Abstract
Pel is a GalNAc-rich bacterial polysaccharide that contributes to the structure and function of Pseudomonas aeruginosa biofilms. The pelABCDEFG operon is highly conserved among diverse bacterial species, and Pel may therefore be a widespread biofilm determinant. Previous annotation of pel gene clusters has helped us identify an additional gene, pelX, that is present adjacent to pelABCDEFG in >100 different bacterial species. The pelX gene is predicted to encode a member of the short-chain dehydrogenase/reductase (SDR) superfamily, but its potential role in Pel-dependent biofilm formation is unknown. Herein, we have used Pseudomonas protegens Pf-5 as a model to elucidate PelX function as Pseudomonas aeruginosa lacks a pelX homologue in its pel gene cluster. We found that P. protegens forms Pel-dependent biofilms; however, despite expression of pelX under these conditions, biofilm formation was unaffected in a ΔpelX strain. This observation led us to identify a pelX paralogue, PFL_5533, which we designate here PgnE, that appears to be functionally redundant to pelX. In line with this, a ΔpelX ΔpgnE double mutant was substantially impaired in its ability to form Pel-dependent biofilms. To understand the molecular basis for this observation, we determined the structure of PelX to 2.1 Å resolution. The structure revealed that PelX resembles UDP-GlcNAc C4-epimerases. Using 1H NMR analysis, we show that PelX catalyzes the epimerization between UDP-GlcNAc and UDP-GalNAc. Our results indicate that Pel-dependent biofilm formation requires a UDP-GlcNAc C4-epimerase that generates the UDP-GalNAc precursors required by the Pel synthase machinery for polymer production.
Keywords: polysaccharide, biofilm, Pseudomonas protegens, X-ray crystallography, epimerase, PelX, PgnE, short-chain dehydrogenase/reductase, Pseudomonas aeruginosa, bacterial adhesion, microbiology, enzyme, Pseudomonas
Exopolysaccharides are a critical component of bacterial biofilms. The opportunistic pathogen Pseudomonas aeruginosa is a model bacterium for studying the contribution of exopolysaccharides to biofilm architecture because biofilms formed by this organism use exopolysaccharides as a structural scaffold (1). P. aeruginosa synthesizes the exopolysaccharides alginate, Psl, and Pel, and each have been shown to contribute structural and protective properties to the biofilm matrix under various conditions (2). Although these polysaccharides differ in their chemical composition and net charge, the synthesis of all three polymers requires sugar-nucleotide precursors. Genes encoding enzymes required for precursor generation are often found within or adjacent to the gene cluster responsible for the production of their associated polysaccharide. For example, Psl requires GDP-mannose precursors, which are generated from mannose-1-phosphate by the enzyme PslB (3). Similarly, alginate requires the precursor GDP-mannuronic acid and the alg locus encodes two of the three enzymes, AlgA and AlgD, required to synthesize this activated sugar (4, 5). The third enzyme, AlgC, is not found within the alg operon and is also involved in synthesizing precursors for Psl and B-band lipopolysaccharide (6).
In Gram-negative bacteria, the pelABCDEFG operon encodes seven gene products that are required for pellicle (Pel) biofilm formation (7). These biofilms form at the air–liquid interface of standing P. aeruginosa cultures (8). In contrast to the Psl and alginate gene clusters, none of the P. aeruginosa pel genes are predicted to be involved in sugar-nucleotide precursor production, indicating that, like AlgC, these functions are encoded by genes elsewhere on the chromosome. Analyses of Pel have demonstrated that it is a cationic polysaccharide rich in GalNAc residues and that the putative Pel polymerase, PelF, preferentially interacts with the nucleotide UDP (9). Additionally, functional characterization of PelA has demonstrated that it is a bifunctional enzyme with both polysaccharide deacetylase and α-1,4-N-acetylgalactosaminidase activities, which further supports the hypothesis that the precursor required for the biosynthesis of Pel is an acetylated sugar (10, 11). Together, these data suggest that a key sugar-nucleotide precursor involved in Pel biosynthesis is UDP-GalNAc, the high-energy precursor needed for the biosynthesis of GalNAc-containing glycans.
We recently made the observation that many bacteria possess an additional ORF in their pel biosynthetic gene clusters that is predicted to encode a member of the short-chain dehydrogenase/reductase (SDR) enzyme superfamily (12, 13). The SDR superfamily is an ancient enzyme family whose members share a common structural architecture and are involved in the synthesis of numerous metabolites, including sugar-nucleotide precursors used for the generation of bacterial cell surface glycans (14). In several species of bacteria, such as the plant-protective pseudomonad Pseudomonas protegens, the SDR encoding gene pelX is found directly upstream of the pel genes, whereas other bacteria such as P. aeruginosa lack this gene within their pel locus.
Plant root colonization by P. protegens Pf-5 requires the formation of biofilms. This process has been shown to require the biofilm adhesin LapA (15). In addition to LapA, biofilms produced by this strain also contain undefined exopolysaccharides (16, 17). Besides Pel, P. protegens Pf-5 has the genetic capacity to synthesize the exopolysaccharides Psl, alginate, and poly-β-1,6-GlcNAc; however, little is known about the role these polymers play in P. protegens biofilm formation (17).
In the present study, we show that P. protegens Pf-5 forms Pel-dependent biofilms at air–liquid interfaces, and using P. protegens PelX as a representative Pel polysaccharide-linked SDR enzyme, we find that this enzyme functions as a UDP-GlcNAc C4-epimerase. We find that the pelX gene is not essential for Pel polysaccharide–dependent biofilm formation because P. protegens possesses a paralogue of this gene, PFL_5533. Deletion of both of these genes was found to substantially impair Pel-dependent biofilm formation. Based on our analyses we designate PFL_5533 as a polysaccharide UDP-GlcNAc epimerase (pgnE) and propose that the production of UDP-GalNAc by UDP-GlcNAc C4-epimerases is a critical step in the biosynthesis of the Pel polysaccharide.
Results
Identification of an SDR family enzyme associated with pel gene clusters
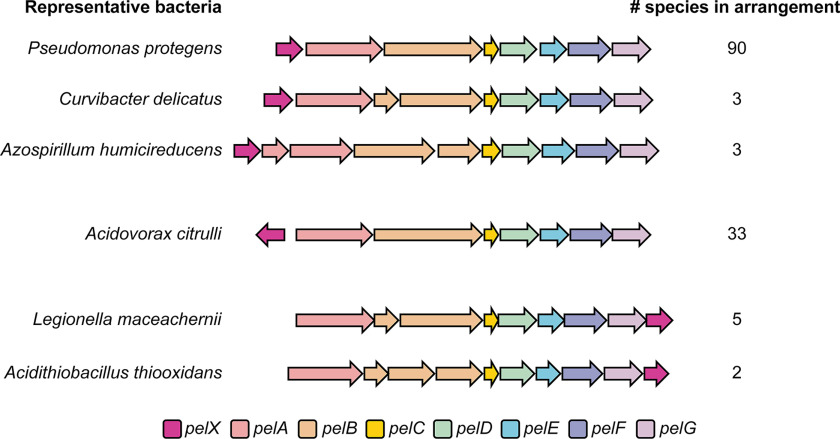
In a previous study, we used the sequence of PelC, a protein required for Pel polysaccharide export, to identify pel biosynthetic loci in a wide range of Proteobacteria (12). In addition to the conserved pelABCDEFG genes, several of these loci contained an additional ORF. We observed several genomic arrangements containing this gene (Fig. 1). In 70% of these genomes, the additional gene is located directly upstream of pelA and may be transcribed together with the pel genes. In 24% of cases, the gene is located upstream of pelA but is divergently transcribed, whereas 5% of the time the gene is encoded downstream of pelG (Fig. 1). Sequence and structure-based analyses of the protein product of this gene, PelX, using BLAST and Phyre2 suggest that it likely encodes an SDR family enzyme (18, 19). In total, we identified 136 pel loci containing a pelX gene (Fig. 1 and Dataset S1).
Figure 1.
A pelX homologous gene is found adjacent to pel biosynthetic gene clusters in three different arrangements. Each gene is shown as an arrow, where each arrow indicates the direction of transcription. The predicted function of each gene is indicated by its color as per the legend (bottom). One representative bacterial species is shown per gene arrangement. The total number of species identified with each arrangement is indicated (right). The full list of species with pelX-containing pel loci can be found in Dataset S1.
To determine whether pelX plays a role in Pel polysaccharide–dependent biofilm formation, we set out to characterize pelX in a species of bacteria for which the regulation of pel gene expression has been studied. In P. protegens, which contains a pelX gene upstream of pelA, the pel gene cluster is under the control of the same Gac/Rsm global regulatory cascade as in P. aeruginosa (17). In addition, two putative recognition sequences for the enhancer-binding protein FleQ are found upstream of pelX (PFL_2971), not pelA, suggesting that in contrast to P. aeruginosa, pelX may be the first gene of the pel operon in this species (20). Given that this operon is likely regulated in a similar manner to the pel locus of P. aeruginosa and that these two species are closely related, we used P. protegens to characterize the role of PelX in biofilm formation.
P. protegens forms pel-dependent biofilms that are enhanced by elevated levels of c-di-GMP
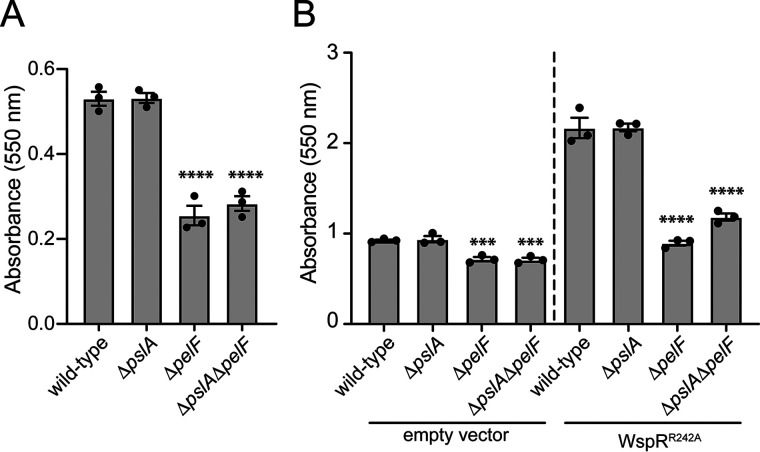
In addition to the pel genes, psl gene expression has been shown to be regulated by the Gac/Rsm pathway in P. protegens and this regulatory cascade is required for P. protegens biofilm formation (17). Interestingly, some strains of P. aeruginosa, including PAO1, use Psl as their predominant biofilm matrix exopolysaccharide whereas others, such as PA14, use Pel (21). Therefore, to determine whether P. protegens biofilms are dependent on Pel and/or Psl, we generated strains lacking pelF or pslA, genes previously shown to be required for Pel- and Psl-dependent biofilm formation, respectively, and examined whether these strains could form biofilms (8, 22). After 5 days of static growth in liquid culture, we found that WT and ΔpslA strains of P. protegens adhered similarly to a polystyrene surface, whereas a strain lacking pelF displayed a marked reduction in surface attachment (Fig. 2A). The level of surface adherence of a ΔpelF ΔpslA double mutant was comparable to that of the ΔpelF strain. Based on these data, we conclude that the Pel polysaccharide is a critical component of P. protegens Pf-5 biofilms.
