Abstract
Background
Using commutable external quality assessment (EQA) materials is important for monitoring successful harmonization efforts. We assessed the commutability of four human serum pool (HSP) preparations to identify candidate EQA materials for alanine aminotransferase (ALT) and aspartate aminotransferase (AST) activity measurement.
Methods
One set each of 85 clinical samples (CSs) was collected for ALT and AST activity measurement. The 15 candidate EQA materials included four types of HSP preparations (A to D): materials A, C, and D contained human original recombinant (HOR) aminotransferases; materials B was mixed leftover samples. The CSs and 15 candidate EQA materials were analyzed using seven routine assays, and the ln-transformed results were analyzed in 21 assay pairs. Commutability was assessed using Deming regression, with a 95% prediction interval (CLSI approach) and the difference in bias with an error component model (International Federation of Clinical Chemistry and Laboratory Medicine [IFCC] approach).
Results
For ALT, all materials were commutable for 14–21 assay pairs according to the CLSI and IFCC approaches. For AST, B01–03 showed commutability for 14–21 assay pairs, and C01–03 and D01–03 showed commutability for no less than 10 assay pairs according to the two approaches. A01–06 were commutable for 9–16 assay pairs according to the CLSI approach, but for 6–9 assay pairs according to the IFCC approach.
Conclusions
Mixed leftover samples showed desirable commutability characteristics as candidate EQA materials for routine aminotransferase activity measurements. Human serum bases supplemented with HOR were commutable for most routine ALT activity measurements.
Keywords: Alanine aminotransferase, Aspartate aminotransferase, Commutability, External quality assessment materials
INTRODUCTION
Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) activity tests are the most common liver chemistry tests, and are, respectively, the second and third most tested among all single items tested in clinical laboratories of 270 hospitals in China [1, 2]. Epidemiological evidence shows that elevated serum ALT and/or AST activity is not only symptomatic of liver injury but can also indicate damage to the myocardium or skeletal muscle [3]. Therefore, accurate measurement of ALT and AST activity is essential for the reasonable interpretation of clinical results and for clinical determination.
External quality assessment (EQA) is used to evaluate the performance of routine assays regularly and monitor the success of harmonization efforts. The application of commutable EQA materials, with these materials behaving like clinical samples (CSs), is recommended to strengthen the reliability of the EQA as a monitoring tool and to avoid peer group evaluation [4–6]. The conventional lyophilized materials, which are most commonly employed in EQA, were sometimes found to be noncommutable with CSs [7–11]. In general, native human serum pool (HSP) is considered as an optimal EQA material but has practical constraints. Hence, there is an urgent need to test the commutability of EQA materials and to use more suitable materials with commutability in EQA. The commonly applied commutability assessment approach was linear regression described in the Clinical and Laboratory Standards Institute (CLSI) EP30-A [12]. Recently, a new approach, difference in bias, is recommended by the International Federation of Clinical Chemistry and Laboratory Medicine (IFCC); it computes the difference between the bias of reference materials and the average bias of CSs to assess commutability [13–15]. Each of these approaches has its own attributes.
For ALT and AST, primary reference measurement procedures using pyridoxal-5-phosphate (P-5′-P) supplementation have been developed, and the IFCC also recommends supplementing P-5′-P in routine aminotransferase assays [16–18]. However, the mainstream assays employed in China for aminotransferase activity measurements lack P-5′-P for various reasons. A national routine chemistry EQA for aminotransferase (dry-chemistry not included) organized by the National Center for Clinical Laboratories in 2018 revealed that only one (i.e., Siemens Dimension) among 93 commercial aminotransferase assays was supplemented with P-5′-P, and laboratories using this assay accounted for only 0.18% (5/2,722) [19]. Therefore, we selected seven routine assays without P-5′-P supplemented for commutability assessment based on the number of user laboratories participating in this EQA for aminotransferase activity measurement.
To our knowledge, the commutabilities of different HSP preparations for ALT and AST have not been compared, especially using difference in bias. To investigate potential EQA commutable materials in China, 15 candidate EQA materials from four types of HSP preparations were analyzed, along with 85 CSs, to assess their commutability for routine ALT and AST activity measurement, according to the CLSI EP30-A guidelines (CLSI approach) [12] and the IFCC recommendations (IFCC approach) [13–15].
MATERIALS AND METHODS
Sample details
Blood samples were collected between February and May 2018 from the clinical laboratories of Beijing Hospital, Beijing Chaoyang Hospital, and Beijing Tongren Hospital, China. CSs were fresh serum stored at 2–8°C within three days of blood collection. One milliliter of each collected CS was transferred into a via, frozen at −80°C immediately, and stored within four months until use. This study was approved by the Ethics Committee of Beijing Hospital, with exemption for obtaining informed consent (approval No. 2018BJYYEC-019–01). In total, 85 CSs from 58 male and 27 female subjects aged 24–90 years were collected for ALT activity measurement (range, 16–318 U/L), and another 85 CSs from 51 male and 34 female subjects aged 18–94 years were collected for AST activity measurement (range, 18–334 U/L). Icteric, lipemic, and hemolyzed samples were excluded, and the activity range of the CSs covered those of the 15 candidate EQA materials for assessment in this study.
