Abstract
Multicellular organisms have multiple genes encoding calponins and calponin-related proteins, some of which are known to regulate actin cytoskeletal dynamics and contractility. However, the functional similarities and differences among these proteins are largely unknown. In the nematode Caenorhabditis elegans, UNC-87 is a calponin-related protein with seven calponin-like (CLIK) motifs and is required for maintenance of contractile apparatuses in muscle cells. Here, we report that CLIK-1, another calponin-related protein that also contains seven CLIK motifs, functionally overlaps with UNC-87 in maintaining actin cytoskeletal integrity in vivo and has both common and different actin-regulatory activities in vitro. We found that CLIK-1 is predominantly expressed in the body wall muscle and somatic gonad in which UNC-87 is also expressed. unc-87 mutation caused cytoskeletal defects in the body wall muscle and somatic gonad, whereas clik-1 depletion alone caused no detectable phenotypes. However, simultaneous clik-1 and unc-87 depletion caused sterility because of ovulation failure by severely affecting the contractile actin networks in the myoepithelial sheath of the somatic gonad. In vitro, UNC-87 bundled actin filaments, whereas CLIK-1 bound to actin filaments without bundling them and antagonized UNC-87–mediated filament bundling. We noticed that UNC-87 and CLIK-1 share common functions that inhibit cofilin binding and allow tropomyosin binding to actin filaments, suggesting that both proteins stabilize actin filaments. In conclusion, partially redundant functions of UNC-87 and CLIK-1 in ovulation are likely mediated by their common actin-regulatory activities, but their distinct actin-bundling activities suggest that they also have different biological functions.
Keywords: Actin, bundling, cytoskeleton, calponin-like (CLIK) motif, ovulation, UNC-87, actomyosin contractility, filament dynamics actin, Caenorhabditis elegans (C. elegans), cofilin, tropomyosin, calponin
Calponins and calponin-related proteins regulate actin filament bundling, actin filament stability, and actomyosin contractility in a wide variety of cell types (1–3). Many of these proteins contain a single calponin-homology (CH) domain and various numbers of calponin-like (CLIK) motifs (also known as CLIK repeats). Although CH domains are recognized as actin-binding domains of several proteins, such as fimbrin, α-actinin, and spectrin (4, 5), the CH domain of calponin is not an actin-binding site (6–9). Instead, CLIK motifs in the C terminus of calponin act as actin-binding sites (6, 9, 10). CLIK motifs are 23-amino acid sequences and present in variable numbers in calponins and calponin-related proteins (9, 11). Vertebrate SM22/transgelin and yeast calponin-related proteins have only one CLIK motif (12–16). Vertebrate calponins have three CLIK motifs (17), whereas calponins in flatworms and mollusks have five CLIK motifs (18–20). UNC-87 in the nematode Caenorhabditis elegans has seven CLIK motifs (21). Importantly, the number of CLIK motifs correlates with the strength of actin-filament binding in vitro (22) and inhibitory effects on actin filament dynamics in cultured cells (10, 23). These observations suggest that CLIK motifs are modular actin-binding sequences with incremental affinity with actin filaments depending on the number of motifs. However, whether all CLIK motifs have the same or different properties remains unknown.
Nematodes have uniquely evolved calponin-related proteins with multiple CLIK motifs and without a CH domain. C. elegans UNC-87, which contains seven CLIK motifs (21), is predominantly expressed in muscular tissues and associated with actin filaments (21, 24, 25). Two isoforms, UNC-87A and UNC-87B, are expressed by two alternative promoters for two separate first exons, and they share all CLIK motifs and similar biochemical properties (21, 24). Mutations in the unc-87 gene cause disorganization of sarcomeric actin filaments in the body wall muscle because of defects in the maintenance of sarcomeres (21, 26, 27). UNC-87 is conserved in other nematode species and plays a critical role in worm motility (28, 29). In vitro, UNC-87 bundles actin filaments (22), inhibits actin-filament severing by actin-depolymerizing factor (ADF)/cofilin (25), and inhibits actomyosin ATPase (24). In addition, the C. elegans genome has three other genes encoding similar calponin-related proteins with various numbers of CLIK motifs: clik-1 (seven motifs), clik-2 (six motifs), and clik-3 (five motifs). A proteomic study reported that CLIK-1, CLIK-2, and CLIK-3 are enriched in the pharyngeal muscle (30). Null mutations of clik-2 and clik-3 cause pumping defects in the pharynx, whereas null mutation of clik-1 causes no detectable phenotypes (31). Therefore, function of clik-1 remains unknown. In this study, we aimed to determine the function of clik-1. Our results indicate that UNC-87 and CLIK-1 are co-expressed in muscle tissues and have overlapping and critical functions in the reproductive system by maintaining cytoskeletal integrity in the somatic gonad. Biochemical characterization showed that UNC-87 and CLIK-1 have common and distinct actin-regulatory activities in vitro.
Results
CLIK-1 and UNC-87 are partially redundant and required for C. elegans reproduction
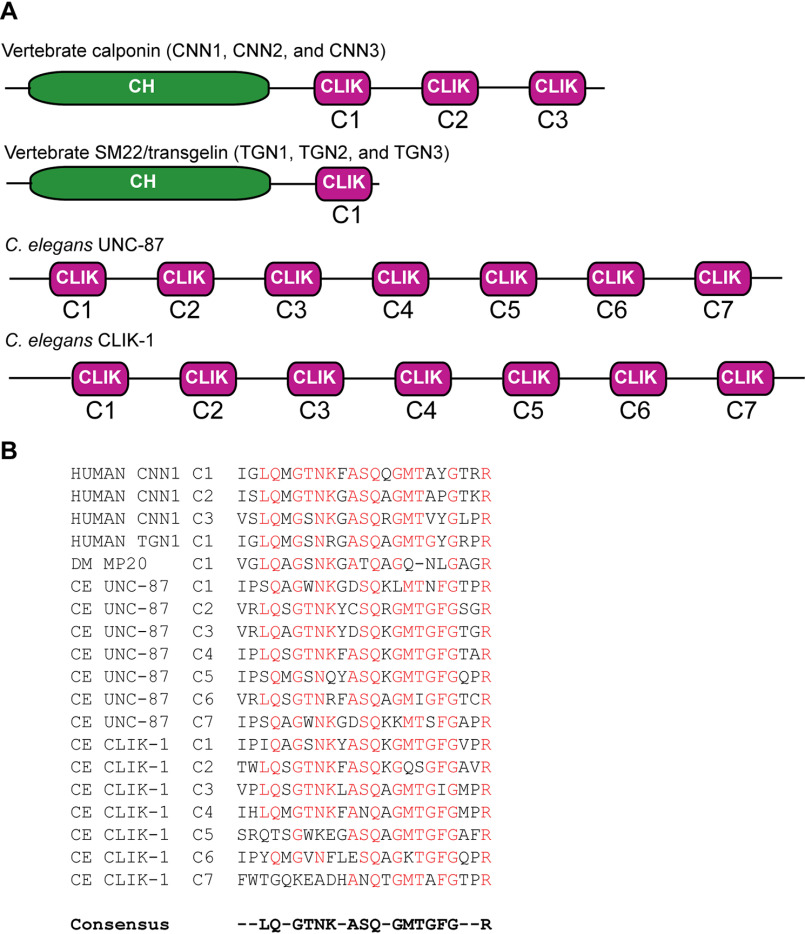
C. elegans has four genes (unc-87, clik-1, clik-2, and clik-3) encoding calponin-related proteins with no CH domains. Among them, primary structures of UNC-87B (hereafter referred to as UNC-87) and CLIK-1 (also known as T25F10.6, GenBankTM accession number CCD74240) are very similar (42% identity) containing seven CLIK motifs (C1–C7) (Fig. 1A). Comparison of CLIK motifs from representative calponins and calponin-related proteins shows that CLIK motifs are highly conserved across species (Fig. 1B). However, sequences of C5, C6, and C7 of CLIK-1 are diverged in their N-terminal halves (Fig. 1B), suggesting that biochemical properties of CLIK-1 are somewhat different from those of UNC-87.
Figure 1.
Domain organizations of calponin and calponin-related proteins. A, schematic representation of domain organizations of vertebrate calponin (CNN1, calponin 1; CNN2, calponin 2; CNN3, calponin 3), vertebrate SM22/transgelin (TGN1, SM22α/transgelin 1; TGN2, transgelin 2; TGN3, transgelin 3), C. elegans UNC-87, and C. elegans CLIK-1. B, sequence alignment of CLIK motifs from calponin and calponin-related proteins: human calponin 1 (CNN1) (NCBI reference sequence NM_001299); human SM22α/transgelin (TGN1) (NCBI reference sequence NM_001001522); Drosophila melanogaster MP20 (NCBI reference sequence NM_057295); C. elegans UNC-87 (NCBI reference sequence NM_001025922); and C. elegans CLIK-1 (GenBankTM accession number CCD74240).
To determine expression pattern and subcellular localization of CLIK-1, GFP was fused to the C terminus of CLIK-1 at the endogenous clik-1 locus using CRISPR/Cas9-mediated genome editing. CLIK-1–GFP was strongly expressed in the body wall muscle (Fig. 2, A, B, C, E, and G) and weakly in the myoepithelial sheath of the somatic gonad (Fig. 2, D, F, and H), where CLIK-1–GFP localized in a sarcomeric pattern in the body wall muscle (Fig. 2C) and in a filamentous pattern in the myoepithelial sheath (Fig. 2D). Staining of fixed worms by tetramethylrhodamine-phalloidin showed that CLIK-1 co-localized with actin filaments in the body wall muscle (Fig. 2, I–K) and myoepithelial sheath (Fig. 2, L–N). However, we did not detect expression of CLIK-1–GFP in the pharynx (Fig. 2, A and B), although we did detect expression of CLIK-1–GFP in the neurons surrounding the pharynx. Thus, our results disagree with the proteomics study reporting enrichment of CLIK-1 in the pharynx (30). Body wall muscle and myoepithelial sheath of the gonad are tissues in which UNC-87 is also expressed and associated with actin filaments (21, 24, 25), suggesting that CLIK-1 and UNC-87 regulate actin filaments in the same cells. Additional characterization of tissue distribution and subcellular localization of CLIK-1–GFP will be reported elsewhere.