Figure 2.
P. protegens forms Pel polysaccharide–dependent biofilms that are enhanced by the overexpression of the diguanylate cyclase, WspR. A and B, biofilm biomass determined using the crystal violet assay for (A) WT P. protegens Pf-5 and the corresponding deletion mutants assayed after 5 days of growth and (B) strains expressing pPSV39 (empty vector) or pPSV39::wspRR242A after 24 h of growth. Error bars indicate the S.E. of the mean of two (A) or three (B) independent trials performed in triplicate. Statistical significance was evaluated using one-way analysis of variance with Bonferroni correction. ***, p < 0.001; ****, p < 0.0001.
Previous analysis of the region upstream of P. protegens pelX identified a FleQ consensus binding sequence (20). FleQ is a bis-(3′,5′)-cyclic dimeric GMP (c-di-GMP)–responsive transcription factor that binds to specific sequences upstream of the pel operon in P. aeruginosa, blocking their transcription (23). When the intracellular concentration of c-di-GMP is high, FleQ switches to an activator and up-regulates transcription of the pel genes (24). Based on these observations, we reasoned that expression of the P. protegens pel operon is likely up-regulated in the presence of elevated levels of c-di-GMP (23). To test this hypothesis, we expressed the well-characterized diguanylate cyclase WspR of P. aeruginosa from an IPTG-inducible plasmid in P. protegens (25). Because WspR activity can be inhibited by c-di-GMP binding to an allosteric site of the enzyme, we inactivated this autoinhibitory site by introducing a previously characterized R242A point mutation into the sequence of the protein (WspRR242A) (26). Upon induction of WspRR242A expression, ∼2.3-fold more P. protegens adhered to polystyrene surfaces compared with a vector control strain (Fig. 2B). Taken together, our data suggest that P. protegens Pel-dependent biofilm formation is enhanced in response to elevated intracellular c-di-GMP levels.
pelX is expressed under biofilm-promoting conditions but is functionally redundant with PFL_5533
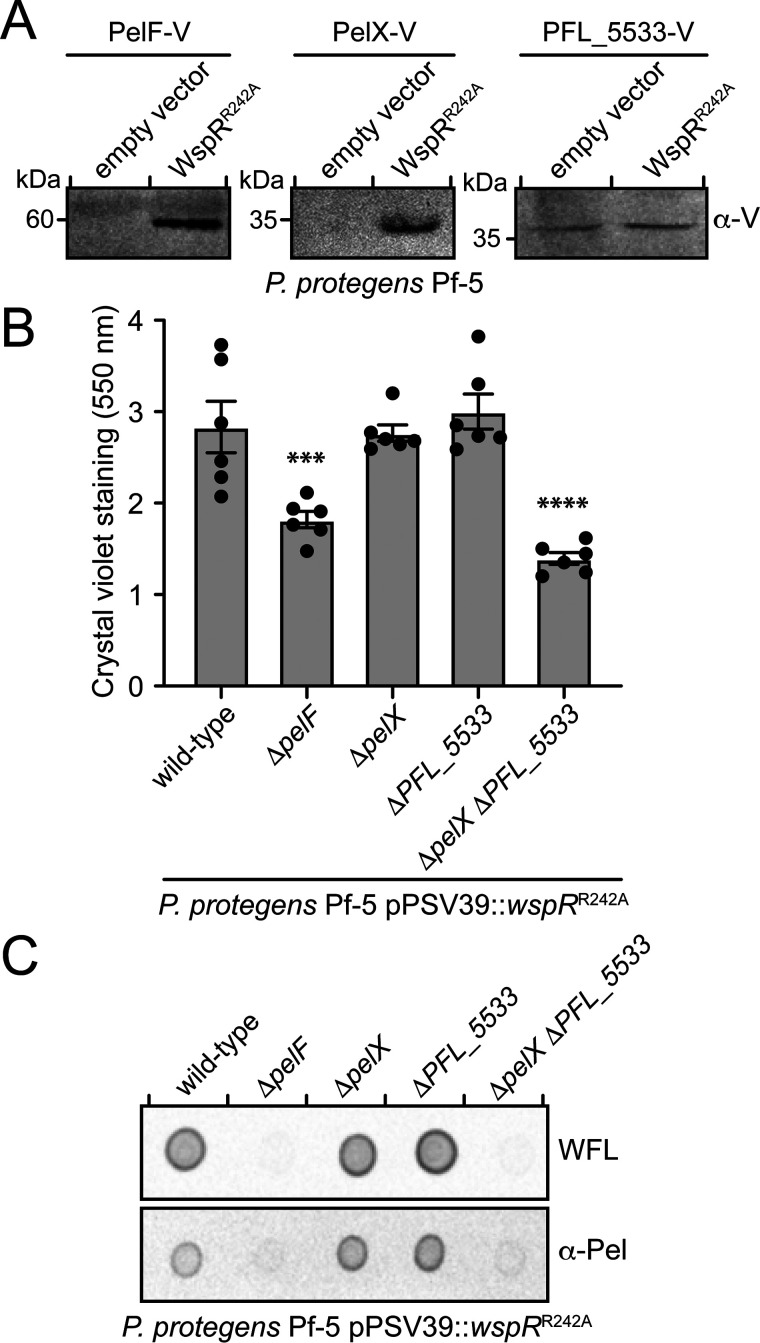
Because Pel-dependent biofilm formation is enhanced in the presence of c-di-GMP and FleQ is predicted to bind upstream of the pelX gene, we reasoned that pelX is most likely expressed in a c-di-GMP–dependent manner along with the rest of the pel genes. To test this, we probed for the expression of PelX by fusing a vesicular stomatitis virus glycoprotein (VSV-G) tag to its C terminus at the native pelX locus on the P. protegens chromosome. To examine expression of the pel operon, a VSV-G tag was similarly added to the C terminus of the putative Pel synthase subunit, PelF (13). Strains expressing either WspRR242A or a vector control were grown under biofilm-conducive conditions and analyzed by Western blotting. In strains lacking WspRR242A, neither PelX nor PelF could be detected; however, in the WspRR242A-expressing strains, both PelX and PelF were detected at their expected molecular mass of 34 and 58 kDa, respectively (Fig. 3A). These data suggest that pelX and pelF expression are positively regulated by c-di-GMP in P. protegens and that PelX is expressed under conditions where the Pel polysaccharide is produced. However, when we deleted pelX, we found that P. protegens biofilm biomass was unaffected, indicating that PelX is not essential for Pel-dependent biofilm formation (Fig. 3B). These findings led us to hypothesize that the P. protegens genome might encode a second SDR enzyme that renders PelX functionally redundant. We queried the PelX amino acid sequence against the P. protegens Pf-5 proteome using BLASTP to identify similar proteins (18). This search identified several proteins from the SDR superfamily (Table 1); however, one protein in particular, PFL_5533, stood out because it shares 68% sequence identity with PelX. To determine whether PFL_5533 is expressed during P. protegens biofilm formation, we fused a C-terminal VSV-G tag to PFL_5533 at its native chromosomal locus and examined its expression in the presence and absence of WspRR242A. We detected similar levels of VSV-G tagged PFL_5533 in both vector control and WspRR242A-expressing strains suggesting that in contrast to pelX, the expression of this gene does not change in response to c-di-GMP (Fig. 3A). The observation that PFL_5533 is expressed during biofilm growth conditions and that it possesses high-sequence homology to pelX led us to probe its potential role in Pel polysaccharide production.
Figure 3.
pelX or PFL_5533 is required for Pel polysaccharide–dependent biofilm formation by P. protegens. A, Western blotting of VSV-G–tagged proteins PelF, PelX, or PFL_5533 in the presence of native c-di-GMP levels (empty vector) or elevated c-di-GMP levels (WspRR242A). B, biofilm biomass as determined using the crystal violet assay after 6 h of static growth. Error bars indicate the S.E. of the mean of two independent trials performed in triplicate. Statistical significance was evaluated using one-way analysis of variance with Bonferroni correction; ***, p < 0.001; ****, p < 0.0001. C, dot blot of culture supernatants of the indicated strains probed using either horseradish peroxidase–conjugated WFL or the Pel antibody (α-Pel) (10, 27).
Table 1.
Short-chain dehydrogenase/reductase enzymes in P. protegens Pf-5
| Pf-5 PFL number | Annotation | Predicted functiona | % Identity to PelX |
|---|---|---|---|
| PFL_2971 | PelX | NAD-dependent epimerase/dehydratase | 100.00 |
| PFL_5533 | NAD-dependent epimerase/dehydratase | 67.97 | |
| PFL_3079 | LspL | UDP-glucuronate 5′-epimerase | 30.54 |
| PFL_5405 | GalE | UDP-Glc 4-epimerase | 29.88 |
| PFL_0305 | rfbB | dTDP-glucose 4,6-dehydratase | 33.60 |
| PFL_5490 | NAD-dependent epimerase/dehydratase | 30.87 | |
| PFL_4822 | 3-β hydroxysteroid dehydrogenase/isomerase | 29.92 | |
| PFL_6133 | NAD-dependent epimerase/dehydratase | 29.37 | |
| PFL_4307 | WbpV | UDP-Glc 4-epimerase WbpV | 29.41 |
| PFL_3045 | arnA | Bifunctional UDP-glucuronic acid decarboxylase/UDP-4-amino-4-deoxy-l-arabinose formyltransferase | 25.98 |
| PFL_4587 | NAD dependent epimerase/dehydratase | 34.09 | |
| PFL_4375 | NAD dependent epimerase/dehydratase | 28.78 | |
| PFL_5491 | Gmd | GDP-mannose 4,6-dehydratase | 28.07 |
| PFL_5106 | WbjB | Trifunctional UDP-d-GlcNAc 4,6-dehydratase/5-epimerase/3-epimerase WbjB | 24.46 |
| PFL_3633 | NAD-dependent epimerase/dehydratase | 22.34 |
a Predicted function based on Pfam analysis (53).
