Abstract
Although the human body appears superficially symmetrical with regard to the left–right (L-R) axis, most visceral organs are asymmetric in terms of their size, shape, or position. Such morphological asymmetries of visceral organs, which are essential for their proper function, are under the control of a genetic pathway that operates in the developing embryo. In many vertebrates including mammals, the breaking of L-R symmetry occurs at a structure known as the L-R organizer (LRO) located at the midline of the developing embryo. This symmetry breaking is followed by transfer of an active form of the signaling molecule Nodal from the LRO to the lateral plate mesoderm (LPM) on the left side, which results in asymmetric expression of Nodal (a left-side determinant) in the left LPM. Finally, L-R asymmetric morphogenesis of visceral organs is induced by Nodal-Pitx2 signaling. This review will describe our current understanding of the mechanisms that underlie the generation of L-R asymmetry in vertebrates, with a focus on mice.
Keywords: chirality, cilia, left–right asymmetry, Nodal
1. Introduction
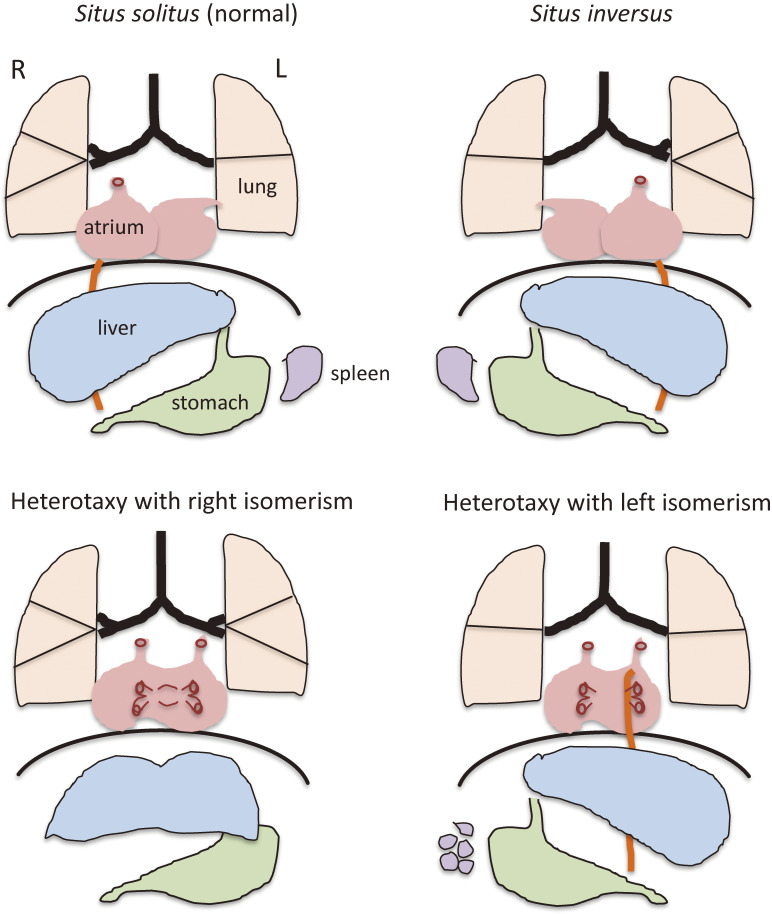
The bodies of bilaterian animals possess three axes: anterior–posterior (A-P), dorso–ventral (D-V), and left–right (L-R). These axes are determined early during development, with the L-R axis being the last to be established. Most visceral organs of vertebrates including humans are L-R asymmetric in terms of their size, shape, or position (Fig. 1). In normal conditions (situs solitus) in humans, the stomach and spleen are located on the left side, whereas the liver is on the right. In addition, the left and right sides of the lung have two and three lobes, respectively. The heart also manifests L-R asymmetries. The apex of the heart is thus located on the left side, and the connection of the great arteries to the ventricles depends on L-R information, with the pulmonary artery and aorta being connected to the right ventricle and left ventricle, respectively. Vertebrates manifest marked diversity in morphological asymmetries of their organs. For instance, there are four lobes of the lung on the right and one on the left in the mouse, female birds have an ovary only on the left side,1) and many snake species possess only a right lung.2)
Figure 1.
Schematic illustration of L-R asymmetry of human visceral organs. Normal L-R asymmetry (situs solitus) and three laterality defects that affect the lung, heart, liver, stomach, and spleen are shown. Heterotaxy with right isomerism is usually associated with a bilateral trilobed lung, a large symmetric liver, and the absence of a spleen. Heterotaxy with left isomerism often manifests as a bilateral bilobed lung, multiple spleens, and a pulmonary vein that drains into both the right and left atria.
The normal laterality pattern is altered in various different ways in pathological conditions (Fig. 1). For organs that exist singly in the viscera such as the heart and stomach, their position may be normal, reversed, or ambiguous. For bilateral organs such as the lung, their morphology may be normal or reversed, or show right isomerism or left isomerism. At the level of the whole organism, the laterality of all organs may be completely reversed in a mirror image (situs inversus). Alternatively, visceral organs might be altered in a discordant manner (heterotaxy), with some organs being unaffected and others showing an ambiguous or reversed position. Two types of heterotaxy are often encountered in humans: heterotaxy with right isomerism and heterotaxy with left isomerism. The former is usually recognized by a bilateral trilobed lung, a large symmetric liver, and the absence of a spleen, whereas the latter is characterized by a bilateral bilobed lung and multiple spleens.
The heart may be the organ most sensitive to laterality defects. Heart anomalies often associated with such defects include transposition of the great arteries, in which the pulmonary artery and aorta are incorrectly connected to the left ventricle and right ventricle, respectively, and double outlet right ventricle, in which both great arteries connect to the right ventricle. Such heart defects in humans are life threatening and usually require immediate medical attention after birth.
2. Historical view
Disorders of L-R asymmetry result in randomization (heterotaxy) or complete reversal (situs inversus) of visceral organs. Humans with laterality defects were described by physicians as long ago as the 16th century. An individual showing the complete mirror-image reversal of visceral organs was recorded in the 18th century.3) Some of the described individuals also manifested bronchiectasis, with their condition being named Kartagener syndrome in the early 20th century4) (Table 1) and later renamed primary ciliary dyskinesia.
Table 1.
Genetic control of L-R asymmetry. Human disorders and mutant animals that provided breakthroughs in our understanding of the mechanisms underlying the development of L-R asymmetry
| Animal | Disorder or mutation | Defects | Year reported | Ref. |
|---|---|---|---|---|
| Human | Kartagener syndrome (ciliary proteins) | Situs inversus/ heterotaxy, chronic respiratory infection, infertility | 1789 | 3 |
| 1933 | 4 | |||
| Mouse | Iv (Dnah11) | L-R is randomized | 1959 | 5 |
| Inv (Inversin) | L-R is reversed | 1993 | 6 | |
| Drosophila | Myo31DF (myosin 1D) | Looping of gut and testes is reversed | 2006 | 13, 14 |
| Snail | Direction of coiling: sinistral vs. dextral | 1923 | 15 | |
The notion that L-R asymmetry is genetically determined became obvious when a mutant mouse (situs inversus viscerum, or iv) with randomized L-R asymmetry was identified in 1959 (Table 1).5) This spontaneously occurring mutation caught the attention of biologists, but the causative gene remained unknown for decades. Another mutant mouse (inv) was described in 1993.6) In contrast to the iv/iv mutant, L-R asymmetry is always reversed in inv/inv mutant mice. In the mid-1990s, the identification of Nodal and Lefty as L-R asymmetrically expressed genes in chick7) and mouse8,9) embryos provided a breakthrough in the study of the molecular and genetic mechanisms underlying the development of L-R asymmetry. The causative gene for the iv mutation was found to encode an axonemal dynein protein in 1997,10) which provided support for the idea that L-R asymmetry requires motile cilia. The gene that harbors the inv mutation was also identified and found to encode a large protein designated Inversin.11) Inversin plays a role in cilia (immotile cilia), but its precise function remains elusive. Although a link between laterality and cilia had been suggested earlier, the discovery of motile cilia and a leftward fluid flow at the node of the mouse embryo in 1998 clearly established the role of cilia in L-R asymmetry for the first time.12)
Unlike many other topics in developmental biology, molecular and genetic analysis of L-R asymmetry was initiated by studies of vertebrates such as the mouse and chick before being expanded to include other vertebrates (Xenopus, zebrafish) and various invertebrates. In particular, the first Drosophila mutant with L-R defects was described in 2006.13,14) Snails, which are characterized by directional coiling of their shell, might be the most beautiful example of L-R asymmetry in animals, and genetics has shown that L-R asymmetry in these animals is determined by a single gene.15–17)
3. Symmetry breaking at the L-R organizer
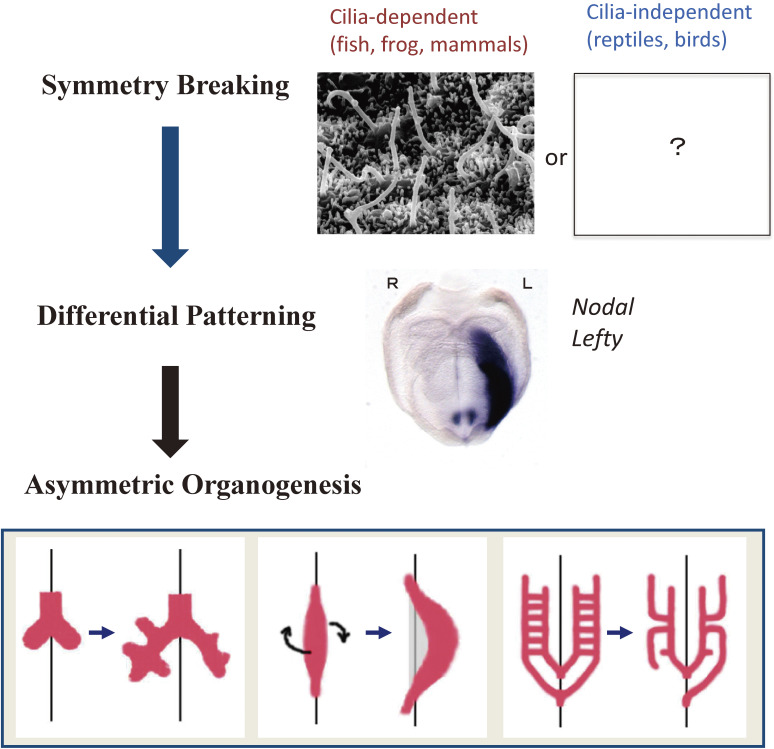
A vertebrate embryo is L-R symmetric when it undergoes gastrulation and forms the primitive streak. The L-R organizer (LRO), at which L-R symmetry is broken, is formed at the anterior tip of the primitive streak and corresponds to the ventral node in the mouse embryo, Hensen’s node in the chick embryo, the gastrocoel roof plate in the clawed frog (Xenopus), and Kupffer’s vesicle in zebrafish.18,19) The breaking of L-R symmetry is followed first by differential patterning of the lateral plate mesoderm (LPM) on the two sides in a manner dependent on signaling by the secretory protein Nodal and subsequently by asymmetric organogenesis (Fig. 2).
Figure 2.
Three steps in the establishment of L-R asymmetry in vertebrates. The three steps include symmetry breaking, patterning, and organogenesis. In the first step, many vertebrates rely on cilia for symmetry breaking, whereas others deploy a cilia-independent, largely unknown mechanism. In the second step, the LPM on the right and left sides undergoes differential patterning as a result of asymmetric Nodal signaling on the left side. In the final step, asymmetric morphogenesis takes place in visceral organs. Modified with permission from Shiratori and Hamada155) and Yoshiba and Hamada.156)
Hereafter, this report will describe L-R symmetry breaking in vertebrates such as mice, frogs, and fish, which is dependent on motile cilia and fluid flow at the LRO (although the description presented is based on studies in mice unless indicated otherwise). Some vertebrates, such as chickens20) and reptiles,21) do not rely on such a mechanism for L-R symmetry breaking. Breaking of L-R symmetry in invertebrates will be described briefly in section 7.
3.1. Two types of cilia at the LRO: motile and immotile.
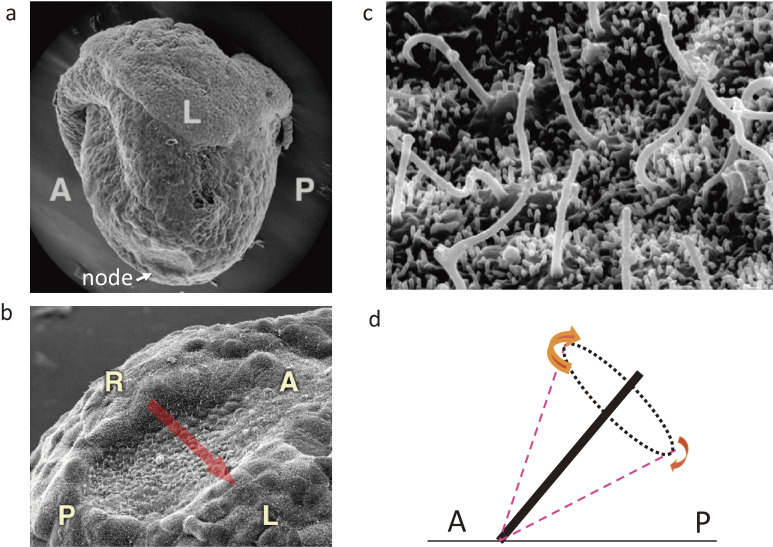
The node of the mouse embryo is a small cavity located at the midline that exists transiently at embryonic day (E) 7.5–8 (Fig. 3). The ventral surface of the node is covered with a layer of epithelial-like cells, each of which possesses a single cilium. However, there are actually two types of ciliated cells at the node. Cells in the central region of the node, often referred to as pit cells, have motile cilia that rotate in a clockwise (when viewed from the ventral side) direction. About 200 motile cilia thus protrude from the central region of the node into the node cavity22) (Fig. 3) and rotate at a speed of 600 rpm12,23) in the mouse embryo. How can motile cilia at the node rotate instead of beating in a planar manner? One reason is that they possess a distinct axonemal structure characterized by a lack of radial spokes.24) Furthermore, the clockwise direction of rotation may be determined by structural chirality of the axoneme, as suggested by mathematical modeling.25)
Figure 3.
