Abstract
Background and Purpose
Irinotecan, used in colorectal cancer therapy, is metabolized by glucuronidation involving different UDP‐glucuronosyltransferase (UGT)1A isoforms leading to facilitated elimination from the body. Individuals homozygous for the genetic variants UGT1A1*28 (Gilbert syndrome) and UGT1A7*3 are more susceptible to irinotecan side effects, severe diarrhoea and leukopenia. The aim of this study was to investigate the protective effects and active constituents of coffee during irinotecan therapy using humanized transgenic (htg)UGT1A‐WT and htgUGT1A‐SNP (carry UGT1A1*28 and UGT1A7*3 polymorphisms) mice.
Experimental Approach
HtgUGT1A mice were pretreated with coffee or caffeic acid (CA) + caffeic acid phenylethyl ester (CAPE) and injected with irinotecan. The effects of coffee and CA + CAPE were investigated using reporter gene assays, immunoblot, TaqMan‐PCR, siRNA analyses and blood counts.
Key Results
Only the combination of the two coffee ingredients, CA and CAPE, mediates protective effects of coffee in a model of irinotecan toxicity by activation of UGT1A genes. Coffee and CA + CAPE significantly increased UGT1A expression and activity along with SN‐38 glucuronide excretion in irinotecan‐injected htgUGT1A mice, resulting in significant improvement of leukopenia, intestinal oxidative stress and inflammation.
Conclusion and Implications
In this study, we identify the compounds responsible for mediating the previously reported coffee‐induced activation of UGT1A gene expression. CA and CAPE represent key factors for the protective properties of coffee which are capable of reducing irinotecan toxicity, exerting antioxidant and protective effects. Provided that CA + CAPE do not affect irinotecan efficacy, they might represent a novel strategy for the treatment of irinotecan toxicity.
Abbreviations
- AhR
aryl hydrocarbon receptor
- Bcl‐xL
B‐cell lymphoma‐extra large
- CA
caffeic acid
- CAPE
caffeic acid phenylethyl ester
- c‐IAP1
cellular inhibitor of apoptosis protein‐1
- c‐IAP2
cellular inhibitor of apoptosis protein‐2
- CRC
colorectal cancer
- HCC
hepatocellular cancer
- NAFLD
non‐alcoholic fatty liver disease
- NRF2/Nrf2
nuclear factor, erythroid 2 like 2
- SNP
single nucleotide polymorphism
- UGT
UDP‐glucuronosyltransferase
- WT
wild type
What is already known
UGT1A polymorphisms are associated with increased risk for irinotecan toxicity including severe leukopenia and diarrhoea.
Coffee has protective effects and induces UGT1A gene expression.
What this study adds
Caffeic acid (CA) and its phenylethyl ester (CAPE) mediate coffee's protective effects on irinotecan toxicity
Protection was also observed in the presence of low function genetic UGT1A variants.
What is the clinical significance
As irinotecan efficacy was unaffected by CA /CAPE they could use to treat irinotecan toxicity.
CA/CAPE may have a high potential in hepatoprotective strategy in other conditions.
1. INTRODUCTION
Irinotecan is a prodrug that is used as a key component in first‐ and second‐line treatment regimens for metastatic colorectal cancer (CRC) and other cancer entities. Ubiquitously expressed carboxylesterases convert irinotecan to its active metabolite SN‐38, which exerts topoisomerase I inhibitor activity (Sanghani et al., 2003). The inactivation of SN‐38 to its inactive glucuronide (SN‐38G) is catalysed by different UDP‐glucuronosyltransferase 1A (UGT1A) enzymes and leads to facilitated excretion of the conjugate in bile (Tukey, Strassburg, & Mackenzie, 2002). A number of studies have provided evidence that patients who are homozygous for the genetic variants UGT1A1*28, UGT1A7*3 and UGT1A9*1b, lack full glucuronidation activity and are susceptible to irinotecan drug side effects, such as severe diarrhoea and leukopenia (Cecchin et al., 2009; Lankisch et al., 2008). In a previous study, we were able to show that 76% of individuals homozygous for the UGT1A1*28 polymorphism are also homozygous for three other genetic variants (UGT1A3‐66T < C, UGT1A6*2 and UGT1A7*3). This complete UGT1A haplotype is present in 10% of the White population in the homozygous form. Consequently, we developed a humanized transgenic (htg) mouse line expressing the human UGT1A gene locus containing all aforementioned single nucleotide polymorphisms (SNPs) and another mouse line expressing human wild‐type (WT) UGT1A genes. Compared to htgUGT1A‐WT mice, htgUGT1A‐SNP mice exhibited a reduced basal expression and a decreased inducibility by common UGT1A inducers (Ehmer et al., 2012).
Coffee has been reported to provide diverse health benefits to its consumers. Among these are a lower risk for the development of hepatocellular cancer (HCC), cirrhosis, non‐alcoholic fatty liver disease (NAFLD) and colorectal cancer (Heath, Brahmbhatt, Tahan, Ibdah, & Tahan, 2017; Ross et al., 2000; Salazar‐Martinez et al., 2004). There is a broad array of compounds in coffee that may be responsible for these protective effects and their identification has a high potential for pharmaceutical intervention or prevention.
Caffeic acid (CA) and its derivate, caffeic acid phenylethyl ester (CAPE) are phenolic degradation products of chlorogenic acid and have been linked to free radical scavenging activity, as well as to anti‐carcinogenic and anti‐inflammatory properties (Chao, Hsu, & Yin, 2009; Rajendra Prasad, Karthikeyan, Karthikeyan, & Reddy, 2011; Sato et al., 2011; Zhang, Tang, Li, Zhu, & Duan, 2014). Different coffee preparations were shown to up‐regulate UGT1A expression in vitro and in a htgUGT1A mouse model (Kalthoff, Ehmer, Freiberg, Manns, & Strassburg, 2010). To this date, the responsible compounds have not been identified.
The aim of this study was therefore to identify the coffee ingredient responsible for the previous reported coffee‐mediated up‐regulation of human UGT1A expression. Moreover, we analysed the protective effects of coffee and coffee ingredients caffeic acid and caffeic acid phenylethyl ester using a model of drug‐induced toxicity. Our hypothesis was that a coffee (caffeic acid + caffeic acid phenylethyl ester)‐mediated induction of UGT1A expression in individuals carrying the UGT1A‐SNP haplotype may prevent or ameliorate irinotecan side effects such as leukopenia and intestinal inflammation.
2. METHODS
2.1. Chemicals
Caffeic acid, caffeic acid phenylethyl ester, Tween 80, RIPA‐buffer and chemicals listed in Table 1 were purchased from Sigma‐Aldrich (Taufkirchen, Germany). Irinotecan was purchased from Actavis (Wien, Austria).
TABLE 1.
Analysed coffee ingredients
| Substance | Concentration in coffee | Reference |
|---|---|---|
| Guaiacol | 4.2 mg·L−1 | (Griffin, 2006) |
| 4‐Ethylguaiacol | 0.16 mg·L−1 | (Griffin, 2006) |
| 2,3‐Butanedione | 50.8 mg·L−1 | (Griffin, 2006) |
| 2,3‐Pentanedione | 39.6 mg·L−1 | (Griffin, 2006) |
| 3‐Hydroxy‐4,5‐dimethyl‐2‐fuarone | 1.47 mg·L−1 | (Griffin, 2006) |
| 4‐Hydroxy‐2.5‐dimethyl‐3(2H)‐fuarone | 109 mg·L−1 | (Griffin, 2006) |
| Vanillin | 4.8 mg·L−1 | (Griffin, 2006) |
| Maltol | 45 mg·L−1 | (Belitz, Grosch, & Schieberle, 2009) |
| Furfurylmercaptane | 1 ppm | (Flament & Bessière‐Thomas, 2002) |
| Trigonelline | 300 mg·L−1 | (Debry, 1994) |
| Methylpyridinium iodide | 72 mg·L−1 | (Preedy, 2014) |
| Ferulic acid | 90 mg·L−1 | (Mattila & Kumpulainen, 2002) |
| p‐coumaric acid | 13.7 mg·L−1 | (Mattila & Kumpulainen, 2002) |
| Hippuric acid | 2.2 mg·L−1 | (Preedy, 2014) |
| 4‐O‐caffeoylquinic acid | 750 mg·L−1 | (Preedy, 2014) |
| Chlorogenic acid | 800 mg·L−1 | (Preedy, 2014) |
| Acetic acid | 225 mg·L−1 | (Griffin, 2006) |
| Quinic acid | 495 mg·L−1 | (Griffin, 2006) |
| Formic acid | 73 mg·L−1 | (Griffin, 2006) |
| Lactic acid | 195 mg·L−1 | (Griffin, 2006) |
| Citric acid | 461 mg·L−1 | (Griffin, 2006) |
| Malic acid | 137 mg·L−1 | (Griffin, 2006) |
| Nicotinic acid | 50 mg·L−1 | (Clarke & Macrae, 1988) |
| Caffeic acid (CA) | 800 mg·L−1 | (Higdon & Frei, 2006; Iwahashi, 2014) |
| Caffeic acid phenylethyl ester (CAPE) | 160 mg·L−1 | ‐ |
Note: Analysed coffee ingredients and their concentration in coffee.
2.2. Standardized preparation of coffee
Detailed coffee preparation method for cell culture treatment was described before (Kalthoff et al., 2010). In short, 6 g of coffee powder (Jacobs Krönung, Kraft Foods, Bremen, Germany) per 150 ml water was used for brewing a stock solution. The filtrate was diluted (12% coffee + 88% medium) for cell culture treatment (Kalthoff et al., 2010). Green coffee brew was prepared using 9.7 g of green coffee powder because the green coffee beans were 1.66‐fold heavier than roasted beans.