To determine whether PFL_5533 contributes to biofilm formation by P. protegens, we generated a strain lacking this gene and examined biofilm formation in our WspRR242A overexpression background. Similar to our ΔpelX strain, we detected no significant difference in biofilm formation between ΔPFL_5533 and WT strains (Fig. 3B). In contrast, a ΔpelX ΔPFL_5533 double mutant exhibited a defect in biofilm formation comparable to that of a ΔpelF strain, which is incapable of producing Pel (Fig. 3B). To confirm that this reduction in biofilm formation was because of decreased Pel polysaccharide secretion, P. protegens culture supernatants were analyzed using a lectin from Wisteria floribunda (WFL) that specifically recognizes terminal GalNAc moieties and Pel-specific antisera generated using P. aeruginosa Pel polysaccharide (10, 27). Culture supernatants from WT P. protegens displayed a strong signal when analyzed by both of these detection methods, whereas a ΔpelF strain exhibited no signal, indicating that these tools can be used to monitor Pel polysaccharide produced by this bacterium (Fig. 3C) (9). In line with our biofilm data, Pel was detected in culture supernatants from ΔpelX and ΔPFL_5533 strains at levels comparable to WT, whereas no Pel polysaccharide was detected in the ΔpelX ΔPFL_5533 double mutant. Taken together, these data indicate that pelX and PFL_5533 have genetically redundant functions in biofilm formation under our experimental conditions, and that the activity of a predicted SDR family enzyme is essential for Pel polysaccharide biosynthesis and Pel-dependent biofilm formation by P. protegens.
PelX is a UDP-GlcNAC C4-epimerase that preferentially epimerizes N-acetylated UDP-hexoses
To gain further insight into PelX function, we initiated structural and functional studies on recombinant PelX protein. Initial efforts to purify His6-tagged PelX overexpressed in Escherichia coli yielded two species consistent with a monomer and dimer of PelX when analyzed by SDS-PAGE. Addition of reducing agent significantly lowered the abundance of the putative PelX dimer, suggesting that this higher molecular weight species likely arose from the formation of an intermolecular disulfide bond. This intermolecular disulfide bond is likely not biologically relevant given that the bacterial cytoplasm is a reducing environment. As sample heterogeneity can be problematic for both the interpretation of biochemical data and protein crystallization, we generated a PelX variant in which the cysteine residue presumed to be involved in disulfide bond formation (Cys-232) was mutated to serine (PelXC232S). This PelXC232S variant appeared as a monomer on SDS-PAGE and its purification to homogeneity was straightforward. When examined by size-exclusion chromatography, PelXC232S had an apparent molecular mass of 64 kDa compared with its expected monomeric molecular mass of 35 kDa, suggesting that like other characterized SDR enzymes, PelX forms noncovalent, SDS-sensitive dimers in solution (Fig. S1) (28).
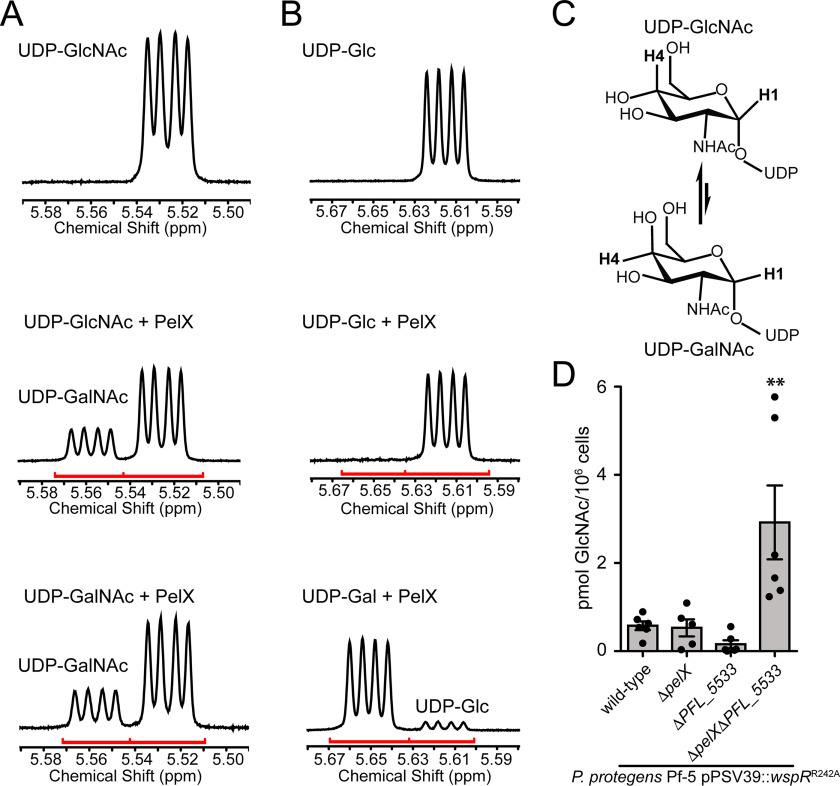
The SDR superfamily of enzymes is known to catalyze numerous chemical reactions, including dehydration, reduction, isomerization, epimerization, dehalogenation, and decarboxylation (14). We hypothesized that PelX likely functions as an epimerase because UDP-GalNAc, the putative precursor for Pel, is typically generated from UDP-GlcNAc by SDR epimerase–catalyzed stereochemical inversion at the C4 position of the hexose ring. Characterized SDR C4-epimerases are classified into three groups based on their substrate preference (29). Group 1 epimerases preferentially interconvert nonacetylated UDP-hexoses, group 2 epimerases are equally able to interconvert nonacetylated and N-acetylated UDP-hexoses, whereas group 3 epimerases preferentially interconvert N-acetylated-UDP-hexoses. Given that the Pel polysaccharide is GalNAc rich, we hypothesized that PelX likely functions as either a group 2 or group 3 epimerase. To examine the potential epimerase activity of PelX, we used 1H NMR to monitor the stereochemistry of UDP-GlcNAc, UDP-GalNAc, UDP-Glc, or UDP-galactose (UDP-Gal) in the presence or absence of purified PelXC232S. Two 1H NMR resonances with characteristic multiplicities in the 5.4–5.7 ppm H-1′′ region allow for the differentiation of UDP-GalNAc/UDP-Gal from UDP-GlcNAc/UDP-Glc, respectively (Fig. 4, A and B). Using these resonances, we found that PelXC232S readily converts UDP-GalNAc to UDP-GlcNAc and vice versa (Fig. 4, A and C). PelXC232S also converted a minor amount of UDP-Gal to UDP-Glc, however, we did not observe significant conversion of UDP-Glc to UDP-Gal (Fig. 4B). Collectively, these data define PelX as a group 3 UDP-hexose C4-epimerase.
Figure 4.
PelX is a Class 3 UDP-GlcNAc C4-epimerase. A and B, 1H NMR focused on the anomeric H-1 region of the spectra (red line) for (A) UDP-GlcNAc incubated without enzyme (top), UDP-GlcNAc with PelX (middle), and UDP-GalNAc with PelX (bottom) and (B) UDP-Glc incubated without enzyme (top), UDP-Glc with PelX (middle), and UDP-Gal with PelX (bottom). C, PelX catalyzes the epimerization of UDP-GlcNAc to UDP-GalNAc by inversion of the hydroxyl group at position C4. D, LC-MS/MS quantification of GlcNAc from cell extracts of the indicated strains. Error bars represent the S.E. of the mean of six independent biological replicates. Statistical significance was evaluated using unpaired t test. *, p < 0.05.
To corroborate our biochemical data, we next performed absolute quantification of cellular GalNAc and GlcNAc levels in our WspRR242A-expressing P. protegens WT, ΔpelX, ΔPFL_5533, and ΔpelX ΔPFL_5533 strains. Although GalNAc levels were below the limit of our detection methods, we found that GlcNAc levels were significantly elevated in the epimerase-deficient background compared with both WT and the individual epimerase mutant strains (Fig. 4D). Taken together with our 1H NMR results, these data suggest that PelX and its homologue PFL_5533 function to generate pools of UDP-GalNAc precursors for polymerization into Pel polysaccharide.
PelX resembles members of the SDR enzyme superfamily
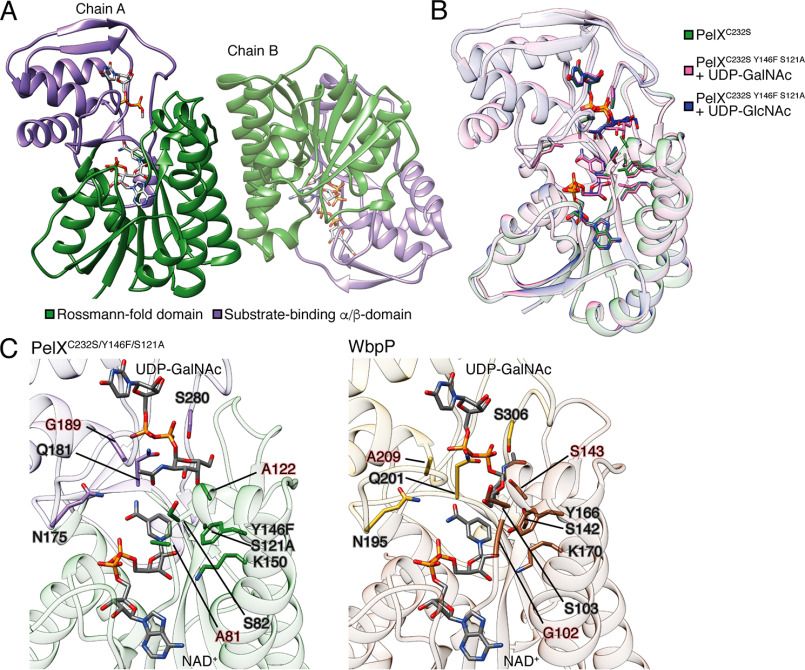
Having established that PelX is a UDP-GlcNAc C4-epimerase, we next sought to determine its structure to obtain further insight into substrate recognition by this enzyme. Despite its straightforward purification and homogenous oligomeric state, we found PelXC232S to be recalcitrant to crystallization. We next attempted to crystallize PelXC232S in complex with its confirmed substrate UDP-GlcNAc. Crystals of PelXC232S incubated with UDP-GlcNAc appeared within 3 days and the structure of the complex was solved to 2.1 Å resolution using molecular replacement with the SDR family member WbpP (PDB ID: 1SB8) as the search model (28). PelX crystallized in space group P21212 and contains a dimer in the asymmetric unit, an arrangement observed for many other structurally characterized SDR family members (Fig. 5A) (30). The dimer interface of PelXC232S is similar to that observed in the WbpP crystal structure where each protomer contributes two α-helices to a four-helix bundle.
