Abstract
The trimethylamine methyltransferase MttB is the first described member of a superfamily comprising thousands of microbial proteins. Most members of the MttB superfamily are encoded by genes that lack the codon for pyrrolysine characteristic of trimethylamine methyltransferases, raising questions about the activities of these proteins. The superfamily member MtcB is found in the human intestinal isolate Eubacterium limosum ATCC 8486, an acetogen that can grow by demethylation of l-carnitine. Here, we demonstrate that MtcB catalyzes l-carnitine demethylation. When growing on l-carnitine, E. limosum excreted the unusual biological product norcarnitine as well as acetate, butyrate, and caproate. Cellular extracts of E. limosum grown on l-carnitine, but not lactate, methylated cob-(I)alamin or tetrahydrofolate using l-carnitine as methyl donor. MtcB, along with the corrinoid protein MtqC and the methylcorrinoid:tetrahydrofolate methyltransferase MtqA, were much more abundant in E. limosum cells grown on l-carnitine than on lactate. Recombinant MtcB methylates either cob(I)alamin or Co(I)-MtqC in the presence of l-carnitine and, to a much lesser extent, γ-butyrobetaine. Other quaternary amines were not substrates. Recombinant MtcB, MtqC, and MtqA methylated tetrahydrofolate via l-carnitine, forming a key intermediate in the acetogenic Wood–Ljungdahl pathway. To our knowledge, MtcB methylation of cobalamin or Co(I)-MtqC represents the first described mechanism of biological l-carnitine demethylation. The conversion of l-carnitine and its derivative γ-butyrobetaine to trimethylamine by the gut microbiome has been linked to cardiovascular disease. The activities of MtcB and related proteins in E. limosum might demethylate proatherogenic quaternary amines and contribute to the perceived health benefits of this human gut symbiont.
Keywords: bacterial metabolism, microbiology, energy metabolism, folate, microbiome, enzyme catalysis, cobalamin, one-carbon metabolism, carnitine, acetogenesis
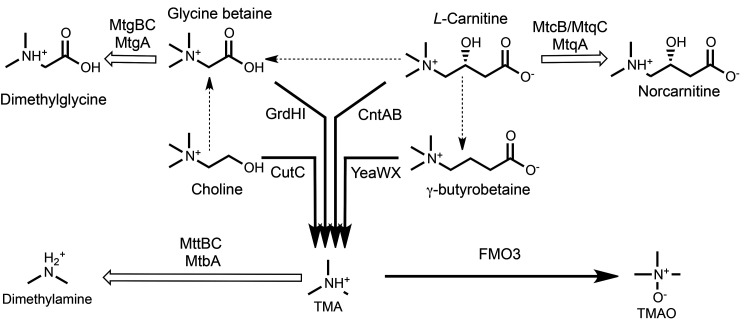
At present, ∼10,000 representatives of the MttB protein superfamily can be found in nearly 2000 different archaeal and bacterial genomes maintained at the National Center for Biotechnology. The first-described member of this large and well-distributed superfamily is the trimethylamine methyltransferase MttB, which catalyzes the corrinoid-dependent demethylation of trimethylamine (TMA) (1, 2). MttB is one of the few proteins known to possess the rare genetically encoded amino acid pyrrolysine (1, 3, 4). However, the genes encoding the vast majority of the superfamily lack the amber codon necessary for co-translational insertion of the pyrrolysine residue that is characteristic of verified TMA methyltransferases (5), leaving the function of their gene products an open question. This conundrum was in part resolved by the discovery of MtgB, a nonpyrrolysine MttB homolog from nitrite-respiring Desulfitobacterium hafniense Y51 (5). MtgB initiates the corrinoid-dependent demethylation of glycine betaine to dimethylglycine as part of a multicomponent glycine betaine:THF methyltransferase system that also requires MtgC, a corrinoid-binding protein, and MtgA, a methylcorrinoid:THF methyltransferase (see Fig. 1 for a schematic of reactions involved in quaternary amine and TMA metabolism). Highly similar homologs of MtgB, MtgC, and MtgA were recently implicated in glycine betaine demethylation catalyzed by Acetobacterium woodii, an acetogen (6). The notable sequence divergence among MttB superfamily members led Ticak et al. (5) to hypothesize that different members of the family may have evolved specificity for other quaternary amines beyond glycine betaine. If so, the impact of the MttB superfamily could be significant in environments where organisms encoding nonpyrrolysine MttB family members are found. One such environment is the human intestine (5), where the metabolism of quaternary amines by members of the microbiome is now known to have a significant effect on human health (7).
Figure 1.
Microbial quaternary amine metabolism emphasizing reactions known to lead toward (black arrows) or away from (white arrows) TMA and TMAO production as described in the introduction. Pathways with two or more enzymes converting one quaternary amine into another are indicated by dashed arrows. The demethylation of l-carnitine to form norcarnitine is demonstrated for the first time here. Notably, members of the MttB superfamily are involved in all reactions known to lead away from TMA production.
In this work, we demonstrate for the first time that an MttB family member is an l-carnitine methyltransferase. l-Carnitine is widely used by eukaryotic cells for transport of fatty acids into the mitochrondria. As a result, l-carnitine is commonly found in many foodstuffs as a component of an omnivorous diet. Following ingestion, l-carnitine, as well as other quaternary amines such as choline and glycine betaine (8, 9), enter the intestines, where they are either absorbed by the host or converted by members of the gut microbiota into TMA (10). In the gut, l-carnitine is primarily dehydrated and reduced to γ−butyrobetaine, which is then converted to TMA (11, 12). Once in the bloodstream, TMA is transported to the liver and converted into trimethylamine N-oxide (TMAO), predominantly by flavin monooxygenase 3 (13). High serum levels of TMAO have been shown to promote formation of atherosclerotic plaques in a mouse model (8). Furthermore, serum levels of TMAO were significantly correlated with the incidence of heart attack, stroke, and death in a clinical population (14). TMAO, as well as choline and l-carnitine, are further associated with increased risk for atherosclerosis (9). Loss-of-function mutations of FMO3 itself can lead to trimethylaminuria, whose sufferers emit the odor of unmetabolized TMA (15).
The health effects of TMA and TMAO have led to renewed interest in understanding quaternary amine degradation by members of the gut microbiome. Choline and l-carnitine have long been known to be converted to glycine betaine (16, 17), which can be then cleaved by betaine reductase to form TMA (18). It is indicative of our relative lack of knowledge concerning the potential for methylamine metabolism by the microbiota that only recently have other enzymes been found that act directly on quaternary amines to generate TMA. Choline was shown to be directly converted to TMA by CutC, a glycyl radical enzyme that acts as a choline-TMA lyase (19). TMA may be produced from l-carnitine in a single step by the l-carnitine monoxygenase CntAB (20). l-Carnitine can be converted to γ−butyrobetaine, which the oxygenase YeaWX can convert to TMA (11). YeaWX also has some activity with l-carnitine and choline. Recent work has indicated that an anoxic uncharacterized pathway for TMA production from γ-butyrobetaine also exists in the gut microbiome (12). Inhibitors of the major enzymes of quaternary amine degradation have been proposed as drugs to potentially decrease the net production of TMA in the gastrointestinal tract (21, 22).
Microbes have also been proposed to control net gut TMA production, and thus net TMAO levels, in humans. Supporting the idea that microbiota influence TMAO levels, introduction of “humanized” gut microbiomes into germ-free mice led to differential production of TMAO (23), and atherosclerosis susceptibility could be transmitted via transplantation of gut microbiota (24). Different microbes thus might contribute to or interfere with the net synthesis of TMA (25). Only one group of microbes that might diminish TMA production has been previously identified (26, 27); the methanogenic archaea inhabiting the gut, whose genomes encode the pyrrolysyl-protein MttB, the TMA methyltransferase (1, 2). Another route that might conceivably limit TMA production would be competition for the quaternary amines that are precursors to TMA. However, such routes of quaternary amine degradation that would not eventually yield TMA under anaerobic conditions have been unknown, save for one, the demethylation of glycine betaine by the nonpyrrolysine (nonPyl) MttB family member, MtgB (5).
Here we show that Eubacterium limosum ATCC 8486, an acetogenic and butyrogenic human gut isolate, consumes l-carnitine to produce norcarnitine. The latter is, to our knowledge, a novel biological product. The nonPyl MttB family member MtcB, along with a corrinoid protein and a corrinoid-dependent THF methyltransferase, were significantly more abundant in cells grown on l-carnitine than on lactate. These three proteins together catalyzed the methylation of THF with l-carnitine, thus providing a key intermediate toward the catabolic synthesis of acetate, butyrate, and carproate. MtcB initiates THF methylation by methylation of an abundant corrinoid protein specifically with l-carnitine. These results expand the known substrates of the MttB superfamily to include a proatherogenic dietary component and reveal a novel anoxic mechanism of l-carnitine degradation via demethylation.