Cilia at the node of a mouse embryo as revealed by scanning electron microscopy. a. Lateral view of a mouse embryo at E7.5. The arrow indicates the location of the node. b. Ventral view of the mouse node. The arrow indicates the direction of fluid flow at the node. c. Higher magnification of the node showing the presence of motile cilia. d. Posterior tilt of motile cilia. Clockwise rotation of posteriorly tilted cilia generates fluid flow (leftward) most efficiently when the cilia are farthest from the surface. A, anterior; L, left; P, posterior; R, right. Modified with permission from Shiratori and Hamada.155)
Cells at the periphery of the node, often referred to as crown cells, possess immotile cilia. Such cilia (also known as primary cilia) are solitary protrusions that are found at the surface of nearly all cells, and they are rendered immotile by the lack of dynein arms in their axonemes. Primary cilia function as antennae that are able to sense the surrounding environment, and they are able to detect not only molecules such as the signaling proteins Hedgehog and Wnt but also mechanical stimuli.26) In particular, evidence suggests that primary cilia are essential for Hedgehog signaling during development. In kidney epithelial cells, primary cilia also respond to mechanical force generated by fluid flow by activating the Ca2+ channel Pkd2.27,28) On the other hand, a recent study suggested that most primary cilia, including those at the node of the mouse embryo, do not function as mechanosensors that generate a Ca2+ signal.29) This issue may need further investigation, however, given that intraciliary Ca2+ oscillations were detected at the LRO of zebrafish embryos.30)
How crown cells and pit cells are specified during development remains unknown. Specification of crown cells in the mouse embryo requires Notch signaling.31) In the zebrafish embryo, anisotropic mechanical strain graded along the medial–lateral axis specifies the identity of cilia at the LRO.32) Short immotile cilia are thus generated at the medial region of the LRO where mechanical strain is low, whereas long motile cilia are generated at the lateral region where mechanical strain is high. It remains to be seen whether a similar mechanism specifies motile and immotile cilia at the LRO of other vertebrates such as in mice. Nevertheless, the spatial distribution of motile and immotile cilia at the LRO is conserved between zebrafish and mice.
3.2. Unidirectional fluid flow generated by rotational movement of cilia breaks L-R symmetry.
The rotational movement of the motile cilia at the node generates a leftward laminar flow of extraembryonic fluid present in the node cavity12,33,34) (Fig. 3), which occurs at a speed of ∼15–20 µm/s. This leftward fluid flow at the node, known as nodal flow,12,33) is responsible for L-R symmetry breaking. The loss of nodal flow as a result of the lack of motile cilia or the loss of their motility thus results in aberrant L-R patterning of the LPM.12,35,36) Furthermore, L-R patterning of the mouse embryo was reversed when the direction of the flow was experimentally reversed,37) providing direct evidence that the direction of the fluid flow determines L-R asymmetry. Moreover, humans with laterality defects have been found to harbor mutations in genes that are required for ciliary motility.38) There are more than 20 genes (http://www.informatics.jax.org/disease/DOID:0110620), including those for dynein proteins (such as Dnah5 and Dnah11), proteins required for cytoplasmic assembly of dynein complexes, proteins that contribute to the transport of these complexes from the cytoplasm to cilia, and proteins necessary for docking of the complexes to axonemal microtubules.
How can unidirectional fluid flow be generated by rotational movement of the cilia? Evidence from modeling39) and in vivo observations40,41) suggests that the clockwise rotation of node cilia generates a leftward flow because the cilia are tilted toward the posterior side.
3.3. L-R symmetry breaking by translation of preexisting information.
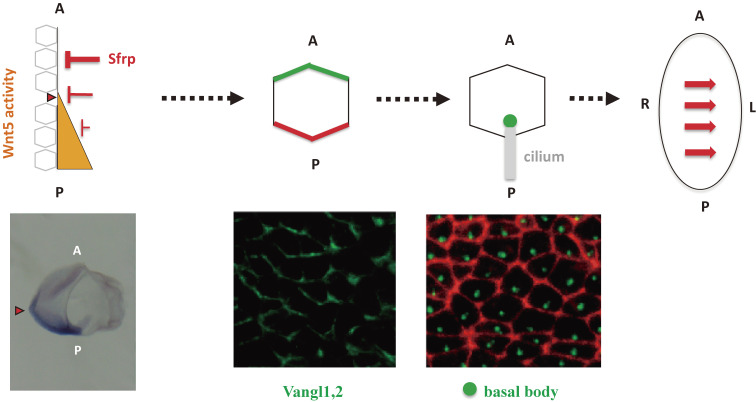
Given that the L-R axis is the last axis to be determined during development, L-R symmetry breaking is thought to be achieved in a manner dependent on preexisting positional cues. Indeed, two preexisting positional cues are represented in the cilia of node pit cells: The A-P and D-V axes are thus represented by the posterior tilt and ventral protrusion of the cilia, respectively (Fig. 4). A third cue is provided by the apparent chirality of the cilia that is attributable to a distinctive arrangement of microtubules and dynein arms and which is thought to allow the cilia to rotate in the clockwise direction. The node cilia thus generate the leftward flow by making use of the preexisting A-P and D-V positional cues and their structural chirality.
Figure 4.
Polarization of node cells with motile cilia along the A-P axis. Asymmetric expression of Wnt5 and its inhibitor Sfrp along the A-P axis generates a graded distribution of Wnt5a/b activity (orange). This will induce polarized localization of PCP core proteins (such as Vangl1, 2 in green and Dvl in red) in node cells with motile cilia (pit cells) along the A-P axis of the mouse embryo. Finally, the basal body will be localized to the posterior side of pit cells. Modified with permission from Minegishi et al.42)
How is A-P information translated into the posterior tilt of the node cilia? Node cells are polarized along the A-P axis of the embryo by the planar cell polarity (PCP) pathway (Fig. 4), with PCP core proteins such as Prickle2 and Vangl1 being localized to the anterior side of node cells and another PCP core protein, Dvl, being localized to the posterior side. This polarized distribution of PCP proteins results in positioning of the basal body (an organelle located at the base of the cilium that serves as the template for ciliogenesis) at the posterior side of each node cell and subsequently confers a posterior tilt on the cilium.
The positional cue that polarizes node cells along the A-P axis is a gradient of Wnt5 activity along this axis42) (Fig. 4). Two noncanonical Wnt ligands, Wnt5a and Wnt5b, are expressed in the posterior region of the node, whereas Sfrps (secreted Frizzled-related proteins), inhibitors of Wnt signaling, are expressed anterior to the node. The reciprocal expression of Wnt5a/b and their inhibitors may be expected to generate a gradient of Wnt5 activity along the A-P axis, with the activity being highest on the posterior side of the node. Overall, evidence suggests that a gradient of Wnt5 activity polarizes node cells so that PCP core proteins become localized to either anterior or posterior sides of the cells.
The basal body is initially localized at the center of pit cells of the node before its positioning at the posterior side of the cells. It remains unknown how the basal body changes its position within individual node cells. However, actomyosin may provide mechanical force capable of shifting the position of the basal body, given that Rho-associated kinase (Drok) links PCP signaling to the actin cytoskeleton in Drosophila43) and that the polarized distribution of PCP proteins such as Celrs1 and Shroom3 regulates cytoskeletal dynamics and apical actomyosin contractility.44,45) Of note, the basal body of ependymal ciliated epithelial cells is not correctly positioned and the beating pattern of airway cilia is disrupted, whereas laterality defects are not apparent, in a rat mutant lacking myosin 1d, an unconventional myosin.46) In contrast, myosin 1d is required for leftward fluid flow and L-R asymmetric gene expression in Xenopus.47) Whether actomyosin contractility contributes to positioning of the basal body of LRO cells such as pit cells in the mouse embryo remains to be demonstrated.
3.4. Sensing of unidirectional fluid flow by immotile cilia.
The precise action of nodal flow remains unknown, but there are two prevailing hypotheses. The flow may transport small molecules or vesicular particles to the left side,33,48) where they may then act as an L-R determinant. Alternatively, mechanical stimuli generated by the flow may be sensed by the embryo.49,50)
Genetic evidence has indicated that it is the immotile cilia at the periphery of the node that sense the fluid flow and that such sensing triggers activation of the Ca2+ channel Pkd2.51) However, it remains unknown what the immotile cilia sense for L-R symmetry breaking—a chemical determinant, mechanical force, or some other signal? A chemical L-R determinant transported by the flow has not been identified in the more than 20 years since the discovery of nodal flow. Mathematical modeling favors either chemical sensing52) or mechanical sensing.39) A recent study failed to detect Ca2+ influx in primary cilia in response to fluid flow, and it was suggested that immotile cilia at the node are not Ca2+-responsive mechanosensors.29) Further investigation is thus required to clarify how nodal flow is sensed by the embryo.
3.5. Role of Ca2+ in flow sensing.
Several lines of evidence suggest that Ca2+ plays a role in the sensing of nodal flow. First, a Ca2+ channel composed of Pkd253) and Pkd1l154–56) is required for L-R patterning (Fig. 5). Given that the L-R defects of Pkd2−/− mutant mice can be rescued by expression of a Pkd2 transgene specifically in crown cells,51) Pkd2 in these cells is sufficient for flow sensing. Pkd2, together with Pkd1l1, also likely functions specifically in the cilium of crown cells, given that a mutation of Pkd2 that disrupts the ciliary localization of the encoded protein gives rise to L-R defects similar to those of Pkd2−/− embryos.51,54) Pkd2 encodes a Ca2+ channel with a short extracellular domain (although, to be more precise, Pkd2 is regarded as a nonselective cation channel and may not be specific for Ca2+). The Pkd1l1 protein possesses a much larger extracellular domain at its amino terminus, suggesting that Pkd1l1 may be responsible for sensing the flow signal. Genetic evidence57) also suggests that Pkd1l1 may inhibit the channel activity of Pkd2 in condition with no flow, and that flow may relieve this inhibition and thereby activate the Pkd2 channel.
Figure 5.
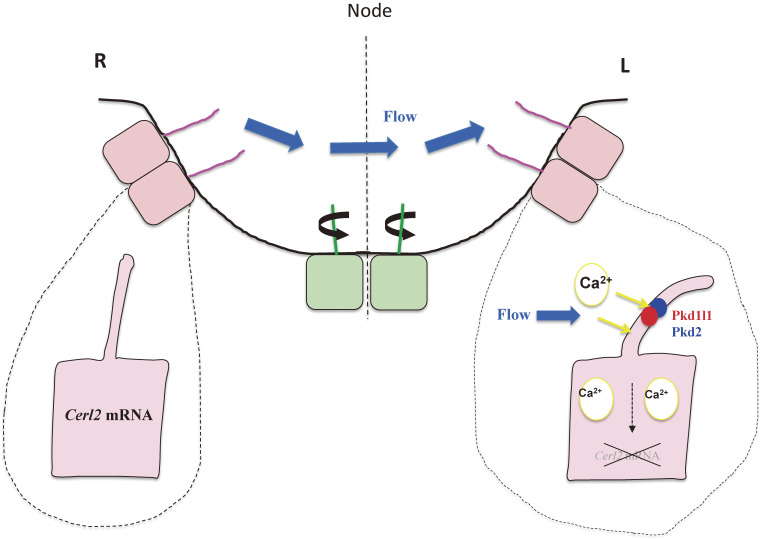
Immotile cilia at the periphery of the node sense nodal flow. Ciliated cells located in the central region of the node of the mouse embryo (green) possess motile cilia that rotate and generate nodal flow, whereas those located peripherally (pink) possess immotile cilia that sense the flow. Sensing of the flow requires a Pkd2-Pkd1l1 complex with Ca2+ channel activity that is localized to the cilium. The flow-mediated signal, the identity of which remains unknown, triggers the degradation of Cerl2 mRNA preferentially on the left side.
