Abstract
Disorders that disrupt myelin formation during development or in adulthood, such as multiple sclerosis and peripheral neuropathies, lead to severe pathologies, illustrating myelin's crucial role in normal neural functioning. However, although our understanding of glial biology is increasing, the signals that emanate from axons and regulate myelination remain largely unknown. To identify the core components of the myelination process, here we adopted a microarray analysis approach combined with laser-capture microdissection of spinal motoneurons during the myelinogenic phase of development. We identified neuronal genes whose expression was enriched during myelination and further investigated hepatoma-derived growth factor-related protein 3 (HRP3 or HDGFRP3). HRP3 was strongly expressed in the white matter fiber tracts of the peripheral (PNS) and central (CNS) nervous systems during myelination and remyelination in a cuprizone-induced demyelination model. The dynamic localization of HPR3 between axons and nuclei during myelination was consistent with its axonal localization during neuritogenesis. To study this phenomenon, we identified two splice variants encoded by the HRP3 gene: the canonical isoform HRP3-I and a newly recognized isoform, HRP3-II. HRP3-I remained solely in the nucleus, whereas HRP3-II displayed distinct axonal localization both before and during myelination. Interestingly, HRP3-II remained in the nuclei of unmyelinated neurons and glial cells, suggesting the existence of a molecular machinery that transfers it to and retains it in the axons of neurons fated for myelination. Overexpression of HRP3-II, but not of HRP3-I, increased Schwann cell numbers and myelination in PNS neuron–glia co-cultures. However, HRP3-II overexpression in CNS co-cultures did not alter myelination.
Keywords: myelin, oligodendrocyte, Schwann cells, alternative splicing, nuclear translocation, spinal motoneuron, hepatoma-derived growth factor-related protein 3 (HRP3), HDGFRP3, axon, neuritogenesis
The myelin sheath is a lipid-rich insulating substance that enables fast propagation of an action potential over the axon while maintaining the continuity of the motor, sensory and cognitive functions of the nervous system in vertebrates (1, 2). Myelination is a sequential, multistep process in which a myelinating glia (an oligodendrocyte in the CNS and a Schwann cell in the PNS) adheres to an axon, ensheathes it, and wraps it multiple times.
The timing, specificity, rate, and amount of myelination are controlled by signals from neurons, astrocytes, and endocrine cells (3–7). For example, neuronal electrical activity modulates myelination by oligodendrocytes and Schwann cells (8–12). Increased neuronal cAMP induces glia to increase myelination (13–15). Additionally, synaptic vesicle release regulates myelination (15). Neuregulin-1 (NRG1) on the axon membranes plays a critical role in the development, differentiation and survival of the glia (16, 17). In NRG1 up-regulated transgenic animals, myelination via Schwann cells increases (18). NRG1/ErbB signaling modulates axon selection and myelin thickness in the PNS (19). In the CNS, the NRG1/ErbB pathway regulates oligodendrocytes' response to neuronal activity, hence also modulating myelination (20). Many factors such as NT-3 (neurotrophin 3), nerve growth factor, brain-derived neurotrophic factor (BDNF), platelet-derived growth factor, and IGF-1 (insulin-like growth factor 1) modulate myelination by regulating glia (12, 21–23). Even with all of this information, we are still far from a comprehensive understanding of the molecular mechanisms underlying the complex cellular interactions that culminate in the formation of myelin.
Furthermore, there are no therapies for many myelination diseases such as multiple sclerosis, Pelizaeus–Merzbacher disease, and Charcot–Marie–Tooth disease (24, 25). Promotion of remyelination by existing glial progenitor cells or myelination-capable cell transplantation is the best strategy to reverse the neural damage. However, developing both of these strategies requires a comprehensive knowledge of the myelination process, including the neuronal factors that regulate myelination (25–27).
To identify novel neuronal molecules that play roles in myelination, we analyzed gene expression profiles of spinal motoneurons (MNs) during and after the completion of myelination. A great majority of the MNs are in a promyelin form between the first and third day postnatum (P1–P3) in the rat spinal cord; they are fully myelinated by P21 (28, 29). Due to their large body size, we were able to isolate rat spinal MNs at P1 and P21 using laser-capture microdissection to compare their gene expression profiles. We compiled a comprehensive list of genes enriched in MNs that were being myelinated.
Hepatoma-derived growth factor–related protein 3 (HRP3) is one of the genes that are enriched during myelination. Previously, HRP3 was implicated in neurite outgrowth, survival of neurons, and angiogenesis in the retina (30–32). We showed that peak HRP3 expression and axonal localization coincided with the period of myelination in the white matter fiber tracts during developmental myelination and during remyelination after injury. Intriguingly, HRP3 localizes to axons during neurite growth, where it regulates microtubules and is restricted to nuclei afterward (30). To explain the dynamic subcellular localization of HRP3, we identified that alternative splicing of HRP3 created two isoforms: HRP3-I, the canonical isoform, and HRP3-II, which contains a functional nuclear export signal (NES) motif that is required for axonal targeting. Finally, in myelinating co-cultures of dorsal root ganglion (DRG) neurons, increased HRP3-II but not HRP3-I resulted in increased Schwann cell numbers and myelination. HRP3-II overexpression in CNS or DRG neurons did not increase myelination by oligodendrocytes.
Results
Gene expression profiling during myelination of spinal MNs
To uncover novel factors critical to myelination, we analyzed genes whose expression changed significantly in MNs during the early phase of myelination and after the completion of myelination. Laser-capture microdissection (LCM) was used to isolate single MNs from the lumbar spinal cord of P1 and P21 Wistar albino rats. We focused on the lumbar level because it is the most MN-dense region of the spinal cord. A rostro-caudal gradient of myelination exists in the developing postnatal spinal cord. In the lumbar region, 60–70% of rat spinal MN fibers are either in a promyelin or early myelinated form between the first and third day postnatum (28, 29). All spinal MNs are fully myelinated by P21.
To verify our isolation procedure, we evaluated the expression of well-established cell type–specific markers, including those of neurons, astrocytes, and oligodendrocytes, by real-time PCR analysis. A significant enrichment of neuronal markers was observed in the MN LCM samples, as compared with whole spinal cord tissue, which exhibited minimal astrocytic and oligodendroglial contaminants (Fig. S1).
To identify the genes that were differentially expressed between MNs of P1 and P21 animals, we extracted total RNA from these two populations and carried out cDNA microarray using the Rat Genome 230 2.0 array (Affymetrix). Animals from four separate litters were divided into P1 and P21 experimental groups, resulting in four biological replicates for each age that were processed independently for microarray analysis using Affymetric RAE230A/B rat genome arrays (between 1,000 and 2,000 cells per chip array; GEO number GSE133744). A total of 211 genes were up-regulated at the onset of myelination (i.e. in P1 samples relative to P21 samples (Table S1)), and 223 genes were up-regulated after the completion of myelination (i.e. in P21 samples relative to P1 samples (Table S2)) at 95% confidence or higher.
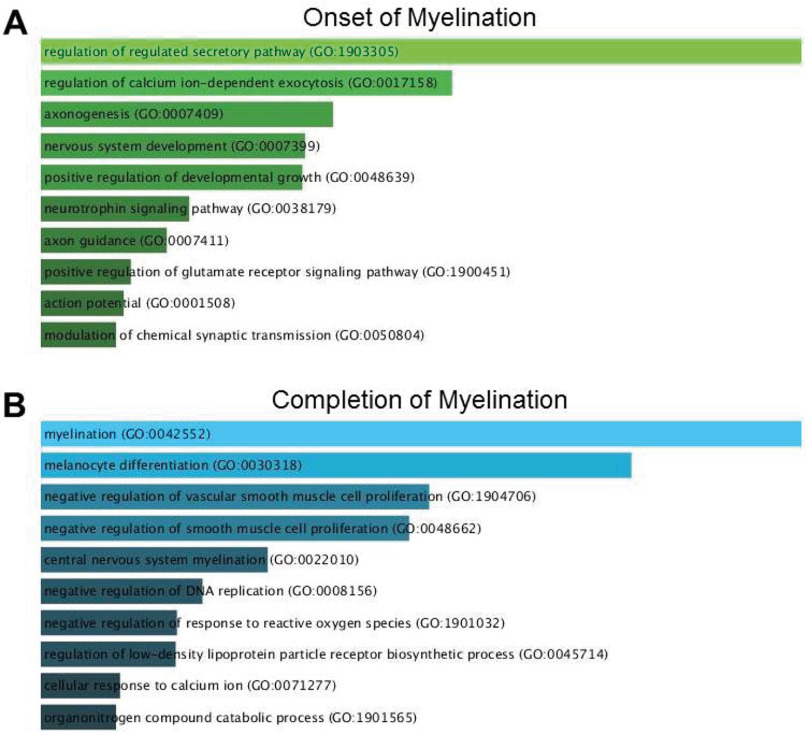
Genes involved in neural development, axon morphogenesis, and regulation of secretion and Ca2+ were enriched among the transcripts up-regulated during myelination (Fig. 1A). For example, neurotrophin-3, which is up-regulated over 1,000 times, plays a critical role in MN survival during the early perinatal period (33). Myosin heavy chain Myr 8 is an actin-based motor protein that may be involved in the process of neuronal cell migration (34). Finally, the severalfold up-regulation of doublecortin, a marker primarily expressed in migrating and differentiating neurons, indicates the yet immature stage of MNs by the time of birth (35). Not surprisingly, genes involved in myelination were highly enriched among the genes up-regulated after myelination was complete (Fig. 1B) because myelin was absent at P1 and any minor contaminants would inevitably be highly enriched.
Figure 1.
GO analysis of differentially expressed genes for enriched biological processes. A, at the onset of myelination, genes involved in neural development, axon morphogenesis, and regulation of secretion and Ca2+ were enriched. B, at P21, among other biological processes, myelination dominated because myelin was absent at P1, and any minor contaminants were inevitably highly enriched.
We then asked whether any of the genes that were up-regulated during myelination (Table S1) were expressed in the white matter tracts of the nervous system, which would make them attractive candidates for regulating the onset and progression of myelination. Several genes were selected based on literature, expression data, and resource availability. A series of immunohistochemical analyses on spinal cord, sciatic nerve, and various brain areas of WT animals at different stages of development and on cultured spinal MNs were performed for the selected genes. HRP3, Reg2, cannabinoid receptor, PACAP-38, DKK3, Npn1, PTPRO, agrin, FGF13, VEGF, and Npr1 were found to be expressed within the white matter tracts or in the processes of cultured spinal MNs (data not shown). We continued our analysis with HRP3, as it was found to be correlated with development of myelination in all the areas of peripheral nerves, spinal cord, and brain that we analyzed in vivo (Figs. 2 and 3; see below).
