Abstract
Infectious diseases are the ever-present threats to public health and the global economy. Accurate and timely diagnosis is crucial to impede the progression of a disease and break the chain of transmission. Conventional diagnostic techniques are typically time-consuming and costly, making them inefficient for early diagnosis of infections and inconvenient for use at the point of care. Developments of sensitive, rapid, and affordable diagnostic methods are necessary to improve the clinical management of infectious diseases. Quartz crystal microbalance (QCM) systems have emerged as a robust biosensing platform due to their label-free mechanism, which allows the detection and quantification of a wide range of biomolecules. The high sensitivity and short detection time offered by QCM-based biosensors are attractive for the early detection of infections and the routine monitoring of disease progression. Herein, the strategies employed in QCM-based biosensors for the detection of infectious diseases are extensively reviewed, with a focus on prevalent diseases for which improved diagnostic techniques are in high demand. The challenges to the clinical application of QCM-based biosensors are highlighted, along with an outline of the future scope of research in QCM-based diagnostics.
Keywords: Quartz crystal microbalance, Biosensor, Infectious disease, Diagnosis, Virus, Influenza
1. Introduction
Infectious diseases are health disorders caused by pathogenic microorganisms such as bacteria, viruses, fungi, and parasites. These diseases can be transmitted from one organism to another via direct or indirect contact, causing various illnesses that can lead to death. Despite significant advances in the prevention and treatment measures, infectious diseases continue to be prevalent and pose a constant threat to public health and the global economy. Infections of tuberculosis and malaria consistently rank among the leading causes of death worldwide, imposing steady yet substantial burdens (Bloom and Cadarette, 2019). Meanwhile, seasonal outbreaks of influenza and coronaviruses have resulted in pandemics that have claimed numerous human lives and devastated the world economy within a short period (Zambon, 2014). On the whole, infectious diseases account for 15 million deaths each year, with the dominant proportion of these occurrences from low-to middle-income nations (Pashchenko et al., 2018). In the fight against infectious diseases, early and accurate diagnosis is the most effective way to break the chain of transmission and mitigate the impacts of these diseases.
Infectious diseases are normally diagnosed by the detection of disease biomarkers in the biological samples of infected patients. Typical disease biomarkers include whole infectious agents (e.g. bacteria and viruses), residues from the infectious agents (e.g. nucleic acids and proteins), and antibodies against the pathogens. Conventional diagnostic techniques such as microscopy, enzyme-linked immunosorbent assay (ELISA), and polymerase chain reaction (PCR) are well established gold standards for the diagnosis of many infectious diseases. However, these assays involve tedious procedures, skilled operators, and expensive instrumentation, all of which translate into high assay costs and significant delays between sample collection and medical diagnosis (Giamblanco et al., 2015; Ragavan et al., 2018). These shortcomings become especially prominent in resource-limited settings, where many infections are undiagnosed due to the poor access to diagnostic services (Sharma et al., 2015; Sin et al., 2014).
In line with the efforts to improve disease diagnostics, World Health Organization (WHO) has introduced the Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free, and Deliverable to end-users (ASSURED) criteria as a benchmark for in vitro diagnostic tests (Kosack et al., 2017). The emphasis on the development of sensitive, rapid, and affordable diagnostic techniques that can be used at or near the point of care has contributed to the rapid growth in biosensor technologies. Biosensors are analytical devices that detect target analytes and convert the molecular recognition events into measurable signals (Sin et al., 2014). A biosensor consists of two elements: the receptors that capture the target molecules, and the transducer mechanism that produces electrical signals in response to target recognition (Srinivasan and Tung, 2015). Compared to conventional assays, biosensors provide an inexpensive platform for detection, with simpler operating procedures that can be conducted at the point of care (Sin et al., 2014); these attributes make biosensors ideal for use as a rapid diagnostic device for infectious diseases.
The quartz crystal microbalance (QCM) is a biosensor platform that incorporates a mechanical transducer, which operates on the principle of mass detection. QCM-based biosensors have gained significant interest in the field of pathogen detection due to their ability to detect virtually any type of biomolecule via a label-free method (Afzal et al., 2017). Moreover, the rapid detection process and the high sensitivity of QCM systems are particularly attractive for the development of novel diagnostic tools. Herein, the detection strategies employed for disease biosensing in QCM platforms are critically discussed, and the prospects of QCM-based biosensors as a rapid diagnostic technique for infectious diseases are evaluated. This review focuses on the development of QCM-based biosensors for the diagnosis of six prevalent infectious diseases that are global health burdens, in which the notable advancements in the field and the shortcomings of reported works are highlighted. The research gaps and opportunities in QCM-based biosensing are also discussed in line with the current trend towards point-of-care diagnostics.
2. Quartz crystal microbalance: Theory for biosensing
The key component of QCM is a thin disc of quartz with electrodes that serve as sensing surfaces on both sides of the crystal. QCM operates on the principle of the piezoelectric effect, in which the application of an external electric field on quartz produces mechanical stresses in the crystal. Imposing an alternating voltage on the crystal causes it to oscillate in the direction perpendicular to the plate surface (Chen et al., 2018). At series resonance, crystal oscillation occurs at a characteristic resonant frequency that is influenced by the mass per unit area at the crystal surface, as described by the Sauerbrey relation (Eq. (1)) (Qiao et al., 2016).
| (1) |
where is the frequency shift due to the change in surface mass (Hz); is the fundamental resonant frequency of the crystal (Hz); is the change in the mass per unit area at the crystal surface (g cm-2); (g cm-3) and (g cm-1 s-2) are the density and shear modulus of the quartz crystal, respectively.
In a liquid medium, the density and viscosity of the liquid contribute to the frequency variation, as described by the theory for liquids (Eq. (2)) (Afzal et al., 2017).
| (2) |
where is the frequency variation due to the density-viscosity effect of the liquid; and are the density and viscosity of the liquid, respectively. For a rigid film immersed in a Newtonian fluid, observed frequency responses can be correlated to the changes in surface mass with consideration of the acoustic properties of the liquid (Qiao et al., 2016).
In recent years, QCM with dissipation monitoring (QCM-D) has been introduced to the biosensing application to monitor the dissipative losses in crystal oscillation alongside the changes in resonant frequency. Viscoelastic and soft films, which are commonly formed upon the binding of biomolecules to the sensor surface, dampen the oscillations and do not fully couple to the crystal oscillation (Cooper and Singleton, 2007). As such, the simultaneous monitoring of frequency and dissipation provides additional information on the structural changes at the crystal surface (Fogel et al., 2016; Karczmarczyk et al., 2017).
The mass-based detection principle of QCM has many inherent advantages for disease biosensing. QCM systems can detect virtually any type of molecule because mass is an intrinsic property of all substances, making it a versatile platform for detecting the diverse types of disease biomarkers. Molecular recognition events at the crystal surface are instantaneously reflected in the frequency response without the need for labelling procedures, resulting in a short detection time, typically between 30 min to 1 h. Moreover, QCM-based biosensors are highly sensitive to changes in surface mass to the order of nanogram per cm2, enabling it to achieve a low detection limit comparable to that of an ELISA (Afzal et al., 2017; Srinivasan and Tung, 2015). These attributes make QCM an invaluable tool for the early detection of infectious diseases, where disease biomarkers are usually present in very low concentrations in clinical samples during the initial stages of an infection. Furthermore, the ability of QCM-based biosensors to quantify analytes extends their application scope beyond screening for infections; this advantageous feature allows the routine monitoring of disease progression and treatment efficacy. Combined with the portable instrument, QCM-based biosensors have tremendous potential to be used at the point of care (Sin et al., 2014; Yao and Fu, 2014).
3. Receptors for target recognition
For QCM-based biosensors to function effectively as a diagnostic device, the sensitivity and selectivity of the receptors to the targeted disease biomarkers are of paramount importance. In biosensors, sensitivity is indicated by the detection limit, which refers to the minimum concentration of analyte that can be reliably detected (Ballantine et al., 1996). A low detection limit is desirable as it enables infections to be detected at the early stages. Target selectivity refers to the ability of the biosensor to respond selectively to a specific target. A high target selectivity minimises non-specific interactions of interferents in a sample with the sensing surface; this in turn reduces the occurrences of false-positive results and high noise levels that will adversely impact the sensitivity of the biosensor (Palladino et al., 2018). As QCM is a mass-based detection technique, any particle that interacts with the surface of the crystal will produce a frequency response. Thus, the QCM surface needs to be functionalised with receptors that can effectively capture the target molecules onto the crystal surface while minimising the adsorption of non-targeted species. Here, the different types of receptors that are commonly employed for target recognition in QCM-based biosensors are described, along with the advantages and challenges associated with each approach.
3.1. Antibodies and antigens
Antibodies are immunoglobulins produced by the immune system to protect the body against foreign molecules, termed as antigens. Each antibody has regions known as the antigen-binding sites, which have a selective affinity to specific molecules, and through which the antibody recognises and binds to its target molecule to form an antibody-antigen complex. The high selectivity and affinity to targets make antibodies a useful sensing material in the development of sensitive assays with reproducible results (Ragavan et al., 2018). Moreover, specific antibodies are available for many types of protein biomarkers, e.g. surface proteins of bacteria and viruses (Hiatt and Cliffel, 2012; Shen et al., 2011), non-structural proteins secreted by the pathogens (Sharma et al., 2011; Tai et al., 2005), and cell signalling proteins that serve as indicators of infections (Zhou et al., 2019). Antibody-based detection is well-established in the field of disease detection, with techniques such as ELISA and haemagglutination-inhibition assay being regarded as the standard diagnostic procedures for many infectious diseases.
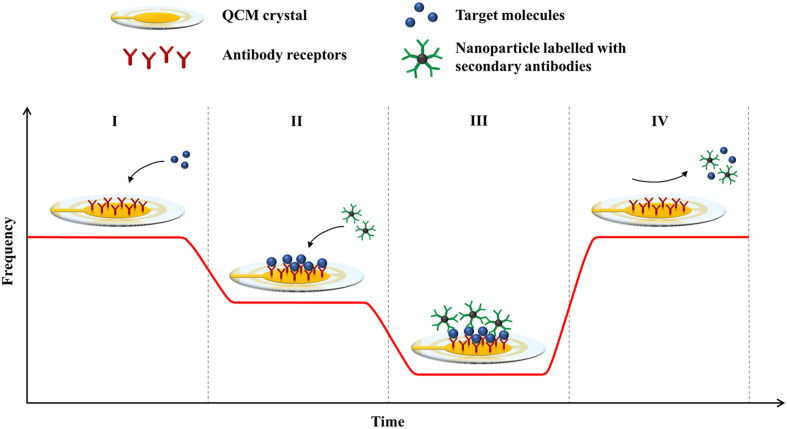
Based on the same principle, antibodies can be used in QCM-based biosensors to capture target molecules onto the crystal surface to induce a frequency response (Fig. 1 ). A variety of methods for immobilising antibodies onto the QCM surface are available, including physical adsorption (Xu et al., 2011), silanisation (Xie et al., 2015), and the ordered attachment of antibodies via self-assembled monolayers (e.g. thiols and cysteamine) (Sharma et al., 2011; Thies et al., 2017) and cross-linker molecules (e.g. Proteins A and G) (Chen et al., 2011; Hiatt and Cliffel, 2012). The process of immobilising antibodies onto the sensor surface is complicated, and a significant loss of bioactivity due to the denaturation and random orientation of antibodies is common. Hence, a fair amount of research on QCM-based biosensors has focused on improving antibody immobilisation by using a combination of methods or novel ligands for antibody attachment. In particular, self-assembled monolayers and Protein A linkers are the popular methods for fabricating QCM-based immunosensors due to their ability to preserve the biological activity of antibodies by producing an organised layer that exposes the binding sites of antibodies (Chauhan et al., 2015; Steinem and Janshoff, 2007).