Second, L-R asymmetric Ca2+ signaling has been detected at the node of mouse embryos,50,58) because they have L-R asymmetric Ca2+ oscillations at the LRO of the zebrafish embryo.30) Third, several Ca2+ blockers have been found to disrupt asymmetric gene expression in crown cells.51) Thus, GdCl3 (an inhibitor of stretch-sensitive transient receptor potential [TRP] channels), 2-aminoethyl diphenylborinate (an inhibitor of the inositol 1,4,5-trisphosphate [IP3] receptor), and thapsigargin (an inhibitor of Ca2+-dependent ATPase activity in the endoplasmic reticulum) impair L-R asymmetric expression of a flow-responsive transgene.51,59) Finally, inhibition of fibroblast growth factor (FGF) signaling, which is essential for normal L-R asymmetry, was found to attenuate the Ca2+ signal on the left side of the ventral node of the mouse embryo without disturbing the leftward fluid flow.48)
These observations suggest a possible route for Ca2+ within node cells. First, Ca2+ can enter crown cells via a stretch-sensitive TRP channel (such as Pkd2) located in the membrane of immotile cilia. This intraciliary Ca2+ would then reach the cell body, act at the IP3 receptor directly or indirectly, and induce the release of more Ca2+ from the endoplasmic reticulum into the cytosol.
3.6. Cerl2/Dand5 mRNA as a target of flow-mediated signaling.
What are the molecular events that take place in crown cells when they receive the flow-mediated signal? Cerl2 (also known as Dand5) mRNA is the target of this signal.60,61) The Cerl2 protein inhibits the activity of the transforming growth factor β (TGFβ)-related protein Nodal, likely by directly interacting with it. The activity of Nodal is greatly increased by its formation of a heterodimer with another TGFβ-related protein, Gdf1 (growth differentiation factor 1),62) and Cerl2 may inhibit formation of the Nodal-Gdf1 heterodimer. The level of Cerl2 mRNA is R > L asymmetric (higher on the right side than on the left side) among crown cells, and Cerl2 mutant mice manifest randomization of L-R decision-making.63)
The abundance of Cerl2 mRNA is initially symmetric (R = L) at the early headfold stage of mouse embryogenesis, but it becomes R > L asymmetric as the velocity of nodal flow increases, with the amount on the left side being down-regulated.59,61) The L-R asymmetry of Cerl2 expression is determined not at the level of transcription but rather posttranscriptionally,64) in particular, by the decay of Cerl2 mRNA in a manner dependent on its 3′ untranslated region. The degradation of Cerl2 mRNA preferentially on the left is triggered by the leftward fluid flow (Fig. 5) and is further enhanced by the operation of complex Wnt-Cerl2 interlinked feedback loops, in which Wnt3 increases Wnt3 expression and promotes Cerl2 mRNA decay whereas Cerl2 promotes the degradation of Wnt3. Both mathematical modeling and experimental data64) suggest that these feedback loops constitute a bistable switch that is capable of amplifying in a noise-resistant manner a small L-R bias conferred by nodal flow.61)
The precise mechanism of Cerl2 mRNA decay in response to nodal flow remains unknown. However, it may be relevant that Bicc1, a putative RNA-binding protein specifically expressed at the node of mouse embryos, is essential for L-R asymmetry.65) Furthermore, Bicc1 interacts with ANKS3 and ANKS6,66,67) and ANKS3 mutations have been identified in patients with laterality defects.68) Finally, Bicc1 also binds to Cerl2 mRNA in zebrafish and thereby inhibits its translation.69) It remains to be determined whether a Bicc1-Anks3-Anks6 complex plays a role in the degradation of Cerl2 mRNA and how Ca2+ might be involved in this process.
3.7. The mechanism of L-R symmetry breaking is not conserved among vertebrates: cilia- and flow-independent symmetry breaking in chick and reptile embryos.
The chick embryo deploys a mechanism of L-R symmetry breaking that is independent of motile cilia and fluid flow.20) Motile cilia are absent at Hensen’s node of the chick embryo. Furthermore, the avian talpid2 mutant, in which the gene for C2CD3, a protein essential for ciliogenesis, is disrupted, manifests a ciliopathy phenotype, including polydactyly and facial clefting. However, it does not show laterality defects,70) indicating that cilia are not required for L-R symmetry breaking in the chick. Instead, asymmetric cell rearrangement, in particular a leftward movement of cells around Hensen’s node, is responsible for L-R symmetry breaking in this species. This rearrangement results in the relative displacement of cells expressing Sonic hedgehog (Shh) and those expressing FGF8, and it thereby gives rise to asymmetric expression domains. It has also recently been shown that reptile embryos develop L-R asymmetry without motile cilia and fluid flow at the LRO.21) It will thus be of interest to learn the origin of L-R asymmetry in reptiles and birds.
Although it is generally believed that the mechanism of L-R symmetry breaking is conserved among mammals, this conclusion requires further evidence. LRO morphology varies substantially among mammals,71) and it has been suggested20) that the LRO of the pig embryo does not have sufficient space for motile cilia to generate fluid flow.
4. Signaling from the LRO to the lateral plate
4.1. Asymmetric signals at the node.
Three signaling molecules expressed in crown cells play a key role in setting up molecular L-R asymmetry at the node in mouse embryos (Fig. 6). First, Nodal is expressed bilaterally in node crown cells in a manner dependent on a crown cell-specific enhancer (NDE), the activity of which requires binding sequences for Rbpjk, a downstream mediator of Notch signaling, suggesting that such expression is induced by Notch.31) The expression of Nodal in crown cells is followed by that in the left LPM, with the former being essential for the latter, given that specific ablation of NDE results in the loss of Nodal expression in the LPM.
Figure 6.
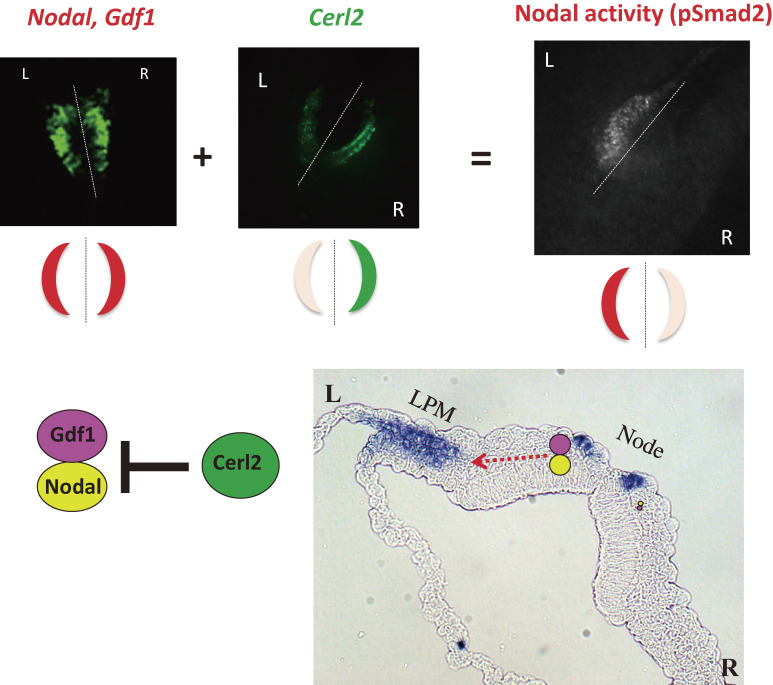
Generation of molecular asymmetries at the node. Whereas Nodal mRNA and Gdf1 mRNA are present at similar levels on both sides of the node of a mouse embryo, Cerl2 mRNA shows an asymmetric (R > L) distribution (top panels). Nodal and Gdf1 form a heterodimer that constitutes an active form of Nodal. Given that Cerl2 is an inhibitor of Nodal (bottom left panel), the level of Nodal activity, which is reflected by the abundance of phosphorylated Smad2/3 (pSmad2), shows a R ≪ L pattern (top panels). The Nodal-Gdf1 heterodimer produced by and secreted from perinodal crown cells is thought to be transported to the LPM on the left side via an intraembryonic route (red dotted arrow in the bottom right panel). On reaching the LPM, the Nodal-Gdf1 heterodimer is thought to activate expression of Nodal (indicated by purple staining in the bottom right panel), which is responsive to Nodal signaling. Modified with permission from Shiratori and Hamada,155) Yoshiba and Hamada,156) and Shiratori and Hamada.157)
Second, Gdf1 is also bilaterally expressed in node crown cells. Furthermore, as mentioned above, the activity of Nodal complexed with Gdf1 is much greater (∼100-fold) than that of the Nodal homodimer. In a Gdf1 mutant mouse, Nodal activity in or near crown cells is greatly attenuated59) and the expression of Nodal in the LPM is absent.72) Collectively, these observations suggest that Gdf1 itself is not active at physiological concentrations but that it instead acts as a co-ligand for Nodal at the node.
The third important signaling molecule expressed in crown cells is Cerl2 (Dand5), a member of the Cerberus/Dan family of proteins that inhibit Nodal signaling.63) As mentioned above, the level of Cerl2 mRNA in crown cells is symmetric before the development of nodal flow, but becomes R > L asymmetric in response to the flow. This asymmetric (R > L) expression of Cerl2 ensures that Nodal activity in crown cells is higher on the left side (Fig. 6). The Cerl2-generated asymmetry (R < L) of Nodal activity at the node closely correlates with the asymmetry of Nodal expression in the LPM.59) It thus appears that Cerl2 selectively inhibits Nodal-Gdf1 heterodimer activity in crown cells on the right side, which then leads to induction of Nodal expression in the left LPM. This mechanism is conserved at least in zebrafish,73) Xenopus, and mouse.
4.2. Transfer of the Nodal signal from the node to the LPM.
Nodal produced in crown cells is required for subsequent Nodal expression in the left LPM.74,75) Evidence suggests that active Nodal protein (presumably, the Nodal-Gdf1 heterodimer) produced in crown cells on the left side is transported to the left LPM, where it directly activates Nodal expression. First, a transcriptional enhancer (ASE) required for Nodal expression in the LPM is responsive to Nodal. Second, Cryptic, a co-receptor of Nodal essential for left-sided Nodal expression in the lateral plate, is required only in the LPM for correct L-R patterning.76)
Available data also suggest that active Nodal protein (Nodal-Gdf1 heterodimer) synthesized in crown cells is transported from the node to the LPM via an intraembryonic route (Fig. 6). First, Nodal expression in the LPM remained unaffected when mouse embryos were incubated with recombinant Nodal in culture medium. Second, Nodal interacts with sulfated glycosaminoglycans that are localized specifically to the basement membrane between the node and lateral plate. Furthermore, inhibition of sulfated glycosaminoglycan synthesis abolished Nodal expression in the LPM.76)
5. Differential patterning of the lateral plate on both sides
5.1. Role of the TGFβ-related factors Nodal and Lefty.
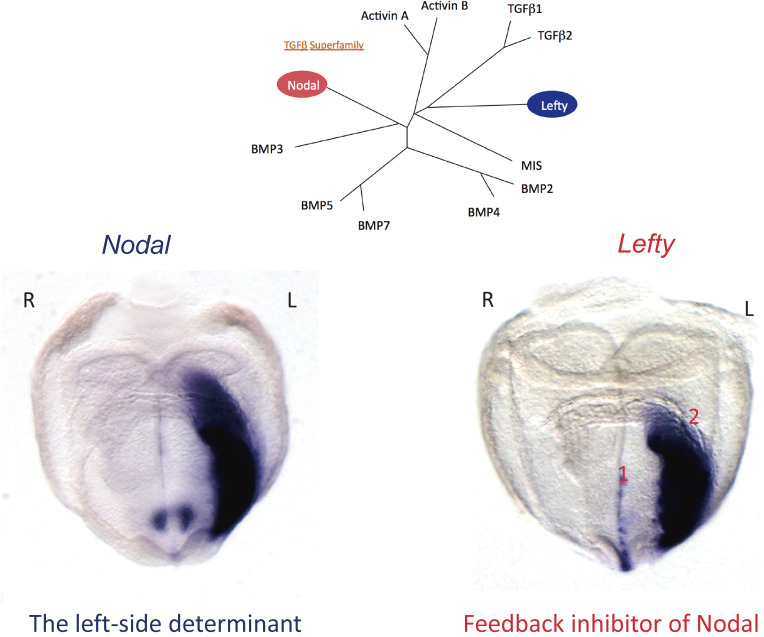
Two TGFβ-related proteins, Nodal and Lefty, are expressed asymmetrically in the LPM (Fig. 7) and play a major role in its patterning.7–9,77) Genetic and biochemical studies have established that Nodal and Lefty have opposite functions. Nodal functions as a left-side determinant,74) whereas Lefty is an antagonist of Nodal that restricts Nodal activity to the left side.78) Mutant mice lacking Nodal expression in the LPM thus show right isomerism of bilateral organs such as the lung, whereas mutant mice that express Nodal on both sides of the LPM manifest left isomerism. On the other hand, the absence of Lefty results in bilateral Nodal expression in the LPM and subsequent left isomerism.78,79) Vertebrates possess two Lefty genes, Lefty1 and Lefty2, both of which are expressed in the left LPM and at the midline.80) Given that Lefty1 and Lefty2 have indistinguishable activities and show similar expression patterns, they will be referred to collectively as Lefty in this review.
Figure 7.