Results
Demethylation of l-carnitine during growth of E. limosum
E. limosum strains have previously been reported to grow utilizing glycine betaine or choline (28). We found that E. limosum ATCC 8486 can also utilize l-carnitine as a growth substrate. Growth with l-carnitine was best in a medium supplemented with yeast extract and casamino acids with a doubling time of ∼6–7 h. Little or no growth was observed when E. limosum was inoculated into the same medium not supplemented with l-carnitine (Fig. 2A). E. limosum grew more slowly on l-carnitine when in a completely defined medium with a doubling time of ∼16–18 h, indicating that yeast extract and casamino acids were stimulatory but not necessary for growth with l-carnitine (Fig. S1).
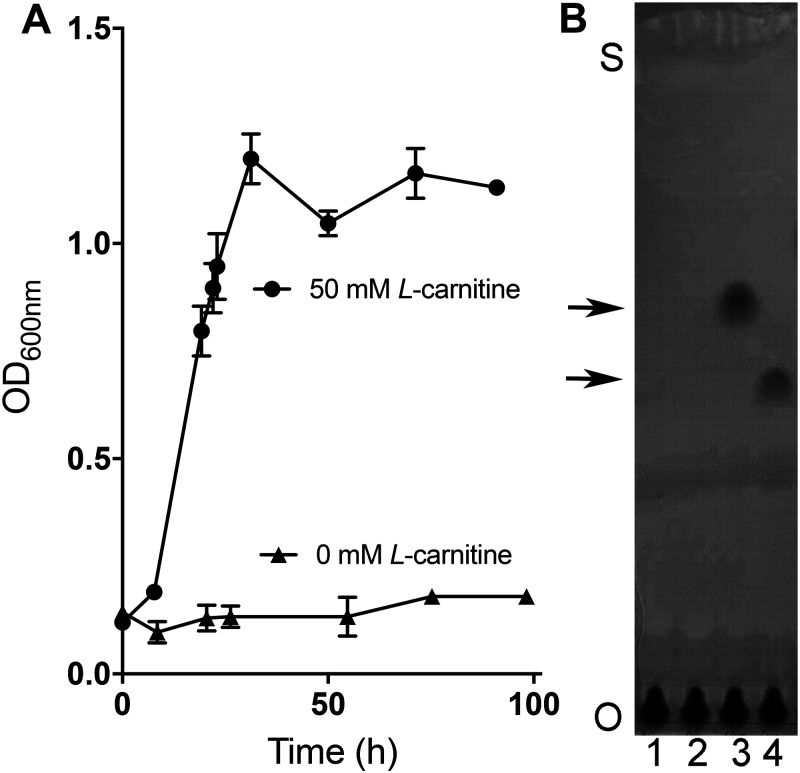
Figure 2.
E. limosum cultures accumulate norcarnitine during growth on l-carnitine. A, growth dependence of E. limosum upon the addition of 50 mm l-carnitine in LS medium supplemented with 0.1% yeast extract, 0.2% casamino acids, and 10 mm sodium acetate. Error bars, S.D. from an average of three different cultures. B, TLC of culture supernatants demonstrate that l-carnitine is consumed and norcarnitine is produced during growth of E. limosum. Cultures were inoculated into medium containing either no substrate (lanes 1 and 2) or 50 mm l-carnitine (lanes 3 and 4) as described for A. Samples were removed just after inoculation (lanes 1 and 3) and again 24 h after growth had ceased in the l-carnitine–supplemented culture (lanes 2 and 4). Samples were applied at the origin (O) and developed until solvent reached near the top of the plate (S), and then the plates stained were stained with bromocresol green. The top and bottom arrows indicate the migration positions of l-carnitine and norcarnitine standards, respectively (not shown).
E. limosum strains were reported to demethylate glycine betaine and choline during growth (28). Therefore, we examined l-carnitine–grown cultures to determine whether l-carnitine was also demethylated during growth. Culture supernatants taken before and after growth were analyzed by TLC, followed by staining with bromocresol green (Fig. 2B). l-Carnitine was not detectable after growth. Instead, a compound was present that co-migrated to a position identical to that of a norcarnitine standard. To confirm this presumptive identification, the scraped spot was extracted with solvent and submitted to mass spectral analysis. The m/z value observed for the compound eluted from TLC plates was within 3 ppm of the theoretical value for norcarnitine (Table S1). MS/MS analysis of the parent ion revealed ions with m/z values predicted for fragmentation of norcarnitine. Additionally, the supernatants from l-carnitine cultures before and after growth were analyzed by MS following chromatography on an anion-exchange cartridge, which confirmed the presence of norcarnitine after growth (Fig. S2).
Product stoichiometry of E. limosum growth on l-carnitine
E. limosum is capable of synthesizing acetyl-CoA from two one-carbon units (29) and will also produce butyrate and (in some strains) caproate from acetyl-CoA (30). Demethylation of l-carnitine did indeed support methylotrophic short-chain fatty acid production, as evidenced by the stoichiometry of l-carnitine degradation in defined medium in which l-carnitine and CO2 were the only carbon sources (aside from the defined vitamin mixture). In 10-ml cultures (n = 6), E. limosum demethylated 476 ± 51 µmol of l-carnitine to 534 ± 25 µmol of norcarnitine. In the process, 102 ± 1 µmol of CO2 were consumed to produce 77 ± 18 µmol of acetate, 78 ± 4 µmol of butyrate, and 10 ± 2 µmol of caproate. Any TMA produced was below the detection limit (∼50 μm TMA) by GC of stationary phase cultures. Total carbon recovery was 108 ± 11%. MS of the supernatant before and after growth provided no evidence of dehydration or further demethylation of l-carnitine beyond norcarnitine (Fig. S3). Given this and assuming a 1:1 stoichiometry between the l-carnitine consumed and norcarnitine produced, the carbon recovery of the methyl group of l-carnitine and CO2 in acid products was 91 ± 11%. Overall, our data support the following idealized equation for l-carnitine metabolism by E. limosum: 64 l-carnitine + 17 CO2 → 64 norcarnitine + 12 acetate + 12 butyrate + 1.5 caproate.
Corrinoid-dependent methyltransferase activities in cell extracts
Acetogens such as E. limosum form methyl-THF as an obligate step in the synthesis of acetate (29, 31). During growth on methylated substrates, methyl-THF is synthesized by substrate-specific multicomponent methyltransferase systems (32–35). Methyl transfer to THF consists of three components: 1) a methylated substrate:corrinoid protein methyltransferase, 2) a corrinoid-binding protein homologous to the cobalamin-binding domain of methionine synthase (36, 37), and 3) a methylcorrinoid protein:THF methyltransferase. During catalysis, the corrinoid protein cofactor undergoes multiple cycles, alternating between the unmethylated Co(I) state and the methyl-Co(III) form. Adventitious oxidation of the Co(I)-corrinoid necessitates reactivation by an ATP-dependent reductive activation protein (sometimes referred to as a fourth component), which reduces the Co(II)-corrinoid back to the Co(I) state (38–40). However, not all corrinoid proteins require the activating protein for in vitro activity and instead can be reduced to Co(I) using a low potential chemical reductant (32, 41).
To determine whether a corrinoid-dependent methyltransferase system might underlie l-carnitine demethylation by E. limosum, we first examined l-carnitine–grown cell extracts for l-carnitine:THF methyltransferase activity and found an l-carnitine–dependent rate of 82.3 ± 4.9 nmol of methyl-THF min−1 mg of protein−1 (average of three preparations). Methylation of THF depended on the addition of extract, THF, and Ti(III)citrate, but ATP did not stimulate the reaction. The requirement of Ti(III)citrate for THF methylation by extracts is consistent with involvement of Co(I)-corrinoid–dependent methyltransferase in the reaction, so we further tested extracts for this activity by using UV-visible spectroscopy (Fig. 3A). Extracts catalyzed l-carnitine–dependent methylation of Ti(III)citrate-reduced cob(I)alamin at a rate of 94 ± 27 nmol min−1 mg of protein−1 (average of three preparations) when supplemented with l-carnitine (Fig. 3B). We did not observe methylation of cob(I)alamin when l-carnitine was omitted (Fig. 3B). Activity with choline, γ-butyrobetaine, glycine betaine, or tetramethylammonium ion was not detectable when tested as described under “Experimental procedures.” Additionally, we did not detect l-carnitine:cob(I)alamin methyltransferase activity in the extracts of cells grown on lactate (Fig. 3B).