Fig. 1.
Principle of detection in a QCM-based immunosensor. The graph shows the frequency responses of the biosensor at different stages of the assay: (I) frequency baseline of a QCM crystal with immobilised antibody receptors; (I-II) binding of target molecules to the receptors; (II-III) binding of nanoparticles labelled with secondary antibodies; (IV) regeneration of the sensor surface.
In many QCM-based assays, secondary antibodies are commonly used as mass enhancers for signal amplification. After the binding of target molecules to the immobilised antibodies on the crystal surface, the secondary antibodies are added to bind to the captured analytes to cause a further frequency shift (Fig. 1). The secondary antibodies can be used without any modification (Tai et al., 2005), or they are conjugated to nanoparticles for added mass (Li et al., 2011; Zhang et al., 2016). The secondary binding event increases the resolution of the frequency response, which effectively lowers the detection limit of the biosensor. Additionally, the amplification step can verify the identity of bound particles and improve the specificity of the QCM assay (Chen et al., 2009).
The major challenge in using antibodies as the receptors for biosensing is their poor chemical and physical stability. The fragile three-dimensional (3D) structures of antibodies are highly sensitive to the properties of the surrounding medium (e.g. pH, temperature, and ionic strength). As the specificity of antibodies relies on their unique 3D conformations, structural changes may result in a loss of function. The application of antibody-based biosensors is further limited in the case where the biological samples such as blood and serum in which contain enzymes that may denature these bioreceptors (Afzal et al., 2017; Srinivasan and Tung, 2015). The poor stability of antibodies typically results in low reusability of QCM-based immunosensors; sensitivity tends to deteriorate after several cycles of detection and regeneration (Chauhan et al., 2015; Li et al., 2011). Additionally, the supply and storage of antibody-based biosensors may prove to be a significant challenge in resource-limited regions due to the limited lifespan of antibodies, which ranges from 6 to 12 months under refrigeration (Whitcombe et al., 2011).
An alternative approach to target recognition based on antibody-antigen interactions is through the presentation of epitopes of infectious agents on the crystal surface. Here, the epitopes serve as the receptors to which antibodies generated by the host immune response bind. Epitopes are smaller in size compared to antibodies, and their interactions are less dependent on orientation. As such, the process of functionalising sensor surfaces with epitopes is usually less tedious (Gerdon et al., 2005). However, this method is less useful for early detection due to the existence of a window period of up to several months before a detectable level of antibodies is produced in the body (Ly et al., 2016; Su et al., 2003).
3.2. Nucleic acid probes
Nucleic acids are the genetic materials of organisms, found either as ribonucleic acid (RNA) or deoxyribonucleic acid (DNA) in the nucleus, nucleocapsid or cytoplasm of pathogens. In many infectious diseases, nucleic acids are among the earliest biomarkers to reach a detectable level in the host (Allain and Opare-Sem, 2016; Gillespie et al., 2019). The level of foreign nucleic acids is often used as an indication of viral replication, which is essential in characterising the phase of an infection. Besides, nucleic acids can provide information on the genotype of pathogens and the emergence of variants resistant to antibiotics, vaccines, and drugs (Kotha et al., 2018).
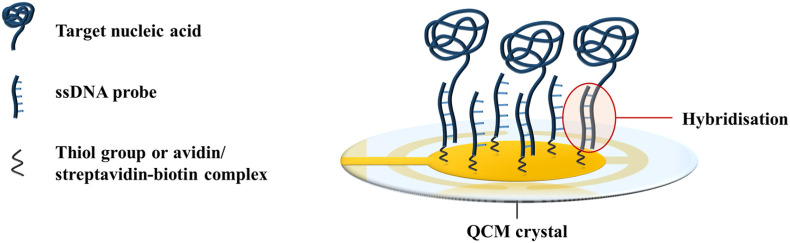
DNA probes are the most commonly employed receptors for nucleic acid detection in QCM-based biosensors. The sequence complementarity between two DNA strands forms the basis for target recognition in QCM-based biosensors. Single-stranded DNA (ssDNA) probes that are complementary to the target gene of the infectious agent can be immobilised onto the crystal surface via Au–S or avidin/streptavidin-biotin bonds (Afzal et al., 2017). When a sample of the infectious agent is added to the biosensor, hybridisation of the target sequences of the pathogen to the ssDNA probes occurs via complementary base pairing (Fig. 2 ). The binding event results in a mass loading at the crystal surface, and creates a frequency response. The recognition mechanism is highly specific as the DNA probes will only bind to their complementary sequences. As such, nucleic acid biosensors can detect up to single-base differences in genetic sequences (Yao et al., 2013, 2008), which is useful for distinguishing the pathotypes of viruses (Cattoli et al., 2011).
Fig. 2.
Hybridisation of target nucleic acids to ssDNA probes on the QCM surface. Illustration adapted from Afzal et al. (2017).
Similar to antibodies, the 3D structures of nucleic acid probes are sensitive to environmental conditions. Hence, their sensing performance may vary according to the solution used (Afzal et al., 2017). A more stable alternative to DNA is the peptide nucleic acid (PNA), which is a DNA mimic produced by replacing the sugar-phosphate backbone with synthetic peptide bonds (Saadati et al., 2019). In recent years, novel bio-recognition elements known as aptamers have been increasingly popular in replacing antibodies as the receptors in biosensors. Aptamers are short-stranded nucleic acids or peptides with a molecular weight below 25 kDa, and a unique 3D structure that confers high selectivity to target molecules. Highly specific aptamers can be selected via the systematic evolution of ligands by exponential enrichment (SELEX) process. Aptamers can target a wide range of molecules such as nucleic acids, antigens, and proteins with affinities comparable to those of monoclonal antibodies (Ragavan et al., 2018). Additionally, they are easy to functionalise, non-aggregating, and more resistant to thermal degradation (Wang and Li, 2013).
A significant drawback of nucleic acid biosensors is their inability to detect the analyte directly from an intact cell containing the genetic material (Steinem and Janshoff, 2007). Thus, samples need to be pre-treated to extract the nucleic acids from the cells prior to detection. Once released from the cells, the nucleic acids are often amplified via PCR to obtain a sufficient concentration of analyte that can be detected by the QCM-based biosensor (Hao et al., 2011). The reliance on laboratory procedures is highly inconvenient for the on-site application of these biosensors. Furthermore, PCR is a costly procedure that requires a thermal cycler, an expensive instrument that is not readily affordable by many laboratories in developing and underdeveloped countries. To eliminate the dependence on thermal cyclers, isothermal amplification methods (e.g. loop-mediated isothermal amplification and rolling circuit amplification) have been explored, although the design of primers for these techniques is relatively intricate as compared to PCR (Hao et al., 2011; Shojaei et al., 2015).
The design of a nucleic acid-based QCM biosensor requires a thoughtful consideration of the properties of the detection probes and the target genes. In general, DNA and aptamers have better thermal stability than antibodies. However, they are susceptible to degradation by the nucleases that are commonly present in clinical samples. The most straightforward approach to circumvent this problem is by pre-treating the samples to remove or inactivate the circulating nucleases. Another option is to modify the DNA or aptamer probes (e.g. by including local modifications to the ribose 2’ sites) to improve their resistance to nucleases (Afzal et al., 2017). As many target genes are shared among different pathogen strains, thus probe sequences need to be designed carefully to avoid cross-reactivity. For the screening of aptamers in the SELEX process, it is essential to use a very pure target sample to select for highly specific aptamers (Menger et al., 2016).
3.3. Molecularly-imprinted polymers
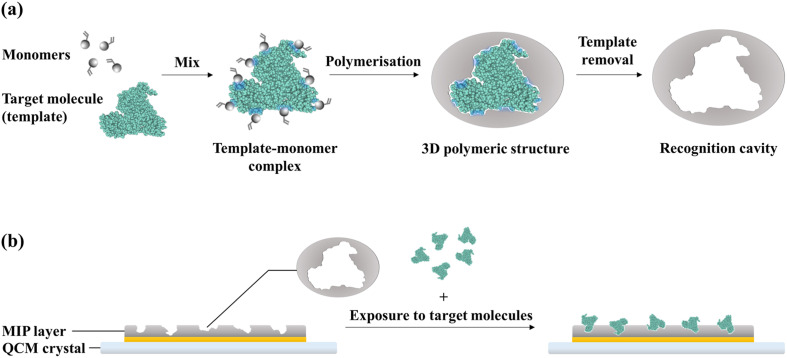
Molecularly-imprinted polymers (MIPs) are synthetic receptors that mimic the recognition properties of natural antibodies. The principle of formation of MIPs is illustrated in Fig. 3 a. MIPs are synthesised by performing a polymerisation reaction in the presence of the target molecules, which serve as the templates. The monomers assemble around the target molecules via functional group interactions; consequently, the polymerisation reaction produces a 3D polymeric structure around the templates. The subsequent elution of the templates from the polymer matrix leaves behind recognition cavities that are complementary to the target molecules in terms of shape and position of functional groups. As a result, the target molecules can selectively bind to the MIP via a combination of non-covalent interactions. In this manner, whole molecules or specific epitopes can be imprinted onto polymers to create binding sites for these targets (Schirhagl, 2013; Whitcombe et al., 2011).
Fig. 3.
Principle of recognition in a MIP-based biosensor: (a) formation of recognition cavities in MIPs; (b) binding of target molecules to the recognition cavities on the MIP-functionalised crystal surface.
MIPs were introduced in biosensor platforms as an alternative to the less stable natural receptors (e.g.antibodies and DNA probes). A QCM crystal functionalised with a MIP layer as a synthetic receptor for capturing target molecules is illustrated in Fig. 3b. QCM crystals can be functionalised by forming the polymer directly on the sensor surface via in situ polymerisation or electropolymerisation. Alternatively, MIPs can be synthesised separately as nanoparticles for immobilisation onto the sensor surface (Whitcombe et al., 2011). The former approach is preferred in QCM systems as it creates thin films on the crystal surface that minimises the swelling effect of functional layers, which can mask binding events and introduce signal drifts (Menger et al., 2016; Phan et al., 2014).
To date, MIPs have been applied in QCM-based biosensors to detect a variety of targets, including proteins (Ma et al., 2017; Tai et al., 2005), whole viruses (Wangchareansak et al., 2013), and small-molecule drugs (Eren et al., 2015). The primary advantage of using MIPs for biosensing is their high stability compared to antibodies and nucleic acids. Polymers are resistant to thermal and chemical degradations, and their functionality is less dependent on environmental conditions, enabling the detection process to be performed over a broad range of conditions. Unlike antibodies, MIPs can be stored under ambient conditions without experiencing a significant loss of activity (Afzal et al., 2017; Eren et al., 2015). MIP-based biosensors have a higher reuse potential as the receptors are less likely to be degraded during washing procedures to regenerate the surface (Steinem and Janshoff, 2007). Besides that, costly and tedious immobilisation procedures can be avoided by the direct polymerisation of MIPs on the crystal surface (Whitcombe et al., 2011). Polymerisation reactions can be completed within a few hours, as compared to the production of antibodies, which takes months and involves the use of animals (Srinivasan and Tung, 2015; Steinem and Janshoff, 2007). As such, the cost of production of MIPs is typically two orders of magnitude lower than that of antibodies, in the range of $0.1–0.5 per mg (Afzal et al., 2017).
