Abstract
Advances in cancer research have revolutionized the way cancer is diagnosed and treated. Any cancer is now known to be an amalgamation of many subtypes, each carrying its specific cancer-causing gene or oncogene. It is also evident that a given oncogene is often present across a wide range of cancer subtypes, albeit at different frequencies. These lines of information have brought cancer genomic medicine (CGM) to the clinic, where genetic information is used to optimize therapeutic intervention. In 2017, the Expert Meeting for Cancer Genomic Medicine Promotion Consortium in the Ministry of Health, Labour and Welfare (MHLW) of Japan submitted a blueprint for the CGM platform in Japan. Accordingly, the MHLW designated a total of 206 hospitals that conduct cancer gene panel testing under the national health insurance system and established the Center for Cancer Genomics and Advanced Therapeutics to store genomic/clinical information of cancer patients. Since June 2019, the CGM officially started in Japan.
Keywords: molecularly targeted drug, cancer genomic medicine, cancer gene panel test
Introduction
Cancer is a disorder of immortal cells that clonally expand through the relentless acquisition of genomic and epigenomic anomalies. A given cancer cell may carry tens of thousands of somatic genomic alterations, and some of them directly contribute to the malignant phenotypes of cancer. International research initiatives involving large-scale sequencing projects for cancer genomes1,2) have identified a plethora of cancer-causing mutations, such as those producing activated kinases or differentiation blockers. Along with these discoveries, molecularly targeted drugs have been developed against growth driver proteins and have successfully prolonged the survival of patients with some cancer subtypes.
Cancer genome analyses have also revealed that such abnormal growth drivers may be shared across distinct cancer subtypes, albeit at differing frequencies.3) Therefore, it became apparent that the choice of molecularly targeted drugs should be optimized to the genome profiles, not the origin of the tumor for each patient. Together with the advent of next-generation sequencing (NGS), cancer genomic medicine (CGM) was thus realized where genomic alterations in tumors are simultaneously examined for genes with matching drugs available. Such momentum was further accelerated by the Precision Medicine Initiative (https://obamawhitehouse.archives.gov/precision-medicine) declared by Mr. Obama, the former president of the U.S.A.
Since June 1, 2019, Japan has started CGM by approving cancer gene panel tests covered by the national health insurance system. As of April 2020, a total of 206 institutes can conduct gene panel tests for cancer care, and the number of such hospitals will likely increase gradually. This review will summarize how the national platform of the CGM has been built and the direction that Japanese CGM is moving toward.
The rise of CGM
Chronic myeloid leukemia (CML) is caused by a reciprocal chromosome translocation between chromosomes 9 and 22 that produces an activated, fusion-type tyrosine kinase BCR-ABL1. Imatinib, a small molecule targeting ABL1, is highly effective in suppressing the growth of CML cells.4) Likewise, amplification of the ERBB2 gene is observed in approximately one-third of breast cancer cases, and trastuzumab, a monoclonal antibody targeting ERBB2, was shown to be effective in inducing apoptosis in ERBB2-amplified breast cancer.5) These studies and clinical trials have led the era of molecularly targeted drugs and companion diagnostics.
It became apparent, however, that such growth-promoting, aberrant proteins may be present in the other cancer subtypes as well. ERBB2 amplification, for instance, can also be found in gastric cancer,6) and the treatment of such tumors with trastuzumab indeed demonstrated clinical efficacy.7) Similarly, in 4–5% of non-small cell lung cancers (NSCLCs), we discovered the EML4-ALK fusion-type oncogene that produces an activated tyrosine kinase.8) Treatment with ALK-specific tyrosine kinase inhibitors (TKIs) for such tumors resulted in marked therapeutic effects. Of note, ALK also becomes fused to other partner genes in different cancers: NPM1 in anaplastic large cell lymphoma, TPM3/4 in inflammatory myofibroblastic tumors, VCL in renal medullary carcinoma, and FN1 in ovarian sarcoma.9) A clinical trial of such ALK-rearranged tumors with an ALK inhibitor had shown promising results.10) Thus, it was proposed that such ALK-dependent tumors be collectively called “ALKoma”, because all these tumors can be effectively treated using the same ALK-TKIs, irrespective of the pathological classification and the organs from which the cancers arise.9)
Given these findings, it is no longer appropriate that molecularly targeted drugs are approved only in a cancer subtype for specific organs. Rather, any cancer subtypes should be treated with targeted drugs if they share the same targets. On the other hand, the development of NGS has enabled the simultaneous assessment of mutations in hundreds of genes. Taking advantage of this technology, cancer gene panel tests (or cancer genome profiling tests) were made to examine the mutation profiles of genes linked to drug selection or to therapeutic intervention, and the CGM era had begun.