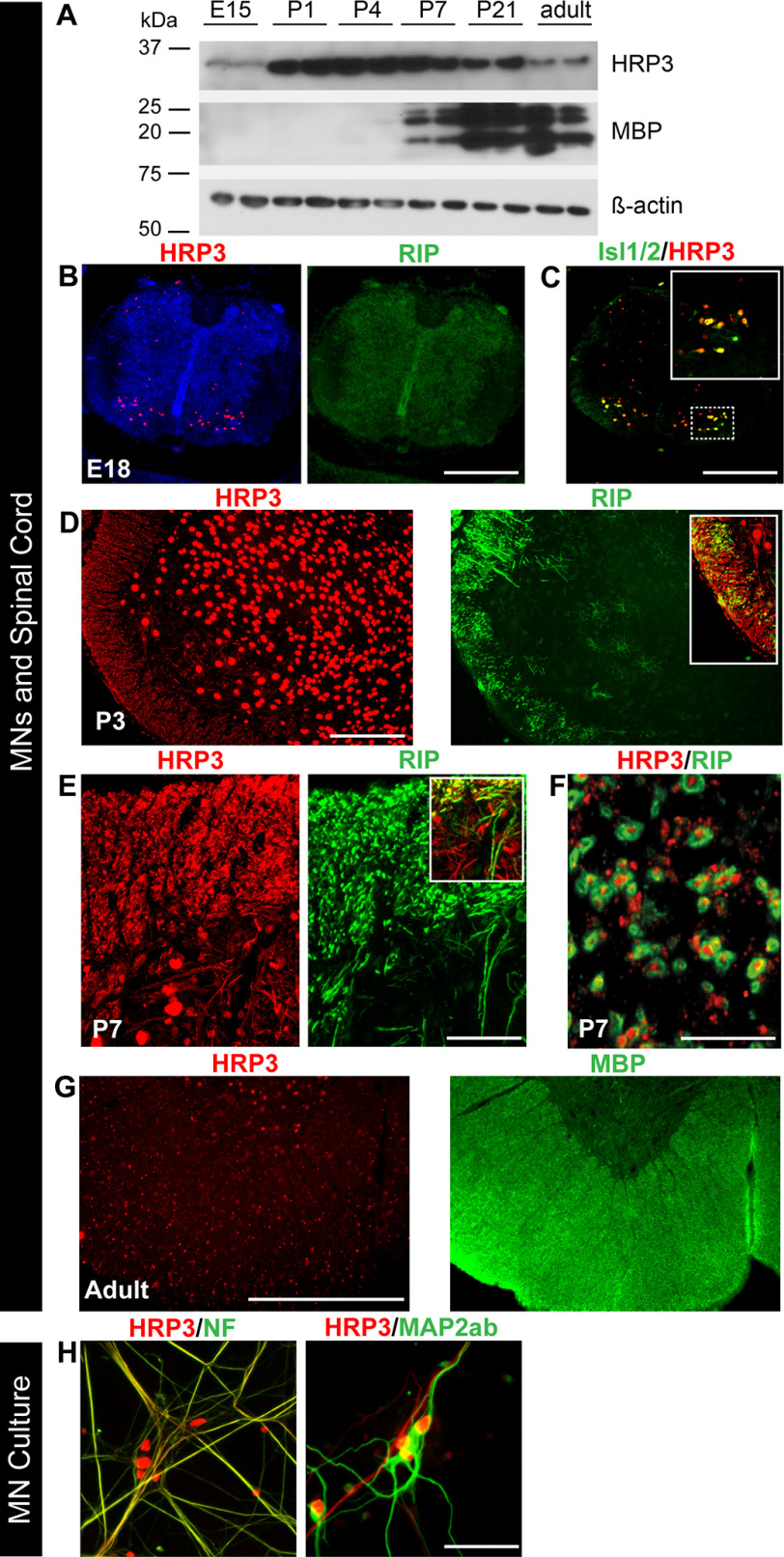
Figure 2.
HRP3 was up-regulated at the onset of myelination, with significant neuronal body and axonal expression in the developing MNs and white matter tract of spinal cord. A, Western blotting showing peak expression of HRP3 at the onset of and during myelination (P1–P7). Positions of the relevant molecular weight markers are indicated. B and G, immunohistochemistry with antibodies to HRP3 (red) in the rat spinal cord at E18 (premyelination), P3–P7 (active myelination), and adult (myelination completed). Myelin is identified by Rip (an early marker of myelination; green) and MBP (a later marker of myelination; green). B and C, during embryogenesis, at the premyelination stage identified by Rip (green), HRP3 (red) (B) was detected in the nuclei of developing MNs, marked with Isl1/2 (green) (C). The boxed region is shown at higher magnification in the top right corner of the image. Scale bars (B and C), 25 μm. D and E, during active myelination, HRP3 (red) localized to neuronal cell nuclei (D) and white matter tracts identified by Rip (green; arrows) (E). The boxed region shown in D and E highlights the appearance of HRP3 in the presumptive white matter tract before the actual formation of myelin. Scale bars, 100 μm (D) and 40 μm (E). F, the cross-sections of the ventral funiculus of spinal cord confirm the specific expression of HRP3 in axons. Scale bar, 5 μm. G, after completion of myelination, as identified by MBP (green), HRP3 remained in the nucleus. Scale bar, 100 μm. H, in spinal MN cultures HRP3 localized to the axons (NF, green), but not to the dendrites (MAP2ab, green). Scale bar, 40 μm.
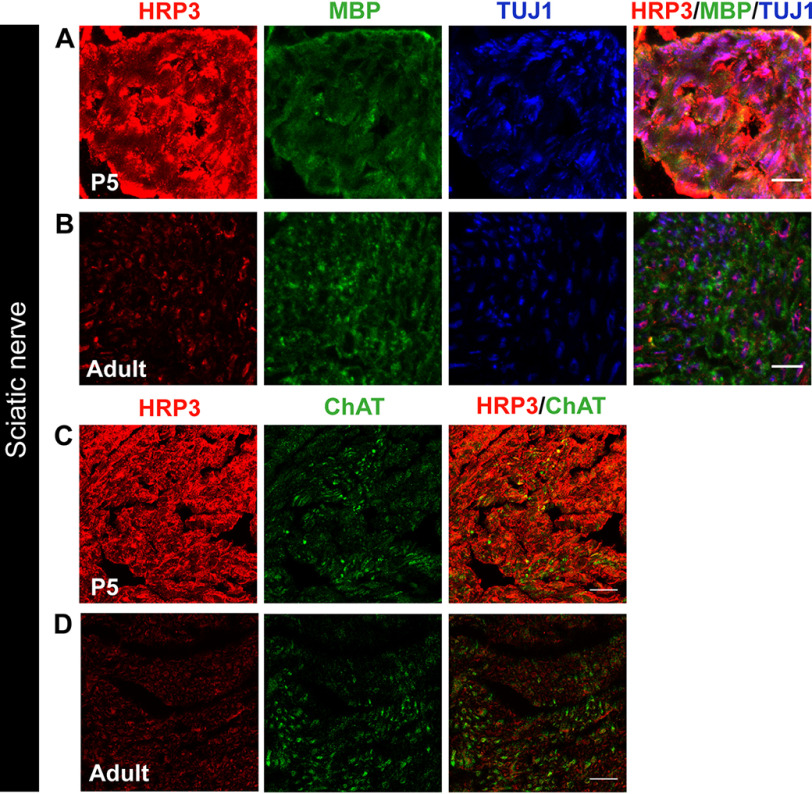
Figure 3.
HRP3 expression in PNS nerve fibers is transiently up-regulated during the myelination period. A and B, immunofluorescence analysis of HRP3 (red) expression was high and diffused in mouse sciatic nerve fibers (TUJ1; blue) during the period of myelination (MBP, green) compared with the adult nerve fibers. Images were acquired and visualized using the same parameters. Representative images are shown for comparison. Scale bars (A and B), 10 μm. E and F, HRP3 (red) levels were not different between motor neuron fibers (ChAT, green) and other fibers running in the sciatic nerve. HRP3 was acquired and visualized using the same parameters. Representative images are shown for comparison. Scale bars (C and D), 20 μm.
HRP3 levels and localization are temporally controlled during myelination
First, we examined the spinal cord from embryogenesis to adulthood to confirm that HRP3 expression parallels microarray data. Western blotting analysis revealed that HRP3 expression was low in the embryo; its expression increased 1 or 2 days before myelination and stayed high during early myelination, then decreased during late myelination, as marked by increased myelin basic protein (MBP) expression, and finally was lower in the adult animals (Fig. 2A). At an early stage of spinal cord development (E18), lack of Rip expression was an indication that myelination had not yet started and, at this stage, HRP3 expression was restricted to the nuclei of MNs marked by Isl1/2 (Fig. 2, B and C). During the most active period of spinal cord myelination (P1–P7), HRP3 expression was high both in the cell nuclei and in the developing white matter tracts (Fig. 2, D and E). HRP3 was present in the axons, wrapped by Rip-positive myelin membrane in the ventral funiculus of the spinal cord at P7 (Fig. 2F). Similar to previous observations with cortical neurons (30), in cultures of spinal MNs, HRP3 was detected in the axons but not in the dendrites (Fig. 2H). Finally, after the completion of myelination, HRP3 localized to the nuclei again (Fig. 2G).
We then examined HRP3's localization in rat spinal cord using subcellular fractionation. A hypotonic detergent-free lysis method resulted in three distinct fractions: nuclear, cytosolic, and membranous. Immunoblots revealed that HRP3 was clearly present in the membrane fraction along with the membrane-bound ATPase (Fig. S2).
Next, we examined HRP3 expression in the sciatic nerve. The level and localization of HRP3 within the axons were dependent upon the developmental stage. At P5, which is within the myelination period for mice, HRP3 staining was brighter (Fig. 3, A and C) than in the adult sciatic nerve (Fig. 3, B and D) when imaged under the same conditions. Moreover, HRP3 was present throughout the entire axon diameter during myelination (Fig. 3, A and C). HRP3 was concentrated in patches after myelination was complete (Fig. 3, B and D). A similar trend was observed in the rat sciatic nerve fibers (data not shown). Next, we asked whether HRP3 expression was different in MN axons compared with other nerve fibers within the sciatic nerve. No differences between choline acetyltransferase (ChAT)-positive MN axons and other axons were observed during or after myelination (Fig. 3, C and D). Coincidingly, HRP3 was observed in the cytoplasm and fibers of developing DRG neurons (36). These observations suggest that HRP3 may play a role in myelination of sensory neurons as well as the MNs.
