Abstract
Background
Somatic mutations are important biomarkers for selecting an optimal targeted therapy and predicting outcomes for non-small-cell lung cancer (NSCLC) patients that are often detected from tissue samples. However, tissue samples are not always readily available from these patients. The exploration of using circulating tumor DNA (ctDNA) to identify somatic mutations offers an alternative source that should be explored.
Methods
In this retrospective study, we included 280 patients diagnosed with adenocarcinoma between 2017 and 2018 in a hospital in eastern China. Tissue or ctDNA was collected, and a wide spectrum of somatic mutations was analyzed by targeted next-generation sequencing platforms. Associations among the mutation status, biomarkers, screening methods, disease stages, and interaction with treatment with overall survival (OS) were investigated.
Results
We found that the EGFR L858R mutation was the most frequently identified mutation in adenocarcinoma in this population by both methods, followed by KRAS (p=3.7e-09), PIK3CA (p=5e-04), and HER2 mutations (p=6.3e-03). We observed that EGFR mutations were significantly mutually exclusive with KRAS, HER2, and MET. FGFR1 mutations were significantly more abundantly detected in the ctDNA group. We found an interaction effect between EGFR mutation and target therapies. The ability of the targeted therapy to improve OS in patients with a single EGFR mutation (HR=0.069, p=0.07) approached significance, but this was not the case for the patients with more than one EGFR mutation or without an EGFR mutation (HR=0.813, p=0.725). Furthermore, the effect of chemotherapy was more predominant in the EGFR group in comparison to the control group.
Conclusion
These findings provide useful information on the distribution of somatic mutations via different screening methods and how this related to the optimal treatment selection in Chinese patients with NSCLC.
Keywords: non-small-cell lung cancer, somatic mutation, Chinese patients
Introduction
In China, lung cancer, as the most common cancer, is a major public health problem, and non-small-cell lung cancer (NSCLC) accounts for the vast majority of lung cancer patients.1 Most lung cancer patients are diagnosed with advanced NSCLC, and there are few satisfactory and effective treatment options. However, recent developments in targeted therapies for a subgroup with specific molecularly activated oncogenes dramatically changes the survival rate and lifespan for NSCLC patients.2
Nevertheless, targeted therapies are fundamentally different from chemotherapy in the sense that clinicians need to detect the relevant driver genes before selecting the genotype-directed targeted drugs. Therefore, uncovering the mutational profile of NSCLC is an important tool for tailoring therapy to patients, and this has a substantial impact on prognosis.3,4 Recent studies demonstrate that East Asian patients have specific mutation profiles, and a large proportion harbor EGFR mutations.5 In particular, those more likely to harbor these mutations are females who have never smoked and who were diagnosed with adenocarcinoma.6 Hence, it is of significant clinical value to better understand the molecular epidemiology in a Chinese population.
Mutation profiling is often performed using the tumor tissue obtained after surgery. However, for advanced NSCLC patients, obtaining this type of tissue is not always possible, because it is often difficult to access sufficient tumor tissue for molecular testing because such an invasive treatment might be harmful for these patients. In an effort to overcome this limitation, recently, highly sensitive screening methods that utilize the cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA), which are found in the blood circulation, have emerged as valuable cancer indications with great potential for molecular diagnosis and monitoring cancer progression.7 Importantly, clinical evidence indicates that there is a substantial discordance in the genomic alterations and efficacy between the tumor tissue and blood ctDNA samples from the same patient.8,9 Thus, it is important to carefully evaluate these different profiling methods in order to provide more information about whether ctDNA is useful for molecular diagnostics.
In this retrospective study, we analyzed 280 NSCLC patients who were diagnosed with adenocarcinoma in a hospital in eastern China between 2017 and 2018. We profiled their somatic mutations from blood or tumor sampling using targeted next-generation sequencing (NGS) platforms. We investigated the relationship between the patterns of somatic mutations and a wide spectrum of factors, with regard to the screening methods, and the stage at onset, treatment response, and survival rate. We detected differential profiles of somatic mutations tested using blood and tumor tissue. Mutually exclusive events and the co-occurrence of certain oncogenes were observed. Furthermore, we further found that targeted therapy only had a significant effect when the patients harbored a single EGFR mutation, and the effect of chemotherapy was also significantly stronger when the patients harbored the single EGFR mutation. Our findings provide useful information and guidance for selecting more appropriate mutation profiling method and a combination of treatments.
Materials and Methods
Patient and Sample Collection
Between January 2017 and December 2018, a total of 400 NSCLC patients provided informed consent to participate in this retrospective study. Fresh tumor tissue was extracted from 300 patients during a surgery or a needle aspiration, and blood samples (10–20 mL) were collected from the remaining 100 patients who did not undergo surgery. Cell-Free DNA BCT® tubes (STRECK) were used to collect the peripheral blood. The cfDNA and genomic DNA (gDNA) are stable for up to 14 days at 6°C to 37°C in this type of tube. The samples were centrifuged at 2,000 g for 15 min to separate the plasma and blood cells, and this was followed by a secondary centrifugation at 16000 g for 10 min. The resulting plasma samples were frozen and stored at −80°C until DNA isolation. The Hwa Mei Hospital, University of Chinese Academy of Sciences approved this study. All of the samples and clinical data included in this retrospective study were irrevocably anonymized. This study was conducted in accordance with the Declaration of Helsinki.