For mouse treatment, 3 g (instead of 6 g) of coffee powder was used for coffee preparation (because of a better drinking acceptance).
2.3. Cell culture
The human hepatocarcinoma cell line HepG2 (RRID:CVCL_0027) and human oesophageal squamous cell carcinoma cell line KYSE70 (RRID:CVCL_1356) were grown in RPMI 1640 (Fisher Scientific, Schwerte, Germany) supplemented with 10% FBS at 37°C in a humidified atmosphere containing 5% CO2.
2.4. Plasmids
A 500‐bp (UGT1A1), a 258‐bp (UGT1A3 and a 530‐bp (UGT1A7 and UGT1A9) DNA fragments of each UGT1A 5′ upstream sequence were amplified by PCR from healthy blood donors (known for WT/SNP sequence of the respective 5′ upstream region). The PCR fragments were cut by Xho I and Nhe I and ligated into pGL3 vector (Promega, Mannheim, Germany). All inserts were sequenced in full using the Dye Terminator Cycle Sequencing Kit 1.1 (Applied Biosystems, Darmstadt, Germany) and the ABI 310 automated sequencer (Applied Biosystems, Darmstadt, Germany) (SNPs: UGT1A1*28; UGT1A3‐66T > C, UGT1A7‐57T > G and UGT1A9*1b) (Erichsen et al., 2010; Kalthoff et al., 2010).
2.5. Luciferase assays
Luciferase assays were performed as described before (Kalthoff et al., 2010). Cells were treated with coffee (12% + 88% medium) for 48 h. For the analysis of coffee ingredients listed in Table 1, concentrations in regular coffee were ascertained by literature inquiry. Chemicals were dissolved in vehicle in the determined concentration and—analogous to coffee—diluted with RPMI medium (12% dissolved chemical + 88% medium). All experiments were performed in five independent experiments analysing one treatment group consisting of three individual cell culture wells (in total 15 wells). Results were analysed using Microsoft Excel software. Columns represent mean ± SD. Results are shown as fold induction (in comparison to vehicle control) in order to assure a better comparability between the inducibilities of different reporter gene constructs. The absolute expression levels of the different constructs are highly variable so that a combined presentation within one graph appears inappropriate.
2.6. Transfection of siRNA
For siRNA experiments, siRNA (MWG Biotech, Ebersberg, Germany) against Nrf2 (AAGAGUAUGAGCUGGAAAAACTT), aryl hydrocarbon receptor (AAGCGGCAUAGAGACCGACUUTT), or non‐silencing control (UAAUGUAUUGGAACGCAUATT) was transfected together with respective reporter gene constructs in OPTI‐MEM (Invitrogen, Karlsruhe, Germany) into cells using Lipofectamine 2000 (Invitrogen, Karlsruhe, Germany). The final concentration of siRNA was 100 nM. Cells were treated with caffeic acid + caffeic acid phenylethyl ester on the next day for further 48 h. Knockdown efficiency of siRNAs was determined by Western blot analysis.
2.7. Compliance with requirements for studies using animals
The humanized transgenic UGT1A mouse model has been used in several other studies investigating the regulation of human UGT1A genes (Ehmer et al., 2012; Kalthoff et al., 2010; Kalthoff, Landerer, Reich, & Strassburg, 2017; Kalthoff, Winkler, Freiberg, Manns, & Strassburg, 2013). Furthermore, the coffee‐induced up‐regulation by coffee was shown in this mouse model (Kalthoff et al., 2010). Mice have been widely used in pharmacological research and therefore, the humanized transgenic UGT1A mice were an appropriate model for this study. In this study, we used only male mice because we were able to show that coffee induces UGT1A gene expression in a gender specific manner. In contrast to female mice, coffee treatment leads to an overall induction of UGT1A expression in the liver, jejunum and colon in males (Kalthoff et al., 2013). Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny, Browne, Cuthill, Emerson, & Altman, 2010) and with the recommendations made by the British Journal of Pharmacology.
2.8. Treatment of humanized transgenic mice
All experiments were performed in accordance to the “German Animal‐Protection Law” and the relevant guidelines of the Local Institutional Animal Care unit of our university (Haus für experimentelle Therapie, Bonn, Germany) and authorized by the relevant North Rhine‐Westphalian state agency for Nature, Environment and Consumer Protection (LANUV, Germany).
For this study, experimentally naive male 8‐ to 12‐week‐old htgUGT1A‐WT and htgUGT1A‐SNP (containing 10 UGT1A SNPs: UGT1A1*28, UGT1A3‐66T > C, UGT1A3 V47A, UGT1A3 W11R, UGT1A6*2a [S7A/T181A/R184S], UGT1A7*3 [N129K/R131K/W208R/−57T > G]) mice weighing 20–25 g at the starting point of the experiment were used. The mice of each mouse line were randomized (by picking numbers out of a hat) into five groups consisting of each 10 male mice (in total 50 mice per mouse line).
One group was treated with 200 mg·kg−1 of irinotecan (i.p.) and another with vehicle (NaCl 0.9% = control). For the analysis of protective effects of coffee and caffeic acid + caffeic acid phenylethyl ester on irinotecan toxicity, one group was pretreated with coffee (3 g per 150 ml in drinking bottle), another with caffeic acid + caffeic acid phenylethyl ester (400 mg·L−1 per 80 mg·L−1 in 5% Tween 80 in drinking bottle) and a last group was treated with vehicle (5% Tween 80 in drinking bottle) for 14 days followed by irinotecan injection (200 mg·kg−1, i.p.). For mouse treatment, we used half as much caffeic acid + caffeic acid phenylethyl ester compared to the in vitro assays, because coffee concentration was also halved for a better drinking acceptance.
Before irinotecan/NaCl treatment, mice were housed in individually ventilated cages with aspen wood bedding (two to five mice per cage) and fed standard irradiated rodent chow (19% protein rodent diet V1534‐000, 3.3% fat; Sniff, Soest, Germany) and water/coffee ad libitum. For the collection of urine, animals were set into a metabolic cage (one mouse per cage) without bedding after irinotecan injection for 20 h.
After 20 h of urine collection, blood counts (from blood taken from Vena facialis) were generated using a Vet abc blood counter (Scil animal care company) and animals were killed by cervical dislocation. Liver, stomach and intestine were extracted and immediately shock‐frozen in liquid nitrogen and stored at −80°C until use.
2.9. Randomization and blinding
All cell culture plate wells for the in vitro experiments and mice for the in vivo experiments were randomized into different treatment groups. The analyses of the in vitro and in vivo experiments were not performed under blinded conditions because the data acquisition and analyses were standardized and automated with automatic routines. This standardized and automated approach reduced possible operator bias and the need for blinding.
2.10. Western blot
50 mg of liver/colon tissue of five individual mice per treatment group (n = 5) was individually homogenized in 1600‐μl RIPA‐buffer with proteinase inhibitor and incubated for 1 h at 4°C on a shaker. After centrifugation (15870 x g; 10 min; 4°C), supernatant was collected into new tubes. 40 μg of isolated protein was boiled for 5 min in Laemmli sample buffer and separated by SDS‐PAGE prior to electrotransfer onto a nitrocellulose membrane. Incubation with primary antibody (UGT1Aall [RRID:AB_972125], Strassburg, Nguyen, Manns, & Tukey, 1999) was carried out in 10% dry milk. After incubation with appropriate secondary antibodies (Millipore, Schwalbach, Germany), protein was visualized by chemiluminescence (Biorad, München, Germany) using ChemiDoc MP imaging system (Bio‐Rad, München, Germany). Staining with β‐actin antibody (Santa Cruz Biotechnology Cat# sc‐69879, RRID:AB_1119529) was used as loading control. Western blot analysis for each treatment group was performed in five individual experiments using five individual mice per group. Data were quantitatively assessed and additionally depicted in graphs. Values represent mean ± SD. The immuno‐related procedures used comply with the recommendations made by the British Journal of Pharmacology (Alexander et al., 2018).
2.11. Gene expression analysis
RNA was isolated from mouse tissue using Trizol (Invitrogen, Karlsruhe, Germany) according to the manufacturer's instructions. 5‐μg RNA isolated from five individual mice was incubated with DNase I (Thermo Fisher Scientific, Schwerte, Germany) at room temperature for 15 min, followed by inactivation at 65°C for 10 min. DNase I‐treated RNA was then used as template in oligo (dT)‐primed cDNA synthesis reaction using the SuperScript III First‐Strand Synthesis System (Thermo Fisher Scientific, Schwerte, Germany). For gene expression analysis, cDNA concentrations were determined by qPCR relative to mouse β‐actin. qPCR reactions with gene‐specific primers and probes were performed in a CFX96 real‐time PCR detection system (Bio‐Rad) using qPCR MasterMix (Eurogentec). For p65, IL‐6 and TNFα gene expression analysis, TaqMan® Gene Expression Assays from Thermo Fisher Scientific were used (p65: Mm00712720_m1; IL‐6: Mm00446190_m1; TNFα: Mm00443260_g1). Expression relative to mouse β‐actin was calculated using Bio‐Rad CFX Manager 3.0 Software. Columns represent mean ± SD from five individual mice per group.
2.12. Hydrogen peroxide assay
10 mg of liver/colon tissue of five individual mice per group (n = 5) was homogenized and analysed according to the manufacturer's instructions (OxiSelectTMHydrogen Peroxide Assay Kit). Samples were analysed using Multiskan Go Reader (ThermoScientific). The hydrogen peroxide assay for each treatment group was performed using five individual mice per group. Values represent mean ± SD.
