Abstract
Retromer orchestrates the selection and export of integral membrane proteins from the endosome via retrograde and plasma membrane recycling pathways. Long-standing hypotheses regarding the retromer sorting mechanism posit that oligomeric interactions between retromer and associated accessory factors on the endosome membrane drives clustering of retromer-bound integral membrane cargo prior to its packaging into a nascent transport carrier. To test this idea, we examined interactions between components of the sorting nexin 3 (SNX3)–retromer sorting pathway using quantitative single particle fluorescence microscopy in a reconstituted system. This system includes a supported lipid bilayer, fluorescently labeled retromer, SNX3, and two model cargo proteins, RAB7, and retromer-binding segments of the WASHC2C subunit of the WASH complex. We found that the distribution of membrane-associated retromer is predominantly comprised of monomer (∼18%), dimer (∼35%), trimer (∼24%), and tetramer (∼13%). Unexpectedly, neither the presence of membrane-associated cargo nor accessory factors substantially affected this distribution. The results indicate that retromer has an intrinsic propensity to form low order oligomers on a supported lipid bilayer and that neither membrane association nor accessory factors potentiate oligomerization. The results support a model whereby SNX3-retromer is a minimally concentrative coat protein complex adapted to bulk membrane trafficking from the endosomal system.
Keywords: cell biology, trafficking, single particle analysis, membrane bilayer, oligomerization, biochemical reconstitution, retromer, WASHC2C
Retromer is an evolutionarily conserved protein complex that orchestrates sorting and export of integral membrane proteins from the endosome. Loss of retromer function, which is implicated in a variety of disease conditions, results in increased rates of turnover of plasma membrane proteins and retrograde cargo proteins in the lysosome, with broad consequences to cell and organism physiology (1–3). Many integral membrane proteins have been identified that are sorted by retromer, but little is known regarding the underlying protein-sorting mechanisms.
Retromer is composed of three proteins VPS26, VPS29, and VPS35 that form a stable, soluble heterotrimer (4–7) that is recruited to the endosome by binding to sorting signals of integral membrane protein cargo and to membrane-associated accessory proteins, including sorting nexins and RAB7 network components (2, 8). Genetic and structural analyses of retromer trimer complexed with different sorting nexins suggest that retromer is a modular sorting device that associates with different sorting nexins (e.g. SNX-BARs, SNX3, SNX27) to constitute distinct coat protein assemblies on the endosome membrane (2). Retromer exhibits an inherent structural plasticity and general low ordered oligomeric behavior when not associated with a membrane (9, 10). Recent structural advances suggest that on SNX-BAR–coated tubules a retromer dimer engages two membrane-associated SNX-BAR proteins via the VPS26 subunits, stabilizing the loosely packed SNX-BAR lattice (11). The sorting nexins SNX3 and SNX27 also associate with retromer via the VPS26 subunit, but if and how these interactions influence retromer coat protein function is unknown.
We discovered that the yeast (Saccharomyces cerevisiae)-sorting nexin Snx3/Grd19 functions as a cargo-selective retromer adapter that associates with retromer on the endosome membrane and aids in cargo recognition (12). Studies of cultured human cells and other model organisms confirmed the existence of an SNX3-retromer sorting pathway in metazoans (13–15), where cargo-sorting signal is recognized via the SNX3-retromer interface (16). Using bulk biochemical reconstitution, we previously discovered that multi-valent interactions among retromer, SNX3, RAB7, and an integral membrane cargo confer recruitment of retromer to the surface of small unilamellar vesicles (17). Interestingly, a study of the homologous yeast proteins suggested that yeast Snx3 possesses membrane remodeling activity that is potentiated by retromer and a soluble fragment of a retromer cargo protein (18). In that study, morphologically diverse patches of the tagged proteins were observed on the surface of giant unilamellar vesicles (GUVs), raising the possibility that oligomerization of these components on the endosome membrane may underlie cargo-protein sorting.
In this study we examined reconstituted components of the human SNX3-retromer sorting system on supported lipid bilayers by quantitative fluorescence microscopy. We find that when associated with a membrane, both in the presence and in the absence of cargo and accessory proteins, retromer exists mainly as monomers and lower order oligomers (dimer to tetramer). The results suggest that cargo is modestly concentrated by retromer prior to export from the endosome by the SNX3-retromer pathway, prompting a revision in long-standing models of retromer-mediated cargo sorting.
Results and Discussion
As a coat protein of endosome-derived transport carriers that recognizes retrograde cargo-sorting signals, retromer has been proposed to concentrate integral membrane protein cargo prior to transport carrier formation (1, 7, 19, 20). To test this, we first sought to determine the oligomeric state of retromer when it is associated with a membrane. Accordingly, a supported lipid bilayer (SLB) was constructed to mimic the relatively planar surface geometry of the vacuolar domain of the sorting endosome, where retromer sorting domains are formed. The SLB contained physiological endosomal lipids, phosphatidylcholine, phosphatidylserine, and PtdIns-3-P, and nonphysiological Ni-NTA–DGS, with a nickel ion–containing head group that is recognized by poly-histidine sequences, and trace amounts of rhodamine-phosphatidylethanolamine (Rh-PE) or NBD-phosphatidylethanolamine (NBD-PE), used to assess lipid mobility and the quality of the bilayer (Fig. S1A). Fluorescence recovery after photobleaching (FRAP) analyses confirmed free diffusion of lipids within the supported bilayer; bilayers that were not fluid were excluded from analysis. Experiments also confirmed that association of a fluorescent His10-tagged peptide with the SLB depends upon the presence of Ni-NTA–DGS lipid and that the lipid-associated polypeptide is mobile (Fig. S1, B and C).
Retromer exists as monomers and low order oligomers on a membrane
Retromer was assembled in lysates of bacterial cells expressing individual retromer proteins, as we have used previously (17). To directly visualize retromer on the SLB by fluorescence microscopy, VPS26 was produced with N-terminal His10 and SNAP tags (Fig. 1), which facilitated binding to the SLB and labeling with a single fluorophore (Alexa Fluor 488). The His10-SNAP-VPS26 fusion protein was incorporated into the retromer trimer at the same stoichiometry as that of untagged VPS26 (1:1:1), judged by comparison of Coomassie Blue–stained preparations, indicating that fusion appendages do not perturb retromer trimer assembly. Although the effective concentration of retromer in cells is not known, two quantitative shotgun proteomics studies estimated the abundance of retromer subunits to correspond ≤100 nm in the cytosol in HeLa and U2-OS cells (21, 22). Hence, retromer was incubated with SLBs over a range of concentrations (75 pm–100 nm investigated, 1 nm nominal retromer concentration is shown in Fig. S1). After incubating labeled retromer with the SLB for 2 h, the SLB was washed to remove unbound proteins and total internal reflection fluorescence microscopy (TIRFM) was used to visualize SLB-associated proteins (23). The results show that retromer is distributed homogenously on the SLB (Fig. S1C), suggesting that it does not self-organize into clusters over this range of concentrations.
Figure 1.