Candidate EQA materials
The 15 candidate EQA materials for commutability assessment were grouped into materials A-D based on the four types of frozen HSP preparations. Materials A were obtained from BIO-CRM Corp. (Jiaxing, China) [20] and comprised two lots, with each lot containing three concentrations (A01–03: lot 1 in 2017; A04–06: lot 2 in 2018). A01–06 were prepared using HSPs supplemented with human original recombinant (HOR) ALT and AST developed at BIO-CRM. Materials B (B01–03), prepared using the mixed leftover samples, contained natural concentrations of aminotransferase. In total, 93 leftover samples (from 44 male and 49 female subjects aged 18–89 years) were collected in vials daily between February and May 2018 at Beijing Hospital and frozen at −80°C. The frozen serum aliquots were grouped into three sets according to the original aminotransferase concentrations, thawed, and pooled to prepare B01–03 (about 20 U/L, 60 U/L, 250 U/L for ALT and 20 U/L, 70 U/L, 270 U/L for AST). Base materials C (C01–03) were leftover samples collected from healthy individuals under 30 years of age and stored at 2–8°C. The duration for leftover sample collection lasted for three days. Base materials D (D01–03) were sampled from frozen human plasma pools supplemented with mannitol-adenine-phosphate red blood cell preservation solution [21, 22] collected from blood banks. The frozen plasma pools were thawed and centrifuged at 2,205×g for 20 minutes to obtain the serum base material. Both materials C and D were supplemented with the same two HOR aminotransferases (i.e., one for ALT and one for AST) prepared at the Beijing Institute of Geriatrics through expression in Escherichia coli cells [23]. Materials C and D were adjusted to three concentrations, as close as possible to those for materials B. All materials were filtered through a 0.22-μm membrane filter using a Corning bottle-top vacuum filter in a sterile cabinet to minimize microbial contamination, mixed thoroughly by inverting, aliquoted at 1 mL/vial, and stored at −80°C until use within a month.
ALT and AST activity measurement procedures and experimental design
Detailed information on the seven P-5′-P-non-supplemented assays used is provided in Table 1. All assays are based on kinetic spectrophotometry (continuous-monitoring method) and were performed on a Hitachi 7180 analyzer (Hitachi High-Technologies Corp., Tokyo, Japan) in four runs, using the parameters’ setting and calibration recommended by the manufacturers for each assay. Materials A-D were interspersed among the CSs during evaluation. The test samples were thawed, mixed, and analyzed according to ALT and AST activity measurement procedures in triplicate in three phases (the first phase started with the first triplicate measurement of each sample, measurements in the second phase were performed in the reverse order, and the order in the third phase was the same as that in the first phase).
Table 1.
Details of the seven routine assays for ALT and AST activity measurement
| Assays | Manufacturer | Country/city | Reagent lot (ALT/AST) | Calibrator lot | Traceability of calibrator |
|---|---|---|---|---|---|
| BioSino | BioSino Bio-Technology & Science | China/Beijing | 170771/170721 | 170,113 | IFCC primary reference procedure |
| BSBE | Beijing Strong Biotechnologies | China/Beijing | 17–0726/17–0717 | 877UE* | JSCC TS01 and ERM AD457† |
| DiaSys | DiaSys Diagnostic Systems | Germany /Holzheim | 3783/3763 | 21,658 | modified IFCC procedure without P-5′-P |
| KHB | Shanghai Kehua Bio-Engineering | China/Shanghai | 2017051R/2017061R | 18,642,601* | modified IFCC procedure without P-5′-P |
| LEADMAN | Beijing Leadman Biochemistry | China/Beijing | 611292K/611161K | 708101H | ERM AD454 and ERM AD457† |
| Maccura | Maccura Biotechnology | China/Chengdu | 617031/617021 | 717,031 | modified IFCC procedure without P-5′-P |
| Wako | Wako Pure Chemical Industries | Japan/Osaka | TP935, 936/TN182, 183 | TN225 | IFCC primary reference procedure |
Randox and Roche calibrators are specified for BSBE and KHB, respectively;
Certified reference materials for ALT and AST.
Abbreviations: IFCC, International Federation of Clinical Chemistry and Laboratory Medicine; JSCC, Japan Society of Clinical Chemistry; P-5′-P, pyridoxal-5-phosphate.
Statistical analysis
The 85 CSs covering the activity range of the candidate EQA materials were used to assess the commutability of materials for ALT and AST activity. The results from the seven assays were analyzed in 21 assay pairs. Ln-transformed concentrations were used to obtain the consistent scatter of the difference plot over the concentrations [14]. Commutability was assessed according to the two approaches mentioned above and are described in detail below. Data were analyzed using Microsoft Excel 2016 (Microsoft Corporation, Redmond, Washington, USA).
CLSI approach based on linear regression analysis
As recommended in the CLSI EP30-A guidelines [12], Deming regression was performed using the ln-transformed data for the CSs after identifying outliers in the difference plot and visually inspecting linearity by ordinary linear regression. The 95% prediction interval was calculated for the commutability assessment.
IFCC approach based on difference in bias analysis
Commutability assessment of candidate EQA materials
The outliers were excluded by inspecting the precision profile and difference plot. The mean bias between assay x and assay y for all CSs was denoted as BCS, and the SD of all biases for the CSs was denoted as sCS. The standard uncertainty for the CSs, μ(BCS), can be expressed using Eq. 1, where N is the number of CSs.
| (1) |
For the candidate EQA materials, bias was calculated as the difference between two assays and was denoted as BRM. The associated uncertainty was calculated using Eq. 2, where sx(RM) and sy(RM) were estimated by pooling the SDs of the 15 candidate EQA materials’ k replicates.
| (2) |
The difference in bias, DRM, was estimated using Eq. 3, with the expanded uncertainty U(DRM) expressed by Eq. 4, in which a coverage factor of 1.9 was used to achieve a coverage of approximately 90%. The values of DRM and U(DRM) were compared with the commutability criterion C, which was set at 6.7% for ALT and 5.6% for AST based on the biological variation [24].
| (3) |
| (4) |
Three commutability assessment outcomes are possible: (1) when the uncertainty interval DRM±U(DRM) is within 0±C, commutable; (2) when DRM±U(DRM) is outside 0±C, noncommutable; (3) when DRM±U(DRM) and 0±C are overlapping, inconclusive.