Presently, the predominant challenge in developing MIP-based biosensors is to achieve the levels of sensitivity and target specificity comparable to those of antibody-based biosensors. The incomplete removal of templates during the elution step and the entrapment of proteins beneath the surface of the polymer often result in a low density of accessible binding sites (Lu et al., 2012; Ma et al., 2017). Furthermore, the imprinting of disease biomarkers is particularly challenging due to the fragile and large structure of biomolecules, which tend to create wide cavities that enhance non-specific binding (Whitcombe et al., 2011). For this reason, specific epitopes rather than whole proteins are the preferred templates in developing MIPs for biosensing applications (Ma et al., 2017). Hydrophilic monomers and cross-linkers are desirable for the synthesis of MIPs because the resultant polymer film has a greater compatibility with aqueous environments, which improves the molecular recognition efficiency (Mattiasson and Ye, 2015; Zhang, 2014). At present, the application of MIPs as receptors for QCM-based detection of diseases is still relatively limited, and a wide possibility of polymer systems for use in QCM-based biosensing remains to be explored.
4. QCM-based biosensors for prevalent infectious diseases
In the last two decades, applications of QCM-based biosensors for disease detection have steadily increased, owing to an improved understanding of QCM behaviour in liquid media and the continuous improvements in target recognition. The high sensitivity and rapid detection offered by QCM-based biosensors are particularly advantageous in the cases of (1) highly contagious diseases that need to be speedily detected and contained; (2) disorders that can be improved by early treatments; and (3) chronic infections that require the routine monitoring of disease progression. In this section, the detection strategies and the performances of QCM-based biosensors that have been reported for six prevalent infectious diseases, which represent global burdens to public health, are highlighted (Table 1 ) and discussed.
Table 1.
Selected examples of QCM-based biosensors for the detection of prevalent infectious diseases.
| Disease | Target | Receptor | Detection limit | Detection time | Signal amplification strategy | Reference |
|---|---|---|---|---|---|---|
| Malaria | PfHRP-2 | Anti-PfHRP-2 antibody | 12 ng mL-1 | – | – | Sharma et al. (2011) |
| P. falciparum and P. vivax | DNA probe | – | 4 h | Target sequences amplified by PCR | Wangmaung et al. (2014) | |
| Hepatitis B | HBV DNA | Peptide nucleic acid probe | 8.6 ng mL-1 | 50 min | Mass enhanced using RecA proteins coated with DNA probes complementary to HBV DNA | Yao et al. (2008) |
| HBsAg | Anti-HBs antibody | 0.53 μg mL-1 | 30 min | Surface modified with HBPs to increase the amount of receptors immobilised | Shen et al. (2011) | |
| HBV DNA | DNA probe | 104 copies mL-1 | 1 h | Target sequences amplified by RCA | Yao et al. (2013) | |
| HBsAg | Anti-HBs antibody | 2 ng mL-1 | 1 h | Mass enhanced using HBPs labelled with antibodies | Zhang et al. (2016) | |
| HBcAg | Anti-HBc antibody | 0.6 μg mL-1 | 25 min | Mass enhanced using the hydrogel swelling effect | Lim et al. (2017) | |
| Influenza | Influenza A and B viruses | Anti-M1 antibody | 103 PFU mL-1 | 1 h | Mass enhanced using AuNPs labelled with antibodies | Hewa et al. (2009) |
| H5N1 virus | Polyclonal antibody against HA glycoprotein | 0.0128 HAU | 2 h | Mass enhanced using magnetic nanobeads labelled with antibodies | Li et al. (2011) | |
| H5N1 virus | DNA aptamer | 0.0128 HAU | 30 min | Mass enhanced using the hydrogel swelling effect | Wang and Li (2013) | |
| H5N1, H5N3, H1N1, H1N3, and H6N1 viruses | Polymer imprint of whole viruses | 105 particles mL-1 | 40 min | – | Wangchareansak et al. (2013) | |
| H5N1 virus | DNA aptamer | 1.25 HAU mL-1 | 10 min | Surface modified with a nanowell pattern to increase the surface area for the immobilisation of receptors | Wang et al. (2017) | |
| HA glycoprotein | SA | 0.26 μg mL-1 | 30 min | – | Diltemiz et al. (2013) | |
| Dengue | DENV | Monoclonal antibodies against the envelope and NS1 proteins | 0.05 μg mL-1 | 30–60 min | – | Su et al. (2003) |
| NS1 protein | Polymer imprint of the NS1 epitope | 5 ng mL-1 | 50 min | Mass enhanced using detection antibodies | Tai et al. (2005) | |
| DNA sequences reverse-transcribed from DENV-2 genome | DNA probe | 2 PFU mL-1 | 1.5 h | Mass enhanced using AuNPs modified with oligonucleotide probes | Chen et al. (2009) | |
| NS1 protein | Immunoglobulin G antibody | 0.1 μg mL-1 | 15–25 min | Surface modified with cellulose nanocrystals | Pirich et al. (2017) | |
| HIV infection | gp41 glycoprotein | Polymer imprint of the gp41 epitope | 2 ng mL-1 | 10 min | – | Lu et al. (2012) |
| p24 antigen | Polyclonal antibody | 1 ng mL-1 | >2 h | Mass enhanced using detection antibodies and AuNPs | Ly et al. (2016) | |
| Tuberculosis | Mtb | Anti-tuberculosis antibody | 15 cells mL-1 | 30 min | – | He et al. (2002) |
| Mtb | α-LAM and anti-H37Rv antibodies | 8.7 × 105 cells mL-1 | <20 min | – | Hiatt and Cliffel (2012) | |
| IFN-γ, TNF-α, and IL-2 | Antibodies against IFN-γ, TNF-α, and IL-2 | 6.3 fg mL-1 (IFN-γ); 7.3 fg mL-1 (TNF-α); 7.8 fg mL-1 (IL-2) | >2 h | Surface modified with AuNPs, and mass enhanced using soluble silver nanoparticles labelled with antibodies | Zhou et al. (2019) |
4.1. Influenza
Commonly known as the flu, influenza is a respiratory disease caused by viruses from the Orthomyxoviridae family. Influenza types A and B are in circulation among humans, and are often responsible for the seasonal epidemics and occasional pandemics. Influenza A virus is divided into several subtypes based on the haemagglutinin (HA) and neuraminidase (NA) glycoproteins on the viral surface (Vemula et al., 2016). Highly contagious in nature, influenza A subtypes have been behind four major pandemics: the 1918 Spanish flu; 1957 Asian flu; 1968 Hong Kong flu; and the 2009 swine flu (Saunders-Hastings and Krewski, 2016). With many circulating strains and a history of outbreaks, influenza has attracted a substantial amount of QCM research in the past decade.
Initial studies for QCM-based biosensors used in the detection of influenza were based on antibodies as the receptors. Hewa et al. (2009) developed an ELISA to detect influenza A and B viruses using antibodies that target the matrix protein 1 (M1) of these viruses. The authors then translated the technique into a QCM immunoassay, using Protein A to orientate the antibodies for the maximal binding of antigens. Following the capture of virus particles onto the crystal surface, gold nanoparticles (AuNPs) labelled with anti-M1 antibodies were introduced to amplify the frequency response. The resultant immunosensor had a detection range of 103–107 PFU mL-1, which coincides with the viral titers in nasal wash samples from symptomatic patients. Notably, the QCM immunosensor had a higher sensitivity compared to the rapid diagnostic test (RDT) kit and the conventional diagnostic techniques such as cell culture, shell vial assay, and ELISA. However, the direct detection of viruses from nasal wash samples was not feasible due to the high background noise caused by the sample matrix.
In another work, Li et al. (2011) detected H5N1 viruses using magnetic nanobeads labelled with anti-H5 antibodies as the mass enhancers. The nanobead amplification step reduced the detection limit by two orders of magnitude in phosphate buffered saline and allowed the detection of virus concentrations above 0.128 HAU in tracheal swab samples. The immunosensor demonstrated the ability to distinguish between avian influenza virus (AIV) subtypes, as the frequency response to H5N1 virus was recorded to be five times greater than that observed for H3N2, H4N2, and H4N8 viruses.
Later on, Wang and Li (2013) generated an aptamer that was specific to the surface proteins of AIV H5N1 via the SELEX process. When studied using a Dot-ELISA, the aptamer proved to be highly specific to AIV H5N1, whereas the conventionally-used anti-H5 antibodies showed cross-reactivity to other H5 subtypes (e.g. H5N2, H5N3, and H5N9). Instead of immobilising the aptamer directly onto the QCM crystal surface, the aptamer was incorporated into a hydrogel containing complementary ssDNA . Hybridisation between the aptamer and ssDNA formed cross-links in the hydrogel structure, which caused the hydrogel to assume a shrunken state. Upon exposure to the target DNA, the aptamer preferentially bound to the target molecules, resulting in the dissolution of the aptamer-ssDNA cross-links, causing the gel to swell. When the hydrogel aptasensor was compared to a QCM-based immunosensor with a similar density of receptors, a larger frequency shift was recorded by the aptasensor for the same viral titer. Consequently, the detection limit of the hydrogel aptasensor was one order of magnitude lower than that of the immunosensor.
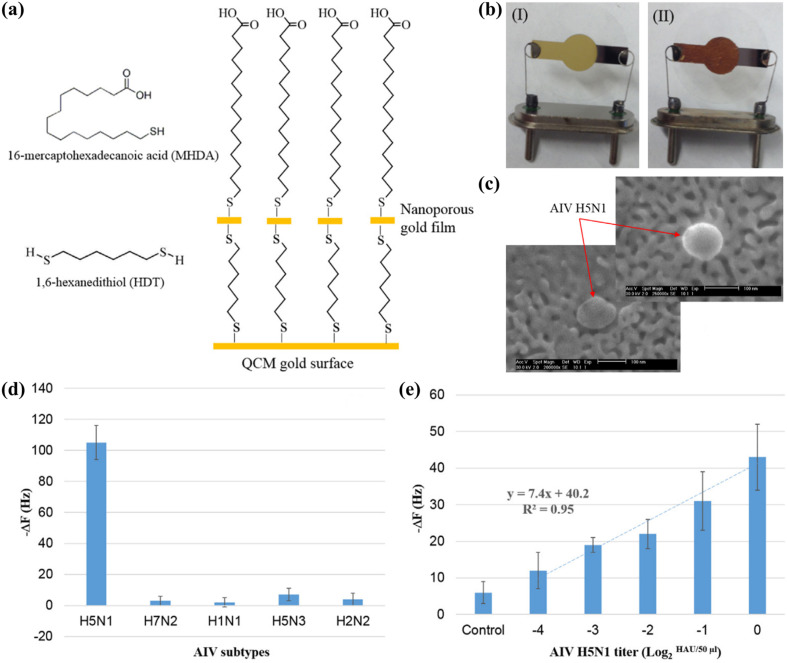
The higher sensitivity of the aptasensor was attributed to the signal amplification caused by the swelling effect of the hydrogel, as well as the higher stability of the aptamer compared to antibodies, which tend to lose a considerable proportion of their biological activity upon immobilisation (Afzal et al., 2017; Wang and Li, 2013). Moreover, detection could be completed within 30 min, as compared to 1 h using the anti-H5 immunosensor developed by Li et al. (2011), since the addition of mass enhancers was not necessary. The same research group then reported a more straightforward method to improve the sensitivity of the aptasensor, which involved the use of an inexpensive metallic corrosion method to create a nanowell pattern on the QCM surface (Fig. 4 a, b). The nanostructuring approach increased the surface area of the electrode by 13 times, resulting in a 5-fold increase in the density of the immobilised aptamer. The nanowell aptasensor had a remarkable detection time of 10 min (Wang et al., 2017), matching the assay time of commercial RDTs for influenza, which typically ranges between 10–30 min (Cho et al., 2013).