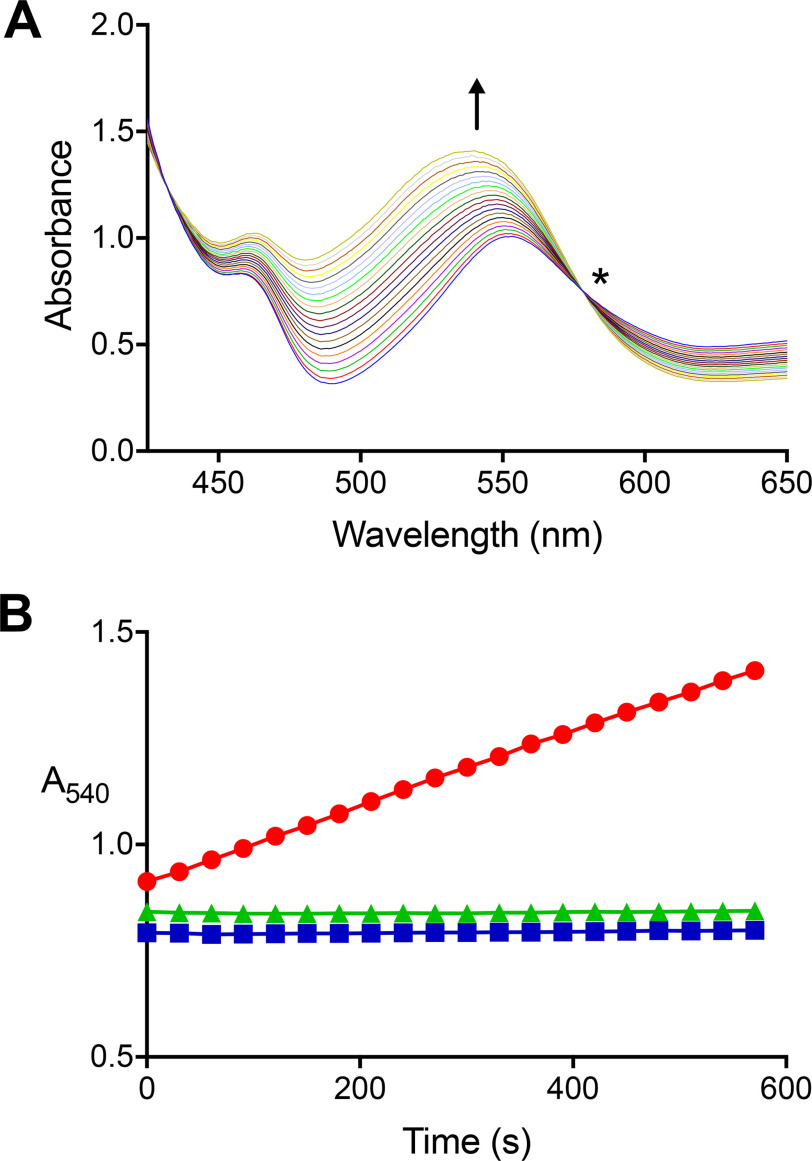
Figure 3.
A carnitine:cob(I)alamin methyltransferase activity is present in extracts of carnitine-grown, but not lactate-grown, E. limosum. A, UV-visible spectra were collected every 30 s during a single carnitine:cob(I)alamin methyltransferase reaction initiated by the addition of extract from carnitine-grown cells. The arrow indicates the direction of increased absorbance at 540 nm with time indicative of methylcob(IIII)alamin formation. The sharp isosbestic point (*) at 578 nm indicates that other cobalamin species did not accumulate appreciably during the reaction. The complete UV-visible spectrum is not shown due to the intense absorbance of Ti(III)citrate and cob(I)alamin below 425 nm. B, absorbance changes at 540 nm (●) in a single reaction containing l-carnitine, cob(I)alamin, and extract of l-carnitine–grown cells. No reaction was observed if l-carnitine was omitted (■) or if l-carnitine–grown cell extract was replaced with lactate-grown cell extract (▴).
Candidates for mediating l-carnitine:THF methyl transfer in E. limosum
Cell extracts from l-carnitine– but not lactate–grown cells catalyzed formation of methyl-cob(III)alamin with l-carnitine. Therefore, to identify candidate proteins that might mediate this activity and couple it to THF methylation, we identified proteins found in cells grown with l-carnitine using label-free proteomic analysis (see Tables S2–S4). We compared this data set with one we described previously from cells growing on lactate (42), focusing on proteins potentially involved in catabolism (Table S2). For each substrate, four separate cultures were grown to mid-log phase prior to individual harvest, lysis, tryptic digestion, and peptide analysis by LC–MS/MS using the same methodology. The relative abundances of proteins in each sample were determined from the percentage of summed emPAI values (43).
Catabolic proteins are generally present at high levels in anaerobes, and high-abundance proteins involved in acetate and butyrate formation were readily identified in cells grown on either lactate or l-carnitine. Abundant catabolic proteins specific to growth on each substrate were also identified (Table S2). For example, in lactate-grown cells, a lactate dehydrogenase and associated proteins (44, 45) were abundant among the ∼1630 proteins identified. These same proteins were at very low abundance or not detectable among the ∼1400 proteins identified from cells grown on l-carnitine. Conversely, homologs of the two methyltransferases and corrinoid proteins involved in multicomponent systems for THF methylation were abundant when cells were grown on l-carnitine, but much less so when grown on lactate. Therefore, these proteins were considered as candidates for an l-carnitine:THF methyltransferase system. The proteins included an MttB superfamily member (WP_038351887.1) that we designated MtcB, which constituted 2.9 ± 0.87 mol % of detected proteins from l-carnitine–grown cells. MtcB was not detectable in lactate-grown cells, and a t test indicated that the difference in MtcB abundance between lactate– and l-carnitine–grown cells had a low probability of occurring randomly (p = 0.00058). Of the genes located near mtcB in the E. limosum genome (Fig. S3), only the product of the adjacent downstream gene (WP_038351886.1) was detectable in cells grown on l-carnitine, but not on lactate (Table S2). This protein, a member of the major facilitator family (MFS) of transporters, was present at low abundance in the l-carnitine proteome. It should be noted that membrane proteins are generally underrepresented by the proteomic protocol employed here.
The corrinoid cofactor methylated by MttB superfamily members is bound to a discrete corrinoid-binding protein. Three homologs of methylotrophic corrinoid proteins were detected in l-carnitine–grown cells. Two of these proteins were not abundant (<0.001 mol %). The third (WP_038352545.1) was present at a relative abundance of 0.64 ± 0.38 mol % but was only 0.0083 ± 0.004 mol % of protein in lactate-grown cells, indicating a 76-fold increase (p = 0.016) increase in abundance when l-carnitine was the growth substrate. We have designated this corrinoid protein MtqC. Additionally, an ATP-dependent reductive activation protein belonging to a superfamily defined largely as COG3894 was identified (WP_038351871.1, designated RamQ) was 14-fold (p = 0.0001) more abundant during growth on l-carnitine (0.13 ± 0.02% of protein) when compared with growth on lactate (0.009 ± 0.003% of protein).
The E. limosum ATCC 8486 genome (46) encodes five proteins homologous to the MetH domain that carries out the methylcobalamin:THF methyltransferase subreaction of methionine synthase (47). We detected two of these enzymes in our proteomic data sets. The first (WP_013381869.1) is encoded in a gene cluster that contains the components of carbon monoxide dehydrogenase/acetyl-CoA synthase and is therefore most likely the corrinoid iron-sulfur protein methyltransferase (AcsE) in E. limosum. Although quite abundant in l-carnitine–grown cells (3.5± 0.7% of detected protein), its abundance was also high during growth on lactate (6.2 ± 1.6% of detected protein). The second MetH-like enzyme detected in l-carnitine–grown cells is WP_038351870.1, which we have designated MtqA. MtqA is 0.10 ± 0.032% of detected protein in lactate-grown cells but increased to 3.5 ± 0.65 mol %, in l-carnitine–grown cells, representing a 35-fold (p = 0.00010) increase.
MtcB is an l-carnitine–dependent corrinoid methyltransferase
Due to its abundance in l-carnitine–grown cells, we hypothesized that the MttB family member MtcB might be an l-carnitine:corrinoid methyltransferase. Therefore, we expressed the mtcB gene in Escherichia coli and purified recombinant MtcB (Fig. S4), which we found did carry out a robust l-carnitine:cob(I)alamin–dependent methyltransferase reaction with spectral changes identical to those observed for the same reaction mediated by cell extract. We monitored the reaction at 540 nm and found that the reaction was completely dependent on l-carnitine and MtcB (Fig. 4A). The enzyme displayed saturation kinetics with both substrates with varying l-carnitine and cob(I)alamin concentrations (Fig. 4B). A series of double reciprocal plots (1/v against 1/[l-carnitine]) at different cob(I)alamin concentrations (Fig. S5) generated a pattern typical of a sequential enzyme mechanism. The enzyme had a Vmax of 2.10 (95% CI, 1.98–2.24) µmol min−1 mg−1 MtcB and apparent Km values for l-carnitine and cob(I)alamin corresponding to 0.40 (95% CI, 0.20–0.60) mm and 1.34 (95% CI, 1.24–1.45) mm, respectively.