Taken together, these data demonstrate that HRP3 levels and localization were temporally controlled in neurons, with HRP3 levels increasing during myelination in an axon-specific manner.
A novel alternatively spliced variant of HRP3 is targeted to the axon
To investigate the potential importance of HRP3 in myelination, we aimed to overexpress HRP3 in rat DRG neurons and MNs in vitro. Surprisingly, we found that overexpressed HRP3 (fused to GFP or Myc to distinguish the overexpressed form from the endogenous protein) failed to localize to the axons of DRG neurons and MNs in vitro or in vivo (Fig. 4A and Fig. S3A), unlike endogenous HRP3. We therefore sought to identify the mechanism of HRP3's localization to different cellular compartments. Eukaryotic proteins are usually transported in and out of the nucleus either through passive transport (efficient for small-size proteins) or by means of active transport, utilizing the cellular nucleo-cytoplasmic shuttling machinery. Proteins carrying a nuclear localization signal (NLS) are actively transported into the nucleus, whereas proteins with a NES sequence are transported out of the nucleus. Because HRP3 contains an NLS but no NES, we reasoned that HRP3 could be alternatively spliced into a new variant containing a NES. The NCBI database of expressed sequence tags was searched for sequences related to HRP3. A cDNA clone (accession no. Q9JMG7) was identified that was predicted to encode a polypeptide very similar to HRP3, but with a distinct C-terminal end. A motif search through the NetNES 1.1 prediction server, a web interface for an algorithm designed to predict the presence of NESs (37), predicted a leucine-rich NES. We named this novel gene product HRP3-II (Fig. 4D and Fig. S3C); the canonical variant, HRP3-I, contains an NLS without NES (Fig. 4C and Fig. S3C). Henceforth, HRP3 will refer to both isoforms. Although there is a relatively high diversity of leucine-rich NES, HRP3-II NES conformed to the consensus, in which the presence of regularly spaced, hydrophobic amino acids such as leucine, isoleucine, valine, phenylalanine, and methionine seemed to be the most important feature of the signal (Fig. S3D) (38).
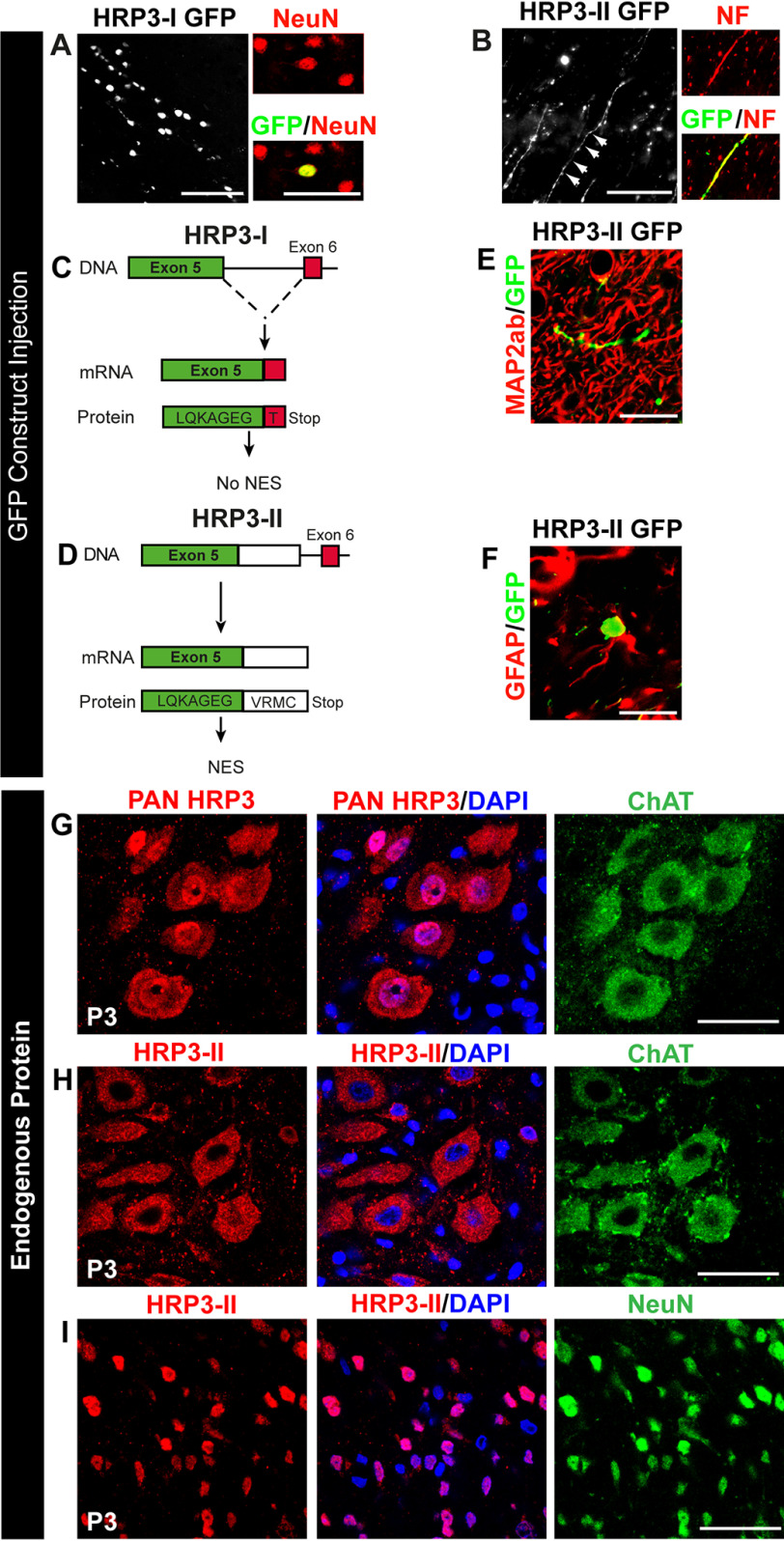
Figure 4.
Identification of the axonal variant of HRP3 (HRP3-II) and distinct subcellular localization of HRP3-II in neurons to be myelinated. A, the canonical isoform of HRP3, HRP3-I, localized to the neuronal nuclei (NeuN, red) in vivo as demonstrated by injection of its GFP fusion construct, HRP3-I–GFP (green), into adult rat brains. Scale bar, 100 μm. B, GFP construct of the new isoform HRP3-II–GFP (green) localized to the axons identified by neurofilament (red). Scale bar, 100 μm. C and D, schematic representations of the genomic organization of the rat HRP3-I (C) and HRP3-II (D). Note that HRP3-II is generated by an alternative 5′ splice site choice and contained at the C-terminal domain a unique region of 4 amino acids. E and F, HRP3-II GFP (green) was not found in dendrites (MAP2ab, red) (E) or outside nuclei of astrocytes (GFAP, red) (F). Scale bars, 35 μm (E) and 20 μm (F). G, a pan-HRP3 antibody (red) recognizing both isoforms revealed nuclear (DAPI, blue) and cytoplasmic (ChAT, green) localization of endogenous HRP3-I and HRP3-II in spinal MNs at the onset of myelination. Scale bar, 20 μm. H, our HRP3-II–specific antibody (red) showed that the endogenous HRP3-II localized to the cytoplasm in the MNs (ChAT, green). Scale bar, 20 μm. I, HRP3-II (red) localized to the nuclei of the dorsal neurons (NeuN, green). Scale bar, 20 μm.
Using cDNAs prepared from P7 spinal cord, we confirmed by RT-PCR that HRP3-II was expressed at the mRNA level, and its sequence was verified by DNA sequencing (Fig. S3E). The stop codon identified in HRP3-II was followed by a 3′-UTR and a consensus polyadenylation sequence (AATAAA) located 690 nucleotides upstream of the poly(A) tail, thereby indicating that the 3′ coding sequence was complete (not shown). Putative orthologs in rat, human, and horse were aligned to demonstrate the high level of conservation of the C-terminal end of HRP3-II and HRP3-I (Fig. S3F). Note that we identified a third and shorter HRP3 splice variant, but its C-terminal end was not evolutionarily conserved (Fig. S3F).
To test the functionality of HRP3-II NES, N-terminal GFP fusions of HRP3-I and HRP3-II were cloned into lentiviral vectors. After injection into the striatum of adult mice, HRP3-II–GFP, as predicted, was localized in cell nuclei and axonal processes of some neurons (Fig. 4B and Fig. S3B) in contrast to HRP3-I–GFP, which remained solely in the cell nuclei (Fig. 4A and Fig. S3A). Interestingly, HRP3-II–GFP was found neither in dendrites (Fig. 4E) nor outside the nuclei of astrocytes (Fig. 4F). Hence, we uncovered the presence of a NES that was functionally restricted to neuronal cells in the alternatively spliced intron 5 of HRP3. The novel isoform, HRP3-II, was exported out of the nucleus and targeted to the axons in a cell type–specific manner.
HRP3-II is exported from the nuclei of developing neurons and mature neurons to be myelinated
To examine HRP3-II independent of HRP3-I, we developed a polyclonal antibody against the C-terminal end of HRP3-II. The antibody recognized HRP3-II but not HRP3-I expressed by transfected HEK293T cells (Fig. S4A) and a band at a similar molecular weight from the crude spinal cord extracts (Fig. S4B) via Western blotting analysis. Similarly, HRP3-II but not HRP3-I was visualized by immunofluorescence on transfected HEK293T cells (Fig. S4C).
We next examined HRP3-II expression at different developmental stages. HRP3-II remained entirely localized outside the nuclei of MNs from E18 to adult stage, confirming the functionality of its NES (Fig. S4, D and E). The decreased presence of HRP3-II in the cytoplasm of adult MNs parallels low levels of axonal HRP3 observed in the adult sciatic nerve (Fig. S4E and Fig. 3 (B and D)). During myelination, a pan-HRP3 antibody identified HRP3 in both nuclei and cytoplasm of neurons (Fig. 4G). Surprisingly, HRP3-II localized to the nuclei of the neurons in the dorsal spinal cord, unlike MNs marked by ChAT, a cholinergic neuron-specific marker (Fig. 4, H and I). Because cholinergic neurons are myelinated, these data suggested that only neurons that were fated for myelination might be capable of translocating HRP3-II outside the nucleus (Fig. 4, H and I). The isoform-specific antibody failed to detect HRP3-II directly in the axon, suggesting that the C-terminal epitope recognized by the peptide antibody might be masked by posttranslational modifications. Thus, our results confirmed the identification of a novel HRP3 splice variant at the protein level and further indicated that alternative splicing of HRP3 induced profound changes in the subcellular localization and possibly the neuronal function of the protein. The temporal pattern of HRP3-II presence in the neuronal cytoplasm matched the temporal pattern detected by pan-HRP3 antibodies, with a peak of expression during the myelinogenic phase of MNs in the developing spinal cord. Interestingly, HRP3-II export out of the nucleus was specifically observed in neuron subtypes undergoing myelination.
