined that, similar to other cell types, MEF2 activity may be regulated by class IIA HDACs in osteoclasts. To determine the significance of MEF2 activity during osteoclast differentiation, we analyzed the skeletal phenotype of mice null for MEF2 during osteoclast differentiation. We focused our analysis on the role of MEF2A and D in regulating osteoclast differentiation because they were the MEF2 family members most highly expressed during osteoclast differentiation. In vitro osteoclast differentiation and activity were impaired in the MEF2A or MEF2D cultures, while the AD double KO cultures displayed almost a complete lack of TRAP positive cells. These observations suggest that MEF2A and D both make necessary contributions to osteoclast lineage determination and differentiation. Feng et al. performed the only other study characterizing the role of MEF2 in osteoclasts[15]. They determined that MEF2C cooperates with MITF and NFATc1 to activate the Atp6v0d2 promoter in RAW264.7 macrophage cell line, which can undergo osteoclast differentiation with RANKL stimulation. This cooperation might suggest a positive role for MEF2 in osteoclasts, consistent with our findings, although their study did not actually test this further. Their data and ours also reveal differences in the pattern of of Mef2 gene expression between primary BMM culture and RAW264.7 cells, most notably concerning expression of Mef2c, which was well expressed in RAW264.7 cells but had much lower expression in wild type BMMs or osteoclasts. In this study, we did not assess the role of MEF2C; however, we are currently breeding mice to evaluate the role of MEF2C in regulating osteoclast differentiation.
In macrophages MEF2A and D have been shown to regulate macrophage terminal differentiation [14]. In macrophages MEF2A and D have been shown to heterodimerize and interact with HDAC1 and HDAC7 or p300 histone acetyltransferase to regulate c-jun expression [14]. Interestingly, we did not measure any change in c-Jun expression by western blot in any of our MEF2 null osteoclasts suggesting that MEF2A and D may regulate other targets in osteoclasts (Fig 4). In retinal photoreceptor cells, MEF2D binds and regulates retina-specific enhancers by interacting with a retinal specific transcription factor CRX [16]. In osteoclasts it could be that MEF2A and/or D regulate genes different from genes they regulate in macrophages due to their potential interactions with osteoclast specific transcription factors such as NFATc1. In the future ChIP and RNA SEQ experiments will be performed to characterize MEF2 targets and binding partners in osteoclasts.
From our in vitro data, we anticipated seeing impaired osteoclasts in vivo, probably leading to reduced bone resorption and increased bone parameters. When we examined mice at 3 months age, surprisingly only A-KO females had the expected osteopetrotic phenotype, Mef2D KO had little phenotype, while and AD KO mice showed decreased trabecular bone, the opposite of what we had predicted. Interestingly, examination of AD-KO male males showed little phenotype at 1month age. AD-KO female mice at 1month did show indications of increased bone measures compared to their controls, which matched our expected phenotype. Thus, both male and female AD KO mice showed negative balance between bone formation and bone resorption as they aged from one to three months. We propose that this difference in phenotypes between in vitro culture and in vivo differences between male and female mice of the same genotype indicates extrinsic factors influence osteoclasts and the net balance between bone formation and resorption.
One possible explanation for the difference between the in vitro and in vivo phenotypes involves interactions between osteoclasts and resting T-cells, which have been shown to blunt osteoclast differentiation [26, 27]. The c-Fms-Cre that we used in our study has been shown to affect neutrophils, dendritic and T-cell lineages as well as macrophages [21]. MEF2 is expressed in and regulates T cell development through regulation of Nur77 expression downstream of the T-cell receptor [28]. In future studies, we will perform FACS analysis on our AD-KO mice to determine if changes in the T-cell population or other cells of the bone marrow environment account for the differences between the in vitro and in vivo phenotypes of our MEF2 mice.
We hypothesize that interactions between estrogens and MEFs in vivo influence the phenotype of the female A-KO mice. Multiple studies have determined that estrogen is the major hormone regulator of the skeleton in both men and women [29–31]. In previous studies looking at the role of MEF2 and class IIA HDACs in cardiac cells where estrogen has a cardioprotective effect, estrogen receptor alpha was shown to be a direct target of MEF2 [32]. We observe an osteopetrotic phenotype in our A-KO female but not male mice which suggests that in osteoclasts expression and or activity of the estrogen receptor may be regulated by MEF2A. Additionally, we measured a 3-fold increase in Mef2c expression in A-KO cultures (Fig 8A, last panel). In cardiac cells, estrogen receptor expression was shown to be activated by MEF2C [32]. The female-specific phenotype in the A-KO mice may be a result of the increased Mef2c expression. Additionally, it was demonstrated in cardiac cells that HDAC4, 5 and 9 interact with and HDAC5 and 9 repress the estrogen receptor’s transcriptional activation [32]. There is no reported sex specific effect in the skeletal phenotype of HDAC9- and HDAC5-KO mice; however, any estrogen effect may be masked by the global nature of the models [33–35]. Studies are currently underway to determine if MEF2A directly or indirectly regulates expression and/or activity of the estrogen receptor through its interaction with class IIA HDACs.