Figure 2.
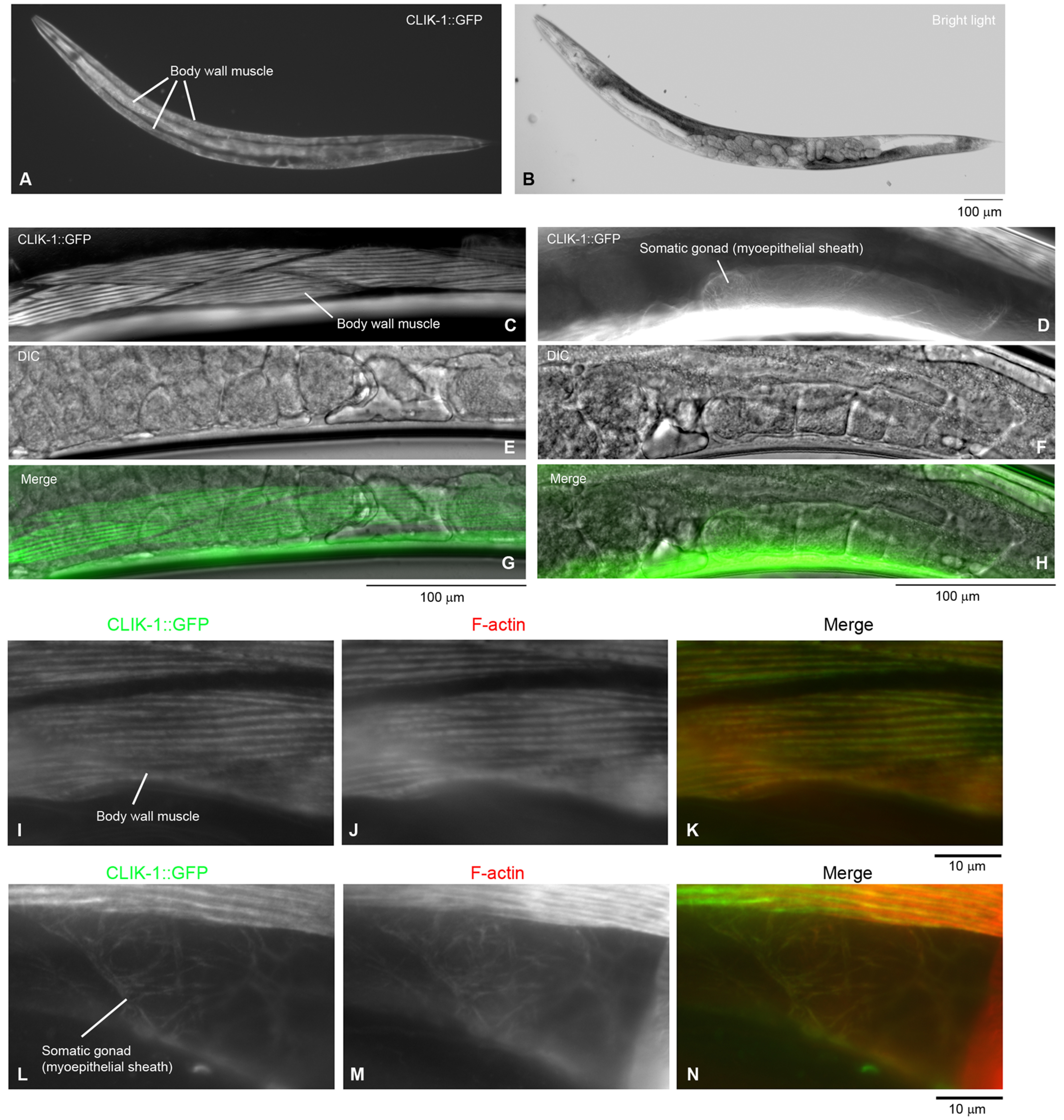
Expression and localization of CLIK-1 in C. elegans. GFP was fused to the C terminus of CLIK-1 using CRISPR/Cas9-mediated genome editing. A–H, expression and localization of CLIK-1–GFP were characterized in live worms. Expression pattern of CLIK-1–GFP in an entire adult hermaphrodite (A) and a bright-light image of the same animal (B) are shown. Bar, 100 μm. Localization of CLIK-1–GFP (C and D) and differential interference contrast images (E and F) were examined in live adult hermaphrodites at a high magnification. Merged images are shown in E and F. Bars, 100 μm. Expression of CLIK-1–GFP in the body wall muscle (C, E, and G) and the myoepithelial sheath of the somatic gonad (D, F, and H) is highlighted. I–N, the worms were fixed and stained with tetramethylrhodamine-phalloidin to visualize actin filaments (F-actin). Images of CLIK-1–GFP (I and L) and F-actin (J and M) were merged in K and N (CLIK-1–GFP in green and F-actin in red). The body wall muscle (I–K) and the myoepithelial sheath of the somatic gonad (L–N) are highlighted. Bar, 10 μm.
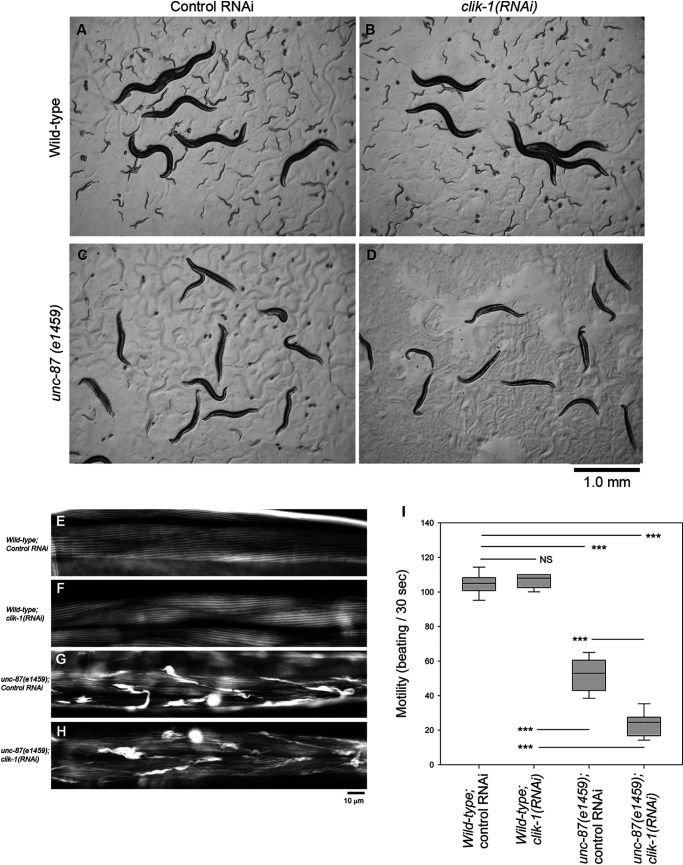
In a recent study, Wang et al. (31) reported that clik-1 gene knockout did not cause any obvious phenotypes, and we confirmed their observations using a separately isolated clik-1–null strain (clik-1(ok2355)) that contains a 1-kb deletion in the clik-1 gene. However, we found that knockdown of clik-1 by RNAi in an unc-87 null mutant (unc-87(e1459)) (21, 24) caused sterility with severe cytoskeletal defects in the reproductive system (Figs. 3 and 4). When worms were treated with control RNAi, WT worms actively moved and produced many progeny (Fig. 3, A and I), whereas unc-87(e1459) worms were slow-moving and produced much less progeny (Fig. 3, C and I). RNAi of clik-1 did not affect movement and fecundity in WT background (Fig. 3, B and I) but worsened worm motility nearly to paralysis (Fig. 3, D and I) and caused sterility in unc-87(e1459) (Fig. 3D, see the absence of eggs and small worms on the culture plate). In the body wall muscle, actin was organized into a striated sarcomeric pattern in WT with control RNAi (Fig. 3E) or clik-1(RNAi) (Fig. 3F). Sarcomeric actin in unc-87(e1459) with control RNAi was disorganized with accumulation in aggregates (Fig. 3G), and the actin disorganization was not apparently enhanced by clik-1(RNAi) in unc-87(e1459) (Fig. 3H), suggesting that enhancement of paralysis by clik-1(RNAi) is not due to structural defects in the sarcomeres. Currently, we do not know why the motility of unc-87(e1459); clik-1(RNAi) worms is severely impaired.
Figure 3.
Simultaneous depletion of unc-87 and clik-1 causes impaired worm motility and sterility. WT or unc-87(e1459) worms were treated with control RNAi or clik-1(RNAi). A–D, live worms on the culture plates are shown. The large worms are the RNAi-treated worms. The small worms and eggs are F1 progeny from the treated worms. Bar, 1.0 mm. E–H, organization of actin filaments in the body wall muscle as observed by staining with tetramethylrhodamine-phalloidin. Bar, 10 μm. I, worm motility was quantified as number of beats per 30 s. n = 10. Boxes represent the range of the 25th and 75th percentiles, with the medians marked by solid horizontal lines, and whiskers indicate the 10th and 90th percentiles. NS, not significant. ***, p < 0.001.
Figure 4.