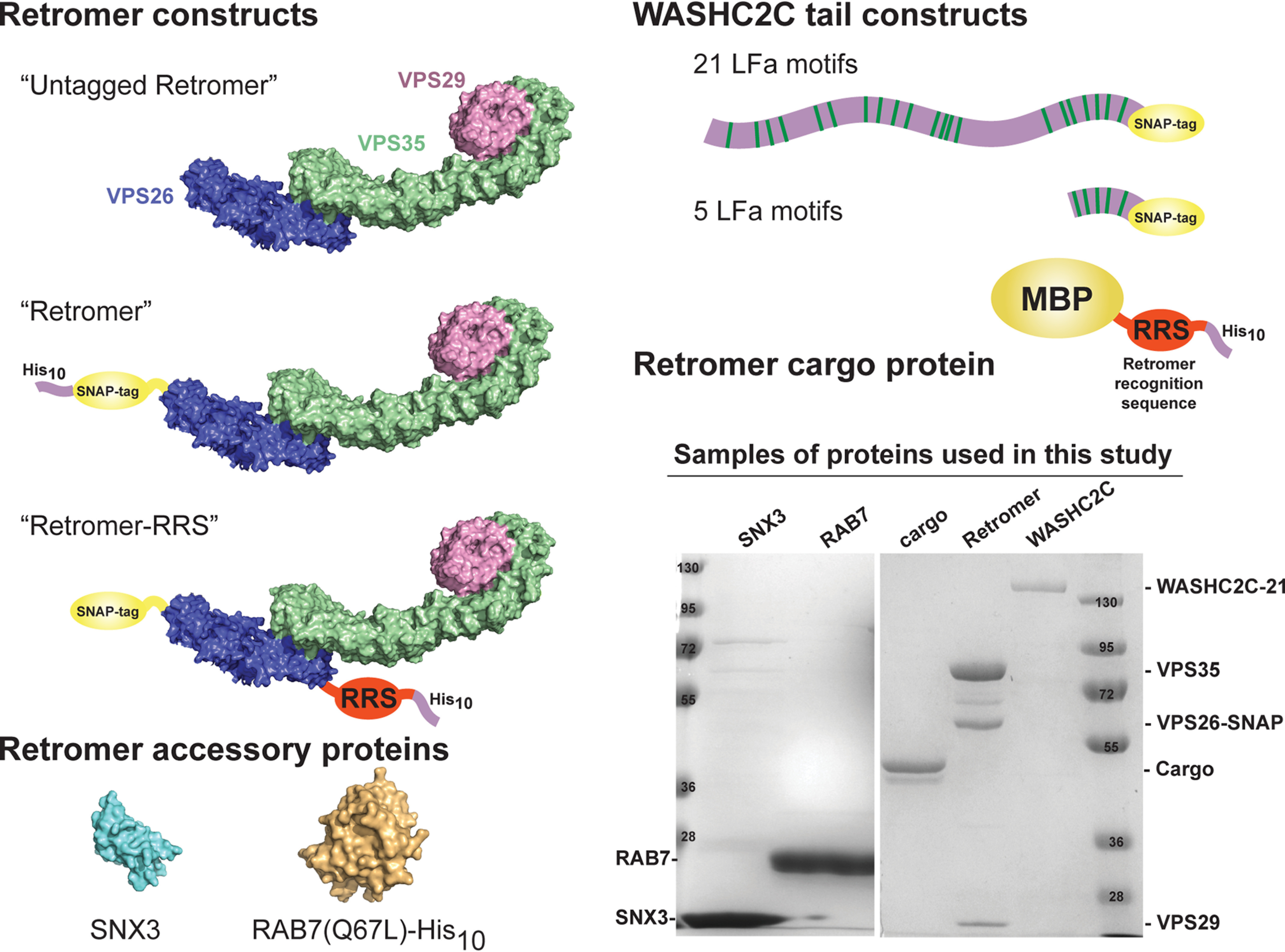
Engineered proteins used in this study. A diagram of each of the salient features of each of the proteins used in this study is shown. “Untagged Retromer” refers to recombinant VPS26/VPS35/VPS29 retromer trimer. “Retromer” refers to a trimer where VPS26 carries a SNAP tag (for labeling with Alexa Fluor 488 dye) and a His10 tag for attachment to supported bilayers containing Ni-NTA–DGS lipids. “Retromer-RRS” contains a VPS26 subunit with a SNAP tag at the N terminus and the sequence of the DMT1-II retromer sorting signal (sequence: TAQPELYLMNTMDADSLVRGL) and a His10 tag fused at its C terminus. In this construct, stoichiometric retromer-cargo interactions are enforced artificially. SNX3, RAB7 (Q67L)-His10 (a GTP-locked, constitutively active mutant), WASHC2C constructs contain either 21 or 5 LFa motifs. “Retromer cargo protein” is composed of an N-terminal maltose-binding protein with a sorting signal and a His10 tag fused at its C terminus. An example of an SDS-PAGE Coomassie Blue–stained gel shows the purity and stoichiometry of complexes. The following structure files from the Protein Data Bank were used to prepare the figure: RAB7A, 1T91; SNX3, 2YPS; retromer, 6H7W.
Next, we established a single molecule fluorescence microscopy assay to determine retromer oligomeric state on the SLB. These experiments used lower protein concentrations (∼75 pm), resulting in a lower protein density on the SLB, such that there are ∼1000–2000 labeled retromer particles in a 100-μm2 region (with ∼0.3-μm separation between fluorescence-labeled particles) to facilitate single-particle analysis. We monitored intensity of Alexa Fluor 488–labeled retromer puncta continuously over time (stream acquisitions with 17- to 18-ms exposure per frame) to capture single fluorophore bleaching steps until most puncta bleached away (Fig. 2A). Fluorescent particles photobleached in either single or multi-step bleaching profiles, indicative of either single or multiple fluorophores in the particle (Fig. 2B). We used the decrease in fluorescence intensity because of the last bleaching event in individual puncta to estimate the intensity of single Alexa Fluor 488 fluorophores (Fig. 2B). This was then used to calculate retromer copy number per particle at the beginning of the movie, before bleaching. Importantly, in this approach, each image stack provides its own single-molecule intensity calibration for the cluster copy number estimation, enhancing robustness of the approach against experiment-to-experiment variations.
Figure 2.
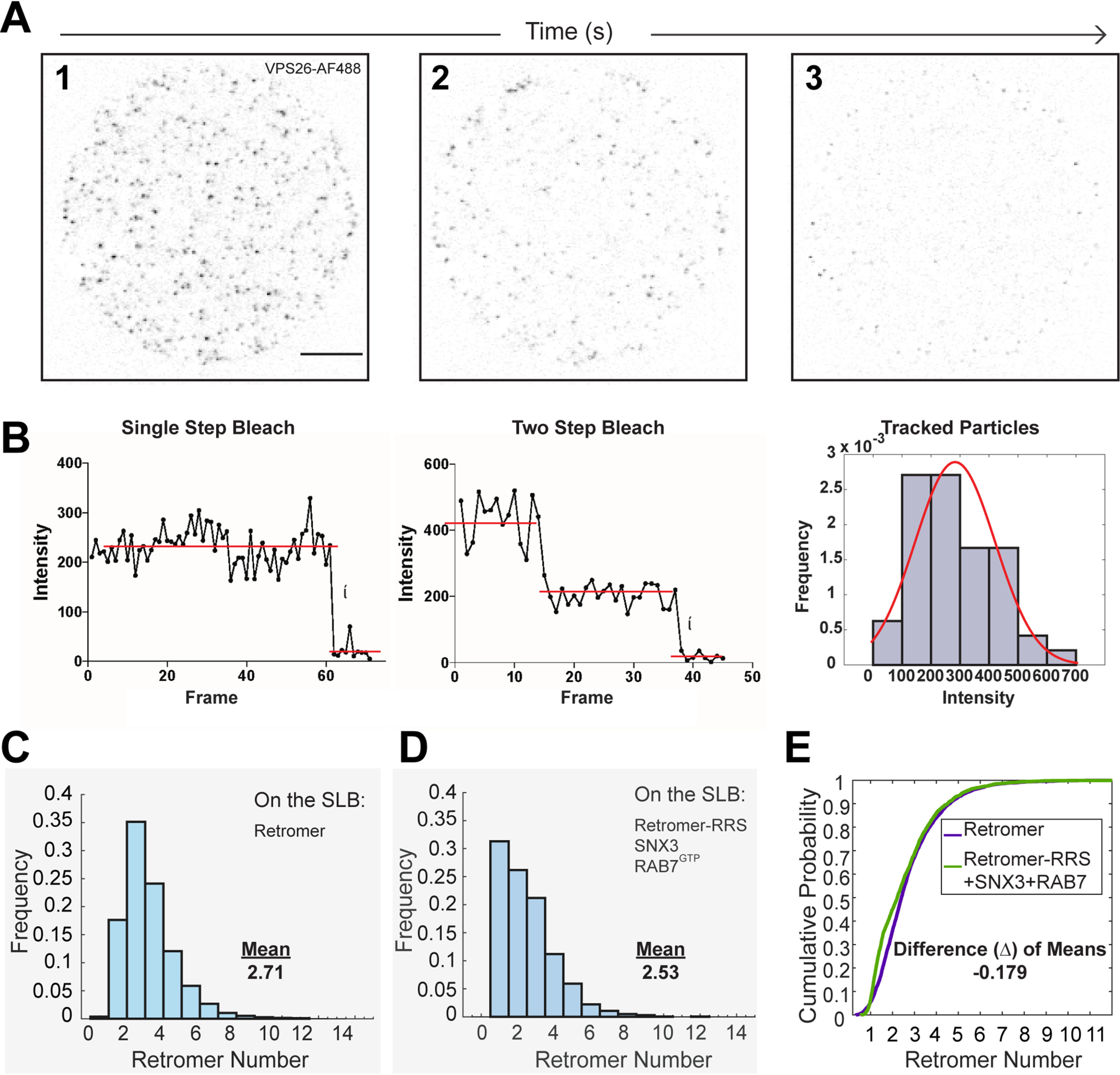
Retromer forms low order oligomers on supported bilayers. Total internal reflection fluorescence microscopy and single-particle analysis were used to determine florescence values of single fluorophores and to track and characterize fluorescent retromer clusters on the SLB. A, three frames from a TIRFM image stack of Alexa Fluor 488–labeled retromer are shown (solution concentration 75 pm; gray values inverted). The retromer clusters observed in the first frame (image 1) bleach over time (images 2 and 3), allowing single-molecule fluorescence bleaching events to be identified. Scale bar: 10 μm. B, example fluorescence intensity profiles of single-particle tracks and distribution of single-fluorophore intensities. Left: the mean pixel value from a 3 × 3 pixel region centered around the tracked position of a spot as a function of time (frames). A distribution of fluorescence intensity drops during the last bleaching events is shown on the right overlaid with a best-fit Gaussian distribution (red line). C, frequency distribution of retromer cluster size on a SLB. The number of retromer complexes in single fluorescent puncta (>150 per image) in the first frame of TIRFM image stacks was calculated using the single-fluorophore intensity calibration (obtained as in B) and accounting for labeling efficiency. The distribution of cluster sizes is plotted for a bilayer containing only retromer. The mean of the distribution is indicated. D, frequency distribution of retromer-RRS cluster size on a SLB containing SNX3 and RAB7. His-tagged, Alexa Fluor 488–labeled retromer-RRS (solution concentration 75 pm), SNX3 (3.25 nm), and RAB7 (Q67L)-His10 (1 nm) were attached to the SLB and analyzed by single-particle TIRFM. The number of retromer complexes in single fluorescent puncta (>150 per image) in the first frame of TIRFM image stacks was calculated. The mean of the distribution is indicated. E, retromer cluster sizes plotted as empirical cumulative distribution functions for both retromer alone and retromer-RRS. The differences between the means of the two distributions is indicated.