HRP3-II overexpression increased myelination in DRG neuron–Schwann cell co-cultures
Myelination may be sensitive to alterations in the levels of axonal proteins. For example, the amount of membrane-bound neuregulin-1 type III isoform expressed on axons has dramatic effects on glial cell differentiation, with a threshold level triggering Schwann cell myelination (39). In contrast, the threshold level of type III NRG1 is not sufficient to initiate oligodendrocyte myelination (39). Therefore, we investigated the possible effects of overexpression and knockdown of the HRP3 splice variants on myelination in an in vitro model of PNS myelination.
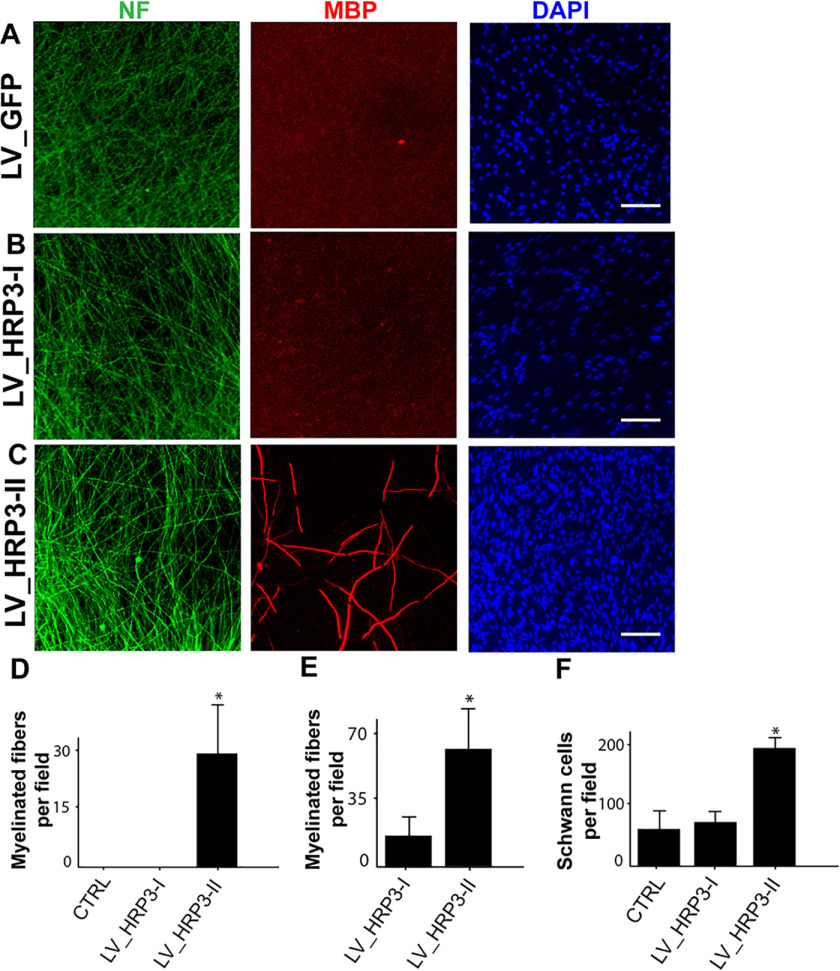
First, we examined the effects of HRP3 overexpression in DRG neuron–Schwann cell co-cultures by immunofluorescence analysis (Fig. 5). We used lentiviruses to overexpress HRP3-I, HRP3-II, or GFP in rat DRG neurons. We found that HRP3-II overexpression in DRG neurons resulted in a significant increase in myelination, as compared with GFP– and HRP3-I–overexpressing neurons in co-cultures with suboptimal Schwann cell density (Fig. 5D). In addition, HRP3-II overexpression in DRG neurons increased myelination under optimal Schwann cell density (Fig. 5E). The increase in myelination may be the result of increased Schwann cell numbers in HRP3-II–overexpressing co-cultures compared with GFP and HRP3-I–overexpressing co-cultures (Fig. 5F). To check whether any residual HRP3-II lentivirus might have infected Schwann cells indirectly and mediated this effect, we examined proliferation of immunopurified cultured Schwann cells after HRP3-II overexpression and found no significant increase in Schwann cell numbers (data not shown).
Figure 5.
HRP3-II, but not HRP3-I, promoted myelination in DRG neuron–Schwann cell co-cultures. A and C, lentivirus-mediated overexpression of GFP (A), HRP3-I (B), or HRP3-II (C) in DRG neurons (NF, green) co-cultured with primary Schwann cells (100,000 cells/coverslip) counterstained with DAPI (blue). D and E, quantification of myelination in the co-cultures. D, suboptimal Schwann cell seeding density did not trigger myelination in the GFP control and HRP3-I–expressing co-cultures, but myelination was triggered by HRP3-II–overexpressing neurons. E, similar increase in myelination was observed with higher Schwann cell seeding density. F, HRP3-II overexpression in the DRG neurons increased the number of Schwann cells, as judged by an increase in the number of DAPI-positive cells at suboptimal seeding density. Error bars, S.D. *, p < 0.001, unpaired Student's t test. Scale bars, 40 μm.
Next, we asked whether knocking down HRP3 in DRG neuron–Schwann cell co-cultures would impair myelination. We used lentiviruses to express HRP3 or scrambled siRNAs in rat DRG neurons. Interestingly, reduced HRP3 levels did not alter myelination (Fig. S5).
In conclusion, these findings suggest that the axonal HRP3-II variant can regulate myelin formation by Schwann cells, possibly indirectly via modulating glial cell metabolism, such as by increasing Schwann cell numbers.
HRP3 is expressed in the CNS white matter fiber tracts during myelination and remyelination
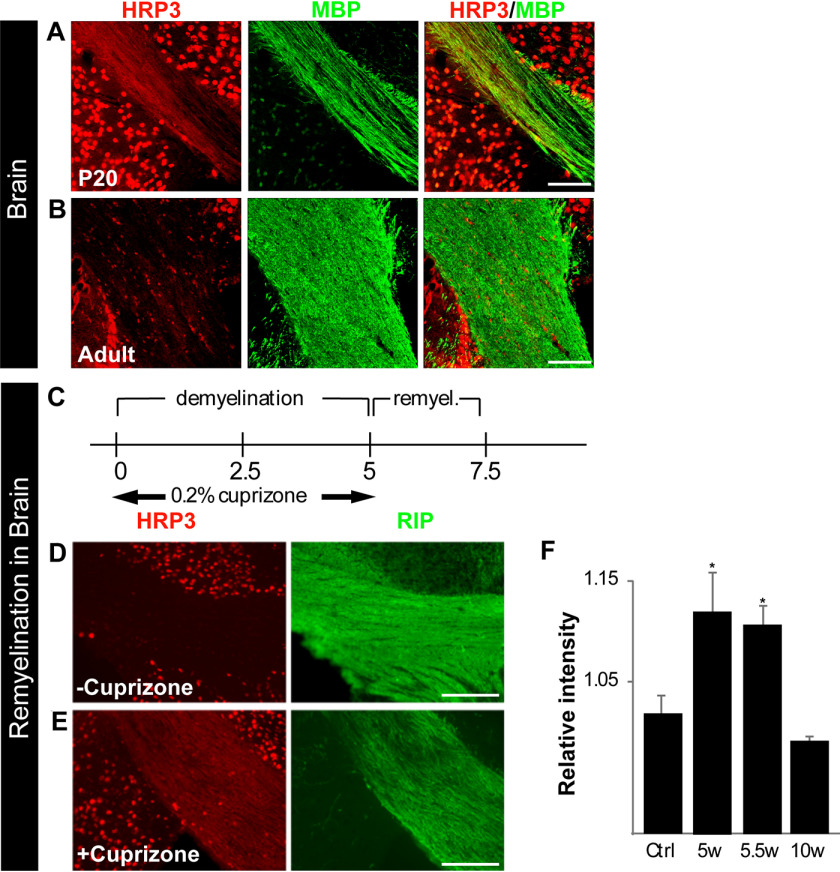
Encouraged by the correlation between HRP3 localization in the MNs and the sciatic nerve and myelination, we examined HRP3 expression in the corpus callosum of the brain during myelination and remyelination. P10–P30 is the most active period for myelination in the corpus callosum. At P20, HRP3 expression was high at the myelinating axons and the surrounding nuclei (Fig. 6A). After myelination was complete, it was localized almost completely to the nuclei (Fig. 6B). Interestingly, HRP3's localization followed a similar pattern during remyelination (Fig. 6, D and E). In the cuprizone experimental model, demyelination can be reproducibly induced in large areas of mouse brain, and an almost complete remyelination takes place in a matter of weeks (Fig. 6C). This process follows a characteristic pattern of cellular events, including phagocytosis of myelin debris, oligodendrocyte precursor cell proliferation, and oligodendrocyte differentiation (40). We examined HRP3 expression in the corpus callosum after cuprizone-induced demyelination. HRP3 levels increased during remyelination compared with controls and declined after the completion of remyelination (Fig. 6F).
Figure 6.
HRP3 expression in defined CNS white matter tracts consistently peaks with the period of myelination and during remyelination. A, HRP3 (red) transiently localized to the white matter tracts in the developing corpus callosum during myelination (P20) (MBP, green). Scale bar, 45 μm. B, similar to spinal MNs, HRP3 remained in the neuronal nuclei of adult brain. Scale bar, 30 μm. C and F, HRP3 was transiently up-regulated in white matter tracts during remyelination. C, a timeline of the experimental demyelination and remyelination. D and E, HRP3 (red) localized to the white matter tracts (Rip, green) in the remyelinating corpus callosum of the animals treated with cuprizone (E), whereas it remained in the nuclei in untreated control animals (D). Scale bars (D and E), 100 μm. F, quantification of staining intensity confirmed increased HRP3 protein expression in the corpus callosum of cuprizone-treated mice during remyelination (unpaired t test; *, p < 0.005). Error bars, S.D.
