Abstract
PCBP1, a member of the poly(C)-binding protein (PCBP) family, has the capability of binding heavily oxidized RNA and therefore participates in the cellular response to oxidative conditions, helping to induce apoptosis. There are four other members of this family, PCBP2, PCBP3, PCBP4, and hnRNPK, but it is not known whether they play similar roles. To learn more, we first tested their affinity for an RNA strand carrying two 8-oxoguanine (8-oxoG) residues at sites located in close proximity to each other, representative of a heavily oxidized strand or RNA with one 8-oxoG or none. Among them, only PCBP2 exhibited highly selective binding to RNA carrying two 8-oxoG residues similar to that observed with PCBP1. In contrast, PCBP3, PCBP4, and hnRNPK bound RNA with or without 8-oxoG modifications and exhibited slightly increased binding to the former. Mutations in conserved RNA-binding domains of PCBP2 disrupted the specific interaction with heavily oxidized RNA. We next tested PCBP2 activity in cells. Compared with WT HeLa S3 cells, PCBP2-KO cells established by gene editing exhibited increased apoptosis with increased caspase-3 activity and PARP1 cleavage under oxidative conditions, which were suppressed by the expression of WT PCBP2 but not one of the mutants lacking binding activity. In contrast, PCBP1-KO cells exhibited reduced apoptosis with much less caspase-3 activity and PARP cleavage than WT cells. Our results indicate that PCBP2 as well as PCBP1 bind heavily oxidized RNA; however, the former may counteract PCBP1 to suppress apoptosis under oxidative conditions.
Keywords: oxidative stress, 8-oxoguanine, RNA-binding protein, apoptosis, apoptosis, oxygen radicals, gene knockout
Almost all cellular components, such as sugars, lipids, proteins, and nucleic acids, are always at high risk of being oxidized by one-electron oxidants and reactive oxygen species (ROS), including hydroxyl radical (·OH) and singlet oxygen (1O2). ROS are inevitable byproducts of electron transport in the mitochondria or other normal metabolic pathways and are also generated during various biological processes, such as host defense, neurotransmission, vasodilation, and signal transduction. The production of ROS is enhanced under various pathophysiological conditions or environmental exposure to ionizing radiation and chemicals, and increased accumulation of oxidatively generated damage during aging is likely to be a major cause of various types of cellular dysfunction, causing degenerative disorders and neoplasms (1).
Among the various types of oxidative damage to cellular macromolecules, the oxidation of nucleobases seems to play pivotal roles in mutagenesis, carcinogenesis, neurodegeneration, and aging (2–4). Among the nucleobases, guanine is the most susceptible to oxidation by hydroxyl radicals or one-electron oxidants and is modified to 8-oxo-7,8-dihydroguanine (8-oxoG). 8-OxoG can take both syn and anti conformations and thus pair with adenine as well as cytosine. Once 8-oxoG is present opposite cytosine in DNA, GC to TA transversion mutation may be induced through DNA replication. In addition, 8-oxoG present in DNA also mispairs with adenine during RNA synthesis, thereby leading to the alteration of the gene expression (5).
Guanine bases in RNA are more susceptible to oxidation by ROS than in dsDNA, in which the bases are protected by hydrogen bonds and by various bound proteins, because RNA molecules are mostly single-stranded. Moreover, ROS also oxidize GTP in the nucleotide pool and generate 8-oxo-GTP, which can be incorporated into RNA (6). Indeed, the level of 8-oxoG in RNA is 14–25 times higher than that in DNA after exposure to hydrogen peroxide (H2O2) (7). The persistence of 8-oxoG–containing RNA induces alteration in the gene expression, and the abnormal protein thus formed may be hazardous to the cell (8).
It is well-known that RNA oxidation is a prominent feature in the brains of Alzheimer's disease patients (9). More recently, we showed that there is a causal relationship between RNA oxidation and β-amyloid (Aβ) peptide formation using Chinese hamster ovary cells to which 8-oxoGTP was externally supplied (10). Cells carrying 8-oxoG in mRNA secreted 28 types of previously unknown Aβ-derived peptides, in addition to a greater amount of pathogenic Aβ peptides that have previously been characterized. These results suggest that organisms must have some mechanisms to eliminate the deleterious effects of RNA oxidation (6).
Recently, we found two protein factors that bind oxidized RNA. One of them, AUF1 protein, recognizes 8-oxoG–containing oligo-RNA and participates in selective mRNA decay under oxidative conditions (11). Another binding protein, PCBP1, a member of the poly(C)-binding proteins (PCBPs), might be involved in the induction of apoptosis via complex formation with 8-oxoG–containing RNA (12). In the present study, we examined all members of the PCBP protein family to investigate whether some of them can bind 8-oxoG–containing RNA and control apoptosis under oxidative conditions, compared with PCBP1. Our results indicate that PCBP2 may counteract PCBP1 to suppress apoptosis induced under oxidative stress by binding to heavily oxidized RNA.
Results
PCBP family proteins bind heavily oxidized RNA
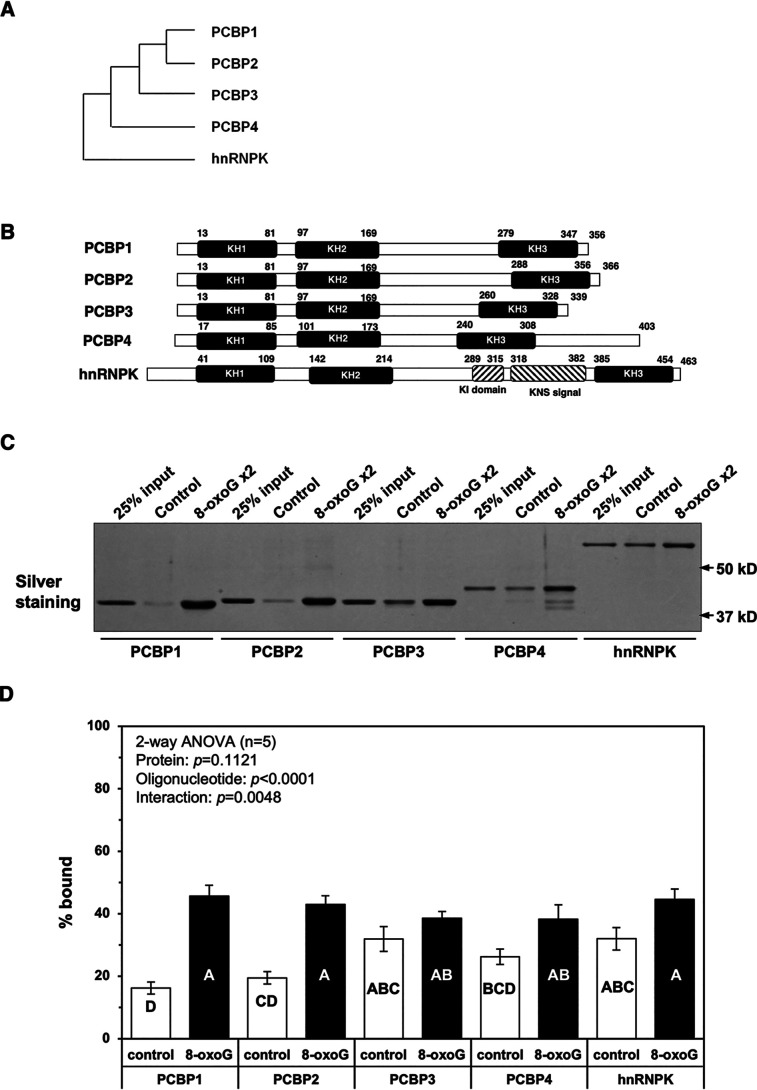
We previously reported that PCBP1, which was originally identified as poly(C)-binding protein belonging to a subfamily of K homology (KH) domain RNA-binding proteins (13, 14), specifically binds heavily oxidized RNA (12). The PCBP family includes five proteins, PCBP1, PCBP2, PCBP3, PCBP4, and hnRNPK. Among them, PCBP1 shares the highest homology with PCBP2, as shown in Fig. 1A and Table 1.
Figure 1.
PCBP family proteins bind heavily oxidized RNA. A, phylogenic tree of the five members of the PCBP protein family. Among the five members of the PCBP protein family, PCBP2 shares the highest homology with PCBP1. B, structural comparison of the five members of the PCBP protein family. All PCBP family proteins contain three KH domains, KH1, KH2, and KH3, each consisting of ∼70 amino acid residues that enable RNA and DNA binding with high affinity toward polycytosine tracts. hnRNPK carries a nuclear localization signal and a nuclear shuttling domain (KNS) together, which allows it to translocate between the cytoplasm and nucleus, and also contains a segment called the K protein interactive (KI) region, located between the KH2 and KH3 domain, which has an intrinsically disordered structure. The unique K protein interactive domain and nuclear shuttling domain signal between the KH2 and KH3 domains. C, binding capacities of PCBP family proteins to oligoribonucleotides carrying no (control) or two 8-oxoG residues (8-oxoG x2). Control and 8-oxoG x2 oligoribonucleotides were conjugated to magnetic beads and used as probes. These RNA beads were mixed with a purified preparation of each recombinant protein of the five PCBP family members and incubated for 90 min at 4 °C for binding. The beads were then washed with HNTG buffer three times, and the retained materials were subjected to SDS-PAGE, followed by silver staining. 25% input, a quarter of each recombinant protein applied to binding was also subjected to SDS-PAGE. D, a quantitative comparison of the binding capacities of PCBP family proteins with oligoribonucleotides carrying no (control) or two 8-oxoG residues (8-oxoG). The band intensities shown in C were measured by ImageQuant TL, and the percentage bound was determined. Data from five independent experiments are shown as the mean ± S.E. (error bars). A two-way ANOVA was performed as a statistical analysis. Bars without the same letter showed statistically significant differences in a post hoc Tukey's HSD test, p < 0.05.