2.13. UGT‐Glo‐Assay
UGT1A enzyme activity was analysed by UGT‐Glo‐Assay Kit (Promega, Mannheim, Germany). For microsome isolation, 10 mg of liver/colon tissue of five individual mice per group (n = 5) was shredded using a QiagenTissue Lyser followed by resuspension in 1‐ml buffer (50 mmol·L−1 of Tris–HCl [pH 7.4] plus 10 mmol·L−1 of MgCl2) and homogenization with a Potter–Elvehjem tissue grinder with 10 strokes at 875 x g on ice. The tissue homogenate was centrifuged at 10,000 g for 5 min at 4°C and the supernatant collected. The pellet was resuspended in 0.5 ml of buffer and centrifuged at 10,000 g for 5 min and the supernatant was collected. The combined supernatants were centrifuged at 150,000 g for 60 min at 4°C and the pellet was resuspended in 0.2 ml of buffer. Protein concentrations were determined by the method of Bradford. Microsomal protein was stored at −80°C. 1 μg of mouse tissue isolated microsomes was used per reaction. Activity was analysed after 90 min of incubation according to the manufacturer's instructions using 25‐μM UGT Multienzyme Substrate. The activity assay for each treatment group was performed using five individual mice per group. Values represent mean ± SD.
2.14. Determination of SN‐38 glucuronides (G) in urine
LC‐MS/MS measurements were performed on a Waters Xevo TQ‐S or Waters Xevo TQ‐MS mass spectrometer equipped with a Waters Acquity UPLC system and analzsed with MassLynx and TargetLynx software packages. Urine samples were diluted 1:100 (H2O), vortexeand centrifuged (15,000 g, 5 min). 40 μl of the supernatant was mixed with 80‐μl acetonitrile, 40‐μl methanol, 40‐μl methanolic d3‐SN‐38‐glucuronide (Toronto Research Chemicals, Canada, 2.30 mg·L−1) solution and 200‐μl acetic acid (2% in H2O), subsequently vortexed and measured. For calibration, eight standard samples were prepared using pooled urine of untreated mice and calibration solutions containing known concentrations of SN‐38‐glucuronide (Toronto Research Chemicals, Canada, 10–4,126 μg·L−1). LC conditions: biphenyl column (Restek Raptor Biphenyl 2,7 μm 2,1 × 100 mm), column temperature 30°C, 0.1% formic acid in H2O as mobile phase A (A), 0.1% formic acid in acetonitrile as mobile phase B (B), flow rate 0.4 ml·min−1. Elution steps: 90% A (1 min), linear gradient towards 65% A (5 min), linear gradient towards 5% A (1 min), 5% A (2 min), linear gradient towards 90% A (1 min), 90% A (2 min). Recorded MRM transitions (m/z) for MS/MS detection were as follows: SN‐38‐glucuronide 569.2 to 393, 569.2 to 349, d3‐SN‐38‐glucuronide 572.2 to 396, 572.2 to 352. Integrals of standard samples were fitted with a second‐degree polynomial fit. For normalization, urinary creatinine levels were determined using Creatinine (urinary) Coloremetric Assay Kit (Cayman) according the manufacturer's instructions. SN‐38‐G and creatinine levels were determined for each mouse (group size n = 10). Values represent mean ± SD.
2.15. Data and statistical analysis
The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology (Curtis et al., 2018).
For both the in vitro and in vivo experiments, the group sizes for each experiment are provided within the respective methods sections and figure legends. For all in vivo experiments, 10 animals per group were used. The group size was calculated based on the results of previous experiments using an online power calculator (http://www.clinical‐trials.de/de/Werkzeuge/Online_Hilfsmittel/online_hilfsmittel.html).
In vitro assays using cell culture were performed in five independent repeats each containing three individual cell culture wells (in total 15 wells).
For determination of statistical significance, Student's t‐test was used. P < 0.05 was considered significant. All the comparisons were made with the respective vehicle treatment. The results are expressed as the mean ± SD.
2.16. Nomenclature of ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander, Kelly, et al., 2019; Alexander, Fabbro, et al., 2019; Alexander, Kelly, et al., 2019).
3. RESULTS
3.1. Effect of green coffee and different coffee ingredients on UGT1A transcription in KYSE70 cells
We first studied whether unroasted green coffee is capable of inducing previously shown (Kalthoff et al., 2010) transcriptional UGT1A up‐regulation. In Figure 1a, the induction of an UGT1A3 reporter gene construct (8.8‐fold) by regular coffee in KYSE70 cells is shown. In contrast, green coffee brew was not able to induce UGT1A3 transcription.
FIGURE 1.
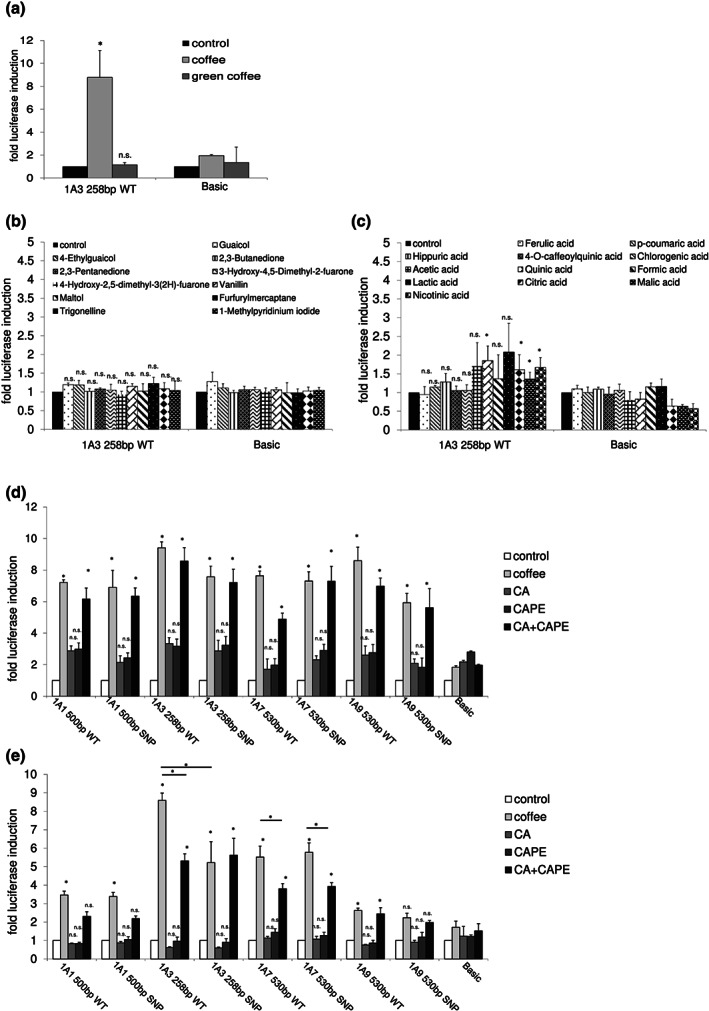
In vitro UGT1A regulation by coffee and different coffee ingredients. A UGT1A3 reporter gene construct was tested for inducibility by green coffee (a), coffee aromatics, trigonelline (b), and coffee acids (c) in KYSE70 cells. Different UGT1A reporter gene constructs were induced by coffee and a combination of caffeic acid (CA) and caffeic acid phenylethyl ester (CAPE) in KYSE70 cells (d) and HepG2 cells (e). Columns represent mean ± SD from five independent experiments analysing one treatment group consisting of three individual cell culture wells (in total 15 wells). Significance was determined in relation to basic control vector. *P < 0.05; n.s., non‐significant
Because roasting appeared to produce compounds capable of UGT1A gene activation, we analysed different aromatic coffee and trigonelline compounds. None of these led to increased UGT1A3 transcription in KYSE70 cells (Figure 1b). Among different acids tested for UGT1A inducibility, quinic, lactic, malic and nicotinic acid showed a significant up‐regulation of UGT1A3 expression, but luciferase expression did not come close to the expression levels elicited by coffee exposure (between 1.3‐ and 1.9‐fold vs. coffee: 8.8‐fold) (Figure 1c).
3.2. Transcriptional UGT1A regulation by coffee, caffeic acid, caffeic acid phenylethyl ester and combination of caffeic acid + caffeic acid phenylethyl ester in cell culture
The caffeic acid derivate caffeic acid phenylethyl ester is present in a broad array of plants including the coffee bean (Macías‐Pérez, Beltrán‐Ramírez, & Villa‐Treviño, 2012). The concentration of caffeic acid in coffee is assumed to amount to approximately 35–250 mg per cup (Higdon & Frei, 2006; Iwahashi, 2014). In the luciferase assays of this study, we used 800 mg·L−1 of caffeic acid. Although caffeic acid phenylethyl ester is also a coffee ingredient, its concentration in the beverage coffee has not been reported. Therefore, after studying different concentrations, we chose 160 mg·L−1, which was not cytotoxic but led to highest UGT1A inducibility (Figure S1).
In KYSE70 cells, UGT1A1, UGT1A3, UGT1A7 and UGT1A9 reporter gene constructs were significantly up‐regulated by coffee treatment (5.5‐ to 8.3‐fold) (Figure 1d). The examined SNP containing (UGT1A1*28, UGT1A3‐66T > C, UGT1A7‐57T > G and UGT1A9*1b) reporter gene constructs did not significantly affect the coffee‐mediated induction of luciferase expression. Interestingly, caffeic acid and caffeic acid phenylethyl ester used individually were not able to induce UGT1A expression, whereas the combination of both (caffeic acid + caffeic acid phenylethyl ester) led to a significant induction (4.8‐ to 8.8‐fold) comparable to that observed with coffee.