We first attached His10-tagged, Alexa Fluor 488–labeled retromer to the SLB and analyzed its number distribution in fluorescent spots (retromer clusters) using single-particle fluorescence microscopy. The distributions of four distinct preparations of retromer were determined and evaluated for technical and experimental consistency using estimation statistics and “effect size” analysis, an alternative analytic statistical method to null hypothesis significance testing that is appropriate for evaluating large datasets (24–26). This analysis (Table S1) showed that mean retromer number per cluster varied by less than one retromer complex in both technical replicate experiments (same retromer preparation on different SLBs) and between different retromer preparations (experimental replicates). Hence, data from technical and experimental replicate experiments were pooled in all subsequent analyses. The results are plotted as a histogram showing the frequency distribution of retromer monomer and oligomers on the SLB (Fig. 2C). The predominant species of retromer on the SLB are monomers (∼18%), dimers (∼35%), trimers (∼24%), and tetramers (∼13%), but pentamers and rare higher order oligomers up to 10 are also observed (∼10%). Notably, this distribution of retromer oligomers on SLBs is similar to that for untagged solution phase retromer reported in a cryo-EM study where dimers and tetramers were the prevalent retromer species in vitrified ice (9). The prevalence of retromer dimers possibly reflects two different retromer-to-retromer binding modes, where one is the 2-fold symmetric dimer observed in solution phase described by Kendall and colleagues (9), and the second mode is mediated by the VPS35-VPS35 dimerization interface observed in the retromer-SNX-BAR coat (11) and in solution (16). A distinct binding mode is needed to explain the small proportion of higher order oligomers observed in our study, which might consist of chains of retromer complexes in solution phase observed by cryo-EM (9), or the patches of yeast Snx3 and yeast retromer observed by Purushothaman and Ungermann (18). These results suggest that membrane association per se does not influence retromer oligomeric state.
Retromer accessory proteins do not influence oligomeric state
Retromer association with the endosome membrane is conferred by binding to integral membrane cargo proteins, sorting nexins, and RAB7 (17, 18, 27, 28); therefore, we next sought to determine whether these accessory factors influence retromer oligomerization on an SLB. His-tagged, Alexa Fluor 488–labeled model cargo protein was homogeneously dispersed on the SLB (Fig. S2A) and incubation with “untagged retromer” (recombinant, not His10-tagged or labeled) did not affect this appearance (not shown). However, the effect of retromer may not be apparent because of the low affinity with which retromer binds sorting signals. Accordingly, we added the sorting nexin, SNX3, which facilitates retromer membrane recruitment and forms part of the DMT1-II cargo-binding site (16), onto the SLB. After confirming that purified, fluorescently labeled SNX3 associates with the SLB by binding to PtdIns-3-P (Fig. S1D), unlabeled SNX3 and untagged retromer were sequentially added in stochiometric excess of the nominal cargo concentration and the distribution of cargo fluorescence was monitored for any changes occurring with the addition of SNX3 and soluble retromer (Fig. S2). At all times examined, cargo fluorescence was homogenously distributed on the surface of the SLB (Fig. S2B). Thus, at the protein concentrations accessible in our experimental system, a model retromer cargo is not clustered by SNX3-retromer (Fig. S2, B and C).
We next measured retromer particle size at low protein density on the SLB using quantitative single-particle TIRFM. For these experiments we used a modified system because, at the low densities of proteins on the SLB required for single-particle analysis, only a small proportion of cargo molecules will be bound by retromer. Accordingly, we constructed a nondissociable model of the retromer-cargo complex, termed retromer-RRS, that was inspired by crystallographic studies of Lucas et al. (16) where a similar VPS26-DMT1-II fusion protein facilitated elucidation of the cargo-binding site on SNX3-retromer (16). The effect of an additional retromer accessory protein, RAB7, was also examined because RAB7 is proposed to aid in retromer recruitment to the endosome (17, 27). His-tagged, Alexa Fluor 488–labeled (via the SNAP tag) retromer-RRS fusion protein, SNX3, and RAB7 (Q67L)-His10 (a mutant form of RAB7 in the GTP-bound conformation) were incubated together with the SLB, and single-particle fluorescence data were collected to determine the distribution of retromer-RRS complexes present on the SLB (Fig. 2D), as before. Retromer-RRS shows a higher proportion of monomer in the presence of SNX3 and RAB7 at the expense of dimer (Fig. 2D), although the effect size is small (mean = 2.53 retromer complexes/cluster in the presence of SNX3 and RAB7 versus 2.71 for retromer alone; difference of means, Δ = −0.18 retromer complexes/cluster) (Fig. 2E), and we consider it unlikely to be of physiological significance. This analysis indicates that cargo occupancy, SNX3 and RAB7 (Q67L)-His10 do not promote retromer oligomerization on a SLB. We note that this conclusion contrasts with that of a study of yeast retromer and engineered accessory proteins (GFP-, His6-tagged ySNX3, tagged-retromer, and a soluble retromer cargo peptide) which were observed to form patches on the surface of a GUV (18). This difference might be attributed to the different experimental approaches used (SLB versus GUV, differing lipid compositions, different cargo proteins used, yeast versus human proteins, etc.); however, we note that clustering of yeast SNX3-retromer depended on the presence of a second, nonphysiological membrane-binding site on ySNX3 (His6 tag) and its lipid ligand (Ni-NTA–DGS) in the GUV membrane (18).
WASHC2C disordered segment does not influence retromer oligomeric state
In metazoans, retromer recruits the WASH protein complex from the cytosol to the endosome membrane where it promotes ARP2/3-dependent actin polymerization and retromer-dependent sorting (8, 29–32). Binding of WASH to retromer is conferred by an ∼1100 amino acid–long unstructured segment of the WASHC2C/FAM21C subunit containing 21 LFa (Leu-Phe-(Asp/Glu)3–10-Leu-Phe) shown to constitute retromer-binding sites by solution-phase binding assays (31, 33). On the basis of the multi-valences of WASHC2C-retromer interaction, Jia et al. (31) speculated that recruitment of WASH to the endosome membrane would result in clustering of membrane-associated retromer-cargo complexes (31). We tested this hypothesis using our single-particle analysis platform.