Table 1.
Homology between PCBP1 and PCBP family members
| Protein | PCBP1 | PCBP2 | PCBP3 | PCBP4 | hnRNPK |
|---|---|---|---|---|---|
| Amino acid residues | 356 | 366 | 339 | 403 | 463 |
| Homology to PCBP1 (%) | 100 | 82.83 | 66.48 | 52.69 | 28.63 |
| KH1 (amino acid residues) | 69 | 69 | 69 | 69 | 69 |
| Homology to PCBP1 (%) | 100 | 95.65 | 86.96 | 73.91 | 30.77 |
| KH2 (amino acid residues) | 73 | 73 | 73 | 73 | 73 |
| Homology to PCBP1 (%) | 100 | 93.15 | 83.56 | 75.34 | 41.27 |
| KH3 (amino acid residues) | 69 | 69 | 69 | 69 | 69 |
| Homology to PCBP1 (%) | 100 | 91.18 | 83.58 | 65.15 | 52.38 |
| KH2-KH3 linker (amino acid residues) | 109 | 118 | 90 | 66 | 170 |
| Homology to PCBP1 (%) | 100 | 64.96 | 44.29 | 23.94 | 45.45 |
To examine whether each of the PCBP family proteins has a similar capacity to bind heavily oxidized RNA, we prepared recombinant human PCBP2, PCBP3, PCBP4, and hnRNPK proteins that also possessed three KH domains (KH1–KH3) (15, 16) (Fig. 1B and Table 1). As shown in Fig. 1C, recombinant PCBP2 and, to a lesser extent, PCBP3, PCBP4, and hnRNPK efficiently bound a 30-mer oligoribonucleotide containing two 8-oxoG residues at positions 9 and 15 (8-oxoG x2) in close proximity to each other. However, a quantitative analysis revealed that only PCBP1 and PCBP2 exhibited highly selective binding to the oligoribonucleotide containing two 8-oxoG residues compared with the control oligoribonucleotide without 8-oxoG residue (Fig. 1D). PCBP3, PCBP4, and hnRNPK exhibited slightly increased binding to the oligoribonucleotide containing two 8-oxoG residues, but the difference was not statistically significant.
PCBP1 and PCBP2 proteins have the capacity to selectively bind heavily oxidized RNA
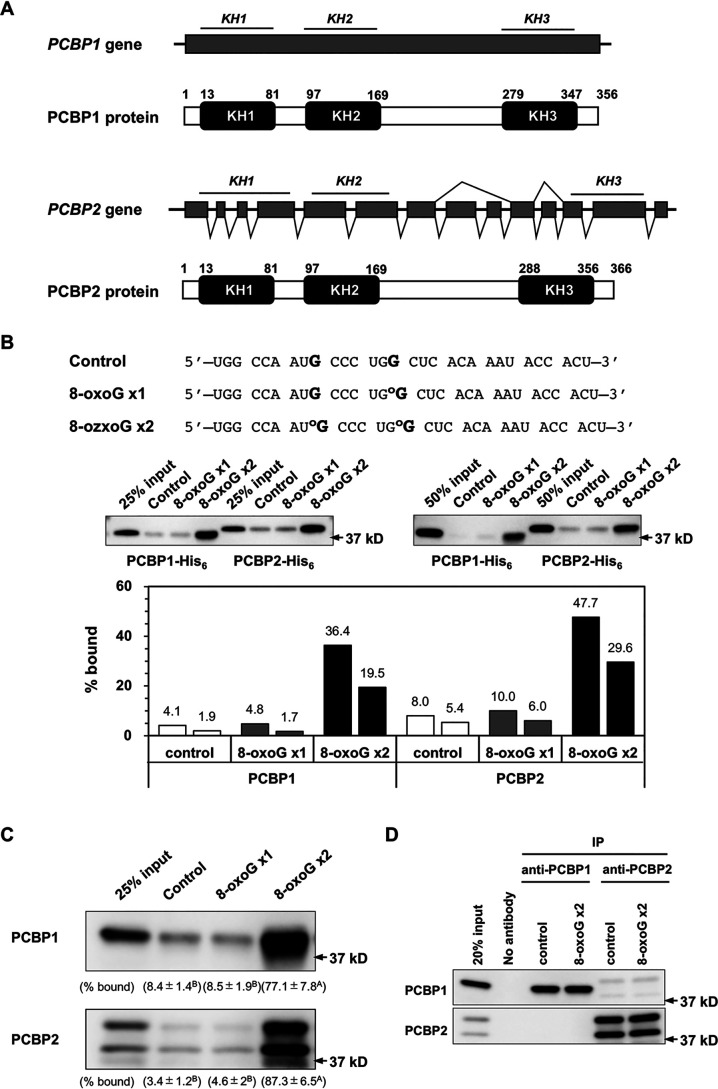
The PCBP1 gene is known to be a retrotransposed derivative of the PCBP2 gene with the highest homology among all PCBP family members (Fig. 2A and Table 1), and these two paralogs essentially serve the same cellular functions (15, 17–19). The two proteins share >80% of amino acid residues (82.8%), and the other three proteins of the PCBP family have much less homology (Table 1). To examine whether PCBP2 binds selectively to heavily oxidized RNA similarly to PCBP1, we compared binding to oligoribonucleotides containing one 8-oxoG at position 9 (8-oxoG x1) or two 8-oxoG residues at positions 9 and 15 (8-oxoG x2) in vitro (Fig. 2B). Two independent binding assays revealed that PCBP2 selectively bound to the oligoribonucleotide containing two 8-oxoG residues, and binding to the oligoribonucleotide containing one 8-oxoG residue was almost the same as that of the control oligonucleotide without 8-oxoG (Fig. 2B). These results indicate that among the five members of the PCBP protein family, PCBP1 and PCBP2 have the capacity to selectively bind to heavily oxidized RNA.
Figure 2.
PCBP1 and PCBP2 proteins have capacities to selectively bind heavily oxidized RNA. A, structures of the PCBP1 and PCBP2 genes and their coding proteins. The PCBP1 gene is a retrotransposed derivative of the PCBP2 gene and thus lacks an intron. B, the binding of PCBP1 and PCBP2 to two types of 8-oxoG–containing oligoribonucleotide probes. Each purified recombinant protein with a His6 tag was applied to beads carrying oligoribonucleotides with no (control), one 8-oxoG residue (8-oxoG x1), and two 8-oxoG residues (8-oxoG x2), and the materials were processed as described above. Western blotting was performed using an anti-His6 antibody. The band intensities shown in the middle panels were measured by ImageQuant TL, and the percentage bound was determined. Data from two independent experiments are shown. C, binding of authentic PCBP1 and PCBP2 to two types of 8-oxoG–containing oligoribonucleotide probes. Crude extract prepared from HeLa S3 cells was applied to beads carrying oligoribonucleotides with no (control), one 8-oxoG residue (8-oxoG x1), and two 8-oxoG residues (8-oxoG x2), and the materials were processed as described above. Western blotting was performed using anti-PCBP1 and anti-PCBP2 antibodies. The band intensities shown in the left panels were measured by ImageQuant TL, and the percentage bound was determined. Data from three independent experiments are shown in parentheses as the mean ± S.E. A two-way ANOVA was performed as a statistical analysis. Data without the same letter showed statistically significant differences in a post hoc Tukey's HSD test, p < 0.05. D, immunoprecipitation of PCBP1 or PCBP2 in the presence of control or 8-oxoG x2 probes. 8-OxoG x2 complex precipitated by anti-PCBP1 contained only PCBP1, and that precipitated by anti-PCBP2 contained only PCBP2, as did control complex.
To confirm the binding activities of authentic PCBP1 and PCBP2 proteins in human cells, extracts prepared from HeLa S3 were applied to control, 8-oxoG x1, and 8-oxoG x2 probes, and pulldown products were subjected to Western blotting with anti-PCBP1 and anti-PCBP2 antibodies. As shown in Fig. 2C, both PCBP1 and PCBP2 were only detected in the pulldown fractions with 8-oxoG x2 probes, indicating that the authentic PCBP1 and PCBP2 selectively bound to the heavily oxidized RNA. We immunoprecipitated PCBP1 or PCBP2 in the presence of control or 8-oxoG x2 probes, and the latter bound >77% of PCBP1 and 87% of PCBP2 in the extracts. We then detected PCBP1 and PCBP2 in the complex by Western blotting. As shown in Fig. 2D, the complex precipitated by anti-PCBP1 contained only PCBP1, and that precipitated by anti-PCBP2 contained only PCBP2, indicating that PCBP1 and PCBP2 bound 8-oxoG x2 probes in a mutually exclusive manner with no interaction.