Estimates of the error component from the CSs
The sCS mentioned above is the estimate of the observed variation in differences. According to the IFCC recommendations [14], sMSSD, excluding the variability from a possible trend in differences, is used as an estimate of random error. In this study, the random error was separated into two components: the variation from replicate measurements (sE) and the sample-specific effect (sd). The sE was estimated as the pooled SDs for all CSs from replicates. To test the null hypothesis of a no sample-specific effect, the test statistic was calculated using Eq. 6, where F has an F-distribution. If sd existed, it was estimated as the residual variation between assays not accounting for other error components and expressed by Eq. 7.
| (6) |
| (7) |
RESULTS
Commutability assessment using the CLSI approach
The between-assay correlations of 21 assay pairs are detailed in Supplemental Data Table S1. The commutability results for the candidate EQA materials for the 21 assay pairs in the Deming regression analysis are summarized in Table 2 and detailed in Supplemental Data Tables S2 and S3. Representative commutability assessment plots based on Deming regression are shown in Fig. 1. For ALT activity measurement, out of 21 assay pairs, 15–21 commutable assay pairs were observed for materials A, and 16–21 commutable assay pairs were observed for materials B to D (Table 2). For AST activity measurement, materials A showed commutability for 9–16 out of 21 assay pairs, whereas materials B to D showed commutability for 15–21 assay pairs (Table 2).
Table 2.
Summary of commutability assessment results for 15 candidate EQA materials
| EQA materials | ALT | AST | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Concentration (U/L) | Commutable pairs in CLSI | Commutable pairs in IFCC | Concentration (U/L) | Commutable pairs in CLSI | Commutable pairs in IFCC | |
| A01 | 94.1 | 21/21 | 21/21 | 34.5 | 13/21 | 9/21 |
|
| ||||||
| A02 | 172.5 | 21/21 | 21/21 | 43.5 | 9/21 | 6/21 |
|
| ||||||
| A03 | 257.0 | 21/21 | 21/21 | 62.8 | 9/21 | 7/21 |
|
| ||||||
| A04 | 39.2 | 15/21 | 14/21 | 31.8 | 16/21 | 7/21 |
|
| ||||||
| A05 | 130.6 | 21/21 | 21/21 | 95.2 | 10/21 | 6/21 |
|
| ||||||
| A06 | 210.9 | 20/21 | 20/21 | 191.1 | 11/21 | 9/21 |
|
| ||||||
| B01 | 20.6 | 20/21 | 15/21 | 20.8 | 21/21 | 14/21 |
|
| ||||||
| B02 | 58.0 | 21/21 | 21/21 | 66.6 | 21/21 | 18/21 |
|
| ||||||
| B03 | 256.0 | 21/21 | 21/21 | 272.0 | 21/21 | 19/21 |
|
| ||||||
| C01 | 20.9 | 16/21 | 16/21 | 25.5 | 18/21 | 10/21 |
|
| ||||||
| C02 | 57.9 | 21/21 | 21/21 | 61.4 | 15/21 | 15/21 |
|
| ||||||
| C03 | 280.5 | 21/21 | 21/21 | 279.7 | 15/21 | 10/21 |
|
| ||||||
| D01 | 22.5 | 16/21 | 21/21 | 20.4 | 15/21 | 10/21 |
|
| ||||||
| D02 | 62.7 | 21/21 | 21/21 | 55.2 | 15/21 | 13/21 |
|
| ||||||
| D03 | 307.1 | 21/21 | 19/21 | 265.7 | 15/21 | 10/21 |
Abbreviations: CLSI, Clinical and Laboratory Standards Institute; EQA, external quality assessment; IFCC, International Federation of Clinical Chemistry and Laboratory Medicine.
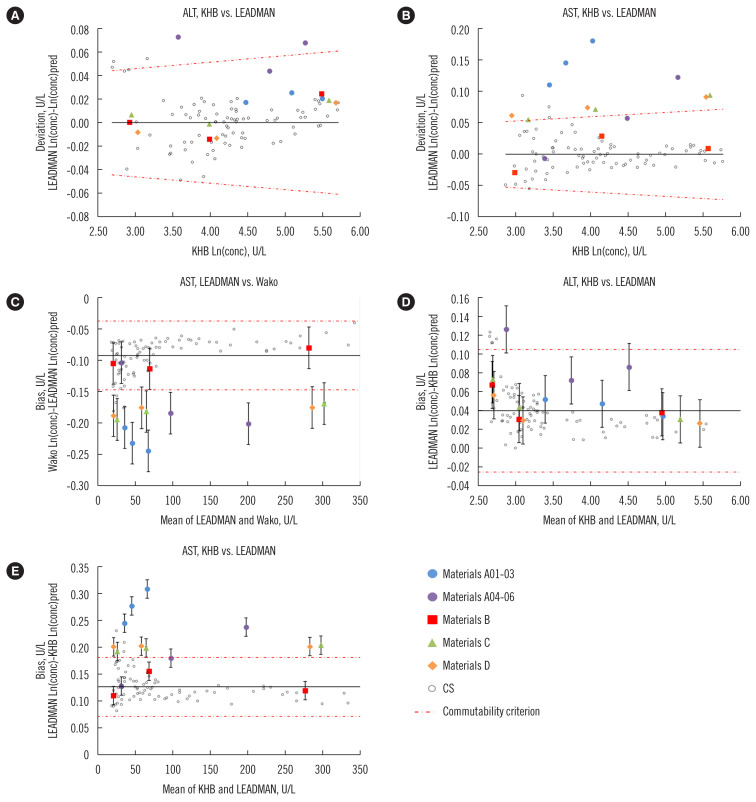
Fig. 1.
Five representative commutability assessment plots based on the ALT and AST activity measurements of 15 materials according to the CLSI and IFCC approaches. Panels A–B: CLSI approach; panels C–E: IFCC approach; error bars: expanded uncertainty U(DRM); solid black line: 0 line in panels A–B, mean bias line in panels C–E.