L-R asymmetric expression of Nodal and Lefty in the mouse embryo. Cluster analysis (top) shows the relations among members of the TGFβ superfamily of proteins. In situ hybridization (bottom) reveals the L-R asymmetric expression of Nodal (which encodes a left-side determinant) and Lefty (which encodes a feedback inhibitor of Nodal) in the E8.0 mouse embryo. Note that Nodal and Lefty are expressed in the LPM on the left side, whereas Lefty is also expressed at the midline (left side of the floor plate). There are actually two Lefty genes, with Lefty2 being preferentially expressed in the LPM and Lefty1 at the midline. Modified with permission from Shiratori and Hamada.157)
Nodal activates intracellular signaling through interaction with type I (ALK4 and ALK7) and type II (ActRIIa and ActRIIb) TGFβ receptors81) (Fig. 8). As described above, Nodal forms a heterodimer with Gdf1 (Vg1 in frog), with Gdf1 serving as a co-ligand that increases Nodal activity.62,82) Unlike other TGFβ family members, however, Nodal requires an EGF-CFC (epidermal growth factor–Cripto-FRL1-Cryptic) family protein (such as Cryptic or Cripto) as a co-receptor. Smad2 and Smad3, together with Smad4, are intracellular components of the Nodal signaling pathway. FoxH1 is the major transcription factor that interacts with Smad2/3 and transduces the Nodal signal. Lefty inhibits Nodal signaling by interacting in a competitive manner with the EGF-CFC co-receptor83) and the type II TGFβ receptor chain.84)
Figure 8.
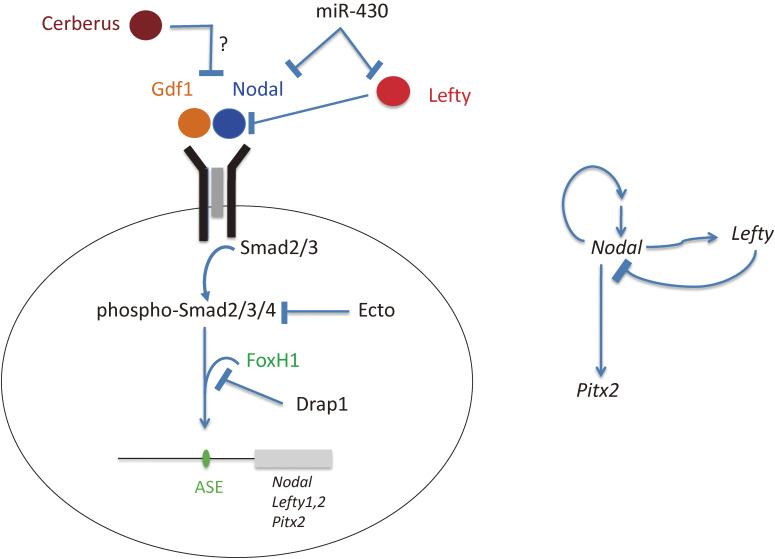
Control of Nodal signaling by various regulators in the developing vertebrate embryo. The Nodal signaling pathway in vertebrate embryos (Nodal → receptor → Smad2/3/4 → FoxH1 → target genes) is regulated in a spatiotemporal manner at multiple levels by various inhibitors including Lefty, Cerberus (Cerl2), Ectodermin (Ecto), Drap1, and miR-430 (left panel). Nodal and Lefty also participate in positive and negative regulatory loops (right panel).
Nodal signaling is precisely regulated both positively and negatively at several levels by various molecules during development (Fig. 8). Drap1 is a transcriptional repressor that interacts with FoxH1 and inhibits its DNA binding activity.85) Ectodermin negatively controls Nodal signaling by inhibiting Smad4 activity.86) Mice lacking Drap1 or Ectodermin thus manifest increased Nodal signaling. Nodal signaling is also subject to translational repression,87) at least three different mechanisms have been identified in the zebrafish embryo: (1) repression of Nodal mRNA translation by Ybx1, (2) repression of Cr1 (Cripto) mRNA translation, and (3) repression of Nodal mRNA translation by the microRNA miR-430.88) Nodal signaling is essential not only for L-R patterning but also for A-P patterning as well as mesoderm induction and patterning.89) These various negative regulators of Nodal signaling have been shown to be essential for mesoderm formation, but it is unknown whether they play a role in L-R patterning.
5.2. Transcriptional regulation of Nodal and Lefty and formation of a self-enhancement lateral inhibition system by the encoded proteins.
L-R asymmetric expression of Nodal and Lefty is transient, with a duration of only several hours in the mouse embryo, from the two- to six-somite stage. This transient expression is established by positive and negative regulatory loops connecting these genes (Fig. 8). Asymmetric expression of Nodal and Lefty in the LPM relies on the Nodal-responsive, FoxH1-dependent enhancer ASE.90) Nodal expression is thus positively regulated by Nodal itself and negatively regulated by the feedback inhibitor Lefty. Interaction of Nodal protein with target cells triggers the transcription of both Nodal and Lefty. The Nodal thereby produced amplifies expression of Nodal and Lefty, whereas the Lefty produced eventually terminates the expression of both genes. The Lefty-mediated negative loop is therefore expected to restrict the duration and area of Nodal signaling in a highly precise manner, accounting for the transient nature of asymmetric Nodal and Lefty expression. The activities of Nodal and Lefty proteins and this transcriptional regulatory relation between the corresponding genes suggest that the two proteins may constitute a self-enhancement lateral inhibition system,91) a type of Turing reaction-diffusion system.92) Such a system comprising an activator and an inhibitor requires in principle that the inhibitor diffuses faster than the activator, and this is indeed the case for Nodal-Lefty at least in the zebrafish embryo.93,94) Computational simulations of gradient formation indicate that diffusivity, extracellular interactions, and selective ligand destruction collectively shape the Nodal morphogen gradient.94) The Nodal-Lefty network may contribute to various patterning events during development including scaling.95)
Asymmetric gene expression is also subject to epigenetic regulation. Depletion of DNA methyltransferase enzymes (Dnmt1 or Dnmt3bb.1) in zebrafish embryos thus gives rise to hypomethylation of the Lefty2 enhancer, resulting in up-regulation of Lefty2 expression, attenuation of Nodal signaling, down-regulation of Nodal expression in the LPM, and disruption of organ laterality.96) Lefty2 expression in the postimplantation mouse embryos is also regulated by the DNA demethylase Tet1.97)
6. Asymmetric organogenesis
6.1. Asymmetric organs.
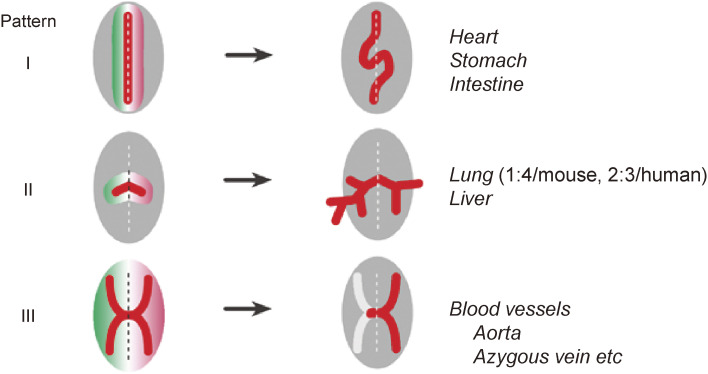
Various visceral organs begin to develop anatomic asymmetries after asymmetric Nodal expression in the LPM has disappeared. Theoretically, a symmetric structure can become asymmetric in at least three ways (Fig. 9). First, a primordial organ that is present in just one copy in an embryo could initially be located at the midline but subsequently undergo a directional morphological change such as looping that would result in positioning of the organ on one side. Examples of such morphogenesis by directional looping include the heart and gut. Second, a primordial organ could be initially formed as a bilaterally symmetric structure, but the two sides might subsequently undergo differential growth or branching. Examples of this type of morphogenesis include the lung, which has one lobe on the left and four lobes on the right in the adult mouse (two lobes on the left and three lobes on the right in human). The third type is unilateral regression, as exemplified during remodeling of the vascular system. The embryonic vascular system is thus initially bilaterally symmetric, but some parts subsequently undergo regression specifically on one side.
Figure 9.
Generation of morphological asymmetries. Three different patterns for generation of morphological asymmetries rely on (I) directional looping, (II) differential branching, or (III) one-sided regression. Examples of anatomic structures generated by each mechanism are shown. Modified with permission from Shiratori and Hamada155) and courtesy of Yukio Saijoh (University of Utah).
6.2. Genetic basis of the asymmetric morphogenesis: role of Nodal signaling and the transcription factor Pitx2.
The main intracellular mediator of asymmetric organogenesis is the transcription factor Pitx2,98–101) the left-sided expression of which is induced by Nodal.102) Like Nodal and Lefty, Pitx2 is expressed asymmetrically in the LPM, but its left-sided expression persists until much later stages of development (Fig. 10). Mice deficient in Pitx2 (to be more precise, Pitx2c, the isoform that is asymmetrically expressed) manifest laterality defects in most visceral organs.103) Pitx2c is responsible for the generation of left-side morphology. Thus, in its absence, bilateral organs such as the lung exhibit right isomerism.
Figure 10.
L-R asymmetric expression of Nodal, Lefty, and Pitx2 in the mouse embryo. In situ hybridization reveals that, whereas L-R asymmetric expression of Nodal and Lefty is transient (detectable around E8), L-R asymmetric expression of Pitx2 persists much longer. Asymmetric expression of Nodal and Lefty is controlled by a Nodal-responsive enhancer (ASE), which renders their asymmetric expression transient. Pitx2, however, contains both ASE and an additional enhancer that maintains its expression at later stages. Modified with permission from Shiratori and Hamada.157)
Whereas the development of most L-R asymmetric organs requires Pitx2, some asymmetric morphogenic events are independent of this protein. One such example is heart looping in mice, which is under the control of left-sided Nodal signaling but not of Pitx2. In mice specifically lacking Pitx2c, heart looping and the stomach thus remain normal even though other asymmetric organs show laterality defects.103,104) Similarly, asymmetric looping of the heart and gut remains normal in a zebrafish Pitx2 mutant (most likely a null mutant).105) The direction of heart looping is also largely normal in a zebrafish mutant lacking southpaw (Nodal in zebrafish), suggesting that heart looping is independent of Nodal signaling in this organism.106) In support of this notion, a recent study suggests that heart looping in zebrafish and chick is controlled by a bone morphogenetic protein (BMP)–Prrx1a axis on the right side.107) The Nodal-Pitx2 pathway in the left LPM and the BMP-Prrx1a pathway in the right LPM seem to operate in parallel and to engage in mutual repression. Whereas Prrx1 or Prrx1/Prrx2 mutant mice show normal heart looping,108,109) the role of Prrx1 in chicks may be filled by Snail1 in mice.107) The precise role of right-side-specific signaling in asymmetric organogenesis awaits further clarification.
6.3. Heart and outflow tract.
Heart formation is under the control of correct L-R patterning,110) which determines the position of the atria relative to nearby organs (such as the stomach and spleen), the orientation of ventricular looping, and the position of the great vessels. In fact, screening of a large number of chemically mutagenized mice for congenital heart disease (CHD) identified 61 genes that can give rise to this condition.111) Mutations in 30 of these 61 genes gave rise to CHD with laterality defects, and 23 of these 30 genes belong to the ciliome family of genes that are required for the motility of rotating cilia or for the sensing of fluid flow by immotile cilia at the mouse node. Similarly, among 28 CHD genes identified in humans, 11 are cilia related.112,113) The L-R symmetry-breaking event mediated by cilia thus has profound effects on heart formation.
Heart development begins with the specification of bilateral precardiogenic mesoderm, which subsequently converges at the midline to generate a linear heart tube. The heart is symmetric until this stage, but as the heart tube elongates along the A-P axis it starts to bulge on its ventral side and to bend rightward. The developing heart thus changes its shape from a simple linear tube to a structure with a rightward helical loop. This process, known as heart looping, is crucial for the subsequent formation of the atrial and ventricular chambers. In addition to heart looping, other events that take place during heart formation also depend on L-R asymmetry including (1) spiral septum formation, defects in which result in transposition of the great arteries and double outlet right ventricle, and (2) formation of the pulmonary vein by angiogenic sprouting from the left atrium.
How does the linear heart tube undergo rightward (dextral) looping? Three theoretical mechanisms have been postulated: differential growth within the heart tube, oriented growth within the heart tube, and buckling.110) The heart tube elongates as a result of ingression of cardiac progenitor cells from the surrounding tissues at both ends of the tube, which are known as the venous and arterial poles. Observation of the developing mouse heart at this stage by high-resolution microscopy has identified two types of asymmetry at these poles.114) First, the arterial pole manifests rotational asymmetry and is also positioned asymmetrically across the midline. This rotational asymmetry present before heart looping is apparent not only in the mouse but also in other animals such as chick and zebrafish.115,116) Second, the venous pole exhibits asymmetric ingression and proliferation of cardiac progenitor cells, with more cells ingressing through the right half of the venous pole than through the left half.114) A computational model suggested that biomechanical forces generated by the asymmetric rotation and asymmetric cell ingression can drive rightward heart looping.114) Indeed, cells that surround and will undergo ingression into the heart show mechanical stress, and this epithelial tension contributes to elongation and looping of the developing heart.117) However, the molecular and cellular mechanisms underlying the asymmetric rotation and asymmetric cell ingression remain unknown.