The structure–function relationship of PCBP2
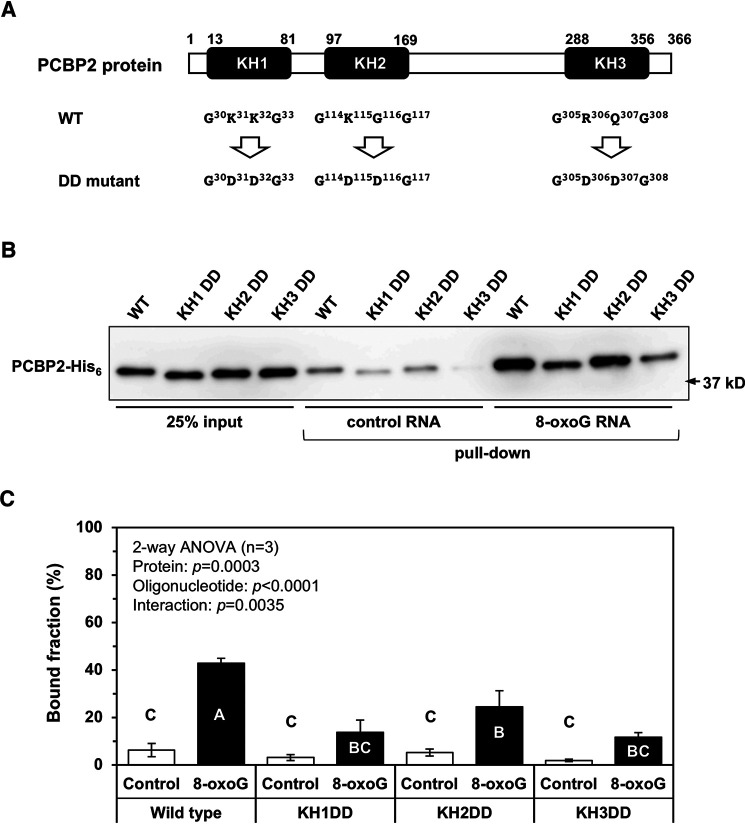
PCBP2, as well as PCBP1, contains three KH domains, each consisting of ∼70 amino acid residues with homologies >91% (Table 1). It is implied that these domains are essential for multiple functions of the PCBP family proteins (13–15, 20–22), including the binding of PCBP1 to heavily oxidized RNA (12). We therefore introduced amino acid substitutions to each of the three domains to determine which domains were important for specific binding to oxidized RNA. Essential basic amino acid residues in the GXXG loop for nucleic acid binding, Lys or Arg, and the adjacent residues in each KH domain were substituted with the Asp-Asp (DD) sequence (Fig. 3A). As shown in Fig. 3 (B and C), DD mutation at the KH1 and KH3 domains significantly reduced the binding capacity for 8-oxoG–containing RNA to almost 30% of the level of the WT protein, whereas 60% of the selective binding level of the WT protein was retained in the presence of DD mutation at the KH2 domain.
Figure 3.
The structure–function relationships of PCBP2. A, structure of PCBP2 and its mutation sites. The numbers of amino acid residues are shown above the three KH domains of PCBP2. Below each KH domain, the amino acid substitution sites in the GXXG loop essential for nucleic acid binding are shown. B, the binding capacity of mutant forms of PCBP2 to 8-oxoG–containing oligoribonucleotide. Each purified KH domain mutant protein was mixed with beads attached to a 30-mer oligoribonucleotide carrying no (control) or two residues of 8-oxoG (8-oxoG x2). After incubation, the beads were washed with HNTG buffer and then boiled in SDS sample buffer. The samples were separated using SDS-PAGE and analyzed by Western blotting using an anti-His6 antibody. C, a quantitative comparison of the binding capacities of mutant PCBP2 to oligoribonucleotides carrying no (control) or two 8-oxoG residues (8-oxoG). Band intensities shown in B were measured by ImageQuant TL, and percentage bound were determined. Data from three independent experiments are shown as the mean ± S.E. (error bars). A two-way ANOVA was performed as a statistical analysis. Bars without the same letter showed statistically significant differences in a post hoc Tukey's HSD test, p < 0.05.
Apoptosis-related reactions in PCBP2-deficient cells
To observe the biological significance of PCBP2-associated events, we constructed PCBP2-null cell lines via gene editing (23), as shown in Fig. 4A. Based on the results of Western blotting, two PCBP2-knockout (KO) cell lines (PCBP2-KO(2-14) and PCBP2-KO(1-4)) were isolated (Fig. 4B). Both PCBP2-KO cells were viable but exhibited slower proliferation than the parental HeLa S3 (WT) cells (Fig. 4C), similarly to PCBP1-KO cells (12). A cell cycle analysis revealed that PCBP2-KO cells had an increased population in the G0/G1 phase and decreased population in the G2/M phase, with lower rates of S phase than WT and PCBP1-KO cells, the latter of which exhibited a decreased population in the G0/G1 phase and increased population in the G2/M phase compared with WT cells (Fig. 4D). Because both PCBP2 and PCBP1 are suggested to be involved in various cellular processes, the slower proliferation rate may be attributed to one deficiency or a combination of deficiencies in these processes, and our data indicate that PCBP1 and PCBP2 deficiencies affect cell proliferation in different manners.
Figure 4.
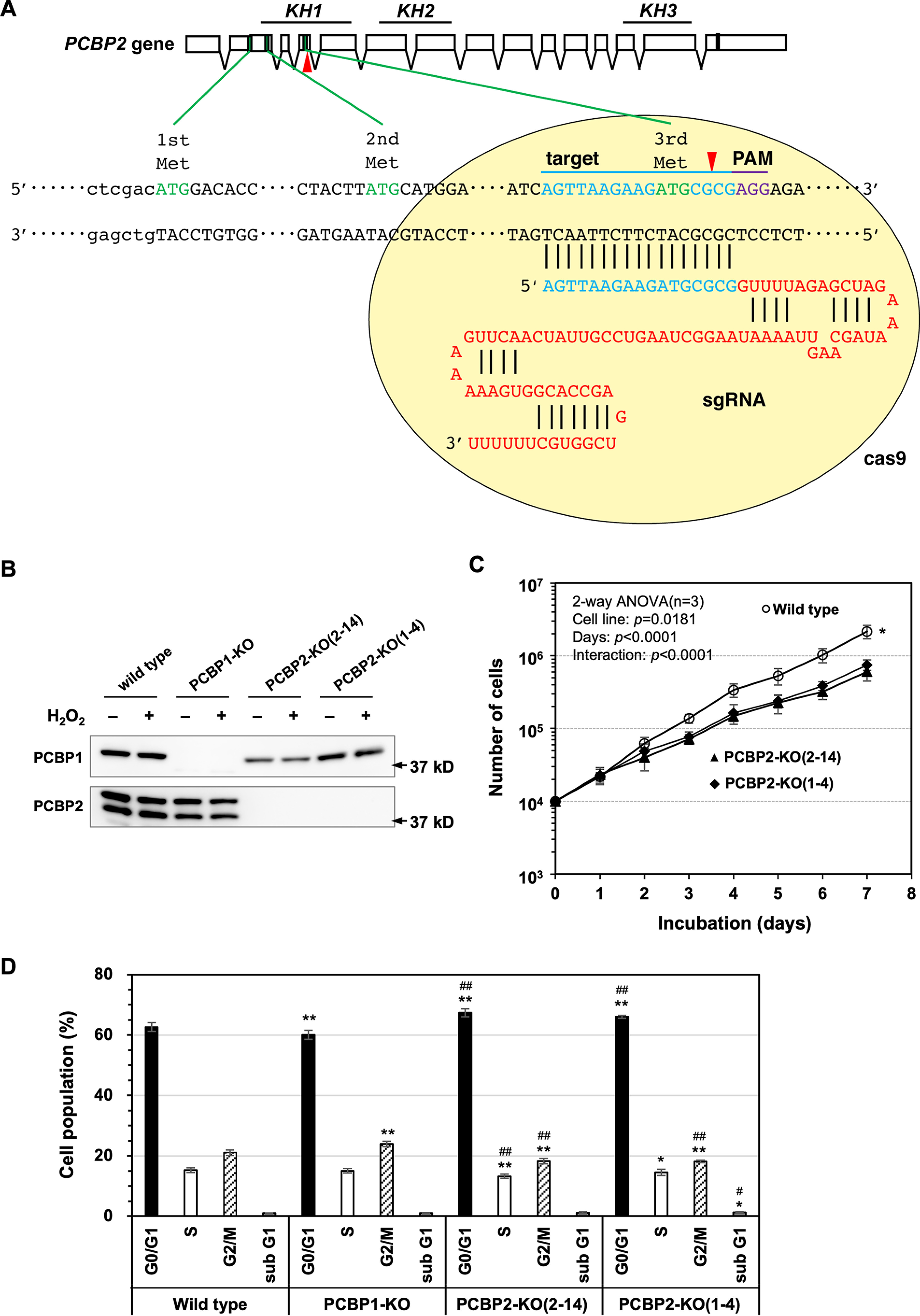
PCBP2-knockout cells exhibit growth retardation. A, schematic illustration of the process of PCBP2 gene editing using the CRISPR/Cas9 system. Single guide RNA designed for genome editing was introduced into HeLa S3 cells. The red arrowheads indicate the site of cutting by Cas9 nuclease. Cleavage of genomic DNA causes an insertion-deletion mutation by inaccurate DNA repair and results in frameshift in the coding region. Boxes, exons of the PCBP2 gene. The green boxes represent the methionine codon, and the black box represents the stop codon. Blue characters indicate the target sequence; violet characters indicate the PAM sequence in the CRISPR/Cas9 system. B, Western blotting of PCBP1-KO and two PCBP2-KO cell lines. HeLa S3 (WT) and HeLa S3-derived PCBP1-KO, PCBP2-KO(2–14), and PCBP2-KO(1–4) cells were lysed in SDS sample buffer and subjected to SDS-PAGE. The expression of PCBP1 and PCBP2 protein was determined by Western blotting using anti-PCBP1 and anti-PCBP2 antibodies. C, growth curves of WT and PCBP2-KO cells. 10,000 WT, PCBP2-KO(2–14), and PCBP2-KO(1–4) cells were plated and incubated under normal conditions. At the indicated times, the cells were detached by trypsin and counted. The data represent the mean ± S.E. (error bars) of three independent experiments. A two-way ANOVA was performed as a statistical analysis. *, p < 0.05 versus PCBP2-KO(2–14) and PCBP2-KO(1–4). D, distribution of cell cycle phases. A cell cycle analysis was performed using flow cytometry. n = 3; more than 30,000 cells were analyzed for each sample. Fisher's exact test in the categorical analysis: *, p < 0.05 versus WT; **, p < 0.00001 versus WT; #, p < 0.05 versus PCBP1-KO; ##, p < 0.00001 versus PCBP1-KO.