Abbreviations: CLSI, Clinical and Laboratory Standards Institute; CS, clinical sample; IFCC, International Federation of Clinical Chemistry and Laboratory Medicine; Ln(conc), ln-transformed concentrations; Ln(conc)pred, predicted ln-transformed concentrations in Deming analysis.
Commutability assessment using the IFCC approach
The results of commutability assessment based on difference in bias of the ln-transformed concentrations are summarized in Table 2 and detailed in the Supplemental Data Tables S4 and S5. Representative commutability assessment plots created using the difference in bias approach are shown in Fig. 1. The commutability assessment results for ALT activity demonstrated that all materials A showed commutability for 14–21 out of 21 assay pairs and materials B to D showed commutability for 15–21 assay pairs (Table 2). In the AST activity commutability assessment, materials A showed commutability for 6–9 out of 21 assay pairs, materials B for 14–19 assay pairs, and materials C and D for 10–15 assay pairs (Table 2).
Error components contributing to the differences among the 21 assay pairs are presented in Table 3. For ALT activity measurement, the values of sCS for the 21 assay pairs ranged from 0.011 to 0.035, whereas those of sE, the variation from replicates, ranged from 0.005 to 0.009, and those of sd, the sample-specific effect, ranged from 0.008 to 0.018. For AST activity measurement, the sCS was 0.011–0.038, sE was 0.009–0.016, and sd was 0.007–0.029 for the 21 assay pairs.
Table 3.
Estimate of error components for routine assay pairs of clinical samples
| Assay pairs (X–Y) | ALT | AST | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| sCS | sE | sd | P* | sCS | sE | sd | P* | |
| Biosino-BSBE | 0.017 | 0.005 | 0.013 | <0.05 | 0.024 | 0.015 | 0.014 | <0.05 |
|
| ||||||||
| Biosino-DiaSys | 0.024 | 0.006 | 0.017 | <0.05 | 0.018 | 0.012 | 0.008 | <0.05 |
|
| ||||||||
| Biosino-KHB | 0.011 | 0.008 | 0.015 | <0.05 | 0.018 | 0.011 | 0.009 | <0.05 |
|
| ||||||||
| Biosino-LEADMAN† | 0.029 | 0.006 | 0.015 | <0.05 | 0.029 | 0.012 | 0.023 | <0.05 |
|
| ||||||||
| Biosino-Maccura | 0.026 | 0.006 | 0.013 | <0.05 | 0.017 | 0.013 | 0.014 | <0.05 |
|
| ||||||||
| Biosino-Wako | 0.016 | 0.006 | 0.011 | <0.05 | 0.024 | 0.015 | 0.009 | >0.05 |
|
| ||||||||
| BSBE-DiaSys | 0.021 | 0.006 | 0.015 | <0.05 | 0.017 | 0.013 | 0.011 | <0.05 |
|
| ||||||||
| BSBE-KHB | 0.014 | 0.008 | 0.012 | <0.05 | 0.020 | 0.012 | 0.010 | <0.05 |
|
| ||||||||
| BSBE-LEADMAN† | 0.027 | 0.005 | 0.015 | <0.05 | 0.038 | 0.013 | 0.029 | <0.05 |
|
| ||||||||
| BSBE-Maccura | 0.026 | 0.006 | 0.015 | <0.05 | 0.011 | 0.013 | 0.008 | >0.05 |
|
| ||||||||
| BSBE-Wako | 0.015 | 0.006 | 0.010 | <0.05 | 0.027 | 0.016 | 0.015 | <0.05 |
|
| ||||||||
| DiaSys-KHB | 0.024 | 0.008 | 0.013 | <0.05 | 0.018 | 0.009 | 0.014 | <0.05 |
|
| ||||||||
| DiaSys-LEADMAN† | 0.035 | 0.006 | 0.016 | <0.05 | 0.017 | 0.010 | 0.017 | <0.05 |
|
| ||||||||
| DiaSys-Maccura | 0.024 | 0.007 | 0.012 | <0.05 | 0.013 | 0.011 | 0.010 | <0.05 |
|
| ||||||||
| DiaSys-Wako | 0.014 | 0.008 | 0.009 | <0.05 | 0.023 | 0.013 | 0.008 | >0.05 |
|
| ||||||||
| KHB-LEADMAN† | 0.027 | 0.009 | 0.018 | <0.05 | 0.029 | 0.009 | 0.025 | <0.05 |
|
| ||||||||
| KHB-Maccura | 0.027 | 0.008 | 0.015 | <0.05 | 0.014 | 0.009 | 0.007 | <0.05 |
|
| ||||||||
| KHB-Wako | 0.016 | 0.008 | 0.011 | <0.05 | 0.016 | 0.012 | 0.013 | <0.05 |
|
| ||||||||
| LEADMAN-Maccura† | 0.021 | 0.006 | 0.017 | <0.05 | 0.024 | 0.010 | 0.024 | <0.05 |
|
| ||||||||
| LEADMAN-Wako† | 0.025 | 0.007 | 0.013 | <0.05 | 0.034 | 0.014 | 0.022 | <0.05 |
|
| ||||||||
| Maccura-Wako | 0.017 | 0.006 | 0.008 | <0.05 | 0.023 | 0.015 | 0.011 | <0.05 |
P in the hypothesis test for the no sample-specific effect;
Routine assay pairs, including LEADMAN.
Abbreviations: sCS, observed variation for clinical samples; sd, sample-specific effect; sE, replicate measurements variation.