cfDNA and Tissue gDNA Extraction
The gDNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tissues using the TIANamp FFPE DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions, and the gDNA was eluted into a volume of 50 μL. The purity of the gDNA was assessed by electrophoresis using a 1% agarose gel, and the DNA concentration was determined using a Qubit Fluorometer and the QubitTM dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). The cfDNA was extracted from 500 µL aliquots of serum using the QIAamp circulating nucleic acid kit (Qiagen, Hilden, Germany), equipped with the QIAvac 24 Plus vacuum manifold, according to the manufacturer. The purity of the cfDNA was assessed using an Agilent High Sensitivity DNA Kit and the Bioanalyzer 2100 instrument (Agilent Technologies, Santa Clara, CA). When necessary, an additional purification step was performed using Agencourt AMPure XP (Beckman Coulter, Brea, CA) on order to remove any larger contaminating nucleic acid. The quantification of the cfDNA was done using a Qubit 2.0 Fluorometer with the Agilent High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA, USA).
Targeted Next-Generation Sequencing and Sequencing Data Analysis
The targeted sequencing of the extracted samples was performed using one of the following platforms. Briefly, each sample was sequenced by either a panel (Longshi) targeting all coding exons of 13 genes (Table S1) or another panel (Longtong) targeting 59 genes (Table S2). The panels also contained probes for copy variation detection in the genes. The gDNA was sheared, end repaired, ligated with barcoded Illumina sequencing adapters, amplified, and size selected. The resulting libraries were quantified using qPCR, and then, they were pooled and sequenced with 150 base paired-end reads using NextSeq 500 sequencers (Illumina). The raw data was processed using automated custom clinical bioinformatics pipelines. For each sample, the variant frequency was selected to be > 0.1%. The Multiplex I cfDNA Reference Standard Set (Horizon Discovery, Cambridge, MA) was used to assess the accuracy and the minimum variant frequency threshold.
Accordingly, the NCCN Guidelines outline the best practices for variants screening assessment. The variants were annotated with ANNOVAR (2016–02-01) based on the genomic coordinates GRCh37.75, and the complex variants were further annotated with SnpEff (v4.3). Next, we determined if the variants were present in the dbSNP (v147) common database. The variants that were not found in the dbSNP database were filtered again using the ClinVar (20181028) database, Cosmic (70), 1000g_EAS, ExAC_ALL, ExAC_EAS. The pathogenic variants were annotated as “likely-pathogenic,” “pathogenic,” or “drug response” by the ClinVar database.
The Raindrop digital PCR System (Raindrop, USA) was used to assess the mutant allele frequency, according to the manufacturer’s instructions.
Chemotherapy and Targeted Therapy
In general, most standard chemotherapy drugs are designed to kill hyperproliferative cells in the body. Thus, since cancer cells divide quickly, these drugs inhibit cancer. However, chemotherapy drugs can also affect other non-cancer cells in the body that have the capacity to divide rapidly, and this can sometimes cause serious side effects. On the other hand, targeted therapy drugs are different because they target specific regions of the actual cancer cells that permit their abnormal proliferative capacity. Cancer cells have a variety of drug target regions. Targeted drugs are meant to do the following: (1) inhibit chemical signals that allow cancer cells to grow and divide; (2) alter proteins within the cancer cells so the cells will undergo apoptosis; (3) inhibit angiogenesis; (4) stimulate the immune system to attack the cancer cells; or (5) transport toxins to the cancer cells to kill them but not the normal cells.
Assessment of Disease Stage
The disease progression at diagnosis of each patient was first assessed according to the TNM staging system10 based on biopsy and clinical data. The extent of the spread of cancer was assessed using the TNM Classification of Malignant Tumors, which is recognized worldwide. This classification system was done as follows: T was used to describe the size of the original (primary) tumor and whether there was tissue invasion; N was used to describe whether there were any nearby (regional) lymph nodes involved; and M was used to describe any distant metastasis. The 8th clinical staging system11 was used to group the anatomic stage/prognostic grouping from the TNM staging system.
Statistical Analysis
The mutation data was processed using the “maftools” R package.12 The tests for the differential mutated genes between the tissue and ctDNA groups and the co-occurrence test of the mutated genes were performed using the same R package. The survival analysis was conducted using the Cox proportional hazard models using the “survival” R package.13 The duration of the overall survival (OS) was calculated using the number of months between the diagnosis and death. The age at diagnosis, disease stage, and chemotherapy were adjusted in the model. The Chi-square test of homogeneity was performed using R.
Results
Patient Characteristics
Table 1 presents the basic characteristics from a total of 280 adenocarcinoma patients that were included in this retrospective study. The patients diagnosed with stage I made up more than half of the population, while the patients diagnosed with stages III and IV each made up ~17% of the population. The average age of the patients in each stage was ~63 at diagnosis. The vast majority (~95%) of the patients were non-smokers. Almost all of the stages I or II patients underwent resection to remove the tumor, while only 31.25% of the stage IV patients had surgery. Chemotherapy was performed for most patients following the removal of the tumor in stages II, III and IV. Around half of the stage IV patients received a targeted therapy, while few patients in stages I and II did. The average follow-up time was 10.5 (±8.5) months. At the end of the follow-up period, the OS rates were 99.37%, 91.67%, 87.5%, and 68.09% for the patients in stages I, II, III, and IV, respectively.
Table 1.