Figure 5.
PelX is a member of the short-chain dehydrogenase/reductase superfamily of enzymes. Cartoon representations of PelXC232S. A, PelXC232S is displayed as found in the asymmetric unit with its N-terminal Rossmann fold domain shown in green, and its C-terminal substrate-binding α/β-domain in purple. B, overlay of PelXC232S/Y146F/S121A substrate complexes with the WT PelXC232S colored according to the legend in the figure. C, comparison of the active site of PelXC232S/Y146F/S121A–UDP-GalNAc complex, and WbpP–UDP-GalNAc complex (PDB ID: 1SB8). UDP-GalNAc and NADH (NAD+), and active site residues are shown in stick representation.
The overall structure of PelXC232S shows that it possesses the characteristic domains associated with the SDR family, which includes an N-terminal NAD+-binding Rossmann fold (residues 1–172 and 218–243) and a C-terminal α/β-domain involved in substrate-binding (residues 173–217 and 244–310) (Fig. 5A). PelXC232S contains the GXXGXXG motif required for binding NAD+ that is found in all SDR family members as well as the active site catalytic triad SX24YX3K (31). Although NAD+ was not exogenously supplied in the purification or crystallization buffers, electron density for this cofactor was clearly observed, suggesting it was acquired during PelXC232S overexpression in E. coli. Although the addition of UDP-GlcNAc was essential for the formation of crystals, we were unable to model the GlcNAc moiety of this molecule because of the poor quality of the electron density (Fig. S3). We speculate that the sugar moiety may be disordered because PelXC232S is catalytically active and converting a portion of the UDP-GlcNAc to UDP-GalNAc. Modeling UDP alone rather than UDP-GlcNAc improved the refinement statistics of the overall model and resulted in ligand B-factors comparable to the surrounding protein atoms (Table 2).
Table 2.
Data collection and refinement statistics
| PelXC232S + UDP-GlcNAca | PelXC232S/Y146F/S121A + UDP-GlcNAc | PelXC232S/Y146F/S121A + UDP-GalNAc | |
|---|---|---|---|
| Data Collection | |||
| Wavelength (Å) | 1.075 | 1.075 | 1.075 |
| Space group | P21212 | P21212 | P21212 |
| Cell dimensions | |||
| a, b, c (Å) | 123.0, 75.5, 79.2 | 124.2, 75.5, 79.3 | 123.6, 75.3, 79.3 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.00 – 2.1 (2.18-2.10) | 50.00 - 2.1 (2.18-2.10) | 50.00 -2.1 (2.18-2.10) |
| Total no. of reflections | 676002 | 598870 | 589555 |
| Total no. of unique reflections | 43495 | 43637 | 44099 |
| Rmerge (%)b | 8.0 (58.0) | 12.2 (72.2) | 10.6 (78.7) |
| I/σI | 38.8 (5.3) | 23.6 (3.6) | 26.5 (3.1) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 99.5 (98.6) |
| Redundancy | 11.4 (14.6)c | 7.6 (12.5) | 7.6 (12.2) |
| Refinement | |||
| Rwork/Rfree (%)d | 16.7/19.7 | 15.6/19.5 | 15.6/19.5 |
| Average B-factors (Å2) | |||
| Protein | 41.29 | 31.23 | 35.80 |
| NAD | 32.25 | 21.64 | 25.42 |
| UDP-sugar | 40.82 | 33.13 | 33.19 |
| Water | 41.52 | 37.69 | 40.39 |
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.006 | 0.006 | 0.006 |
| Bond angles (°) | 0.881 | 0.863 | 0.897 |
| Ramachandran plot (%)e | |||
| Total favored | 97.87 | 97.55 | 97.71 |
| Total allowed | 2.13 | 2.45 | 2.13 |
| Coordinate error (Å)f | 0.21 | 0.2 | 0.22 |
| PDB code | 6WJB | 6WJ9 | 6WJA |
a Although this structure was crystallized with UDP-GlcNAc, density for the sugar was not visible and therefore only UDP was modeled and refined.
b Rmerge = Σ Σ |I(k) − |/Σ I(k), where I(k) and represent the diffraction intensity values of the individual measurements and the corresponding mean values. The summation is over all unique measurements.
c Values in parentheses correspond to the highest resolution shell.
d Rwork = Σ ‖Fobs| − k|Fcalc‖/|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is the sum extended over a subset of reflections (5%) excluded from all stages of the refinement.
e As calculated using MOLPROBITY (54).
f Maximum-Likelihood Based Coordinate Error, as determined by PHENIX (52).
Previous studies on a catalytically inactive variant of the UDP-Gal 4-epimerase GalE from E. coli allowed for the co-crystallization and modeling of UDP-Glc and UDP-Gal in the active site of this enzyme (32). In their study, these authors targeted the serine and tyrosine residues of the consensus Sx24Yx3K active site motif. Guided by this approach, we generated a variant of PelXC232S with S121A and Y146F mutations and confirmed that this variant is catalytically inactive (Fig. S2). PelXC232S/S121A/Y146F crystallized readily with either UDP-GlcNAc or UDP-GalNAc, and both structures were solved to a resolution of 2.1 Å using molecular replacement (Table 2). The final models of PelXC232S/S121A/Y146F in complex with UDP-GlcNAc or UDP-GalNAc were both refined to an Rwork/Rfree of 15.6%/19.5% (Table 2). In these structures, the electron density for the sugar moieties was well-defined compared with the PelXC232S–UDP-GlcNAc co-crystal structure and allowed for the unambiguous modeling of the expected sugar-nucleotides (Fig. S3). Given that both structures showed improved ligand density for their respective substrates, these structures substantiate our biochemical data showing that UDP-GlcNAc and UDP-GalNAc are substrates for PelX. Examination of the active site of our PelXC232S/S121A/Y146F-substrate complexes did not show any significant differences in the positions of active site residues, suggesting that both sugar-nucleotides are recognized by the enzyme in a similar manner (Fig. 5B). We next compared our substrate-bound PelXC232S/S121A/Y146F structures to the UDP-GalNAc–bound structure of the aforementioned UDP-hexose C4-epimerase WbpP from P. aeruginosa. WbpP shares 32% sequence identity to PelX and also catalyzes the epimerization of UDP-GlcNAc to UDP-GalNAc (28). The overall structure of WbpP is highly similar to PelXC232S/S121A/Y146F (PDB code 1SB8, root mean square deviation 1.9 Å over 306 Cα) except that WbpP possesses an additional N-terminal α-helix not found in PelX. The active site residues identified as being important for sugar-nucleotide interaction in WbpP are invariant in PelX (Fig. 5C) with the exception of Ala-81, Ala-122 and Gly-189 in PelX, which correspond to residues Gly-102, Ser-143 and Ala-209 in WbpP, respectively (28). These differences are not predicted to impair specificity toward the UDP-GlcNAc/GalNAc substrate. Rather, Demendi et al. (33) found that bulkier residues (G102K, A209N), and mutation of S143A actually displayed enhanced specificity toward acetylated substrates. However, while the positions of the PelXC232S/S121A/Y146F and WbpP active site residues and NAD+ cofactor are highly similar, comparison of the bound UDP-GalNAc substrate between the two structures reveals distinct differences in the conformations of the GalNAc moiety (Fig. 5C). We suspect that this difference in conformation may be a result of the co-crystallization of UDP-GalNAc with WT WbpP, whereas to observe electron density for the GalNAc moiety of UDP-GalNAc in complex with PelX we had to mutate two active site residues, S121A and Y146F. The residues equivalent to Ser-121 and Tyr-146 in WbpP make contact with the C4 hydroxyl group of GalNAc and thus are likely involved in substrate orientation. These observations suggest that the conformation of UDP-GalNAc in our mutant PelX co-crystal structure may not represent a state adopted during catalysis, but demonstrate a high degree of conformational freedom of the sugar moiety within the relatively large substrate-binding pocket. The GlcNAc moiety of UDP-GlcNAc in our PelXC232S/S121A/Y146F–UDP-GlcNAc co-crystal structure was also found in a similar orientation as in our UDP-GalNAc–containing structure. Taking these considerations into account and given that WbpP and PelX share a high degree of sequence similarity and interconvert identical substrates with similar preference, we speculate that the epimerization of N-acetylated UDP-hexoses by PelX most likely occurs via a similar catalytic mechanism as proposed for WbpP (28, 33). In summation, our structural data support our biochemical studies showing that PelX belongs to the group 3 family of UDP-N-acetylated hexose C4-epimerases.
Discussion
In this study, we report the characterization of the Pel polysaccharide precursor-generating enzyme PelX. Using P. protegens Pf-5 as a model bacterium, we found that pelX is required for Pel polysaccharide–dependent biofilm formation in a strain that also lacks the pelX paralogue PFL_5533. Guided by our 1H NMR analyses and multiple crystal structures, we have shown that PelX functions as a UDP-GlcNAc C4-epimerase and that it preferentially interconverts UDP-GlcNAc/UDP-GalNAc over UDP-Glc/UDP-Gal, defining it as a group 3 UDP-N-acetylhexose C4-epimerase. Based on these observations and the data presented herein we propose naming PFL_5533 polysaccharide UDP-GlcNAc epimerase (pgnE).