In HepG2 cells, coffee treatment increased the expression of all UGT1A (except for UGT1A9 530‐bp SNP) reporter gene constructs (2.6‐ to 8.6‐fold) (Figure 1e). Only the UGT1A3‐66T > C SNP significantly reduced coffee‐mediated induction. None of the tested constructs led to a significant induction by caffeic acid or caffeic acid phenylethyl ester alone. After combined treatment with caffeic acid + caffeic acid phenylethyl ester UGT1A3 WT/SNP, UGT1A7 WT/SNP and UGT1A9 WT constructs showed a significant induction, which was overall lower than that observed with coffee (1.9 to 5.3‐fold). UGT1A1‐WT and SNP constructs were induced twofold by caffeic acid + caffeic acid phenylethyl ester (not significant) compared to a 3.5‐fold induction by coffee.
3.3. Effect of siRNA‐mediated knockdown of aryl hydrocarbon receptor and Nrf2 on UGT1A regulation by caffeic acid + caffeic acid phenylethyl ester
In a previous study of our laboratory, we identified aryl hydrocarbon receptor (AhR) and nuclear factor, erythroid 2 like 2 (NRF2 or Nrf2) as mediators of coffee‐induced UGT1A up‐regulation (Kalthoff et al., 2010). In order to examine if aryl hydrocarbon receptor and Nrf2 are also responsible for the CA + caffeic acid phenylethyl ester‐mediated UGT1A induction, we treated KYSE70 and HepG2 cells with either siRNA against aryl hydrocarbon receptor or Nrf2 and analysed UGT1A reporter gene expression. In agreement with our previously reported findings, caffeic acid + caffeic acid phenylethyl ester‐mediated UGT1A up‐regulation was abolished when KYSE70 and HepG2 cells were co‐transfected with either aryl hydrocarbon receptor or Nrf2 siRNA, exemplary shown for the UGT1A1 and UGT1A3 reporter gene constructs in Figure 2a,b. In conclusion, the presence of both transcription factors is essential for coffee and caffeic acid + caffeic acid phenylethyl ester‐induced up‐regulation of UGT1A expression.
FIGURE 2.
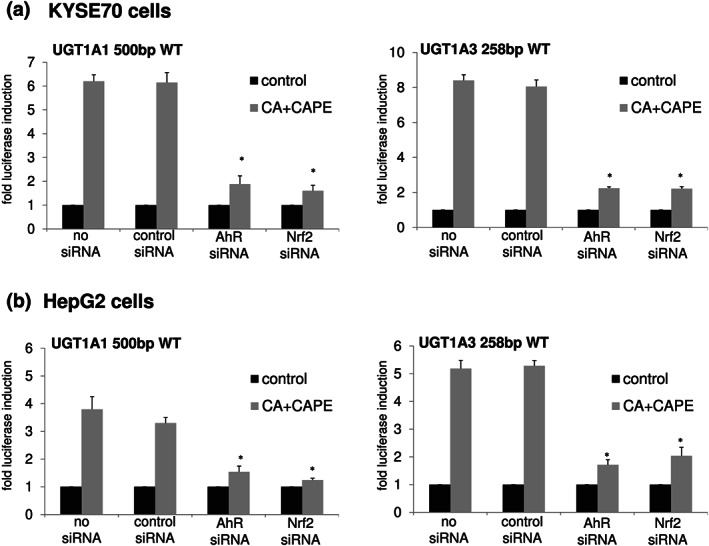
siRNA‐mediated knockdown of aryl hydrocarbon receptor and Nrf2. Both treatments with aryl hydrocarbon receptor (AhR) and Nrf2 siRNA (100 nM) led to a significant reduction of caffeic acid (CA) + caffeic acid phenylethyl ester (CAPE)‐induced UGT1A up‐regulation in KYSE70 cells (a) and HepG2 cells (b). Columns represent mean ± SD from of five independent experiments analysing one treatment group consisting of three individual cell culture wells (in total 15 wells). Significance was determined in relation to constructs treated with control siRNA. *P < 0.05
3.4. In vivo regulation of UGT1A mRNA expression in htgUGT1A‐mice treated with irinotecan in combination with coffee or caffeic acid + caffeic acid phenylethyl ester
In the liver of irinotecan‐treated htgUGT1A‐WT mice, transcription of UGT1A mRNA was highly up‐regulated by both coffee and caffeic acid + caffeic acid phenylethyl ester pretreatment (Figure 3a). A significant induction of UGT1A1 expression was observed after coffee pretreatment and caffeic acid + caffeic acid phenylethyl ester exposure. Similarly, UGT1A3, UGT1A4 and UGT1A6 expression was significantly increased after coffee and caffeic acid + caffeic acid phenylethyl ester pretreatment. A significant up‐regulation of UGT1A1, UGT1A3, UGT1A4 and UGT1A6 by coffee and caffeic acid + caffeic acid phenylethyl ester was also observed in the liver of irinotecan‐treated htgUGT1A‐SNP mice, although absolute expression levels remained under those detected in htgUGT1A‐WT mice. In the colon, UGT1A1, UGT1A3 and UGT1A4 expression was significantly increased by coffee and caffeic acid + caffeic acid phenylethyl ester pretreatment in both htgUGT1A‐WT and SNP mice (Figure 3b) although absolute expression levels of htgUGT1A‐SNP mice were lower than those observed in WT mice.
FIGURE 3.
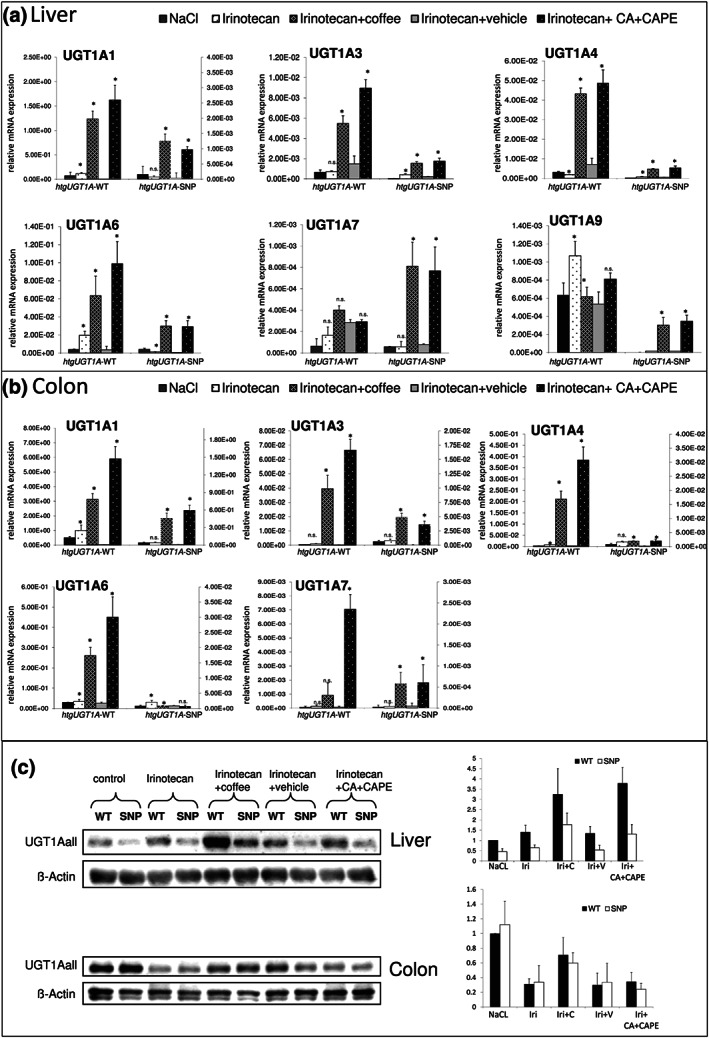
Inducing effects of coffee and caffeic acid (CA) + caffeic acid phenylethyl ester (CAPE) treatment on UGT1A mRNA and protein expression. (a,b) UGT1A mRNA expression was highly up‐regulated by coffee and CA + CAPE pretreatment in the liver (a) and the colon (b) of htgUGT1A‐WT and single nucleotide polymorphism (SNP) mice. Columns represent mean ± SD from five individual mice. Significance of irinotecan group was determined in comparison to NaCl group; coffee group was compared to irinotecan group; CA + CAPE group was compared to irinotecan + vehicle group. Values for htgUGT1A‐WT mice refer to the left y‐axis; values for htgUGT1A‐SNP mice refer to the right y‐axis (colon). (c) Induction of UGT1A protein expression in irinotecan‐injected htgUGT1A mice pretreated with coffee or CA + CAPE. Western blot analysis for each treatment group (using five individual mice [n = 5] of every treatment group) was performed. A representative Western blot is shown. Data were quantitatively assessed and additionally depicted in graphs. Values represent mean ± SD. Iri, Irinotecan; c, coffee; v, vehicle; *P < 0.05; n.s., non‐significant
3.5. In vivo regulation of UGT1A protein expression in htgUGT1A mice treated with irinotecan in combination with coffee or caffeic acid + caffeic acid phenylethyl ester
UGT1A protein levels were determined using an antibody against all UGT1A isoforms (anti‐UGT1Aall). In the liver, UGT1A protein was up‐regulated by pretreatment with coffee and caffeic acid + caffeic acid phenylethyl ester in htgUGT1A‐WT and SNP mice (Figure 3c). Protein levels in htgUGT1A‐SNP mice were slightly reduced compared to WT counterparts. In the colon, overall UGT1Aall protein expression was reduced by irinotecan treatment which was reversed by coffee but not by caffeic acid + caffeic acid phenylethyl ester pretreatment (Figure 3c).