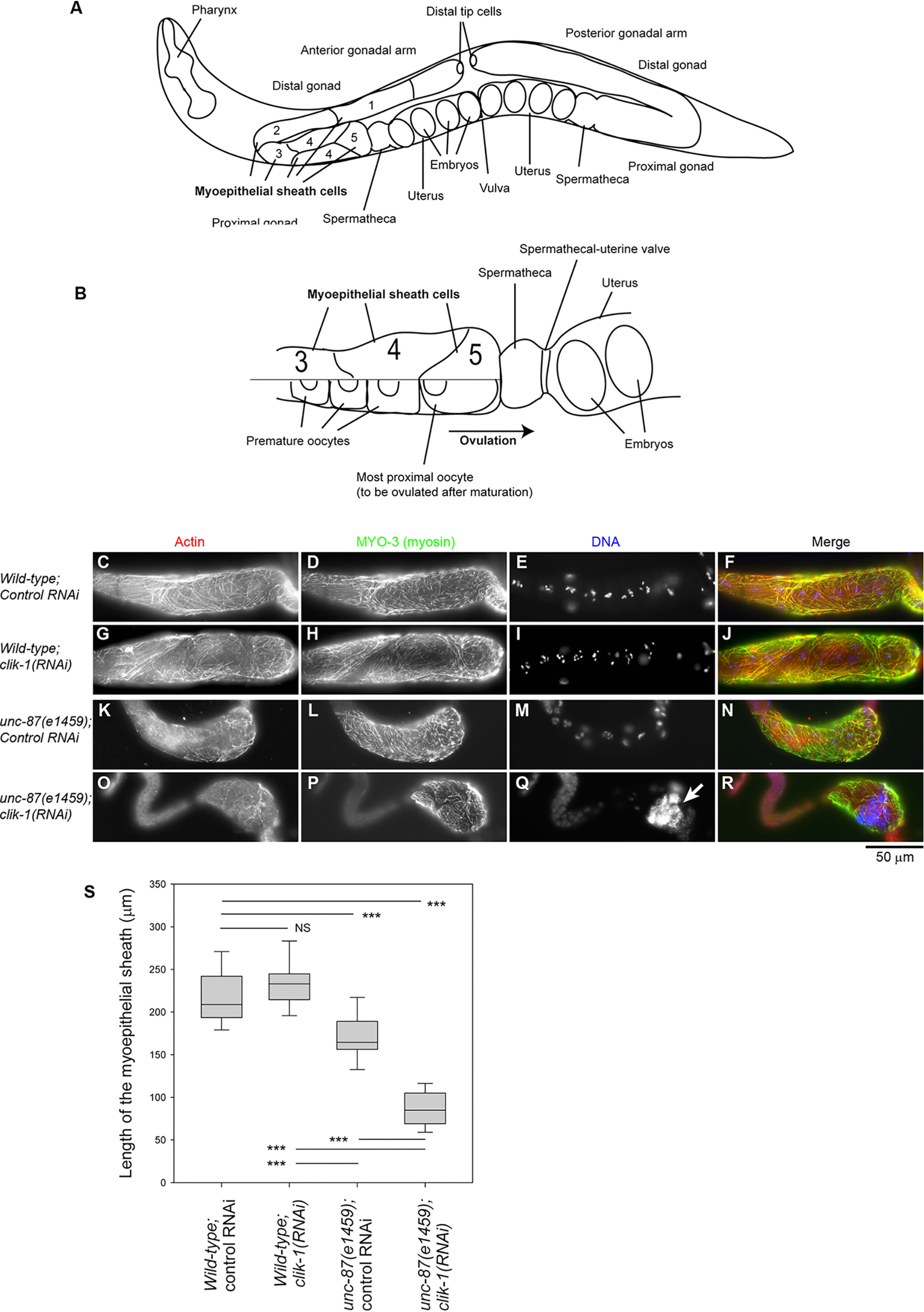
Simultaneous depletion of unc-87 and clik-1 causes severe disorganization of actin filaments in the somatic gonad. WT or unc-87(e1459) worms were treated with control RNAi or clik-1(RNAi). A and B, anatomy of the C. elegans gonad (adapted from Ref. 37). A hermaphrodite has two (anterior and posterior) gonadal arms (A). Each arm has five pairs of myoepithelial sheath cells (only shown in the anterior arm), and pairs 3–5 surround oocytes (B). The top half of B shows the myoepithelial sheath, and the bottom half of B shows the oocytes. C–R, dissected gonads were stained for F-actin (C, G, K, and O), MYO-3 (myosin) (D, H, L, and P), and DNA (E, I, M, and Q). The proximal sides of the gonads are oriented to the right. Merged images (F-actin in red, MYO-3 in green, and DNA in blue) are shown in the right column (F, J, N, and R). Bar, 50 μm. S, lengths of the myoepithelial sheath as marked by the MYO-3 staining were measured. Sample sizes (n) were 29 (WT; control RNAi), 28 (WT; clik-1(RNAi)), 26 (unc-87(e1459); control RNAi), and 27 (unc-87(e1459); clik-1(RNAi)). Boxes represent the range of the 25th and 75th percentiles, with the medians marked by solid horizontal lines, and whiskers indicate the 10th and 90th percentiles. NS, not significant. ***, p < 0.001.
Simultaneous depletion of unc-87 and clik-1 disrupted cytoskeletal integrity in the somatic gonads, which explains why worm reproduction was strongly impaired (Fig. 4). In C. elegans hermaphrodites, oocytes are surrounded by the myoepithelial sheath in the gonad that provides contractile forces during ovulation of mature oocytes (32, 33) (Fig. 4, A and B). The myoepithelial sheath is a smooth muscle–like somatic tissue that contains nonstriated actomyosin networks (34–36). The MYO-3 myosin heavy chain was used as a marker for the myoepithelial sheath (34, 36, 37). In WT with control RNAi, the myoepithelial sheath contained networks of actin and MYO-3 myosin and covered multiple oocytes (as shown by clusters of condensed chromosomes) (Fig. 4, C–F). RNAi of clik-1 in WT did not cause alteration in the actomyosin networks in the myoepithelial sheath (Fig. 4, G–J). In unc-87(e1459) with control RNAi, actin filaments were somewhat disorganized, and the myoepithelial sheath was not as extended as WT with control RNAi (Fig. 4, K–N and S). Strikingly, in unc-87(e1459) with clik-1(RNAi), actin filaments were only sparsely distributed, and the myoepithelial sheath was shortened and nearly collapsed to one side of the gonad (Fig. 4, O–S). The oocytes in these worms contained large accumulations of DNA (Fig. 4Q, arrow) as a result of endomitotic DNA replication during ovulation failure (38). These phenotypes indicate that UNC-87 and CLIK-1 have partially redundant function to maintain actin cytoskeletal integrity in the myoepithelial sheath and to execute proper ovulation, which is an essential process in worm reproduction.
CLIK-1 competes with UNC-87 for actin filament binding and inhibits filament bundling
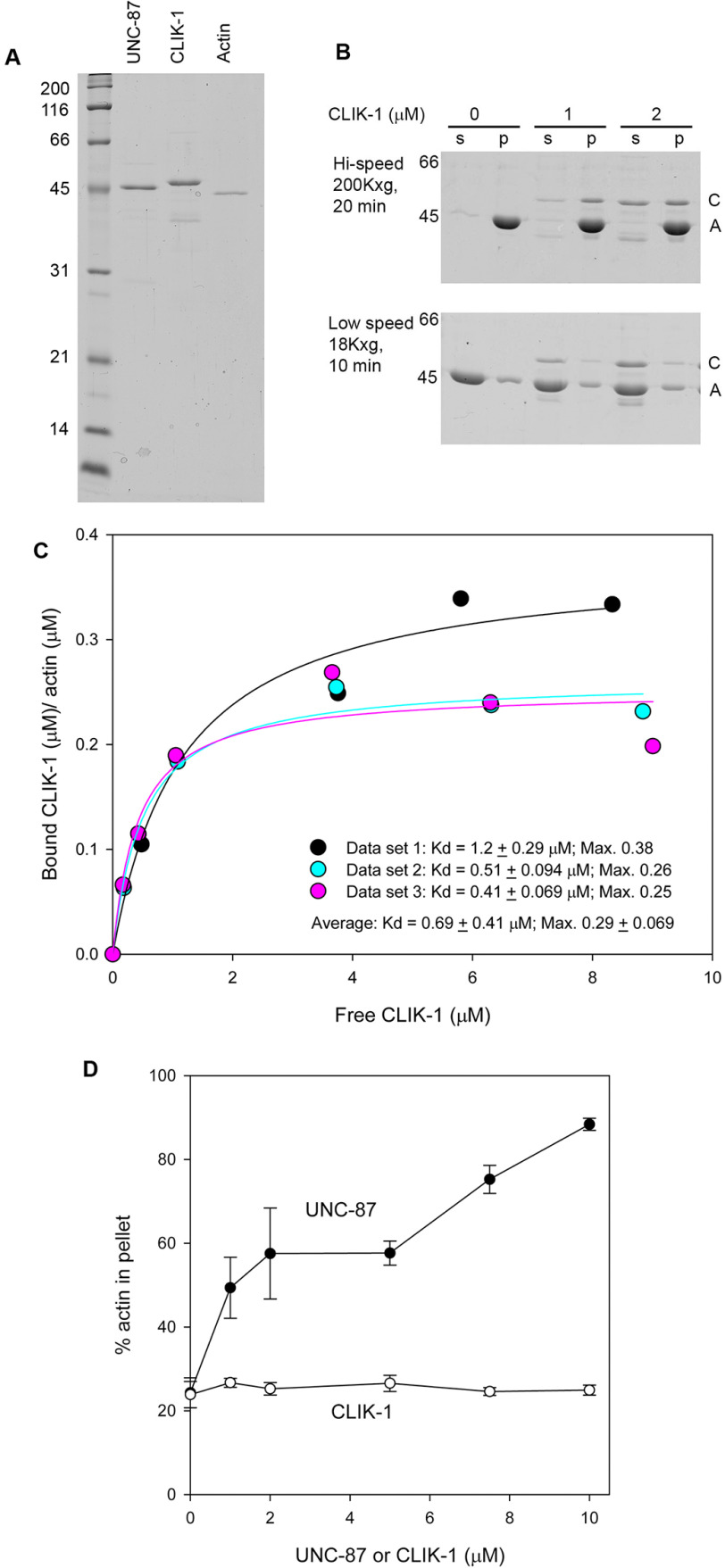
To characterize the biochemical properties of CLIK-1 and compare them with those of UNC-87, we prepared recombinant CLIK-1 protein with no extra tag sequence (Fig. 5A). When CLIK-1 was incubated with filamentous (F-) actin, CLIK-1 co-sedimented with F-actin after ultracentrifugation (Fig. 5B), indicating that CLIK-1 bound to F-actin. Quantitative analysis of the F-actin co-sedimentation assays at various CLIK-1 concentrations demonstrated that CLIK-1 bound to F-actin with a dissociation constant (Kd) of 0.69 ± 0.41 μm (n = 3), which is higher than the estimated Kd (∼0.15 μm) for UNC-87 binding to F-actin (25). Binding was saturated at ∼0.29 (mol CLIK-1/mol actin), indicating that CLIK-1 binds to actin filaments with a stoichiometry of 1:3 or 1:4 (Fig. 5C). Under low-speed centrifugation (18,000 × g, 10 min), CLIK-1 did not increase actin in the pellets (Fig. 5, B and D), whereas UNC-87 increased actin in the pellets by bundling filaments (22) (Fig. 5D). Therefore, CLIK-1 binds to actin filaments but, unlike UNC-87, lacks filament bundling activity.