We first determined if SLB-associated retromer can recruit WASHC2C-21 (Fig. 1), composed of an ∼1100 amino acid–long segment of WASHC2C containing all 21 LFa motifs and a C-terminal SNAP tag, to the SLB (Fig. 3). Labeled WASHC2C-21 was incubated alone or co-incubated with labeled His10-tagged retromer, with an SLB. We confirmed that WASHC2C-21 binds to the SLB via retromer by imaging both proteins on an area of the SLB with enriched retromer using multiple fluorescence channels (Fig. 3). We then asked if segments of the WASHC2C unstructured segment can influence retromer cluster size on the SLB. His10-tagged, Alexa Fluor 488–labeled retromer was attached to the SLB via binding to Ni-NTA–DGS and then incubated with varying amounts of WASHC2C-21 to cover stoichiometries ranging from retromer excess to WASHC2C excess (50:1, 20:1, 5:1, 1:1, and 1:13) (Fig. 4, WASHC2C-21), noting that in HeLa cells retromer is estimated to be ∼10 times as abundant as WASHC2C (22). A truncated version, WASHC2C-5, containing just the last five LFa motifs was also examined (Fig. 4, WASHC2C-5). We observed that the mean number of complexes in single retromer clusters varied by less than one retromer complex for both WASHC2C-5 and WASHC2C-21 over a broad range of retromer:WASH ratios. Empirical cumulative distribution plots show all datasets to converge at the same range of retromer oligomeric states (Fig. 4C). Although statistical analysis indicates that some of these small differences are significant (Table S2), they can be explained by variability in technical replicates (Table S1). Taken together, the data indicate that WASHC2C does not promote retromer oligomerization on the SLB. Curiously, incubation of retromer with WASHC2C-21 resulted in a trend toward smaller retromer oligomers (by less than one retromer complex), but the physiological significance is unclear, especially in consideration of the WASHC2C peptide fragment, not the complete WASH complex, that was used. Together, the results indicate that the WASHC2C unstructured segment does not elicit clustering of retromer under the conditions tested.
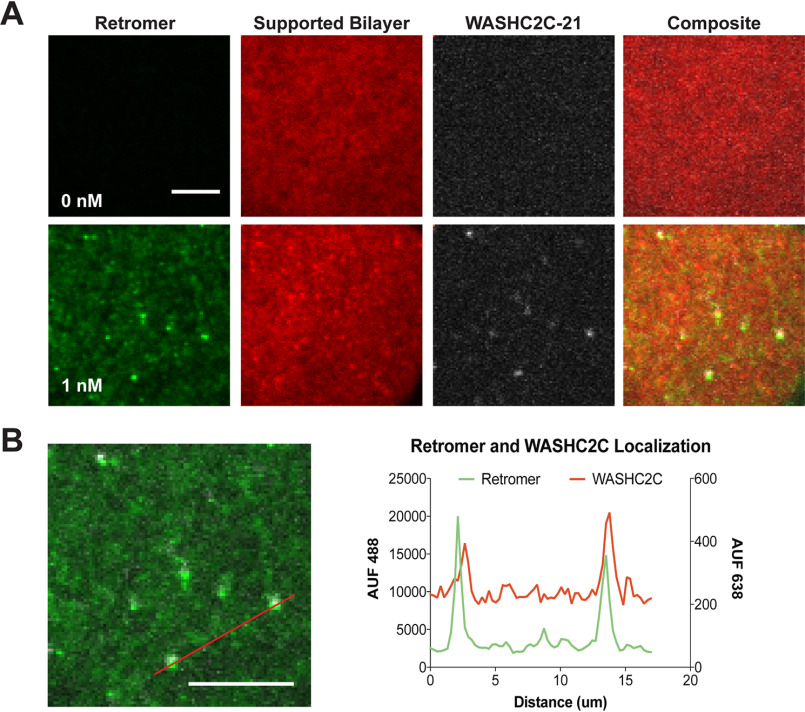
Figure 3.
WASHC2C is recruited to the supported bilayer by retromer. A, TIRFM images are shown of SLBs incubated with Alexa Fluor 647–labeled WASHC2C-21 (solution concentration 1 nm) (top row), or with Alexa Fluor 488–labeled retromer (solution concentration 1 nm) (bottom row). After incubation, unbound material was removed, the SLB was washed, and then retromer, WASHC2C, and the SLB (rhodamine-PE) were imaged on the SLB surface by TIRFM. WASHC2C-21 fluorescence is only observed on SLBs when retromer is present. B, retromer and WASHC2C are associated on the SLB. An enlarged view of the overlay of the retromer and WASHC2C TIRFM channels from (A) is shown. The line scan documents co-localization of retromer and WASHC2C-21. Scale bar: 10 μm.
Figure 4.
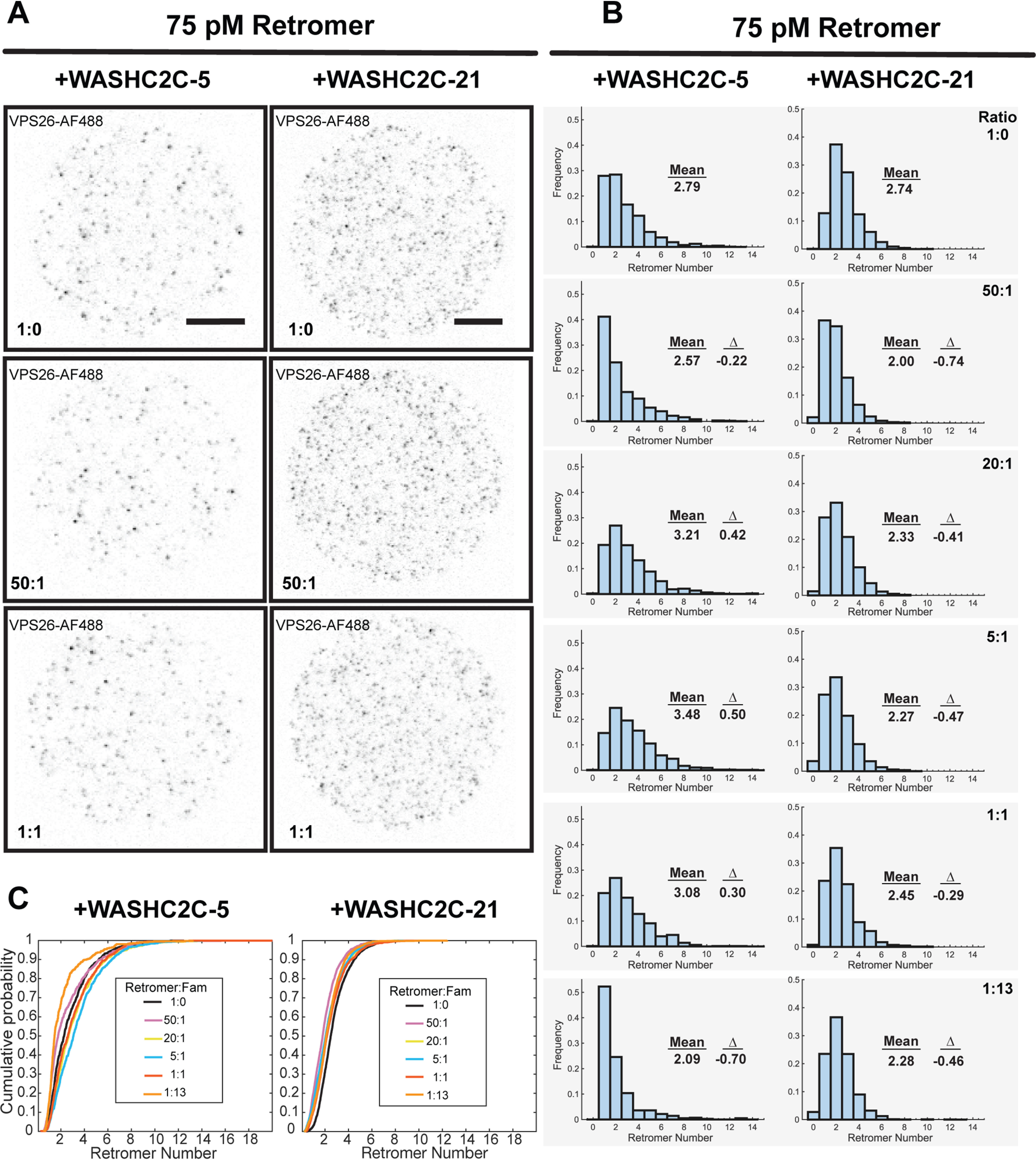
WASHC2C does not influence Retromer oligomeric state on a supported bilayer. Single particle TIRFM analysis was used to measure retromer cluster size in the presence of varying amounts of WASHC2C proteins. His-tagged, Alexa Fluor 488–labeled retromer was attached to a SLB (solution concentration 75 pm) and the indicated Alexa Fluor 647–labeled WASHC2C protein was then added to the indicated retromer:WASHC2C ratio. After incubation, the SLB was washed and retromer was imaged. A, the left column of images shows retromer fluorescence after incubation with WASHC2C-5 (containing 5 LFa motifs) and the right column shows incubations with WASHC2C-21 (containing 21 LFa motifs). The number of retromer complexes in single puncta was calculated and the distribution was plotted. Retromer fluorescence is shown (gray values inverted). Scale bar: 10 μm. B, distributions of retromer oligomer size on the SLB after incubation with WASHC2C-5 or -21 at the indicated ratios. The mean for each distribution is indicated. In addition, the difference of the mean with the distribution obtained in the absence of WASHC2C fragments (1:0), Δ, is shown in each panel. The presence of WASHC2C shifted the mean by less than one retromer per cluster. Small shifts in the monomer-to-oligomer ratios are likely insignificant as well, because such differences are observed among technical or biological replicates (e.g. compare the first panels with 1:0, in the absence of WASHC2C fragments) C, distributions shown in B, were plotted as empirical cumulative distribution functions.