Basic Characteristics of the Patients Included in the Study
| Stage | N | Per (%) | Smoking (%) | Age | Surgery (%) | Chemo (%) | Targeted (%) | Survival (%) |
|---|---|---|---|---|---|---|---|---|
| I | 158 | 56.43 | 0.63 | 63.05063 | 96.20 | 27.22 | 1.27 | 99.37 |
| II | 12 | 4.29 | 0 | 62.75 | 91.67 | 91.67 | 0 | 91.67 |
| III | 49 | 17.50 | 0 | 63.5625 | 69.39 | 69.39 | 22.45 | 87.5 |
| IV | 48 | 17.14 | 4.17 | 63.65957 | 31.25 | 52.08 | 43.75 | 68.09 |
| NA | 13 | 4.64 | 0 | NA | 100 | 0 | 0 | NA |
Abbreviations: N, the number of patients; Per, the percentage of the patients diagnosed with this stage; Age, average age at diagnosis; Survival, percentage of patients alive by the end of follow-up.
EGFR L858R is the Most Common Mutation in Adenocarcinoma
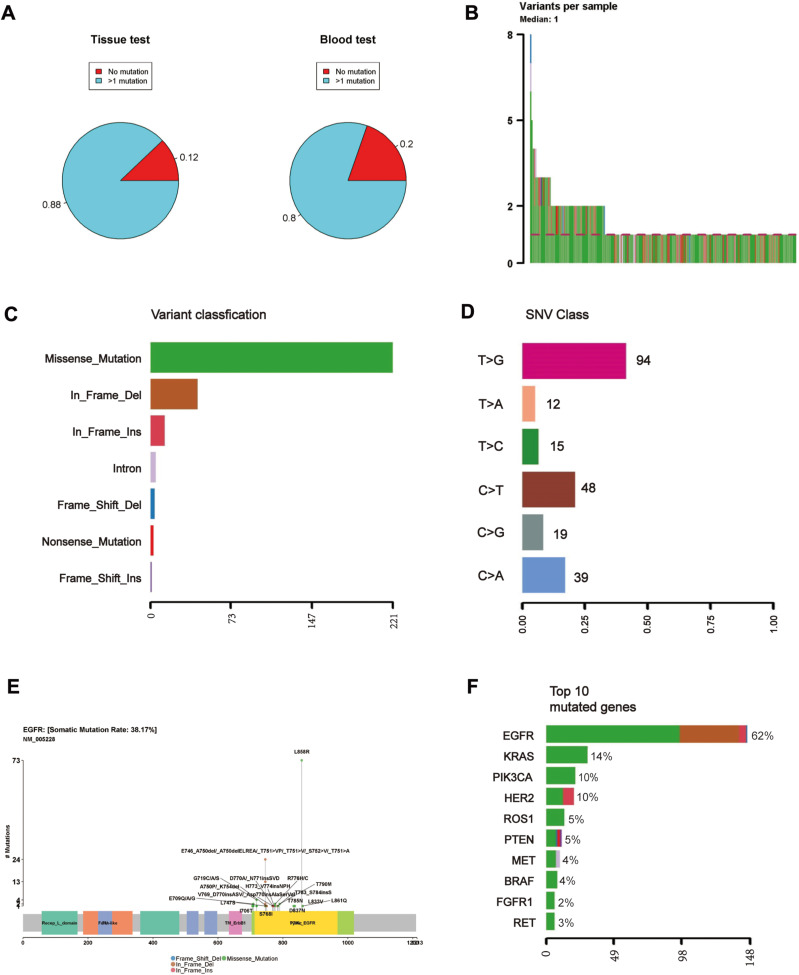
Among the 280 adenocarcinoma patients, a targeted genetic mutation screening test was performed using the tumor tissue or the ctDNA from the blood for 239 and 41 patients, respectively. Most patients who underwent the ctDNA test were diagnosed with stage IV, because surgery was not an optimal option and thus, it was difficult to obtain fresh tissue for the biopsy. A total of 88% of the patients tested using their tumor tissue showed somatic mutations, and this was compared with a detection rate of only 80% for the patients who were tested using their ctDNA (Figure 1A), suggesting that ctDNA-based tests potentially had a lower sensitivity than the test using tumor tissue. The Chi-square test of homogeneity yielded a p-value of 0.16. The non-significant result may be due to the limited sample size of the ctDNA group.
Figure 1.
(A) Percentage of the patients for whom at least one somatic mutation was identified based on a genetic test from tumor tissue or ctDNA from blood. (B) A local plot showing the mutation spots on the protein structure of EGFR and the number of patients harboring those mutations. (C–F) A summary report of the detected mutations, including (C) variant classification, (D) variant type, (E) SNV class, (F) the number of detected variants per sample.
More than half of the patients had only one detected mutation (Figure 1B). The largest proportion of detected mutations (>2/3) were missense mutations, followed by in-frame deletions, and insertions (Figure 1C). Most of the mutations were single nucleotide polymorphisms (SNPs), and the most abundant single nucleotide variation was the transition from thymine (T) to guanine (G) (Figure 1D), which was attributed to the well-known mutation L858R in exon 21 of EGFR (Figure 1E). We found that EGFR was the most frequently mutated gene, accounting for almost 50% of all the detected mutations regardless of the source of biopsy (Figures 1F, 2A and B). In addition, KRAS, PIK3CA and HER2 were among the most frequently mutated genes (Figure 1F). We observed that EGFR had the highest variant allelic frequency (VAF), while ROS1 and FGFR1 had substantially lower (VAF) compared to the other genes despite the small sample size (Figure 2C).