Figure 5.
CLIK-1 binds to actin filaments without bundling them. A, purity of recombinant UNC-87 (0.5 µg), recombinant CLIK-1 (0.5 µg), and rabbit muscle actin (0.2 µg) was examined by SDS-PAGE (12% acrylamide gel). Molecular mass markers in kDa are shown on the left. B, F-actin sedimentation assays at high speed (200,000 × g, 20 min) (top) and low speed (18,000 × g, 10 min) (bottom). F-actin (10 μm) was incubated with 0–2 μm CLIK-1 for 1 h and centrifuged at the indicated conditions. Supernatants (s) and pellets (p) were separated and examined by SDS-PAGE. Positions of CLIK-1 (C) and actin (A) are indicated on the right. C, quantitative analysis of the high-speed F-actin sedimentation assays. The high-speed F-actin sedimentation assays were performed using 5 μm F-actin and 0–10 μm CLIK-1. Molar ratios of CLIK-1 to actin in the pellets (actin-dependent sedimentation of CLIK-1 was determined by subtracting nonspecific sedimentation of CLIK-1) (bound CLIK-1 (µm)/actin (µm)) are plotted as a function of free CLIK-1. Data from three independent experiments are plotted. A dissociation constant (Kd) and maximal (Max.) bound CLIK-1 (µm)/actin (µm) was determined from each data set (mean ± S.E. of mean from regression analysis). Averages of Kd (± standard deviation) and Max. (± standard deviation) are shown on the lower right. D, quantitative analysis of the low-speed F-actin sedimentation assays. The low-speed F-actin sedimentation assays were performed with 10 μm F-actin and 0–10 μm UNC-87 or CLIK-1. Percentages of actin in the pellets (bundled actin filaments) are plotted as a function of concentration of UNC-87 or CLIK-1. The data are means ± standard deviation from three independent experiments.
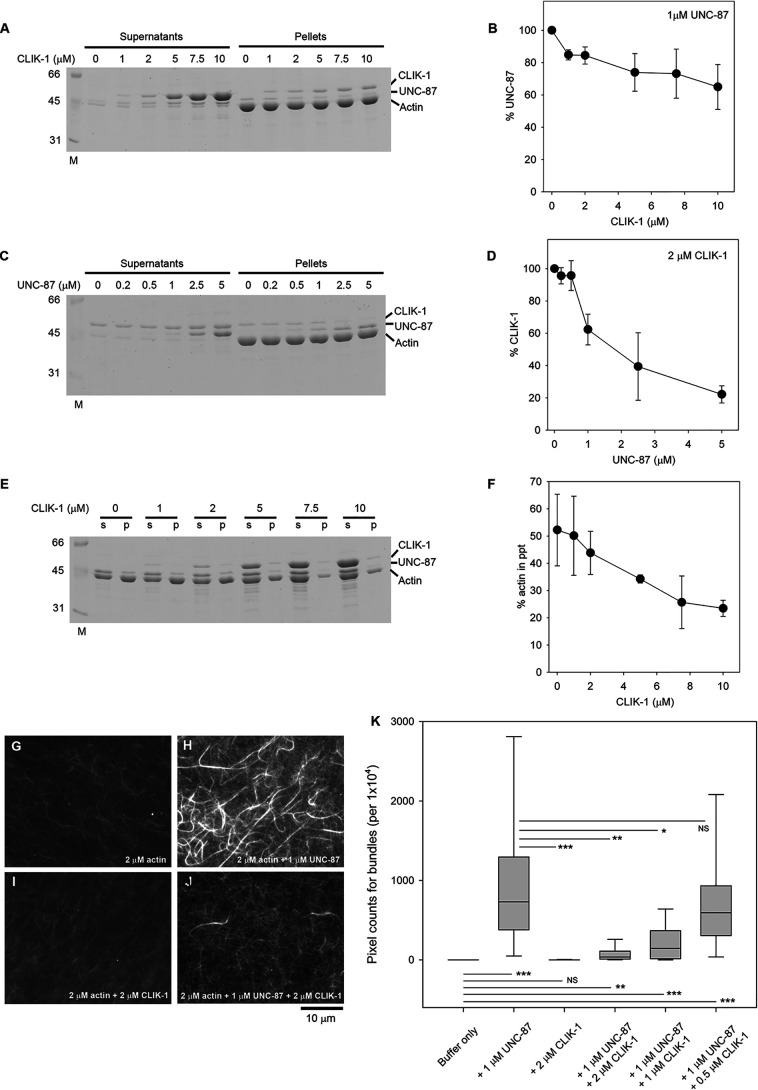
Because CLIK-1 and UNC-87 localize to actin filaments in the same cells, we examined their functional relationship when the two proteins are both present. Not surprisingly, given their sequence similarity, CLIK-1 and UNC-87 competed for F-actin binding (Fig. 6, A–F). In the F-actin co-sedimentation assays at high speed (200,000 × g, 20 min) in the presence of a constant UNC-87 concentration, increasing concentrations of CLIK-1 reduced UNC-87 in the pellets (Fig. 6, A and B). In the opposite experiments with a constant CLIK-1 concentration, increasing concentrations of UNC-87 reduced CLIK-1 in the pellets (Fig. 6, C and D). UNC-87 was more resistant to dissociation than CLIK-1 and more effective to dissociate the counterpart than CLIK-1, which is consistent with the higher affinity of UNC-87 with actin than that of CLIK-1. Interestingly, the competition between UNC-87 and CLIK-1 resulted in antagonistic modulation of actin filament bundling. UNC-87 bundled actin filaments and increased actin in the pellets under low-speed centrifugation (Fig. 6, E and F). However, increasing concentrations of CLIK-1 decreased actin in the pellets even in the presence of UNC-87 (Fig. 6, E and F). Their antagonistic effects on actin bundling were further demonstrated by direct observation of fluorescently labeled actin filaments. Actin filament bundles were formed in the presence of UNC-87 (Fig. 6, compare G and H), but not CLIK-1 (Fig. 6I). When both UNC-87 (1 μm) and CLIK-1 (2 μm) were present, very few actin bundles were formed (Fig. 6, J and K). The concentration of CLIK-1 needed to be higher than that of UNC-87 to cause significant reduction of actin bundles (Fig. 6K). Thus, the balance of UNC-87 and CLIK-1 can determine the extent of actin filament bundling in vitro.
Figure 6.
Competitive binding of UNC-87 and CLIK-1 to actin filaments. A and B, F-actin (10 μm) was incubated with 1 μm UNC-87 and 0–10 μm CLIK-1 and examined by high-speed co-sedimentation assays. Supernatants and pellets were analyzed by SDS-PAGE (A). Relative amounts of UNC-87 in the pellets (100% in the absence of CLIK-1) were quantified and plotted as a function of CLIK-1 concentration (B). The data are means ± standard deviation from three independent experiments. C and D, F-actin (10 μm) was incubated with 2 μm CLIK-1 and 0–5 μm UNC-87 and examined by high-speed co-sedimentation assays. Supernatants and pellets were analyzed by SDS-PAGE (C). Relative amounts of CLIK-1 in the pellets (100% in the absence of UNC-87) were quantified and plotted as a function of UNC-87 concentration (D). The data are means ± standard deviation from three independent experiments. E and F, F-actin (10 μm) was incubated with 2.5 μm UNC-87 and 0–10 μm CLIK-1 and examined by low-speed sedimentation assays. Supernatants (s) and pellets (p) were analyzed by SDS-PAGE (E). Percentages of actin in the pellets (bundled actin filaments) were plotted as a function of CLIK-1 concentration. The data are means ± standard deviation from four independent experiments. G–K, direct observation of actin bundling by fluorescence microscopy. DyLight 549–labeled actin (2 μm) was incubated with buffer only (G) or buffer with 1 μm UNC-87 (H), 2 μm CLIK-1 (I), or 1 μm UNC-87 and 2 μm CLIK-1 (J) and observed with fluorescence microscopy. Bar, 10 μm. K, actin bundle formation was quantified from randomly selected regions of interest (100 × 100 pixels) (n = 25). Boxes represent the range of the 25th and 75th percentiles, with the medians marked by solid horizontal lines, and whiskers indicate the 10th and 90th percentiles. NS, not significant. *, 0.01 < p < 0.05; **, 0.001 < p < 0.01; ***, p < 0.001.