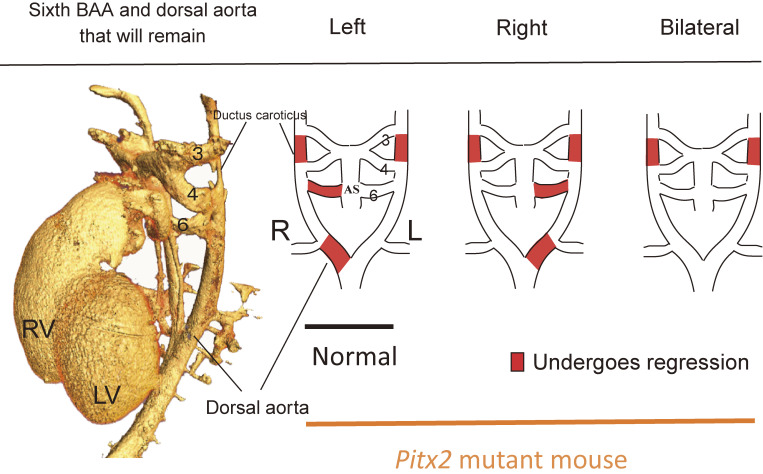
The developing heart and outflow tract also undergo asymmetric remodeling. For example, the aortic arch, which is connected to the left ventricle and arches toward the left side, is derived from the dorsal aorta and branchial arch arteries (BAAs) at embryonic stages. The dorsal aorta and BAAs are initially formed symmetrically but subsequently undergo asymmetric remodeling. The left sixth BAA thus persists and gives rise to the aortic arch, whereas the right sixth BAA regresses (Fig. 11). A dynamic morphological change in the outflow tract of the heart, which is under the control of Pitx2, results in provision of an asymmetric blood supply to the left sixth BAA.118) This uneven distribution of blood flow results in differential signaling by both the platelet-derived growth factor receptor and vascular endothelial growth factor receptor 2. The consequent stabilization of the left sixth BAA and regression of the right sixth BAA underpin left-sided formation of the aortic arch. Hemodynamics generated by a Pitx2-induced morphological change in the outflow tract are thus responsible for the asymmetric remodeling of the great arteries.
Figure 11.
L-R asymmetric remodeling of the sixth branchial arch artery in the mouse embryo. In a normal embryo, the right side of the sixth branchial arch artery (BAA) and the right side of the dorsal aorta regress as development proceeds, eventually resulting in arching of the aorta toward the left side. In Pitx2 mutant mice; however, this pattern of remodeling is impaired. AS, aortic sac; RV and LV, right and left ventricle.
6.4. Gut.
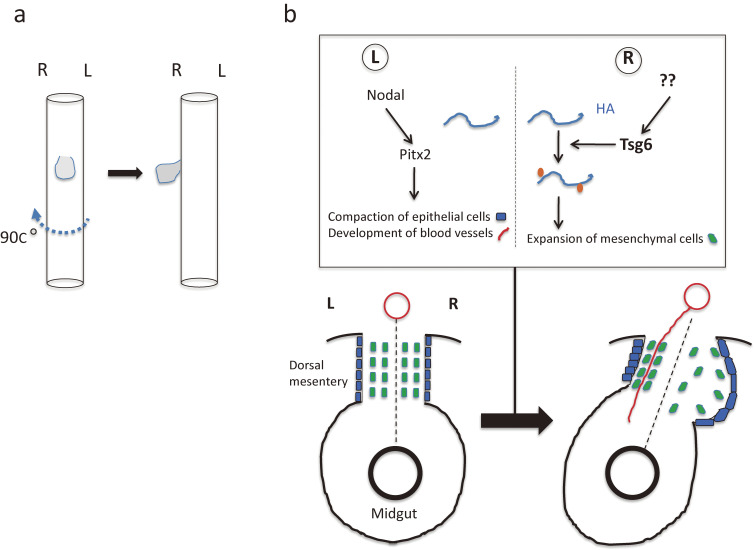
The developing gut consists of the foregut, midgut, and hindgut, from anterior to posterior. The foregut will give rise to the anterior portions of the digestive tract including the esophagus, stomach, and duodenum, as well as to adjacent organs such as the pancreas and liver. The liver is an asymmetric organ in that it is located asymmetrically in the body cavity. A 90-degree clockwise rotation (looking down from the rostral side) of the foregut results in translocation of the liver primordium to the right side (Fig. 12a). The liver also manifests morphological asymmetries in its size and shape. In humans, for example, the right side of the liver is five to six times as large as is the left side, and it shows a distinct morphology and more complex lobation pattern. Similar morphological asymmetries are present in the liver of other vertebrates, although the precise morphologies differ among species. During formation of the liver in Xenopus embryos, endoderm cells on the right side become columnar and apically constricted, whereas those on the left side become rounder and rearrange into a compact structure.119) The development of such cellular asymmetries is regulated by Pitx2c and is responsible for the generation of future morphological asymmetries of the adult liver. Of interest, the cellular response to Pitx2c in the developing liver is opposite to that apparent in the embryonic midgut, in which endoderm cells on the left side of the dorsal mesentery remain columnar and those on the right side become compact.
Figure 12.
Rotation and looping of the developing gut. a. Clockwise rotation of the foregut results in translocation of the liver primordium (gray) to the right side of the body cavity. b. Cellular changes that initiate L-R asymmetry in the midgut tube of the chick embryo.120,123) Asymmetries arise in the dorsal mesentery between Hamburger–Hamilton (HH) stages 20 and 22. Left side-specific Nodal-Pitx2 expression drives compaction of mesenchymal cells (green rectangles) within the left dorsal mesentery and promotes retention of a columnar morphology of epithelial cells (blue rectangles) on the left side of the dorsal mesentery. In the right dorsal mesentery, hyaluronan (HA) undergoes modification by Tsg6, which catalyzes the covalent attachment of a heavy chain (orange) of inter-α-trypsin inhibitor. Signals that induce Tsg6 expression in the right half of the dorsal mesentery remain unknown. The modified form of hyaluronan is more stable and accumulates in the right dorsal mesentery, resulting in expansion of mesenchymal cells and exclusion of blood vessels (red line) in the right dorsal mesentery. Together, these cellular asymmetries drive leftward tilting of the gut tube. Left (L) and right (R) sides of the dorsal mesentery are indicated together with the midline (broken line). Modified with permission from Hamada.158)
The midgut is the future small intestine. Given that the gut is a long tubular organ that far exceeds the length of the body, it must be packaged to fit within the limited abdominal space. This feat is accomplished by a process known as gut looping, which takes place early in development. At the early embryonic stage, the gut is a simple tube composed of endodermal epithelium with mesenchyme recruited from the LPM. The mesenchyme portion becomes the dorsal mesentery, which will connect the dorsal edge of the gut to the body wall along its entire length. As the gut elongates rapidly along the A-P axis, the midgut undergoes a 270-degree anticlockwise rotation. The symmetry-breaking event that initiates this rotation and will result in midgut looping is a leftward tilt of the dorsal mesentery (Fig. 12b).120,121) It has been suggested that two types of cellular changes in and around the dorsal mesentery are responsible for this leftward tilt.121,122) First, mesenchymal cells of the right half of the dorsal mesentery become dispersed, whereas those of the left half become compacted. Second, epithelial endoderm cells on the right side of the dorsal mesentery expand and flatten, whereas those on the left side retain a narrow columnar shape. Together, these cellular asymmetries drive the leftward tilting of the midgut. These cellular changes are under the control of Pitx2, which is expressed in the left half of the dorsal mesentery. The genes for N-cadherin and Daam2, which are targets of Pitx2, are also expressed on the left side of the dorsal mesentery, and their expression may confer adhesive and condensed features to cells on the left side. These observations suggest that left side-specific events governed by Nodal-Pitx2 determine the direction of midgut looping. However, a right side-specific pathway may also exist. Covalent modification of the extracellular matrix component hyaluronan by Tsg6 takes place on the right side of chick and mouse embryos, and this modification is essential for midgut looping.123) Tsg6 is expressed in the dorsal mesentery on the right side of chick embryos, and Tsg6 mutant mice manifest aberrant rotation of the midgut.123) The precise relation between the Nodal-Pitx2 pathway and the Tsg6-hyaluronan pathway remains to be clarified.
Additional loops form as the gut elongates, bends, and twists. Association of the gut and dorsal mesentery is required for formation of the mature loops in the gut.124) The looping morphology can thus be maintained after dissection of the intestine together with the dorsal mesentery away from all other embryonic tissues. In contrast, separation of the intestine from the dorsal mesentery results in uncoiling of the gut and further coiling of the dorsal mesentery. These observations suggest that the gut tube is under compression whereas the dorsal mesentery is under tension. Such isotropic forces arise from differential growth between the faster-growing gut and the slower-growing dorsal mesentery.124)
6.5. Brain.
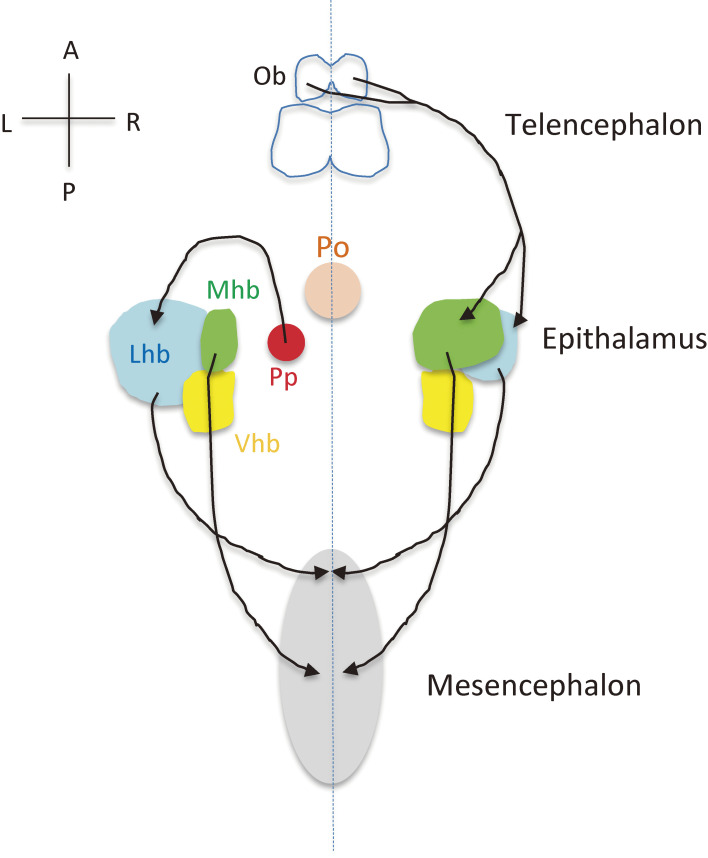
Most insight into the development of brain asymmetry has come from studies of zebrafish, which shows a pronounced anatomic asymmetry in the epithalamus.125) The epithalamus consists of bilateral habenular nuclei and an unpaired pineal complex (Fig. 13). The habenular nuclei form a conserved limbic conduction system linking the telencephalon (forebrain) to the mesencephalon (midbrain). The pineal is situated at the midline and functions as a photosensitive clock that secretes melatonin. The parapineal is located left of the midline and projects exclusively to the dorsal habenula on the left side.126) As a result, the left habenula and right habenula receive asymmetric inputs, with such asymmetric neural circuits being responsible for certain behaviors. For example, the response to fear depends on the projection from the left habenula to the interpeduncular nucleus of the midbrain.127) Epithalamic asymmetries are established by collective migration of parapineal cells to the left side of the brain, which is controlled by several signaling pathways including asymmetric Nodal signaling and bilateral FGF signaling.128) Nodal is thus expressed on the left side of the epithalamus, whereas Fgf8 is expressed bilaterally in the habenular nuclei. In an Fgf8 mutant, the parapineal fails to migrate and remains at the midline. If Nodal expression is lost or becomes symmetric, the parapineal migrates in a random manner, either to the left or right side, suggesting that asymmetric Nodal signaling determines the direction of parapineal migration.129) FGF signaling is activated in a few cells located at the leading edge of the parapineal during its migration, and asymmetric Nodal signaling contributes to a leftward bias of FGF pathway activation.130)
Figure 13.
The epithalamic system and L-R asymmetric neural projection in zebrafish. The habenula of zebrafish is divided into a dorsal component and a ventral component (Vhb), with the dorsal habenula being constituted by the lateral (Lhb) and medial (Mhb) subnuclei of unequal size. Further components of the zebrafish epithalamus are the photosensitive pineal (Po) and the asymmetrically organized parapineal organ (Pp). The Lhb and Mhb project to the mesencephalon. The olfactory bulb (Ob) projects to the Lhb and Mhb on the right side. Bilaterally symmetric neural projections are not shown.
Anatomic and functional asymmetries also exist between the left and right cortical hemispheres of the human brain.131,132) Various cognitive functions are thus lateralized, with the most pronounced asymmetries being apparent in the language system. Other asymmetrically organized cognitive systems include visuospatial processing, auditory processing, and behaviors such as hand or foot preference. However, the neuroanatomic asymmetries underlying these functional differences remain largely unknown. Although genes that are asymmetrically expressed in the human cortex have been identified,133–136) the biological relevance of such asymmetry remains to be seen. Linkage analysis has also identified candidate genes for the control of cerebral asymmetry such as PCSK6137) and LRRTM1.138) Specific alleles of such genes were found to be linked to handedness in limited family groups, but the associations failed to achieve statistical significance in genome-wide analysis of samples from the general population.