Functional redundancy of sugar-nucleotide synthesizing enzymes in biofilm producing bacteria is not unprecedented. For example, in P. aeruginosa PAO1, PslB and WbpW both catalyze the synthesis of GDP-mannose, a precursor molecule required for Psl polysaccharide and A-band lipopolysaccharide (LPS). Like PelX and PgnE, PslB and WbpW have been shown to be genetically redundant as a defect in Psl polysaccharide, or A-band LPS is only observed when both pslB and wbpW are deleted (22). Although P. aeruginosa PAO1 has another paralogue of PslB and WbpW, AlgA, the algD promoter responsible for transcription of the algA gene is not significantly activated in nonmucoid strains such as PAO1 (34). Psl biosynthesis, like Pel, is also regulated by c-di-GMP through FleQ (23), whereas being an integral component of the P. aeruginosa outer membrane, the genes responsible for A-band LPS synthesis are constitutively expressed (35). Although at present what additional glycans PgnE may be involved in producing is unknown, it is clear that the existence of paralogous sugar-nucleotide synthesizing enzymes may be a means of keeping up with metabolic demand during the synthesis of multiple cell-surface polysaccharides.
We previously reported the isolation of Pel polysaccharide from P. aeruginosa PAO1 and carbohydrate composition analyses showed that it is rich in GalNAc (9). Therefore, the co-regulation of a UDP-GlcNAc C4-epimerase with the pel genes likely ensures that adequate quantities of UDP-GalNAc are available for Pel biosynthesis when a biofilm mode of growth is favored. In contrast to P. protegens Pf-5, P. aeruginosa PAO1 does not contain a pelX gene in its Pel biosynthetic gene cluster, yet this bacterium is also capable of producing Pel polysaccharide (36). In the PAO1 genome, the poorly characterized PA4068 gene is found in the same genomic context as pgnE whereby both genes are part of a two-gene operon, with the second gene predicted to encode a dTDP-4-dehydrorhamnose reductase (PA4069/PFL_5534) (37). In addition, the protein encoded by PA4068 shares 76% identity with PgnE, suggesting that this gene may function analogously to pgnE and by extension pelX. A ΔPA4068 mutant was found to display a surface attachment defect during secretin-induced stress, suggesting a role for this gene in surface glycan production (37). However, it has been established that Psl is the primary polysaccharide required for P. aeruginosa PAO1 biofilm formation even though this strain is genetically capable of synthesizing Pel (36). Consequently, studies characterizing Pel polysaccharide production by PAO1 have relied on an engineered strain that lacks the ability to produce Psl and expresses the pel genes from an arabinose-inducible promoter. It may be that only low levels of UDP-GalNAc are required to sustain Pel polysaccharide production by WT PAO1 and thus a second UDP-GlcNAc C4-epimerase that is dedicated to Pel production is not required. In contrast, Pel polysaccharide appears to be a major biofilm matrix constituent in P. protegens Pf-5 and thus the higher levels of Pel production in this organism may necessitate the need for increased synthesis of UDP-GalNAc precursors.
The epimerization of UDP-Gal to UDP-Glc by PelX occurs much less efficiently than its N-acetylated counterpart. Creuzenet and colleagues noted a similar trend for WbpP, a UDP-GlcNAc C4-epimerase involved in P. aeruginosa PAK O-antigen biosynthesis, and hypothesized that the poor efficiency displayed by this enzyme toward nonacetylated substrates means that this reaction is unlikely to occur in vivo (38). The equilibrium of the PelX-catalyzed epimerization between UDP-GalNAc and UDP-GlcNAc in vitro is skewed toward the more thermodynamically stable UDP-GlcNAc epimer. A similar balance for this equilibrium has been documented for other epimerases (38, 39). We speculate that the continuous polymerization of UDP-GalNAc by the putative Pel polysaccharide polymerase, PelF, would keep the cellular concentration of UDP-GalNAc low and thus drive the equilibrium toward its production.
In conclusion, this work demonstrates the involvement of a Pel polysaccharide precursor generating enzyme required for biofilm formation in P. protegens. Our data linking the production of UDP-GalNAc to Pel polysaccharide production lend genetic and biochemical support to the chemical analyses that showed Pel is a GalNAc-rich carbohydrate polymer (9). Furthermore, the identification of a new Pel polysaccharide–dependent biofilm forming bacterium provides an additional model system that can be used for the characterization of this understudied polysaccharide secretion apparatus.
Experimental Procedures
Bacterial strains, microbiological media, and physiological buffers
All bacterial strains and plasmids used in this study are listed in Table S1. Jensen's medium contained per liter of MilliQ water: 5 g NaCl, 2.51 g K2HPO4, 13.46 g glutamic acid, 2.81 g l-valine, 1.32 g l-phenylalanine, 0.33 g/liter MgSO4•7H2O, 21 mg CaCl2•2H2O, 1.1 mg FeSO4•7H2O, 2.4 mg ZnSO4•7H2O, and 1.25% d-glucose. Semi-solid agar medium in Petri dishes was prepared by adding 1.0% noble agar to Jensen's medium. A 10× solution of PBS was purchased from Amresco and diluted, as required, in sterile MilliQ water. King's B medium contained per liter of MilliQ water: 10 g proteose peptone no. 2 (DIFCO), 1.5 g anhydrous K2HPO4, 15 g glycerol, and 5 ml MgSO4. Lysogeny broth (LB) contained per liter of MilliQ water: 10 g tryptone, 10 g NaCl, and 5 g yeast extract. E. coli strains were grown with shaking at 37°C. P. protegens strains were grown at 30°C. The following concentration of antibiotics were used: gentamicin 15 μg ml−1 (E. coli); gentamicin 30 μg ml−1 (P. protegens); kanamycin, 25 μg ml−1. Plasmids were maintained in DH5a(λpir).
Bioinformatic identification of PelX among pel gene clusters in sequenced bacterial genomes
We previously constructed a database of genomes containing pel gene clusters using the Geneious platform (12, 13, 40). Briefly, identification of pel gene clusters was made via BLASTP (18) searching of the National Center for Biotechnology Information, Pseudomonas (41), and Burkholderia (42) databases (as of May 6, 2018) using P. aeruginosa PAO1 PelC (NP_251752.1) as the query sequence. Annotated genomes encoding PelC orthologues were downloaded from the databases and manually binned according to synteny of the pel operon. Conserved domains encoded by open reading frames (ORFs) linked to pel loci were queried by searching the Conserved Domain Database (43). Visualizations of pel gene clusters were drawn to scale using Geneious Prime 2020 and Adobe Illustrator.
Sequence analysis of PelX and PgnE orthologues
According to the Pseudomonas Genome database, PFL_2971 and PFL_5533 belong to the Pseudomonas orthologue groups (POGs) POG020331 and POG001617, respectively. Prior to this study, the POGs were unnamed; therefore, based on our observations, we have named these POGs as pelX and pgnE. PelX primary amino acid sequences were aligned using MUSCLE (44) to identify highly conserved amino acid residues. Additionally, the P. protegens PelX sequence was submitted to Phyre2 to determine the predicted fold of the protein (19). The PelX and PgnE protein sequences from P. protegens Pf-5 were obtained from the Pseudomonas Genome Database (41). Comparison of the PelX structure to previously determined structures was performed using the DALI pairwise comparison server (45).
Construction of P. protegens chromosomal mutations
In-frame, unmarked pslA (PFL_4208), pelF (PFL_2977), pelX (PFL_2971), and PFL_5533 gene deletions in P. protegens Pf-5 were constructed using an established allelic replacement strategy (46). Flanking upstream and downstream regions of the ORFs were amplified and joined by splicing-by-overlap extension PCR (primers are listed in Table S1). The pslA, pelF, and pelX, alleles were generated using forward upstream and downstream reverse primers tailed with EcoRI and XbaI, restrictions sites, respectively (Table S1). The PFL_5533 allele was generated using forward upstream and downstream reverse primers tailed with EcoRI and HindIII restriction sites, respectively (Table S1). This PCR product was gel purified, digested, and ligated into pEXG2, and the resulting constructs, pLSM33, pLSM34, and pLSM35, pLSM36 were identified and sequenced as described above.
The VSV-G–tagged pelF, pelX, and PFL_5533 constructs were generated by amplifying flanking upstream and downstream regions surrounding the stop codon of the ORFs of each gene. The reverse upstream and forward downstream primers (Table S1) were tailed with complementary sequences encoding the VSV-G peptide immediately before the stop codon. Amplified upstream and downstream fragments were joined by splicing-by-overlap extension PCR using forward upstream and reverse downstream primers tailed with EcoRI and HindIII restriction sites, respectively (Table S1). These PCR products were gel purified, digested, and ligated into pEXG2, as described above. Clones with positive inserts were verified by Sanger sequencing to generate pLSM37, pLSM38, and pLSM39.
The aforementioned pEXG2-based plasmids were introduced into P. protegens Pf-5 via biparental mating with donor strain E. coli SM10 (47). Merodiploids were selected on LB containing 60 μg ml−1 gentamicin (Gen) and 25 μg ml−1 Irgasan. SacB-mediated counter-selection was carried out by selecting for double crossover mutations on no-salt LB agar containing 5% (w/v) sucrose. Unmarked gene deletions were identified by PCR with primers targeting the outside, flanking regions of pslA, pelF, pelX, and PFL_5533 (Tables S1 and S2). These PCR products were Sanger sequenced using the same primers to confirm the correct deletion.
Generation of WspR overexpression strains
The wspR nucleotide sequence from P. aeruginosa PAO1 was obtained from the Pseudomonas Genome Database and used to design primers specific to full-length wspR (Table S1). The forward primer encodes an EcoRI restriction site and a ribosomal-binding site, whereas the reverse primer encodes a HindIII restriction site. The amplified PCR products were digested with EcoRI and HindIII restriction endonucleases and subsequently cloned into the pPSV39 vector (Table S1). Confirmation of the correct nucleotide sequence of wspR was achieved through DNA sequencing (The Center for Applied Genomics, The Hospital for Sick Children). Arg-242 was mutated to an alanine to prevent allosteric inhibition of WspR using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies), as described previously. The resulting expression vector (pLSM-wspRR242A) encodes residues 1–347 of WspR. Introduction of the pPSV39 empty vector or pSLM-wspRR242A into P. protegens was carried out by electroporation. Positive clones were selected for LB agar containing 30 μg ml−1 Gen.