Figure 2.
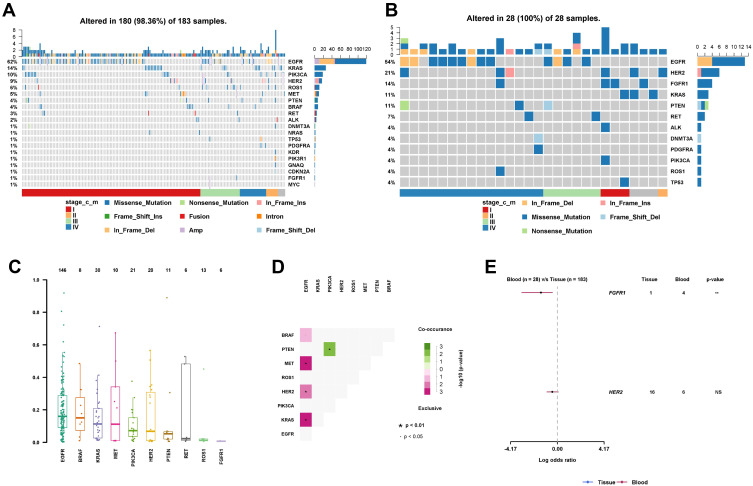
(A) VAF of the top 10 most frequent mutated genes. (B) A co-occurrence matrix showing which pair of gene mutations were mutually exclusive or coexistent. (C) The differential mutation rate between the tissue and ctDNA. (D) An oncoplot showing the scattering of the somatic mutations with regard to cancer stage and variant type in the tissue samples. (E) An oncoplot showing the scattering of somatic mutations with regard to cancer stage and variant type in the ctDNA samples.
EGFR is Mutually Exclusive When Compared to KRAS, HER2, and MET
Next, we explored whether these mutations were mutually exclusive or likely to co-occur. We found that the EGFR mutations were most significantly (p=3.7e-09) mutually exclusive when compared to the KRAS mutations (Figure 2D), supporting previous findings.14 We further discovered that the EGFR mutations were significantly mutually exclusive with HER2 and MET (p=5e-04 and 6.3e-03, respectively), but not with PIK3CA or PTEN. We also observed the co-existence of PTEN and PIK3CA mutations, consistent with previous reports.15
ctDNA Test Detected More FGFR1 Mutations
When comparing the mutations detected from tumor tissue with those detected from the ctDNA in blood, we found that the mutations in FGFR1 were significantly more abundantly detected in the ctDNA group (p<0.01) (Figure 2E). The enrichment of the mutations in FGFR1 for the ctDNA test was not attributed to the larger number of patients with Stage IV in that group, as the mutations were scattered over all the cancer stages (Figure 2A and B). In addition, EGFR amplification was detected in ~10% of the samples with EGFR mutations in the tissue group, while no amplification was detected in the ctDNA group (Figure 2A and B). Across all the detected mutations, we tested and found no mutations that were more enriched for a specific cancer stage. We also found no enrichment of certain mutations for a specific age group.
Targeted Therapy is Effective for Patients with EGFR Mutations
We investigated whether the mutations impacted the OS of the adenocarcinoma patients over the course of the follow-up period. In particular, the focus was on the effect of the targeted therapy on the patients carrying EGFR mutations in the advanced stages. This was because, for stage I and II patients, targeted therapy is rarely applied, and consequently our retrospective cohort was not able to achieve enough statistical power to detect the effect. Thus, we performed a survival analysis using the 97 patients diagnosed with Stage IIIA or higher. We found that the hazard rate was significantly associated with the stage of cancer and the use of chemotherapy in the total samples (Table 2). Stage IIIB and IVA substantially increased the hazard (p=0.03 and 0.05) while receiving chemotherapy dramatically improved the prognosis (p=1.6e-03). However, age was not associated with the hazard rate, probably because the range of age in this study was narrow. The targeted therapy increased the survival rate in the total samples albeit not significant (p=0.4).
Table 2.
Effects of Stage, Chemotherapy and Targeted Therapy on Overall Survival in the Total Samples, the EGFR Group and the Control Group
| Predictor | Hazard Ratio | 95% CI | p |
|---|---|---|---|
| All Samples | |||
| Targeted | 0.67 | 0.26–1.71 | 0.399 |
| Chemotherapy | 0.192 | 0.069–0.534 | 0.0016** |
| IIIB | 6.60 | 1.18–36.84 | 0.0316** |
| IVA | 4.22 | 0.95–18.79 | 0.0587* |
| EGFR group | |||
| Targeted | 0.069 | 0.0034–1.37 | 0.0796* |
| Chemotherapy | 0.014 | 5.57e-04-0.34 | 0.0087** |
| IIIB | 37.5 | 1.28–1095.69 | 0.0353** |
| IVA | 3.96 | 0.17–90.35 | 0.388 |
| Control group | |||
| Targeted | 0.813 | 0.26–2.57 | 0.725 |
| Chemotherapy | 0.424 | 0.13–1.36 | 0.149 |
| IIIB | 2.862 | 0.18–46.30 | 0.459 |
| IVA | 8.001 | 1.02–62.94 | 0.048** |
Notes: *0.05<P<0.1; **P<0.05.