Fig. 4.
Hydrogel aptasensor for the detection of AIV H5N1: (a) chemical modification of the QCM surface with a nanoporous gold film; (b) QCM electrode before (I) and after (II) the surface modification; (c) scanning electron microscopy (SEM) image of an AIV H5N1 bound to the nanowell structure on the crystal surface; (d) frequency shifts of the aptasensor to 2 HAU of different AIV subtypes; (e) frequency responses to tracheal swab samples spiked with 2-4 to 20 HAU of AIV H5N1. Reprinted with permission from Wang et al. (2017). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
During an infection, the HA glycoprotein on the viral envelope interacts with molecules containing sialic acid (SA) on the surface of host cells to facilitate cytoplasmic invasion (Hai et al., 2017). Several studies have exploited this interaction for the detection of influenza viruses in QCM platforms. Diltemiz et al. (2013) immobilised SA molecules onto the crystal surface and demonstrated the ability of the system to detect the HA glycoprotein. In another work, Hai et al. (2017) modified the QCM crystal by grafting a conducting polymer and 2,6-sialyllactose onto the surface. The biosensor did not rely on antibodies in the detection mechanism, and it was able to detect H1N1 viruses to a detection limit of 0.12 HAU.
4.2. Hepatitis B
Hepatitis B is a viral infection affecting approximately 290 million people worldwide, with more than 600,000 deaths annually caused by the end-stage liver complications resulting from chronic hepatitis B (CHB). The disease is caused by infection of the hepatitis B virus (HBV), which induces liver inflammation triggered by the host immune response against the infected liver cells. Chronic liver inflammation leads to liver diseases (e.g. fibrosis and cirrhosis), and heightens the risk of developing hepatocellular carcinoma (HCC), which is the primary cancer of the liver. The progression of liver disease and HCC can be prevented through timely diagnosis, vaccination, and administration of antiviral drugs against CHB. Thus, early detection and routine monitoring of hepatitis B biomarkers in the blood and serum of patients are crucial in providing an effective therapy to prevent irreversible liver damage (Fourati and Pawlotsky, 2016; Kotha et al., 2018; Peeling et al., 2017).
HBV DNA levels in serum are a measure of viral replication and serve as a risk indicator of HCC. The viral DNA is also a critical biomarker as it allows monitoring of the antiviral response (Kotha et al., 2018). Yao et al. (2008) constructed two different biosensors using PNA and DNA probes, respectively, to detect HBV DNA. In addition to having a higher biological stability, the PNA-based biosensor was found to have a higher specificity and a shorter reaction time in detecting the viral DNA. For an increased signal resolution, RecA proteins coated with ssDNA probes were used as the mass enhancers. The resulting biosensor had a low detection limit (8.6 ng mL-1), and the detection could be completed within 50 min.
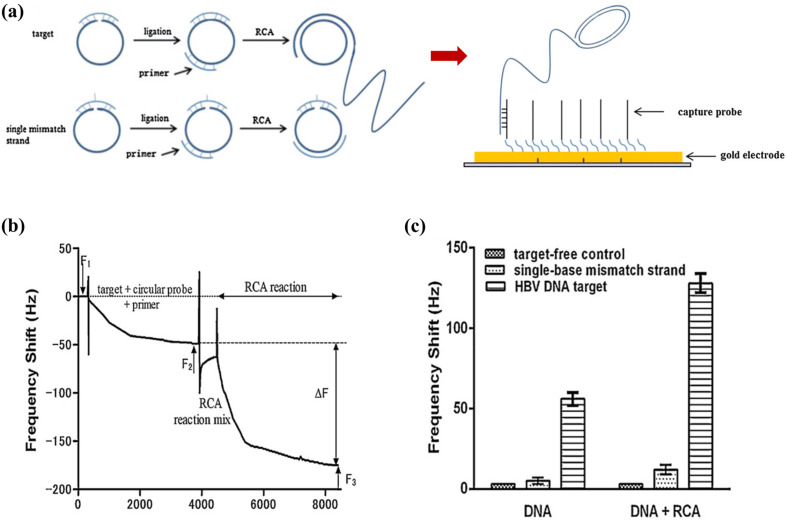
At present, the heavy reliance of nucleic acid-based biosensors on PCR for the amplification of target DNA to the detectable levels significantly increases the overall cost, duration, and complexity of these assays. In their subsequent work, Yao et al. (2013) designed a rolling circuit amplification (RCA) process to replace PCR for the amplification of HBV DNA in their detection procedure (Fig. 5 a). Amplification of target DNA was conducted via the RCA reaction in the QCM detection cell, with the process monitored in real-time (Fig. 5b). The RCA process resulted in a 10-fold magnification in the frequency shift, allowing as low as 104 copies mL-1 of HBV DNA to be detected. The RCA-QCM system completed the amplification and detection of targets within 60 min without the need for thermal cyclers. Furthermore, the unique design of the circular probe used in the amplification process enabled the single-base discrimination of sequences (Fig. 5c). The linear range of the biosensor (103–108 copies mL- 1 of HBV DNA) provided an appropriate working range for detection as it coincides with serum levels of the biomarker during an infection (Luckenbaugh et al., 2015). When tested on HBV-positive clinical samples, the RCA-QCM biosensor showed a 96% correlation with a real-time PCR assay. The excellent outcomes of that study proved the strategy of integrating isothermal amplification methods with QCM-based detection to be a promising approach for overcoming the inconveniences of conventional techniques for nucleic acid amplification.
Fig. 5.
RCA-QCM biosensor: (a) hybridisation of a circular probe with a target strand results in the circularisation and ligation of the probe, whereas RCA does not occur for a single-mismatch strand. The RCA product then binds to the crystal surface via the complementary capture probes; (b) real-time frequency response of the biosensor during the RCA reaction; (c) frequency shifts of the biosensor (with and without RCA reaction) in response to the negative control, HBV DNA target, and single-mismatch strand. Reprinted with permission from Yao et al. (2013).
Another clinically-significant biomarker of HBV is the hepatitis B surface antigen (HBsAg). The level of HBsAg in serum reflects the balance between the viral activity and host immune responses. Thus, an accurate quantitation of HBsAg will aid in determining the phase of infection, the severity of liver damage, the risk of HCC, and the response to antiviral treatments (Fourati and Pawlotsky, 2016). Shen et al. (2011) and Zhang et al. (2016) both used anti-HBsAg antibodies (anti-HBs) as the receptors for HBsAg detection. Both studies employed hyperbranched polymers (HBPs) for signal amplification, albeit through different approaches. Shen et al. (2011) modified the QCM surface with HBPs to provide a 3D structure for the immobilisation of more anti-HBs receptors. Conversely, Zhang et al. (2016) immobilised anti-HBs directly onto the QCM surface, and used the HBPs labelled with detection antibodies as the mass enhancers. While the former method achieved a signal amplification of 34%, the latter approach produced a 5-fold enhancement in frequency shift that resulted in a lower detection limit of 2 ng mL- 1. Furthermore, the quantitation of HBsAg in blood serum samples using the nanoparticle amplification strategy showed a good agreement with ELISA measurements (Zhang et al., 2016). A comparison of these studies suggests that by exploiting the mass of the HBPs for signal enhancement was more effective for increasing the signal resolution in a mass-based detection system such as QCM.
In another work, Lim et al. (2017) quantified hepatitis B core antigens (HBcAg) by the swelling effect of a hydrogel immobilised with anti-HBcAg antibodies (anti-HBc) and the pendant HBcAg. The hydrogel-based biosensor was able to detect HBcAg in the spiked serum samples within 25 min, and no cross-reactivity was observed with HBsAg. Additionally, the hydrogel showed a consistent swelling and deswelling behaviour upon several cycles of intermittent exposure to HBcAg samples and blank buffer, demonstrating that the biosensor could be regenerated and reused. The biosensor also had a wide detection range of 2–2000 μg mL- 1; however, this detection range was above the serum levels of HBcAg (50–3000 ng mL- 1) (Luckenbaugh et al., 2015). This shortcoming would limit its applicability for the direct detection of HBcAg from serum samples, as a pre-treatment step would be required to concentrate the target protein.
4.3. Dengue
Dengue is a vector-borne disease transmitted by the mosquitoes Aedes aegypti and Aedes albopictus. The disease is prevalent in tropical and sub-tropical climates, which promote the reproduction and incubation of dengue virus (DENV) in these mosquitoes. Currently, the virus is classified into four serotypes, namely DENV-1, DENV-2, DENV-3, and DENV-4. Because of the antigenic distinction among serotypes, infection with one serotype does not provide cross-protective immunity against the other serotypes. While primary infections of dengue may be inapparent, a second infection by another serotype can progress into a harsher condition, such as dengue hemorrhagic fever or dengue shock syndrome (DSS). DSS is associated with circulatory collapse, and it has a mortality rate of 1–5% (Darwish et al., 2015).
The non-structural 1 (NS1) protein is the most common biomarker for dengue detection. A high level of NS1 protein is present in the blood during the viremia phase, with concentrations in the range of 1–10 μg mL-1 the day after infection (Pirich et al., 2017). An early work by Su et al. (2003) used monoclonal antibodies against the viral envelope and NS1 proteins as the receptors. The QCM immunosensor showed great promise for the early detection of dengue as its sensitivity surpassed the 5 μg mL-1 detection limit of a commercial ELISA. In efforts to overcome the stability limitations of QCM immunosensors, the research group then used a linear epitope of the NS1 protein to fabricate a MIP film on the QCM surface to serve as artificial receptors for these analytes (Tai et al., 2006, 2005). The MIP biosensor based on the NS1 protein had a detection limit of 1–10 μg mL-1, and its sensitivity was retained after one month of storage (Tai et al., 2006).
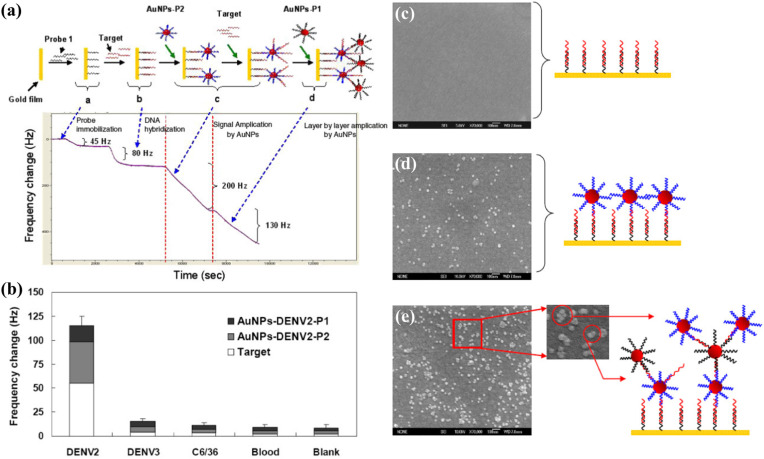
In another work, Chen et al. (2009) used DNA capture probes to detect sequences related to the DENV-2 genome, with two steps of signal amplification using AuNPs labelled with oligonucleotide probes (Fig. 6 ). The two-step amplification significantly lowered the detection limit from 100 PFU mL-1 to 2 PFU mL-1, matching the sensitivity of a real-time PCR assay and reducing the detection time. The low detection limit allowed the DNA-QCM biosensor to detect the viral genome in the initial phase of infection. Furthermore, the biosensor showed a high specificity to DENV-2, producing a significantly larger response to DENV-2-positive blood samples compared to DENV-3-positive and uninfected blood samples (Fig. 6b). The ability to distinguish between DENV serotypes makes it highly useful in the diagnosis of dengue. More recently, Pirich et al. (2017) created a nanoporous structure on QCM crystals by coating the surfaces with cellulose nanocrystals. The increase in surface roughness allowed a greater amount of antibody receptors to be immobilised, producing a detection limit of 0.1 μg mL-1, which is the lowest for QCM-based detection of NS1 protein by far.