A molecular asymmetry detected in the mouse brain is the distribution of the NMDA subtype of glutamate receptor in the hippocampus.139,140) The synaptic distribution of the NMDA receptor subunit GluRε2 (NR2B) in the adult hippocampus is thus asymmetric between the left and right hemispheres as well as between the apical and basal dendrites of individual neurons. L-R asymmetry in hippocampal circuits may be required for spatial learning and memory,141) and it is disrupted in the iv/iv mutant mouse, which lacks motile cilia and thus does not develop nodal flow at the LRO,142) suggesting that such asymmetry depends on the action of motile cilia. It remains unclear, however, where and when hippocampal asymmetry is generated during development. Although it may originate from ciliary function at the node, it may alternatively be dependent on motile cilia present in the neural tube. The origin of hippocampal asymmetry is an important issue for future studies.
6.6. Other visceral organs.
In most birds, unlike other animals, adult females have only one functional gonad (ovary) and a single oviduct, both of which are located on the left side. A pair of gonads is initially formed in a female embryo, but subsequently only the left ovary remains while the right ovary degenerates. Such asymmetric gonadal development is regulated by Pitx2, which is expressed in the left gonad.143,144) The absence of Pitx2 in the right gonad allows expression of retinaldehyde dehydrogenase 2 (RALDH2), which catalyzes the synthesis of retinoic acid, on the right side. Retinoic acid suppresses expression of the nuclear receptor Ad4BP (also known as Sf-1) and estrogen receptor α on the right side, resulting in degeneration of the right gonad.
7. L-R asymmetry in invertebrates
There is considerable diversity in mechanisms for the development of L-R asymmetry among animals (Fig. 14). Like vertebrates, chordates such as amphioxus145,146) and ascidians147) deploy cilia and the Nodal-Pitx2 signaling pathway. Sea urchins also rely on motile cilia and the Nodal-Pitx2 pathway,148) but, unlike vertebrates, Nodal specifies the future right side, whereas BMP acts on the left side.149,150) Snails17,151,152) and Drosophila153,154) do not depend on cilia but instead rely on cytoskeletal proteins for the generation of cellular chirality, which serves as the symmetry-breaking event. Drosophila even lacks the Nodal-Pitx2 pathway, which is conserved in all other species examined.
Figure 14.
Mechanisms of L-R asymmetry establishment in various animals. Note that some steps (such as the Nodal-Pitx2 pathway) are conserved, whereas others are divergent.
8. Conclusions
There has been rapid progress in our understanding of L-R asymmetry in animals over the last 20 years or so, since the first discovery of L-R asymmetrically expressed genes. Systematic screening for L-R mutants in various model animals and genetic studies of human patients with laterality defects have identified a large number of genes that are required for establishment of L-R asymmetry. We also now largely understand the key role of cilia in symmetry breaking in many animals, the role and regulation of asymmetric Nodal signaling, and the cellular basis of asymmetric morphogenesis for some visceral organs. However, despite this recent progress, many challenging questions remain to be answered. For example, how does fluid flow achieve its effects, and how does an embryo sense the flow? What is the origin of L-R asymmetry during embryogenesis? If directional fluid flow breaks L-R symmetry, as in mice, what determines the direction of ciliary rotation? Can it be ascribed to structural chirality of dynein arms in the axoneme? How does brain asymmetry develop in humans? Finally, the evolutionary conservation and diversity of mechanisms for the establishment of L-R asymmetry among animals are still largely unexplored.
Acknowledgments
I thank former and current members of my lab for insightful discussion. Recent work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (nos. 24113001 and 24113004) and from Core Research for Evolutional Science and Technology of the Japan Science and Technology Agency. The author declares no conflicts of interest.
Profile
Hiroshi Hamada was born in Kagawa prefecture, Japan, in 1950 and graduated from Okayama University School of Medicine in 1975. After 5 years as a post-doc at the NIH, he became an Assistant Professor at Memorial University in Newfoundland, Canada, and started working on the regulation of embryonal carcinoma cell differentiation. This moved his interests toward developmental biology. He returned to Japan as an Associate Professor in The University of Tokyo, and subsequently a Professor at the Institute for Molecular & Cellular Biology, The University of Tokyo/Graduate School of Frontier Biosciences, Osaka University for 20 years. After retirement from Osaka University, he moved to RIKEN Center for Developmental Biology as the Director in 2015. He is currently Team Leader of RIKEN Center for Biosystems Dynamics Research. His laboratory used the mouse system to study embryonic patterning and organogenesis. Prompted by the finding of Lefty, a left–right asymmetrically expressed TGFβ member, his group investigated how body axes, left–right and anterior–posterior axes, are established during development. His current interests are the mechanism of left–right symmetry breaking and its diversity among various animals. He was elected as an Associate Member of EMBO and received the Keio Medical Science Prize in 2014.
References
- 1).Smith C.A., Sinclair A.H. (2004) Sex determination: Insights from the chicken. Bioessays 26, 120–132. [DOI] [PubMed] [Google Scholar]
- 2).van Soldt B.J., Metscher B.D., Poelmann R.E., Vervust B., Vonk F.J., Müller G.B., et al. (2015) Heterochrony and early left-right asymmetry in the development of the cardiorespiratory system of snakes. PLoS One 10, e116416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Baillie M. (1789) An account of a remarkable transposition of the viscera in the human body. Lond. Med. J. 10, 178–197. [PMC free article] [PubMed] [Google Scholar]
- 4).Kartagener M. (1933) Zur pathogenese der bronchiektasen. 1. Mitteilung: Bronchiektasien bei situs viscerum inversus. Beitr. Klin. Tuberk. 83, 489–501. [Google Scholar]
- 5).Hummel K.P. (1959) Developmental anomalies in mice resulting from action of the gene, disorganization, a semi-dominant lethal. Pediatrics 23, 212–221. [PubMed] [Google Scholar]
- 6).Yokoyama T., Copeland N.G., Jenkins N.A., Montgomery C.A., Elder F.F., Overbeek P.A. (1993) Reversal of left-right asymmetry: A situs inversus mutation. Science 260, 679–682. [DOI] [PubMed] [Google Scholar]
- 7).Levin M., Johnson R.L., Stern C.D., Kuehn M., Tabin C. (1995) A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell 82, 803–814. [DOI] [PubMed] [Google Scholar]
- 8).Meno C., Saijoh Y., Fujii H., Ikeda M., Yokoyama T., Yokoyama M., et al. (1996) Left-right asymmetric expression of the TGFβ-family member lefty in mouse embryos. Nature 381, 151–155. [DOI] [PubMed] [Google Scholar]
- 9).Collignon J., Varlet I., Robertson E.J. (1996) Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 381, 155–158. [DOI] [PubMed] [Google Scholar]
- 10).Supp D.M., Witte D.P., Potter S.S., Brueckner M. (1997) Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice. Nature 389, 963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Mochizuki T., Saijoh Y., Tsuchiya K., Shirayoshi Y., Takai S., Taya C., et al. (1998) Cloning of inv, a gene that controls left/right asymmetry and kidney development. Nature 395, 177–181. [DOI] [PubMed] [Google Scholar]
- 12).Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., et al. (1998) Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837. [DOI] [PubMed] [Google Scholar]
- 13).Hozumi S., Maeda R., Taniguchi K., Kanai M., Shirakabe S., Sasamura T., et al. (2006) An unconventional myosin in Drosophila reverses the default handedness in visceral organs. Nature 440, 798–802. [DOI] [PubMed] [Google Scholar]
- 14).Speder P., Adam G., Noselli S. (2006) Type ID unconventional myosin controls left-right asymmetry in Drosophila. Nature 440, 803–807. [DOI] [PubMed] [Google Scholar]
- 15).Sturtevant A.H. (1923) Inheritance of direction of coilling in Limnaea. Science 58, 269–270. [DOI] [PubMed] [Google Scholar]
- 16).Ueshima R., Asami T. (2003) Evolution: Single-gene speciation by left-right reversal. Nature 425, 679. [DOI] [PubMed] [Google Scholar]
- 17).Abe M., Kuroda R. (2019) The development of CRISPR for a mollusc establishes the formin Lsdia1 as the long-sought gene for snail dextral/sinistral coiling. Development 146, dev175976. [DOI] [PubMed] [Google Scholar]
- 18).Blum M., Feistel K., Thumberger T., Schweickert A. (2014) The evolution and conservation of left-right patterning mechanisms. Development 141, 1603–1613. [DOI] [PubMed] [Google Scholar]
- 19).Nakamura T., Hamada H. (2012) Left-right patterning: Conserved and divergent mechanisms. Development 139, 3257–3262. [DOI] [PubMed] [Google Scholar]
- 20).Gros J., Feistel K., Viebahn C., Blum M., Tabin C.J. (2009) Cell movements at Hensen’s node establish left/right asymmetric gene expression in the chick. Science 324, 941–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Kajikawa E., Horo U., Ide T., Mizuno K., Minegishi K., Hara Y., et al. (2020) Nodal paralogues underlie distinct mechanisms for visceral left-right asymmetry in reptiles and mammals. Nat. Ecol. Evol. 4, 261–269. [DOI] [PubMed] [Google Scholar]
- 22).Sulik K., Dehart D.B., Iangaki T., Carson J.L., Vrablic T., Gesteland K., et al. (1994) Morphogenesis of the murine node and notochordal plate. Dev. Dyn. 201, 260–278. [DOI] [PubMed] [Google Scholar]
- 23).Hirokawa N., Tanaka Y., Okada Y. (2009) Left-right determination: Involvement of molecular motor KIF3, cilia, and nodal flow. Cold Spring Harb. Perspect. Biol. 1, a000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Shinohara K., Chen D., Nishida T., Misaki K., Yonemura S., Hamada H. (2015) Absence of radial spokes in mouse node cilia is required for rotational movement but confers ultrastructural instability as a trade-off. Dev. Cell 35, 236–246. [DOI] [PubMed] [Google Scholar]
- 25).Chen D., Zhong Y., Shinohara K., Nishida T., Hasegawa T., Hamada H. (2014) The dynein-triggered ciliary motion in embryonic nodes: An exploratory study based on computational models. Biomed. Mater. Eng. 24, 2495–2501. [DOI] [PubMed] [Google Scholar]
- 26).Goetz S.C., Anderson K.V. (2010) The primary cilium: A signalling centre during vertebrate development. Nat. Rev. Genet. 11, 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Nauli S.M., Alenghat F.J., Luo Y., Williams E., Vassilev P., Li X., et al. (2003) Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33, 129–137. [DOI] [PubMed] [Google Scholar]
- 28).Praetorius H.A. (2015) The primary cilium as sensor of fluid flow: New building blocks to the model. A review in the theme: Cell signaling: Proteins, pathways and mechanisms. Am. J. Physiol. Cell Physiol. 308, C198–C208. [DOI] [PubMed] [Google Scholar]
- 29).Delling M., Indzhykulian A.A., Liu X., Li Y., Xie T., Corey D.P., et al. (2016) Primary cilia are not calcium-responsive mechanosensors. Nature 531, 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Yuan S.L., Zhao L., Brueckner M., Sun Z.X. (2015) Intraciliary calcium oscillations initiate vertebrate left-right asymmetry. Curr. Biol. 25, 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Krebs L.T., Iwai N., Nonaka S., Welsh I.C., Lan Y., Jiang R., et al. (2003) Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 17, 1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Chien Y.H., Srinivasan S., Keller R., Kintner C. (2018) Mechanical strain determines cilia length, motility, and planar position in the left-right organizer. Dev. Cell 45, 316–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Hirokawa N., Tanaka Y., Okada Y., Takeda S. (2006) Nodal flow and the generation of left-right asymmetry. Cell 125, 33–45. [DOI] [PubMed] [Google Scholar]
- 34).Hirokawa N., Tanaka Y., Okada Y. (2012) Cilia, KIF3 molecular motor and nodal flow. Curr. Opin. Cell Biol. 24, 31–39. [DOI] [PubMed] [Google Scholar]
- 35).Takeda S., Yonekawa Y., Tanaka Y., Okada Y., Nonaka S., Hirokawa N. (1999) Left-right asymmetry and kinesin superfamily protein KIF3A: New insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J. Cell Biol. 145, 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Okada Y., Nonaka S., Tanaka Y., Saijoh Y., Hamada H., Hirokawa N. (1999) Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol. Cell 4, 459–468. [DOI] [PubMed] [Google Scholar]
- 37).Nonaka S., Shiratori H., Saijoh Y., Hamada H. (2002) Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature 418, 96–99. [DOI] [PubMed] [Google Scholar]