3.6. Effect of coffee and caffeic acid + caffeic acid phenylethyl ester pretreatment on UGT enzyme activity in the liver and colon of irinotecan‐treated htgUGT1A mice
To investigate whether UGT enzyme activity was up‐regulated by irinotecan, coffee and caffeic acid + caffeic acid phenylethyl ester, we measured UGT activity using the UGT‐Glo™ Assay (Promega). Irinotecan treatment alone did not increase UGT catalytic activity in comparison to control mice (Figure 4a). However, coffee and caffeic acid + caffeic acid phenylethyl ester pretreatment led to a significant induction of UGT activity in liver and colon of htgUGT1A‐WT and SNP mice compared to control animals. In general, the UGT activity measured in colon was lower compared to that detected in the liver of htgUGT1A mice. The level of UGT activity induction by coffee and caffeic acid + caffeic acid phenylethyl ester was comparable between htgUGT1A‐WT and SNP mice. Moreover, the UGT activity levels observed in the liver and colon of htgUGT1A‐SNP mice were only slightly lower compared to those in WT mice.
FIGURE 4.
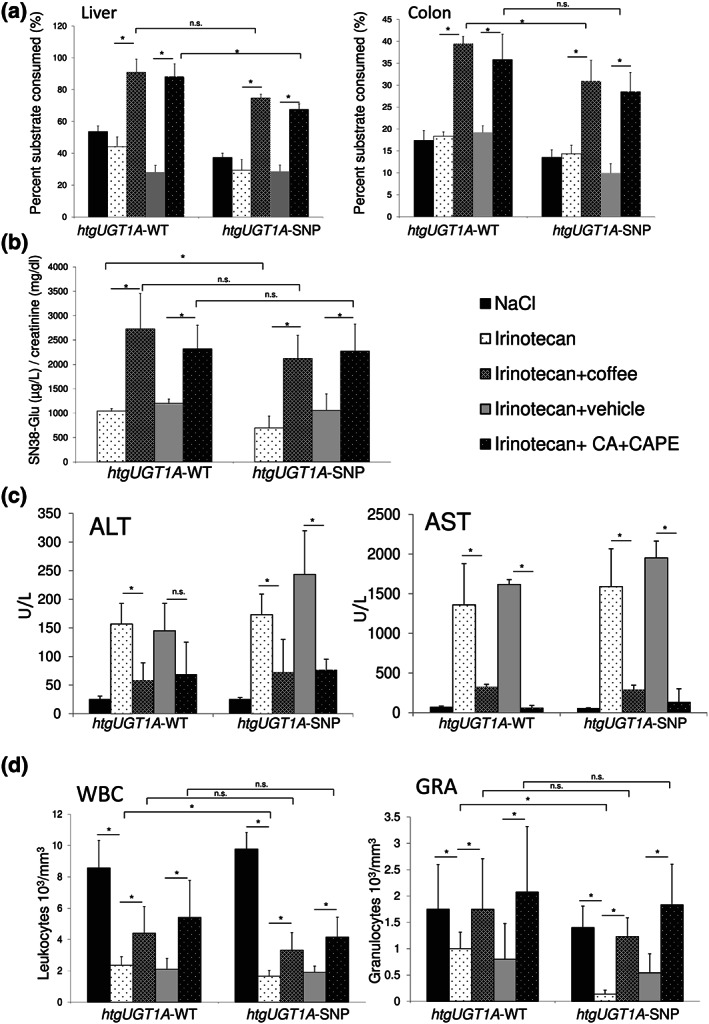
Effects of coffee and caffeic acid (CA) + caffeic acid phenylethyl ester (CAPE) pretreatment on UGT activity, aminotransferases and leukopenia. (a) UGT enzyme activity against a proluciferin substrate was determined using UGT‐Glo™ Assay (Promega). UGT activity was significantly induced by coffee and CA + CAPE pretreatment in htgUGT1A‐WT and single nucleotide polymorphism (SNP) mice. Columns represent mean ± SD from five individual mice per group. (b) SN‐38 glucuronide formation analysed by LC‐MS/MS measurements (urine) was significantly increased by coffee and CA + CAPE pretreatment in both mouse lines (values represent mean of n = 10 mice per group ±SD). (c,d) Protective effects of coffee and CA + CAPE pretreatment on irinotecan‐induced elevation of aminotransferases and leukopenia (values represent mean of n = 10 mice per group ±SD). (c) ALT and AST levels were reduced by coffee and CA + CAPE pretreatment in irinotecan‐injected htgUGT1A mice. (d) Irinotecan‐induced leukopenia was reversed by pretreatment with coffee and CA + CAPE. Significance was determined as indicated by lines. *P < 0.05; n.s., non‐significant
These data suggest that coffee and caffeic acid + caffeic acid phenylethyl ester can significantly increase the enzymatic activity of UGT enzymes in the liver and the colon of both htgUGT1A‐WT and SNP mice.
3.7. Determination of SN‐38 glucuronides in urine of irinotecan‐injected htgUGT1A mice pretreated with coffee or caffeic acid + caffeic acid phenylethyl ester
To examine the effect of coffee and caffeic acid + caffeic acid phenylethyl ester on SN‐38 glucuronide formation, urine of irinotecan‐injected and coffee/caffeic acid + caffeic acid phenylethyl ester‐pretreated htgUGT1A mice were collected for 20 h after irinotecan injection. SN‐38G in urine was quantified by LC‐MS/MS measurements. SN‐38G levels normalized against creatinine were significantly higher in urine of irinotecan‐treated htgUGT1A‐WT mice compared to those of SNP mice (Figure 4b). Coffee and caffeic acid + caffeic acid phenylethyl ester pretreatment led to a significant increase of SN‐38G formation in both htgUGT1A‐WT and SNP mice. There were no significant differences between SN‐38G levels between coffee or caffeic acid + caffeic acid phenylethyl ester‐pretreated htgUGT1A‐WT and SNP mice. These data demonstrate that coffee and its constituents caffeic acid + caffeic acid phenylethyl ester activate the glucuronidation of the irinotecan metabolite SN‐38.
3.8. Pretreatment with coffee or caffeic acid + caffeic acid phenylethyl ester improved irinotecan‐mediated elevation of serum alanine (ALT) and aspartate (AST) aminotransferases
Irinotecan treatment led to a significant elevation of ALT and AST serum levels in htgUGT1A‐WT and SNP mice (Figure 4c). After pretreatment with coffee or caffeic acid + caffeic acid phenylethyl ester, ALT and AST levels were reduced in both mouse lines. The ALT decrease by caffeic acid + caffeic acid phenylethyl ester in htgUGT1A‐SNP mice was significant but not in htgUGT1A‐WT mice. No significant differences in AST and ALT levels were detectable between htgUGT1A‐WT and SNP mice suggesting that the UGT1A haplotype (containing 10 different SNPs) does not affect the protective effects of coffee and caffeic acid + caffeic acid phenylethyl ester.
3.9. Pretreatment with coffee or caffeic acid + caffeic acid phenylethyl ester reverses irinotecan‐mediated leukopenia
Twenty hours after irinotecan injection, the number of leukocytes was determined in order to measure irinotecan‐induced bone marrow toxicity. The mice receiving irinotecan showed a significant reduction in leukocyte and granulocyte numbers (Figure 4d). Furthermore, leukocyte levels but especially granulocyte numbers were significantly reduced in htgUGT1A‐SNP mice compared to their WT counterparts. This in accordance with the observation that patients homozygous for the Gilbert's syndrome related UGT1A1*28 allele are more likely to develop neutropenia following irinotecan therapy (Liu, Cheng, Kuang, Liu, & Xu, 2013). Pretreatment with coffee or caffeic acid + caffeic acid phenylethyl ester significantly reversed irinotecan‐induced reduction of leukocyte and granulocyte number in both htgUGT1A‐WT and SNP mice. No significant differences between both mouse lines pretreated with coffee or caffeic acid + caffeic acid phenylethyl ester were observed indicating a protective effect of coffee and caffeic acid + caffeic acid phenylethyl ester also in the presence of the UGT1A haplotype.
3.10. Pretreatment with coffee or caffeic acid + caffeic acid phenylethyl ester decreased irinotecan‐induced oxidative stress and inflammatory response
The administration of antineoplastic agents is accompanied by the production of ROS contributing to many side effects that are common to many anticancer drugs including gastrointestinal toxicity (Touchefeu et al., 2014). Therefore, we sought to analyse the levels of hydrogen peroxide in the liver and colon of irinotecan‐treated mice. Hepatic and colonic H2O2 levels were significantly increased by irinotecan treatment in htgUGT1A‐WT and SNP mice (Figure 5a). Coffee and caffeic acid + caffeic acid phenylethyl ester pretreatment resulted in a significant reduction of H2O2 levels in the colon compared to single irinotecan treatment. In the liver, the reduction of ROS by coffee and caffeic acid + caffeic acid phenylethyl ester was only significant in htgUGT1A‐SNP mice although there was a trend (not significant) in their WT counterparts. Irinotecan‐treated as well as coffee and caffeic acid + caffeic acid phenylethyl ester‐pretreated htgUGT1A‐WT mice exhibited significantly lower H2O2 levels in the liver and the colon compared to htgUGT1A‐SNP mice.
FIGURE 5.