Fig. 6.
DNA biosensor for dengue detection: (a) frequency responses of the biosensor at each stage of the detection process; (b) frequency shifts for spiked blood samples. SEM images of the QCM crystal surface: (c) after the hybridisation of target DNA; (d) after the binding of the first layer of AuNPs; (e) after the binding of the second layer of AuNPs. Reprinted with permission from Chen et al. (2009). Copyright IOP 2009.
4.4. Malaria
Malaria is caused by Plasmodium spp. parasites, which are transmitted by female Anopheles mosquitoes. The symptoms of malaria include fever, chills, headache, vomiting, muscle pain, and fatigue. Plasmodium falciparum causes a severe neurological complication known as cerebral malaria, whereas other species (e.g. Plasmodium vivax) cause milder forms of the disease (Ragavan et al., 2018). Early diagnosis and prompt administration of antimalarial drugs are critical in the treatment of malaria, as it can develop into a severe illness if not treated within the first 24 h of infection (Zarei, 2018).
Presently, the gold standard for malaria diagnosis is the identification of parasites in blood smears using microscopy. However, microscopy is known to be inaccurate due to its inability to detect low parasite densities and mixed infections (Ittarat et al., 2013; Ragavan et al., 2018). By designing primers of different lengths, Wangmaung et al. (2014) amplified the genes of P. falciparum and P. vivax using PCR. The amplified sequences were then detected using a QCM-based biosensor with DNA probes specific to a common gene of both species. As different lengths of target DNA were selected for P. falciparum and P. vivax, different magnitudes of frequency shifts were produced for each species, enabling single and mixed infections to be distinguished. Target DNA bands observed in agarose gel electrophoresis proved the accuracy of the QCM-based diagnosis. The QCM-based biosensor was found to be more sensitive and accurate compared to routine microscopy, which was prone to false negatives and misdiagnoses of P. vivax as P. falciparum. However, the drawback of this method was the lengthy detection procedure, which took up to 4 h due to the need for a PCR amplification step prior to QCM detection. In this regard, the development of PCR alternatives that can be integrated with the biosensor could expedite the detection procedure, as demonstrated by the RCA-QCM biosensor developed by Yao et al. (2013) for HBV detection.
In another approach, Sharma et al. (2011) developed a QCM immunosensor to diagnose P. falciparum infections via detection of the histidine-rich protein 2 (PfHRP-2). PfHRP-2 is a unique biomarker secreted by P. falciparum, and it can be detected in the blood, serum, plasma, and urine (Ragavan et al., 2018). The QCM method reported a low detection limit (12 ng mL- 1). The results of this biosensor-based diagnosis using the infected serum samples were in a good agreement with that of a commercial immunochromatographic test (ICT) kit. Notably, the QCM-based biosensor demonstrated a significant advantage over the ICT kit, in which the former was able to quantify the concentration of PfHRP-2 in samples, whereas the latter provided only colourimetric indications.
4.5. Human immunodeficiency virus infection
The human immunodeficiency virus (HIV) attacks the immune cells of an infected person and weakens the immune system. The acute phase of infection manifests in the form of a flu-like illness that is often not recognised as HIV infection due to the non-specific symptoms. The acute stage is followed by a prolonged asymptomatic period that may last up to 10 years, during which CD4 immune cells decline steadily (Gillespie et al., 2019). As the host immune system is compromised, the body becomes susceptible to opportunistic infections such as tuberculosis, cryptococcal meningitis, and cancer (Zarei, 2018). End-stage HIV infection is known as acquired immunodeficiency syndrome (AIDS), in which a patient who does not receive appropriate treatment may succumb to opportunistic infections within two years (Gillespie et al., 2019).
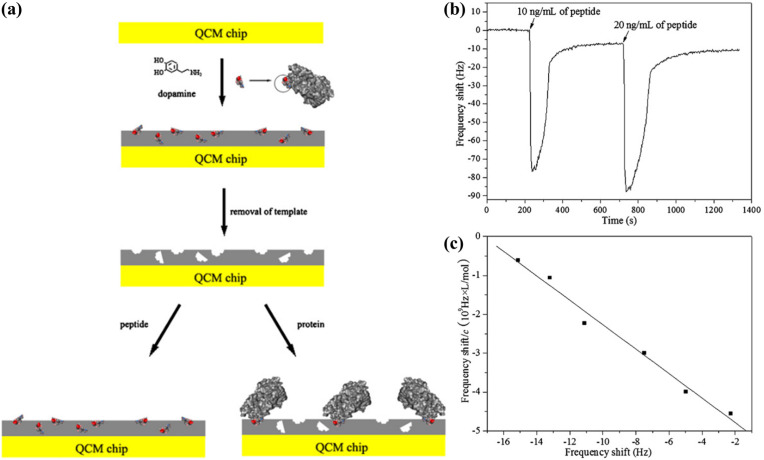
Lu et al. (2012) presented a MIP-based biosensor to detect the gp41 glycoproteins of HIV-1 for monitoring the viral infection (Fig. 7 ). The highlight of this research was the use of polydopamine, a novel bio-inspired polymer, to form a MIP film. The film thickness was optimised by varying the concentration of monomer used in the polymerisation process. The ability to adjust the film thickness provides a simple method to overcome the long-standing challenge in MIP-based recognition, which is the low density of recognition sites on the crystal surface. As a result, the biosensor achieved a low detection limit (2 ng mL-1) that was comparable to that of an ELISA. Furthermore, the recovery of gp41 proteins from diluted urine samples was up to 94.1%, showing that the biosensor had the potential to handle complex matrices. It is worth noting that the procedures for QCM functionalisation and target detection in this work are among the simplest that have been reported so far. The molecularly-imprinted polydopamine film was synthesised on the crystal surface via a simple step involving the self-oxidation of dopamine monomers incubated in an alkaline buffer. An optimised MIP film removed the need for signal amplification procedures; consequently, the detection process was markedly less complicated and could be completed within 10 min (Fig. 7b).
Fig. 7.
Polydopamine-based biosensor for the detection of HIV gp41 glycoproteins: (a) schematic representation of the epitope imprinting process; (b) real-time frequency responses of the biosensor to injections of the target peptide; (c) a Scatchard plot showing the linear range of the biosensor to the target peptide. Reprinted with permission from Lu et al. (2012).
Another recognised approach for detecting HIV infections is to target the p24 capsid protein of the virus, which is a virological marker that appears earlier than the anti-HIV antibodies by more than a week (Gillespie et al., 2019; Kirsch et al., 2013). Ly et al. (2016) demonstrated the feasibility of detecting p24 using a QCM immunosensor coupled with AuNPs for signal enhancement. The immunosensor had a wide detection range (1–107 ng mL-1). However, the detection process was slightly lengthy, taking more than 2 h due to the stepwise addition of detection antibodies and AuNPs. Nevertheless, the QCM immunosensor was a promising approach for the early detection of HIV infections, and for replacing the existing methodsthat depend mainly on the detection of anti-HIV antibodies in serum samples (Kirsch et al., 2013).
4.6. Tuberculosis
Tuberculosis is a bacterial infection caused by Mycobacterium tuberculosis (Mtb). The disease is airborne and typically affects the lungs. Tuberculosis is known to be the leading cause of death by a single infectious agent, and it is a common cause of death in patients with AIDS. Timely diagnosis and treatment with antibiotics can effectively cure and reduce the spread of the disease (World Health Organization, 2019).
Thus far, QCM immunosensors have been developed to detect whole cells of Mtb (He et al., 2002), the bacterial antigens (Hiatt and Cliffel, 2012; Montoya et al., 2016), and tuberculosis-related cytokines (Zhou et al., 2019). Hiatt and Cliffel (2012) constructed an immunosensor using lipoarabinomannan (LAM), a surface antigen of Mtb, as the target biomarker. The biosensor was able to detect both Mtb and LAM to the levels of 8.7 × 105 cells mL- 1 and 60 nM, respectively. Although the sensitivity of the QCM immunosensor for the detection of whole Mtb did not match that of a conventional ELISA, which had a detection limit that was one order of magnitude lower, the biosensor proved to be a more rapid technique because the detection could be completed within 20 min.
In a recent work, Zhou et al. (2019) reported an innovative approach for the continuous monitoring of three diagnostic biomarkers of latent tuberculosis infection (LTBI), namely interferon-gamma (IFN-γ), tumour necrosis factor-alpha (TNF-α), and interleukin-2 (IL-2). During the detection process, IFN-γ, TNF-α, and IL-2 molecules from the sample were captured onto the QCM surface by the immobilised antibody receptors. Silver nanoparticles labelled with specific antibodies against IFN-γ were then loaded to amplify the frequency shift caused by the bound IFN-γ molecules. After the signal had stabilised, hydrogen peroxide solution was injected into the detection cell to dissolve the silver nanoparticles and restore the frequency baseline. The signal amplification step was then repeated by loading silver nanoparticles labelled with specific antibodies against TNF-α and IL-2, respectively, into the detection cell. In this way, the concentration of each biomarker in a mixed sample could be determined, with detection limits in the fg mL- 1 range for all three molecules. Notably, the QCM-based quantitation in the spiked human serum samples showed a high recovery of LTBI biomarkers (97.8–108%) and an excellent agreement with the results of an ELISA. To date, there have been few reports on multi-analyte detection by QCM-based biosensors. The abovementioned work demonstrated the feasibility of performing the differential detection of multiple biomarkers in QCM platforms, which may promote future developments towards a more robust diagnostic system.
5. Challenges to clinical implementation
So far, QCM-based biosensors have demonstrated the ability to achieve detection limits that are applicable to the circulating levels of biomarkers (Pirich et al., 2017; Yao et al., 2013). Diagnostic results of these biosensors have shown excellent agreements with those of gold standard techniques such as ELISA (Zhou et al., 2019) and PCR (Wangmaung et al., 2014; Yao et al., 2013). Furthermore, QCM-based detection has been shown to outperform microscopy (Wangmaung et al., 2014), RDTs (Hewa et al., 2009; Sharma et al., 2011), dot-blot assays (Wang and Li, 2013) and shell vial assays (Hewa et al., 2009). Additionally, QCM-based biosensors have been reported to have a better target selectivity compared to surface plasmon resonance (Diltemiz et al., 2013; Tombelli et al., 2005) and potentiometric (Hai et al., 2017) biosensors developed using similar procedures. Although the reported results have been promising, QCM-based devices have yet to be commercialised for the clinical diagnosis of infectious diseases due to several challenges, namely the complex matrices of clinical samples and the low accessibility of QCM-based assays.
5.1. Complex matrices of clinical samples
The major hurdle to the clinical application of QCM-based biosensors is their poor detection performance in clinical samples. Clinical samples such as whole blood, serum, and urine exist as complex matrices, as they consist of various biomolecules that may interact with the sensor surface to compromise the sensitivity of the biosensor and produce inaccurate results (Palladino et al., 2018; Sin et al., 2014). As such, the detection limit may be raised by an order of magnitude in clinical samples to account for the high noise levels caused by the non-specific interactions (Li et al., 2011; Wang et al., 2017). Although numerous works have demonstrated the successful detection of analytes in clinical samples, additional procedures such as DNA extraction (Yao et al., 2008, 2013), PCR (Ittarat et al., 2013; Wangmaung et al., 2014), swab preparation (Li et al., 2011; Wang et al., 2017), and centrifugation (Lu et al., 2012) are often necessary for the extraction of target analytes and removal of interferents prior to detection.