Crystal violet assay
Overnight cultures grown in King's B media (KBM), were diluted to a final OD of 0.005 in 1 ml of KBM in a 24-well VDX plate (Hampton Research) and left undisturbed at 30°C for 120 h. Nonattached cells were removed and the wells were washed thoroughly with water and stained with 1.5 ml 0.1% (w/v) crystal violet. After 10 min, the wells were washed again and the stain solubilized using 2 ml of 95% (v/v) ethanol for 10 min. 200 μl was transferred to a fresh 96-well polypropylene plate (Nunc) and the absorbance measured at 550 nm. For strains containing empty pPSV39 or pLSM-wspRR242A, the above protocol was modified slightly. As c-di-GMP significantly up-regulated biofilm formation, crystal violet staining for these strains was performed as described previously using 96-well polypropylene plates that were incubated statically for 6 or 24 h at 30°C. All strains were grown in KBM containing 30 μg ml−1 Gen and 30 μm IPTG.
Dot blots
Pel antisera were obtained as described in Colvin et al. (10) from P. aeruginosa PA14 PBADpel. The adsorption reaction was conducted as described by Jennings et al. (9). Culture supernatants containing secreted Pel were harvested by centrifugation (16,000 × g for 2 min) from 1 ml aliquots of P. protegens grown overnight at 30°C in LB containing 30 μg ml−1 Gen and 30 μm IPTG, and treated with proteinase K (final concentration, 0.5 mg ml−1) for 60 min at 60°C, followed by 30 min at 80°C to inactivate proteinase K.
Pel immunoblots were performed as described by Colvin et al. (10) and Jennings et al. (9). 5 μl of secreted Pel, prepared as described above, was pipetted onto a nitrocellulose membrane and left to air dry for 10 min. The membrane was blocked with 5% (w/v) skim milk in TBS (10 mm Tris-HCl, pH 7.5, 150 mm NaCl) containing 0.1% (v/v) Tween 20 (TBS-T) for 1 h at room temperature and probed with adsorbed α-Pel at a 1:60 dilution in 1% (w/v) skim milk in TBS-T overnight at 4°C with shaking. Blots were washed three times for 5 min each with TBS-T, probed with goat α-rabbit HRP-conjugated secondary antibody (Bio-Rad) at a 1:2000 dilution in TBS-T for 45 min at room temperature with shaking, and washed again. All immunoblots were developed using SuperSignal West Pico (Thermo Scientific) following the manufacturer's recommendations.
For WFL-HRP immunoblots, 5 μl of secreted Pel, prepared as described above, was pipetted onto a nitrocellulose membrane and left to air dry for 10 min. The membrane was blocked with 5% (w/v) BSA in TBS-T for 1 h at room temperature and probed with 10 μg/ml of WFL-HRP (EY Laboratories) in 2% (w/v) BSA in TBS-T with 0.2 g/liter CaCl2 overnight at room temperature with shaking. Membranes were washed twice for 5 min and once for 10 min with TBS-T, then developed as described above.
Western blotting sample preparation and analysis
For analysis of protein levels from WspRR242A overexpressing strains containing VSV-G–tagged PelF, PelX, or PFL_5533, 5 ml of LB media containing 30 μm IPTG and 30 μg ml−1 Gen was inoculated with the appropriate strain and allowed to grow overnight at 30°C with shaking. Culture density was normalized to an A600 = 1 and 1 ml of cells was centrifuged at 5000 × g for 5 min to pellet cells. The cell pellet was resuspended in 100 μl of 2× Laemmli buffer, boiled for 10 min at 95°C, and analyzed by SDS-PAGE followed by Western blotting. For Western blot analysis, a 0.2-μm PVDF membrane was wetted in methanol and soaked for 5 min in Western transfer buffer (25 mm Tris-HCl, 150 mm glycine, 20% (v/v) methanol) along with the SDS-PAGE gel to be analyzed. Protein was transferred from the SDS-PAGE gel to the PVDF membrane by wet transfer (25 mV, 2 h). The membrane was briefly washed in TBS-T before blocking in 5% (w/v) skim milk powder in TBS-T for 2 h at room temperature with gentle agitation. The membrane was briefly washed again in TBS-T before incubation overnight with α–VSV-G antibody in TBS-T with 1% (w/v) skim milk powder at 4°C. The next day, the membrane was washed four times in TBS-T for 5 min each before incubation for 1 h with secondary antibody (1:2000 dilution of Bio-Rad Affinity-purified mouse α-rabbit IgG conjugated to alkaline phosphatase) in TBS-T with 1% (w/v) skim milk powder. The membrane was then washed three times with TBS-T for 5 min each before development with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium chloride (BioShop ready-to-use BCIP/NBT solution). Developed blots were imaged using a Bio-Rad ChemiDoc imaging system.
Cloning and mutagenesis
The pelX nucleotide sequence from P. protegens Pf-5 (PFL_2971) was obtained from the Pseudomonas Genome Database (41) and used to design primers specific to full-length pelX (Table S1). The amplified PCR products were digested with NdeI and XhoI restriction endonucleases and subsequently cloned into the pET28a vector (Novagen). Confirmation of the correct nucleotide sequence of pelX was achieved through DNA sequencing (ACGT DNA Technologies Corporation). The resulting expression vector (pLSM-PelX) encodes residues 1–309 of PelX fused to a cleavable N-terminal His6 tag (His6-PelX) for purification purposes (Table S2). To prevent aggregation of PelX in solution, a nonconserved cysteine (Cys-232) was mutated to a serine with the aid of the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) and confirmed with DNA sequencing (ACGT DNA Technologies Corporation). The PelXC232S active site mutant (S121A/Y146F) was generated analogously.
Expression and purification of PelX
The expression of PelXC232S was achieved through the transformation of the PelXC232S expression vector into E. coli BL21 (DE3) competent cells, which were then grown in 2 liters LB containing 50 μg ml−1 kanamycin at 37°C. The cells were grown to an A600 of 0.6 whereupon IPTG was added to a final concentration of 1.0 mm to induce expression. The induced cells were incubated for 20 h at 25°C prior to being harvested via centrifugation at 6260 × g for 20 min at 4°C. The resulting cell pellet was stored at −20 °C until required.
The cell pellet from 2 liters of bacterial culture was thawed and resuspended in 80 ml of Buffer A (50 mm Tris-HCl pH 8.0, 300 mm NaCl, 5% (v/v) glycerol, and 1 mm tris(2-carboxyethyl)phosphine (TCEP)) containing 1 SIGMAFAST Protease Inhibitor EDTA-free mixture tablet (Sigma). Because of the presence of two remaining cysteines in PelXC232S, TCEP was included to prevent intermolecular crosslinking of the protein. These cysteines are not predicted to be involved in disulfide bond formation given their poor conservation and the cytoplasmic localization of PelXC232S. The resuspension was then lysed by homogenization using an Emulsiflex-C3 (Avestin, Inc.) at a pressure between 15,000–20,000 psi, until the resuspension appeared translucent. Insoluble cell lysate was removed by centrifugation for 25 min at 25,000 × g at 4°C. The supernatant was loaded onto a 5 ml Ni2+–nitrilotriacetic acid column pre-equilibrated with Buffer A containing 5 mm imidazole to reduce background binding. To remove contaminating proteins, the column was washed with 5 column volumes of Buffer A containing 20 mm imidazole. Bound protein was eluted from the column with 3 column volumes of Buffer A containing 250 mm imidazole. SDS-PAGE analysis revealed that the resulting His6-PelXC232S was ∼99% pure and appeared at its expected molecular mass of 35 kDa. Fractions containing PelXC232S were pooled and concentrated to a volume of 2 ml by centrifugation at 2200 × g at a temperature of 4°C using an Amicon Ultra Centrifugal filter device (Millipore) with a 10-kDa molecular weight cut-off. PelXC232S was purified and buffer exchanged into Buffer B (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5% (v/v) glycerol, 1 mm TCEP) by size-exclusion chromatography using a HiLoad 16/60 Superdex 200 gel-filtration column (GE Healthcare). PelXC232S eluted as a single Gaussian-shaped peak, and all PelXC232S containing fractions were pooled and concentrated by centrifugation, as above, to 8 mg ml−1 and stored at 4°C. PelXC232S/Y146F/S121A was purified similarly.
Determination of the PelX oligomerization state by gel filtration analysis
Oligomerization of PelXC232S was determined using a Superdex 200 10/300 GL column (GE Life Sciences). The column was equilibrated in Buffer B. Molecular weight standards (Sigma, 12–200 kDa) were applied to the column as directed. PelXC232S was applied to the column at 7.5 mg ml−1 (100 μl) and protein elution was monitored at 280 nm.
NMR activity assay
The following method has been adapted from Wyszynski et al. (39). Enzymatic reactions were performed in 30-mm sodium phosphate, pH 8.0, with 50 μg of freshly purified PelXC232S and 10 mm UDP-GlcNAc, UDP-Glc, UDP-Gal, or 5 mm UDP-GalNAc in a total reaction volume of 220 μl. After incubation at 37°C for 1 h, the mixture was flash frozen and lyophilized. The resulting material was dissolved in 220 μl of D2O and analyzed by 1H NMR. As control experiments, the same procedures were applied to samples lacking PelX or UDP-GlcNAc. Data were collected on a Varian 600 MHz NMR spectrometer.
Intracellular metabolite extraction
P. protegens Pf-5 WT, ΔpelX, ΔPFL_5533, and ΔpelX ΔPFL_5533 strains that had been transformed with a plasmid expressing WspRR242A (pLSM21) were streaked out twice in succession on Jensen's agar containing 30 μg/ml gentamicin, and these first and second subcultures were grown for 48 h at 30°C. For each biological replicate, cells from the second subcultures were collected using a polyester swab and suspended in 30 ml of Jensen's medium to match an optical density at 600 nm (A600) of 0.6. Subsequently, two 10 ml aliquots of this standardized culture were each passed through a syringe filter (0.45 μm, PVDF, Millipore) to collect the bacteria. These filters were placed face up on Jensen's agar using flame-sterilized tweezers and were then incubated at 30°C for 3 h.