HRP3-positive nuclei were observed in the corpus callosum (Fig. 6, B and D) as well as in the white matter of the spinal cord (Fig. 2G), where very few neuronal nuclei are present. Previously, it was shown that HRP3 was expressed in the oligodendrocytes of adult animals (41). Similarly, we also observed HRP3 colocalized with an oligodendrocyte marker, GST-Pi (Fig. S6A), only in the adult. At earlier stages, neither the cytoplasmic nor the nuclear HRP3 colocalized with another oligodendrocyte marker, GalC (Fig. S6B). In addition, previous reports on HRP3 expression during embryogenesis did not describe expression in the early oligodendrocytes (36, 42).
HRP3-II overexpression did not increase myelination by oligodendrocytes
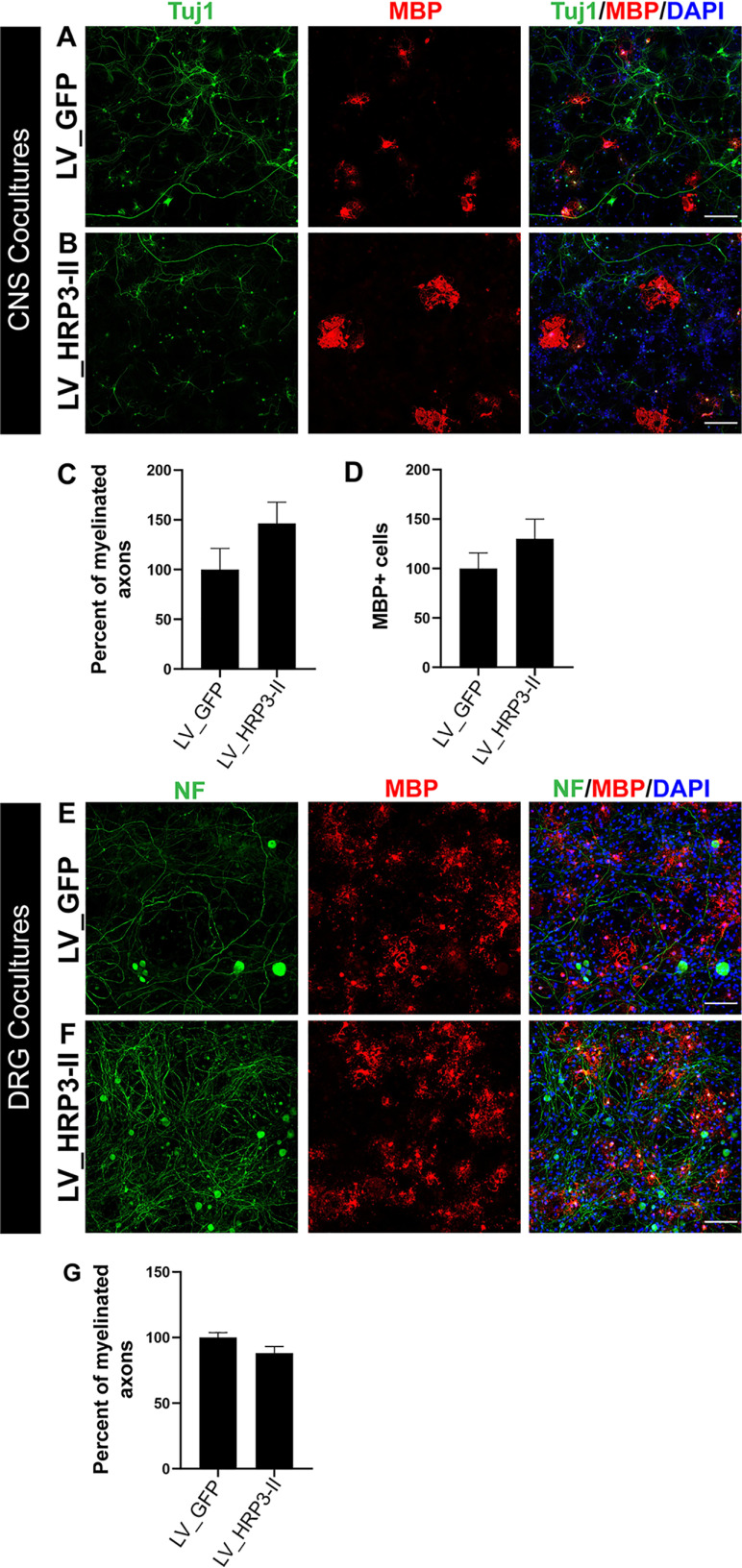
Next, to determine the possible role of HRP3 in CNS myelination, we used an in vitro CNS myelination model in which both neurons and oligodendrocytes were generated from mouse embryonic stem cells (mESCs) (43). HRP3-II GFP was overexpressed in mESC-derived CNS neurons and localized to neurites, some of which were myelinated (Fig. S7). However, HRP3-II variant overexpression in CNS neurons (Fig. 7, A and B) did not significantly increase myelination or numbers of MBP-positive (MBP+) mature oligodendrocytes compared with the GFP-expressing controls (Fig. 7, C and D). Finally, we asked whether overexpression of HRP3-II in DRG neurons could regulate myelination by oligodendrocytes. mESC-derived oligodendrocytes were co-cultured with mouse DRG neurons overexpressing HRP3-II or GFP (Fig. 7, E and F). HRP3-II overexpression did not increase myelination by oligodendrocytes (Fig. 7G).
Figure 7.
HRP3-II and did not increase myelination or number of mature oligodendrocytes in CNS co-cultures. A and B, mESC-derived CNS neurons (TUJ1, green) were infected with lentiviruses expressing GFP (A) or HRP3-II (B) and co-cultured with mESC-derived oligodendrocytes (MBP, red). Scale bars, 200 μm (A and B). C and D, percentage of myelinated fibers (C) and MBP+ oligodendrocyte number (D) did not increase significantly in cortical neurons overexpressing HRP3-II, compared with GFP-expressing cortical neurons. Error bar, S.E.; not significant, Mann–Whitney U test. E and F, lentivirus-mediated overexpression of GFP (E) or HRP3-II (F) in mouse primary DRG neurons (NF, green) co-cultured with mESC-derived oligodendrocytes (MBP, red). Scale bars (C and D), 100 μm. G, percentages of myelinated fibers were similar in both groups of DRG neuron-oligodendrocyte co-cultures. Error bars, S.E.
These findings suggest that the axonal HRP3-II variant overexpression in neurons is inadequate to regulate myelin formation by oligodendrocytes. This difference between oligodendrocytes and Schwann cells may reflect an intrinsic difference between CNS and PNS myelination processes.
Discussion
In the present study, by comparing gene expression profiles of MNs isolated both during and after myelination, we identified novel neuronal genes whose expression correlates with the onset and progression of myelination. Understanding the neuron-derived signals that regulate myelination is crucial for studying demyelinating diseases. Indeed, at least one of the genes up-regulated during myelination, HRP3, is observed at both mRNA and protein levels in multiple sclerosis plaques (44). HRP3's presence in these plaques may reflect its role. Therefore, the strategy described here and the gene expression data sets (GSE133744) are important to the researchers who study MNs, myelin, and related diseases.
In addition, we performed a series of immunohistochemical analyses with the top candidate genes and showed that HRP3 was highly expressed in the white matter fiber tracts precisely during the myelination of neurons. Therefore, we further analyzed HRP3, which is a member of the hepatoma-derived growth factor–related proteins. HRP3 is also an interesting candidate to modulate myelination because it regulates neurite growth in cortical neurons (30). HRP3 localizes to the growing neurites, where it promotes tubulin polymerization and stabilizes and bundles microtubules. After the neuritogenesis is complete, HRP3 leaves the axon and concentrates at the nucleus. Here, we described the mechanism behind this dynamic localization as the alternative splicing of HRP3. Whereas both HRP3-I and HRP3-II isoforms contain NLS, only the isoform we discovered, HRP3-II, has a NES. In parallel with the previous observations of HRP3 localizing only in the axons of growing cortical neurons (30), the NES-carrying HRP3-II was translocated specifically down the axons of MNs but not into their dendrites, suggesting that sequestration by an anchoring axon-specific protein might be working together with the NES to direct HRP3-II to the axon.
Interestingly, HRP3-II's NES was functional only in the neuron subtypes that were myelinated, whereas HRP3-II remained in the nucleus of nonmyelinated neurons in the spinal cord and the glial cells. Hence, we asked whether neuronal HRP3-II could regulate myelination in vitro. When HRP3-II was overexpressed specifically in the DRG neurons, their myelination by Schwann cells but not by oligodendrocytes was increased. The former was accompanied with increased Schwann cell numbers. In addition, overexpression of HRP3-II in the CNS neurons did not increase the number of myelinating oligodendrocytes or myelination significantly. Based on these results, we hypothesize that HRP3-II overexpression by DRG neurons is increasing myelination by increasing the availability of the Schwann cell pool to interact with, and myelinate, a given axon. In the PNS, Schwann cell proliferation is critical for matching axon and Schwann cell numbers during radial sorting, a prerequisite to myelination in peripheral nerves (45). Schwann cell proliferation has been shown to be axonally induced in in vitro studies and depends on direct contact with neurites and neurite fractions (46). In addition, sciatic nerve transection in newborn rats impairs Schwann cell proliferation (47), consistent with the idea that axons provide a mitogenic signal for developing Schwann cells. Neuregulin 1 is likely to play a major role in this process, as neuronal overexpression of the neuregulin type III-β3 isoform (also known as SMDF, for sensory and motoneuron-derived factor) leads to Schwann cell hyperproliferation and myelination of Remak bundles (48). However, the increased proliferation in the transgenic nerves is transient, with no persistence after P14, although expression of the SMDF transgene continues after P14 through adulthood.
The observed difference between PNS and CNS in vitro co-cultures may reflect an intrinsic difference between oligodendrocytes and Schwann cells. Indeed, the effect of NRG signaling on myelination by oligodendrocytes appears to be less straightforward (20). NRG switches oligodendrocytes from a neuronal activity–independent myelination mode to an activity-dependent myelination mode. Moreover, this switch is redundant, as BDNF signaling can replace NRG/ErbB signaling (20). These complexities and redundancies may explain the difference we observed by HRP3-II overexpression in vitro. It is possible that oligodendrocytes' response to CNS neurons overexpressing HRP3-II was dampened by other factors either in the co-culture medium or released by the neurons, and/or an additional unknown factor might have been the rate-limiting step in oligodendrocyte myelination that remains to be elucidated. In addition, DRG neurons might not express these factors, resulting in no change in oligodendrocyte myelination.