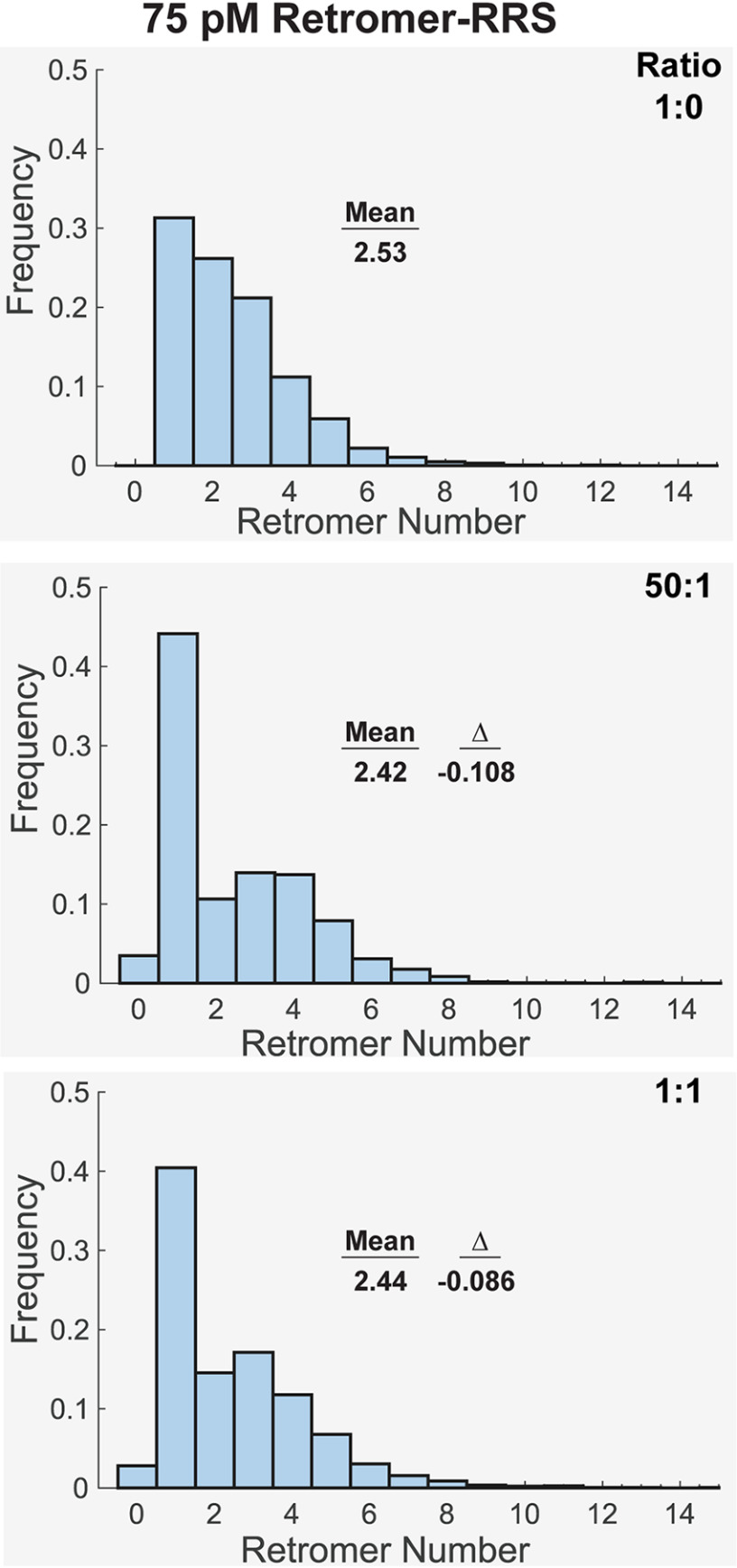
Finally, we asked if retromer, cargo, SNX3, RAB7, and WASHC2C act synergistically to influence retromer clustering at low protein densities (Fig. 5). As before, retromer-RRS was used to enforce cargo occupancy. We had retromer-RRS on the SLB in the presence of SNX3 and RAB7GTP. Next, WASHC2C-21 was added to the reactions to examine the effect of sub-stoichiometric (50:1) and stoichiometric (1:1) WASHC2C-21. Single-particle analyses revealed increases in the proportions of retromer-RRS monomer at both retromer-RRS:WASHC2C-21 ratios, similar to the effect of sub-stoichiometric amounts of WASHC2C (Fig. 4), although more pronounced (Fig. 5). These results indicate that binding of retromer and WASHC2C on the SLB is (or is close to) stoichiometric, and further suggest that WASHC2C does not exert an effect (e.g. allosteric) to promote retromer oligomerization. Consistent with these interpretations, Jia and colleagues (31) reported that only the last two of the 21 LFa motifs (LFa20-21), which are those bound by VPS35 with the highest affinity, are essential for WASH-dependent sorting of integral membrane cargo. These findings suggest that WASH does not exert its role in the retromer pathway by clustering retromer or retromer-cargo complexes.
Figure 5.
Cargo, SNX3, RAB7, and WASHC2C do not influence Retromer oligomeric state on a supported lipid bilayer. Retromer-RRS-His10 (Alexa Fluor 488–labeled; solution concentration 75 pm), SNX3 (1 nm), and Rab7 (Q67L)-His10 (1 nm) were incubated with a SLB. Following binding and washing to remove unbound protein, WASHC2C-21 was added to the reaction cell at the indicated retromer:WASHC2C-21 ratios. Single particle analysis of retromer TIRFM image stacks was used to measure the number of retromer complexes in individual puncta. At both ratios tested, the presence of WASHC2C-21 skews the distribution to favor retromer monomers, chiefly at the expense of dimers. The mean for each distribution, and the difference (Δ) from the reference mean (1:0 distribution in top panel) are indicated. Note that the data in the top panel is also presented in Fig. 2D.
Implications for retromer-sorting mechanism
When associated with a supported lipid bilayer, human retromer has an intrinsic propensity to form low order clusters (<5 monomers) and neither membrane association, nor the presence of modified forms of membrane-associated accessory factors—cargo, SNX3, RAB7, and WASHC2C—potentiates retromer oligomerization on an SLB. The sizes of retromer clusters observed in this study agree well with biophysical and structural studies of untagged, recombinant retromer in solution-phase, where retromer monomer and dimer were the most prevalent species (9, 10, 16). Only retromer dimers are present in atomic structures of SNX-BAR–retromer–coated tubules (yeast Vps5 and retromer) (11); the close agreement with our observations suggests that a retromer dimer is the functional protomer of the SNX3-retromer coat, as previously suggested (16).
Retromer has been proposed to constitute a coat protein complex for endosome-derived transport carriers that, by analogy to better-characterized conventional vesicle coats (e.g. clathrin), polymerize on the membrane to enrich nascent carriers in particular integral membrane cargo (7, 20). A key feature of the conventional transport vesicle paradigm is the small area of densely coated membrane enriched in integral membrane cargo that gives rise to small (<100 nm diameter) transport carriers. In contrast, sorting of retrograde and recycling integral membrane in the endosomal system is weakly concentrative, relying instead on the large surface area of endosome-derived carriers to mediate bulk export of proteins and lipid from the endosome (34). Results presented here suggest that retromer concentrates cargo minimally, although this study was necessarily limited so it is possible that conditions or factors not examined, such as different membrane topologies, rigidities, posttranslational modifications, and/or WASH-mediated actin polymerization, influence retromer oligomerization. We note, however, that the structure of yeast VPS5-retromer–coated membrane tubules that show the coat to be highly heterogenous with limited long-range order, which is consistent with a minimally concentrative sorting mechanism (2, 11). Collectively, data do not support long-standing hypotheses of retromer sorting that invoke oligomerization as a driving force for sorting of integral membrane proteins in the endosomal system. Rather, low order oligomerization of retromer and associated factors is likely an adaptation of bulk membrane trafficking pathways characteristic of the endosomal system that ensure membrane homeostasis of the plasma membrane and endo-lysosome organelles.