5. Conclusion
Up to date there has been little information characterizing the role of MEF2 in regulating osteoclast gene expression. We present data that MEF2A and MEF2D are expressed during osteoclast differentiation and that osteoclasts null for either MEF2A or MEF2D are decreased in their ability to form multinuclear cells while osteoclasts null for both MEF2A and MEF2D are almost completely inhibited in their ability to undergo osteoclast differentiation. However, in vivo only female MEF2A mice have the predicted osteopetrotic skeletal phenotype. Our data together suggests a significant role for the MEF2 family of transcription factors in regulating osteoclast differentiation.
Supplementary Material
Highlights.
MEF2A and D are the most abundant MEF2 family members in osteoclasts
Loss of MEF2A and D result in a complete loss of osteoclast differentiation in vitro
MEF2A null female mice have an osteopetrotic phenotype in vivo
MEF2A and D null mice are osteopenic in vivo
Acknowledgements
We would like to thank the other members of the Mansky lab for their thoughtful comments during the writing of this manuscript. We would also like to thank thank Ms. Ashley Gunderson (Division of Oral and Maxillofacial Pathology, University of Minnesota, Minneapolis, MN) for embedding and sectioning of the MEF2 bones.
Funding Sources
This work was supported by are supported by NIH R01 AR061352 (K.M and E.J), T32AR050938 and Ray C. Anderson Fellowship (N.B.) and bridge funds by the University of Minnesota School of Dentistry (K.C.M.).
Footnotes
Conflict of Interest
The authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Crockett JC, Rogers MJ, Coxon FP, Hocking LJ, Helfrich MH, Bone remodelling at a glance, J Cell Sci 124(Pt 7) (2011) 991–8. [DOI] [PubMed] [Google Scholar]
- [2].Khosla S, Westendorf JJ, Modder UI, Concise review: Insights from normal bone remodeling and stem cell-based therapies for bone repair, Stem Cells 28(12) (2010) 2124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Raggatt LJ, Partridge NC, Cellular and molecular mechanisms of bone remodeling, J Biol Chem 285(33) (2010) 25103–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Boyle WJ, Simonet WS, Lacey DL, Osteoclast differentiation and activation, Nature 423(6937) (2003) 337–42. [DOI] [PubMed] [Google Scholar]
- [5].Feng X, McDonald JM, Disorders of bone remodeling, Annu Rev Pathol 6 (2011) 121–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Novack DV, Mbalaviele G, Osteoclasts-Key Players in Skeletal Health and Disease, Microbiol Spectr 4(3) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stemig M, Astelford K, Emery A, Cho JJ, Allen B, Huang TH, Gopalakrishnan R, Mansky KC, Jensen ED, Deletion of histone deacetylase 7 in osteoclasts decreases bone mass in mice by interactions with MITF, PLoS One 10(4) (2015) e0123843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pham L, Kaiser B, Romsa A, Schwarz T, Gopalakrishnan R, Jensen ED, Mansky KC, HDAC3 and HDAC7 have opposite effects on osteoclast differentiation, J Biol Chem 286(14) (2011) 12056–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jin Z, Wei W, Dechow PC, Wan Y, HDAC7 inhibits osteoclastogenesis by reversing RANKL-triggered beta-catenin switch, Mol Endocrinol 27(2) (2013) 325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dressel U, Bailey PJ, Wang SC, Downes M, Evans RM, Muscat GE, A dynamic role for HDAC7 in MEF2-mediated muscle differentiation, J Biol Chem 276(20) (2001) 17007–13. [DOI] [PubMed] [Google Scholar]
- [11].Potthoff MJ, Olson EN, MEF2: a central regulator of diverse developmental programs, Development 134(23) (2007) 4131–40. [DOI] [PubMed] [Google Scholar]
- [12].McKinsey TA, Zhang CL, Olson EN, MEF2: a calcium-dependent regulator of cell division, differentiation and death, Trends Biochem Sci 27(1) (2002) 40–7. [DOI] [PubMed] [Google Scholar]
- [13].Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S, Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7, J Biol Chem 276(50) (2001) 47496–507. [DOI] [PubMed] [Google Scholar]
- [14].Aude-Garcia C, Collin-Faure V, Bausinger H, Hanau D, Rabilloud T, Lemercier C, Dual roles for MEF2A and MEF2D during human macrophage terminal differentiation and c-Jun expression, Biochem J 430(2) (2010) 237–44. [DOI] [PubMed] [Google Scholar]