DISCUSSION
To propel the harmonization of aminotransferase activity measurements, the commutability of HSP preparations was assessed in this study, and two commutability assessment approaches were compared. Results of the CLSI approach for ALT activity showed no obvious difference when compared with those of the IFCC approach (Table 2; and Supplemental Data Tables S2 and S4). For AST activity, more commutability was observed for the CLSI approach than for the IFCC approach, although the overall conclusions were largely the same between the two approaches (Table 2; and Supplemental Data Tables S3 and S5). These observations suggested that the commutability criteria in these two approaches had similar stringencies for the ALT assay pairs, whereas the criterion for the AST assay pairs in the CLSI approach was less stringent than that in the IFCC approach. A possible explanation is that the criterion in the IFCC approach is fixed and based on medical requirements, whereas the CLSI criterion, which uses prediction intervals, is related to the random errors in each assay pair [14, 25, 26]. The magnitude of the random errors in this study is reflected in the following analysis based on the estimation of the error components.
Based on the error model in the IFCC approach, the possible error components that led to the differences between CS assay pairs could be estimated and analyzed. The results of error component estimation (Table 3) indicated that sd, the sample-specific effect, was one of the main sources of random error for both ALT and AST activity measurements. For AST, sE, the variation from replicate measurements (0.009–0.016) also substantially contributed to the disagreement between routine assay pairs, consistent with the less stringent CLSI commutability criterion. For six assay pairs related to LEADMAN, the sd was 0.017–0.029 (P<0.05), whereas the sd of the other assay pairs was 0.007–0.015 (Table 3). The LEADMAN assay tended to show a larger residual standard deviation (Sy.x) based on a correlation analysis, in which the results of each assay and the mean of the seven assays were compared (Supplemental Data Table S6). Therefore, the LEADMAN assay could have a limitation in terms of measurement selectivity. In addition, for assay pairs related to LEADMAN, there tended to be more non-commutability in the two approaches than for other assay pairs (Supplemental Data Tables S3 and S5). A possible reason is the above-mentioned limitation in the LEADMAN assay or the non-commutability of those materials.
Collecting blood from donors and rigorously preparing fresh HSPs according to the CLSI C37-A guidelines [27] are the most effective approaches to obtain commutable materials [5, 28]. Another promising source of commutable materials is leftover samples, which are easier to obtain. Considering the complex preparation of multienzyme EQA materials and the higher concentrations of EQA materials, except those for mixed leftover samples, materials with supplementation of HOR in the human serum base were also prepared in this study. Thus, four different types of HSP preparations were analyzed to identify their commutability and select candidate EQA materials. For ALT, all materials showed desirable commutability based on the CLSI and IFCC approaches. B01–03 also showed satisfying commutability for AST (Table 2; CLSI: 21–21 out of 21, IFCC: 14–19 out of 21). These observations suggested desirable commutability of the mixed leftover samples for both ALT and AST. For other HSP preparations with HORs in AST activity measurements, A01–06 showed undesirable commutability (6–9), whereas materials C–D were commutable for 10–15 assay pairs based on the IFCC approach (Table 2). The major difference between materials A and C–D is the supplemented HORs, which are from different origins. It is difficult to determine whether the commutability of the materials C–D for AST is satisfactory, since it remains inconclusive whether measurement selectivity limitations are related to the LEADMAN assay or the materials themselves resulted in noncommutability in the current experimental design. Nevertheless, human serum base supplemented with HOR showed great potential as a commutable EQA material for ALT and AST activity measurement [29, 30]. However, the HOR source affects the commutability of candidate EQA materials for AST activity measurement and should therefore be considered.
Commutability assessment is a continuing task for institutions and EQA manufacturers. Since the commutability conclusion from one lot or one concentration cannot be applied arbitrarily to others [7–10], we investigated multiple lots and concentrations of candidate EQA materials. Considering the wide-spread use of the heterogeneous system, which accounts for 61% of all measurements (1652/2722 from EQA data in 2018) in Chinese laboratories, we used the Hitachi 7180 analyzer, with combinations of seven mainstream reagents in the current study. In the future, other systems, including, Beckman AU5800, Siemens Advia 2400, and Roche Cobas 8000, should be investigated.
The plasma processing for the base materials D might produce precipitation despite a few steps of centrifugation or filtering. However, we did not observe coagulation during the preparation and testing of these materials. Considering the stability of ALT and AST activity, it is difficult to use fresh CSs in a large-scale commutability study using seven routine assays. Therefore, CSs were frozen until analysis according to previous enzyme commutability studies and the EP30-A guidelines [12, 25, 31]. In the current study design, with three measurements in three phases, the position effect (variation from measurement order) for the IFCC approach was lost in the replicate measurements variation (sE) due to nonadjacent replicates [25]. However, the relatively small values of sE for ALT indicated that position effects may not have been a major variation source for ALT.
In conclusion, mixed leftover samples showed desirable commutability characteristics as candidate EQA materials for routine aminotransferase activity measurements. Human serum bases supplemented with HOR were also commutable for most routine ALT activity measurements.
Supplementary Information
Supplemental Data Table S1.