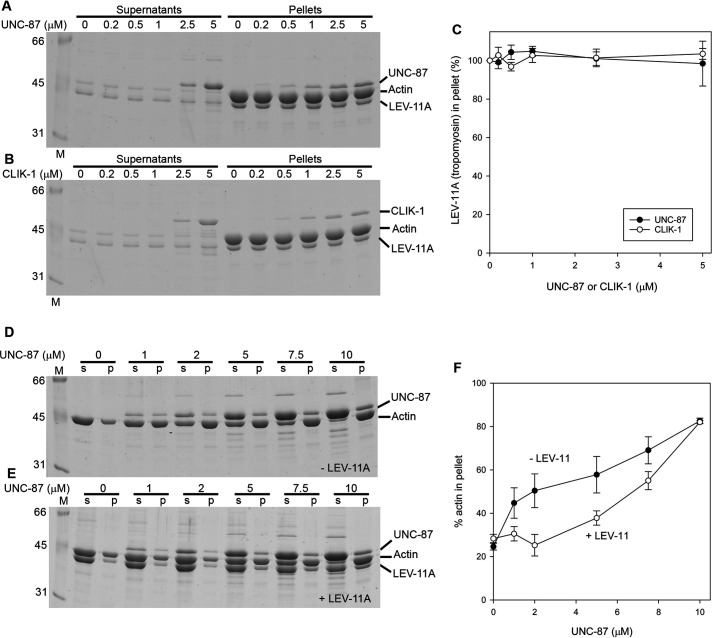
Tropomyosin binds to actin filaments simultaneously with CLIK-1 or UNC-87 and partially inhibits filament bundling
Although the biochemical experiments suggest that CLIK-1 is an inhibitor of actin filament bundling, we did not detect excessive actin filament bundling in the body wall muscle or myoepithelial sheath of clik-1(RNAi) worms or clik-1 knockout worms. Therefore, we reasoned that other actin-binding protein(s) can inhibit actin filament bundling by UNC-87. Because tropomyosin is a major F-actin–binding protein, we examined how tropomyosin interacts with actin filaments in the presence of UNC-87 or CLIK-1. The lev-11 gene encodes at least six tropomyosin isoforms, and LEV-11A is a major high-molecular-weight isoform in muscle cells (39–42). LEV-11A bound to actin filaments in the presence of either UNC-87 or CLIK-1 (Fig. 7, A–C). In addition, in the presence of a saturating concentration of LEV-11A, actin bundling, as determined by low-speed actin sedimentation, was inhibited when the UNC-87 concentrations were low (Fig. 7, D–F). At high UNC-87 concentrations, actin filament bundling was no longer inhibited by LEV-11A (Fig. 7, D–F). Therefore, tropomyosin, in addition to CLIK-1, can also inhibit UNC-87–induced actin filament bundling under certain conditions.
Figure 7.
Tropomyosin binds to F-actin in the presence of UNC-87 or CLIK-1 and partially inhibits UNC-87–mediated actin filament bundling. A–C, F-actin (10 μm) was incubated with 1 μm LEV-11A (tropomyosin) and 0–10 μm UNC-87 (A) or CLIK-1 (B) and examined by high-speed co-sedimentation assays. Supernatants and pellets were analyzed by SDS-PAGE. Relative amounts of LEV-11A in the pellets (100% in the absence of UNC-87 or CLIK-1) were quantified and plotted as a function of UNC-87 or CLIK-1 concentration (C). The data are means ± standard deviation from three independent experiments. D–F, F-actin (10 μm) was incubated with 0–10 μm UNC-87 in the absence (D) or presence of 2 μm LEV-11A (E) and examined by low-speed sedimentation assays. Supernatants (s) and pellets (p) were analyzed by SDS-PAGE. Percentages of actin in the pellets (bundled actin filaments) were plotted as a function of UNC-87 concentration. The data are means ± standard deviation from three independent experiments.
CLIK-1 and UNC-87 prevent ADF/cofilin from F-actin binding
UNC-87 stabilizes actin filaments by preventing biding of ADF/cofilin to the filaments (25). Similarly, CLIK-1 inhibited ADF/cofilin binding to actin filaments. UNC-60B, a muscle-specific ADF/cofilin isoform in C. elegans, severs actin filaments, and UNC-60B–bound actin filaments can be sedimented by ultracentrifugation (43–46). UNC-60B, which co-sedimented with F-actin, decreased when increasing concentrations of UNC-87 (Fig. 8, A and C) or CLIK-1 (Fig. 8, B and C) were included. UNC-87 was more efficient to prevent actin binding of UNC-60B than CLIK-1 (Fig. 8C), which is consistent with the difference in their affinity with actin. Thus, UNC-87 and CLIK-1 share a common actin-stabilizing function by antagonizing ADF/cofilin.
Figure 8.
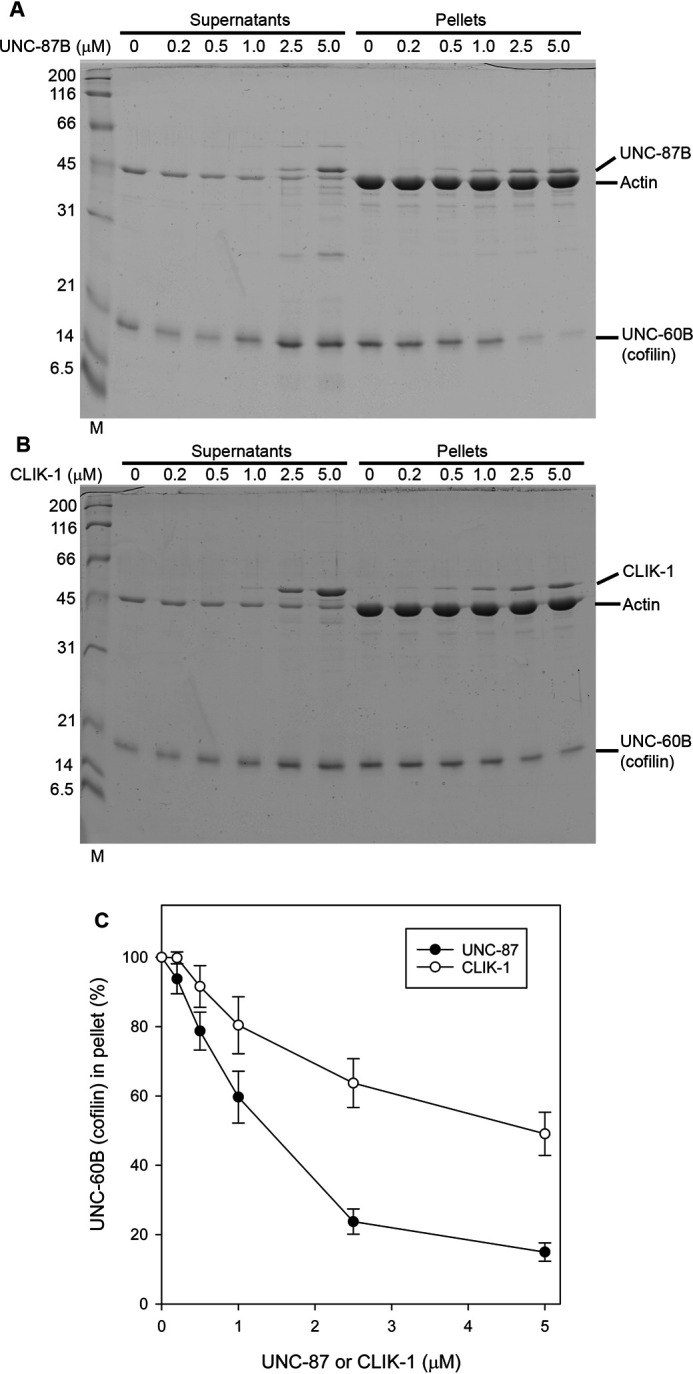
UNC-87 and CLIK-1 inhibit F-actin binding of ADF/cofilin. A and B, F-actin (10 μm) was incubated with 10 μm UNC-60B (ADF/cofilin) and 0–5 μm UNC-87 (A) or CLIK-1 (B) and examined by high-speed co-sedimentation assays. Supernatants and pellets were analyzed by SDS-PAGE. C, relative amounts of UNC-60B in the pellets (100% in the absence of UNC-87 or CLIK-1) were quantified and plotted as a function of UNC-87 or CLIK-1 concentration. The data are means ± standard deviation from three independent experiments.
Discussion
Here, we demonstrate that two calponin-related actin-binding proteins, UNC-87 and CLIK-1, are critical for cytoskeletal integrity in the C. elegans reproductive system. Depletion of both UNC-87 and CLIK-1 causes severe disorganization of actin filaments in the myoepithelial sheath of the somatic gonad, which results in sterility. Single depletion of either UNC-87 or CLIK-1 caused much milder or no phenotypes, indicating that UNC-87 and CLIK-1 have partially redundant functions in vivo. A previous single-cell proteomic study reported that CLIK-1 is enriched in the pharyngeal muscle cells (30). However, we do not detect strong expression of CLIK-1 in the pharynx. Instead, CLIK-1 expression is strong in other contractile tissues, including body wall muscle and myoepithelial sheath and overlaps with UNC-87 expression. Interestingly, biochemical analysis reveals both common and distinct actin-regulatory properties of UNC-87 and CLIK-1. Both proteins bind to actin filaments and prevent binding of ADF/cofilin to actin filaments. However, UNC-87 bundles actin filaments, but CLIK-1 does not. Their competitive binding to actin filaments modulates the extent of filament bundling when the two proteins co-exist. Thus, our current study uncovers novel functional aspects of calponin-related proteins in the regulation of the actin cytoskeleton.
The myoepithelial sheath of the somatic gonad (ovary) is highly sensitive to depletion of UNC-87 and CLIK-1. The myoepithelial sheath is a monolayer of smooth muscle–like cells that contain nonstriated contractile actin networks (34–36) (see Ref. 36 for structural details). Its contraction is tightly coupled with oocyte maturation and pushes only one mature oocyte out of the ovary for subsequent fertilization (32, 33). Contraction of the myoepithelial sheath is regulated by both troponin-tropomyosin complex (47–49) and phosphorylation of myosin light chain (37). UNC-87 has an inhibitory effect on actomyosin contractility and may facilitate relaxation of the myoepithelial sheath (24). Severe disorganization of the actin filaments in the myoepithelial sheath in double depletion of UNC-87 and CLIK-1 suggests that the two calponin-related proteins have redundant roles to stabilize actin filaments. CLIK-1 shares a common function with UNC-87 to protect actin filaments from ADF/cofilin (Fig. 8). ADF/cofilin and actin-interacting protein 1 are required for assembly of actin networks in the myoepithelial sheath (50, 51). However, excessive activities of these proteins are expected to destabilize actin filaments. Either UNC-87 or CLIK-1 is sufficient to stabilize actin filaments, but loss of both proteins can cause severe actin disorganization such that the myoepithelial sheath is no longer functional. In addition, ovulatory contraction of the myoepithelial sheath involves extensive morphological changes and mechanical stress (32, 33), and these calponin-related proteins may have a role in providing mechanical stability to the actin filaments as demonstrated biophysically for mammalian calponin (52, 53).