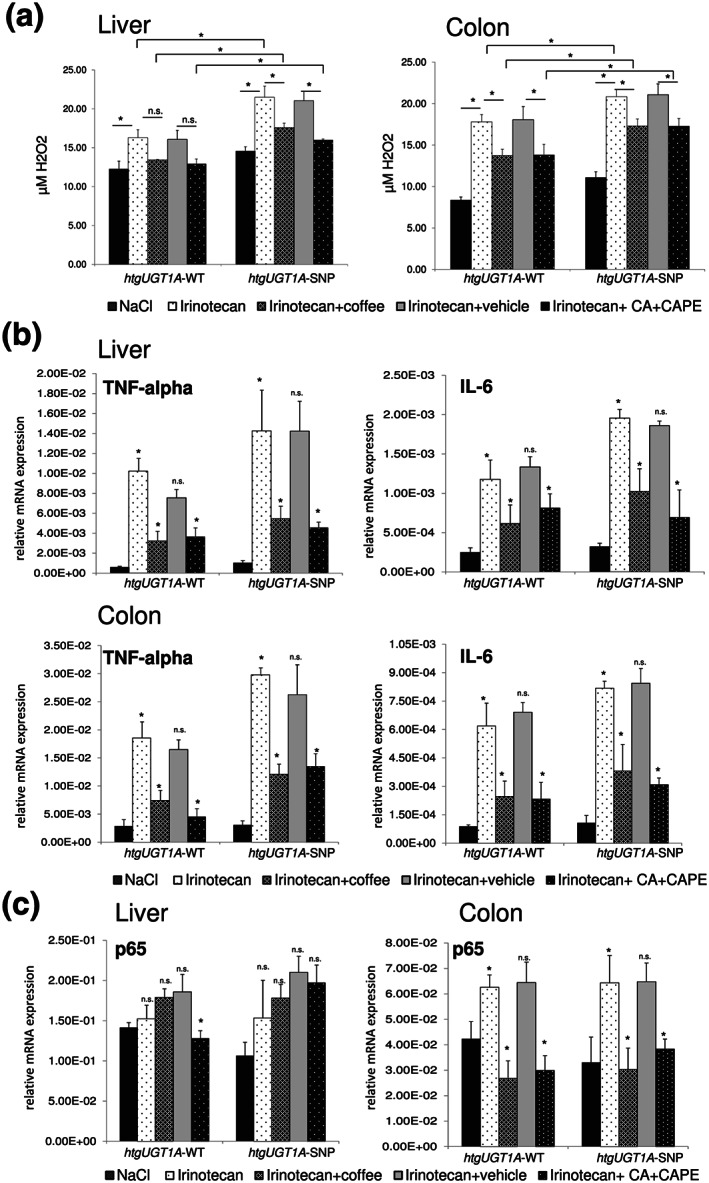
Protective effects of coffee and caffeic acid (CA) + caffeic acid phenylethyl ester (CAPE) pretreatment on irinotecan‐induced oxidative stress, inflammatory response and p65 up‐regulation. (a) Pretreatment with coffee and CA + CAPE led to a significant reduction of hydrogen peroxide levels in the liver and in the colon. Significance was determined as indicated by lines. Columns represent mean ± SD from five individual mice per group. Significance was determined as indicated by lines. (b) Irinotecan‐induced up‐regulation of TNFα and IL‐6 was significantly decreased in the liver and the colon of htgUGT1A‐WT and single nucleotide polymorphism (SNP) mice. (c) In the colon, pretreatment with coffee and CA + CAPE abolished the irinotecan‐induced up‐regulation of p65 expression in htgUGT1A‐WT and single nucleotide polymorphism SNP mice. Columns represent mean ± SD from five individual mice per group (b,c). Significance of irinotecan group was determined in comparison to NaCl group; coffee group was compared to irinotecan group; CA + CAPE group was compared to irinotecan + vehicle group (b,c). *P < 0.05; n.s., non‐significant
In order to examine the inflammatory response after irinotecan treatment, we measured mRNA expression levels of the pro‐inflammatory cytokines IL‐6 and TNFα. In the liver and the colon of htgUGT1A‐WT and SNP mice, IL‐6 and TNFα levels were significantly increased by irinotecan treatment (Figure 5b). This effect was significantly reduced by coffee and caffeic acid + caffeic acid phenylethyl ester pretreatment.
3.11. Pretreatment with coffee or caffeic acid + caffeic acid phenylethyl ester abolished the irinotecan‐induced up‐regulation of p65 mRNA expression in the colon of htgUGT1A mice
Irinotecan treatment has been shown to lead to the activation of NF‐κB in a variety of human CRC cell lines (Xu & Villalona‐Calero, 2002). Since it is known that siRNA‐mediated down‐regulation of the NF‐κB subunit p65 can effectively enhance in vitro and in vivo sensitivity to irinotecan (Guo, Verma, Gaynor, Frenkel, & Becerra, 2004), we analysed the p65 mRNA expression levels in the liver and the colon of irinotecan‐treated and coffee/caffeic acid + caffeic acid phenylethyl ester‐pretreated htgUGT1A mice. In contrast to the liver, colonic p65 expression was significantly increased by irinotecan in htgUGT1A‐WT and SNP mice (Figure 5c). This induction was completely abolished by pretreatment with coffee and caffeic acid + caffeic acid phenylethyl ester. Moreover, p65 levels in coffee and caffeic acid + caffeic acid phenylethyl ester‐pretreated htgUGT1A‐WT mice were even lower than those detected in NaCl‐treated mice. These data suggest that coffee and the coffee ingredients caffeic acid + caffeic acid phenylethyl ester can reduce p65 expression potentially leading to an increased chemosensitivity to irinotecan.
4. DISCUSSION
Although coffee was initially believed to represent a health hazard potentially promoting cancer and heart disease, it has been realized in recent years that coffee consumption in fact confers substantial health benefits. Numerous coffee components are candidates for these beneficial effects and require identification in order to develop druggable candidates for therapeutic interventions.
Since roasting appeared to be the critical step for the protective effects of coffee, we looked into possible candidates. A substantial amount of chlorogenic acid is degraded into caffeic acid and quinic acid during roasting. In addition, caffeic acid phenylethyl ester is a derivative of caffeic acid. Used individually, both caffeic acid and caffeic acid phenylethyl ester were not capable of inducing UGT1A reporter gene constructs. Only the combined exposure of caffeic acid + caffeic acid phenylethyl ester led to a significant up‐regulation of UGT1A expression comparable to that observed by coffee in cell culture experiments. This inducing effect was confirmed in irinotecan‐treated htgUGT1A mice. Most hepatic and intestinal UGT1A isoforms were highly up‐regulated by coffee and to the same degree by the combined exposure to the coffee‐constituents caffeic acid and caffeic acid phenylethyl ester.
Irinotecan is known for its narrow therapeutic window with feared complications and toxicity such as severe myelosuppression and massive diarrhoea (Rajendra Prasad et al., 2011; Sato et al., 2011). In humans, these side effects are most commonly the dose‐limiting toxicities in cancer chemotherapy and limit the anti‐tumour activity of irinotecan (Iwahashi, 2014). Our hypothesis was that irinotecan‐induced side effects may be ameliorated by coffee (caffeic acid + caffeic acid phenylethyl ester)‐mediated up‐regulation of mRNA expression and activity of irinotecan detoxifying UGT1A enzymes.
HtgUGT1A‐SNP mice solely treated with irinotecan exhibited a more severe leukopenia and granulopenia compared to their WT counterparts. These findings are consistent with the fact that patients carrying the UGT1A haplotype have a higher risk to develop a more severe leukopenia after irinotecan administration (Lankisch et al., 2008). In accordance with the results obtained from the analysis of the UGT1A mRNA expression and enzyme activity, pretreatment with coffee or caffeic acid + caffeic acid phenylethyl ester led to an improvement of leukopenia and serum liver enzymes. Surprisingly, no significant differences between the two mouse lines were observed after pretreatment with coffee or caffeic acid + caffeic acid phenylethyl ester concerning myelosuppression and serum transaminases indicating a compensation of the SNP associated increased toxicity. These data are in line with the examined SN‐38 glucuronide formation showing similar coffee and caffeic acid + caffeic acid phenylethyl ester‐mediated absolute induction levels of enzymatic activity comparing htgUGT1A‐WT and SNP mice. This is the first study showing a pharmacologically induced protective effect on irinotecan‐induced leukopenia and elevation of transaminases in presence of the human UGT1A SNP haplotype.
Some discrepancies were observed concerning differences between coffee and caffeic acid + caffeic acid phenylethyl ester inducing effects on protein level in the colon (colonic UGT1A1all protein expression was reduced by irinotecan treatment, which was reversed by coffee but not by caffeic acid + caffeic acid phenylethyl ester pretreatment) and differences between protein expression and UGT enzyme activity (in contrast to colonic protein expression, enzyme activity was not significantly inhibited by irinotecan but significantly increased by coffee and caffeic acid + caffeic acid phenylethyl ester in htgUGT1A‐WT and SNP mice). Obviously, there are differences between UGT1A mRNA, protein expression and UGT enzyme activity. Genome‐wide correlation between expression levels of mRNA and protein are notoriously poor, hovering around 40% explanatory power across many studies. The discrepancy is typically attributed to different regulation mechanisms of the transcript and the protein product (Maier, Guell, & Serrano, 2009). However, studies investigating direct effects on enzyme activity have to be specific for one enzyme and are rare in respect to UGT1A enzymes. Some drugs such as atazanavir and indinavir are known to inhibit UGT enzyme activity (Strassburg, Kalthoff, & Ehmer, 2008). Further studies are required to examine a potential activating mechanism of caffeic acid + caffeic acid phenylethyl ester on UGT enzyme activity.