Experimental Procedures
Molecular biology
cDNAs encoding human SNX3, RAB7, VPS35, VPS26, VPS29, VPS26-RRS, WASHC2C-21 (WASHC2C-357-1318), and WASHC2C-5 (WASHC2C-921-1318), and model cargo protein were amplified by PCR and cloned into bacterial expression vectors. Retromer, RAB7, and SNX3 were prepared as described in Harrison et al. (17). Fluorescence-tagged retromers, retromer-RRS, model cargo protein, and WASHC2C fragments were prepared as described here.
Model cargo protein was constructed from the cytoplasmic tail of the retromer cargo DMT1-II (15, 17). Construct contained a His10 tag followed by an 18 amino acid linker containing a cysteine (used for fluorescence labeling) (35), a 37 amino acid fragment of the divalent metal transporter DMTII-1 containing a retromer recognition sequence (amino acids 532–568) (15, 17), followed by maltose binding protein. The DMT sequence used LGMSFLDSGHTSHLGLTAQPELYLMNTMDADSLVRGL.
Retromer-RRS used a VPS26 construct in which the last four residues of VPS26 were removed, followed by a further truncated DMT1-II sequence, followed by a His10 tag: TAQPELYLMNTMDADSLVRGLHHHHHHHHHH.
Protein expression and purification
Proteins were prepared fresh on the first or second day of each 3-day experiment. All proteins were expressed by auto-induction (36) from BL21(DE3) E. coli cells. The cells were pelleted from culture medium and re-suspended in lysis buffer. Cells were lysed by three passages through a cell disruptor (Avestin) at >10,000 psi. The lysate was clarified by 30,000 × g centrifugation. Lysis buffers were supplemented with 0.1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 1 mm DTT, and cOmpleteTM Protease Inhibitor Mixture tablet (Roche Diagnostics).
GST-tagged proteins (retromer, SNX3, WASHC2C-5, and WASHC2C-21) were purified by incubating clarified lysate with GSH Sepharose 4B (GE Healthcare) beads. The beads were washed with lysis buffer and the proteins released from the beads with TEV protease. If the protein was to be fluorescently labeled, the labeling was carried out before the proteins were released from the beads. Purified proteins were quantified by Bradford assay (Thermo Scientific).
Retromer
Retromer subunit VPS35 was GST-TEV tagged. The other retromer subunits (untagged VPS-29, untagged VPS26, His-SNAP–tagged VPS26, and His-SNAP–tagged VPS26-RRS fusion) were not GST tagged. Cell pellets of separate cultures expressing VPS35, VPS29, and one of the VPS26 constructs were combined and resuspended together in lysis buffer no. 1 (20 mm HEPES, pH 8, and 150 mm NaCl). After cell lysis and clarification, retromer was purified using the GST-tagged protein protocol, where the VPS35-conjugated beads were washed with lysis buffer no. 1. Unpartnered VPS26 and VPS29 subunits were washed away during purification. Normal assembly of heterotrimeric retromer complexes in lysis buffer was confirmed by SDS-PAGE.
SNX3
GST-TEV-SNX3 was purified by incubating clarified lysate with GSH Sepharose 4B (GE Healthcare) beads. The beads were washed with lysis buffer no. 1 and the proteins released from the beads with TEV protease as described for retromer.
WASHC2C-21
GST-TEV-WASHC2C-357-SNAP was purified by incubating clarified lysate with GSH Sepharose 4B (GE Healthcare) beads. The beads were washed with lysis buffer no. 2 (25 mm HEPES, 500 mm NaCl, 20 mm imidazole, 10% glycerol, 1 mm TCEP, pH 7.8) and the proteins released from the beads with TEV protease.
WASHC2C-5
His6-SUMO-WASHC2C-5-SNAP was purified using lysis buffer no. 2 and Ni-NTA–agarose beads (Qiagen). The protein was incubated with Ni-NTA beads and washed with 50 mm imidazole. The His6-SUMO-WASHC2C-5-SNAP was released from the Qiagen beads with His6-SUMO protease.
RAB7
His10-RAB7 (Q67L)-His10, a constitutively active form of the protein (37), was purified using an ÄKTAprime plus FPLC system equipped with a 1-ml His-Trap HP column (GE Healthcare-Amersham Biosciences) and the high salt buffer (25 mm HEPES, 500 mm NaCl, 20 mm imidazole, 10% glycerol, 1 mm TCEP, pH 7.8). Immediately before use, RAB7 was incubated on ice with a 5× excess of 100 mm GTP disodium salt solution (Sigma) for 30 min. In all experiments with RAB7-GTP, the RAB7-GTP is His-tagged and attaches to SLBs by binding to nickel lipids.
Cargo
The model cargo protein, with a His10 tag, was purified by the same procedure as RAB7 using the high-salt buffer.
Fluorescence labeling of recombinant proteins
Proteins were labeled on the second day of a 3-day experiment. SNAP-tagged proteins were fluorescence labeled while still bound to purification beads by incubation with a 2× molar excess of SNAP Surface Alexa Fluor 488 or 647 as marked overnight at 4°C with constant mixing. After 16-18 h, the unreacted dyes were removed by thoroughly washing the beads with a minimum of 60 bead-volumes of lysis buffer prior to protein release. After free dye removal, all proteins clarified with a 100,000 × g centrifugation step and immediately quantified by Bradford assay. The concentration of dye was determined with a spectrophotometer. The labeling efficiency was calculated by e = Mdye/Mprotein, where Mdye and Mprotein are the molar concentration of dye and protein. Proteins with labeling efficiency below 75% were not used.
Model cargo protein was labeled via the engineered single cysteine in the linker between the His-tag and the cargo protein sequence. The protein was labeled with a 5× excess of Alexa Fluor 488 C5 maleimide dye (ThermoScientific) in the dark and tumbled overnight at 4°C. Unreacted dye was quenched with the addition of 5 mm 2-mercaptoethanol (Sigma-Aldrich). Free dye was removed with three sequential 2-ml Zeba desalting columns (Thermo Scientific) that had been equilibrated with liposome buffer A (25 mm HEPES, 200 mm NaCl, 1 mm MgCl2, pH 7.5) with 10% glycerol. To label SNX3, a single cysteine (L11C, C140S) mutant of GST-SNX3 was labeled overnight on the GSH Sepharose beads (GE Lifesciences) at 4°C with a 5× excess of Alexa Fluor 546 C5 maleimide dye (Thermo Scientific). Unreacted dye was quenched and removed as described.
Liposomes
Liposomes were prepared generally by the protocol of Su et al. (38) with some adjustments. Liposomes were made from pure synthetic lipids (Avanti Polar Lipids, and Echelon Biosciences (PtdIns-3-P)) by combining 92 mol% dioleoyl-phosphatidylcholine, 5 mol% dioleoyl-phosphatidylserine, 2 mol% Ni-NTA–DGS, and 1 mol% di-palmitoyl phosphatidylinositol 3-phosphate in a new 4-ml glass vial. Trace amounts (0.01 μl) of either 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (Rh-PE) or 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (ammonium salt) (NBD-PE) (Avanti) were added to visualize the SLB. For a typical total volume of ∼80 μl of lipids in the vial, 1 ml of 9:1 chloroform:methanol was added to the vial and the contents were gently swirled to mix the different lipids together. The contents of the vial were dried to a lipid film with nitrogen and residual solvent removed for 2 h under vacuum. The lipid film was rehydrated in the vial to 0.62 mm with 400 μl of buffer A by shaking for 30 min and was then transferred to an Eppendorf tube. The lipid solution was then frozen at −80°C until the day of the experiment. On the day of imaging, lipid-buffer mix was sonicated on ice with a microtip sonicator (Misonex, Qsonica, S-4000, 417A) for 20 min at an amplitude of 30 and 50% duty cycle. The sonicated liposomes were centrifuged for 20 min at 100,000 × g to pellet any larger liposomes or aggregates. The supernatant was used to make the SLB within 30 min of clarification.
Supported lipid bilayers
SLBs were made by the protocol of Su et al. (38) with some adjustments. A 96-well, black, glass-bottomed plate that has low background fluorescence (MatriPlate MGB096-1-2LG-L) was cleaned in an overnight soak in 5% Hellmanex III heated to 50°C. After thoroughly rinsing with Milli-Q filtered water and drying the slide, the clean wells were sealed with PCR sealing foil sheets (Thermo Scientific). To form an SLB, an individual well was opened and cleaned with two 1-h incubations at 50°C of freshly prepared, sterile filtered 5 m NaOH. The well was rinsed twice with 0.5 ml Milli-Q water and twice with buffer A. The rinsed well was then filled with 0.2 ml of buffer A and 10 μl of the liposomes were added. After incubation for 1 h at 37°C, the well was washed three times with buffer A to remove any nonadhered liposomes. The SLB was then blocked with 0.1% casein in buffer A for 20 min at 37°C. We checked the mobility of the SLB by FRAP or visual inspection (described below) for each experiment; immobile membranes were discarded.