The components required for CGM
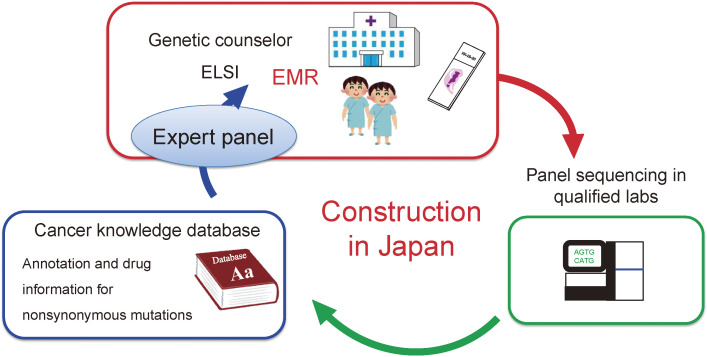
Japan is unique in that almost every Japanese person is covered by the national health insurance system. If Japan integrates CGM into its healthcare system, it will be necessary to build a CGM platform that can accommodate tens of thousands of cancer patients. Furthermore, the total CGM system should have several important components in addition to cancer gene panel tests. As shown in Fig. 1, physicians and co-medical staff must have sufficient literacy about CGM and should be capable of explaining CGM to their patients. Moreover, pathology departments have to prepare high-quality specimens that are suitable for NGS analyses.
Figure 1.
Components required for CGM. To sustain CGM within the national health insurance system, Japan has to build a large infrastructure for CGM. EMR, electric medical record; ELSI, ethical, legal and social implications.
In quality-assured laboratories, cancer specimens (and often with paired peripheral blood samples) are processed for NGS sequencing with gene panel tests. The raw sequencing data are then mapped to the human reference genome, and genomic alterations are calculated for each specimen. When paired normal samples are analyzed together with the tumors, somatic mutations in the tumors can be called by subtracting germline polymorphisms. Then, amino acid changes are predicted based on the mutation profiles and referred to the cancer knowledge database (CKDB) for the clinical annotation of each mutation. The CKDB is similar to an encyclopedia and contains clinical trial databases matched to each mutation as well as an extensive list of pathogenicity for gene mutations/variants. By referring to the CKDB, mutation profiles can be translated to clinical advice for the optimal selection of approved drugs and/or clinical trials for every patient.
Such reports are then sent back to hospitals and used for discussion about the therapeutic intervention for a given patient by an “expert panel” or “cancer board” that consists of experts of different expertise. If a patient has a germline pathogenic variant in a hereditary cancer syndrome gene, a genetic counselor should explain to the patient the causative genetic background and assist the patient in selecting therapeutic interventions.
If Japan is going to start CGM as part of the national health insurance system, it has to construct this CGM platform in a nationwide manner.
Construction of the national platform of CGM in Japan
To actualize CGM in Japan, the Ministry of Health, Labour and Welfare (MHLW) held an Expert Meeting for Cancer Genome Medicine Promotion Consortium composed of 12 representatives of various academic societies and patient advocacy groups. This meeting was set to discuss how the Japanese CGM should be constructed/organized and what infrastructures are necessary to support the Japanese CGM. After several rounds of discussion, the meeting submitted an official report to the Minister of the MHLW (https://www.mhlw.go.jp/file/05-Shingikai-10901000-Kenkoukyoku-Soumuka/0000169236.pdf, in Japanese) on June 27, 2017. The report contains two important proposals for the Japanese platform of CGM.