To our knowledge, CLIK-1 is the only calponin-related protein lacking actin filament bundling activity. Because only a limited number of calponin-related proteins have been characterized biochemically, there may be other calponin-related proteins with variable functions. Vertebrate calponin bundles actin filaments in vitro (54, 55) and plays important roles in cytoskeletal structures containing actin bundles (10, 56). Vertebrate SM22/transgelin (57, 58) and yeast calponin-related proteins (12–14, 59) have only one CLIK motif and still bundles actin filaments in vitro. A calponin-related protein with no actin-bundling activity, such as CLIK-1 or an equivalent protein, may compete with conventional calponin and/or SM22/transgelin to modulate the extent of actin bundle formation. Although such an antagonistic relationship was not clearly detected between UNC-87 and CLIK-1 in the body wall muscle and myoepithelial sheath in vivo, it may be of functional significance in other cell types or other actin-binding proteins, such as tropomyosin, may participate in the modulation of actin bundle formation.
The differences in the actin-regulatory activities of UNC-87 and CLIK-1 suggest that not all CLIK motifs behave in the same manner. Previous studies suggested that CLIK motifs are functionally similar modular motifs because the number of CLIK motifs correlates with the effectiveness in stabilizing actin filaments in cultured cells (10, 23). The difference between UNC-87 and CLIK-1 in actin-bundling activity indicate that some of the CLIK motifs in UNC-87 specifically mediate actin bundling. Because UNC-87 is monomeric (22), a single UNC-87 molecule needs to bind to multiple actin filaments to bundle them, suggesting that some of the CLIK motifs preferentially bind to a second filament. By contrast, CLIK-1 does not bundle actin filaments. Therefore, all the CLIK motifs in CLIK-1 bind to the same filament. There may be a difference in cooperativity among actin-binding sites. In UNC-87, a first actin-binding site may prevent a second actin-binding site from binding to the same filament, whereas in CLIK-1, a first actin-binding site may promote binding of a second actin-binding site to the same filament. Alternatively, the structure of UNC-87 may be suited to cross-bridge multiple filaments, whereas that of CLIK-1 may be optimal to bind to a single filament. Further dissection of biochemical properties of CLIK motifs should inform us about how different CLIK motifs contribute to functional differentiation of calponin-related proteins.
Experimental procedures
C. elegans strains and culture
The worms were cultured following standard methods (60). The following strains were obtained from the Caenorhabditis Genetics Center and used in this study: N2 WT, CB1459 unc-87(e1459), and RB1820 clik-1(ok2355). clik-1(ok2355) was outcrossed three times and used in this study (outcrossed strain: ON225).
CRISPR/Cas9 genome editing
The GFP sequence was inserted in the genome in-frame at the 3´-end of the clik-1 coding region by CRISPR/Cas9-mediated genome editing using vectors and protocols described by Dickinson et al. (61). A single-guide RNA target sequence (GATGTAATTACCACATACGA) was cloned into pDD162 (Addgene Plasmid # 47549) for expression of both single-guide RNA and Cas9 nuclease. Homology arms of ∼500 bp each were fused with the GFP::self-excising cassette region from pDD282 (Addgene plasmid 66823) by fusion PCR using Q5 high-fidelity DNA polymerase (New England Biolabs) and used as a homologous repair template. Hygromycin-resistant worms with a roller phenotype were isolated as knock-in worms. They were treated at 34 °C for 4 h to induce excision of the self-excising cassette. In the next generation, nonroller GFP-positive worms were isolated, and three independent clik-1::GFP strains, ON352 clik-1(kt1 [clik-1::gfp]), ON353 clik-1(kt2 [clik-1::gfp]), and ON354 clik-1(kt3 [clik-1::gfp]), were established.
RNAi experiments
RNAi experiments were performed by feeding with Escherichia coli HT115(DE3) expressing dsRNA as described previously (62). The RNAi experiments were started by treating L1 larvae, and phenotypes were observed when they grew to adult worms after 3 days. Control RNAi experiments were performed using the unmodified L4440 plasmid vector (kindly provided by Andrew Fire, Stanford University) (63). An RNAi clone for clik-1 (V-4N16) was obtained from Source BioScience. Images of worms on agar plates were captured using a Nikon Coolpix 995 digital camera mounted on a Zeiss Stemi 2000 stereo microscope.
Fluorescence microscopy
Live worms were anesthetized in M9 buffer containing 0.1% tricaine and 0.01% tetramisole for 30 min, mounted on 2% agarose pads, and observed by epifluorescence and differential interference contrast optics using a Nikon Eclipse TE2000 inverted microscope (Nikon Instruments, Tokyo, Japan).
Staining of whole worms with tetramethylrhodamine–phalloidin (Sigma–Aldrich) was performed as described previously (64). To preserve GFP fluorescence in phalloidin staining in Fig. 2 (I–N), adult worms expressing CLIK-1–GFP were anesthetized by 0.1% tricaine and 0.5 mm levamisole and cut in half using a pair of 28-gauge needles on polylysine-coated glass slides. Glass coverslips were overlaid on cut worms, frozen by dry ice, and then removed while they were frozen using a razor blade. They were immediately fixed by 4% paraformaldehyde in 1× cytoskeleton buffer (10 mm MES-KOH, pH 6.1, 138 mm KCl, 3 mm MgCl2, 2 mm EGTA) containing 0.32 m sucrose for 20 min at room temperature, permeabilized by PBS containing 0.5% Triton X-100 and 30 mm glycine (PBS-TG) for 10 min, and stained with 0.2 µg/ml tetramethylrhodamine–phalloidin in PBS-TG for 1 h at room temperature. They were washed three times with PBS-TG (5 min each).
Gonads were dissected from adult hermaphrodites on polylysine-coated glass slides as described previously (36). They were fixed by methanol at −20°C for 5 min and washed with PBS and primary antibodies in PBS containing 1% BSA. After being washed with PBS, they were treated with fluorophore-labeled secondary antibodies, followed by washing with PBS.
Primary antibodies used were rabbit anti-actin polyclonal antibody (Cytoskeleton, Denver, CO) and mouse anti–MYO-3 monoclonal antibody (clone 5–6) (65). The secondary antibodies used were Alexa 488–labeled goat anti-mouse IgG from Life Technologies and Cy3-labeled goat anti-mouse IgG from Jackson ImmunoResearch (West Grove, PA).
Fixed samples were mounted with ProLong Gold (Life Technologies) and observed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope with a CFI Plan Fluor ELWD 40× (dry; NA 0.60) or Plan Apo 60× (oil; NA 1.40) objective. Images were captured by a SPOT RT monochrome charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI) and processed by IPLab imaging software (BD Biosciences) and Photoshop CS3 (Adobe, San Jose, CA) or by ORCA Flash 4.0 LT monochrome scientific complementary metal–oxide–semiconductor camera (Hamamatsu Photonics, Shizuoka, Japan) and processed by Nikon NIS-Elements and Adobe Photoshop CS3. Quantification of the myoepithelial sheath lengths was performed using ImageJ.
Protein preparation
Actin was prepared from rabbit muscle acetone powder (Pel-Freeze Biologicals) as described by Pardee and Spudich (66). Recombinant LEV-11A (tropomyosin) (41), UNC-87B (22), and UNC-60B (ADF/cofilin) (45) were expressed in E. coli and purified as described previously. Unstained molecular weight markers (Nacalai USA, catalog no. 29458-24) were used in SDS-PAGE.
The ORF of clik-1 cDNA was amplified with reverse-transcriptase–PCR and cloned at the NdeI–BamHI sites of pET-3a (Novagen) with no extra tag sequence. E. coli BL21(DE3) pLysS was transformed with the expression vector and cultured in M9ZB medium containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37 °C until A600 reached 0.6 cm−1. Then protein expression was induced by adding 0.1 mm isopropyl β-d-thiogalactopyranoside for 3 h at 37 °C. The cells were harvested by centrifugation at 5000 × g for 10 min and disrupted by sonication in a buffer containing 0.1 m KCl, 1 mm EDTA, 20 mm Tris-HCl (pH 7.5), 0.2 mm DTT, and 1 mm phenylmethanesulfonyl fluoride. The homogenates were centrifuged at 25,000 × g for 30 min, and the supernatants were fractionated by 35% saturated ammonium sulfate (194 g/liter). Insoluble proteins were separated by centrifugation at 20,000 × g for 20 min, dissolved in a small volume of buffer A (0.1 m NaCl, 0.2 mm DTT, and 20 mm Tris-HCl, pH 7.5), and dialyzed overnight against buffer A. The dialyzed proteins were cleared by centrifugation at 20,000 × g for 20 min, applied to a Mono-Q 4.6/100PE column (GE Healthcare) and eluted with a linear gradient of NaCl from 0.1 to 0.5 m. Fractions containing CLIK-1 were applied to HiLoad 16/60 Superdex 75 (GE Healthcare) and eluted with a buffer containing 0.1 m KCl, 0.2 mm DTT, 20 mm HEPES-KOH, pH 7.5. Pure CLIK-1 fractions were pooled and concentrated by a MacroSep centrifugal device (molecular weight cutoff at 10,000) (Pall Corporation) and stored at −80 °C. Protein concentrations were determined by a BCA protein assay kit (Thermo Scientific).