Remarkably, HRP3 can be secreted by neurons (31). The soluble HRP3 functions as a neuronal survival factor, whereas when bound to the surface of the cell culture dish, HRP3 promotes neurite growth, similar to the intracellular protein. Abouzied et al. (31) showed that HRP3 released from neurons to the cell culture medium protects neurons from death. To test whether HRP3 had a similar effect on the glial cells, we overexpressed HRP3 in either DRG neurons or mESC-derived CNS neurons and fed Schwann cells and oligodendrocytes with these media, respectively. No effect was observed on Schwann cells or oligodendrocytes (data not shown). These findings suggest that the effect of HRP3 on glial cells and myelination occurs possibly through a bound form of HRP3 or is indirect via its effect on the neurons. It is possible that neuronal overexpression of HRP3-II promotes survival, neuritogenesis, and/or an unknown property of neurons that leads to increased myelination.
Experimental procedures
Laser-capture microdissection, RNA purification, and gene chip hybridization
All experimental protocols involving rats and mice were approved by the Institutional Animal Care and Use Committee of the Salk Institute or the Ethics Committee of the Istanbul Medipol University.
Cryostat sections (12 µm thick) were stained with cresyl violet, rapidly dehydrated through xylenes, and stored until use in a vacuum dessicator. An Arcturus PixCell II machine (Arcturus, Mountain View, CA, USA) was used to laser-capture between 1,500 and 2,000 MNs per gene chip at the lumbar level of the spinal cord. Total RNA was purified using the Absolutely RNA Nanoprep Kit (Stratagene, La Jolla, CA, USA), and yield was assessed using RiboGreen RNA Quantitation Reagent (Molecular Probes, Inc, Eugene, OR, USA). Around 50 ng of RNA was obtained per pool of four rats. RNA (50 ng) was amplified in two rounds using the MessageAmp aRNA kit (Ambion Inc., Austin, TX, USA). This method utilizes reverse transcription with T7-oligo(dT) primers followed by in vitro transcription with T7 RNA polymerase. Biotinylated aRNA (15 µg) was fragmented and hybridized to Affymetrix Rat Genome RAE230A/B arrays according to the manufacturer's protocol. GeneChips were performed in quadruplicates, with each of four litters being split into a P1 and P21 sample. A total of 53–65% of the 31,000 probe sets were identified in MNs at P1 and P21 (GEO number GSE133744). Analysis and normalization were performed using two methods: the dChip 1.3 (49) and Drop Method (50) software packages. For the dChip analysis, after invariant set normalization and model fitting, a -fold change threshold (≥2) for each P1/P21 litter pair was used. Genes needed to surpass this threshold for each pair to be considered. The Drop Method analysis was unpaired, and genes had to have a 95% confidence value to be considered significant. Genes were required to be significant in both methods for consideration in this study. Values for average -fold changes that are reported here were calculated in dChip.
Gene ontology analysis
Before gene ontology (GO) analysis, probes targeting multiple genes and unknown genes were removed. The -fold changes were averaged for genes targeted by multiple probes. Genes differentially expressed during myelination and after myelination were tested for enrichment against the GO biological process using the EnrichR tool (51, 52).
RT-PCR and Q-PCR analysis
Total RNA was extracted from spinal cord tissue from 4–10-day-old rats using RNAzol. Four µg of total RNA was reverse-transcribed using the Superscript preamplification system (Invitrogen). First-strand cDNA was diluted 3-fold, and 2 µl of cDNA was used for PCR. Quantitative PCR analysis was performed using nonamplified mRNA from laser-captured motoneuron samples and compared with mRNA extracted from whole-lumbar spinal cord tissue. For each reaction, 2 µl of cDNA was placed in a 28-µl reaction mixture containing 15 µl of SYBR Green I PCR Master Mix (Applied Biosystems) and a 70 nm concentration of each primer (IDT). The following primers were used for PCR: HRP3 sense (5′-TTCAGCAACAGAGCTCTTCAG), HRP3-I antisense (5′-AGCATTCATTACGGTAGTTAGG), HRP3-II antisense (5′-TAACATTAGGCCATGGACAC), NF (5′-GCCGCCTACTCAAGGCTAAG; 5′-GCTTCGCAGCTCATTCTCCAG), transferrin (5′-AATTCCACCCTCTGTGACCTGTGT; 5′-TTCCAGGACAGTCTGGTGCTTCA), GFAP (5′-GTGGTATCGGTCCAAGTTTGC; 5′-CGATAGTCATTAGCCTCGTGCTT), β-actin (5′-TTGCTGACAGGATGCAGAAG; 5′-TAGAGCCACCAATCCACACA).
DRG neuron/Schwann cell myelinating co-cultures
Purified DRG neurons were isolated from E15 rat embryos and established on Matrigel-coated glass coverslips. Neuronal media were supplemented with 100 ng/ml nerve growth factor (Serotec). Neurons were infected with lentiviral particles and maintained for 2–3 weeks prior to seeding of Schwann cells. Primary Schwann cells were isolated from 3-day-old rat pups and maintained in Dulbecco's modified Eagle's medium, 10% fetal bovine serum, 2 mm l-glutamine until used. Pure Schwann cells were seeded directly onto DRGs (6-well plates) at low density and high density, 100,000 and 200,000 cells/coverslip, respectively. Myelination was initiated after 1 week by the addition of ascorbic acid (50 µg/ml). Quantification was performed after blinding the experimenter. Each condition was performed in triplicate wells, and data from four independent experiments were averaged.
CNS neuron/oligodendrocyte myelinating co-cultures
The myelinating co-cultures of mESC-derived neurons and oligodendrocytes were prepared as described previously (43). Briefly, neurons were differentiated toward a cortical phenotype (53). Neurons were infected with lentiviral particles and were maintained prior to co-culture establishment. Oligodendrocytes were generated in parallel cultures, and myelinating co-cultures were established by plating oligodendrocytes on neurons (43). Co-cultures were kept in myelination medium, which contained 20 ng/ml glial cell–derived neurotrophic factor (R&D Systems), 500 µg/ml cAMP (Sigma), 0.2 μm ascorbic acid (Sigma), and 40 ng/ml T3 (Sigma–Aldrich).
DRG/oligodendrocyte myelinating co-cultures
Oligodendrocytes were generated from mESCs as described above. DRGs were prepared from 6–8-week-old BALB/c mice by following a previously published protocol that enables selective isolation of the neurons (54). DRG neurons were seeded on 1.8-mg/cm2 poly-l-lysine (Sigma) and then 40 ng/mm2 laminin (Sigma)–coated 8-chamber slides (SPL). Then DRG neurons were infected with lentiviral particles and were kept for 4 days in Neurobasal A medium supplemented with 2% B27 supplement containing 2 mm Glutamax-I, antibiotic/antimycotic (1:100, v/v). On day 4, oligodendrocytes (100,000 cells/well) at day 15 of the differentiation protocol were seeded onto DRG neurons. Co-cultures were kept for 15 days in the 1:1 Dulbecco's modified Eagle's medium/F-12 and RPMI 1640 medium mix supplemented with 2 mm Glutamax-I, antibiotic/antimycotic (1:100, v/v), B27 supplement (1:50, v/v), N2 supplement (1:100, v/v), and 20 ng/ml triiodothyronine (Sigma). Medium was renewed every other day.
Lentiviral particle production
High-titer VSV-pseudotyped lentiviral vector stocks were produced in 293T as described previously, ranging from 5 × 109 to 1 × 1010 multiplicity of infection (55, 56). The GFP fusion proteins were expressed under the control of the CMV promoter.
Intracranial virus injections
Lentiviral particles were injected intracranially for in vivo HRP3-I and HRP3-II expressions. Injections were done into the motor cortex (M2), and coordinates were determined according to the Mouse Brain Atlas (57). Injection coordinates according to the bregma were as follows: x: −1.5; y: 1.5; z: −1.4. Two-month-old WT BALB/c mice were anesthetized with isoflurane and placed in the stereotaxic instrument (David Kopf Instruments, Tujunga, CA, USA). The scalp was incised, and a small hole was opened by drilling at the injection coordinates. 1–2 µl of lentivirus preparations was injected intracranially with a pulled glass pipette (Drummond Scientific, Wiretrol, Broomall, PA, USA), 50-µm tip diameter, and with a 30 nl/min injection speed set by a micromanipulator (Narishige, East Meadow, NY, USA). A waiting time of 10 min was given for lentiviral particles to diffuse, and the injection pipette was slowly withdrawn. The scalp was stitched, and animals were housed for 20 days for transgene expression.
Experimental demyelination and remyelination
Male C57BL/6 WT mice were fed ad libitum 0.2% cuprizone (oxalic bis(cyclohexylidenehydrazide)) (Sigma–Aldrich) mixed into milled chow. Mice were treated for 5 consecutive weeks with cuprizone and then were returned to a diet of normal pellet chow for 1 or 2 weeks (6 or 7 weeks of total treatment) to study the remyelination process. Untreated mice were maintained on a diet of normal pellet chow.
Immunohistochemistry
Rats and mice were anesthetized with a ketamine/xylazine mixture and transcardially perfused with 0.9% NaCl solution followed by 4% paraformaldehyde in 0.1 m phosphate buffer, pH 7.4. Spinal cords, brains, and sciatic nerves were postfixed overnight in 4% paraformaldehyde, transferred into 30% sucrose solution, and embedded in OCT. 10- or 20-µm frozen sections were thaw-mounted onto Superfrost slides (Mettler) and air-dried. Brains from adult animals were cut in 40-µm sections using a sliding microtome (Leica, Nussloch, Germany). 50-µm brain sections were cut with a vibratome. The following antibodies were used in various combinations: rabbit (rb) anti-HRP3 (gift from Dr. S. Franken), 1:300; rat anti-MBP (Serotech), 1:100; rb anti-MBP (Chemicon), 1:300; mouse (mo) anti-Rip (Hybridoma Bank), 1:20; mo anti-NeuN (Chemicon), 1:100; rb anti-neurofilament 200 kDa (Chemicon), 1:300; guinea pig anti-ChAT (Chemicon), 1:200; goat anti-ChAT (Abcam), 1:100; chicken anti-βIII tubulin, 1:400; mo anti-galactocerebroside (Millipore), 1:200; mo anti-MAP2ab (Sigma), 1:250; chicken and rb anti-GFP (Aves laboratory), 1:300; guinea pig anti-Isl1/2 (gift from Dr. S. L. Pfaff), 1:1000; and mo anti-GST-Pi (BD Biosciences), 1:1000. For indirect immunofluorescence, the following secondary antibodies were used for the study: 647 goat anti-rat IgG (1:750, Biotium), chicken anti-goat IgG (H + L) Alexa Fluor 594 (1:400), goat anti-chicken IgG (H + L) Alexa Fluor 568 (1:400), 488 goat anti-rabbit IgG (1:750, Biotium), 633 goat anti-mouse IgG (1:250, Biotium), and donkey-raised secondary antibodies conjugated to either cy3, FITC, or cy5 (Jackson) used at a 1:250 dilution.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde-PBS for 15 min, blocked, and permeabilized with horse serum (10%) and Triton X-100 (0.1%) in PBS and then incubated overnight with a combination of primary antibodies (see above and 1:1,000 mouse or rabbit anti-TUJ1 (Covance), rat anti-MBP (1:100, Bio-Rad), and chicken anti-neurofilament heavy polypeptide (1:100,000, Abcam). On the next day, they were washed with PBS and incubated with secondary antibodies: 633 goat anti-chicken IgG (1:750, Molecular Probes) and 555 goat anti-rat IgG (1:750, Biotium).