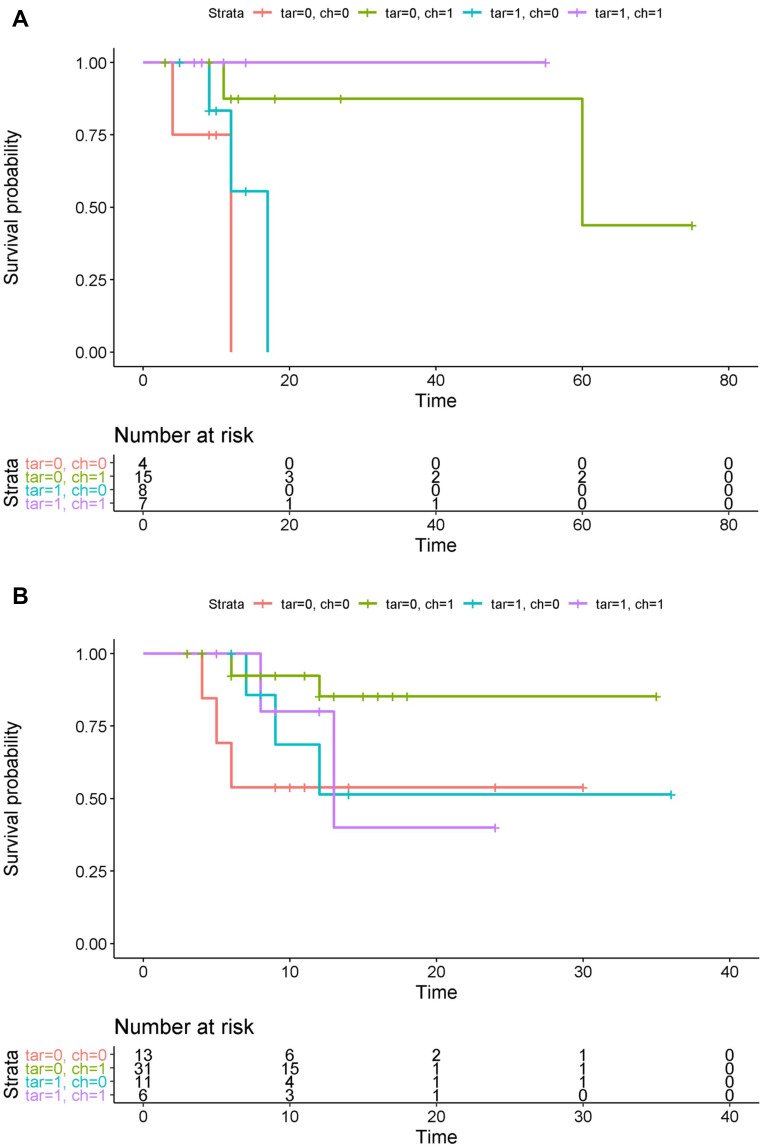
To check whether the targeted therapy had an effect on the patients with EGFR mutations, we split the samples into two groups as follows: (1) an EGFR group that was comprised of 34 samples with EGFR mutations and no other mutations and (2) a control group that consisted of the remaining 63 samples. The patients who did not receive any treatment died most quickly for both groups (Figure 3A and B). We observed that the effect of the targeted therapy in the EGFR group neared significance (p=0.07) (Table 2, Figure 3A), and the effect size was much stronger than that in the control group (Figure 3B). In addition, the effect of chemotherapy was also predominant for the EGFR group relative to the control group.
Figure 3.
Survival curves of the patients according to whether chemotherapy or targeted therapy was applied in (A) the EGFR group and (B) the control group. Tar/ch=0 represents no targeted or chemotherapy, respectively. The unit of time is a month.
Discussion
Recently, treatment paradigms for NSCLC have shifted. Traditionally, the treatments were based on the stratification of patients using histological findings alone. Now, they are aimed at molecularly classifying patients using the presence of genetic alterations within “driver” oncogenes. For NSCLC patients who harbor these alterations, the mechanism typically hinges upon one oncogenic pathway that drives cell survival, which is known as oncogene addiction.16 In fact, it was recently demonstrated that about 50% of pulmonary adenocarcinomas have, at minimum, one genetic mutation within an oncogene driver, and the frequencies are higher among those who have no history of smoking.17,18 Traditionally, standard somatic mutation analyses are done using postoperative material or biopsy tissue. However, more recently, new tumor DNA sources, including blood, have provided a noninvasive approach to identify ctDNA and monitor tumor changes and genetic mutations when the cancer is at advanced stages.19,20
A meta-analysis revealed that circulating cell-free DNA (cfDNA) served as a prognostic and predictive biomarker for NSCLC patients, especially with respect to the EGFR mutation detection.21 Thus, liquid biopsies are proposed as a potential approach to monitor somatic mutations in real time in contrast to tissue biopsies, which represent a single snap-shot in time. Recently, a large study was conducted using specimens from surgery or biopsies from NCSLC patients in a Chinese population, revealing that there were unique EGFR mutations within this population.22 However, determining whether these types of mutation can also be identified using ctDNA in a Chinese NSCLC population has not been done. Thus, in this study, we aimed to assess both tumor tissue samples and ctDNA from blood using NGS to identify somatic mutations in a Chinese NSCLC population.