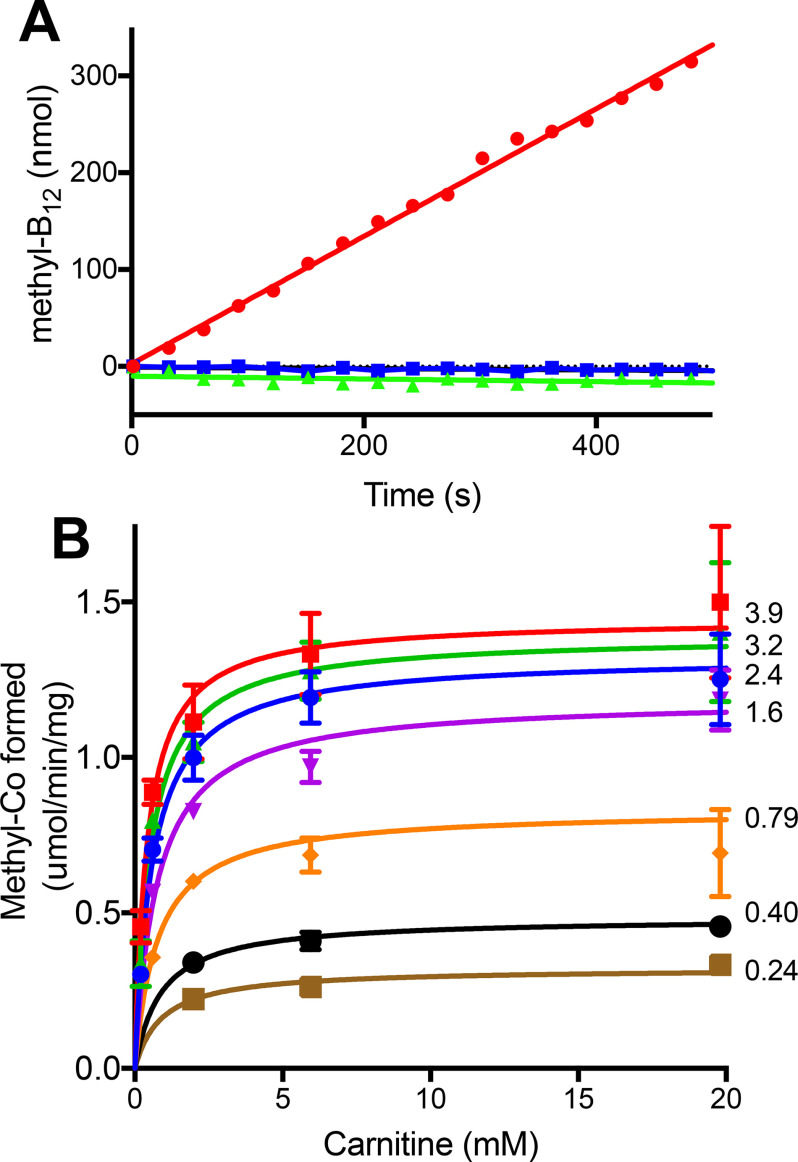
Figure 4.
Purified recombinant MtcB-dependent methylation of cob(I)alamin with l-carnitine. A, methylation was followed by the increase at 540 nm in the complete reaction (●) as described under “Experimental procedures.” Methylation was not observed in the absence of MtcB (▴) or l-carnitine (■). B, kinetic analysis of MtcB in which the l-carnitine concentration was varied at different set cob(I)alamin concentrations. The numbers beside each curve are the millimolar concentration of cob(I)amin used for that data set. Each point is the average of three determinations. Error bars, S.D. Some error bars are not visible due to size of data point markers.
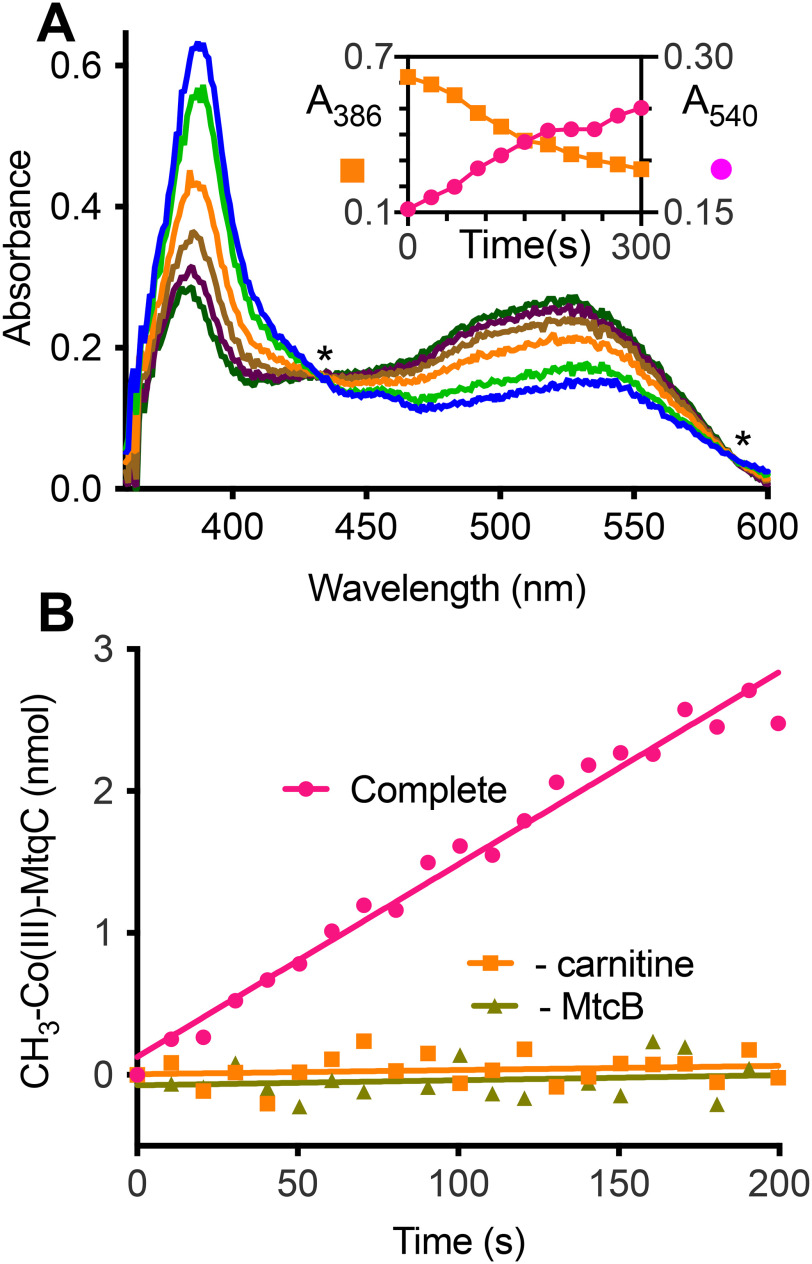
We next tested to see whether MtcB could use l-carnitine to methylate MtqC, the corrinoid protein of increased abundance in l-carnitine–grown cells. Recombinant MtqC was produced as an apoprotein from E. coli; therefore, we reconstituted the purified protein (Fig. S4) with cobalamin prior to further purification. Incubation of the MtqC holoprotein with recombinant RamQ (Fig. S4), Ti(III)citrate, and ATP generated a stable pool of Co(I)-MtqC. Upon subsequent addition of l-carnitine and MtcB, methylation of Co(I)-MtqC occurred as indicated by the disappearance of the 387 nm peak corresponding to Co(I)-MtqC and appearance of a peak at 532 nm corresponding to methyl-Co(III)-MtqC (Fig. 5A). The reaction depended on both l-carnitine and MtcB (Fig. 5B). During the reaction, we observed an isosbestic point near 585 nm indicating that the conversion of Co(I)-MtqC to methyl-Co(III)-MtqC occurred without accumulation of an appreciable amount of a spectrally distinct intermediate. MtcB methylated MtqC with 4 mm l-carnitine at an average rate of 14 µmol min−1 mg−1 (n = 2).
Figure 5.
MtcB methylates MtqC with l-carnitine. A, UV-visible spectral changes associated with methylation of 50 μm Co(I)-MtqC by MtcB with 40 mm l-carnitine. Spectra shown were taken immediately after initiation (light blue spectrum) of a single reaction and every 60 s thereafter until termination (dark green spectrum). Asterisks indicate locations of isosbestic points for the Co(I)-MtqC and methyl-Co(III)-MtqC spectra. Inset, the progress of the reaction was monitored by increase in absorbance at 540 nm accompanied by the decrease in absorbance at 386 nm. B, methyl-Co(III)-MtqC formation during a complete reaction described under “Experimental procedures” was determined using the calculated extinction coefficient for methyl-Co(III)-MtqC. MtqC methylation was not detectable in the absence of l-carnitine or MtcB. Each curve represents a single representative reaction.
No MtcB-dependent methylation of Co(I)-MtqC was detectable with 100 mm choline, glycine betaine, proline betaine, TMA, or tetramethylammonium as methyl donors. However, we found that the purified enzyme did use γ-butyrobetaine (an analog of l-carnitine in which the β-hydroxy group is absent) as a substrate for methylation of MtqC. However, the rate of methylation of MtqC by MtcB with 65 mm γ-butyrobetaine was 0.5 µmol min−1 mg−1, a rate considerably lower than the rate of the enzyme with l-carnitine.