Between-assay correlations of 21 assay pairs for clinical samples based on Deming regression
| Assay pairs (X–Y) | ALT | AST | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Outliers | r2 | DR slope (95% CI) | DR intercept (95% CI) | Outliers | r2 | DR slope (95% CI) | DR intercept (95% CI) | |
| Biosino-BSBE | 1/85 | 0.9994 | 1.00 (0.99–1.00) | 0.05 (0.04 to 0.07) | 1/85 | 0.998 7 | 0.99 (0.99–1.00) | −0.03 (−0.05 to 0.00) |
|
| ||||||||
| Biosino-DiaSys | 1/85 | 0.9989 | 0.98 (0.97–0.98) | 0.08 (0.05 to 0.10) | 3/85 | 0.999 2 | 1.00 (0.99–1.00) | 0.03 (0.01 to 0.05) |
|
| ||||||||
| Biosino-KHB | 0/85 | 0.9990 | 1.00 (1.00–1.01) | −0.11 (−0.13 to −0.09) | 3/85 | 0.999 4 | 1.00 (1.00–1.01) | −0.07 (−0.09 to −0.06) |
|
| ||||||||
| Biosino-LEADMAN | 2/85 | 0.9986 | 0.98 (0.98–0.99) | 0.02 (0.00 to 0.05) | 2/85 | 0.997 8 | 0.99 (0.98–1.00) | 0.10 (0.07 to 0.13) |
|
| ||||||||
| Biosino-Maccura | 0/85 | 0.9958 | 0.99 (0.98–0.99) | 0.03 (−0.01 to 0.07) | 1/85 | 0.999 0 | 1.00 (0.99–1.00) | 0.01 (−0.01 to 0.03) |
|
| ||||||||
| Biosino-Wako | 1/85 | 0.9991 | 0.98 (0.98–0.98) | 0.06 (0.04 to 0.08) | 2/85 | 0.998 8 | 1.01 (1.01–1.02) | −0.09 (−0.11 to −0.06) |
|
| ||||||||
| BSBE-DiaSys | 0/85 | 0.9991 | 0.98 (0.98–0.98) | 0.03 (0.01 to 0.05) | 1/85 | 0.998 8 | 1.01 (1.00–1.01) | 0.04 (0.02 to 0.07) |
|
| ||||||||
| BSBE-KHB | 2/85 | 0.9993 | 1.01 (1.00–1.01) | −0.15 (−0.17 to −0.14) | 2/85 | 0.999 1 | 1.01 (1.01–1.02) | −0.05 (−0.07 to −0.03) |
|
| ||||||||
| BSBE-LEADMAN | 3/85 | 0.9990 | 0.99 (0.98–0.99) | −0.03 (−0.05 to −0.01) | 0/85 | 0.996 7 | 1.00 (0.99–1.01) | 0.13 (0.09 to 0.16) |
|
| ||||||||
| BSBE-Maccura | 0/85 | 0.9984 | 0.98 (0.97–0.98) | 0.07 (0.04 to 0.09) | 0/85 | 0.999 2 | 1.00 (1.00–1.01) | 0.04 (0.02 to 0.05) |
|
| ||||||||
| BSBE-Wako | 1/85 | 0.9993 | 0.98 (0.98–0.99) | 0.01 (−0.01 to 0.03) | 1/85 | 0.998 5 | 1.02 (1.02–1.03) | −0.06 (−0.09 to −0.04) |
|
| ||||||||
| DiaSys-KHB | 2/85 | 0.9990 | 1.03 (1.02–1.03) | −0.18 (−0.21 to −0.16) | 2/85 | 0.999 3 | 1.01 (1.00–1.01) | −0.11 (−0.12 to −0.09) |
|
| ||||||||
| DiaSys-LEADMAN | 3/85 | 0.9989 | 1.01 (1.00–1.01) | −0.06 (−0.08 to −0.03) | 1/85 | 0.998 7 | 1.00 (0.99–1.00) | 0.07 (0.05 to 0.10) |
|
| ||||||||
| DiaSys-Maccura | 1/85 | 0.9990 | 0.99 (0.99–1.00) | 0.05 (0.03 to 0.07) | 1/85 | 0.999 1 | 1.00 (0.99–1.00) | −0.01 (−0.03 to 0.01) |
|
| ||||||||
| DiaSys-Wako | 0/85 | 0.9995 | 1.00 (1.00–1.01) | −0.02 (−0.03 to 0.00) | 2/85 | 0.999 3 | 1.02 (1.01–1.02) | −0.11 (−0.13 to −0.09) |
|
| ||||||||
| KHB-LEADMAN | 2/85 | 0.9985 | 0.98 (0.97–0.99) | 0.13 (0.10 to 0.15) | 2/85 | 0.997 8 | 0.99 (0.98–1.00) | 0.17 (0.14 to 0.20) |
|
| ||||||||
| KHB-Maccura | 1/85 | 0.9980 | 0.97 (0.96–0.98) | 0.21 (0.19 to 0.24) | 2/85 | 0.999 5 | 0.99 (0.99–1.00) | 0.09 (0.07 to 0.10) |
|
| ||||||||
| KHB-Wako | 2/85 | 0.9989 | 0.98 (0.97–0.98) | 0.16 (0.14 to 0.19) | 2/85 | 0.999 1 | 1.01 (1.01–1.01) | −0.01 (−0.02 to 0.01) |
|
| ||||||||
| LEADMAN-Maccura | 2/85 | 0.9985 | 0.99 (0.98–0.99) | 0.11 (0.08 to 0.13) | 1/85 | 0.997 3 | 1.00 (0.99–1.01) | −0.08 (−0.12 to −0.05) |
|
| ||||||||
| LEADMAN-Wako | 3/85 | 0.9992 | 1.00 (0.99–1.00) | 0.05 (0.03 to 0.07) | 1/85 | 0.998 1 | 1.02 (1.02–1.03) | −0.18 (−0.21 to −0.15) |
|
| ||||||||
| Maccura-Wako | 1/85 | 0.9994 | 1.01 (1.00–1.01) | −0.05 (−0.06 to −0.03) | 0/85 | 0.998 9 | 1.02 (1.01–1.02) | −0.10 (−0.12 to −0.08) |
Abbreviations: DR, Deming regression; r2, determination of coefficient; CI, confidence interval.
Supplemental Data Table S2.