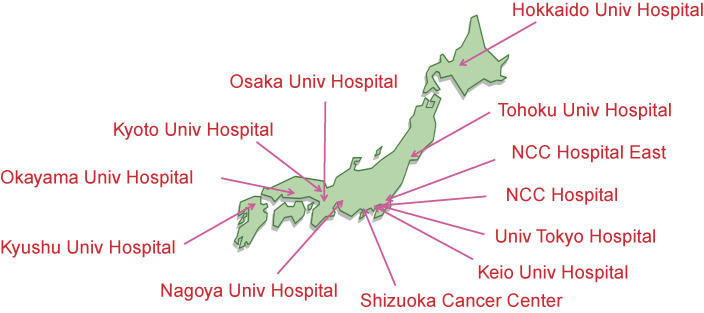
First, CGM should be conducted in a group of certified CGM-ready hospitals only, and the number of such hospitals shall be increased gradually. The report says that such hospitals should (i) have a certified infrastructure and plenty of experience for clinical trials, (ii) have an expert group for genetic counseling for hereditary cancer syndrome, and (iii) have an expert panel for assessing the clinical utility of each gene mutation. The MHLW accordingly chose 11 designated core hospitals for CGM in February 2018. As of April 2020, Japan has 12 designated core hospitals (Fig. 2), 33 designated hospitals and 161 cooperative hospitals, for a total of 206 institutes eligible for CGM under the national health insurance system.
Figure 2.
The twelve designated core hospitals for CGM.
Second, the report suggested that Japan should have a central datacenter to aggregate the genomic data and clinical information of the patients receiving cancer gene panel tests. The MHLW thus established the Center for Cancer Genomics and Advanced Therapeutics (C-CAT) in June 2018. C-CAT and the 206 hospitals above constitute the large network for CGM.
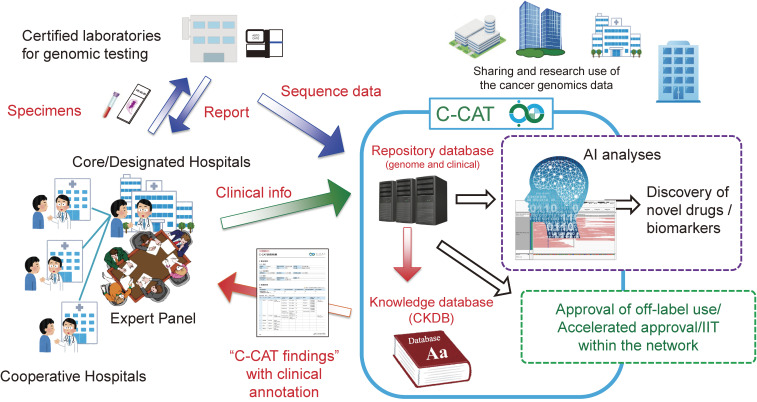
Figure 3 summarizes the national platform of CGM in Japan. If patients provide consent for gene panel testing at either designated core hospitals, designated hospitals, or cooperative hospitals, their appropriate specimens are sent to certified sequencing laboratories that then send back the test reports to the hospitals. The sequencing laboratories, upon patient consent, transfer nucleotide sequencing data such as VCF and FASTQ files to C-CAT (for some gene panel tests, the sequencing data are transferred to C-CAT from the corresponding hospitals). The CGM hospitals also input the clinical information of the patients into the C-CAT repository database. C-CAT thus stores the genomic data and clinical information of patients who receive cancer gene panel tests under the national health insurance system.
Figure 3.
The national platform of CGM in Japan. The figure is modified from the one used in the press conference held in C-CAT on June 1, 2018.