- 38).Fliegauf M., Benzing T., Omran H. (2007) When cilia go bad: Cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880–893. [DOI] [PubMed] [Google Scholar]
- 39).Omori T., Sugai H., Imai Y., Ishikawa T. (2017) Nodal cilia-driven flow: Development of a computational model of the nodal cilia axoneme. J. Biomech. 61, 242–249. [DOI] [PubMed] [Google Scholar]
- 40).Okada Y., Takeda S., Tanaka Y., Belmonte J.C., Hirokawa N. (2005) Mechanism of nodal flow: A conserved symmetry breaking event in left-right axis determination. Cell 121, 633–644. [DOI] [PubMed] [Google Scholar]
- 41).Nonaka S., Yoshiba S., Watanabe D., Ikeuchi S., Goto T., Marshall W.F., et al. (2005) De novo formation of left-right asymmetry by posterior tilt of nodal cilia. PLoS Biol. 3, e268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Minegishi K., Hashimoto M., Ajima R., Takaoka K., Shinohara K., Ikawa Y., et al. (2017) A Wnt5 activity asymmetry and intercellular signaling via PCP proteins polarize node cells for left-right symmetry breaking. Dev. Cell 40, 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Winter C.G., Wang B., Ballew A., Royou A., Karess R., Axelrod J.D., et al. (2001) Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105, 81–91. [DOI] [PubMed] [Google Scholar]
- 44).McGreevy E.M., Vijayraghavan D., Davidson L.A., Hildebrand J.D. (2015) Shroom3 functions downstream of planar cell polarity to regulate myosin II distribution and cellular organization during neural tube closure. Biol. Open 4, 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Nishimura T., Honda H., Takeichi M. (2012) Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell 149, 1084–1097. [DOI] [PubMed] [Google Scholar]
- 46).Hegan P.S., Ostertag E., Geurts A.M., Mooseker M.S. (2015) Myosin Id is required for planar cell polarity in ciliated tracheal and ependymal epithelial cells. Cytoskeleton (Hoboken) 72, 503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Tingler M., Kurz S., Maerker M., Ott T., Fuhl F., Schweickert A., et al. (2018) A conserved role of the unconventional myosin 1d in laterality determination. Curr. Biol. 28, 810–816. [DOI] [PubMed] [Google Scholar]
- 48).Tanaka Y., Okada Y., Hirokawa N. (2005) FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature 435, 172–177. [DOI] [PubMed] [Google Scholar]
- 49).Tabin C.J., Vogan K.J. (2003) A two-cilia model for vertebrate left-right axis specification. Genes Dev. 17, 1–6. [DOI] [PubMed] [Google Scholar]
- 50).McGrath J., Somlo S., Makova S., Tian X., Brueckner M. (2003) Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell 114, 61–73. [DOI] [PubMed] [Google Scholar]
- 51).Yoshiba S., Shiratori H., Kuo I.Y., Kawasumi A., Shinohara K., Nonaka S., et al. (2012) Cilia at the node of mouse embryos sense fluid flow for left-right determination via Pkd2. Science 338, 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Ferreira R.R., Vilfan A., Julicher F., Supatto W., Vermot J. (2017) Physical limits of flow sensing in the left-right organizer. eLife 6, e25078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Pennekamp P., Karcher C., Fischer A., Schweickert A., Horst J., Blum M., et al. (2002) The ion channel polycystin-2 is required for left-right axis determination in mice. Curr. Biol. 12, 938–943. [DOI] [PubMed] [Google Scholar]
- 54).Field S., Riley K.L., Grimes D.T., Hilton H., Simon M., Powles-Glover N., et al. (2011) Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2. Development 138, 1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Kamura K., Kobayashi D., Uehara Y., Koshida S., Iijima N., Kudo A., et al. (2011) Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left-right axis. Development 138, 1121–1129. [DOI] [PubMed] [Google Scholar]
- 56).Vetrini F., D’Alessandro L.C., Akdemir Z.C., Braxton A., Azamian M.S., Eldomery M.K., et al. (2016) Bi-allelic mutations in PKD1L1 are associated with laterality defects in humans. Am. J. Hum. Genet. 99, 886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Grimes D.T., Keynton J.L., Buenavista M.T., Jin X., Patel S.H., Shinohara K., et al. (2016) Genetic analysis reveals a hierarchy of interactions between polycystin-encoding genes and genes controlling cilia function during left-right determination. PLoS Genet. 12, e1006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Takao D., Nemoto T., Abe T., Kiyonari H., Kajiuara-Kobayashi H., Shiratori H., et al. (2013) Asymmetric distribution of dynamic calcium signals in the node of mouse embryo during left-right axis formation. Dev. Biol. 376, 23–30. [DOI] [PubMed] [Google Scholar]
- 59).Kawasumi A., Nakamura T., Iwai N., Yashiro K., Saijoh Y., Belo J.A., et al. (2011) Left-right asymmetry in the level of active Nodal protein produced in the node is translated into left-right asymmetry in the lateral plate of mouse embryos. Dev. Biol. 353, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Schweickert A., Vick P., Getwan M., Weber T., Schneider I., Eberhardt M., et al. (2010) The nodal inhibitor Coco is a critical target of leftward flow in Xenopus. Curr. Biol. 20, 738–743. [DOI] [PubMed] [Google Scholar]
- 61).Shinohara K., Kawasumi A., Takamatsu A., Yoshiba S., Botilde Y., Motoyama N., et al. (2012) Two rotating cilia in the node cavity are sufficient to break left-right symmetry in the mouse embryo. Nat. Commun. 3, 622. [DOI] [PubMed] [Google Scholar]
- 62).Tanaka C., Sakuma R., Nakamura T., Hamada H., Saijoh Y. (2007) Long-range action of Nodal requires interaction with GDF1. Genes Dev. 21, 3272–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Marques S., Borges A.C., Silva A.C., Freitas S., Cordenonsi M., Belo J.A. (2004) The activity of the Nodal antagonist Cerl-2 in the mouse node is required for correct L/R body axis. Genes Dev. 18, 2342–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Nakamura T., Saito D., Kawasumi A., Shinohara K., Asai Y., Takaoka K., et al. (2012) Fluid flow and interlinked feedback loops establish left-right asymmetric decay of Cerl2 mRNA. Nat. Commun. 3, 1322. [DOI] [PubMed] [Google Scholar]
- 65).Maisonneuve C., Guilleret I., Vick P., Weber T., Andre P., Beyer T., et al. (2009) Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development 136, 3019–3030. [DOI] [PubMed] [Google Scholar]
- 66).Yakulov T.A., Yasunaga T., Ramachandran H., Engel C., Müller B., Hoff S., et al. (2015) Anks3 interacts with nephronophthisis proteins and is required for normal renal development. Kidney Int. 87, 1191–1200. [DOI] [PubMed] [Google Scholar]
- 67).Rothe B., Leettola C.N., Leal-Esteban L., Cascio D., Fortier S., Isenschmid M., et al. (2018) Crystal structure of Bicc1 SAM polymer and mapping of interactions between the ciliopathy-associated proteins Bicc1, ANKS3, and ANKS6. Structure 26, 209–224.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Shamseldin H.E., Yakulov T.A., Hashem A., Walz G., Alkuraya F.S. (2016) ANKS3 is mutated in a family with autosomal recessive laterality defect. Hum. Genet. 135, 1233–1239. [DOI] [PubMed] [Google Scholar]
- 69).Zhang Y., Cooke A., Park S., Dewey C.N., Wickens M., Sheets M.D. (2013) Bicaudal-C spatially controls translation of vertebrate maternal mRNAs. RNA 19, 1575–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Chang C.F., Schock E.N., O’Hare E.A., Dodgson J., Cheng H.H., Muir W.M., et al. (2014) The cellular and molecular etiology of the craniofacial defects in the avian ciliopathic mutant talpid2. Development 141, 3003–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Schröder S.S., Tsikolia N., Weizbauer A., Hue I., Viebahn C. (2016) Paraxial nodal expression reveals a novel conserved structure of the left-right organizer in four mammalian species. Cells Tissues Organs 201, 77–87. [DOI] [PubMed] [Google Scholar]
- 72).Rankin C.T., Bunton T., Lawler A.M., Lee S.J. (2000) Regulation of left-right patterning in mice by growth/differentiation factor-1. Nat. Genet. 24, 262–265. [DOI] [PubMed] [Google Scholar]
- 73).Montague T.G., Gagnon J.A., Schier A.F. (2018) Conserved regulation of Nodal-mediated left-right patterning in zebrafish and mouse. Development 145, dev171090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74).Brennan J., Norris D.P., Robertson E.J. (2002) Nodal activity in the node governs left-right asymmetry. Genes Dev. 16, 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75).Saijoh Y., Oki S., Ohishi S., Hamada H. (2003) Left-right patterning of the mouse lateral plate requires nodal produced in the node. Dev. Biol. 256, 160–172. [DOI] [PubMed] [Google Scholar]
- 76).Oki S., Hashimoto R., Okui Y., Shen M.M., Mekada E., Otani H., et al. (2007) Sulfated glycosaminoglycans are necessary for Nodal signal transmission from the node to the left lateral plate in the mouse embryo. Development 134, 3893–3904. [DOI] [PubMed] [Google Scholar]
- 77).Lowe L.A., Supp D.M., Sampath K., Yokoyama T., Wright C.V., Potter S.S., et al. (1996) Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature 381, 158–161. [DOI] [PubMed] [Google Scholar]
- 78).Meno C., Shimono A., Saijoh Y., Yashiro K., Mochida K., Ohishi S., et al. (1998) lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell 94, 287–297. [DOI] [PubMed] [Google Scholar]
- 79).Meno C., Takeuchi J., Sakuma R., Koshiba-Takeuchi K., Ohishi S., Saijoh Y., et al. (2001) Diffusion of nodal signaling activity in the absence of the feedback inhibitor Lefty2. Dev. Cell 1, 127–138. [DOI] [PubMed] [Google Scholar]
- 80).Meno C., Ito Y., Saijoh Y., Matsuda Y., Tashiro K., Kuhara S., et al. (1997) Two closely-related left-right asymmetrically expressed genes, lefty-1 and lefty-2: Their distinct expression domains, chromosomal linkage and direct neuralizing activity in Xenopus embryos. Genes Cells 2, 513–524. [DOI] [PubMed] [Google Scholar]
- 81).Schier A.F., Shen M.M. (2000) Nodal signalling in vertebrate development. Nature 403, 385–389. [DOI] [PubMed] [Google Scholar]
- 82).Montague T.G., Schier A.F. (2017) Vg1-Nodal heterodimers are the endogenous inducers of mesendoderm. eLife 6, e28183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83).Cheng S.K., Olale F., Brivanlou A.H., Schier A.F. (2004) Lefty blocks a subset of TGFβ signals by antagonizing EGF-CFC coreceptors. PLoS Biol. 2, E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84).Sakuma R., Ohnishi Y., Meno C., Fujii H., Juan H., Takeuchi J., et al. (2002) Inhibition of Nodal signalling by Lefty mediated through interaction with common receptors and efficient diffusion. Genes Cells 7, 401–412. [DOI] [PubMed] [Google Scholar]
- 85).Iratni R., Yan Y.T., Chen C., Ding J., Zhang Y., Price S.M., et al. (2002) Inhibition of excess nodal signaling during mouse gastrulation by the transcriptional corepressor DRAP1. Science 298, 1996–1999. [DOI] [PubMed] [Google Scholar]
- 86).Morsut L., Yan K.P., Enzo E., Aragona M., Soligo S.M., Wendling O., et al. (2010) Negative control of Smad activity by ectodermin/Tif1γ patterns the mammalian embryo. Development 137, 2571–2578. [DOI] [PubMed] [Google Scholar]
- 87).Sampath K., Robertson E.J. (2016) Keeping a lid on nodal: Transcriptional and translational repression of nodal signalling. Open Biol. 6, 150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Choi W.Y., Giraldez A.J., Schier A.F. (2007) Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318, 271–274. [DOI] [PubMed] [Google Scholar]
- 89).Robertson E.J. (2014) Dose-dependent Nodal/Smad signals pattern the early mouse embryo. Semin. Cell Dev. Biol. 32, 73–79. [DOI] [PubMed] [Google Scholar]
- 90).Saijoh Y., Adachi H., Sakuma R., Yeo C.Y., Yashiro K., Watanabe M., et al. (2000) Left-right asymmetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Mol. Cell 5, 35–47. [DOI] [PubMed] [Google Scholar]
- 91).Nakamura T., Mine N., Nakaguchi E., Mochizuki A., Yamamoto M., Yahsiro K., et al. (2006) Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Dev. Cell 11, 495–504. [DOI] [PubMed] [Google Scholar]
- 92).Meinhardt H., Gierer A. (2000) Pattern formation by local self-activation and lateral inhibition. Bioessays 22, 753–760. [DOI] [PubMed] [Google Scholar]
- 93).Müller P., Rogers K.W., Jordan B.M., Lee J.S., Robson D., Ramanathan S., et al. (2012) Differential diffusivity of Nodal and Lefty underlies a reaction-diffusion patterning system. Science 336, 721–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94).Wang Y., Wang X., Wohland T., Sampath K. (2016) Extracellular interactions and ligand degradation shape the nodal morphogen gradient. eLife 5, e13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95).Almuedo-Castillo M., Bläßle A., Mörsdorf D., Marcon L., Soh G.H., Rogers K.W., et al. (2018) Scale-invariant patterning by size-dependent inhibition of Nodal signalling. Nat. Cell Biol. 20, 1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Wang L., Liu Z., Lin H., Ma D., Tao Q., Liu F. (2017) Epigenetic regulation of left-right asymmetry by DNA methylation. EMBO J. 36, 2987–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97).Khoueiry R., Sohni A., Thienpont B., Luo X., Velde J.V., Bartoccetti M., et al. (2017) Lineage-specific functions of TET1 in the postimplantation mouse embryo. Nat. Genet. 49, 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98).Logan M., Pagan-Westphal S.M., Smith D.M., Paganessi L., Tabin C.J. (1998) The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell 94, 307–317. [DOI] [PubMed] [Google Scholar]