F-actin sedimentation assays
Filamentous actin (5 or 10 μm) was incubated with proteins of interest in F-buffer (0.1 m KCl, 2 mm MgCl2, 20 mm HEPES-KOH, pH 7.5, 1 mm DTT) at room temperature. To examine F-actin binding of the proteins of interest by high-speed co-sedimentation, the samples were ultracentrifuged at 42,000 rpm (200,000 × g) for 20 min using a Beckman 42.2Ti rotor. To examine F-actin bundling by low-speed sedimentation, the samples were centrifuged at 15,000 rpm (18,000 × g) for 10 min using a Hettich MIKRO 200 centrifuge. Supernatants and pellets were separated, adjusted to the same volumes, and examined by SDS–PAGE (12% acrylamide gel). For quantitative analysis of high-speed F-actin co-sedimentation, control experiments were performed without actin, and nonspecific sedimentation of CLIK-1 was determined and subtracted from sedimentation of CLIK-1 in the presence of actin. The gels were stained with Coomassie Brilliant Blue R-250 (National Diagnostics) and scanned by an Epson Perfection V700 scanner at 300 dpi. Band intensity was quantified with ImageJ. Bound CLIK-1 (µm)/actin (µm) was plotted as a function of free CLIK-1 (µm). The data were fitted to the following equation using SigmaPlot v13.0 (Systat Software, San Jose, CA),
| (Eq. 1) |
where b = bound CLIK-1 (µm)/actin (µm), Max = maximal bound CLIK-1 (µm)/actin (µm), Kd = dissociation constant, and [CLIK-1] = [CLIK-1]free.
Microscopic observation of actin bundles
Direct observation of actin bundles by fluorescent microscopy was performed as described previously (24, 67) using DyLight 549–labeled actin (68). Briefly, DyLight 549–labeled actin filaments (2 μm actin, 20% labeled) were incubated with proteins of interest for 5 min at room temperature in F-buffer, and 2 µl of the samples were applied to nitrocellulose-coated coverslips (22 × 40 mm) and mounted with noncoated coverslips (18 × 18 mm). They were immediately observed with epifluorescence using a Nikon Eclipse TE2000 inverted microscope with a Plan Apo 60 × (oil; NA 1.40) objective. Images were captured by a SPOT RT monochrome charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI) and processed by IPLab imaging software (BD Biosciences) and Photoshop CS3 (Adobe, San Jose, CA). For quantification of actin bundles, regions of interest of 12.7 μm × 12.7 μm (100 × 100 pixels) were randomly selected. Then using ImageJ, a threshold was set to eliminate single filaments, and the number of pixels above the threshold regardless the intensity was counted.
Statistical analysis
The data in Figs. 3I, 4S, and 6K were analyzed by one-way analysis of variance with the Holm–Sidak method of pairwise multiple comparison procedures using SigmaPlot, version 13.0 (Systat Software, San Jose, CA, USA).
Data availability
All data are contained in the article. Raw data are available from S. O. upon request.
Acknowledgments
This work is dedicated to the memory of Kanako Ono, who passed away during the preparation of this article. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by Grant P40 OD010440 from the National Institutes of Health Office of Research Infrastructure Programs. mAb 5–6 (developed by Henry Epstein, University of Texas Medical Branch, Galveston, TX) was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences of the University of Iowa (Iowa City, IA).
Author contribution—S. O. conceptualization; S. O. and K. O. data curation; S.O. and K. O. formal analysis; S. O. supervision; S. O. funding acquisition; S. O. validation; S. O. and K. O. investigation; S. O. and K. O. methodology; S. O. writing-original draft; S. O. project administration; S. O. writing-review and editing.
Funding and additional information—This work was supported by National Institutes of Health Grant R01 AR048615 (to S. O.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Ono, S., and Ono, K., unpublished observations.
- CH
- calponin homology
- CLIK
- calponin-like
- ADF
- actin-depolymerizing factor
- F-actin
- filamentous actin.
References
- 1. Wu K. C., and Jin J. P. (2008) Calponin in non-muscle cells. Cell Biochem. Biophys. 52, 139–148 10.1007/s12013-008-9031-6 [DOI] [PubMed] [Google Scholar]
- 2. Carmichael J. D., Winder S. J., Walsh M. P., and Kargacin G. J. (1994) Calponin and smooth muscle regulation. Can. J. Physiol. Pharmacol. 72, 1415–1419 10.1139/y94-204 [DOI] [PubMed] [Google Scholar]
- 3. Winder S. J., and Walsh M. P. (1996) Calponin. Curr. Topics Cell. Reg. 34, 33–61 10.1016/S0070-2137(96)80002-1 [DOI] [PubMed] [Google Scholar]
- 4. Gimona M., Djinovic-Carugo K., Kranewitter W. J., and Winder S. J. (2002) Functional plasticity of CH domains. FEBS Lett. 513, 98–106 10.1016/S0014-5793(01)03240-9 [DOI] [PubMed] [Google Scholar]
- 5. Korenbaum E., and Rivero F. (2002) Calponin homology domains at a glance. J. Cell Sci. 115, 3543–3545 10.1242/jcs.00003 [DOI] [PubMed] [Google Scholar]
- 6. Gimona M., and Mital R. (1998) The single CH domain of calponin is neither sufficient nor necessary for F-actin binding. J. Cell Sci. 111, 1813–1821 [DOI] [PubMed] [Google Scholar]
- 7. Gimona M., and Winder S. J. (1998) Single calponin homology domains are not actin-binding domains. Curr. Biol. 8, R674–R675 [DOI] [PubMed] [Google Scholar]
- 8. Galkin V. E., Orlova A., Fattoum A., Walsh M. P., and Egelman E. H. (2006) The CH-domain of calponin does not determine the modes of calponin binding to F-actin. J. Mol. Biol. 359, 478–485 10.1016/j.jmb.2006.03.044 [DOI] [PubMed] [Google Scholar]
- 9. Stradal T., Kranewitter W., Winder S. J., and Gimona M. (1998) CH domains revisited. FEBS Lett. 431, 134–137 10.1016/S0014-5793(98)00751-0 [DOI] [PubMed] [Google Scholar]
- 10. Gimona M., Kaverina I., Resch G. P., Vignal E., and Burgstaller G. (2003) Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol. Biol. Cell 14, 2482–2491 10.1091/mbc.e02-11-0743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin R. M., Chilton N. B., Lightowlers M. W., and Gasser R. B. (1997) Echinococcus granulosus myophilin–relationship with protein homologues containing “calponin-motifs.” Int. J. Parasit. 27, 1561–1567 10.1016/S0020-7519(97)00146-X [DOI] [PubMed] [Google Scholar]
- 12. Goodman A., Goode B. L., Matsudaira P., and Fink G. R. (2003) The Saccharomyces cerevisiae calponin/transgelin homolog Scp1 functions with fimbrin to regulate stability and organization of the actin cytoskeleton. Mol. Biol. Cell 14, 2617–2629 10.1091/mbc.e03-01-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Winder S. J., Jess T., and Ayscough K. R. (2003) SCP1 encodes an actin-bundling protein in yeast. Biochem. J. 375, 287–295 10.1042/bj20030796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakano K., Bunai F., and Numata O. (2005) Stg 1 is a novel SM22/transgelin-like actin-modulating protein in fission yeast. FEBS Lett. 579, 6311–6316 10.1016/j.febslet.2005.10.011 [DOI] [PubMed] [Google Scholar]
- 15. Pearlstone J. R., Weber M., Lees-Miller J. P., Carpenter M. R., and Smillie L. B. (1987) Amino acid sequence of chicken gizzard smooth muscle SM22 alpha. J. Biol. Chem. 262, 5985–5991 [PubMed] [Google Scholar]
- 16. Prinjha R. K., Shapland C. E., Hsuan J. J., Totty N. F., Mason I. J., and Lawson D. (1994) Cloning and sequencing of cDNAs encoding the actin cross-linking protein transgelin defines a new family of actin-associated proteins. Cell Motil. Cytoskeleton 28, 243–255 10.1002/cm.970280307 [DOI] [PubMed] [Google Scholar]
- 17. Takahashi K., and Nadal-Ginard B. (1991) Molecular cloning and sequence analysis of smooth muscle calponin. J. Biol. Chem. 266, 13284–13288 [PubMed] [Google Scholar]
- 18. Yang W., Zheng Y. Z., Jones M. K., and McManus D. P. (1999) Molecular characterization of a calponin-like protein from Schistosoma japonicum. Mol. Biochem. Parasit. 98, 225–237 10.1016/S0166-6851(98)00171-6 [DOI] [PubMed] [Google Scholar]
- 19. Matusovsky O. S., Dobrzhanskaya A. V., Pankova V. V., Kiselev K. V., Girich U. V., and Shelud'ko N. S. (2017) Crenomytilus grayanus 40kDa calponin-like protein: cDNA cloning, sequence analysis, tissue expression, and post-translational modifications. Comp. Biochem. Physiol. D Genomics Proteomics 22, 98–108 10.1016/j.cbd.2017.02.006 [DOI] [PubMed] [Google Scholar]
- 20. Wang J., Gao J., Xie J., Zheng X., Yan Y., Li S., Xie L., and Zhang R. (2016) Cloning and mineralization-related functions of the calponin gene in Chlamys farreri. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 201, 53–58 10.1016/j.cbpb.2016.06.008 [DOI] [PubMed] [Google Scholar]
- 21. Goetinck S., and Waterston R. H. (1994) The Caenorhabditis elegans muscle-affecting gene unc-87 encodes a novel thin filament-associated protein. J. Cell Biol. 127, 79–93 10.1083/jcb.127.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kranewitter W. J., Ylanne J., and Gimona M. (2001) UNC-87 is an actin-bundling protein. J. Biol. Chem. 276, 6306–6312 10.1074/jbc.M009561200 [DOI] [PubMed] [Google Scholar]
- 23. Lener T., Burgstaller G., and Gimona M. (2004) The role of calponin in the gene profile of metastatic cells: inhibition of metastatic cell motility by multiple calponin repeats. FEBS Lett. 556, 221–226 10.1016/S0014-5793(03)01401-7 [DOI] [PubMed] [Google Scholar]
- 24. Ono K., Obinata T., Yamashiro S., Liu Z., and Ono S. (2015) UNC-87 isoforms, Caenorhabditis elegans calponin-related proteins, interact with both actin and myosin and regulate actomyosin contractility. Mol. Biol. Cell 26, 1687–1698 10.1091/mbc.E14-10-1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamashiro S., Gimona M., and Ono S. (2007) UNC-87, a calponin-related protein in C. elegans, antagonizes ADF/cofilin-mediated actin filament dynamics. J. Cell Sci. 120, 3022–3033 10.1242/jcs.013516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goetinck S., and Waterston R. H. (1994) The Caenorhabditis elegans UNC-87 protein is essential for maintenance, but not assembly, of bodywall muscle. J. Cell Biol. 127, 71–78 10.1083/jcb.127.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Waterston R. H., Thomson J. N., and Brenner S. (1980) Mutants with altered muscle structure of Caenorhabditis elegans. Dev. Biol. 77, 271–302 10.1016/0012-1606(80)90475-3 [DOI] [PubMed] [Google Scholar]