In C-CAT, the mutation profiles of individual patients are referred to the CKDB to annotate the clinical significance of each mutation. Such annotation information (for instance, matching approved drugs and matching clinical trials in Japan) is summarized as a “C-CAT findings” report that is sent back to an expert panel in the hospital to discuss the optimal therapeutic intervention for each patient.
The C-CAT database is further utilized to increase drug access for Japanese cancer patients. The number of approved drugs for cancer treatments is fewer than that in the U.S.A. We thus aim to use the C-CAT data to try/accept the off-label use of cancer drugs within the national CGM platform. From October 2019, a new official scheme for off-label use started within the 11 designated core hospitals. As of April 2020, three pharmaceutical companies have donated compounds to this scheme for compassionate use.
Future direction of Japanese CGM
This national initiative of CGM will markedly improve patient care in Japan in many ways. First, “C-CAT findings” are obtained for every patient, which facilitates patient recruitment into clinical trials regardless of where the patients are treated. The C-CAT findings provide a list of matching clinical trials via the CKDB that is curated and constantly updated by Japanese clinical oncologists. As of June 2020, the CKDB of C-CAT stores information for >700 cancer clinical trials in Japan.
Second, the C-CAT data are an essential resource for investigator-initiated and company-driven clinical trials in Japan. Furthermore, the data may be a driving force for foreign pharmaceutical companies to choose Japan to test their compounds in Asian countries because only Japan has a national database for mutations in cancer.
Third, as the official scheme for off-label use described above, the entire CGM network consisting of C-CAT and CGM hospitals will be a platform to maximize drug access for Japanese cancer patients. Sunami et al. reported that, in a clinical research, NGS analyses of cancer specimens with a 114 gene-panel test led to treatments with matching targeted drugs only in 13.3% of patients.11) This low drug accessibility is partly due to the small number of available clinical trials in Japan. We thus hope that such a situation for drug accessibility in Japan will change in the future by virtue of the CGM network.
Fourth, the C-CAT data will be utilized for a variety of purposes. C-CAT is constructing a search portal site for the C-CAT database for CGM hospitals. By using the search engine, any hospital within the CGM network can search for patients with a given gene mutation and obtain information on the drugs given to and/or clinical outcome of those patients. A similar search engine will also be widely provided to academia/industry outside the CGM network. If, for instance, a pharmaceutical company develops a new molecularly targeted drug, they can readily identify how many eligible patients there are in Japan who carry the mutated targets. Furthermore, C-CAT is developing a portal site for the complete set of anonymized C-CAT data. Here, the clinical and genomic information of individual patients can be accessed and analyzed for research use. Large-scale examination of C-CAT data will enable the discovery of new biomarkers/therapeutic targets. We hope that the C-CAT data will become a novel foundation for next-generation cancer care. It should be noted that such complete data can be accessed by approved users, only provided that the patient gave consent for industry usage of the anonymized data.
Finally, the C-CAT data should play an essential role for the government to improve patient care in Japan. Nearly complete mutation profiles (both somatic mutations and, in some cancer panels, germline variants) of Japanese cancer patients are stored in the C-CAT repository database. The Japanese government can, for instance, use the data to assess the number of hereditary cancer syndrome carriers and to scientifically estimate the cost-benefit of given cancer drugs.
Since the approval of cancer gene panel tests in June 2019 in Japan, more than 4,000 patients deposited their clinical and genomic information into C-CAT within the 2019 fiscal year, and more than 10,000 patients are expected to do so in the 2020 fiscal year. The Japanese CGM platform is thus growing steadily, and is a unique and ambitious program that we have harnessed and are going to continuously develop.
Acknowledgments
Thanks go to all members in my research group involved in the discovery of the EML4-ALK oncogene and who have swiftly brought such a discovery to the clinic. I am also grateful to all C-CAT staff for their dedication to actualize cancer genomic medicine in Japan.