Irinotecan and its metabolites cause acute damage to the intestinal mucosa leading to increased oxidative stress, epithelial damage and severe diarrhoea (Sakai, Diener, Gartmann, & Takeguchi, 1995),(Rtibi et al., 2017). In this study, we detected an irinotecan‐induced elevation of H2O2 levels not only in the colon but also in the liver. Moreover, this effect was significantly attenuated by pretreatment with coffee and caffeic acid + caffeic acid phenylethyl ester in the liver of htgUGT1A‐SNP mice and in the colon of both htgUGT1A mouse lines. ROS can directly induce tissue injury and trigger a cascade of inflammatory pathways where NF‐κB is thought to be pivotal in this process. Once activated by chemotherapy and ROS, NF‐κB acts to induce gene expression and production of pro‐inflammatory cytokines such as TNFα, IL‐1β and IL‐6, which in turn lead to tissue injury and apoptosis (Lee, Ryan, & Doherty, 2014). In line with these data, we observed a significant increase of TNFα and IL‐6 expression in the liver and colon of irinotecan‐treated htgUGT1A mice that was attenuated by coffee and caffeic acid + caffeic acid phenylethyl ester pretreatment. In a recent study, we were able to show that UGT1A activation in benzo(a)pyrene exposure‐associated oxidative stress is protective and that this effect is directly linked to the presence of UGT1A enzymes (Kalthoff et al., 2017). Therefore, we think that the protective effects of coffee/caffeic acid + caffeic acid phenylethyl ester are mediated by both elevated UGT1A catalysed SN38 glucuronidation and increased UGT1A mediated detoxification of cell damage originated reactive metabolites leading to decreased levels of oxidative stress.
Apart from the caffeic acid phenylethyl ester‐mediated inhibition of NF‐κB activation by prevention of DNA binding described in other studies (Marquez et al., 2004; Natarajan, Singh, Burke, Grunberger, & Aggarwal, 1996), we observed an inhibition of p65 mRNA transcription by caffeic acid phenylethyl ester in combination with caffeic acid. In a study from Guo et al. (2004), an enhanced chemosensitivity to irinotecan was described after RNA interference mediated down‐regulation of p65 mRNA expression. Activation of the NF‐κB pathway constitutes a potential mechanism of inducible resistance by malignant cells exposed to irinotecan (Baldwin, 2001; Wang, Mayo, Korneluk, Goeddel, & Baldwin, 1998). The underlying mechanism is thought to include the NF‐κB‐activated gene expression of different inhibitors of apoptosis such as c‐IAP1, c‐IAP2 and Bcl‐xL (Chen, Kandasamy, & Srivastava, 2003 ; Wang et al., 1998). Thus, reducing NF‐κB‐mediated activation by caffeic acid + caffeic acid phenylethyl ester might help to prevent irinotecan‐induced resistance to cell killing.
Although caffeic acid + caffeic acid phenylethyl ester were shown to attenuate irinotecan‐induced leukopenia, intestinal oxidative stress and inflammatory cytokine expression, an effect on the efficacy of irinotecan cannot be excluded. The increased hepatic and intestinal glucuronidation activity of UGT1A enzymes might decrease the systemic exposure to the anti‐cancer metabolite SN‐38, which would limit the potential clinical utility. Therefore, further studies are needed to examine the effect of caffeic acid + caffeic acid phenylethyl ester on the irinotecan‐mediated tumour shrinkage.
In conclusion, in this study, we identify the compounds responsible for mediating the previously reported coffee‐associated activation of UGT1A gene expression. Using a model of drug‐induced toxicity, coffee and a combination of the coffee ingredients caffeic acid and caffeic acid phenylethyl ester were shown to significantly improve leukopenia and to decrease oxidative stress and inflammatory response in htgUGT1A‐WT and SNP mice. Moreover, coffee and caffeic acid + caffeic acid phenylethyl ester were shown to reverse irinotecan‐induced intestinal p65 up‐regulation, which is associated with an inhibition of chemotherapy‐induced apoptosis. However, to estimate the clinical utility, further studies are necessary to investigate the effect of caffeic acid + caffeic acid phenylethyl ester‐induced UGT1A activity on irinotecan efficacy.
AUTHOR CONTRIBUTIONS
S.K. and C.P.S. designed the study and experiments. S.K. and S.P. did experimental work and analysed the data. A.R. and S.H. developed the LC‐MS/MS method and determined SN‐38 glucuronides in urine. S.K., G.H. and C.P.S interpreted and discussed data. S.K. and C.P.S. wrote the manuscript. C.P.S attracted funding.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design & Analysis, Immunoblotting and Immunochemistry, and Animal Experimentation and as recommended by funding agencies, publishers and other organizations engaged with supporting research.
Supporting information
Figure S1. Increasing concentrations of CAPE were not able to induce UGT1A3 reporter gene expression significantly in comparison to Basic control vector in KYSE70 cells. The screen was performed in three independent experiments each analyzing three wells.
ACKNOWLEDGEMENT
This work was funded by Deutsche Forschungsgemeinschaft (DFG) project STR493/9‐1 (to C.P.S.).
Kalthoff S, Paulusch S, Rupp A, Holdenrieder S, Hartmann G, Strassburg CP. The coffee ingredients caffeic acid and caffeic acid phenylethyl ester protect against irinotecan‐induced leukopenia and oxidative stress response. Br J Pharmacol. 2020;177:4193–4208. 10.1111/bph.15162
REFERENCES
- Alexander, S. P. H. , Roberts, R. E. , Broughton, B. R. S. , Sobey, C. G. , George, C. H. , Stanford, S. C. , … Ahluwalia, A. (2018). Goals and practicalities of immunoblotting and immunohistochemistry: A guide for submission to the British Journal of Pharmacology . British Journal of Pharmacology, 175, 407–411. 10.1111/bph.14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , … CGTP Collaborators . (2019). The concise guide to pharmacology 2019/20: Introduction and Other Protein Targets. British Journal of Pharmacology, 176: S1‐S120. 10.1111/bph.14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , … CGTP collaborators . (2019). The concise guide to pharmacology 2019/20: Enzymes. British Journal of Pharmacology, 176, S297–S396. 10.1111/bph.14572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , … GTP collaborators . (2019). The concise guide to pharmacology 2019/20: Transporters. British Journal of Pharmacology, 176, S397–S493. 10.1111/bph.14753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, A. S. Jr. (2001). Series introduction: The transcription factor NF‐κB and human disease. The Journal of Clinical Investigation, 107(1), 3–6. 10.1172/JCI11891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitz, H.‐D. , Grosch, W. , & Schieberle, P. (2009). Food chemistry: Individual aroma compounds. Berlin: Springer. [Google Scholar]
- Cecchin, E. , Innocenti, F. , D'Andrea, M. , Corona, G. , De Mattia, E. , Biason, P. , … Toffoli, G. (2009). Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. Journal of Clinical Oncology, 27(15), 2457–2465. 10.1200/JCO.2008.19.0314 [DOI] [PubMed] [Google Scholar]
- Chao, P. C. , Hsu, C. C. , & Yin, M. C. (2009). Anti‐inflammatory and anti‐coagulatory activities of caffeic acid and ellagic acid in cardiac tissue of diabetic mice. Nutrition & Metabolism (London), 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Kandasamy, K. , & Srivastava, R. K. (2003). Differential roles of RelA (p65) and c‐Rel subunits of nuclear factor κB in tumor necrosis factor‐related apoptosis‐inducing ligand signaling. Cancer Research, 63(5), 1059–1066. [PubMed] [Google Scholar]
- Clarke, R. J. , & Macrae, R. (1988). Coffee In Physiology (Vol. 3). London, New York: Elsevier Applied Science. [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , … MacEwan, D. J. (2018). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175, 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debry, G. (1994). Coffee and health. Montrouge, France: John Libbey Eurotext. [Google Scholar]
- Ehmer, U. , Kalthoff, S. , Fakundiny, B. , Pabst, B. , Freiberg, N. , Naumann, R. , … Strassburg, C. P. (2012). Gilbert syndrome redefined: A complex genetic haplotype influences the regulation of glucuronidation. Hepatology, 55(6), 1912–1921. 10.1002/hep.25561 [DOI] [PubMed] [Google Scholar]
- Erichsen, T. J. , Aehlen, A. , Ehmer, U. , Kalthoff, S. , Manns, M. P. , & Strassburg, C. P. (2010). Regulation of the human bile acid UDP‐glucuronosyltransferase 1A3 by the farnesoid X receptor and bile acids. Journal of Hepatology, 52(4), 570–578. 10.1016/j.jhep.2010.01.010 [DOI] [PubMed] [Google Scholar]
- Flament, I. , & Bessière‐Thomas, Y. (2002). Coffee flavor chemistry. New Jersey: New York Wiley. [Google Scholar]
- Griffin, M. (2006). Coffee chemistry: Coffee acidity. from http://www.coffeeresearch.org/science/sourmain.htm