For instance, the QCM immunosensor developed by Hewa et al. (2009) for influenza A and B viruses demonstrated a low detection limit and a fitting detection range when tested with dilutions of viruses in phosphate buffered saline. However, the viruses could not be detected in nasal wash samples due to the significant noise caused by other biomolecules in the samples. A positive detection from the nasal wash samples was finally achieved by passaging the samples in cell cultures overnight before analysis. Consequently, the entire detection procedure took two days, which was even longer than the detection by ELISA and reverse transcriptase PCR. Similarly, the reliance of DNA biosensors on PCR for the amplification of target sequences typically prolongs the detection process by several hours (Wangmaung et al., 2014). Sample pre-treatment involving laboratory procedures contributes to significant time delays and additional costs (Allain and Opare-Sem, 2016). Moreover, the need for laboratory facilities considerably reduces the field applicability of QCM-based biosensors. Hitherto, the challenges of matrix effects and sample processing have presented a significant difficulty in translating many emerging biosensor technologies from research laboratories to clinical applications (Sin et al., 2014).
Presently, most of the QCM-based biosensors rely on various signal amplification strategies, including mass enhancement, hydrogel swelling effect, and surface nanostructuring to achieve satisfactory levels of sensitivity and specificity, especially when performing the detection in biological matrices (Table 1). Of these methods, mass enhancement using AuNPs is the most common, and it is capable of producing large signal amplification effects. Nanomaterials are frequently used to enhance the sensitivity of assays due to their low cost, excellent thermal stability, and their ability to provide large surface areas (Dultsev and Tronin, 2015). However, the multi-step detection procedure may be inconvenient for point-of-care detection because it increases the detection time, reagent consumption, and complexity of the assay (Chen et al., 2009; Zhou et al., 2019).
5.2. Affordability and accessibility of QCM assays
At present, RDTs based on dipsticks and lateral flow immunoassays are popular methods for disease detection, especially for conducting mass screenings and for applications in low-resource settings. Compared to RDTs, QCM-based biosensors provide quantitative results, superior sensitivity, and a lower tendency for misdiagnosis, particularly in the case of mixed infections (Wangmaung et al., 2014). However, RDTs are highly affordable and deliverable to end-users; these characteristics make RDTs a tough market competitor for QCM-based biosensors, especially for point-of-care applications. Thus, QCM-based biosensors will need to emulate the performances of RDTs in these aspects to emerge as a competitive diagnostic tool.
Currently, antibody-based approaches predominate in QCM research for disease biosensing. However, the fabrication of QCM immunosensors is tedious and expensive. For this reason, the development of simpler and more economical methods to functionalise QCM crystals for biosensing is imperative. On the same note, cheaper options for electrode materials may also be considered. For instance, Wangmaung et al. (2014) used QCM crystals with silver electrodes, which were estimated to be 10 times cheaper than the conventional gold-coated crystals. However, this resulted in a trade-off in reusability as the silver-coated QCM crystals were less chemically stable.
In regards to reducing the cost of QCM assays, another parameter that warrants further study is the stability of the developed biosensors. Stability of a biosensor is characterised by the ability to retain its sensitivity and specificity under storage and reuse; a high stability translates to less stringent requirements for storage and a longer shelf life of the biosensor. Unlike RDTs, the functionalised QCM crystals may be regenerated for subsequent uses, which presents a potential approach to lowering the unit cost of QCM assays. Nonetheless, concerns pertaining to the reuse of these sensors, e.g. the risk of infection by the residual biomarkers and the additional operations required for regeneration, will need to be considered. At present, there are insufficient studies on the storage stability and reusability of QCM-based biosensors; moreover, available studies have reported a considerable variation in the performances of biosensors in these aspects. Efforts to increase the stability of receptors for regeneration is keyed to improving the reusability of QCM-based biosensors. The use of MIPs may provide the desired improvements in stability; however, studies on MIP-based biosensors for disease biosensing are currently lacking.
6. Research opportunities
In this section, research gaps in the field of QCM-based diagnostics are outlined, along with several areas of research that may be explored to circumvent the challenges mentioned and improve the applicability of QCM-based biosensors as a rapid diagnostic device for disease detection.
6.1. Improved methods for crystal functionalisation
Although many types of receptors and methods for surface functionalisation have been introduced to QCM platforms for biosensing, thorough studies on the properties of receptors and their recognition mechanisms are lacking. A better understanding of the recognition behaviour at the crystal surface and the optimisation of processes will aid efforts to improve the performances of the developed biosensors. A prominent example is in the construction of antibody- and DNA-based biosensors, in which most of the procedures aimed at achieving the maximum coverage of the QCM surface with receptors. However, several studies have suggested that a high density of receptors may not always correspond to increased sensitivity. Giamblanco et al. (2015) showed that the hybridisation efficiency and selectivity of a DNA biosensor towards the target sequences could be improved via optimisation of the DNA probe density on the QCM surface. The authors reported that the efficiency of target recognition decreased at a high density of immobilised DNA probes due to an overcrowding effect. Upon the optimisation of probe density, the desired sensitivity was achieved without the need for signal amplification. Sharma et al. (2011) observed a similar behaviour in a QCM immunosensor, in which the overloading of antibody receptors led to steric effects that resulted in a lower sensor response. Thus, the optimisation of the fabrication processes may inherently improve biosensor performance and simplify the detection process.
In the fabrication of MIP-based biosensors, the use of novel materials and the rational design of polymer compositions may improve the biosensing performance (Lu et al., 2012; Wangchareansak et al., 2013; Whitcombe et al., 2011). As an example, Wangchareansak et al. (2013) found that the addition of N-vinylpyrrolidone monomers to a co-polymer system consisting of acrylamide, methacrylic acid, and methyl methacrylate produced a dramatic increase in the selectivity of the biosensor towards different subtypes of influenza A virus. Optimisation of the ratio between monomers and cross-linkers further improved the sensitivity and specificity of the MIP-based biosensor. The extensive ranges of monomers and cross-linkers provide the possibility of tuning the material properties of MIPs via rational design to achieve the desired performances. Additionally, combinatorial and computational analysis may be utilised to design MIPs with improved recognition properties for the biosensing of disease biomarkers (Whitcombe et al., 2011).
6.2. Integration with microfluidic platforms
With small channel sizes in the nano- and micrometre ranges, microfluidic systems offer the advantages of low power and reagent consumptions, better flow manipulation, rapid diffusion, and short assay times (Sin et al., 2014). Microfluidic devices and lab-on-a-chip (LOC) platforms can perform many laboratory functions such as target extraction, nucleic acid amplification, as well as mixing and washing steps in the microscale (Sharma et al., 2015). These features make microfluidic devices an ideal platform for sample preparation for use in line with analytical techniques (Sin et al., 2014; Soares et al., 2016). Additionally, on-chip sample preparation can achieve the gentle and consistent sample processing that may enhance diagnosis results (Toner and Irimia, 2005). As such, microfluidic devices have gained recognition as the future technology for point-of-care medical diagnosis (Sharma et al., 2015). On that note, the many advantages of microfluidic and LOC platforms can be exploited to expedite and automate QCM-based detection processes.
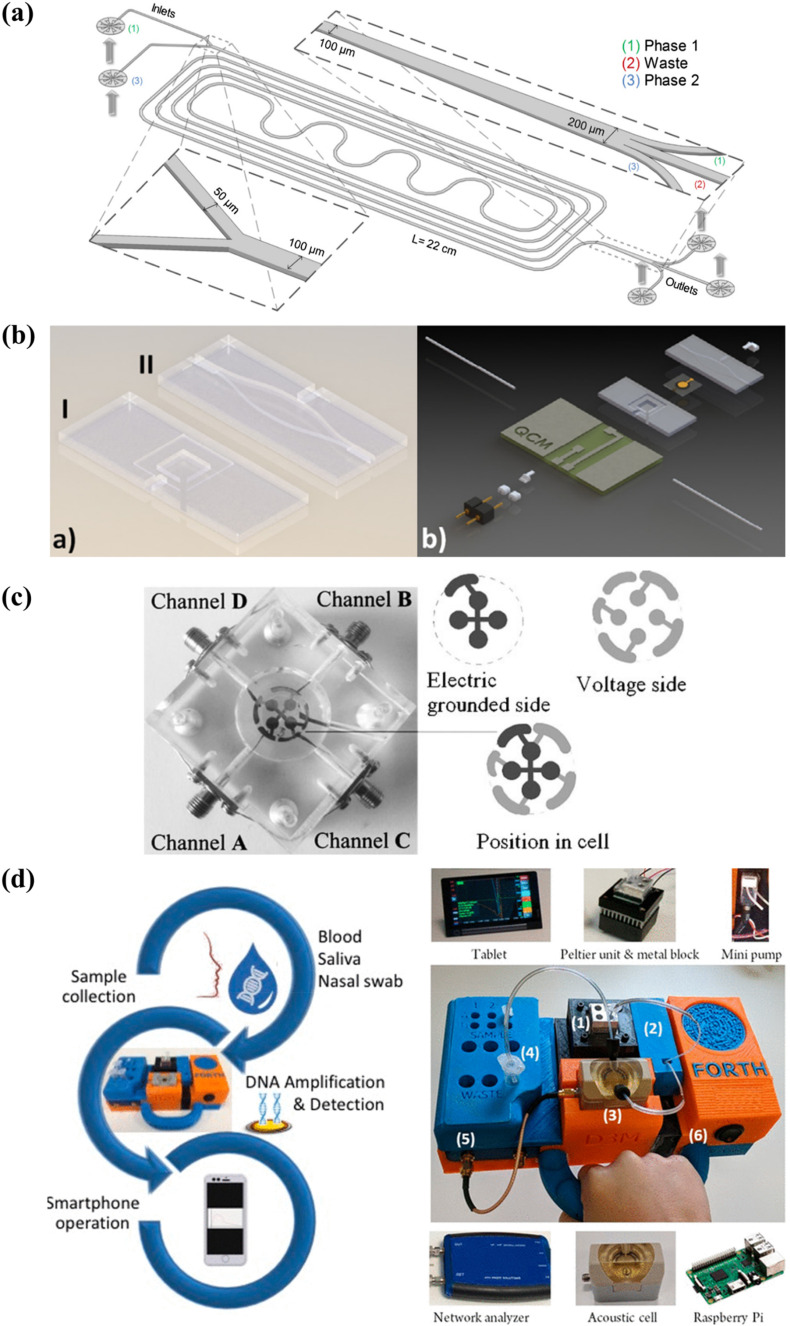
To date, several separation techniques such as the aqueous two-phase systems (ATPS) (Jacinto et al., 2015; Soares et al., 2017) and magnetic bead separation (Foudeh et al., 2012) have been successfully integrated into microfluidic devices for the processing of complex samples. The design of a microfluidic ATPS used in a selective extraction process is illustrated in Fig. 8 a. In a past study, a microfluidic ATPS was coupled to a lateral flow immunoassay to extract and concentrate mycotoxins from red wine samples for detection. The pre-treatment step, which could be completed in 20 min, lowered the detection limit of the assay from 100 ng mL- 1 to 0.26 ng mL- 1 (Soares et al., 2017). Microfluidic sample preparation has indeed proven to be capable of reducing matrix effects, consequently improving the performance of detection assays.