In our study, 88% of the patients that we tested using their tumor tissue were identified to have somatic mutations, and the detection rate was only 80% for the patients tested using their ctDNA. This discrepancy is to be expected due to the limitation of the size of the ctDNA sample population compared to that of the tumor tissue samples. Furthermore, the accuracy of many of the current technologies to detect specific mutations among the total cfDNA can be a limitation, because the ctDNA copy numbers are low compared to the sizable amount of wild-type cfDNA that is present.23 In addition, it is also worth considering that the ctDNA released by the tumor cells may be genetically distinctive when it is compared to the most of the cells within the primary tumor because of tumor heterogeneity.24
However, even though there was a disparity in the detection of the somatic mutation percentages between the sample types, when we examined the identity of the mutations, we found that there were little differences in the findings when the mutations were identified, regardless of the DNA source. Indeed, EGFR was the most frequently identified mutated gene, and it accounted for almost 50% of all of the detected mutations irrespective of the source of biopsy. These finding are supported by another study that examined newly diagnosed advanced lung adenocarcinoma patients and found that 65% of the patients with harbored an EGFR mutation in their tissue samples and had concordant EGFR mutation results using liquid biopsy. The concordant results were associated with significantly poorer PFS in stage IV patients treated with EGFR tyrosine kinase inhibitors (TKIs).26 The importance of the detecting EGFR mutations for NSCLC is clear, and now there is intriguing evidence that these mutations in particular can be monitored non-invasively, which will be beneficial for treatments, predictions, and survival.
We also identified KRAS as one of the most commonly mutated genes. With regard to human cancer, the RAS genes are considered some of the most frequently mutated genes and are comprised of three members, namely HRAS, KRAS and NRAS. The enzymes encoded by the RAS family genes enzymes trigger numerous signaling pathways, including RAF-MEK-ERK and PI3K-AKT-mTOR. Consequently, these genes are crucial for regulating cell proliferation, differentiation, and survival.27 With regard to NSCLC, KRAS mutations are among the most rampant oncogenic driver mutations. The KRAS mutation status in NSCLC is associated with the activation of different signaling pathways, which might lead to an altered prognostic significance and response to therapy.28,29 Thus, it is important to identify this mutation with accuracy. With respect the clinical utility of determining KRAS mutations in a liquid biopsy as a marker of sensitivity to EGFR-TKIs in NSCLC, the current studies have not shown significant differences in terms of the overall survival.30 However, one study did reveal that mutant KRAS patients had a worse PFS than wild type subjects.31 Thus, the prognostic and predictive value of KRAS mutations in cfDNA as a biomarker still requires further investigation.
The PI3K pathway is essential for cell metabolism and proliferation.32 The PIK3CA gene encodes the class I PI3K p110α, and mutations within this gene are frequently identified in an assortment of cancers.33 We found a high level of PIK3CA mutations in our samples. For NSCLC, PIK3CA mutations are considered oncogenic and targetable.34 However, it is questionable whether the PIK3CA mutation alone is enough to function as an oncogenic driver in tumor formation for NSCLC patients.35 Studies are currently examining this notion. For example, PIK3CA-mutated NSCLC is identified clinically and genetically as a heterogeneous subgroup among adenocarcinomas, showing no deleterious effect on survival after surgery or systemic therapy. However, for lung cancer, PIK3CA mutations occur in patients with prior malignancies.36 Thus, further examination into the PIK3CA mutational status in NSCLC patients is worth exploring. To the best of our knowledge, this has not been done using cfDNA for NSCLC. However, PIK3CA, as a driver mutation, was detected using ctDNA in breast cancer and head and neck squamous cell carcinoma patients.37,38 Thus, this finding in NSCLC should be further explored in a larger cohort.
Due to the significance of the EGFR mutations in NSCLC, we further explored whether it existed mutually exclusive or was likely to co-occur with the other mutations. Our data revealed that the EGFR mutations were most significantly mutually exclusive with the KRAS mutations. Traditionally, EGFR and KRAS mutations have been deemed mutually exclusive. Thus, the occurrence of KRAS mutations is considered a characteristic of anti-EGFR TKI therapy resistance. Nevertheless, data attained using highly sensitive technologies indicate that in a heterogeneous tumor cell population, certain tumors do share KRAS and EGFR mutations.39 Thus, given the importance of these 2 onco-drivers in NSCLC, it will be important to examine these relationships further in the future. With respect to the detection of EGFR and KRAS mutations in cfDNA as predictive and prognostic biomarkers of NSCLC, the EGFR mutation clearly shows a relationship to the acquired resistance to EGFR-TKIs.30 However, as mentioned above, for KRAS mutations in cfDNA as a biomarker, their prognostic and predictive values are still under investigation.
Interestingly, we found that FGFR1 mutations were significantly more abundantly detected in the ctDNA group compared to the tumor tissue. FGFR1 shows oncogenic characteristics in squamous cell carcinomas using preclinical models, as such FGFR1 amplification usually occurs via the activation the signal transducer and activator of transcription (STAT), AKT, and mitogen-activated protein kinase pathways, paving the way for the prospective therapeutic targeting in squamous cell carcinomas.40 However, the use of inhibitors targeting FGFR1 for this subtype is less than effective.41 Thus, in lung cancer, focus has turned to adenocarcinoma. The amplification of FGFR1 is observed in 1% to 3% of lung adenocarcinomas, and evidence suggests that the mRNA and protein expressions of FGFR1 may be superior prognosticators of FGFR inhibition ability.42 Interestingly, the dual inhibition of EGFR and FGFR in FGFR1-overexpressing, EGFR-activated models demonstrates combinatorial results on tumor growth, indicating that patients whose tumors bear these features may have improved outcomes with combined EGFR/FGFR inhibitory treatments.43 A better exploration of these two mutations would be beneficial in future studies. FGFR mutations are considered less common mutations in NSCLC, because they are found in only 1–2% of cases. However, it is found in ctDNA from these patients.44 Thus, using cfDNA to explore FGFR mutations may provide more information compared to what is known from tumor biopsies.