- 99).Yoshioka H., Meno C., Koshiba K., Sugihara M., Itoh H., Ishimaru Y., et al. (1998) Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell 94, 299–305. [DOI] [PubMed] [Google Scholar]
- 100).Piedra M.E., Icardo J.M., Albajar M., Rodriguez-Rey J.C., Ros M.A. (1998) Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell 94, 319–324. [DOI] [PubMed] [Google Scholar]
- 101).Ryan A.K., Blumberg B., Rodriguez-Esteban C., Yonei-Tamura S., Tamura K., Tsukui T., et al. (1998) Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature 394, 545–551. [DOI] [PubMed] [Google Scholar]
- 102).Shiratori H., Sakuma R., Watanabe M., Hashiguchi H., Mochida K., Sakai Y., et al. (2001) Two-step regulation of left-right asymmetric expression of Pitx2: Initiation by nodal signaling and maintenance by Nkx2. Mol. Cell 7, 137–149. [DOI] [PubMed] [Google Scholar]
- 103).Liu C., Liu W., Lu M.F., Brown N.A., Martin J.F. (2001) Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development 128, 2039–2048. [DOI] [PubMed] [Google Scholar]
- 104).Shiratori H., Yashiro K., Shen M.M., Hamada H. (2006) Conserved regulation and role of Pitx2 in situs-specific morphogenesis of visceral organs. Development 133, 3015–3025. [DOI] [PubMed] [Google Scholar]
- 105).Ji Y., Buel S.M., Amack J.D. (2016) Mutations in zebrafish pitx2 model congenital malformations in Axenfeld-Rieger syndrome but do not disrupt left-right placement of visceral organs. Dev. Biol. 416, 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106).Noël E.S., Verhoeven M., Lagendijk A.K., Tessadori F., Smith K., Choorapoikayil S., et al. (2013) A Nodal-independent and tissue-intrinsic mechanism controls heart-looping chirality. Nat. Commun. 4, 2754. [DOI] [PubMed] [Google Scholar]
- 107).Ocana O.H., Coskun H., Minguillon C., Murawala P., Tanaka E.M., Galceran J., et al. (2017) A right-handed signalling pathway drives heart looping in vertebrates. Nature 549, 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108).Bergwerff M., Gittenberger-de Groot A.C., Wisse L.J., DeRuiter M.C., Wessels A., Martin J.F., et al. (2000) Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 436, 12–19. [DOI] [PubMed] [Google Scholar]
- 109).Martin J.F., Bradley A., Olson E.N. (1995) The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 9, 1237–1249. [DOI] [PubMed] [Google Scholar]
- 110).Desgrange A., Le Garrec J.F., Meilhac S.M. (2018) Left-right asymmetry in heart development and disease: Forming the right loop. Development 145, dev162776. [DOI] [PubMed] [Google Scholar]
- 111).Li Y., Klena N.T., Gabriel G.C., Liu X., Kim A.J., Lemke K., et al. (2015) Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 521, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112).Yuan S., Zaidi S., Brueckner M. (2013) Congenital heart disease: Emerging themes linking genetics and development. Curr. Opin. Genet. Dev. 23, 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113).Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J.D., et al. (2013) De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114).Le Garrec J.F., Dominguez J.N., Dedsgrange A., Ivanovitch K.D., Raphael E., Bangham J.A., et al. (2017) A predictive model of asymmetric morphogenesis from 3D reconstructions of mouse heart looping dynamics. eLife 6, 28951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115).Smith K.A., Chocron S., von der Hardt S., de Pater E., Soufan A., Bussmann J., et al. (2008) Rotation and asymmetric development of the zebrafish heart requires directed migration of cardiac progenitor cells. Dev. Cell 14, 287–297. [DOI] [PubMed] [Google Scholar]
- 116).Chen J.N., van Eeden F.J., Warren K.S., Chin A., Nusslein-Volhard C., Haffter P., et al. (1997) Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 124, 4373–4382. [DOI] [PubMed] [Google Scholar]
- 117).Francou A., De Bono C., Kelly R.G. (2017) Epithelial tension in the second heart field promotes mouse heart tube elongation. Nat. Commun. 8, 14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118).Yashiro K., Shiratori H., Hamada H. (2007) Haemodynamics determined by a genetic programme govern asymmetric development of the aortic arch. Nature 450, 285–288. [DOI] [PubMed] [Google Scholar]
- 119).Womble M., Amin N.M., Nascone-Yoder N. (2018) The left-right asymmetry of liver lobation is generated by Pitx2c-mediated asymmetries in the hepatic diverticulum. Dev. Biol. 439, 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120).Huycke T.R., Tabin C.J. (2018) Chick midgut morphogenesis. Int. J. Dev. Biol. 62, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121).Davis N.M., Kurpios N.A., Sun X., Gros J., Martin J.F., Tabin C.J. (2008) The chirality of gut rotation derives from left-right asymmetric changes in the architecture of the dorsal mesentery. Dev. Cell 15, 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122).Kurpios N.A., Ibanes M., Davis N.M., Lui W., Katz T., Martin J.F., et al. (2008) The direction of gut looping is established by changes in the extracellular matrix and in cell: Cell adhesion. Proc. Natl. Acad. Sci. U.S.A. 105, 8499–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123).Sivakumar A., Mahadevan A., Lauer M.E., Narvaez R.J., Ramesh S., Demler C.M., et al. (2018) Midgut laterality is driven by hyaluronan on the right. Dev. Cell 46, 533–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124).Savin T., Kurpios N.A., Shyer A.E., Florescu P., Liang H., Mahadevan L., et al. (2011) On the growth and form of the gut. Nature 476, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125).Concha M.L., Bianco I.H., Wilson S.W. (2012) Encoding asymmetry within neural circuits. Nat. Rev. Neurosci. 13, 832–843. [DOI] [PubMed] [Google Scholar]
- 126).Gamse J.T., Kuan Y.S., Macurak M., Brosamie C., Thisse B., Thisse C., et al. (2005) Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development 132, 4869–4881. [DOI] [PubMed] [Google Scholar]
- 127).Duboue E.R., Hong E., Eldred K.C., Halpern M.E. (2017) Left habenular activity attenuates fear responses in larval zebrafish. Curr. Biol. 27, 2154–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128).Signore I.A., Palma K., Concha M.L. (2016) Nodal signalling and asymmetry of the nervous system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129).Concha M.L., Burdine R.D., Russell C., Schier A.F., Wilson S.W. (2000) A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron 28, 399–409. [DOI] [PubMed] [Google Scholar]
- 130).Roussigne M., Wei L., Tsingos E., Kuchling F., Alkobtawi M., Tsalavouta M., et al. (2018) Left/right asymmetric collective migration of parapineal cells is mediated by focal FGF signaling activity in leading cells. Proc. Natl. Acad. Sci. U.S.A. 115, E9812–E9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131).Corballis M.C. (2014) Left brain, right brain: Facts and fantasies. PLoS Biol. 12, e1001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132).Gunturkun O., Ocklenburg S. (2017) Ontogenesis of Lateralization. Neuron 94, 249–263. [DOI] [PubMed] [Google Scholar]
- 133).Karlebach G., Francks C. (2015) Lateralization of gene expression in human language cortex. Cortex 67, 30–36. [DOI] [PubMed] [Google Scholar]
- 134).Ocklenburg S., Schmitz J., Moinfar Z., Moser D., Klose R., Lor S., et al. (2017) Epigenetic regulation of lateralized fetal spinal gene expression underlies hemispheric asymmetries. eLife 6, 22784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135).Sun T., Patoine C., Abu-Khalil A., Visvader J., Sum E., Cherry T.J., et al. (2005) Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science 308, 1794–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136).de Kovel C.G.F., Lisgo S.N., Fisher S.E., Francks C. (2018) Subtle left-right asymmetry of gene expression profiles in embryonic and foetal human brains. Sci. Rep. 8, 12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137).Arning L., Ockenburg S., Schulz S., Ness V., Gerding W.M., Hengstler J.G., et al. (2013) PCSK6 VNTR polymorphism is associated with degree of handedness but not direction of handedness. PLoS One 8, e67251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138).Ludwig K.U., Mattheisen M., Muhleisen T.W., Roeske D., Schmal C., Breuer R., et al. (2009) Supporting evidence for LRRTM1 imprinting effects in schizophrenia. Mol. Psychiatry 14, 743–745. [DOI] [PubMed] [Google Scholar]
- 139).Kawakami R., Shinohara Y., Kato Y., Sugiyama H., Shigemoto R., Ito I. (2003) Asymmetrical allocation of NMDA receptor epsilon2 subunits in hippocampal circuitry. Science 300, 990–994. [DOI] [PubMed] [Google Scholar]
- 140).Ukai H., Kawahara A., Hirayama K., Case M.J., Aino S., Miyabe M., et al. (2017) PirB regulates asymmetries in hippocampal circuitry. PLoS One 12, e0179377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141).Goto K., Kurashima R., Gokan H., Inoue N., Ito I., Watanabe S. (2001) Left-right asymmetry defect in the hippocampal circuitry impairs spatial learning and working memory in iv mice. PLoS One 5, e15468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142).Kawakami R., Dobi A., Shigemoto R., Ito I. (2008) Right isomerism of the brain in inversus viscerum mutant mice. PLoS One 3, e1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143).Guioli S., Lovell-Badge R. (2007) PITX2 controls asymmetric gonadal development in both sexes of the chick and can rescue the degeneration of the right ovary. Development 134, 4199–4208. [DOI] [PubMed] [Google Scholar]
- 144).Ishimaru Y., Komatsu T., Kasahara M., Katoh-Fukui Y., Ogawa H., Toyama Y., et al. (2008) Mechanism of asymmetric ovarian development in chick embryos. Development 135, 677–685. [DOI] [PubMed] [Google Scholar]
- 145).Zhu X., Shi C., Zhong Y., Liu X., Yan Q., Wu X., et al. (2020) Cilia-driven asymmetric Hedgehog signalling determines the amphioxus left-right axis by controlling Dand5 expression. Development 147, dev182469. [DOI] [PubMed] [Google Scholar]
- 146).Soukup V. (2017) Left-right asymmetry specification in amphioxus: Review and prospects. Int. J. Dev. Biol. 61, 611–620. [DOI] [PubMed] [Google Scholar]
- 147).Yamada S., Tanaka Y., Imai K.S., Saigou M., Onuma T.A., Nishida H. (2019) Wavy movements of epidermis monocilia drive the neurula rotation that determines left-right asymmetry in ascidian embryos. Dev. Biol. 448, 173–182. [DOI] [PubMed] [Google Scholar]
- 148).Tisler M., Wetzel F., Mantino S., Kremnyov S., Thumberger T., Schweickert A., et al. (2016) Cilia are required for asymmetric nodal induction in the sea urchin embryo. BMC Dev. Biol. 16, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149).Su Y.H. (2014) Telling left from right: Left-right asymmetric controls in sea urchins. Genesis 52, 269–278. [DOI] [PubMed] [Google Scholar]
- 150).Molina M.D., de Crozé N., Haillot E., Lepage T. (2013) Nodal: Master and commander of the dorsal-ventral and left-right axes in the sea urchin embryo. Curr. Opin. Genet. Dev. 23, 445–453. [DOI] [PubMed] [Google Scholar]
- 151).Abe M., Takahashi H., Kuroda R. (2014) Spiral cleavages determine the left-right body plan by regulating Nodal pathway in monomorphic gastropods, Physa acuta. Int. J. Dev. Biol. 58, 513–520. [DOI] [PubMed] [Google Scholar]
- 152).Grande C., Patel N.H. (2009) Nodal signalling is involved in left-right asymmetry in snails. Nature 457, 1007–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153).Inaki M., Liu J., Matsuno K. (2016) Cell chirality: Its origin and roles in left-right asymmetric development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154).Coutelis J.B., Gonzalez-Morales N., Geminard C., Noselli S. (2014) Diversity and convergence in the mechanisms establishing L/R asymmetry in metazoa. EMBO Rep. 15, 926–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155).Shiratori H., Hamada H. (2006) The left-right axis in the mouse: From origin to morphology. Development 133, 2095–2104. [DOI] [PubMed] [Google Scholar]
- 156).Yoshiba S., Hamada H. (2014) Roles of cilia, fluid flow, and Ca2+ signaling in breaking of left-right symmetry. Trends Genet. 30, 10–17. [DOI] [PubMed] [Google Scholar]
- 157).Shiratori H., Hamada H. (2014) TGFβ signaling in establishing left-right asymmetry. Semin. Cell Dev. Biol. 32, 80–84. [DOI] [PubMed] [Google Scholar]
- 158).Hamada H. (2018) Hyaluronan works on the right for directional gut looping. Dev. Cell 46, 525–526. [DOI] [PubMed] [Google Scholar]