Fig. 8.
Research opportunities in QCM-based diagnostics: (a) design of a microfluidic ATPS for the selective extraction of virus-like particles from cell cultures; (b) 3D design of a microfluidic QCM sensor for the detection of C-reactive protein; (c) a tetra-electrode QCM system connected to four channels for sample delivery; (d) a portable QCM platform for point-of-care genetic testing. Reprinted with permissions from Jacinto et al. (2015), Thies et al. (2017), Latif et al. (2011) (Copyright Springer 2011), and Papadakis et al. (2019) (Copyright ACS 2019), respectively.
In recent years, QCM systems have been miniaturised and integrated into microfluidic chips to perform detection assays (Fig. 8b) (Tao et al., 2015; Thies et al., 2017). The ability of QCM to be operated under continuous or partial flow modes makes it compatible for integration with microfluidic systems. Microfluidic QCM systems can be expanded to include a sample pre-treatment step, thereby creating a stand-alone platform for the direct detection of disease biomarkers from clinical samples. Such developments would eliminate the need for laboratory processes to enable the point-of-care application of QCM-based biosensors. Most importantly, this will provide a more rapid diagnosis and a considerable reduction in assay cost (Allain and Opare-Sem, 2016). Additionally, microfluidic and LOC platforms can aid in the facilitation of multi-step operations involving signal amplification, washing, and regeneration procedures. These procedures can be automated and performed with a better process control to create a more rapid and robust diagnostic system. Moreover, a microfluidic QCM device can provide a closed platform for performing the analysis of clinical samples to reduce the biohazard risk (Yamaguchi et al., 2017).
6.3. QCM-D and multi-electrode QCMs
Thus far, the full capability of QCM systems has yet to be utilised for disease biosensing. The QCM-D technique, which allows the simultaneous monitoring of frequency and dissipation, has been increasingly applied in the studies of cell adhesion and protein interactions. However, the use of this function for disease biosensing is still in its infancy. The combined data of frequency and dissipation can provide information on the rigidity and viscoelasticity of particles adsorbed on the crystal surface (Karczmarczyk et al., 2017); this information will aid in verifying the identity of the bound particles causing the frequency responses. In this way, QCM-D offers a built-in validation that may improve the reliability of QCM-based diagnoses. For biosensing in liquid samples, more accurate results can be obtained by taking advantage of the ability of QCM-D to monitor frequency responses at different overtones. Higher overtones are less sensitive to changes in the bulk solution and may provide better sensitivity and reproducibility in measurements (Bianco et al., 2013; Karczmarczyk et al., 2017; Pirich et al., 2017).
Finally, multi-electrode QCM crystals (Fig. 8c) can be explored for high-throughput sampling and the simultaneous detection of multiple analytes (Afzal et al., 2017). Development of a more robust detection system with such capabilities will be highly useful for mass screening applications and for diagnosing diseases that have many circulating subtypes. Besides, the detection of multiple biomarkers will aid in characterising the phase of infection, since pathogens produce different biomarkers at various stages of their life cycles (Pashchenko et al., 2018).
6.4. Mobile health diagnostics
After sample collection and testing, the compilation and analysis of data are integral to producing a diagnosis. A core feature of the highly successful glucose biosensor is the simple readout that makes the interpretation of test results easy for end-users. Enabling a similar capability in QCM devices would empower end-users and promote the use of these biosensors for point-of-care diagnosis and health monitoring (Ragavan et al., 2018). The increasing adoption of smartphones and the changing consumer attitude towards self-testing are among the factors contributing to the current trend of mobile health, which involves the application of mobile devices (e.g. smartphones and tablets) for healthcare (Wood et al., 2019). QCM-based biosensors are highly suited for this application since the output of these devices is a digital signal that can be processed by computers. Efforts to keep QCM developments up to date with these technological advancements would go a long way in increasing the competitiveness of QCM-based biosensors as a rapid diagnostic device.
Today, many commercial QCM systems can incorporate wireless communication for establishing connections to smart devices for data collection and interpretation (Fig. 8d). The development of accompanying smartphone applications that can interpret frequency responses into the state of infection would aid physicians and patients in monitoring the progression of a disease and the treatment efficacy. Mobile health is forecasted to increase access to the testing, diagnosis, and treatment of infectious diseases (Wood et al., 2019), presenting an attractive future scope for QCM-based diagnostics.
7. Conclusions and future perspectives
Sensitive, rapid, and affordable diagnostic technologies are indispensable in the fight against infectious diseases. QCM technology has evolved significantly over the past decades to emerge as an excellent platform for disease biosensing. Thus far, QCM-based biosensors have proven their ability to detect a broad range of disease biomarkers with low detection limits that are relevant for the early detection of infections. Presently, the bridging of the gap between research and commercialisation of QCM-based biosensors for infectious diseases is hindered by the hurdles of sample preparation and matrix effects, as well as the lower affordability of QCM-based assays relative to RDTs. To bring QCM-based biosensors closer to the stage of clinical application, research should aim towards improving the applicability and the detection performance of these biosensors under clinically-relevant conditions, in efforts to fulfil the ASSURED criteria established by WHO for medical diagnostics. In this regard, enhanced methods for functionalising QCM surfaces for biosensing are essential. Such attempts will involve improving the properties and recognition performances of receptors, e.g. by increasing the stability of antibody and nucleic acid receptors, and by improving the specificity of MIPs. The next research focus is to overcome the matrix effects of clinical samples and the reliance on laboratory sample processing, to realise the point-of-care application of QCM-based biosensors for disease detection. In our opinion, a significant advantage of QCM systems lies in their tremendous potential for an innovative integration of other technologies that can increase the robustness of biosensing. In line with the current trend towards point-of-care healthcare and diagnosis, microfluidic and mobile health technologies will play a key role in boosting the applicability and the competitiveness of QCM-based biosensors for the rapid detection of infectious diseases. Although QCM-based biosensors are yet to reach clinical application, their prospects as a rapid diagnostic device are undoubtedly promising.
Author contributions
Hui Jean Lim: Conceptualisation, Writing - Original Draft. Tridib Saha: Writing - Review & Editing. Beng Ti Tey: Writing - Review & Editing. Wen Siang Tan: Writing - Review & Editing. Chien Wei Ooi: Writing - Review & Editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Tropical Medicine and Biology Platform, Monash University Malaysia. Lim H. J. acknowledges the financial support provided by Monash University Malaysia under the Higher Degree Research Scholarship.
References
- Afzal A., Mujahid A., Schirhagl R., Bajwa S.Z., Latif U., Feroz S. Chemosensors. 2017;5:7. [Google Scholar]
- Allain J.-P., Opare-Sem O. Nat. Rev. Gastroenterol. Hepatol. 2016;13:643–653. doi: 10.1038/nrgastro.2016.138. [DOI] [PubMed] [Google Scholar]
- Ballantine D.S., Jr., White R.M., Martin S.J., Ricco A.J., Zellers E.T., Frye G.C., Wohltjen H. Elsevier; 1996. Acoustic Wave Sensors: Theory, Design and Physico-Chemical Applications. [Google Scholar]
- Bianco M., Aloisi A., Arima V., Capello M., Ferri-Borgogno S., Novelli F., Leporatti S., Rinaldi R. Biosens. Bioelectron. 2013;42:646–652. doi: 10.1016/j.bios.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Bloom D.E., Cadarette D. Front. Immunol. 2019;10:1–12. doi: 10.3389/fimmu.2019.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoli G., Susta L., Terregino C., Brown C. J. Vet. Diagn. Invest. 2011;23:637–656. doi: 10.1177/1040638711407887. [DOI] [PubMed] [Google Scholar]
- Chauhan R., Solanki P.R., Singh J., Mukherjee I., Basu T., Malhotra B.D. Food Contr. 2015;52:60–70. [Google Scholar]
- Chen J.-C., Sadhasivam S., Lin F.-H. Process Biochem. 2011;46:543–550. [Google Scholar]
- Chen J.Y., Penn L.S., Xi J. Biosens. Bioelectron. 2018;99:593–602. doi: 10.1016/j.bios.2017.08.032. [DOI] [PubMed] [Google Scholar]
- Chen S.-H., Chuang Y.-C., Lu Y.-C., Lin H.-C., Yang Y.-L., Lin C.-S. Nanotechnology. 2009;20:215501. doi: 10.1088/0957-4484/20/21/215501. [DOI] [PubMed] [Google Scholar]
- Cho C.H., Woo M.K., Kim J.Y., Cheong S., Lee C.-K., An S.A., Lim C.S., Kim W.J. J. Virol. Methods. 2013;187:51–56. doi: 10.1016/j.jviromet.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Cooper M.A., Singleton V.T. J. Mol. Recogn. 2007;20:154–184. doi: 10.1002/jmr.826. [DOI] [PubMed] [Google Scholar]
- Darwish N.T., Alias Y.B., Khor S.M. TrAC Trends Anal. Chem. 2015;67:45–55. [Google Scholar]
- Diltemiz S.E., Ersöz A., Hür D., Keçili R., Say R. Mater. Sci. Eng. C. 2013;33:824–830. doi: 10.1016/j.msec.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Dultsev F.N., Tronin A.V. Sensor. Actuator. B Chem. 2015;216:1–5. [Google Scholar]
- Eren T., Atar N., Yola M.L., Karimi-Maleh H. Food Chem. 2015;185:430–436. doi: 10.1016/j.foodchem.2015.03.153. [DOI] [PubMed] [Google Scholar]
- Fogel R., Limson J., Seshia A.A. Essays Biochem. 2016;60:101–110. doi: 10.1042/EBC20150011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudeh A.M., Didar T.F., Veres T., Tabrizian M., Fatanat Didar T., Veres T., Tabrizian M. Lab Chip. 2012;12:3249–3266. doi: 10.1039/c2lc40630f. [DOI] [PubMed] [Google Scholar]