- [15].Feng H, Cheng T, Steer JH, Joyce DA, Pavlos NJ, Leong C, Kular J, Liu J, Feng X, Zheng MH, Xu J, Myocyte enhancer factor 2 and microphthalmia-associated transcription factor cooperate with NFATc1 to transactivate the V-ATPase d2 promoter during RANKL-induced osteoclastogenesis, J Biol Chem 284(21) (2009) 14667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Andzelm MM, Cherry TJ, Harmin DA, Boeke AC, Lee C, Hemberg M, Pawlyk B, Malik AN, Flavell SW, Sandberg MA, Raviola E, Greenberg ME, MEF2D drives photoreceptor development through a genome-wide competition for tissue-specific enhancers, Neuron 86(1) (2015) 247–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Andzelm MM, Vanness D, Greenberg ME, Linden DJ, A Late Phase of Long-Term Synaptic Depression in Cerebellar Purkinje Cells Requires Activation of MEF2, Cell Rep 26(5) (2019) 1089–1097 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R, Guidelines for assessment of bone microstructure in rodents using micro-computed tomography, J Bone Miner Res 25(7) (2010) 1468–86. [DOI] [PubMed] [Google Scholar]
- [19].Jepsen KJ, Silva MJ, Vashishth D, Guo XE, van der Meulen MC, Establishing biomechanical mechanisms in mouse models: practical guidelines for systematically evaluating phenotypic changes in the diaphyses of long bones, J Bone Miner Res 30(6) (2015) 951–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pon JR, Marra MA, MEF2 transcription factors: developmental regulators and emerging cancer genes, Oncotarget 7(3) (2016) 2297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Deng L, Zhou JF, Sellers RS, Li JF, Nguyen AV, Wang Y, Orlofsky A, Liu Q, Hume DA, Pollard JW, Augenlicht L, Lin EY, A novel mouse model of inflammatory bowel disease links mammalian target of rapamycin-dependent hyperproliferation of colonic epithelium to inflammation-associated tumorigenesis, Am J Pathol 176(2) (2010) 952–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T, Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts, Dev Cell 3(6) (2002) 889–901. [DOI] [PubMed] [Google Scholar]
- [23].Matsuo K, Galson DL, Zhao C, Peng L, Laplace C, Wang KZ, Bachler MA, Amano H, Aburatani H, Ishikawa H, Wagner EF, Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos, J Biol Chem 279(25) (2004) 26475–80. [DOI] [PubMed] [Google Scholar]
- [24].Sharma SM, Bronisz A, Hu R, Patel K, Mansky KC, Sif S, Ostrowski MC, MITF and PU.1 recruit p38 MAPK and NFATc1 to target genes during osteoclast differentiation, J Biol Chem 282(21) (2007) 15921–9. [DOI] [PubMed] [Google Scholar]
- [25].Carson JA, Manolagas SC, Effects of sex steroids on bones and muscles: Similarities, parallels, and putative interactions in health and disease, Bone 80 (2015) 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li Y, Toraldo G, Li A, Yang X, Zhang H, Qian WP, Weitzmann MN, B cells and T cells are critical for the preservation of bone homeostasis and attainment of peak bone mass in vivo, Blood 109(9) (2007) 3839–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Toraldo G, Roggia C, Qian WP, Pacifici R, Weitzmann MN, IL-7 induces bone loss in vivo by induction of receptor activator of nuclear factor kappa B ligand and tumor necrosis factor alpha from T cells, Proc Natl Acad Sci U S A 100(1) (2003) 125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Youn HD, Sun L, Prywes R, Liu JO, Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2, Science 286(5440) (1999) 790–3. [DOI] [PubMed] [Google Scholar]
- [29].Khosla S, Pathogenesis of age-related bone loss in humans, J Gerontol A Biol Sci Med Sci 68(10) (2013) 1226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Khosla S, Amin S, Orwoll E, Osteoporosis in men, Endocr Rev 29(4) (2008) 441–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Albright F, Post-menopausal osteoporosis, Trans Association American Physicians 55 (1940) 298–305. [Google Scholar]
- [32].van Rooij E, Fielitz J, Sutherland LB, Thijssen VL, Crijns HJ, Dimaio MJ, Shelton J, De Windt LJ, Hill JA, Olson EN, Myocyte enhancer factor 2 and class II histone deacetylases control a gender-specific pathway of cardioprotection mediated by the estrogen receptor, Circ Res 106(1) (2010) 155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Obri A, Makinistoglu MP, Zhang H, Karsenty G, HDAC4 integrates PTH and sympathetic signaling in osteoblasts, J Cell Biol 205(6) (2014) 771–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wein MN, Spatz J, Nishimori S, Doench J, Root D, Babij P, Nagano K, Baron R, Brooks D, Bouxsein M, Pajevic PD, Kronenberg HM, HDAC5 controls MEF2C-driven sclerostin expression in osteocytes, J Bone Miner Res 30(3) (2015) 400–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jin Z, Wei W, Huynh H, Wan Y, HDAC9 Inhibits Osteoclastogenesis via Mutual Suppression of PPARgamma/RANKL Signaling, Mol Endocrinol 29(5) (2015) 730–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.