Finally, in our retrospective analysis, we examined the treatment and survival rates of the patients compared to their EGFR mutation status, which was determined by 2 methods. These data revealed that the targeted therapy was effective for the EFGR mutation patients. It is estimated that more than 60% of NSCLCs show EGFR expression, making EGFR an essential therapeutic target. Clinically active TKIs for EGFR have been developed, and these are successful in patients whose tumors contain activating mutations in the tyrosine kinase domain of the EGFR gene. Thus, mutation analyses are required to pinpoint these patients, because when the selection only utilizes clinicopathologic characteristics, it is insufficient.45 Given that this retrospective study found similar EGFR mutation rates in by the biopsies and the ctDNA, we propose that a future prospective study be conducted to assess the ability of ctDNA to predict outcome and provide treatment guidance.
Conclusions
For NSCLC, the isolation and analysis ctDNA is an efficient and favorable genomic profiling tool. Our study demonstrated a reasonable comparison between the mutational analyses of ctDNA and tumor tissue. Thus, using ctDNA might be specifically valuable when it is not safe to assess the tissue biopsy because of its physical condition. Moreover, the use of ctDNA has the potential to identify oncogenic driver mutations for which targetable therapies might be useful. Our findings support recent research, which establishes that ctDNA testing is a valuable addition to tissue genotyping for NSCLC. Ultimately, the findings described here provide support the application of ctDNA testing as a routine clinical practice for NSCLC.
Ethics Approval and Informed Consent
The Hwa Mei Hospital, University of Chinese Academy of Sciences approved this study.
Disclosure
Wei Chen is an employee of Zhongyuan Union Clinical Laboratory Co., Ltd., Tianjin, China. The authors report no funding and no other potential conflicts of interest for this work.
References
- 1.Zhou C. Lung cancer molecular epidemiology in China: recent trends. Transl Lung Cancer Res. 2014;3(5):270–279. doi: 10.3978/j.issn.2218-6751.2014.09.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–742. doi: 10.1016/S1470-2045(11)70184-X [DOI] [PubMed] [Google Scholar]
- 3.Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet. 2016;387(10026):1415–1426. doi: 10.1016/S0140-6736(16)00004-0 [DOI] [PubMed] [Google Scholar]
- 4.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F, Zhou C. Lung cancer in never smokers-the East Asian experience. Transl Lung Cancer Res. 2018;7(4):450–463. doi: 10.21037/tlcr.2018.05.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 7.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–554. doi: 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Han T, Zhou Y, Mao B, Zhuang W. Comparing the efficacy of targeted next-generation sequencing in the identification of somatic mutations in circulating tumor DNA from different stages of lung cancer. Neoplasma. 2019;66(4):652–660. doi: 10.4149/neo_2018_181130N910 [DOI] [PubMed] [Google Scholar]
- 9.Guo Q, Wang J, Xiao J, et al. Heterogeneous mutation pattern in tumor tissue and circulating tumor DNA warrants parallel NGS panel testing. Mol Cancer. 2018;17(1):131. doi: 10.1186/s12943-018-0875-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobin LH. TNM Classification of Malignant Tumours 7e. 2009. [Google Scholar]
- 11.Lim W, Ridge CA, Nicholson AG, Mirsadraee S. The 8th lung cancer TNM classification and clinical staging system: review of the changes and clinical implications. Quant Imaging Med Surg. 2018;8(7):709–718. doi: 10.21037/qims.2018.08.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasrebi H. SurvJamda: an R package to predict patients’ survival and risk assessment using joint analysis of microarray gene expression data. Bioinformatics. 2011;27(8):1168–1169. [DOI] [PubMed] [Google Scholar]
- 14.Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res. 2013;19(15):4273–4281. doi: 10.1158/1078-0432.CCR-13-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oda K, Stokoe D, Taketani Y, McCormick F. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005;65(23):10669–10673. doi: 10.1158/0008-5472.CAN-05-2620 [DOI] [PubMed] [Google Scholar]
- 16.Weinstein IB, Joe AK. Mechanisms of disease: oncogene addiction–a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3(8):448–457. doi: 10.1038/ncponc0558 [DOI] [PubMed] [Google Scholar]