Following this incubation, the first filter was placed in 2 ml of sterile PBS containing 1 mm purified recombinant PelA, and this filter was incubated for 30 min at room temperature to break up aggregates (11). An established microtiter dilution method for viable cell counting was used to determine the number of bacteria on the filter (48). The second filter was put into a Petri dish (60 × 15 mm) containing 2 ml of cold 80% (v/v) LC-MS grade methanol, which was incubated for 15 min. Afterward, 1 ml of 80% LC-MS grade methanol was used to wash the filter, and then the 3 ml of the methanol extract were transferred to a 5-ml microcentrifuge tube. These tubes were placed in a centrifuge at 7000 × g for 30 min at 4°C, and then 2 ml of the supernatant were transferred to a 2-ml microcentrifuge tube. The methanol was evaporated using a speed vac, and then the dried cell extracts were suspended in 200 μl of 50% (v/v) LC-MS grade methanol. Cell extracts were then stored at −80°C until LC-MS analysis. MS measurements of GalNAc were normalized to viable cell counts.
LC-MS
Mass spectral data collection was done on a Thermo Scientific Q ExactiveTM Hybrid Quadrupole-OrbitrapTM Mass Spectrometer in negative ion full scan mode (70–1000 m/z) at 140,000 resolution, with an automatic gain control target of 1e6, and a maximum injection time of 200 ms. A Thermo Scientific Ion Max-S API source outfitted with a HESI-II probe was used to couple the mass spectrometer to a Thermo Scientific Vanquish Flex UHPLC platform. Heated electrospray source parameters for negative mode were as follows: spray voltage −2500 V, sheath gas 25 (arbitrary units), auxiliary gas 10 (arbitrary units), sweep gas 2 (arbitrary units), capillary temperature 275°C, auxiliary gas temperature 325°C. A binary solvent mixture of 20 mm ammonium formate at pH 3.0 in LC-MS grade water (solvent A) and 0.1% (v/v) formic acid in LC-MS grade acetonitrile (solvent B) were used in conjunction with a SyncronisTM HILIC LC column (Thermo Fisher Scientific 97502-102130) to achieve chromatographic separation of chemical compounds. For these runs the following gradient was used at a flow rate of 600 μl min−1: 0–2 min, 100% B; 2–15 min, 100–80% B; 15–16 min, 80–5% B; 16–17.5 min, 5% B; 17.5–18 min, 5–100% B; 18–21 min, 100% B. For all runs the sample injection volume was 2 μl. Raw data acquisition was carried out using Thermo Xcalibur 4.0.27.19 software. Data analysis was carried out using MAVEN software (49). Compound identification was achieved through matching of high-resolution accurate mass and retention time characteristics to those of authentic standards. Secondary compound confirmation was performed by matching of fragmentation profiles obtained through parallel reaction monitoring.
Crystallization and structure determination
Commercial sparse matrix crystal screens from Microlytic (MCSG1-4) were prepared at room temperature (22°C) with PelXC232S at a concentration of 8 mg ml−1 (0.23 mm). UDP-GlcNAc was added exogenously to a concentration of 2 mm. Trials were set up in 48-well VDX plates (Hampton Research) by hand with 3 μl drops at a ratio of 1:1 protein to crystallization solution over a reservoir containing 200 μl of the crystallization solution. Crystal trays were stored at 22°C. The best crystals were obtained from condition 32 (0.2 m ammonium sulfate, 0.1 m sodium citrate, pH 5.6, 25% (w/v) PEG 4000) from MCSG-1 (Microlytic). This condition yielded stacked flat square plate crystals that took ∼5 days to grow to maximum dimensions of 300 μm × 300 μm × 50 μm. PelX was unable to form crystals in the absence of UDP-GlcNAc.
Crystals of PelXC232S were cryo-protected in well solution supplemented with 20% (v/v) ethylene glycol by briefly soaking the crystal in a separate drop. Crystals were soaked for 2–3 s prior to vitrification in liquid nitrogen and subsequently stored until X-ray diffraction data were collected on beamline X29A at the National Synchrotron Light Source at Brookhaven National Laboratory. A total of 360 images of 1° Δφ oscillation were collected on an ADSC Q315 CCD detector with a 250 mm crystal-to-detector distance and an exposure time of 0.4 s per image. The data were processed using DENZO and integrated intensities were scaled using SCALEPACK from the HKL-2000 program package (50). The data collection statistics are summarized in Table 2. The structure was solved by molecular replacement using WbpP as a model with PHENIX AutoMR wizard. The resulting map was of good quality and allowed manual model building using COOT (51, 52). The model was then refined using PHENIX.REFINE (52) to a final Rwork/Rfree of 16.7 and 19.7%, respectively.
PelXC232S/S121A/Y146F in complex with UDP-GalNAc or UDP-GlcNAc was crystallized under the same conditions as the WT protein, and data collection and refinement were performed as described above. The corresponding statistics can be found in Table 2.
Data availability
All the data described are located within the manuscript and the supplemental information. The coordinates and structure factors for PelXC232S in complex with NAD+ and UDP, PelXC232S/S121A/Y146F UDP-GlcNAc, and PelXC232S/S121A/Y146F UDP-GalNAc have been deposited in the PDB, ID codes 6WJB, 6WJA, and 6WJ9, respectively.
Supplementary Material
Acknowledgments
Crystallization utilized the Structural and Biophysical Core Facility at The Hospital for Children supported in part by the Canadian Foundation for Innovation. Beam line X29 at the National Synchrotron Light Source is supported by the U. S. Department of Energy and the National Institutes of Health National Center for Research Resources. Metabolomics data were acquired by R.A.G. at the Calgary Metabolomics Research Facility (CMRF), which is supported by the International Microbiome Centre.
This article contains supporting information.
Author contributions—L. S. M., M. R. P., and P. L. H. conceptualization; L. S. M., G. B. W., J. J. H., I. A. L., and P. L. H. formal analysis; L. S. M., G. B. W., H. R., R. P., R. J. W, T. E. R., E. R., A. O., R. A. G., and I. A. L., investigation; L. S. M., G. B. W., and P. L. H. writing-original draft; L. S. M., G. B. W., J. C. W., J. J. H., and P. L. H. writing-review and editing; M. N., H. R., M. R. P., and I. A. L. resources; J. J. H, I. A. L., and P. L. H. supervision; P. L. H. funding acquisition; P. L. H. project administration.
Funding and additional information—This work was supported in part by Canadian Institutes of Health Research (CIHR) Grants MOP 43998 and FDN154327 (to P. L. H.), National Institutes of Health Grant 2R01AI077628 (to M. R. P.), and Natural Sciences and Engineering Research Council (NSERC) Grant 435631-2013 (to J. J. H.). P. L. H. and J. J. H. are recipients of Canada Research Chairs. This work has also been supported by Canada Graduate Scholarships from NSERC (to L. S. M., G. B. W., and J. C. W.), the Ontario Graduate Scholarship Program (to L. S. M. and J. C. W.), The Hospital for Sick Children Foundation Student Scholarship Program (to L. S. M.), a Cystic Fibrosis Canada Graduate Scholarship (to G. B.W.), and an Alberta Innovates Translational Health Chair (to I. A. L.). This work was also supported by the Canada Foundation for Innovation CFI-JELF 34986 (to I. A. L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- SDR
- short-chain dehydrogenase/reductase
- Pel
- pellicle
- c-di-GMP
- cyclic dimeric GMP
- IPTG
- isopropyl-β-d-1-thiogalactopyranoside
- VSV-G
- vesicular stomatitis virus glycoprotein
- WFL
- Wisteria floribunda
- UDP-Gal
- UDP-galactose
- LPS
- lipopolysaccharide
- LB
- lysogeny broth
- Gen
- gentamicin
- KBM
- King's B media
- TBS-T
- Tris-buffered saline with Tween 20
- TCEP
- tris(2-carboxyethyl)phosphine
- Buffer A
- 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5% (v/v) glycerol, and 1 mm TCEP
- Buffer B
- 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5% (v/v) glycerol, 1 mm TCEP.