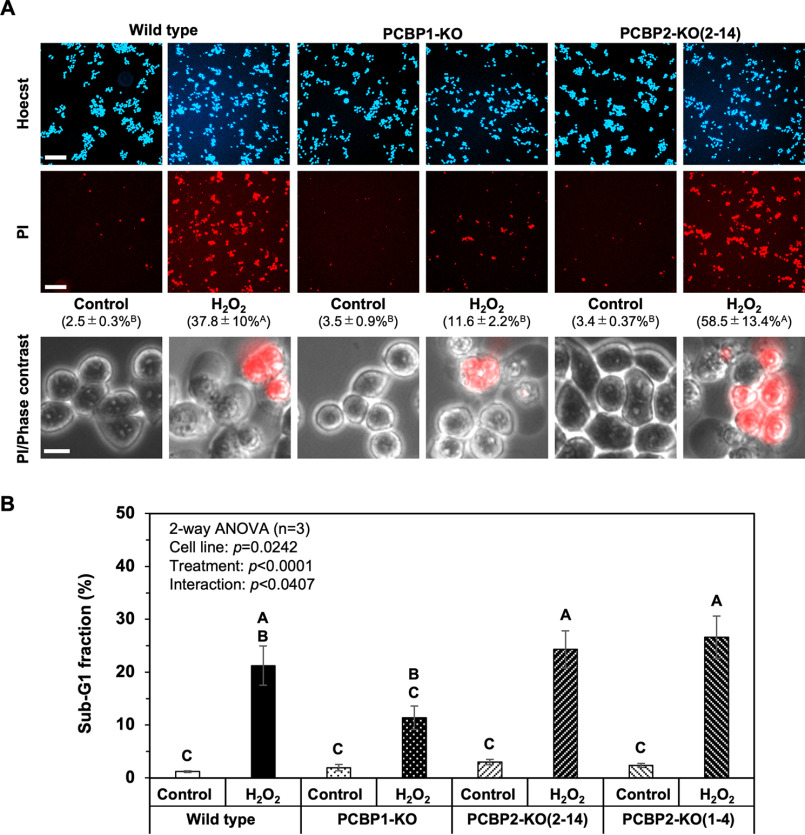
We then examined the cell viability of the three cell lines after exposure to H2O2 (Fig. 5). When WT cells were exposed to 0.2 mm H2O2 (24, 25), dead cells stained by propidium iodide (PI) appeared within 9 h after H2O2 exposure (Fig. 5A). A quantitative examination revealed that PCBP1-KO cells exhibited significantly fewer PI-positive cells (11.6%) compared with WT cells (37.8%), whereas PCBP2-KO cells exhibited significantly more PI-positive cells (58.5%) than WT and PCBP1-KO cells. These results suggest that PCBP2 deficiency increases sensitivity to H2O2-induced cell death, whereas PCBP1 deficiency confers resistance to H2O2-induced cell death. Next, we examined the sub-G1 fraction, which represents the fraction of cells undergoing apoptosis with or without H2O2 exposure (Fig. 5B). The sub-G1 fraction in WT cells increased to 20% within 9 h after H2O2 exposure but significantly decreased to 10% in PCBP1-KO cells. In contrast, the two PCBP2-KO cell lines showed an increased sub-G1 fraction exceeding 20% after H2O2 exposure.
Figure 5.
PCBP2 deficiency increases susceptibility to H2O2-induced apoptosis. A, Cell viability after H2O2 exposure. Appropriate numbers of WT, PCBP1-KO, and PCBP2-KO(2–14) cells were seeded onto plates and incubated overnight. The cells on the dishes were treated with 0.2 mm H2O2 for 9 h at 37 °C, and then the cells were stained with Hoechst 33342 and PI (top two panels; scale bars, 100 μm). PI-positive cells represent damaged or dead cells. Approximately, 2,000–5,000 Hoechst-positive cells were examined in each experiment (n = 3). The percentage of PI-positive cells is shown in parentheses as the mean ± S.E. A two-way ANOVA was performed as a statistical analysis. Data without the same letter showed statistically significant differences in a post hoc Tukey's HSD test, p < 0.05. Magnified phase-contrast images of cells with PI staining are shown in the bottom panels (scale bar, 10 μm). B, percentage of cells in the sub-G1 fraction after H2O2 exposure. Cells (WT, PCBP1-KO, PCBP2-KO(2–14), and PCBP2-KO(1–4) cells) exposed to 0.2 mm H2O2 for 9 h were subjected to flow cytometry. The percentages of cells in the sub-G1 fraction are shown in the bar graph. The data represent the mean ± S.E. (error bars) of three independent experiments. A two-way ANOVA was performed as a statistical analysis. Bars without the same letter showed statistically significant differences in a post hoc Tukey's HSD test, p < 0.05.
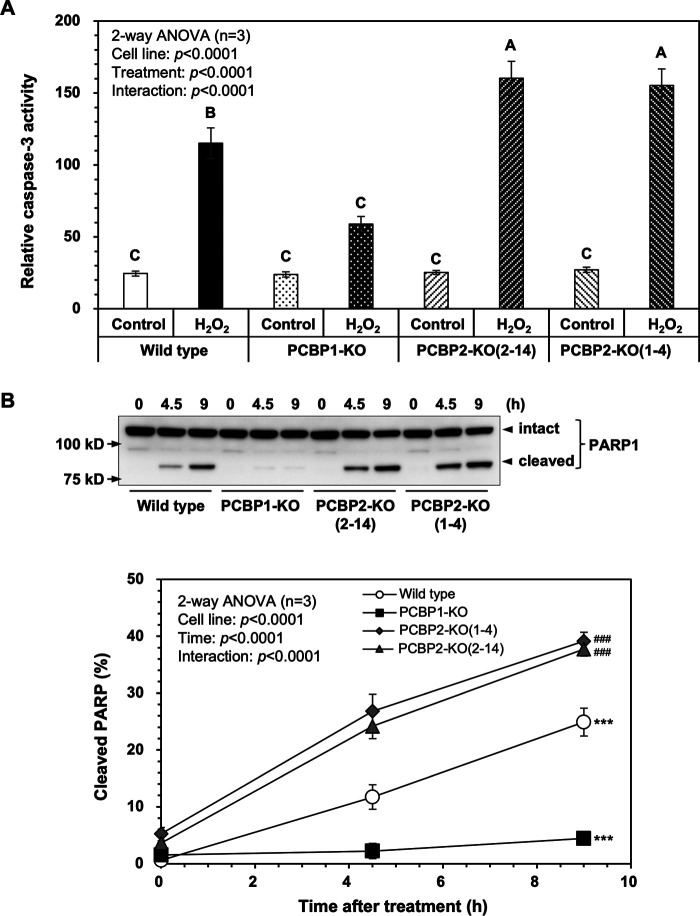
We then measured the apoptosis-related activities of the four cell lines (Fig. 6). As shown in Fig. 6A, the caspase-3 activity was increased more than 4.5-fold in WT cells 9 h after exposure to 0.2 mm H2O2, and PCBP1 deficiency significantly reduced the activity, whereas PCBP2 deficiency significantly increased the activity more than 6-fold, thus confirming that PCBP1 and PCBP2 deficiencies affect H2O2-induced apoptosis differently.
Figure 6.
PCBP2 deficiency enhances H2O2-induced apoptosis. A, relative caspase-3 activity after H2O2 exposure. Cells treated with (H2O2) or without (control) 0.2 mm H2O2 for 9 h were subjected to a caspase-3 assay. The relative caspase-3 activities are shown in the bar graph. The data represent the mean ± S.E. (error bars) of three independent experiments. A two-way ANOVA was performed as a statistical analysis. Bars without the same letter showed statistically significant differences in a post-hoc Tukey's HSD test, p < 0.05. B, cleavage of PARP-1 after H2O2 exposure. WT, PCBP1-KO, PCBP2-KO(2–14), and PCBP2-KO(1–4) cells were incubated with 0.2 mm H2O2. At the indicated times, the cells were recovered and boiled in SDS lysis buffer. The samples were subjected to SDS-PAGE followed by Western blotting using an anti-PARP-1 antibody (top). Band intensities shown in the blots were measured by ImageQuant TL, and the percentage of cleaved PARP1 was determined (bottom). Data from three independent experiments are shown as the mean ± S.E. A two-way ANOVA was performed as a statistical analysis. ***, p < 0.0001 versus all others; ###, p < 0.0001 versus WT.
The amounts of full-length PARP1 and its cleaved forms (following cleavage by caspases) in cells cultured in the presence of 0.2 mm H2O2 were determined by Western blotting (Fig. 6B, top). The cleavage of PARP1 occurred at significantly higher rates in the two PCBP2-KO cell lines than in WT cells, whereas PCBP1-KO cells exhibited a significantly lower rate of PARP1 cleavage (Fig. 6B, bottom).