- Fourati S., Pawlotsky J.-M. F1000Research. 2016;5:2243. doi: 10.12688/f1000research.8983.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdon A.E., Wright D.W., Cliffel D.E. Biomacromolecules. 2005;6:3419–3424. doi: 10.1021/bm050475o. [DOI] [PubMed] [Google Scholar]
- Giamblanco N., Conoci S., Russo D., Marletta G. RSC Adv. 2015;5:38152–38158. [Google Scholar]
- Gillespie S.L., Chinen J., Paul M.E., Shearer W.T. Clinical Immunology. Elsevier; 2019. Human immunodeficiency virus infection and acquired immunodeficiency syndrome; pp. 545–560. [Google Scholar]
- Hai W., Goda T., Takeuchi H., Yamaoka S., Horiguchi Y., Matsumoto A., Miyahara Y. ACS Appl. Mater. Interfaces. 2017;9:14162–14170. doi: 10.1021/acsami.7b02523. [DOI] [PubMed] [Google Scholar]
- Hao R.Z., Song H.-B., Zuo G.M., Yang R.F., Wei H.-P., Wang D.-B., Cui Z.-Q., Zhang Z., Cheng Z.-X., Zhang X.-E. Biosens. Bioelectron. 2011;26:3398–3404. doi: 10.1016/j.bios.2011.01.010. [DOI] [PubMed] [Google Scholar]
- He F., Zhang L., Zhao J., Hu B., Lei J. Sensor. Actuator. B Chem. 2002;85:284–290. [Google Scholar]
- Hewa T.M.P., Tannock G.A., Mainwaring D.E., Harrison S., Fecondo J.V. J. Virol. Methods. 2009;162:14–21. doi: 10.1016/j.jviromet.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt L.A., Cliffel D.E. Sensor. Actuator. B Chem. 2012;174:245–252. doi: 10.1016/j.snb.2012.06.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittarat W., Chomean S., Sanchomphu C., Wangmaung N., Promptmas C., Ngrenngarmlert W. Clin. Chim. Acta. 2013;419:47–51. doi: 10.1016/j.cca.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Jacinto M.J., Soares R.R.G., Azevedo A.M., Chu V., Tover A., Conde J.P., Aires-Barros M.R. Separ. Purif. Technol. 2015;154:27–35. [Google Scholar]
- Karczmarczyk A., Haupt K., Feller K.-H. Talanta. 2017;166:193–197. doi: 10.1016/j.talanta.2017.01.054. [DOI] [PubMed] [Google Scholar]
- Kirsch J., Siltanen C., Zhou Q., Revzin A., Simonian A. Chem. Soc. Rev. 2013;42:8733–8768. doi: 10.1039/c3cs60141b. [DOI] [PubMed] [Google Scholar]
- Kosack C.S., Page A.-L., Klatser P.R. Bull. World Health Organ. 2017;95:639–645. doi: 10.2471/BLT.16.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotha S., Neong S.F., Patel K. Expert Rev. Mol. Diagn. 2018;18:713–722. doi: 10.1080/14737159.2018.1496020. [DOI] [PubMed] [Google Scholar]
- Latif U., Mujahid A., Afzal A., Sikorski R., Lieberzeit P.A., Dickert F.L. Anal. Bioanal. Chem. 2011;400:2507–2515. doi: 10.1007/s00216-011-4927-1. [DOI] [PubMed] [Google Scholar]
- Li D., Wang J., Wang R., Li Y., Abi-Ghanem D., Berghman L., Hargis B., Lu H. Biosens. Bioelectron. 2011;26:4146–4154. doi: 10.1016/j.bios.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Lim S.L., Ooi C.W., Tan W.S., Chan E.S., Ho K.L., Tey B.T. Sensor. Actuator. B Chem. 2017;252:409–417. [Google Scholar]
- Lu C.-H., Zhang Y., Tang S.-F., Fang Z.-B., Yang H.-H., Chen X., Chen G.-N. Biosens. Bioelectron. 2012;31:439–444. doi: 10.1016/j.bios.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Luckenbaugh L., Kitrinos K.M., Delaney IV W.E., Hu J. J. Viral Hepat. 2015;22:561–570. doi: 10.1111/jvh.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly T.N., Park S., Park S.J. Sensor. Actuator. B Chem. 2016;237:452–458. [Google Scholar]
- Ma X.-T., He X.-W., Li W.-Y., Zhang Y.-K. Sensor. Actuator. B Chem. 2017;246:879–886. [Google Scholar]
- Mattiasson B., Ye L. Springer; 2015. Molecularly Imprinted Polymers in Biotechnology. [Google Scholar]
- Menger M., Yarman A., Erdőssy J., Yildiz H.B., Gyurcsányi R., Scheller F.W. Biosensors. 2016;6:35. doi: 10.3390/bios6030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya A., March C., Montagut Y.J., Moreno M.J., Manclus J.J., Arnau A., Jimenez Y., Jaramillo M., Marin P.A., Torres R.A. Curr. Top. Med. Chem. 2016;17:1623–1630. doi: 10.2174/1568026617666161104105210. [DOI] [PubMed] [Google Scholar]
- Palladino P., Minunni M., Scarano S. Biosens. Bioelectron. 2018;106:93–98. doi: 10.1016/j.bios.2018.01.068. [DOI] [PubMed] [Google Scholar]
- Papadakis G., Pantazis A.K., Ntogka M., Parasyris K., Theodosi G.-I., Kaprou G., Gizeli E. ACS Sens. 2019;4:1329–1336. doi: 10.1021/acssensors.9b00264. [DOI] [PubMed] [Google Scholar]
- Pashchenko O., Shelby T., Banerjee T., Santra S. ACS Infect. Dis. 2018;4:1162–1178. doi: 10.1021/acsinfecdis.8b00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., Boeras D.I., Marinucci F., Easterbrook P. BMC Infect. Dis. 2017;17:699. doi: 10.1186/s12879-017-2775-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan N.V.H., Sussitz H.F., Lieberzeit P.A. Biosensors. 2014;4:161–171. doi: 10.3390/bios4020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirich C.L., de Freitas R.A., Torresi R.M., Picheth G.F., Sierakowski M.R. Biosens. Bioelectron. 2017;92:47–53. doi: 10.1016/j.bios.2017.01.068. [DOI] [PubMed] [Google Scholar]
- Qiao X., Zhang X., Tian Y., Meng Y. Appl. Phys. Rev. 2016;3:31106. [Google Scholar]
- Ragavan K.V., Kumar S., Swaraj S., Neethirajan S. Biosens. Bioelectron. 2018;105:188–210. doi: 10.1016/j.bios.2018.01.037. [DOI] [PubMed] [Google Scholar]
- Saadati A., Hassanpour S., de la Guardia M., Mosafer J., Hashemzaei M., Mokhtarzadeh A., Baradaran B. TrAC Trends Anal. Chem. 2019;114:56–68. [Google Scholar]
- Saunders-Hastings P.R., Krewski D. Pathogens. 2016;5:66. doi: 10.3390/pathogens5040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirhagl R. Anal. Chem. 2013;86:250–261. doi: 10.1021/ac401251j. [DOI] [PubMed] [Google Scholar]
- Sharma M.K., Rao V.K., Merwyn S., Agarwal G.S., Upadhyay S., Vijayaraghavan R. Talanta. 2011;85:1812–1817. doi: 10.1016/j.talanta.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Sharma S., Zapatero-Rodríguez J., Estrela P., O'Kennedy R. Biosensors. 2015;5:577–601. doi: 10.3390/bios5030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G., Cai C., Wang K., Lu J. Anal. Biochem. 2011;409:22–27. doi: 10.1016/j.ab.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Shojaei T.R., Tabatabaei M., Shawky S., Salleh M.A.M., Bald D. Mol. Biol. Rep. 2015;42:187–199. doi: 10.1007/s11033-014-3758-5. [DOI] [PubMed] [Google Scholar]
- Sin M.L.Y., Mach K.E., Wong P.K., Liao J.C. Expert Rev. Mol. Diagn. 2014;14:225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares R.R.G., Azevedo A.M., Fernandes P., Chu V., Conde J.P., Aires-Barros M.R. J. Chromatogr. A. 2017;1511:15–24. doi: 10.1016/j.chroma.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Soares R.R.G., Silva D.F.C., Fernandes P., Azevedo A.M., Chu V., Conde J.P., Aires-Barros M.R. Biotechnol. J. 2016;11:1498–1512. doi: 10.1002/biot.201600356. [DOI] [PubMed] [Google Scholar]
- Srinivasan B., Tung S. J. Lab. Autom. 2015;20:365–389. doi: 10.1177/2211068215581349. [DOI] [PubMed] [Google Scholar]
- Steinem C., Janshoff A. first ed. Springer; 2007. Piezoelectric Sensors. [Google Scholar]
- Su C.-C., Wu T.-Z., Chen L.-K., Yang H.-H., Tai D.-F. Anal. Chim. Acta. 2003;479:117–123. [Google Scholar]
- Tai D.-F., Lin C.-Y., Wu T.-Z., Chen L.-K. Anal. Chem. 2005;77:5140–5143. doi: 10.1021/ac0504060. [DOI] [PubMed] [Google Scholar]
- Tai D.-F., Lin C.-Y., Wu T.-Z., Huang J.-H., Shu P.-Y. Clin. Chem. 2006;52:1486–1491. doi: 10.1373/clinchem.2005.064501. [DOI] [PubMed] [Google Scholar]
- Tao W., Xie Q., Wang H., Ke S., Lin P., Zeng X. Sensors. 2015;15:25746–25760. doi: 10.3390/s151025746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies J.-W., Kuhn P., Thürmann B., Dübel S., Dietzel A. Microelectron. Eng. 2017;179:25–30. [Google Scholar]
- Tombelli S., Minunni M., Luzi E., Mascini M. Bioelectrochemistry. 2005;67:135–141. doi: 10.1016/j.bioelechem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Toner M., Irimia D. Annu. Rev. Biomed. Eng. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemula S.V., Zhao J., Liu J., Xue X.W., Biswas S., Hewlett I., Wang X., Biswas S., Hewlett I. Viruses. 2016;8:1–15. [Google Scholar]
- Wang R., Li Y. Biosens. Bioelectron. 2013;42:148–155. doi: 10.1016/j.bios.2012.10.038. [DOI] [PubMed] [Google Scholar]
- Wang R., Wang L., Callaway Z.T., Lu H., Huang T.J., Li Y. Sensor. Actuator. B Chem. 2017;240:934–940. [Google Scholar]
- Wangchareansak T., Thitithanyanont A., Chuakheaw D., Gleeson M.P., Lieberzeit P.A., Sangma C. J. Mater. Chem. B. 2013;1:2190–2197. doi: 10.1039/c3tb00027c. [DOI] [PubMed] [Google Scholar]
- Wangmaung N., Chomean S., Promptmas C., Mas-Oodi S., Tanyong D., Ittarat W. Biosens. Bioelectron. 2014;62:295–301. doi: 10.1016/j.bios.2014.06.052. [DOI] [PubMed] [Google Scholar]
- Whitcombe M.J., Chianella I., Larcombe L., Piletsky S.A., Noble J., Porter R., Horgan A. Chem. Soc. Rev. 2011;40:1547–1571. doi: 10.1039/c0cs00049c. [DOI] [PubMed] [Google Scholar]
- Wood C.S., Thomas M.R., Budd J., Mashamba-Thompson T.P., Herbst K., Pillay D., Peeling R.W., Johnson A.M., McKendry R.A., Stevens M.M. Nature. 2019;566:467–474. doi: 10.1038/s41586-019-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2019. Global Tuberculosis Report 2019. [Google Scholar]
- Xie X., Wang C., Xiao Q., Zheng Y., Li Y., Feng B. Anal. Methods. 2015;7:9018–9025. [Google Scholar]
- Xu T., Miao J., Wang Z., Yu L., Li C.M. Sensor. Actuator. B Chem. 2011;151:370–376. [Google Scholar]
- Yamaguchi N., Tokunaga Y., Goto S., Fujii Y., Banno F., Edagawa A. Sci. Rep. 2017;7:1–6. doi: 10.1038/s41598-017-03293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C.-Y., Fu W.-L. World J. Gastroenterol. 2014;20:12485. doi: 10.3748/wjg.v20.i35.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Xiang Y., Deng K., Xia H., Fu W. Sensor. Actuator. B Chem. 2013;181:382–387. [Google Scholar]
- Yao C., Zhu T., Tang J., Wu R., Chen Q., Chen M., Zhang B., Huang J., Fu W. Biosens. Bioelectron. 2008;23:879–885. doi: 10.1016/j.bios.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Zambon M. Medicine. 2014;42:45–51. doi: 10.1016/j.mpmed.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei M. Biosens. Bioelectron. 2018;106:193–203. doi: 10.1016/j.bios.2018.02.007. [DOI] [PubMed] [Google Scholar]
- Zhang H. Polymer. 2014;55:699–714. [Google Scholar]
- Zhang L.-Q., Shen H.-X., Cheng Q., Liu L.-C. Chem. Pap. 2016;70:1031–1038. [Google Scholar]
- Zhou B., Hao Y., Chen S., Yang P. Microchim. Acta. 2019;186:212. doi: 10.1007/s00604-019-3319-7. [DOI] [PubMed] [Google Scholar]