- 28. Joseph S., Gheysen G., and Subramaniam K. (2012) RNA interference in Pratylenchus coffeae: knock down of Pc-pat-10 and Pc-unc-87 impedes migration. Mol. Biochem. Parasit. 186, 51–59 10.1016/j.molbiopara.2012.09.009 [DOI] [PubMed] [Google Scholar]
- 29. Tan J. A., Jones M. G., and Fosu-Nyarko J. (2013) Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp. Parasit. 133, 166–178 10.1016/j.exppara.2012.11.011 [DOI] [PubMed] [Google Scholar]
- 30. Yuet K. P., Doma M. K., Ngo J. T., Sweredoski M. J., Graham R. L., Moradian A., Hess S., Schuman E. M., Sternberg P. W., and Tirrell D. A. (2015) Cell-specific proteomic analysis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 112, 2705–2710 10.1073/pnas.1421567112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang H., Park H., Liu J., and Sternberg P. W. (2018) An efficient genome editing strategy to generate putative null mutants in Caenorhabditis elegans using CRISPR/Cas9. G3 8, 3607–3616 10.1534/g3.118.200662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCarter J., Bartlett B., Dang T., and Schedl T. (1997) Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 181, 121–143 10.1006/dbio.1996.8429 [DOI] [PubMed] [Google Scholar]
- 33. McCarter J., Bartlett B., Dang T., and Schedl T. (1999) On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205, 111–128 10.1006/dbio.1998.9109 [DOI] [PubMed] [Google Scholar]
- 34. Ardizzi J. P., and Epstein H. F. (1987) Immunochemical localization of myosin heavy chain isoforms and paramyosin in developmentally and structurally diverse muscle cell types of the nematode Caenorhabditis elegans. J. Cell Biol. 105, 2763–2770 10.1083/jcb.105.6.2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strome S. (1986) Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J. Cell Biol. 103, 2241–2252 10.1083/jcb.103.6.2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ono K., Yu R., and Ono S. (2007) Structural components of the nonstriated contractile apparatuses in the Caenorhabditis elegans gonadal myoepithelial sheath and their essential roles for ovulation. Dev. Dyn. 236, 1093–1105 10.1002/dvdy.21091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ono K., and Ono S. (2016) Two distinct myosin II populations coordinate ovulatory contraction of the myoepithelial sheath in the Caenorhabditis elegans somatic gonad. Mol. Biol. Cell 27, 1131–1142 10.1091/mbc.E15-09-0648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iwasaki K., McCarter J., Francis R., and Schedl T. (1996) emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J. Cell Biol. 134, 699–714 10.1083/jcb.134.3.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kagawa H., Sugimoto K., Matsumoto H., Inoue T., Imadzu H., Takuwa K., and Sakube Y. (1995) Genome structure, mapping and expression of the tropomyosin gene tmy-1 of Caenorhabditis elegans. J. Mol. Biol. 251, 603–613 10.1006/jmbi.1995.0459 [DOI] [PubMed] [Google Scholar]
- 40. Anyanful A., Sakube Y., Takuwa K., and Kagawa H. (2001) The third and fourth tropomyosin isoforms of Caenorhabditis elegans are expressed in the pharynx and intestines and are essential for development and morphology. J. Mol. Biol. 313, 525–537 10.1006/jmbi.2001.5052 [DOI] [PubMed] [Google Scholar]
- 41. Barnes D. E., Watabe E., Ono K., Kwak E., Kuroyanagi H., and Ono S. (2018) Tropomyosin isoforms differentially affect muscle contractility in the head and body regions of the nematode Caenorhabditis elegans. Mol. Biol. Cell 29, 1075–1088 10.1091/mbc.E17-03-0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Watabe E., Ono S., and Kuroyanagi H. (2018) Alternative splicing of the Caenorhabditis elegans lev-11 tropomyosin gene is regulated in a tissue-specific manner. Cytoskeleton (Hoboken) 75, 427–436 10.1002/cm.21489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ono S., Baillie D. L., and Benian G. M. (1999) UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 145, 491–502 10.1083/jcb.145.3.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McKim K. S., Matheson C., Marra M. A., Wakarchuk M. F., and Baillie D. L. (1994) The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol. Gen. Genet. 242, 346–357 10.1007/BF00280425 [DOI] [PubMed] [Google Scholar]
- 45. Ono S., and Benian G. M. (1998) Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J. Biol. Chem. 273, 3778–3783 10.1074/jbc.273.6.3778 [DOI] [PubMed] [Google Scholar]
- 46. Yamashiro S., Mohri K., and Ono S. (2005) The two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins differently enhance actin filament severing and depolymerization. Biochemistry 44, 14238–14247 10.1021/bi050933d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ono K., and Ono S. (2004) Tropomyosin and troponin are required for ovarian contraction in the Caenorhabditis elegans reproductive system. Mol. Biol. Cell 15, 2782–2793 10.1091/mbc.e04-03-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Obinata T., Ono K., and Ono S. (2010) Troponin I controls ovulatory contraction of non-striated actomyosin networks in the C. elegans somatic gonad. J. Cell Sci. 123, 1557–1566 10.1242/jcs.065060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Myers C. D., Goh P. Y., Allen T. S., Bucher E. A., and Bogaert T. (1996) Developmental genetic analysis of troponin T mutations in striated and nonstriated muscle cells of Caenorhabditis elegans. J. Cell Biol. 132, 1061–1077 10.1083/jcb.132.6.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ono K., and Ono S. (2014) Two actin-interacting protein 1 isoforms function redundantly in the somatic gonad and are essential for reproduction in Caenorhabditis elegans. Cytoskeleton 71, 36–45 10.1002/cm.21152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ono K., Yamashiro S., and Ono S. (2008) Essential role of ADF/cofilin for assembly of contractile actin networks in the C. elegans somatic gonad. J. Cell Sci. 121, 2662–2670 10.1242/jcs.034215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jensen M. H., Morris E. J., Gallant C. M., Morgan K. G., Weitz D. A., and Moore J. R. (2014) Mechanism of calponin stabilization of cross-linked actin networks. Biophys. J. 106, 793–800 10.1016/j.bpj.2013.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jensen M. H., Watt J., Hodgkinson J. L., Gallant C., Appel S., El-Mezgueldi M., Angelini T. E., Morgan K. G., Lehman W., and Moore J. R. (2012) Effects of basic calponin on the flexural mechanics and stability of F-actin. Cytoskeleton (Hoboken) 69, 49–58 10.1002/cm.20548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kołakowski J., Makuch R., Stepkowski D., and Dabrowska R. (1995) Interaction of calponin with actin and its functional implications. Biochem. J. 306, 199–204 10.1042/bj3060199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lu F. W., Freedman M. V., and Chalovich J. M. (1995) Characterization of calponin binding to actin. Biochemistry 34, 11864–11871 10.1021/bi00037a026 [DOI] [PubMed] [Google Scholar]
- 56. Ciuba K., Hawkes W., Tojkander S., Kogan K., Engel U., Iskratsch T., and Lappalainen P. (2018) Calponin-3 is critical for coordinated contractility of actin stress fibers. Sci. Rep. 8, 17670 10.1038/s41598-018-35948-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shapland C., Hsuan J. J., Totty N. F., and Lawson D. (1993) Purification and properties of transgelin: a transformation and shape change sensitive actin-gelling protein. J. Cell Biol. 121, 1065–1073 10.1083/jcb.121.5.1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Han M., Dong L. H., Zheng B., Shi J. H., Wen J. K., and Cheng Y. (2009) Smooth muscle 22 alpha maintains the differentiated phenotype of vascular smooth muscle cells by inducing filamentous actin bundling. Life Sci. 84, 394–401 10.1016/j.lfs.2008.11.017 [DOI] [PubMed] [Google Scholar]
- 59. Gheorghe D. M., Aghamohammadzadeh S., Smaczynska-de R. II, Allwood E. G., Winder S. J., and Ayscough K. R. (2008) Interactions between the yeast SM22 homologue Scp1 and actin demonstrate the importance of actin bundling in endocytosis. J. Biol. Chem. 283, 15037–15046 10.1074/jbc.M710332200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stiernagle T. (2006) Maintenance of C. elegans. In WormBook (Fay D., ed) pp. 1–11, C. elegans Research Community, Pasadena, CA: 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D., and Goldstein B. (2015) Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200, 1035–1049 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ono S., and Ono K. (2002) Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 156, 1065–1076 10.1083/jcb.200110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Timmons L., Court D. L., and Fire A. (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- 64. Ono S. (2001) The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 152, 1313–1319 10.1083/jcb.152.6.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Miller D. M., Ortiz I., Berliner G. C., and Epstein H. F. (1983) Differential localization of two myosins within nematode thick filaments. Cell 34, 477–490 10.1016/0092-8674(83)90381-1 [DOI] [PubMed] [Google Scholar]
- 66. Pardee J. D., and Spudich J. A. (1982) Purification of muscle actin. Methods Enzymol. 85, 164–181 10.1016/0076-6879(82)85020-9 [DOI] [PubMed] [Google Scholar]
- 67. Ono S. (2016) Basic methods to visualize actin filaments in vitro using fluorescence microscopy for observation of filament severing and bundling. In Cytoskeleton: Methods and Protocols, 3rd Ed., (Gavin R. H., ed.) pp. 187–193, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 68. Liu Z., Klaavuniemi T., and Ono S. (2010) Distinct roles of four gelsolin-like domains of Caenorhabditis elegans gelsolin-like protein-1 in actin filament severing, barbed end capping, and phosphoinositide binding. Biochemistry 49, 4349–4360 10.1021/bi100215b [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained in the article. Raw data are available from S. O. upon request.