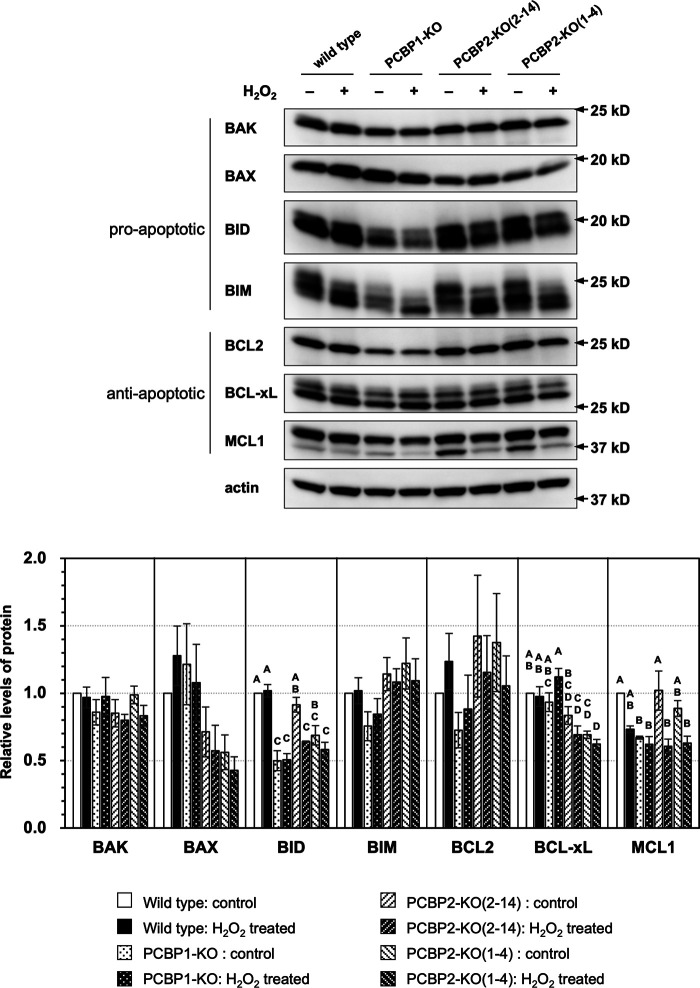
We also examined the expression of pro-apoptotic and anti-apoptotic proteins in WT, PCBP1-KO, and PCBP2-KO cell lines by Western blotting (Fig. 7). Among the four pro-apoptotic proteins, the levels of BAK and BIM proteins were not markedly changed by PCBP1 or PCBP2 deficiency with or without H2O2 exposure (2 h), and the levels of BAX tended to be decreased by PCBP2 deficiency with or without H2O2 exposure, although no statistical significance was seen. We found that the levels of BID were significantly decreased to 50% of those levels seen in WT cells by PCBP1 deficiency, with or without H2O2 exposure, whereas PCBP2 deficiency conversely decreased the levels of BID after exposure to H2O2. Among the three anti-apoptotic proteins, the levels of BCL-xL protein were significantly decreased to <70% of those levels seen in WT cells by PCBP2 deficiency. The expression of MCL1 tended to be decreased to 60% of those levels seen in WT control cells by H2O2 exposure in WT and PCBP2-KO cells, whereas PCBP1 deficiency decreased the levels to 60% of those levels seen in WT cells without H2O2 exposure.
Figure 7.
Expression profiles of pro- and anti-apoptotic proteins in WT, PCBP1-KO, PCBP2-KO(2–14), and PCBP2-KO(1–4) cells. Cells treated with (H2O2) or without (control) 0.2 mm H2O2 for 2 h were subjected to Western blotting using antibodies against BAK, BAX, BID, BIM, BCL2, BCL-xL, MCL1, and actin (top panels). The band intensities shown in the top panels were measured by ImageQuant TL, and the relative level of each protein to WT control was determined after being normalized to the actin level. Data from three independent experiments are shown as the mean ± S.E. (bottom panel). A two-way ANOVA was performed as a statistical analysis. Bars without the same letter showed statistically significant differences in a post-hoc Tukey's HSD test, p < 0.05.
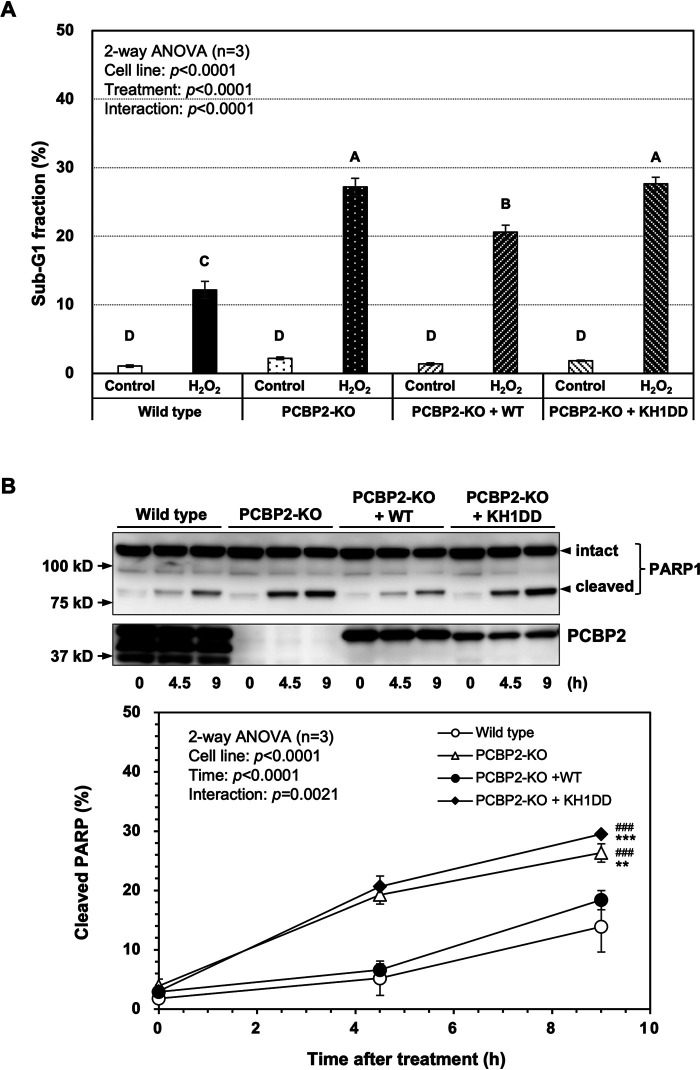
Finally, we introduced WT PCBP2 cDNA (WT) or PCBP2-KH1 mutant (KH1DD) cDNA into PCBP2-KO(2–14) cells and obtained stable cell lines (PCBP2-KO + WT and PCBP2-KO + KH1DD), which were subsequently exposed to H2O2. As shown in Fig. 8A, the sub-G1 fraction in PCBP2-KO + WT cells was significantly decreased compared with that in PCBP2-KO cells after H2O2 exposure, although the level was still higher than that of WT cells. In contrast, PCBP2-KO + KH1DD cells exhibited essentially the same sub-G1 fraction as seen in PCBP2-KO cells. Furthermore, PARP1 cleavage in PCBP2-KO + WT cells was decreased to the level seen in WT cells after H2O2 exposure, whereas in PCBP2-KO + KH1DD cells, a higher level of PARP1 cleavage was maintained, as seen in PCBP2-KO cells after H2O2 exposure (Fig. 8B).
Figure 8.
Enhanced apoptosis of PCBP2-deficient cells after exposure to H2O2 was suppressed by the expression of WT PCBP2 but not KH1DD mutant. A, percentage of cells in the sub-G1 fraction after H2O2 exposure. Cells (WT, PCBP2-KO, PCBP2-KO + WT, and PCBP2-KO + KH1DD) exposed to 0.2 mm H2O2 for 9 h were subjected to flow cytometry. The percentages of cells in the sub-G1 fraction are shown in the bar graph. The data represent the mean ± S.E. (error bars) of three independent experiments. A two-way ANOVA was performed as a statistical analysis. Bars without the same letter showed statistically significant differences in a post-hoc Tukey's HSD test, p < 0.001. B, cleavage of PARP-1 after H2O2 exposure. Cells (WT, PCBP2-KO, PCBP2-KO + WT, and PCBP2-KO + KH1DD) were incubated with 0.2 mm H2O2. At the indicated times, the cells were recovered and boiled in SDS lysis buffer. The samples were subjected to SDS-PAGE followed by Western blotting using an anti-PARP-1 antibody (top). The band intensities shown in the blots were measured by ImageQuant TL, and the percentage of cleaved PARP1 was determined (bottom). Data from three independent experiments are shown as the mean ± S.E. A two-way ANOVA was performed as a statistical analysis. ***, p < 0.0001 versus all others; ###, p < 0.0001 versus WT.
Taken together, these results indicate that PCBP2 has the capacity to suppress the apoptosis induced under oxidative conditions, depending on its binding to oxidized RNA, in contrast to PCBP1, which has the capacity to induce apoptosis under oxidative conditions.
Discussion
In the present study, we demonstrated that among five PCBP family proteins, PCBP2, as well as PCBP1, exhibits highly selective binding to highly oxidized RNA carrying two 8-oxoG residues at sites located in close proximity to each other compared with RNA carrying one 8-oxoG residue or control RNA without 8-oxoG residue and that this binding depended on the KH domains. In contrast, PCBP3, PCBP4, and hnRNPK bind RNAs with or without 8-oxoG and exhibit slightly increased binding to the former. Compared with WT HeLa S3 cells, PCBP2-KO cells exhibited increased apoptosis with increased caspase-3 activity and PARP1 cleavage under oxidative conditions (Figs. 5 and 6), and apoptosis or PARP1 cleavage was suppressed by the expression of WT PCBP2 but not KH1DD mutant lacking binding activity to highly oxidized RNA (Fig. 8). In contrast, PCBP1-KO cells exhibited reduced apoptosis with much less caspase-3 activity and PARP cleavage than WT cells (Figs. 5 and 6). These results indicate that PCBP1 and PCBP2 are involved in the regulation of the early step of apoptosis.