In vitro reconstitution of the l-carnitine:THF methyltransferase reaction with recombinant MtcB, MtqC, and MtqA
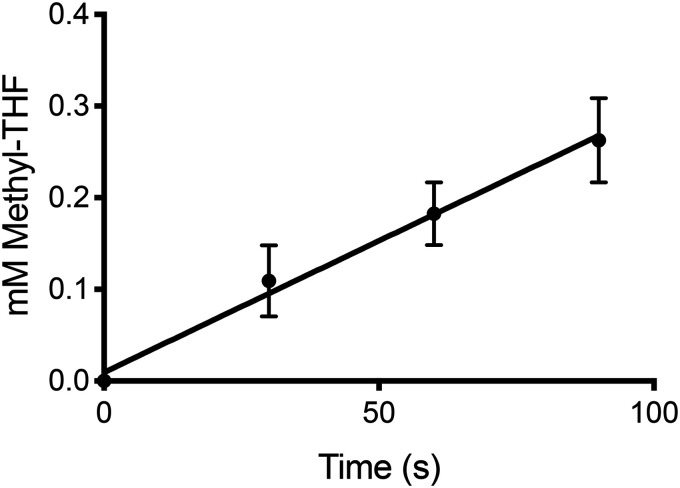
The preceding experiments revealed that MtcB could methylate MtqC in an l-carnitine–dependent manner and that both proteins were more abundant when cells were grown on l-carnitine. The abundance of MtqA, a homolog of the MetH methylcobalamin:THF methyltransferase domain, was also significantly increased when cells were grown on l-carnitine, and therefore we purified recombinantly produced MtqA (Fig. S4) to test whether this protein might participate in methyl transfer between l-carnitine and THF. The addition of MtqA to MtcB and MtqC allowed methylation of THF with carnitine. Equimolar (2 μm) amounts of MtcB and MtqA incubated with a 12.5-fold molar excess of MtqC catalyzed l-carnitine–dependent THF methylation at a rate of 188 ± 38 nmol min−1 mg−1 total protein (Fig. 6). Methyl-THF formation was not detectable in reactions lacking either MtcB, MtqC, or MtqA. As MtcB can methylate MtqC independently of MtqA, the requirement of MtqA for THF methylation indicates that methyl-Co(III)-MtqC is utilized as a substrate to methylate THF at a rate of at least 2.8 µmol min−1 mg−1 MtqA under the conditions of the assay. In contrast to the two methyltransferases and the corrinoid protein, RamQ was not required for the reaction but stimulated the rate of THF methylation. Controls lacking RamQ or ATP produced 38 and 39%, respectively, of the methyl-THF formed in assays supplemented with RamQ. This again indicates that Ti(III)citrate was capable of reducing sufficient MtqC to the active Co(I) form in the absence of RamQ-mediated reduction.
Figure 6.
In vitro reconstitution of l-carnitine:THF methyl transfer using recombinant MtcB, MtqC, and MtqA. Each time point is the average from three independent reactions and determinations. Error bars, S.D.
Discussion
A major stumbling block to precise annotation of microbial genomes and subsequent fuller understanding of the metabolic potential within metagenomes remains our yet incomplete understanding of metabolism. This is especially true for genes encoding members of protein superfamilies that have undergone functional diversification and are therefore sometimes subject to overly specific assignments during annotation that are not justified by retention of essential residues (48). The MttB superfamily is proving to be a case in point. Genes encoding superfamily members are often annotated as TMA methyltransferases due to their similarity to MttB, despite the lack of a pyrrolysine codon that distinguishes the gene encoding MttB, the first member of the superfamily with a described function (1, 3, 4). Many intestinal isolates bear genes encoding nonPyl MttB superfamily members in their genomes. Here we show that a fecal isolate of E. limosum, whose genome encodes 42 nonPyl MttB superfamily members, can grow by demethylation of l-carnitine. MtcB is the only one of these nonPyl MttB homologs that becomes abundant during growth on carnitine, and it acts as an l-carnitine:cobalamin methyltransferase.
MtcB is a strong candidate as a primary entry point of methyl groups from l-carnitine into the acetogenic metabolism of E. limosum. This idea is supported by the relative abundance of MtcB in cells grown on l-carnitine relative to lactate, as well as the favorable kinetics for the l-carnitine–dependent methylation of MtqC by MtcB. Upon the addition of MtqA, MtcB initiates the rapid in vitro methylation of THF relative to the abundance of the methyltransferase (Fig. 7). l-Carnitine–dependent methylation of THF would be crucial for methylotrophic acetogenesis with l-carnitine as the major growth substrate but would not be required for the proposed pathway of acetogenesis from lactate (45) in which lactate is initially catabolized by a regulated lactate dehydrogenase (44). Correspondingly, MtcB is only abundant in cells grown on l-carnitine and not on lactate.
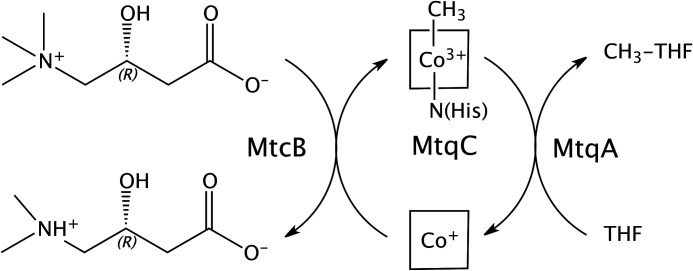
Figure 7.
Schematic of l-carnitine demethylation and methylation of THF by MtcB, MtqC, and MtqA from E. limosum. The ligation of the four nitrogens of the corrin tetrapyrrole ring bound to MtqC is indicated by the square surrounding the cobalt. In the methyl-Co(III) form, the lower axial ligand is a nitrogen provided by a histidine residue that is highly conserved in corrinoid-binding proteins and domains involved in methyltransferase reactions (36).
MtcB is also not abundant in cells grown on another methylotrophic substrate, proline betaine. Recently our laboratory described the ability of MtpB, another E. limosum MttB superfamily member, to demethylate proline betaine (42). MtcB is undetectable in the proteome of proline betaine-grown cells (42). Conversely, MtpB was 3.8 mol % protein in cells grown with proline betaine (42) but 1500-fold less abundant in the l-carnitine–grown cells examined here. MtcB and MtpB also display distinct substrate specificity toward their respective quaternary amines. MtcB's robust activity with l-carnitine contrasts with its inability to detectably demethylate proline betaine. In comparison, MtpB has only trace activity with l-carnitine, ∼0.0005% of the activity with proline betaine as substrate (42). MtcB and MtpB thus superbly illustrate the extent to which members of the MttB superfamily have diversified to utilize distinctive quaternary amines as substrates. Nonetheless, it is of note that MtpB and MtcB both appear to utilize their respective quaternary amines to methylate the same corrinoid protein, MtqC. That these two methyltransferases can utilize the same corrinoid protein is not overly surprising, given that these MttB superfamily members share 45% identity over most of their length and could easily share common determinants for specifically recognizing the same corrinoid protein. The utilization of the same corrinoid protein for both THF methylation systems also would explain why the same activation protein, RamQ, was identified as more abundant when cells were grown on either quaternary amine, relative to growth on lactate.
The utilization of MtqA, MtqC, and RamQ in methylating THF with distinct quaternary amines is reflected in the genomic context of these genes. For example, both ramQ and mtqA are located near a number of genes encoding enzymes involved in the oxidation of methyl-THF (42), an obligate pathway for methylotrophic acetogenesis (49). The methyltransferases are found in other parts of the genome. The mtcB gene encoding the l-carnitine methyltransferase is found among genes with a general role in utilization of cationic amino acids (Fig. S4). Immediately upstream of the mtcB gene is encoded WP_052237179, a homolog of RocR that in Bacillus subtilis controls the catabolism of the cationic amino acid arginine (50). Downstream of mtcB are encoded several proteins involved in transport. Searches of the Transporter Classification Database (51) revealed WP_038351886, encoded immediately adjacent to mtcB, as a member of the MFS, specifically a member of the glycerophosphodiester uptake family, only one of which has a documented substrate, glycerophosphocholine (52). Further downstream are also found encoded WP_13378691 and WP_013378692, homologs of AzlCD involved in the export of branch-chain amino acids, including 4-azaleucine, an alanine homolog bearing a dimethylamino substituent (53, 54). However, as mentioned previously, of the proteins encoded near mtcB, only the MFS family member was detectable in our proteomic database in l-carnitine–grown cells (Table S2).
The majority of the l-carnitine consumed by E. limosum was recoverable as the singly demethylated product, norcarnitine. Excretion of singly demethylated products has been typical of demethylation of quaternary amines by this acetogen (5, 28, 42). However, with l-carnitine, the end product is to our knowledge a novel catabolite. Most references in the literature refer to norcarnitine as a synthetic compound tested as an alternative substrate for l-carnitine–dependent enzymes, such as l-carnitine acetyltransferase (55) or palmityltransferase (56). However, among other l-carnitine derivatives, norcarnitine was shown to serve as sole carbon and nitrogen source for the facultative anaerobe Psuedomonas putida (57), suggesting that the norcarnitine produced by E. limosum during growth on l-carnitine is likely to be degraded by other members of the gut microbiota.