In one three-channel co-localization experiment (Fig. 3), the fluorescence signal was too weak for visualization on the SLB alone because of the low affinity between retromer and WASHC2C. In this experiment, we looked at the bright spots where more lipid and thus protein was concentrated, and signal-to-noise was high enough to detect co-localization.
Protein attachment to supported bilayers
His10-tagged proteins were bound to Ni-NTA–DGS lipids in the SLB. Proteins were added to the well containing 0.2 ml of buffer A with 0.1% casein and 1 mm TCEP. The protein and the bilayer were incubated in the well in the dark for 2 h at 30°C to get a secure attachment to the bilayer. After protein addition, the bilayer was washed three times with 0.1% casein buffer A. Proteins not binding through His10 tags (SNX3, retromer, and WASHC2C) were incubated with the SLB for 30 min in the dark at room temperature and then washed three times with 0.1% casein buffer A. Wells with immobile proteins were not used.
Fluorescence microscopy
TIRF images were collected on a custom-built polarized TIRF microscope with an Olympus microscope body, a 60× objective (oil, PlanApo, NA 1.45), an Andor EM CCD camera (Ixon Ultra), and Micro-Manager software (39). Lasers at 488 nm, 561 nm, and 638 nm were used to excite NBD-PE/Alexa Fluor 488, Rh-PE/Alexa Fluor 546, and Alexa Fluor 647, respectively. Single micrographs were collected with a 100-ms exposure time, and movies were imaged in stream mode (∼60 images/s) with a 17.74-ms frame duration. The 488-nm laser for FRAP measurements with NBD-PE was used at maximum laser power, and data were analyzed as described in Refs. 40 and 41.
Readouts of large-scale clustering events
Some systems, for example the reconstituted adhesion receptor nephrin signaling system (35), may undergo large-scale phase transitions in which very large clusters, networks, or coats of proteins may form. To identify clustering events larger than ∼10 particles we prepared SLBs with high protein densities and examined them by TIRF fluorescence microscopy. These experiments used high protein concentrations in the medium (∼1nm), resulting in high protein densities on the SLB, with separation between fluorescence-labeled proteins smaller than the optical resolution (∼250 nm). Images were inspected visually for localized puncta of intense fluorescence. In some more sensitive quantitative experiments, intensity measurements (line scans) across the illuminated field of view were used to assess fine-scale variation in the uniformity of fluorescence across the surface of the membrane.
Calculation of retromer copies per cluster
Calibration of single fluorophore intensities
Fluorescently labeled proteins bound to a SLB were imaged using TIRF microscopy using continuous stream acquisition until nearly all spots bleached. Imaging started in a virgin region that was never exposed to excitation light. This was achieved by first focusing on a different location, turning on autofocus and moving to a new unilluminated spot. Thus, the fluorescent spots in first frame of a movie represent retromer clusters that have not yet bleached. Laser power was adjusted to achieve stepwise bleaching in the illuminated sample. Each movie was corrected for uneven illumination field and background in a two-step process. First, the final frame in the movie, where almost all the fluorescent particles have bleached, was Gaussian blurred with a width of 2 pixels, then this frame was subtracted from the entire image stack. Individual particles of fluorescence were identified and tracked using the ImageJ plugin SpeckleTrackerJ (42–44). Particles were automatically detected and tracked. Because final bleaching steps could be most confidently identified, we only used the final drop in intensity to estimate single-molecule intensities. Tracking typically started after some spots bleached; a lower spot density facilitated tracking. Tracks stopped the frame before a particle disappeared. This was checked visually, and tracks corrected if necessary. The tracks were saved and further analyzed in MATLAB. The last five frames before bleaching and five subsequent frames at the last position were analyzed by calculating the average pixel intensity in a 3 × 3 pixel region centered around the tracked position of the particle. The drop in intensity, , because of this last bleaching step, was calculated by subtracting the average of the 5 post-bleach intensities from the 5 pre-bleach intensities. The distribution of was plotted and checked that it fitted well with a Gaussian function. The mean single-fluorophore intensity, , was used to estimate the copy numbers of retromer molecules in clusters detected in the first frame, as explained below. Each movie yielded 20–80 single-fluorophore intensity estimates from such bleaching events, and multiple image stacks were analyzed for each individual experiment.
Analysis
We returned to the first frame in every movie and detected particles using SpeckleTrackerJ, employing the same criteria across different conditions. Using MATLAB, we calculated the mean pixel intensity for every spot, I, in a 3 × 3 pixel region around the spot's centroid. Every spot's intensity was divided by the average single-fluorophore intensity for that condition, , and the labeling efficiency, e, to estimate the number N of retromer in that spot (cluster): N = I/. Probability density functions of N are displayed with a bin width of 1. Typically, three movies per condition were tracked and analyzed for single-particle fluorescence, and five movies per condition were used for analysis of retromer cluster size distributions. Two independent protein preparations were used per condition.
Statistical differences between distributions under separate conditions were tested using the two-sample Kolmogorov-Smirnov test (for comparing the distributions in Fig. 2, C and D) and the Kruskal-Wallis or one-way analysis of variance, followed by the multi-comparison tests (using Tukey's Honestly Significant Difference procedure, for comparing distributions in Figs. 4B and 5). These tests yielded low p-values in some cases, indicating the distributions are statistically significantly different. Some differences are indeed obvious by inspection (e.g. Fig. 5, 1:0 versus 50:1), but these differences lie mainly in changes in the monomer-to-oligomer ratios. The mean retromer copies per cluster, and the spans varied within a limited range, 2.5 to 3.1 for the mean and [0–12] to [0–14] for the span. PlotsofDifferences web app (25) was used to calculate estimation statistics, which are based on an estimate of the magnitude of the effect size and the uncertainty (26). In addition, variability among technical and biological replicates could explain the variability among biological conditions tested (Table S1). Thus, we do not find any evidence of biologically meaningful, large shifts in retromer copy numbers across the conditions tested.
WASHC2C titrations
Five wells with identical conditions were set up with the specified protein components. WASHC2C was added to the well at the specified concentration and incubated for 30 min. The well was then washed a minimum of three times with buffer A with 0.1% casein until bright spots of labeled WASHC2C were visible in the TIRF plane of the bilayer. Multiple bleaching movies were collected as described above and changes to the retromer particle distribution were quantified. After analysis of the puncta of fluorescence in the micrographs (Fig. 4A), results were plotted as histograms (Fig. 4, A and B), with the order of the oligomer (i.e. 1 (monomers), 2 (dimers), 3 (tetramers), etc.) on the horizontal axis and relative frequency of the oligomer on the vertical axis. Statistical differences between the distributions were tested as described above.
Data Availability
All data are contained within the manuscript.
Supplementary Material
This article contains supporting information.
Author contributions—C. L. D. and C. G. B. conceptualization; C. L. D., E. K., and C. G. B. funding acquisition; C. L. D. investigation; C. L. D., J. N., E. K., and C. G. B. methodology; C. L. D. and C. G. B. writing-original draft; C. L. D., J. N., and C. G. B. writing-review and editing; E. K. and C. G. B. supervision; C. G. B. resources.
Funding and additional information—This work was supported by the NIGMS and the NINDS, National Institutes of Health Grants R37GM061221 (to C. G. B.), F32GM125120 (to C. L. D.), and R01NS113236 (to E. K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- Buffer A
- 25 mm HEPES, 200 mm NaCl, 1 mm MgCl2, pH 7.5
- FRAP
- fluorescence recovery after photobleaching
- GUV
- giant unilamellar vesicle
- LFa
- leucine-phenylalanine-acidic residue motif
- NBD
- 12-(N-methyl-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl))
- Ni-NTA
- nickel-nitrilotriacetic acid
- Ni-NTA–DGS
- nickel salt of 1,2-dioleoyl-sn-glycero-3-((N-(5-amino-1-carboxypentyl) iminodiacetic acid) succinyl)
- PE
- phosphatidylethanolamine
- Rh-PE
- rhodamine-PE
- RRS
- retromer recognition sequence
- SLB
- supported lipid bilayer
- TCEP
- tris(2-carboxyethyl)phosphine
- TEV
- tobacco etch virus
- TIRFM
- total internal reflection fluorescence microscopy.