Commutability for ALT activity measurements in candidate EQA materials according to the CLSI approach
| Assay pairs (X–Y) | A01 | A02 | A03 | A04 | A05 | A06 | B01 | B02 | B03 | C01 | C02 | C03 | D01 | D02 | D03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biosino-BSBE | C | C | C | C | C | C | C | C | C | NC | C | C | C | C | C |
| Biosino-DiaSys | C | C | C | C | C | C | C | C | C | C | C | C | NC | C | C |
| Biosino-KHB | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-LEADMAN | C | C | C | NC | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-Maccura | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| BSBE-DiaSys | C | C | C | C | C | C | C | C | C | C | C | C | NC | C | C |
| BSBE-KHB | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| BSBE-LEADMAN | C | C | C | NC | C | C | C | C | C | NC | C | C | C | C | C |
| BSBE-Maccura | C | C | C | C | C | C | C | C | C | NC | C | C | C | C | C |
| BSBE-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-KHB | C | C | C | C | C | C | C | C | C | C | C | C | NC | C | C |
| DiaSys-LEADMAN | C | C | C | NC | C | C | C | C | C | C | C | C | NC | C | C |
| DiaSys-Maccura | C | C | C | NC | C | C | C | C | C | NC | C | C | C | C | C |
| DiaSys-Wako | C | C | C | C | C | C | C | C | C | C | C | C | NC | C | C |
| KHB-LEADMAN | C | C | C | NC | C | NC | C | C | C | C | C | C | C | C | C |
| KHB-Maccura | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| KHB-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| LEADMAN-Maccura | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| LEADMAN-Wako | C | C | C | NC | C | C | C | C | C | C | C | C | C | C | C |
| Maccura-Wako | C | C | C | C | C | C | NC | C | C | NC | C | C | C | C | C |
Abbreviations: C, commutable; NC, noncommutable; EQA, external quality assessment.
Supplemental Data Table S3.
Commutability for AST activity measurements in candidate EQA materials according to the CLSI approach
| Assay pairs (X–Y) | A01 | A02 | A03 | A04 | A05 | A06 | B01 | B02 | B03 | C01 | C02 | C03 | D01 | D02 | D03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biosino-BSBE | C | C | C | C | NC | C | C | C | C | C | C | C | C | C | C |
| Biosino-DiaSys | C | C | NC | NC | NC | C | C | C | C | C | C | C | C | C | C |
| Biosino-KHB | C | NC | NC | C | NC | NC | C | C | C | C | C | C | C | C | C |
| Biosino-LEADMAN* | NC | NC | NC | C | C | C | C | C | C | C | NC | NC | NC | NC | NC |
| Biosino-Maccura | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-Wako | C | C | C | C | NC | NC | C | C | C | C | C | C | C | C | C |
| BSBE-DiaSys | NC | NC | NC | C | C | C | C | C | C | C | C | C | NC | C | C |
| BSBE-KHB | C | C | C | C | C | NC | C | C | C | C | C | C | C | C | C |
| BSBE-LEADMAN* | NC | NC | NC | C | NC | C | C | C | C | C | NC | NC | NC | NC | NC |
| BSBE-Maccura | C | C | C | C | NC | C | C | C | C | C | C | C | NC | C | C |
| BSBE-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-KHB | NC | NC | NC | C | C | NC | C | C | C | C | C | C | C | C | C |
| DiaSys-LEADMAN* | C | NC | NC | C | NC | NC | C | C | C | NC | NC | NC | C | NC | NC |
| DiaSys-Maccura | C | NC | NC | NC | NC | C | C | C | C | C | C | C | C | C | C |
| DiaSys-Wako | NC | NC | NC | C | C | NC | C | C | C | C | C | C | C | C | C |
| KHB-LEADMAN* | NC | NC | NC | C | C | NC | C | C | C | NC | NC | NC | NC | NC | NC |
| KHB-Maccura | C | NC | C | NC | NC | NC | C | C | C | C | C | C | C | C | C |
| KHB-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| LEADMAN-Maccura* | NC | NC | NC | NC | C | NC | C | C | C | C | NC | NC | C | NC | NC |
| LEADMAN-Wako* | NC | NC | NC | C | NC | NC | C | C | C | NC | NC | NC | NC | NC | NC |
| Maccura-Wako | C | C | C | NC | NC | C | C | C | C | C | C | C | C | C | C |
Routine assay pairs, including LEADMAN.
Abbreviations: C, commutable; NC, noncommutable; EQA, external quality assessment.
Supplemental Data Table S4.
Commutability for ALT activity measurements in candidate EQA materials according to the IFCC approach
| Assay pairs (X–Y) | A01 | A02 | A03 | A04 | A05 | A06 | B01 | B02 | B03 | C01 | C02 | C03 | D01 | D02 | D03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biosino-BSBE | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-DiaSys | C | C | C | C | C | C | C | C | C | C | C | C | C | C | I |
| Biosino-KHB | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-LEADMAN | C | C | C | NC | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-Maccura | C | C | C | I | C | C | I | C | C | I | C | C | C | C | C |
| Biosino-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| BSBE-DiaSys | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| BSBE-KHB | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| BSBE-LEADMAN | C | C | C | I | C | C | C | C | C | I | C | C | C | C | C |
| BSBE-Maccura | C | C | C | C | C | C | I | C | C | NC | C | C | C | C | C |
| BSBE-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-KHB | C | C | C | C | C | C | I | C | C | C | C | C | C | C | I |
| DiaSys-LEADMAN | C | C | C | I | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-Maccura | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| KHB-LEADMAN | C | C | C | I | C | I | C | C | C | C | C | C | C | C | C |
| KHB-Maccura | C | C | C | I | C | C | I | C | C | I | C | C | C | C | C |
| KHB-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| LEADMAN-Maccura | C | C | C | C | C | C | I | C | C | C | C | C | C | C | C |
| LEADMAN-Wako | C | C | C | I | C | C | C | C | C | C | C | C | C | C | C |
| Maccura-Wako | C | C | C | C | C | C | I | C | C | I | C | C | C | C | C |
Abbreviations: C, commutable; NC, noncommutable; I, inconclusive; EQA, external quality assessment.
Supplemental Data Table S5.