Subcellular fractionation
Nuclear and cytosolic fractions of rat spinal cord were prepared using a hypotonic, detergent-free buffer, essentially according to the manufacturer's instructions (Sigma, CellLytic NuCLEAR Extraction kit). The cytosolic fraction was then centrifuged at 100,000 × g for 1 h to give crude membrane pellet and soluble fractions. Soluble fractions were discarded, and the pellet was washed, centrifuged again, and resuspended into 5 mm Tris, pH 8, 0.6% IGEPAL. Equal amounts of proteins from each fraction (10 µg/lane for nuclear and cytosolic fractions, 100 µg/lane for membrane fractions) were resolved in 4–12% SDS gels.
Antibody production
Antibodies against pan-HRP3 and HRP3 isoform II were raised in rabbits (Covance) against peptides (Sigma) conjugated to KLH (Pierce) and consisting of the following sequences: pan-HRP3, CGDAGNDTRNT; HRP3 isoform II, CAGEGVRM. The antibodies were purified on antigen columns before use.
Image acquisition and analysis
Fluorescent signals were acquired on Zeiss LSM confocal or Zeiss SD confocal microscopes. Images were processed on Zen (Zeiss), ImageJ (58), and Adobe Photoshop and Illustrator (Adobe Systems) software. Staining intensity in the cuprizone experiment was determined for 5–10 sections (2–3 animals for each group) by a blinded experimenter and measured with ImageJ software followed by normalization to staining intensity outside the corpus callosum. The images of myelination by oligodendrocytes were blinded and were quantified using computer-assisted evaluation of myelin formation (43).
MN preparation
MN cultures were prepared from E15 rat spinal cords as described previously (59), except a single step of purification was performed using a 10% (v/v) solution of Optiprep (60%, Nycomed Pharma) in 4.4% glucose/Tricine, 10 mm, pH 7. Mixed MN cultures were maintained in Neurobasal Medium A (Gibco) in the presence of 1 ng/ml BDNF (Sigma) added at the time of cell seeding.
Data availability
The microarray data are deposited in the Gene Expression Omnibus with accession code GSE133744. All other data are contained in this article.
Supplementary Material
Acknowledgments
We gratefully thank Sebastian Franken for generously providing HRP3 antibodies. We also thank Ed Lein, Jonah Chan, Carla Taveggia, and Brigitte Pettmann for advice with the LCM and in vitro cultures and Mary Lynn Gage for editorial comments on the manuscript.
This article contains supporting information.
Author contributions—B. E. K., S. G., and F. H. G. conceptualization; B. E. K., S. G., B. K. V., S. G. G., G. W. Y., J. B. A., and F. H. G. formal analysis; B. E. K. and F. H. G. supervision; B. E. K., S. G., X. X., G. W. Y., and F. H. G. funding acquisition; B. E. K., S. G., B. K. V., S. G. G., and F. H. G. investigation; B. E. K., S. G., and F. H. G. methodology; B. E. K., S. G., S. G. G., and F. H. G., writing-original draft; F. H. G. project administration; B. E. K., S. G., B. K. V., and F. H. G. writing-review and editing; A. M. D. and X. X. resources; B. E. K., S. G., G. W. Y., J. B. A., and F. H. G. data curation.
Funding and additional information—This work was supported by the Christopher and Dana Reeve Foundation, the Roche Foundation, and the Swiss National Science Foundation. Research was also supported in part by the American Heart Association and Paul G. Allen Frontiers Group Grant 19PABHI34610000 (to F. H. G.); Leona M. and Harry B. Helmsley Charitable Trust Grant 2017-PG-MED001; the Grace Foundation; the JPB Foundation; Annette C. Merle-Smith; the Robert and Mary Jane Engman Foundation; Lynn and Edward Streim; and the Ray and Dagmar Dolby Family Fund. G. W. Y. was supported by the Crick-Jacob Center for Theoretical and Computational Biology. X. X. was supported by the Neurosurgery Neuroscience Consortium Fellowship. B. E. K. and B. K. V. were supported by Scientific and Technological Research Council of Turkey Grant 116S059.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- NRG1
- neurogulin
- MN
- motoneuron
- HRP3
- hepatoma-derived growth factor-related protein 3
- HDGF
- hepatoma-derived growth factor
- CNS
- central nervous system
- PNS
- peripheral nervous system
- BDNF
- brain-derived neurotrophic factor
- NES
- nuclear export signal
- DRG
- dorsal root ganglion
- LCM
- laser-capture microdissection
- MBP
- myelin basic protein
- NLS
- nuclear localization signal
- mESC
- mouse embryonic stem cell
- E
- embryonic day
- P
- postnatal day
- ChAT
- choline acetyltransferase
- aRNA
- amplified RNA
- rb
- rabbit
- mo
- mouse
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- DAPI
- 4′,6-diamidino-2-phenylindole
- GO
- gene ontology.
References
- 1. Shimizu T., Osanai Y., and Ikenaka K. (2018) Oligodendrocyte-neuron interactions: impact on myelination and brain function. Neurochem. Res. 43, 190–194 10.1007/s11064-017-2387-5 [DOI] [PubMed] [Google Scholar]
- 2. Nave K. A. (2010) Myelination and the trophic support of long axons. Nat. Rev. Neurosci. 11, 275–283 10.1038/nrn2797 [DOI] [PubMed] [Google Scholar]
- 3. Doyle J. P., and Colman D. R. (1993) Glial-neuron interactions and the regulation of myelin formation. Curr. Opin. Cell Biol. 5, 779–785 10.1016/0955-0674(93)90025-L [DOI] [PubMed] [Google Scholar]
- 4. Nualart-Marti A., Solsona C., and Fields R. D. (2013) Gap junction communication in myelinating glia. Biochim. Biophys. Acta 1828, 69–78 10.1016/j.bbamem.2012.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schumacher M., Hussain R., Gago N., Oudinet J. P., Mattern C., and Ghoumari A. M. (2012) Progesterone synthesis in the nervous system: implications for myelination and myelin repair. Front. Neurosci. 6, 10 10.3389/fnins.2012.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watkins T. A., Emery B., Mulinyawe S., and Barres B. A. (2008) Distinct stages of myelination regulated by γ-secretase and astrocytes in a rapidly myelinating CNS coculture system. Neuron 60, 555–569 10.1016/j.neuron.2008.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Simons M., and Nave K. A. (2015) Oligodendrocytes: myelination and axonal support. Cold Spring Harb. Perspect. Biol. 8, a020479 10.1101/cshperspect.a020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barres B. A., and Raff M. C. (1993) Proliferation of oligodendrocyte precursor cells depends on electrical activity in axons. Nature 361, 258–260 10.1038/361258a0 [DOI] [PubMed] [Google Scholar]
- 9. Shrager P., and Novakovic S. D. (1995) Control of myelination, axonal growth, and synapse formation in spinal cord explants by ion channels and electrical activity. Brain Res. Dev. Brain Res. 88, 68–78 10.1016/0165-3806(95)00081-N [DOI] [PubMed] [Google Scholar]
- 10. Demerens C., Stankoff B., Logak M., Anglade P., Allinquant B., Couraud F., Zalc B., and Lubetzki C. (1996) Induction of myelination in the central nervous system by electrical activity. Proc. Natl. Acad. Sci. U. S. A. 93, 9887–9892 10.1073/pnas.93.18.9887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stevens B., Tanner S., and Fields R. D. (1998) Control of myelination by specific patterns of neural impulses. J. Neurosci. 18, 9303–9311 10.1523/JNEUROSCI.18-22-09303.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zuchero J. B., and Barres B. A. (2013) Intrinsic and extrinsic control of oligodendrocyte development. Curr. Opin. Neurobiol. 23, 914–920 10.1016/j.conb.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malone M., Gary D., Yang I. H., Miglioretti A., Houdayer T., Thakor N., and McDonald J. (2013) Neuronal activity promotes myelination via a cAMP pathway. Glia 61, 843–854 10.1002/glia.22476 [DOI] [PubMed] [Google Scholar]
- 14. Monje P. V., Soto J., Bacallao K., and Wood P. M. (2010) Schwann cell dedifferentiation is independent of mitogenic signaling and uncoupled to proliferation: role of cAMP and JNK in the maintenance of the differentiated state. J. Biol. Chem. 285, 31024–31036 10.1074/jbc.M110.116970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mensch S., Baraban M., Almeida R., Czopka T., Ausborn J., El Manira A., and Lyons D. A. (2015) Synaptic vesicle release regulates myelin sheath number of individual oligodendrocytes in vivo. Nat. Neurosci. 18, 628–630 10.1038/nn.3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quintes S., Goebbels S., Saher G., Schwab M. H., and Nave K. A. (2010) Neuron-glia signaling and the protection of axon function by Schwann cells. J. Peripher. Nerv. Syst. 15, 10–16 10.1111/j.1529-8027.2010.00247.x [DOI] [PubMed] [Google Scholar]
- 17. Wang Z., Colognato H., and Ffrench-Constant C. (2007) Contrasting effects of mitogenic growth factors on myelination in neuron-oligodendrocyte co-cultures. Glia 55, 537–545 10.1002/glia.20480 [DOI] [PubMed] [Google Scholar]
- 18. Michailov G. V., Sereda M. W., Brinkmann B. G., Fischer T. M., Haug B., Birchmeier C., Role L., Lai C., Schwab M. H., and Nave K. A. (2004) Axonal neuregulin-1 regulates myelin sheath thickness. Science 304, 700–703 10.1126/science.1095862 [DOI] [PubMed] [Google Scholar]
- 19. Garratt A. N., Voiculescu O., Topilko P., Charnay P., and Birchmeier C. (2000) A dual role of erbB2 in myelination and in expansion of the Schwann cell precursor pool. J. Cell Biol. 148, 1035–1046 10.1083/jcb.148.5.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lundgaard I., Luzhynskaya A., Stockley J. H., Wang Z., Evans K. A., Swire M., Volbracht K., Gautier H. O., Franklin R. J., Charles F.-C., Attwell D., and Káradóttir R. T. (2013) Neuregulin and BDNF induce a switch to NMDA receptor-dependent myelination by oligodendrocytes. PLos Biol. 11, e1001743 10.1371/journal.pbio.1001743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barres B. A., Lazar M. A., and Raff M. C. (1994) A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development 120, 1097–1108 [DOI] [PubMed] [Google Scholar]