References
- 1. Burd C., and Cullen P. J. (2014) Retromer: A master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol. 6, a016774 10.1101/cshperspect.a016774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen K. E., Healy M. D., and Collins B. M. (2019) Towards a molecular understanding of endosomal trafficking by Retromer and Retriever. Traffic 20, 465–478 10.1111/tra.12649 [DOI] [PubMed] [Google Scholar]
- 3. Wang S., and Bellen H. J. (2015) The retromer complex in development and disease. Development (Camb.) 142, 2392–2396 10.1242/dev.123737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collins B. M., Norwood S. J., Kerr M. C., Mahony D., Seaman M. N., Teasdale R. D., and Owen D. J. (2008) Structure of Vps26B and mapping of its interaction with the retromer protein complex. Traffic 9, 366–379 10.1111/j.1600-0854.2007.00688.x [DOI] [PubMed] [Google Scholar]
- 5. Collins B. M., Skinner C. F., Watson P. J., Seaman M. N., and Owen D. J. (2005) Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nat. Struct. Mol. Biol. 12, 594–602 10.1038/nsmb954 [DOI] [PubMed] [Google Scholar]
- 6. Reddy J. V., and Seaman M. N. (2001) Vps26p, a component of retromer, directs the interactions of Vps35p in endosome-to-Golgi retrieval. Mol. Biol. Cell 12, 3242–3256 10.1091/mbc.12.10.3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seaman M. N., McCaffery J. M., and Emr S. D. (1998) A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142, 665–681 10.1083/jcb.142.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomez T. S., and Billadeau D. D. (2009) A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699–711 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kendall A. K., Xie B., Xu P., Wang J., Burcham R., Frazier M. N., Binshtein E., Wei H., Graham T. R., Nakagawa T., and Jackson L. P. (2020) mammalian retromer is an adaptable scaffold for cargo sorting from endosomes. Structure 28, 393–405.e4 10.1016/j.str.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Norwood S. J., Shaw D. J., Cowieson N. P., Owen D. J., Teasdale R. D., and Collins B. M. (2011) Assembly and solution structure of the core retromer protein complex. Traffic 12, 56–71 10.1111/j.1600-0854.2010.01124.x [DOI] [PubMed] [Google Scholar]
- 11. Kovtun O., Leneva N., Bykov Y. S., Ariotti N., Teasdale R. D., Schaffer M., Engel B. D., Owen D. J., Briggs J. A. G., and Collins B. M. (2018) Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature 561, 561–564 10.1038/s41586-018-0526-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Strochlic T. I., Setty T. G., Sitaram A., and Burd C. G. (2007) Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol. 177, 115–125 10.1083/jcb.200609161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harterink M., Port F., Lorenowicz M. J., McGough I. J., Silhankova M., Betist M. C., van Weering J. R., van Heesbeen R. G., Middelkoop T. C., Basler K., Cullen P. J., and Korswagen H. C. (2011) A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat. Cell Biol. 13, 914–923 10.1038/ncb2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen C., Garcia-Santos D., Ishikawa Y., Seguin A., Li L., Fegan K. H., Hildick-Smith G. J., Shah D. I., Cooney J. D., Chen W., King M. J., Yien Y. Y., Schultz I. J., Anderson H., Dalton A. J., et al. (2013) Snx3 regulates recycling of the transferrin receptor and iron assimilation. Cell Metab. 17, 343–352 10.1016/j.cmet.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tabuchi M., Yanatori I., Kawai Y., and Kishi F. (2010) Retromer-mediated direct sorting is required for proper endosomal recycling of the mammalian iron transporter DMT1. J. Cell Sci. 123, 756–766 10.1242/jcs.060574 [DOI] [PubMed] [Google Scholar]
- 16. Lucas M., Gershlick D. C., Vidaurrazaga A., Rojas A. L., Bonifacino J. S., and Hierro A. (2016) Structural mechanism for cargo recognition by the retromer complex. Cell 167, 1623–1635.e14 10.1016/j.cell.2016.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrison M. S., Hung C. S., Liu T. T., Christiano R., Walther T. C., and Burd C. G. (2014) A mechanism for retromer endosomal coat complex assembly with cargo. Proc. Natl. Acad. Sci. U. S. A. 111, 267–272 10.1073/pnas.1316482111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Purushothaman L. K., and Ungermann C. (2018) Cargo induces retromer-mediated membrane remodeling on membranes. Mol. Biol. Cell 29, 2709–2719 10.1091/mbc.E18-06-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonifacino J. S., and Hurley J. H. (2008) Retromer. Curr. Opin. Cell Biol. 20, 427–436 10.1016/j.ceb.2008.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hierro A., Rojas A. L., Rojas R., Murthy N., Effantin G., Kajava A. V., Steven A. C., Bonifacino J. S., and Hurley J. H. (2007) Functional architecture of the retromer cargo-recognition complex. Nature 449, 1063–1067 10.1038/nature06216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., and Aebersold R. (2011) The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549 10.1038/msb.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., and Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548 10.1038/msb.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Toomre D., and Bewersdorf J. (2010) A new wave of cellular imaging. Annu. Rev. Cell Dev. Biol. 26, 285–314 10.1146/annurev-cellbio-100109-104048 [DOI] [PubMed] [Google Scholar]
- 24. Gardner M. J., and Altman D. G. (1986) Confidence intervals rather than P values: Estimation rather than hypothesis testing. Br. Med. J. 292, 746–750 10.1136/bmj.292.6522.746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goedhart J. (2019) PlotsOfDifferences—a web app for the quantitative comparison of unpaired data. bioRxiv 578575 10.1101/578575 [DOI]
- 26. Ho J., Tumkaya T., Aryal S., Choi H., and Claridge-Chang A. (2019) Moving beyond P values: Data analysis with estimation graphics. Nat. Methods 16, 565–566 10.1038/s41592-019-0470-3 [DOI] [PubMed] [Google Scholar]
- 27. Rojas R., van Vlijmen T., Mardones G. A., Prabhu Y., Rojas A. L., Mohammed S., Heck A. J., Raposo G., van der Sluijs P., and Bonifacino J. S. (2008) Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J. Cell Biol. 183, 513–526 10.1083/jcb.200804048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seaman M. N., Harbour M. E., Tattersall D., Read E., and Bright N. (2009) Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci. 122, 2371–2382 10.1242/jcs.048686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D., and Gautreau A. (2009) The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712–723 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- 30. Duleh S. N., and Welch M. D. (2010) WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton 67, 193–206 10.1002/cm.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jia D., Gomez T. S., Billadeau D. D., and Rosen M. K. (2012) Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol. Biol. Cell 23, 2352–2361 10.1091/mbc.E11-12-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Puthenveedu M. A., Lauffer B., Temkin P., Vistein R., Carlton P., Thorn K., Taunton J., Weiner O. D., Parton R. G., and von Zastrow M. (2010) Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell 143, 761–773 10.1016/j.cell.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harbour M. E., Breusegem S. Y., and Seaman M. N. (2012) Recruitment of the endosomal WASH complex is mediated by the extended 'tail' of Fam21 binding to the retromer protein Vps35. Biochem. J. 442, 209–220 10.1042/BJ20111761 [DOI] [PubMed] [Google Scholar]
- 34. Maxfield F. R., and McGraw T. E. (2004) Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5, 121–132 10.1038/nrm1315 [DOI] [PubMed] [Google Scholar]
- 35. Banjade S., and Rosen M. K. (2014) Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife 3, e04123 10.7554/eLife.04123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 37. Buczynski G., Bush J., Zhang L., Rodriguez-Paris J., and Cardelli J. (1997) Evidence for a recycling role for Rab7 in regulating a late step in endocytosis and in retention of lysosomal enzymes in Dictyostelium discoideum. Mol. Biol. Cell 8, 1343–1360 10.1091/mbc.8.7.1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Su X., Ditlev J. A., Rosen M. K., and Vale R. D. (2017) Reconstitution of TCR signaling using supported lipid bilayers. Methods Mol. Biol. 1584, 65–76 10.1007/978-1-4939-6881-7_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N. (2010) Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. Chapter 14, Unit14.20 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nikolaus J., and Karatekin E. (2016) SNARE-mediated fusion of single proteoliposomes with tethered supported bilayers in a microfluidic flow cell monitored by polarized TIRF microscopy. J. Vis. Exp. 2016, 54349 10.3791/54349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karatekin E., and Rothman J. E. (2012) Fusion of single proteoliposomes with planar, cushioned bilayers in microfluidic flow cells. Nat. Protoc. 7, 903–920 10.1038/nprot.2012.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith M. B., Karatekin E., Gohlke A., Mizuno H., Watanabe N., and Vavylonis D. (2011) Interactive, computer-assisted tracking of speckle trajectories in fluorescence microscopy: Application to actin polymerization and membrane fusion. Biophys. J. 101, 1794–1804 10.1016/j.bpj.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.