Abbreviations
- CGM
cancer genomic medicine
- CKDB
cancer knowledge database
- NGS
next-generation sequencing
- TKI
tyrosine kinase inhibitor
Profile
Hiroyuki Mano was born in 1959, and graduated from the School of Medicine, The University of Tokyo in 1984. After spending his internship at The University of Tokyo Hospital and Jichi Medical University Hospital, he joined the Third Department of Internal Medicine, Faculty of Medicine, The University of Tokyo in 1986, where he became a clinical hematologist and also began to study the intracellular signaling pathways of blood cells. He discovered a novel tyrosine kinase, Tec, in 1990 and revealed that it plays an essential role in B cell development. He moved to the United States in 1989 to join the Department of Biochemistry, St. Jude Children’s Research Hospital in Memphis, Tennessee, as a postdoctoral researcher. He returned to the Third Department of Internal Medicine at The University of Tokyo to become an Assistant Professor in 1991. He then moved to the Department of Molecular Biology at Jichi Medical University, where he was appointed as an Associate Professor in 1993, and he became Professor of the Division of Functional Genomics at the same university in 2001. During his time at Jichi Medical University, Dr. Mano discovered the EML4-ALK oncogene in 2007. He then moved to the Department of Cellular Signaling, Graduate School of Medicine, The University of Tokyo, as a Professor in 2013. Then he moved to National Cancer Center as the Director of Research Institute in 2016, and was also appointed as Director of the Center for Cancer Genomics and Advanced Therapeutics, National Cancer Center in 2018.
For the discoveries of many clinically relevant oncogenes, he was awarded The Medical Award from The Japan Medical Association (2008), The Princess Takamatsu Cancer Research Fund Prize (2009), The Academic Award from the Mochida Memorial Foundation (2010), The Takeda Prize for Medical Science (2010), The Uehara Prize (2010), The Prize for Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (2011), The Asclepios Award from the Bonnie J. Addario Lung Cancer Foundation (2011), The Takamine Memorial Daiichi Sankyo Prize (2011), The Keio Medical Science Prize (2012), the Medal with Purple Ribbon from The Japanese Emperor (2012), The Shiono Prize from The Cell Science Research Foundation (2013), The Bälz Prize from Boehringer Ingelheim (2015), The Yamazaki Teiichi Prize (2019). In 2020, the Japan Academy decided to award him the 110th Japan Academy Prize.
References
- 1).The International Cancer Genome Consortiusm (2010) International network of cancer genome projects. Nature 464, 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Bailey M.H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., et al. (2018) Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385 e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).O’Brien S.G., Guilhot F., Larson R.A., Gathmann I., Baccarani M., Cervantes F., et al. (2003) Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N. Engl. J. Med. 348, 994–1004. [DOI] [PubMed] [Google Scholar]
- 5).Menard S., Pupa S.M., Campiglio M., Tagliabue E. (2003) Biologic and therapeutic role of HER2 in cancer. Oncogene 22, 6570–6578. [DOI] [PubMed] [Google Scholar]
- 6).Gravalos C., Jimeno A. (2008) HER2 in gastric cancer: A new prognostic factor and a novel therapeutic target. Ann. Oncol. 19, 1523–1529. [DOI] [PubMed] [Google Scholar]
- 7).Bang Y.J., Van Cutsem E., Feyereislova A., Chung H.C., Shen L., Sawaki A., et al. (2010) Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 376, 687–697. [DOI] [PubMed] [Google Scholar]
- 8).Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., et al. (2007) Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566. [DOI] [PubMed] [Google Scholar]
- 9).Mano H. (2012) ALKoma: A cancer subtype with a shared target. Cancer Discov. 2, 495–502. [DOI] [PubMed] [Google Scholar]
- 10).Mosse Y.P., Voss S.D., Lim M.S., Rolland D., Minard C.G., Fox E., et al. (2017) Targeting ALK with crizotinib in pediatric anaplastic large cell lymphoma and inflammatory myofibroblastic tumor: A Children’s Oncology Group study. J. Clin. Oncol. 35, 3215–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Sunami K., Ichikawa H., Kubo T., Kato M., Fujiwara Y., Shimomura A., et al. (2019) Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: A hospital-based study. Cancer Sci. 110, 1480–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]