One of the most important implications of this work is that it reveals for the first time the potential of gut microbes having l-carnitine:corrinoid methyltransferase activity to positively affect human health. Work over the past decade has revealed that serum TMAO levels correlate with an increased risk of cardiovascular disease (58) and serum TMAO originates in the TMA excreted by gut microbes metabolizing quaternary amines, such as l-carnitine and its derivative γ-butyrobetaine (9, 11, 12). The demethylation of l-carnitine by MtcB represents the first instance of l-carnitine metabolism producing a catabolic product that cannot be readily converted into TMA. Indeed, quaternary amine demethylation by MtcB and perhaps other MttB superfamily members in different microbes might serve as a natural or therapeutic route to limit the production of TMA from dietary quaternary amines in the gut and thereby limit proatherogenic TMAO accumulation in the serum. It is interesting in this regard that E. limosum itself is widely regarded as a beneficial organism (33, 59, 60) and has been proposed as a marker of human longevity due to its significantly increased abundance in the microbiotia of centenarians relative to younger groups (61). This leads us to speculate that demethylation of proatherogenic quaternary amines by MtcB, and perhaps other MttB family members, may be a factor underlying the perceived health benefits of E. limosum as a human gut symbiont.
Experimental procedures
l-Carnitine demethylation during growth
E. limosum ATCC 8486 was cultured at 37 °C on the E. limosum defined medium supplemented with yeast extract, casamino acids, and sodium acetate as indicated (supporting Experimental procedures). Supernatant from a culture was separated on silica gel 60 TLC plates followed by detection of quaternary amines and demethylation product(s) by bromocresol green as described previously (5). Norcarnitine used as a standard was synthesized via demethylation of l-carnitine with thiophenolate (supporting Experimental procedures). To confirm the identity of demethylation products, chromatographed supernatant was left unstained, and the area with an Rf corresponding to the demethylation product was extracted for mass spectral analysis on a Bruker MaXis ESI Q-TOF mass spectrometer (Bruker, Billerica, MA, USA) as detailed in the supporting Experimental procedures.
Fermentation balance
Cultures in LS defined medium with 50 mm l-carnitine were grown to stationary phase and acidified by injection of 1 ml of 1 m HCl, whereas controls were acidified immediately after inoculation. Net CO2 production was then determined by GC as described previously (62). Supernatant samples taken from the cultures immediately following inoculation and just prior to acidification were analyzed for l-carnitine and norcarnitine content using TLC (5) and densitometry of unknowns and standard curves. GC was used to quantify short-chain fatty acids (63) and trimethylamine (64).
Protein preparations
Cell extracts were prepared from E. limosum grown on either dl-lactate or l-carnitine by lysis with a French pressure cell followed by centrifugation at 48,000 × g. The genes encoding MtcB, MtqC, MtqA, and RamQ were cloned into pSpeed using PIPE techniques (65). MtcB and MtqA were produced by isopropyl 1-thio-β-d-galactopyranoside induction in E. coli BL21(DES3) and purified by nickel affinity column followed by purification on a MonoQ column (GE Healthcare Life Sciences). RamQ was produced anaerobically in E. coli SG13009 (supporting Experimental procedures). The harvested cells were stored anaerobically at −80 °C until needed. RamQ was purified as described for MtcB and MtqA except in a Coy anaerobe chamber with buffers that contained 3 mm DTT for both nickel affinity and monoQ columns. MtqC was produced as an apoprotein in E. coli BL21(DES3) and was reconstituted with hydroxycobalamin following removal of the N-terminal His tag using tobacco etch virus protease. The cleaved MtqC was then incubated under N2/H2 (98:2) at 4 °C for 36 h in a solution containing 3.5 m glycine betaine, 1 mm hydroxocobalamin, 10 mm DTT in 50 mm Tris, pH 7.2. The reconstituted protein was then further purified on phenyl-Sepharose. The cobalamin content was assayed by measuring the absorbance change following dicyano derivatization and using a Δε368 nm of 1.7·104 m−1 cm−1 (66). Further details are available in the supporting Experimental procedures. Protein was assayed using the Bradford method with BSA as a standard.
Enzyme assays
All reactions were carried out under anoxic conditions and dim red light to prevent photolysis of the methyl-Co bond in corrinoid derivatives. All assays were conducted at 37 °C and the indicated concentration of phosphate buffer, pH 7.2. Methylation of cob(I)alamin by l-carnitine or other quaternary amines was measured in 2-mm cuvettes essentially as described previously for the glycine betaine:cob(I)alamin methyltransferase (5) except for use of the 75 mm potassium phosphate buffer, pH 7.2 (supporting Experimental procedures), and the substrate was l-carnitine or other quaternary amines as methylating substrates. Reactions were initiated with either cell extract (0.7-1.0 mg of total protein) or purified MtcB (0.8 μm). When the substrate specificity was tested, the reactions contained 2.3 mm cob(I)alamin, and a 63.4 mm concentration of the potential quaternary amine substrate. Kinetic parameters for MtcB l-carnitine:cob(I)alamin methyltransferase activity were collected by varying l-carnitine concentration between 0.2 and 63.4 mm and the cob(I)alamin concentration between 0.24 and 3.88 mm. Replots of the slopes and y intercepts of the double reciprocal plots of these data were used to calculate Km and Vmax values.
Assays for l-carnitine:THF methyl transfer activity were carried out in a Coy anaerobe chamber having 98% nitrogen and 2% hydrogen under dim red light. Reactions (0.6 ml) containing either cell extract (3.8 mg of total protein) or purified MtcB (2 μm), MtqC (25 μm), MtqA (2 μm), and RamQ (0.4 μm) were conducted in masked 1-ml quartz cuvettes with 1-cm pathlength. Each also contained 6 mm Ti(III) citrate, 1.25 mm MgATP, 2.3 mm tetrahydrofolate in 50 mm potassium phosphate buffer, pH 7.2, and were initiated with 50 mm l-carnitine. At time points, samples (100 μl) were withdrawn and mixed with 20 μl of saturated TCA and centrifuged, and the supernatants were frozen at −80 °C before methyl-THF concentration was determined on a Dionex Ultimate 3000 HPLC system as described previously (5). Methylation of THF with methylcobalamin by MtqA was measured using the spectral assay described previously (5).
MtcB-dependent methylation of MtqC was assayed at 37 °C in 1-cm microcuvettes and initiated with the addition of l-carnitine or other methylated amines. Each 100-μl reaction contained 2 mm Ti(III) citrate, 2 mm ATP, 51 μm MtcC, 0.35 μm RamQ, and 10 nm (with indicated concentrations of L-carnitine) or 100 nm (with 100 mm of other tested substrates) MtcB in 50 mm potassium phosphate buffer, pH 7.2. The Δε540 for the conversion of Co(I)-MtqC to methyl-Co(III) MtqC was determined to be 3032 ± 198 m·cm−1.
Proteomics
The experimental l-carnitine data set obtained here is compared with the same lactate data set described in a previous study (42). Both data sets were obtained with identical procedures. Two sets of four replicate cultures were grown in defined media on either 50 mm dl-lactate or l-carnitine in LS medium to mid-log phase (OD600 ≈ 0.45). The harvested cell pellets were independently subjected to lysis and protein extraction prior to tryptic digestion. Peptides (12 μg) were separated on a two-dimensional liquid chromatography system prior to introduction of the fractionated peptides into an Orbitrap Fusion Mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) for MS/MS peptide sequencing and identification. The merged data were searched against the closed E. limosum ATCC 8486 genome (46) using Mascot Daemon 2.5.1 (Matrix Science, Boston, MA, USA). Scaffold (Proteomic Software, Inc., Portland, OR, USA) was used to compile data and assign emPAI values (43) based on peptide counts for estimation of the mol % of each protein relative to the total set of identified proteins in each proteome. For more details, see the supporting Experimental procedures.
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (67) with the data set identifier PXD013806 (for the lactate data set) and with the identifier PXD013961 (for the l-carnitine data set). All other data cited can be found in the article.
Supplementary Material
Acknowledgments
We express our appreciation to Katherine Huening for development of the protocol for production of recombinant Ram proteins. We also thank Andrew VanSchoiack and Yu Cao from the Ohio State University Campus Chemical Instrument Center for assistance in performing mass spectrometric analysis of demethylation products as well as Jared Ellenbogen and Shane Hendricks for assistance with one of the growth experiments. The Fusion Orbitrap instrument used for the proteomics study was supported by National Institutes of Health Grant S10 OD018056.
This article contains supporting information.
Author contributions—D. J. K., L. Z., and J. A. K. data curation; D. J. K., E. J. B., L. Z., and J. A. K. formal analysis; D. J. K., E. J. B., L. Z., and J. A. K. investigation; D. J. K., L. Z., and J. A. K. visualization; D. J. K., E. J. B., L. Z., and J. A. K. methodology; D. J. K. and J. A. K. writing-original draft; D. J. K., E. J. B., L. Z., and J. A. K. writing-review and editing; E. J. B., L. Z., and J. A. K. resources; L. Z. software; L. Z. and J. A. K. funding acquisition; J. A. K. conceptualization; J. A. K. supervision; J. A. K. validation; J. A. K. project administration.
Funding and additional information—This research was supported by National Institutes of Health Grant 1R01DK109345 (to J. A. K.). The Fusion Orbitrap instrument was supported by National Institutes of Health Grant S10 OD018056 (to L. Z.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- TMA
- trimethylamine
- TMAO
- trimethylamine N-oxide
- nonPyl
- nonpyrrolysine
- THF
- tetrahydrofolate
- emPAI
- exponentially modified protein abundance index
- PIPE
- polymerase incomplete primer extension
- OD
- optical density
- MFS
- major facilitator family
- CI
- confidence interval.