- Guo, J. , Verma, U. N. , Gaynor, R. B. , Frenkel, E. P. , & Becerra, C. R. (2004). Enhanced chemosensitivity to irinotecan by RNA interference‐mediated down‐regulation of the nuclear factor‐κB p65 subunit. Clinical Cancer Research, 10(10), 3333–3341. 10.1158/1078-0432.CCR-03-0366 [DOI] [PubMed] [Google Scholar]
- Heath, R. D. , Brahmbhatt, M. , Tahan, A. C. , Ibdah, J. A. , & Tahan, V. (2017). Coffee: The magical bean for liver diseases. World Journal of Hepatology, 9(15), 689–696. 10.4254/wjh.v9.i15.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon, J. V. , & Frei, B. (2006). Coffee and health: A review of recent human research. Critical Reviews in Food Science and Nutrition, 46(2), 101–123. 10.1080/10408390500400009 [DOI] [PubMed] [Google Scholar]
- Iwahashi, H. (2014). Inhibitory effects of caffeic acid on free‐radical formation. San Diego, United States: Coffee in Health and Disease Prevention. [Google Scholar]
- Kalthoff, S. , Ehmer, U. , Freiberg, N. , Manns, M. P. , & Strassburg, C. P. (2010). Coffee induces expression of glucuronosyltransferases by the aryl hydrocarbon receptor and Nrf2 in liver and stomach. Gastroenterology, 139(5), 1699–1710. 1710 e1691‐1692 [DOI] [PubMed] [Google Scholar]
- Kalthoff, S. , Landerer, S. , Reich, J. , & Strassburg, C. P. (2017). Protective effects of coffee against oxidative stress induced by the tobacco carcinogen benzo[α]pyrene. Free Radical Biology & Medicine, 108, 66–76. [DOI] [PubMed] [Google Scholar]
- Kalthoff, S. , Winkler, A. , Freiberg, N. , Manns, M. P. , & Strassburg, C. P. (2013). Gender matters: Estrogen receptor alpha (ERα) and histone deacetylase (HDAC) 1 and 2 control the gender‐specific transcriptional regulation of human uridine diphosphate glucuronosyltransferases genes (UGT1A). Journal of Hepatology, 59(4), 797–804. 10.1016/j.jhep.2013.05.028 [DOI] [PubMed] [Google Scholar]
- Kilkenny, C. , Browne, W. , Cuthill, I. C. , Emerson, M. , & Altman, D. G. (2010). Animal research: Reporting in vivo experiments: The ARRIVE guidelines. British Journal of Pharmacology, 160, 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankisch, T. O. , Schulz, C. , Zwingers, T. , Erichsen, T. J. , Manns, M. P. , Heinemann, V. , & Strassburg, C. P. (2008). Gilbert's syndrome and irinotecan toxicity: Combination with UDP‐glucuronosyltransferase 1A7 variants increases risk. Cancer Epidemiology, Biomarkers & Prevention, 17(3), 695–701. 10.1158/1055-9965.EPI-07-2517 [DOI] [PubMed] [Google Scholar]
- Lee, C. S. , Ryan, E. J. , & Doherty, G. A. (2014). Gastro‐intestinal toxicity of chemotherapeutics in colorectal cancer: The role of inflammation. World Journal of Gastroenterology, 20(14), 3751–3761. 10.3748/wjg.v20.i14.3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Cheng, D. , Kuang, Q. , Liu, G. , & Xu, W. (2013). Association of UGT1A1*28 polymorphisms with irinotecan‐induced toxicities in colorectal cancer: A meta‐analysis in Caucasians. The Pharmacogenomics Journal, 14(2), 120–129. 10.1038/tpj.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macías‐Pérez, J. R. , Beltrán‐Ramírez, O. , & Villa‐Treviño, S. (2012). Searching for analogues of the natural compound, caffeic acid phenethyl ester, with chemprotective activity. London, UK: A Compendium of Essays on Alternative Therapy. [Google Scholar]
- Maier, T. , Guell, M. , & Serrano, L. (2009). Correlation of mRNA and protein in complex biological samples. FEBS Letters, 583(24), 3966–3973. 10.1016/j.febslet.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Marquez, N. , Sancho, R. , Macho, A. , Calzado, M. A. , Fiebich, B. L. , & Munoz, E. (2004). Caffeic acid phenethyl ester inhibits t‐cell activation by targeting both nuclear factor of activated t‐cells and NF‐κB transcription factors. The Journal of Pharmacology and Experimental Therapeutics, 308(3), 993–1001. [DOI] [PubMed] [Google Scholar]
- Mattila, P. , & Kumpulainen, J. (2002). Determination of free and total phenolic acids in plant‐derived foods by hplc with diode‐array detection. Journal of Agricultural and Food Chemistry, 50(13), 3660–3667. 10.1021/jf020028p [DOI] [PubMed] [Google Scholar]
- Natarajan, K. , Singh, S. , Burke, T. R. Jr. , Grunberger, D. , & Aggarwal, B. B. (1996). Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF‐κB. Proceedings of the National Academy of Sciences of the United States of America, 93(17), 9090–9095. 10.1073/pnas.93.17.9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preedy, V. R. (2014). Coffee in health and disease prevention. San Diego, United States: Elsevier Science. [Google Scholar]
- Rajendra Prasad, N. , Karthikeyan, A. , Karthikeyan, S. , & Reddy, B. V. (2011). Inhibitory effect of caffeic acid on cancer cell proliferation by oxidative mechanism in human ht‐1080 fibrosarcoma cell line. Molecular and Cellular Biochemistry, 349(1–2), 11–19. 10.1007/s11010-010-0655-7 [DOI] [PubMed] [Google Scholar]
- Ross, G. W. , Abbott, R. D. , Petrovitch, H. , Morens, D. M. , Grandinetti, A. , Tung, K. H. , … White, L. R. (2000). Association of coffee and caffeine intake with the risk of parkinson disease. JAMA, 283(20), 2674–2679. 10.1001/jama.283.20.2674 [DOI] [PubMed] [Google Scholar]
- Rtibi, K. , Selmi, S. , Grami, D. , Sebai, H. , Amri, M. , & Marzouki, L. (2017). Irinotecan chemotherapy‐induced intestinal oxidative stress: Underlying causes of disturbed mucosal water and electrolyte transport. Pathophysiology, 24(4), 275–279. 10.1016/j.pathophys.2017.07.002 [DOI] [PubMed] [Google Scholar]
- Sakai, H. , Diener, M. , Gartmann, V. , & Takeguchi, N. (1995). Eicosanoid‐mediated Cl− secretion induced by the antitumor drug, irinotecan (CPT‐11), in the rat colon. Naunyn‐Schmiedeberg's Archives of Pharmacology, 351(3), 309–314. [DOI] [PubMed] [Google Scholar]
- Salazar‐Martinez, E. , Willett, W. C. , Ascherio, A. , Manson, J. E. , Leitzmann, M. F. , Stampfer, M. J. , & Hu, F. B. (2004). Coffee consumption and risk for type 2 diabetes mellitus. Annals of Internal Medicine, 140(1), 1–8. 10.7326/0003-4819-140-1-200401060-00005 [DOI] [PubMed] [Google Scholar]
- Sanghani, S. P. , Quinney, S. K. , Fredenburg, T. B. , Sun, Z. , Davis, W. I. , Murry, D. J. , … Bosron, W. F. (2003). Carboxylesterases expressed in human colon tumor tissue and their role in CPT‐11 hydrolysis. Clinical Cancer Research, 9(13), 4983–4991. [PubMed] [Google Scholar]
- Sato, Y. , Itagaki, S. , Kurokawa, T. , Ogura, J. , Kobayashi, M. , Hirano, T. , … Iseki, K. (2011). In vitro and in vivo antioxidant properties of chlorogenic acid and caffeic acid. International Journal of Pharmaceutics, 403(1–2), 136–138. 10.1016/j.ijpharm.2010.09.035 [DOI] [PubMed] [Google Scholar]
- Strassburg, C. P. , Kalthoff, S. , & Ehmer, U. (2008). Variability and function of family 1 uridine‐5′‐diphosphate glucuronosyltransferases (UGT1A). Critical Reviews in Clinical Laboratory Sciences, 45(6), 485–530. 10.1080/10408360802374624 [DOI] [PubMed] [Google Scholar]
- Strassburg, C. P. , Nguyen, N. , Manns, M. P. , & Tukey, R. H. (1999). UDP‐glucuronosyltransferase activity in human liver and colon. Gastroenterology, 116(1), 149–160. 10.1016/s0016-5085(99)70239-8 [DOI] [PubMed] [Google Scholar]
- Touchefeu, Y. , Montassier, E. , Nieman, K. , Gastinne, T. , Potel, G. , Bruley des Varannes, S. , … de La Cochetière, M. F. (2014). Systematic review: The role of the gut microbiota in chemotherapy‐ or radiation‐induced gastrointestinal mucositis—Current evidence and potential clinical_ applications. Alimentary Pharmacology & Therapeutics, 40(5), 409–421. [DOI] [PubMed] [Google Scholar]
- Tukey, R. H. , Strassburg, C. P. , & Mackenzie, P. I. (2002). Pharmacogenomics of human UDP‐glucuronosyltransferases and irinotecan toxicity. Molecular Pharmacology, 62(3), 446–450. 10.1124/mol.62.3.446 [DOI] [PubMed] [Google Scholar]
- Wang, C. Y. , Mayo, M. W. , Korneluk, R. G. , Goeddel, D. V. , & Baldwin, A. S. Jr. (1998). Nf‐κB antiapoptosis: Induction of TRAF1 and TRAF2 and c‐IAP1 and c‐IAP2 to suppress caspase‐8 activation. Science, 281(5383), 1680–1683. 10.1126/science.281.5383.1680 [DOI] [PubMed] [Google Scholar]
- Xu, Y. , & Villalona‐Calero, M. A. (2002). Irinotecan: Mechanisms of tumor resistance and novel strategies for modulating its activity. Annals of Oncology, 13(12), 1841–1851. [DOI] [PubMed] [Google Scholar]
- Zhang, P. , Tang, Y. , Li, N. G. , Zhu, Y. , & Duan, J. A. (2014). Bioactivity and chemical synthesis of caffeic acid phenethyl ester and its derivatives. Molecules, 19(10), 16458–16476. 10.3390/molecules191016458 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Increasing concentrations of CAPE were not able to induce UGT1A3 reporter gene expression significantly in comparison to Basic control vector in KYSE70 cells. The screen was performed in three independent experiments each analyzing three wells.