It has been established that H2O2 is a signaling molecule in the apoptotic pathways under oxidative stress and during aging (26). H2O2 can cause activation of caspase-9 and -3 accompanied by PARP1 cleavage and cytochrome c release to cytosol; thus, cells exposed to H2O2 execute apoptotic cell death (25, 27). However, the exact mechanism by which H2O2 activates the apoptotic pathway is still unclear.
We previously reported that PCBP1 specifically binds heavily oxidized RNA and that PCBP1 deficiency suppresses the activation of caspase and the apoptosis induced by H2O2 (12). In the present study, we showed that PCBP2 also specifically binds the heavily oxidized RNA, but PCBP2 deficiency enhances caspase-3 activation, PARP1 cleavage by caspase, and the apoptosis induced by H2O2. Because PCBP1 and PCBP2 bind highly oxidized RNA mutually exclusively without interaction (Fig. 2D), it is likely that PCBP2 competes with PCBP1 to bind highly oxidized RNA, thus antagonistically regulating the apoptotic pathway activated by H2O2. It is likely that PCBP1 and PCBP2 act as pro-apoptotic and anti-apoptotic factors, respectively, similar to the BAX and BCL2 families (28, 29). The expression of these pro- and anti-apoptotic factors was marginally affected by either PCBP1 or PCBP2 deficiency or oxidative stress (Fig. 7), suggesting that oxidation of the RNAs encoding these factors may differently influence their translation with or without PCBP1 or PCBP2 under oxidative conditions. It should be clarified in greater detail how oxidation of RNA affects the translation or stability of RNA.
The PCBP protein family consists of five members, PCBP1 to -4 and hnRNPK, with three conserved KH domains (Fig. 1A and Table 1), among which the KH1 and KH3 domains are essential for PCBP1 to bind heavily oxidized RNA. We therefore examined their binding capacity to RNA with or without two 8-oxoG residues at sites located in close proximity to each other. All five members of the PCBP protein family bound RNA with two 8-oxoG residues to a similar extent; however, PCBP3, PCBP4, and hnRNPK also substantially (but slightly less) bound to RNA without 8-oxoG, whereas the binding activity when PCBP2 bound RNA with two 8-oxoG residues was more than 2-fold that observed when it bound RNA with one 8-oxoG residue or RNA without 8-oxoG (as was observed with PCBP1). We therefore concluded that among the PCBP family proteins, PCBP1 and PCBP2 have the capability to selectively bind heavily oxidized RNA and can therefore regulate the apoptotic pathway induced by H2O2.
As the PCBP1 gene is an intron-less, processed gene derived from PCBP2 mRNA, PCBP1 has the highest homology with PCBP2 among all PCBP family proteins (Fig. 1A and Table 1) (13). It has been shown that PCBP2 and PCBP1 have essentially the same capability to bind RNA and share their roles in the regulation of RNA processing (14, 17, 18, 30). Indeed, PCBP2-KO cells exhibited essentially the same growth suppression as observed in PCBP1-KO cells (Fig. 4C) (12). However, the distribution of cell cycle phases in PCBP1-KO and PCBP2-KO cells were differently affected (Fig. 4D), indicating that PCBP1 and PCBP2 regulate cell proliferation in a different manner.
There are many reports indicating that PCBP1 and PCBP2 show differences in RNA binding and have different functions (19, 31–35). PCBP1 and PCBP2 are highly homologous; however, they show some divergence in the linker sequences between KH2 and KH3 domains of the two proteins (Table 1), which are known to be responsible for the different efficiency of poliovirus internal ribosome entry site–mediated translation between PCBP1 and PCBP2 through interaction with poliovirus stem-loop IV RNA (36). It is therefore likely that the linker sequences of PCBP1 and PCBP2 are responsible for their different functions, including the regulation of apoptosis.
It has been reported that the overexpression of PCBP1 triggers caspase-3– and caspase-8–mediated apoptosis of tumor cells; thus, PCBP1 has been considered as a tumor suppressor (37, 38), whereas PCBP2 is highly expressed in cancer tissues, such as esophageal squamous cell carcinoma (ESCC), and the reduced expression of PCBP2 induces apoptosis in ESCC cells (39). PCBP2 is therefore considered to be an oncogene (35). The present findings demonstrated the counteracting potential of PCBP1 and PCBP2 in the regulation of apoptosis; however, it has been reported that PCBP2-specific siRNA-transfected neurons show significantly decreased neuronal apoptosis (40), which suggests that the biological roles of PCBP1 and PCBP2 may differ among cell types. To delineate the differential regulation of apoptosis by PCBP1 and PCBP2, it is essential to identify factors that differentially interact with PCBP1 and PCBP2 and their signaling pathways.
Experimental procedures
Antibodies and chemicals
Anti-PCBP2 and anti-PCBP1 polyclonal antibodies were purchased from MBL (Nagoya, Japan). Anti-PARP1 (F-2) and anti-BCL2 (C2) were purchased from Santa Cruz Biotechnology, Inc. Anti-His6 mAb (9C11) conjugated to peroxidase was purchased from Wako Chemical (Osaka, Japan). Anti-BAK (D4E4), anti-BAX (D2E11), anti-BID (#2002), anti-BIM (C34C5), anti-BCL-xL (54H6), and anti-MCL1 (D35A5) were obtained from Cell Signaling Technology (Danvers, MA, USA), and anti-actin (6D1) was obtained from FUJIFILM Wako Pure Chemical Corp. (Osaka, Japan). Anti-rabbit IgG horseradish peroxidase–linked whole-donkey antibody and anti-mouse IgG horseradish peroxidase–linked whole-sheep antibody were obtained from GE Healthcare. Clarity Western ECL Substrate was supplied by Bio-Rad (Hercules, CA, USA). 8-OxoG–containing oligoribonucleotides and oligodeoxyribonucleotides were synthesized by Tsukuba Oligo Service (Ibaraki, Japan). The RNeasy Mini Kit was purchased from Qiagen (Hilden, Germany). EDTA-free protease inhibitor mixture cOmplete™ and Transcriptor reverse transcriptase were purchased from Roche (Basel, Switzerland). Isopropyl-β-d(−)-thiogalactopyranoside was purchased from Wako Chemical (Osaka, Japan). The HisTALON gravity column purification kit was purchased from Takara Bio Inc. (Shiga, Japan).
Preparation of the recombinant His-PCBP family proteins
RNAs were extracted from HeLa S3, MCF7, and U2OS cells with an RNeasy Mini Kit, and cDNAs were synthesized using total RNAs as the templates. DNA fragments containing ORFs for PCBP family proteins were amplified by PCR using cDNAs (PCBP2 and hnRNPK from HeLa S3, PCBP3 from MCF7, and PCBP4 from U2OS) as templates and cloned into a pET21a vector. Additionally, for the construction of KH mutant clones, base substitutions were introduced to PCBP2 ORF by site-directed mutagenesis. For this, the abovementioned pET21a-PCBP2 construct was used for PCR, with three primer pairs 5′-ATCGGAGACGACGGAGAATCAGTTAAGAAG-3′ and 5′-TTCTCCGTCGTCTCCGATGATACTGCCAAC-3′ for KH1DD mutant, 5′-ATTGGAGACGACGGATGCAAGATCAAGGAA-3′ and 5′-GCATCCGTCGTCTCCAATGAGAGAGCCACA-3′ for KH2DD mutant, and 5′-ATCGGGGACGACGGCGCCAAAATCAATGAG-3′ and 5′-GGCGCCGTCGTCCCCGATTATGCAGCCAAT-3′ for KH3DD mutant). The sequences of these KH mutant clones were validated by nucleotide sequencing. For the expression of recombinant PCBP family proteins, Escherichia coli BL21 (DE3) cells were transformed with pET21a-PCBP family ORF constructs. Transformants were cultured in 2× YT-G medium overnight and diluted with medium for exponential growth. The cells were grown with 1 mm isopropyl-β-d(−)-thiogalactopyranoside for 4 h to express recombinant His-PCBP family proteins and then collected by centrifugation. For the purification of His-PCBP family proteins, a HisTALON Gravity Column Purification Kit (Takara Bio Inc.) was used. Whole-cell extract was prepared from cell pellets suspended in xTractor Buffer, followed by the addition of DNase I. Lysates were incubated on ice for 15 min and centrifuged at 10,000 × g for 20 min at 4 °C. Supernatant was collected and applied to HisTALON gravity columns. After binding, the columns were washed with 8 column volumes of equilibration buffer followed by 7 column volumes of wash buffer. His-PCBP family proteins were then eluted from columns with 5 column volumes of elution buffer containing 150 mm imidazole. Purities of the His-PCBP family proteins were analyzed by SDS-PAGE, and protein concentrations were measured by the Bradford method. To remove excess imidazole, samples were subjected to gel filtration on PD-10 columns.
Preparation of RNA beads
Dynabeads M-270, purchased from Veritas (Tokyo, Japan) were washed three times with 1× BW buffer (5 mm Tris·HCl (pH 7.5), 0.5 mm EDTA, 1 m NaCl, and 0.01% Tween 20). Oligoribonucleotide (50 pmol each) with or without 8-oxoG was mixed with prewashed Dynabeads M-270 (25 µl), followed by rotation of the mixture for 30 min at room temperature. After conjugation, the beads were washed three times with 1× BW buffer. Before use, the beads were washed with the appropriate buffer.