References
- 1. Ferguson D. J. Jr., and Krzycki J. A. (1997) Reconstitution of trimethylamine-dependent coenzyme M methylation with the trimethylamine corrinoid protein and the isozymes of methyltransferase II from Methanosarcina barkeri. J. Bacteriol. 179, 846–852 10.1128/jb.179.3.846-852.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krzycki J. A. (2004) Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 8, 484–491 10.1016/j.cbpa.2004.08.012 [DOI] [PubMed] [Google Scholar]
- 3. Paul L., Ferguson D. J., and Krzycki J. A. (2000) The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol. 182, 2520–2529 10.1128/jb.182.9.2520-2529.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soares J. A., Zhang L., Pitsch R. L., Kleinholz N. M., Jones R. B., Wolff J. J., Amster J., Green-Church K. B., and Krzycki J. A. (2005) The residue mass of l-pyrrolysine in three distinct methylamine methyltransferases. J. Biol. Chem. 280, 36962–36969 10.1074/jbc.M506402200 [DOI] [PubMed] [Google Scholar]
- 5. Ticak T., Kountz D. J., Girosky K. E., Krzycki J. A., and Ferguson D. J. Jr. (2014) A nonpyrrolysine member of the widely distributed trimethylamine methyltransferase family is a glycine betaine methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 111, E4668–E4676 10.1073/pnas.1409642111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lechtenfeld M., Heine J., Sameith J., Kremp F., and Müller V. (2018) Glycine betaine metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 20, 4512–4525 10.1111/1462-2920.14389 [DOI] [PubMed] [Google Scholar]
- 7. Tang W. H. W., Bäckhed F., Landmesser U., and Hazen S. L. (2019) Intestinal microbiota in cardiovascular health and disease: JACC state-of-the-art review. J. Am. Coll. Cardiol. 73, 2089–2105 10.1016/j.jacc.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Z., Klipfell E., Bennett B. J., Koeth R., Levison B. S., Dugar B., Feldstein A. E., Britt E. B., Fu X., Chung Y. M., Wu Y., Schauer P., Smith J. D., Allayee H., Tang W. H., et al. (2011) Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koeth R. A., Wang Z., Levison B. S., Buffa J. A., Org E., Sheehy B. T., Britt E. B., Fu X., Wu Y., Li L., Smith J. D., DiDonato J. A., Chen J., Li H., Wu G. D., et al. (2013) Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Al-Waiz M., Mikov M., Mitchell S. C., and Smith R. L. (1992) The exogenous origin of trimethylamine in the mouse. Metabolism 41, 135–136 10.1016/0026-0495(92)90140-6 [DOI] [PubMed] [Google Scholar]
- 11. Koeth R. A., Levison B. S., Culley M. K., Buffa J. A., Wang Z., Gregory J. C., Org E., Wu Y., Li L., Smith J. D., Tang W. H., DiDonato J. A., Lusis A. J., and Hazen S. L. (2014) γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of l-carnitine to TMAO. Cell Metab. 20, 799–812 10.1016/j.cmet.2014.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koeth R. A., Lam-Galvez B. R., Kirsop J., Wang Z., Levison B. S., Gu X., Copeland M. F., Bartlett D., Cody D. B., Dai H. J., Culley M. K., Li X. S., Fu X., Wu Y., Li L., et al. (2019) l-Carnitine in omnivorous diets induces an atherogenic gut microbial pathway in humans. J. Clin. Invest. 129, 373–387 10.1172/JCI94601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lang D. H., Yeung C. K., Peter R. M., Ibarra C., Gasser R., Itagaki K., Philpot R. M., and Rettie A. E. (1998) Isoform specificity of trimethylamine N-oxygenation by human flavin-containing monooxygenase (FMO) and P450 enzymes: selective catalysis by FMO3. Biochem. Pharmacol. 56, 1005–1012 10.1016/S0006-2952(98)00218-4 [DOI] [PubMed] [Google Scholar]
- 14. Wang Z., Tang W. H., Buffa J. A., Fu X., Britt E. B., Koeth R. A., Levison B. S., Fan Y., Wu Y., and Hazen S. L. (2014) Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur. Heart J. 35, 904–910 10.1093/eurheartj/ehu002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Phillips I. R., and Shephard E. A. (2008) Flavin-containing monooxygenases: mutations, disease and drug response. Trends Pharmacol. Sci. 29, 294–301 10.1016/j.tips.2008.03.004 [DOI] [PubMed] [Google Scholar]
- 16. Gadda G., and McAllister-Wilkins E. E. (2003) Cloning, expression, and purification of choline dehydrogenase from the moderate halophile Halomonas elongata. Appl. Environ. Microbiol. 69, 2126–2132 10.1128/aem.69.4.2126-2132.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meadows J. A., and Wargo M. J. (2015) Carnitine in bacterial physiology and metabolism. Microbiology 161, 1161–1174 10.1099/mic.0.000080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meyer M., Granderath K., and Andreesen J. R. (1995) Purification and characterization of protein PB of betaine reductase and its relationship to the corresponding proteins glycine reductase and sarcosine reductase from Eubacterium acidaminophilum. Eur. J.Biochem. 234, 184–191 10.1111/j.1432-1033.1995.184_c.x [DOI] [PubMed] [Google Scholar]
- 19. Craciun S., and Balskus E. P. (2012) Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. U. S. A. 109, 21307–21312 10.1073/pnas.1215689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu Y., Jameson E., Crosatti M., Schäfer H., Rajakumar K., Bugg T. D., and Chen Y. (2014) Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl. Acad. Sci. U. S. A. 111, 4268–4273 10.1073/pnas.1316569111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuka J., Liepinsh E., Makrecka-Kuka M., Liepins J., Cirule H., Gustina D., Loza E., Zharkova-Malkova O., Grinberga S., Pugovics O., and Dambrova M. (2014) Suppression of intestinal microbiota-dependent production of pro-atherogenic trimethylamine N-oxide by shifting l-carnitine microbial degradation. Life Sci. 117, 84–92 10.1016/j.lfs.2014.09.028 [DOI] [PubMed] [Google Scholar]
- 22. Wang Z., Roberts A. B., Buffa J. A., Levison B. S., Zhu W., Org E., Gu X., Huang Y., Zamanian-Daryoush M., Culley M. K., DiDonato A. J., Fu X., Hazen J. E., Krajcik D., DiDonato J. A., et al. (2015) Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 10.1016/j.cell.2015.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin F. P., Wang Y., Sprenger N., Yap I. K., Lundstedt T., Lek P., Rezzi S., Ramadan Z., van Bladeren P., Fay L. B., Kochhar S., Lindon J. C., Holmes E., and Nicholson J. K. (2008) Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 4, 157 10.1038/msb4100190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gregory J. C., Buffa J. A., Org E., Wang Z., Levison B. S., Zhu W., Wagner M. A., Bennett B. J., Li L., DiDonato J. A., Lusis A. J., and Hazen S. L. (2015) Transmission of atherosclerosis susceptibility with gut microbial transplantation. J. Biol. Chem. 290, 5647–5660 10.1074/jbc.M114.618249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown J. M., and Hazen S. L. (2014) Metaorganismal nutrient metabolism as a basis of cardiovascular disease. Curr. Opin. Lipidol. 25, 48–53 10.1097/MOL.0000000000000036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brugère J. F., Borrel G., Gaci N., Tottey W., O'Toole P. W., and Malpuech-Brugere C. (2014) Archaebiotics: proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes 5, 5–10 10.4161/gmic.26749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramezani A., Nolin T. D., Barrows I. R., Serrano M. G., Buck G. A., Regunathan-Shenk R., West R. E. 3rd, Latham P. S., Amdur R., and Raj D. S. (2018) Gut colonization with methanogenic archaea lowers plasma trimethylamine N-oxide concentrations in apolipoprotein e−/− mice. Sci. Rep 8, 14752 10.1038/s41598-018-33018-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muller E., Fahlbusch K., Walther R., and Gottschalk G. (1981) Formation of N,N-dimethylglycine, acetic acid, and butyric acid from betaine by Eubacterium limosum. Appl. Environ. Microbiol. 42, 439–445 10.1128/AEM.42.3.439-445.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeong J., Bertsch J., Hess V., Choi S., Choi I. G., Chang I. S., and Müller V. (2015) Energy conservation model based on genomic and experimental analyses of a carbon monoxide-utilizing, butyrate-forming acetogen, Eubacterium limosum KIST612. Appl. Environ. Microbiol. 81, 4782–4790 10.1128/AEM.00675-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Genthner B. R., Davis C. L., and Bryant M. P. (1981) Features of rumen and sewage sludge strains of Eubacterium limosum, a methanol- and H2-CO2-utilizing species. Appl. Environ. Microbiol. 42, 12–19 10.1128/AEM.42.1.12-19.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ragsdale S. W., and Pierce E. (2008) Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784, 1873–1898 10.1016/j.bbapap.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Naidu D., and Ragsdale S. W. (2001) Characterization of a three-component vanillate O-demethylase from Moorella thermoacetica. J. Bacteriol. 183, 3276–32781 10.1128/JB.183.11.3276-3281.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen J. X., Deng C. Y., Zhang Y. T., Liu Z. M., Wang P. Z., Liu S. L., Qian W., and Yang D. H. (2016) Cloning, expression, and characterization of a four-component O-demethylase from human intestinal bacterium Eubacterium limosum ZL-II. Appl. Microbiol. Biotechnol. 100, 9111–9124 10.1007/s00253-016-7626-1 [DOI] [PubMed] [Google Scholar]