- 17.Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol. 2011;22(12):2616–2624. doi: 10.1093/annonc/mdr489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, Fang R, Sun Y, et al. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One. 2011;6(11):e28204. doi: 10.1371/journal.pone.0028204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102(45):16368–16373. doi: 10.1073/pnas.0507904102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng M, Chen C, Hulbert A, Brock MV, Yu F. Non-blood circulating tumor DNA detection in cancer. Oncotarget. 2017;8(40):69162–69173. doi: 10.18632/oncotarget.19942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ai B, Liu H, Huang Y, Peng P. Circulating cell-free DNA as a prognostic and predictive biomarker in non-small cell lung cancer. Oncotarget. 2016;7(28):44583–44595. doi: 10.18632/oncotarget.10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng H, Guo X, Sun D, et al. Genomic profiling of driver gene mutations in Chinese patients with non-small cell lung cancer. Front Genet. 2019;10:1008. doi: 10.3389/fgene.2019.01008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arriola E, Paredes-Lario A, Garcia-Gomez R, et al. Comparison of plasma ctDNA and tissue/cytology-based techniques for the detection of EGFR mutation status in advanced NSCLC: Spanish data subset from ASSESS. Clin Transl Oncol. 2018;20(10):1261–1267. doi: 10.1007/s12094-018-1855-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toor OM, Ahmed Z, Bahaj W, et al. Correlation of somatic genomic alterations between tissue genomics and ctDNA employing next-generation sequencing: analysis of lung and gastrointestinal cancers. Mol Cancer Ther. 2018;17(5):1123–1132. doi: 10.1158/1535-7163.MCT-17-1015 [DOI] [PubMed] [Google Scholar]
- 25.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo CY, Lee MH, Tsai MJ, Yang CJ, Hung JY, Chong IW. The factors predicting concordant epidermal growth factor receptor (EGFR) mutation detected in liquid/tissue biopsy and the related clinical outcomes in patients of advanced lung adenocarcinoma with EGFR mutations. J Clin Med. 2019;8(11):1758. doi: 10.3390/jcm8111758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrido P, Olmedo ME, Gomez A, et al. Treating KRAS-mutant NSCLC: latest evidence and clinical consequences. Ther Adv Med Oncol. 2017;9(9):589–597. doi: 10.1177/1758834017719829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricciuti B, Leonardi GC, Metro G, et al. Targeting the KRAS variant for treatment of non-small cell lung cancer: potential therapeutic applications. Expert Rev Respir Med. 2016;10(1):53–68. doi: 10.1586/17476348.2016.1115349 [DOI] [PubMed] [Google Scholar]
- 29.Brady AK, McNeill JD, Judy B, et al. Survival outcome according to KRAS mutation status in newly diagnosed patients with stage IV non-small cell lung cancer treated with platinum doublet chemotherapy. Oncotarget. 2015;6(30):30287–30294. doi: 10.18632/oncotarget.4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garzon M, Villatoro S, Teixido C, et al. KRAS mutations in the circulating free DNA (cfDNA) of non-small cell lung cancer (NSCLC) patients. Transl Lung Cancer Res. 2016;5(5):511–516. doi: 10.21037/tlcr.2016.10.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, An T, Wang J, et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2010;16(4):1324–1330. doi: 10.1158/1078-0432.CCR-09-2672 [DOI] [PubMed] [Google Scholar]
- 32.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7(8):606–619. doi: 10.1038/nrg1879 [DOI] [PubMed] [Google Scholar]
- 33.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502 [DOI] [PubMed] [Google Scholar]
- 34.Oxnard GR, Binder A, Janne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol. 2013;31(8):1097–1104. doi: 10.1200/JCO.2012.42.9829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer. 2006;54(2):209–215. doi: 10.1016/j.lungcan.2006.07.006 [DOI] [PubMed] [Google Scholar]
- 36.Scheffler M, Bos M, Gardizi M, et al. PIK3CA mutations in non-small cell lung cancer (NSCLC): genetic heterogeneity, prognostic impact and incidence of prior malignancies. Oncotarget. 2015;6(2):1315–1326. doi: 10.18632/oncotarget.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chada A, Hoque R. Images: periodic limb movements during sleep noted on ventral thigh surface electromyography in an above-the-knee amputated stump. J Clin Sleep Med. 2019;15(8):1183–1184. doi: 10.5664/jcsm.7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt H, Kulasinghe A, Allcock RJN, et al. A pilot study to non-invasively track PIK3CA mutation in head and neck cancer. Diagnostics. 2018;8(4):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5(8):860–877. doi: 10.1158/2159-8290.CD-14-1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang T, Gao G, Fan G, Li M, Zhou C. FGFR1 amplification in lung squamous cell carcinoma: a systematic review with meta-analysis. Lung Cancer. 2015;87(1):1–7. doi: 10.1016/j.lungcan.2014.11.009 [DOI] [PubMed] [Google Scholar]
- 41.Paik PK, Shen R, Berger MF, et al. A phase Ib open-label multicenter study of AZD4547 in patients with advanced squamous cell lung cancers. Clin Cancer Res. 2017;23(18):5366–5373. doi: 10.1158/1078-0432.CCR-17-0645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wynes MW, Hinz TK, Gao D, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014;20(12):3299–3309. doi: 10.1158/1078-0432.CCR-13-3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quintanal-Villalonga A, Molina-Pinelo S, Cirauqui C, et al. FGFR1 Cooperates with EGFR in lung cancer oncogenesis, and their combined inhibition shows improved efficacy. J Thorac Oncol. 2019;14(4):641–655. doi: 10.1016/j.jtho.2018.12.021 [DOI] [PubMed] [Google Scholar]
- 44.Saarenheimo J, Eigeliene N, Andersen H, Tiirola M, Jekunen A. The value of liquid biopsies for guiding therapy decisions in non-small cell lung cancer. Front Oncol. 2019;9:129. doi: 10.3389/fonc.2019.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206 [DOI] [PubMed] [Google Scholar]