Binding assay
8-OxoG–containing and control oligoribonucleotide beads were added to 200 μl of purified PCBP family protein (100 ng) or crude extracts (100 µg of protein), and the mixtures were rotated at 4 °C for 90 min. After rotation, the beads were washed three times with HNTG buffer (20 mm HEPES-KOH (pH 7.6), 150 mm NaCl, 0.1% (v/v) Triton X-100, 10% (v/v) glycerol), and the bound proteins were analyzed by Western blotting using specific antibodies or silver staining. Band intensities were measured by ImageQuant TL (GE Healthcare).
Immunoprecipitation
To prepare cell extracts, HeLa S3 cells were lysed with HTNG buffer. 8-OxoG or normal G oligoribonucleotide was mixed with the cell extract (100 µg), followed by incubation at 4 °C for 90 min. Anti-PCBP1 or PCBP2 antibody was then added to the reaction solutions, and the mixture was rotated at 4 °C for 90 min. To precipitate interacting proteins against each antibody, 20 μl of 50% slurry Protein G–agarose beads were added to the mixture and incubated at 4 °C for 3 h. After incubation, the beads were washed three times with HTNG buffer, and the bound materials ware analyzed by Western blotting.
Cell culture
HeLa S3 (WT), HeLa S3–derived PCBP1-KO (12), and PCBP2-KO cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) at 37 °C under 5% CO2. For the treatment of cells with H2O2, cells were cultured in DMEM with or without 10% FBS at 37 °C under 5% CO2.
Establishment of the PCBP2-knockout cell line
To perform genome editing for the PCBP2 gene locus, we searched for the target sequence of the CRISPR/Cas9 system using the Optimized CRISPR Design website and selected AGT TAA GAA GAT GCG CG as the target sequence. Two oligonucleotides possessing the target and complementary sequences were inserted into the cloning site of pX330-U6-Chimeric BB-CBh-hSpCas9 vector, purchased from Addgene (Cambridge, MA, USA). The construct was introduced into HeLa S3 cells by electroporation with a NEPA2000 system (Nepa Gene, Tokyo, Japan). The transfected cells were immediately seeded onto a 100-mm dish and cultured with DMEM containing 10% FBS at 37 °C under 5% CO2 for 2 weeks to form colonies. After incubation, candidate colonies were isolated and expanded in medium. To select the PCBP2-KO cell lines, cell extracts were prepared from these candidate clones, and the expression of PCBP2 protein in each cell line was determined by Western blotting using an anti-PCBP2 antibody.
Cloning and sequencing of the chromosomal region of the PCBP2 gene in PCBP2-deficient cell lines
Genomic DNAs were prepared from PCBP2-deficient cells as described previously (12). To amplify the coding region of the PCBP2 gene, genomic DNA was used as a template for PCR, using primers 5′-ATG CAT GGA AAG GTA TGC TCT AGC-3′ and 5′-CTC TTC CAG TTT GTC AAT GAT CAT-3′. The amplified DNA fragment was cloned into the pMD20-T vector (Takara Bio Inc.) using the TA-cloning method. The genomic sequences were determined using M13 reverse primer (5′-CAGGAAACAGCTATGAC-3′) and analyzed for indel mutations in the vicinity of the genome-editing site. DNA sequence analyses revealed that clone 2-14 carried two independent insertion mutations (T or G) at the same site, two-base downstream from the cutting site in exon 4 by Cas9 nuclease (see Fig. 4A, red arrowheads), whereas clone 1-4 carried an insertion mutation (T) at the same site and two different deletions from intron 3 to the cutting site in exon 4 (−70 nucleotides) or from the cutting site to intron 4 (−63 nucleotides). These two clones can thus be regarded as PCBP2-deficient lines.
Cell proliferation assay
Cells (1 × 104) were plated onto 30-mm dishes with 1 ml of medium (DMEM containing 10% FBS) and incubated at 37 °C under 5% CO2. At the indicated times, cells were detached from the dish by brief treatment with trypsin, and the number of cells was counted. Cell counting was performed each day, and the cell numbers of the HeLa S3 (WT), PCBP1-KO, and PCBP2-KO cell lines were compared.
Hoechst 33342/propidium iodide-double staining assay
Cells (1 × 105) were plated onto 30-mm dishes with 1 ml of medium (DMEM containing 10% FBS) and incubated at 37 °C under 5% CO2. The cells were exposed to 0.2 mm H2O2 for 9 h and were stained with 10 μm Hoechst 33342 (Hoechst, Molecular Probes) and 2 µg/ml PI (Sigma–Aldrich) for 30 min at 37 °C under 5% CO2. Cells were observed under a fluorescence microscope (BZ-9000, KEYENCE). The HeLa S3 (WT), PCBP1-KO, and PCBP2-KO cell lines were compared.
Flow cytometry
About 1 × 106 cells were detached from the culture dish with trypsin and collected into a tube. The cells were washed with 1× PBS and suspended in 300 μl of 1× PBS containing 0.1% Triton X-100. To remove any contaminating RNA, 4 μl of RNase A (10 mg/ml) was added to the cell suspension. The sample was then mixed with 100 μl of PI solution (100 µg/ml) and incubated at 37 °C for 30 min in the dark. After incubation, the cells were filtered through a cell strainer tube (352235; Corning Inc., Corning, NY, USA). To identify cell cycle phases G0/G1, S, and G2/M and the sub-G1 fraction, the cell suspensions were analyzed using a flow cytometer, FACSCalibur, and BD CellQuest Pro software program (BD Biosciences).
Establishment of WT PCBP2– and KH1DD mutant–expressing stable cell lines
DNA fragments carrying WT PCBP2 or KH1DD mutant cDNA sequences amplified from pET21a-PCBP2 or pET21a-PCBP2 KH1DD were cloned into the mammalian expression vector pBApo-EF1α (Takara Bio Inc.). The vectors were transfected into the PCBP2-KO(2–14) cell line, and cells harboring the expression vector were screened by drug selection using puromycin. Single-cell clones isolated by screening were expanded and analyzed for the expression of WT PCBP2 or KH1DD mutant protein by Western blotting, and each cell line was designated PCBP2-KO + WT or PCBP2-KO + KH1DD, respectively.
Caspase-3 activity
To measure the caspase-3 activity, an APOPCYTO caspase-3 colorimetric assay kit (MBL) was used as described previously (12).
Cleavage of PARP1
HeLa S3 (WT) and PCBP1-KO, PCBP2-KO cells (1 × 105) were plated onto a 30-mm dish with 1 ml of DMEM containing 10% FBS and incubated for 2 days. After incubation, the cells were washed with fresh medium and incubated with 0.2 mm H2O2 in DMEM containing 10% FBS for 4.5 or 9 h. The cells were then washed with 1× PBS and transferred into a fresh tube and then heated in SDS sample buffer at 96 °C for 10 min. The proteins were applied to SDS-PAGE and examined for PARP1 cleavage by Western blotting using a specific antibody.
Data availability
All data are contained within the article.
Acknowledgments
We thank Dr. Brian Quinn for editing a draft of the manuscript and T. Koizumi for technical assistance.
Author contributions—T. Ishii, T. Igawa, H. H., M. S., and Y. N. conceptualization; T. Ishii, T. Igawa, and Y. N. data curation; T. Ishii, T. Igawa, and Y. N. formal analysis; T. Ishii, H. H., T. F., M. S., and Y. N. supervision; T. Ishii, M. S., and Y. N. funding acquisition; T. Ishii, T. Igawa, and Y. N. validation; T. Ishii, T. Igawa, H. H., and Y. N. investigation; T. Ishii and T. Igawa visualization; T. Ishii, T. Igawa, and Y. N. methodology; T. Ishii, T. Igawa, and Y. N. writing-original draft; H. H., T. F., and M. S. resources; H. H. and M. S. project administration; H. H., T. F., and M. S. writing-review and editing.
Funding and additional information—This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, including the MEXT supported Program for the Strategic Research Foundation at Private Universities (Grant S1411042) and Japan Society for the Promotion of Science (JSPS) Grants-in-aid for Scientific Research 16K07260 and 20K06495 (to T. I.) and 17H01391 (to Y. N).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ROS
- reactive oxygen species
- 8-oxoG
- 8-oxoguanine
- Aβ
- β-amyloid
- PCBP
- poly(C)-binding protein
- KO
- knockout
- PI
- propidium iodide
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- KH
- K homology
- HSD
- honestly significant difference
- ANOVA
- analysis of variance.