- 34. Siebert A., Schubert T., Engelmann T., Studenik S., and Diekert G. (2005) Veratrol-O-demethylase of Acetobacterium dehalogenans: ATP-dependent reduction of the corrinoid protein. Arch. Microbiol. 183, 378–384 10.1007/s00203-005-0001-8 [DOI] [PubMed] [Google Scholar]
- 35. Engelmann T., Kaufmann F., and Diekert G. (2001) Isolation and characterization of a veratrol:corrinoid protein methyl transferase from Acetobacterium dehalogenans. Arch. Microbiol. 175, 376–383 10.1007/s002030100275 [DOI] [PubMed] [Google Scholar]
- 36. Matthews R. G. (2009) Cobalamin- and corrinoid-dependent enzymes. Met. Ions Life Sci. 6, 53–114 10.1039/BK9781847559159-00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drennan C. L., Matthews R. G., and Ludwig M. L. (1994) Cobalamin-dependent methionine synthase: the structure of a methylcobalamin-binding fragment and implications for other B12-dependent enzymes. Curr. Opin. Struct. Biol. 4, 919–299 10.1016/0959-440X(94)90275-5 [DOI] [PubMed] [Google Scholar]
- 38. Ferguson T., Soares J. A., Lienard T., Gottschalk G., and Krzycki J. A. (2009) RamA, a protein required for reductive activation of corrinoid-dependent methylamine methyltransferase reactions in methanogenic Archaea. J. Biol. Chem. 284, 2285–2295 10.1074/jbc.M807392200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nguyen H. D., Studenik S., and Diekert G. (2013) Corrinoid activation by a RACE protein: studies on the interaction of the proteins involved. FEMS Microbiol. Lett. 345, 31–38 10.1111/1574-6968.12178 [DOI] [PubMed] [Google Scholar]
- 40. Hennig S. E., Jeoung J. H., Goetzl S., and Dobbek H. (2012) Redox-dependent complex formation by an ATP-dependent activator of the corrinoid/iron-sulfur protein. Proc. Natl. Acad. Sci. U. S. A. 109, 5235–5240 10.1073/pnas.1117126109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sauer K., and Thauer R. K. (1999) Methanol:coenzyme M methyltransferase from Methanosarcina barkeri—substitution of the corrinoid harbouring subunit MtaC by free cob(I)alamin. Eur. J. Biochem. 261, 674–681 10.1046/j.1432-1327.1999.00355.x [DOI] [PubMed] [Google Scholar]
- 42. Picking J. W., Behrman E. J., Zhang L., and Krzycki J. A. (2019) MtpB, a member of the MttB superfamily from the human intestinal acetogen Eubacterium limosum, catalyzes proline betaine demethylation. J. Biol. Chem. 294, 13697–13707 10.1074/jbc.RA119.009886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., and Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 10.1074/mcp.M500061-MCP200 [DOI] [PubMed] [Google Scholar]
- 44. Schoelmerich M. C., Katsyv A., Sung W., Mijic V., Wiechmann A., Kottenhahn P., Baker J., Minton N. P., and Müller V. (2018) Regulation of lactate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 20, 4587–4595 10.1111/1462-2920.14412 [DOI] [PubMed] [Google Scholar]
- 45. Weghoff M. C., Bertsch J., and Müller V. (2015) A novel mode of lactate metabolism in strictly anaerobic bacteria. Environ. Microbiol. 17, 670–677 10.1111/1462-2920.12493 [DOI] [PubMed] [Google Scholar]
- 46. Song Y., Shin J., Jeong Y., Jin S., Lee J. K., Kim D. R., Kim S. C., Cho S., and Cho B. K. (2017) Determination of the genome and primary transcriptome of syngas fermenting Eubacterium limosum ATCC 8486. Sci. Rep. 7, 13694 10.1038/s41598-017-14123-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Evans J. C., Huddler D. P., Hilgers M. T., Romanchuk G., Matthews R. G., and Ludwig M. L. (2004) Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase. Proc. Natl. Acad. Sci. U. S. A. 101, 3729–3736 10.1073/pnas.0308082100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schnoes A. M., Brown S. D., Dodevski I., and Babbitt P. C. (2009) Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 5, e1000605 10.1371/journal.pcbi.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kremp F., Poehlein A., Daniel R., and Müller V. (2018) Methanol metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 20, 4369–4384 10.1111/1462-2920.14356 [DOI] [PubMed] [Google Scholar]
- 50. Calogero S., Gardan R., Glaser P., Schweizer J., Rapoport G., and Debarbouille M. (1994) RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J. Bacteriol. 176, 1234–1241 10.1128/jb.176.5.1234-1241.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saier M. H. Jr., Reddy V. S., Tsu B. V., Ahmed M. S., Li C., and Moreno-Hagelsieb G. (2016) The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 44, D372–D379 10.1093/nar/gkv1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Großhennig S., Schmidl S. R., Schmeisky G., Busse J., and Stülke J. (2013) Implication of glycerol and phospholipid transporters in Mycoplasma pneumoniae growth and virulence. Infect. Immun. 81, 896–904 10.1128/IAI.01212-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Belitsky B. R., Gustafsson M. C., Sonenshein A. L., and Von Wachenfeldt C. (1997) An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J. Bacteriol. 179, 5448–5457 10.1128/jb.179.17.5448-5457.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kennerknecht N., Sahm H., Yen M. R., Patek M., Saier M. H. Jr., and Eggeling L. (2002) Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J. Bacteriol. 184, 3947–3956 10.1128/jb.184.14.3947-3956.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fritz I. B., and Schultz S. K. (1965) Carnitine acetyltransferase. II. Inhibition by carnitine analogues and by sulfhydryl reagents. J. Biol. Chem. 240, 2188–2192 [PubMed] [Google Scholar]
- 56. Norum K. R. (1964) Palmityl-CoA:carnitine palmityltransferase: purification from calf-liver mitochondria and some properties of the enzyme. Biochim. Biophys. Acta 89, 95–108 [PubMed] [Google Scholar]
- 57. Kleber H. P., Seim H., Aurich H., and Strack E. (1978) Interrelationships between carnitine metabolism and fatty acid assimilation in Pseudomonas putida. Arch. Microbiol. 116, 213–220 10.1007/BF00406039 [DOI] [PubMed] [Google Scholar]
- 58. Brown J. M., and Hazen S. L. (2018) Microbial modulation of cardiovascular disease. Nat. Rev. Microbiol. 16, 171–181 10.1038/nrmicro.2017.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Song Y., and Cho B. K. (2015) Draft genome sequence of chemolithoautotrophic acetogenic butanol-producing Eubacterium limosum ATCC 8486. Genome Announc. 3, 10.1128/genomeA.01564-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kanauchi O., Fukuda M., Matsumoto Y., Ishii S., Ozawa T., Shimizu M., Mitsuyama K., and Andoh A. (2006) Eubacterium limosum ameliorates experimental colitis and metabolite of microbe attenuates colonic inflammatory action with increase of mucosal integrity. World J. Gastroenterol. 12, 1071–1077 10.3748/wjg.v12.i7.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Biagi E., Nylund L., Candela M., Ostan R., Bucci L., Pini E., Nikkïla J., Monti D., Satokari R., Franceschi C., Brigidi P., and De Vos W. (2010) Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5, e10667 10.1371/journal.pone.0010667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cao X., and Krzycki J. A. (1991) Acetate-dependent methylation of two corrinoid proteins in extracts of Methanosarcina barkeri. J. Bacteriol. 173, 5439–5448 10.1128/jb.173.17.5439-5448.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kremer J. D., Cao X., and Krzycki J. (1993) Isolation of two novel corrinoid proteins from acetate-grown Methanosarcina barkeri. J. Bacteriol. 175, 4824–4833 10.1128/jb.175.15.4824-4833.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Burke S. A., and Krzycki J. A. (1995) Involvement of the “A” isozyme of methyltransferase II and the 29-kilodalton corrinoid protein in methanogenesis from monomethylamine. J. Bacteriol. 177, 4410–4416 10.1128/jb.177.15.4410-4416.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Klock H. E., Koesema E. J., Knuth M. W., and Lesley S. A. (2008) Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71, 982–994 10.1002/prot.21786 [DOI] [PubMed] [Google Scholar]
- 66. Grahame D. A. (1991) Catalysis of acetyl-CoA cleavage and tetrahydrosarcinapterin methylation by a carbon monoxide dehydrogenase-corrinoid enzyme complex. J. Biol. Chem. 266, 22227–22233 [PubMed] [Google Scholar]
- 67. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (67) with the data set identifier PXD013806 (for the lactate data set) and with the identifier PXD013961 (for the l-carnitine data set). All other data cited can be found in the article.