References
- 1. Whitfield G. B., Marmont L. S., and Howell P. L. (2015) Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front. Microbiol. 6, 471 10.3389/fmicb.2015.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Franklin M. J., Nivens D. E., Weadge J. T., and Howell P. L. (2011) Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2, 167 10.3389/fmicb.2011.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee H. J., Chang H. Y., Venkatesan N., and Peng H. L. (2008) Identification of amino acid residues important for the phosphomannose isomerase activity of PslB in Pseudomonas aeruginosa PAO1. FEBS Lett. 582, 3479–3483 10.1016/j.febslet.2008.09.013 [DOI] [PubMed] [Google Scholar]
- 4. Roychoudhury S., May T. B., Gill J. F., Singh S. K., Feingold D. S., and Chakrabarty A. M. (1989) Purification and characterization of guanosine diphospho-d-mannose dehydrogenase. A key enzyme in the biosynthesis of alginate by Pseudomonas aeruginosa. J. Biol. Chem. 264, 9380–9385 [PubMed] [Google Scholar]
- 5. Shinabarger D., Berry A., May T. B., Rothmel R., Fialho A., and Chakrabarty A. M. (1991) Purification and characterization of phosphomannose isomerase-guanosine diphospho-d-mannose pyrophosphorylase. A bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J. Biol. Chem. 266, 2080–2088 [PubMed] [Google Scholar]
- 6. Ma L., Wang J., Wang S., Anderson E. M., Lam J. S., Parsek M. R., and Wozniak D. J. (2012) Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ. Microbiol. 14, 1995–2005 10.1111/j.1462-2920.2012.02753.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Friedman L., and Kolter R. (2004) Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690 10.1046/j.1365-2958.2003.03877.x [DOI] [PubMed] [Google Scholar]
- 8. Vasseur P., Vallet-Gely I., Soscia C., Genin S., and Filloux A. (2005) The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151, 985–997 10.1099/mic.0.27410-0 [DOI] [PubMed] [Google Scholar]
- 9. Jennings L. K., Storek K. M., Ledvina H. E., Coulon C., Marmont L. S., Sadovskaya I., Secor P. R., Tseng B. S., Scian M., Filloux A., Wozniak D. J., Howell P. L., and Parsek M. R. (2015) Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 112, 11353–11358 10.1073/pnas.1503058112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colvin K. M., Alnabelseya N., Baker P., Whitney J. C., Howell P. L., and Parsek M. R. (2013) PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol. 195, 2329–2339 10.1128/JB.02150-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le Mauff F., Bamford N. C., Alnabelseya N., Zhang Y., Baker P., Robinson H., Codée J. D. C., Howell P. L., and Sheppard D. C. (2019) Molecular mechanism of Aspergillus fumigatus biofilm disruption by fungal and bacterial glycoside hydrolases. J. Biol. Chem. 294, 10760–10772 10.1074/jbc.RA119.008511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marmont L. S., Rich J. D., Whitney J. C., Whitfield G. B., Almblad H., Robinson H., Parsek M. R., Harrison J. J., and Howell P. L. (2017) Oligomeric lipoprotein PelC guides Pel polysaccharide export across the outer membrane of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 114, 2892–2897 10.1073/pnas.1613606114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whitfield G. B., Marmont L. S., Ostaszewski A., Rich J. D., Whitney J. C., Parsek M. R., Harrison J. J., and Howell P. L. (2020) Pel polysaccharide biosynthesis requires an inner membrane complex comprised of PelD, PelE, PelF, and PelG. J. Bacteriol. 202, e00684–19 10.1128/JB.00684-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kavanagh K. L., Jörnvall H., Persson B., and Oppermann U. (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65, 3895–3906 10.1007/s00018-008-8588-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fox A. R., Soto G., Valverde C., Russo D., Lagares A. Jr., Zorreguieta Á., Alleva K., Pascuan C., Frare R., Mercado-Blanco J., Dixon R., and Ayub N. D. (2016) Major cereal crops benefit from biological nitrogen fixation when inoculated with the nitrogen-fixing bacterium Pseudomonas protegens Pf-5 X940. Environ. Microbiol. 18, 3522–3534 10.1111/1462-2920.13376 [DOI] [PubMed] [Google Scholar]
- 16. Ueda A., and Saneoka H. (2015) Characterization of the ability to form biofilms by plant-associated Pseudomonas species. Curr. Microbiol. 70, 506–513 10.1007/s00284-014-0749-7 [DOI] [PubMed] [Google Scholar]
- 17. Kidarsa T. A., Shaffer B. T., Goebel N. C., Roberts D. P., Buyer J. S., Johnson A., Kobayashi D. Y., Zabriskie T. M., Paulsen I., and Loper J. E. (2013) Genes expressed by the biological control bacterium Pseudomonas protegens Pf-5 on seed surfaces under the control of the global regulators GacA and RpoS. Environ. Microbiol. 15, 716–735 10.1111/1462-2920.12066 [DOI] [PubMed] [Google Scholar]
- 18. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 19. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baraquet C., and Harwood C. S. (2016) FleQ DNA binding consensus sequence revealed by studies of FleQ-dependent regulation of biofilm gene expression in Pseudomonas aeruginosa. J. Bacteriol. 198, 178–186 10.1128/JB.00539-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Colvin K. M., Irie Y., Tart C. S., Urbano R., Whitney J. C., Ryder C., Howell P. L., Wozniak D. J., and Parsek M. R. (2012) The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928 10.1111/j.1462-2920.2011.02657.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Byrd M. S., Sadovskaya I., Vinogradov E., Lu H., Sprinkle A. B., Richardson S. H., Ma L., Ralston B., Parsek M. R., Anderson E. M., Lam J. S., and Wozniak D. J. (2009) Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73, 622–638 10.1111/j.1365-2958.2009.06795.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hickman J. W., and Harwood C. S. (2008) Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389 10.1111/j.1365-2958.2008.06281.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baraquet C., Murakami K., Parsek M. R., and Harwood C. S. (2012) The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40, 7207–7218 10.1093/nar/gks384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hickman J. W., Tifrea D. F., and Harwood C. S. (2005) A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U.S.A. 102, 14422–14427 10.1073/pnas.0507170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De N., Pirruccello M., Krasteva P. V., Bae N., Raghavan R. V., and Sondermann H. (2008) Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 6, e67 10.1371/journal.pbio.0060067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sato T., Tateno H., Kaji H., Chiba Y., Kubota T., Hirabayashi J., and Narimatsu H. (2017) Engineering of recombinant Wisteria floribunda agglutinin specifically binding to GalNAcβ1,4GlcNAc (LacdiNAc). Glycobiology 27, 743–754 10.1093/glycob/cwx038 [DOI] [PubMed] [Google Scholar]
- 28. Ishiyama N., Creuzenet C., Lam J. S., and Berghuis A. M. (2004) Crystal structure of WbpP, a genuine UDP-N-acetylglucosamine 4-epimerase from Pseudomonas aeruginosa: Substrate specificity in UDP-hexose 4-epimerases. J. Biol. Chem. 279, 22635–22642 10.1074/jbc.M401642200 [DOI] [PubMed] [Google Scholar]
- 29. Beerens K., Soetaert W., and Desmet T. (2015) UDP-hexose 4-epimerases: A view on structure, mechanism and substrate specificity. Carbohydr. Res. 414, 8–14 10.1016/j.carres.2015.06.006 [DOI] [PubMed] [Google Scholar]
- 30. Bauer A. J., Rayment I., Frey P. A., and Holden H. M. (1992) The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5 Å resolution. Proteins 12, 372–381 10.1002/prot.340120409 [DOI] [PubMed] [Google Scholar]
- 31. Jörnvall H., Persson B., Krook M., Atrian S., Gonzàlez-Duarte R., Jeffery J., and Ghosh D. (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34, 6003–6013 10.1021/bi00018a001 [DOI] [PubMed] [Google Scholar]
- 32. Thoden J. B., and Holden H. M. (1998) Dramatic differences in the binding of UDP-galactose and UDP-glucose to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry 37, 11469–11477 10.1021/bi9808969 [DOI] [PubMed] [Google Scholar]
- 33. Demendi M., Ishiyama N., Lam J. S., Berghuis A. M., and Creuzenet C. (2005) Towards a better understanding of the substrate specificity of the UDP-N-acetylglucosamine C4 epimerase WbpP. Biochem. J. 389, 173–180 10.1042/BJ20050263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wozniak D. J., Wyckoff T. J., Starkey M., Keyser R., Azadi P., O'Toole G. A., and Parsek M. R. (2003) Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. U.S.A. 100, 7907–7912 10.1073/pnas.1231792100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rocchetta H. L., Burrows L. L., and Lam J. S. (1999) Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63, 523–553 10.1128/MMBR.63.3.523-553.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Colvin K. M., Gordon V. D., Murakami K., Borlee B. R., Wozniak D. J., Wong G. C., and Parsek M. R. (2011) The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7, e1001264 10.1371/journal.ppat.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seo J., Brencic A., and Darwin A. J. (2009) Analysis of secretin-induced stress in Pseudomonas aeruginosa suggests prevention rather than response and identifies a novel protein involved in secretin function. J. Bacteriol. 191, 898–908 10.1128/JB.01443-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Creuzenet C., Belanger M., Wakarchuk W. W., and Lam J. S. (2000) Expression, purification, and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J. Biol. Chem. 275, 19060–19067 10.1074/jbc.M001171200 [DOI] [PubMed] [Google Scholar]
- 39. Wyszynski F. J., Lee S. S., Yabe T., Wang H., Gomez-Escribano J. P., Bibb M. J., Lee S. J., Davies G. J., and Davis B. G. (2012) Biosynthesis of the tunicamycin antibiotics proceeds via unique exo-glycal intermediates. Nat. Chem. 4, 539–546 10.1038/nchem.1351 [DOI] [PubMed] [Google Scholar]
- 40. Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., and Drummond A. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Winsor G. L., Lam D. K., Fleming L., Lo R., Whiteside M. D., Yu N. Y., Hancock R. E., and Brinkman F. S. (2011) Pseudomonas Genome Database: Improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596–D600 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Winsor G. L., Khaira B., Van Rossum T., Lo R., Whiteside M. D., and Brinkman F. S. (2008) The Burkholderia Genome Database: Facilitating flexible queries and comparative analyses. Bioinformatics 24, 2803–2804 10.1093/bioinformatics/btn524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I., Lanczycki C. J., Lu F., Marchler G. H., Song J. S., Thanki N., et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holm L., and Rosenström P. (2010) Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hmelo L. R., Borlee B. R., Almblad H., Love M. E., Randall T. E., Tseng B. S., Lin C., Irie Y., Storek K. M., Yang J. J., Siehnel R. J., Howell P. L., Singh P. K., Tolker-Nielsen T., Parsek M. R., et al. (2015) Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 10, 1820–1841 10.1038/nprot.2015.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. de Lorenzo V., and Timmis K. N. (1994) Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235, 386–405 10.1016/0076-6879(94)35157-0 [DOI] [PubMed] [Google Scholar]
- 48. Harrison J. J., Stremick C. A., Turner R. J., Allan N. D., Olson M. E., and Ceri H. (2010) Microtiter susceptibility testing of microbes growing on peg lids: A miniaturized biofilm model for high-throughput screening. Nat. Protoc. 5, 1236–1254 10.1038/nprot.2010.71 [DOI] [PubMed] [Google Scholar]
- 49. Clasquin M. F., Melamud E., and Rabinowitz J. D. (2012) LC-MS data processing with MAVEN: A metabolomic analysis and visualization engine. Curr. Protoc. Bioinformatics Chapter 14, Unit14 11 10.1002/0471250953.bi1411s37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 51. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 52. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. El-Gebali S., Mistry J., Bateman A., Eddy S. R., Luciani A., Potter S. C., Qureshi M., Richardson L. J., Salazar G. A., Smart A., Sonnhammer E. L. L., Hirsh L., Paladin L., Piovesan D., Tosatto S. C. E., et al. (2019) The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data described are located within the manuscript and the supplemental information. The coordinates and structure factors for PelXC232S in complex with NAD+ and UDP, PelXC232S/S121A/Y146F UDP-GlcNAc, and PelXC232S/S121A/Y146F UDP-GalNAc have been deposited in the PDB, ID codes 6WJB, 6WJA, and 6WJ9, respectively.