References
- 1. Valko M., Leibfritz D., Moncol J., Cronin M. T., Mazur M., and Telser J. (2007) Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 39, 44–84 10.1016/j.biocel.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 2. Sekiguchi M., and Tsuzuki T. (2002) Oxidative nucleotide damage: consequences and prevention. Oncogene 21, 8895–8904 10.1038/sj.onc.1206023 [DOI] [PubMed] [Google Scholar]
- 3. Nakabeppu Y. (2017) Neurodegeneration caused by accumulation of an oxidized base lesion, 8-oxoguanine, in nuclear and mitochondrial DNA: from animal models to human diseases. in The Base Excision Repair Pathway: Molecular Mechanisms and Role in Disease Development and Therapeutic Design (Wilson D. M., III, ed) pp. 523–556, World Scientific, Singapore [Google Scholar]
- 4. Nakabeppu Y., Ohta E., and Abolhassani N. (2017) MTH1 as a nucleotide pool sanitizing enzyme: friend or foe? Free Radic. Biol. Med. 107, 151–158 10.1016/j.freeradbiomed.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 5. Kuraoka I., Suzuki K., Ito S., Hayashida M., Kwei J. S., Ikegami T., Handa H., Nakabeppu Y., and Tanaka K. (2007) RNA polymerase II bypasses 8-oxoguanine in the presence of transcription elongation factor TFIIS. DNA Repair 6, 841–851 10.1016/j.dnarep.2007.01.014 [DOI] [PubMed] [Google Scholar]
- 6. Ishii T., and Sekiguchi M. (2019) Two ways of escaping from oxidative RNA damage: selective degradation and cell death. DNA Repair 81, 102666 10.1016/j.dnarep.2019.102666 [DOI] [PubMed] [Google Scholar]
- 7. Hofer T., Badouard C., Bajak E., Ravanat J. L., Mattsson A., and Cotgreave I. A. (2005) Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol. Chem. 386, 333–337 10.1515/BC.2005.040 [DOI] [PubMed] [Google Scholar]
- 8. Tanaka M., Chock P. B., and Stadtman E. R. (2007) Oxidized messenger RNA induces translation errors. Proc. Natl. Acad. Sci. U. S. A. 104, 66–71 10.1073/pnas.0609737104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nunomura A., Perry G., Pappolla M. A., Wade R., Hirai K., Chiba S., and Smith M. A. (1999) RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer's disease. J. Neurosci. 19, 1959–1964 10.1523/JNEUROSCI.19-06-01959.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dai D. P., Gan W., Hayakawa H., Zhu J. L., Zhang X. Q., Hu G. X., Xu T., Jiang Z. L., Zhang L. Q., Hu X. D., Nie B., Zhou Y., Li J., Zhou X. Y., Li J., et al. (2018) Transcriptional mutagenesis mediated by 8-oxoG induces translational errors in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 115, 4218–4222 10.1073/pnas.1718363115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishii T., Hayakawa H., Sekiguchi T., Adachi N., and Sekiguchi M. (2015) Role of Auf1 in elimination of oxidatively damaged messenger RNA in human cells. Free Radic. Biol. Med. 79, 109–116 10.1016/j.freeradbiomed.2014.11.018 [DOI] [PubMed] [Google Scholar]
- 12. Ishii T., Hayakawa H., Igawa T., Sekiguchi T., and Sekiguchi M. (2018) Specific binding of PCBP1 to heavily oxidized RNA to induce cell death. Proc. Natl. Acad. Sci. U. S. A. 115, 6715–6720 10.1073/pnas.1806912115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leffers H., Dejgaard K., and Celis J. E. (1995) Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur. J. Biochem. 230, 447–453 10.1111/j.1432-1033.1995.tb20581.x [DOI] [PubMed] [Google Scholar]
- 14. Kiledjian M., Wang X., and Liebhaber S. A. (1995) Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J. 14, 4357–4364 10.1002/j.1460-2075.1995.tb00110.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Makeyev A. V., and Liebhaber S. A. (2000) Identification of two novel mammalian genes establishes a subfamily of KH-domain RNA-binding proteins. Genomics 67, 301–316 10.1006/geno.2000.6244 [DOI] [PubMed] [Google Scholar]
- 16. Choi H. S., Hwang C. K., Song K. Y., Law P. Y., Wei L. N., and Loh H. H. (2009) Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem. Biophys. Res. Commun. 380, 431–436 10.1016/j.bbrc.2009.01.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gamarnik A. V., and Andino R. (1997) Two functional complexes formed by KH domain containing proteins with the 5' noncoding region of poliovirus RNA. RNA 3, 882–892 [PMC free article] [PubMed] [Google Scholar]
- 18. Collier B., Goobar-Larsson L., Sokolowski M., and Schwartz S. (1998) Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem. 273, 22648–22656 10.1074/jbc.273.35.22648 [DOI] [PubMed] [Google Scholar]
- 19. Ghanem L. R., Kromer A., Silverman I. M., Chatterji P., Traxler E., Penzo-Mendez A., Weiss M. J., Stanger B. Z., and Liebhaber S. A. (2016) The Poly(C) binding protein Pcbp2 and its retrotransposed derivative Pcbp1 are independently essential to mouse development. Mol. Cell Biol. 36, 304–319 10.1128/MCB.00936-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krecic A. M., and Swanson M. S. (1999) hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11, 363–371 10.1016/S0955-0674(99)80051-9 [DOI] [PubMed] [Google Scholar]
- 21. Napthine S., Treffers E. E., Bell S., Goodfellow I., Fang Y., Firth A. E., Snijder E. J., and Brierley I. (2016) A novel role for poly(C) binding proteins in programmed ribosomal frameshifting. Nucleic Acids Res. 44, 5491–5503 10.1093/nar/gkw480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Asnani M., Pestova T. V., and Hellen C. U. T. (2016) PCBP2 enables the cadicivirus IRES to exploit the function of a conserved GRNA tetraloop to enhance ribosomal initiation complex formation. Nucleic Acids Res. 44, 9902–9917 10.1093/nar/gkw609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Troyano A., Sancho P., Fernández C., de Blas E., Bernardi P., and Aller P. (2003) The selection between apoptosis and necrosis is differentially regulated in hydrogen peroxide-treated and glutathione-depleted human promonocytic cells. Cell Death Differ. 10, 889–898 10.1038/sj.cdd.4401249 [DOI] [PubMed] [Google Scholar]
- 25. Singh M., Sharma H., and Singh N. (2007) Hydrogen peroxide induces apoptosis in HeLa cells through mitochondrial pathway. Mitochondrion 7, 367–373 10.1016/j.mito.2007.07.003 [DOI] [PubMed] [Google Scholar]
- 26. Giorgio M., Trinei M., Migliaccio E., and Pelicci P. G. (2007) Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 8, 722–728 10.1038/nrm2240 [DOI] [PubMed] [Google Scholar]
- 27. Park W. H. (2014) Anti-apoptotic effect of caspase inhibitors on H2O2-treated HeLa cells through early suppression of its oxidative stress. Oncol. Rep. 31, 2413–2421 10.3892/or.2014.3084 [DOI] [PubMed] [Google Scholar]
- 28. Igney F. H., and Krammer P. H. (2002) Death and anti-death: tumour resistance to apoptosis. Nat. Rev. Cancer 2, 277–288 10.1038/nrc776 [DOI] [PubMed] [Google Scholar]
- 29. Youle R. J., and Strasser A. (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59 10.1038/nrm2308 [DOI] [PubMed] [Google Scholar]
- 30. Thakur S., Nakamura T., Calin G., Russo A., Tamburrino J. F., Shimizu M., Baldassarre G., Battista S., Fusco A., Wassell R. P., Dubois G., Alder H., and Croce C. M. (2003) Regulation of BRCA1 transcription by specific single-stranded DNA binding factors. Mol. Cell Biol. 23, 3774–3787 10.1128/mcb.23.11.3774-3787.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blyn L. B., Swiderek K. M., Richards O., Stahl D. C., Semler B. L., and Ehrenfeld E. (1996) Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5' noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 93, 11115–11120 10.1073/pnas.93.20.11115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blyn L. B., Towner J. S., Semler B. L., and Ehrenfeld E. (1997) Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71, 6243–6246 10.1128/JVI.71.8.6243-6246.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yanatori I., Richardson D. R., Imada K., and Kishi F. (2016) Iron export through the transporter ferroportin 1 is modulated by the iron chaperone PCBP2. J. Biol. Chem. 291, 17303–17318 10.1074/jbc.M116.721936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryu M. S., Zhang D., Protchenko O., Shakoury-Elizeh M., and Philpott C. C. (2017) PCBP1 and NCOA4 regulate erythroid iron storage and heme biosynthesis. J. Clin. Invest. 127, 1786–1797 10.1172/JCI90519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo J., and Jia R. (2018) Splicing factor poly(rC)-binding protein 1 is a novel and distinctive tumor suppressor. J. Cell. Physiol. 234, 33–41 10.1002/jcp.26873 [DOI] [PubMed] [Google Scholar]
- 36. Sean P., Nguyen J. H., and Semler B. L. (2008) The linker domain of poly(rC) binding protein 2 is a major determinant in poliovirus cap-independent translation. Virology 378, 243–253 10.1016/j.virol.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang W., Shi H., Zhang M., Liu B., Mao S., Li L., Tong F., Liu G., Yang S., and Wang H. (2016) Poly C binding protein 1 represses autophagy through downregulation of LC3B to promote tumor cell apoptosis in starvation. Int. J. Biochem. Cell Biol. 73, 127–136 10.1016/j.biocel.2016.02.009 [DOI] [PubMed] [Google Scholar]
- 38. Shi H., Li H., Yuan R., Guan W., Zhang X., Zhang S., Zhang W., Tong F., Li L., Song Z., Wang C., Yang S., and Wang H. (2018) PCBP1 depletion promotes tumorigenesis through attenuation of p27Kip1 mRNA stability and translation. J. Exp. Clin. Cancer Res. 37, 187 10.1186/s13046-018-0840-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ye J., Zhou G., Zhang Z., Sun L., He X., and Zhou J. (2016) Poly (C)-binding protein 2 (PCBP2) promotes the progression of esophageal squamous cell carcinoma (ESCC) through regulating cellular proliferation and apoptosis. Pathol. Res. Pract. 212, 717–725 10.1016/j.prp.2016.05.008 [DOI] [PubMed] [Google Scholar]
- 40. Mao X., Liu J., Chen C., Zhang W., Qian R., Chen X., Lu H., Ge J., Zhao C., Zhang D., and Wang Y. (2016) PCBP2 modulates neural apoptosis and astrocyte proliferation after spinal cord injury. Neurochem. Res. 41, 2401–2414 10.1007/s11064-016-1953-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the article.