- 22. McMorris F. A., and McKinnon R. D. (1996) Regulation of oligodendrocyte development and CNS myelination by growth factors: prospects for therapy of demyelinating disease. Brain Pathol. 6, 313–329 10.1111/j.1750-3639.1996.tb00858.x [DOI] [PubMed] [Google Scholar]
- 23. Xiao J., Kilpatrick T. J., and Murray S. S. (2009) The role of neurotrophins in the regulation of myelin development. Neurosignals 17, 265–276 10.1159/000231893 [DOI] [PubMed] [Google Scholar]
- 24. Aydinli F. İ., Çelik E., Vatandaşlar B. K., and Kerman B. E. (2016) Myelin disorders and stem cells: as therapies and models. Turk. J. Biol. 40, 1068–1080 10.3906/biy-1512-83 [DOI] [Google Scholar]
- 25. Pareyson D., Saveri P., and Pisciotta C. (2017) New developments in Charcot-Marie-Tooth neuropathy and related diseases. Curr. Opin. Neurol. 30, 471–480 10.1097/WCO.0000000000000474 [DOI] [PubMed] [Google Scholar]
- 26. Taveggia C., Feltri M. L., and Wrabetz L. (2010) Signals to promote myelin formation and repair. Nat. Rev. Neurol. 6, 276–287 10.1038/nrneurol.2010.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nave K. A., and Werner H. B. (2014) Myelination of the nervous system: mechanisms and functions. Annu. Rev. Cell Dev. Biol. 30, 503–533 10.1146/annurev-cellbio-100913-013101 [DOI] [PubMed] [Google Scholar]
- 28. Fraher J. P., and Kaar G. F. (1985) The development of α and γ motoneuron fibres in the rat. II. A comparative ultrastructural study of their central and peripheral myelination. J. Anat. 141, 89–103 [PMC free article] [PubMed] [Google Scholar]
- 29. Kaar G. F., and Fraher J. P. (1985) The development of α and γ motoneuron fibres in the rat. I. A comparative ultrastructural study of their central and peripheral axon growth. J. Anat. 141, 77–88 [PMC free article] [PubMed] [Google Scholar]
- 30. El-Tahir H. M., Abouzied M. M., Gallitzendoerfer R., Gieselmann V., and Franken S. (2009) Hepatoma-derived growth factor-related protein-3 interacts with microtubules and promotes neurite outgrowth in mouse cortical neurons. J. Biol. Chem. 284, 11637–11651 10.1074/jbc.M901101200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abouzied M. M., El-Tahir H. M., Gieselmann V., and Franken S. (2010) Hepatoma-derived growth factor-related protein-3: a new neurotrophic and neurite outgrowth-promoting factor for cortical neurons. J. Neurosci. Res. 88, 3610–3620 10.1002/jnr.22507 [DOI] [PubMed] [Google Scholar]
- 32. LeBlanc M. E., Wang W., Caberoy N. B., Chen X., Guo F., Alvarado G., Shen C., Wang F., Wang H., Chen R., Liu Z. J., Webster K., and Li W. (2015) Hepatoma-derived growth factor-related protein-3 is a novel angiogenic factor. PLoS One 10, e0127904 10.1371/journal.pone.0127904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Woolley A. G., Sheard P. W., and Duxson M. J. (2005) Neurotrophin-3 null mutant mice display a postnatal motor neuropathy. Eur. J. Neurosci. 21, 2100–2110 10.1111/j.1460-9568.2005.04052.x [DOI] [PubMed] [Google Scholar]
- 34. Patel K. G., Liu C., Cameron P. L., and Cameron R. S. (2001) Myr 8, a novel unconventional myosin expressed during brain development associates with the protein phosphatase catalytic subunits 1α and J. Neurosci. 21, 7954–7968 10.1523/JNEUROSCI.21-20-07954.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M. C., Friocourt G., McDonnell N., Reiner O., Kahn A., McConnell S. K., Berwald-Netter Y., Denoulet P., and Chelly J. (1999) Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron 23, 247–256 10.1016/S0896-6273(00)80777-1 [DOI] [PubMed] [Google Scholar]
- 36. Eltahir H. M. A. (2010) Developmentally-regulated Localization and Possible Functions of HRP-3 in the Murine Nervous Tissue, Ph.D. thesis, Universitäts-und Landesbibliothek Bonn [Google Scholar]
- 37. la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., and Brunak S. (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17, 527–536 10.1093/protein/gzh062 [DOI] [PubMed] [Google Scholar]
- 38. Kutay U., and Güttinger S. (2005) Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 15, 121–124 10.1016/j.tcb.2005.01.005 [DOI] [PubMed] [Google Scholar]
- 39. Taveggia C., Zanazzi G., Petrylak A., Yano H., Rosenbluth J., Einheber S., Xu X., Esper R. M., Loeb J. A., Shrager P., Chao M. V., Falls D. L., Role L., and Salzer J. L. (2005) Neuregulin-1 type III determines the ensheathment fate of axons. Neuron 47, 681–694 10.1016/j.neuron.2005.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stidworthy M. F., Genoud S., Suter U., Mantei N., and Franklin R. J. (2003) Quantifying the early stages of remyelination following cuprizone-induced demyelination. Brain Pathol. 13, 329–339 10.1111/j.1750-3639.2003.tb00032.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. El-Tahir H. M., Dietz F., Dringen R., Schwabe K., Strenge K., Kelm S., Abouzied M. M., Gieselmann V., and Franken S. (2006) Expression of hepatoma-derived growth factor family members in the adult central nervous system. BMC Neurosci. 7, 6 10.1186/1471-2202-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klein K. (2013) Generation and Characterisation of an HRP-3-deficient Mouse Model, Ph.D. thesis, Universitäts-und Landesbibliothek Bonn [Google Scholar]
- 43. Kerman B. E., Kim H. J., Padmanabhan K., Mei A., Georges S., Joens M. S., Fitzpatrick J. A., Jappelli R., Chandross K. J., August P., and Gage F. H. (2015) In vitro myelin formation using embryonic stem cells. Development 142, 2213–2225 10.1242/dev.116517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han M. H., Lundgren D. H., Jaiswal S., Chao M., Graham K. L., Garris C. S., Axtell R. C., Ho P. P., Lock C. B., Woodard J. I., Brownell S. E., Zoudilova M., Hunt J. F., Baranzini S. E., Butcher E. C., et al. (2012) Janus-like opposing roles of CD47 in autoimmune brain inflammation in humans and mice. J. Exp. Med. 209, 1325–1334 10.1084/jem.20101974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Woodhoo A., and Sommer L. (2008) Development of the Schwann cell lineage: from the neural crest to the myelinated nerve. Glia 56, 1481–1490 10.1002/glia.20723 [DOI] [PubMed] [Google Scholar]
- 46. Salzer J. L., and Bunge R. P. (1980) Studies of Schwann cell proliferation. I. An analysis in tissue culture of proliferation during development, Wallerian degeneration, and direct injury. J. Cell Biol. 84, 739–752 10.1083/jcb.84.3.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Komiyama A., and Suzuki K. (1992) Age-related differences in proliferative responses of Schwann cells during Wallerian degeneration. Brain Res. 573, 267–275 10.1016/0006-8993(92)90772-2 [DOI] [PubMed] [Google Scholar]
- 48. Gomez-Sanchez J. A., Lopez de Armentia M., Lujan R., Kessaris N., Richardson W. D., and Cabedo H. (2009) Sustained axon-glial signaling induces Schwann cell hyperproliferation, Remak bundle myelination, and tumorigenesis. J. Neurosci. 29, 11304–11315 10.1523/JNEUROSCI.1753-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li C., and Wong W. H. (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. U. S. A. 98, 31–36 10.1073/pnas.98.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aimone J. B., and Gage F. H. (2004) Unbiased characterization of high-density oligonucleotide microarrays using probe-level statistics. J. Neurosci. Methods 135, 27–33 10.1016/j.jneumeth.2003.11.016 [DOI] [PubMed] [Google Scholar]
- 51. Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R., and Ma'ayan A. (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., McDermott M. G., Monteiro C. D., Gundersen G. W., and Ma'ayan A. (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gaspard N., Bouschet T., Herpoel A., Naeije G., van den Ameele J., and Vanderhaeghen P. (2009) Generation of cortical neurons from mouse embryonic stem cells. Nat. Protoc. 4, 1454–1463 10.1038/nprot.2009.157 [DOI] [PubMed] [Google Scholar]
- 54. Öztürk G., Cengiz N., Erdoğan E., Him A., Oğuz E. K., Yenidünya E., and Ayşit N. (2013) Two distinct types of dying back axonal degeneration in vitro. Neuropathol. Appl. Neurobiol. 39, 362–376 10.1111/j.1365-2990.2012.01295.x [DOI] [PubMed] [Google Scholar]
- 55. Tiscornia G., Singer O., and Verma I. M. (2006) Production and purification of lentiviral vectors. Nat. Protoc. 1, 241–245 10.1038/nprot.2006.37 [DOI] [PubMed] [Google Scholar]
- 56. Consiglio A., Gritti A., Dolcetta D., Follenzi A., Bordignon C., Gage F. H., Vescovi A. L., and Naldini L. (2004) Robust in vivo gene transfer into adult mammalian neural stem cells by lentiviral vectors. Proc. Natl. Acad. Sci. U.S.A. 101, 14835–14840 10.1073/pnas.0404180101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Paxinos G., and Franklin K. B. J. (2001) The Mouse Brain in Stereotaxic Coordinates, pp. 65–69, Academic Press, San Diego [Google Scholar]
- 58. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arce V., Garces A., de Bovis B., Filippi P., Henderson C., Pettmann B., and deLapeyriere O. (1999) Cardiotrophin-1 requires LIFRβ to promote survival of mouse motoneurons purified by a novel technique. J. Neurosci. Res. 55, 119–126 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data are deposited in the Gene Expression Omnibus with accession code GSE133744. All other data are contained in this article.