Commutability for AST activity measurements in candidate EQA materials according to the IFCC approach
| Assay pairs (X–Y) | A01 | A02 | A03 | A04 | A05 | A06 | B01 | B02 | B03 | C01 | C02 | C03 | D01 | D02 | D03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biosino-BSBE | C | I | C | C | NC | C | I | C | C | C | C | I | C | I | I |
| Biosino-DiaSys | C | C | I | I | NC | C | C | C | C | C | C | C | I | C | C |
| Biosino-KHB | I | I | I | I | I | I | C | C | C | C | C | C | C | C | C |
| Biosino-LEADMAN* | NC | NC | NC | I | C | I | C | I | C | I | NC | NC | NC | I | NC |
| Biosino-Maccura | C | C | C | I | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-Wako | I | I | I | I | NC | I | C | I | C | I | C | C | C | C | C |
| BSBE-DiaSys | I | I | I | I | C | C | I | C | C | C | C | C | I | C | C |
| BSBE-KHB | C | C | C | I | I | I | I | C | C | C | C | I | C | C | I |
| BSBE-LEADMAN* | NC | NC | NC | I | NC | I | I | I | C | I | NC | NC | NC | NC | NC |
| BSBE-Maccura | C | C | C | I | NC | C | I | C | C | C | C | C | I | C | C |
| BSBE-Wako | C | C | C | I | C | I | I | C | I | I | C | I | C | I | I |
| DiaSys-KHB | I | NC | NC | C | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-LEADMAN* | I | I | NC | C | I | NC | C | C | C | I | I | NC | I | NC | NC |
| DiaSys-Maccura | C | I | I | NC | NC | C | C | C | C | C | C | C | C | C | C |
| DiaSys-Wako | I | I | I | C | I | I | C | C | C | I | C | I | I | C | I |
| KHB-LEADMAN* | NC | NC | NC | C | I | NC | C | C | C | I | NC | NC | NC | NC | NC |
| KHB-Maccura | C | I | C | NC | NC | C | C | C | C | C | C | C | C | C | C |
| KHB-Wako | C | C | I | C | I | C | I | C | C | I | C | C | C | C | C |
| LEADMAN-Maccura* | NC | NC | NC | NC | C | NC | C | C | C | I | NC | NC | I | NC | NC |
| LEADMAN-Wako* | NC | NC | NC | C | NC | NC | C | C | C | NC | NC | I | NC | I | I |
| Maccura-Wako | I | I | C | NC | NC | I | C | C | I | I | C | I | I | C | I |
Routine assay pairs, including LEADMAN.
Abbreviations: C, commutable; NC, noncommutable; I, inconclusive; EQA, external quality assessment.
Supplemental Data Table S6.
Correlation analysis between seven routine assays and the means of all assays for 85 clinical samples
| Assay | ALT | AST | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| r2 | Slope (95% CI) | Intercept (95% CI) | Sy.x | r2 | Slope (95% CI) | Intercept (95% CI) | Sy.x | |
| Biosino | 0.9999 | 1.04 (1.04–1.05) | −0.88 (−1.11 to −0.66) | 0.63 | 0.9999 | 1.02 (1.02–1.02) | −0.30 (−0.52 to −0.08) | 0.69 |
|
| ||||||||
| BSBE | 0.9999 | 1.07 (1.07–1.08) | −0.58 (−0.87 to −0.28) | 0.83 | 0.9998 | 0.94 (0.94–0.95) | 0.40 (0.06 to 0.74) | 1.06 |
|
| ||||||||
| Diasys | 0.9999 | 0.98 (0.98–0.98) | 1.01 (0.76 to 1.26) | 0.71 | 0.9999 | 1.02 (1.01–1.02) | 0.39 (0.14 to 0.63) | 0.76 |
|
| ||||||||
| KHB | 0.9999 | 0.95 (0.94–0.95) | −0.95 (−1.22 to −0.69) | 0.75 | 1.0000 | 0.96 (0.96–0.96) | −0.34 (−0.51 to −0.17) | 0.52 |
|
| ||||||||
| LEADMAN* | 0.9998 | 0.96 (0.96–0.97) | 0.14 (−0.22 to 0.50) | 1.02 | 0.9997 | 1.06 (1.06–1.07) | 0.75 (0.26 to 1.24) | 1.51 |
|
| ||||||||
| Maccura | 0.9998 | 1.00 (1.00–1.00) | 0.80 (0.43 to 1.17) | 1.04 | 0.9999 | 1.00 (1.00–1.01) | 0.09 (−0.21 to 0.40) | 0.93 |
LEADMAN routine assay.
Abbreviations: r2, determination of coefficient; CI, confidence interval; Sy.x, residual standard deviation.
ACKNOWLEDGMENTS
The authors gratefully acknowledge their colleagues at Beijing Chaoyang Hospital Department of Laboratory Medicine and Beijing Tongren Hospital Department of Laboratory Medicine for their kind assistance in providing residual clinical samples.
Footnotes
AUTHOR CONTRIBUTIONS
ZTJ and WJ conceived and planned the study. QTQ collected samples with the help of WM. HW prepared human original recombinase. QTQ and LQC prepared materials B-D and carried out all the measurements. YY and ZHJ performed the data analysis. LQC wrote the draft of the manuscript. ZJ reviewed and edited the manuscript. ZCB and CWX supervised the study. All authors read and approved the final version of the manuscript.
CONFLICTS OF INTEREST
No potential conflicts of interest relevant to this article were reported.
RESEARCH FUNDING
This research was supported by The National Key Research and Development Program of China (2018YFC1002204).
REFERENCES
- 1.Kwo PY, Cohen SM, Lim JK. ACG clinical guideline: evaluation of abnormal liver chemistries. Am J Gastroenterol. 2017;112:18–35. doi: 10.1038/ajg.2016.517. [DOI] [PubMed] [Google Scholar]
- 2.Zhong K, Wang W, Falin H, Wang Z. Investigation and analysis of current application status of testing items of clinical laboratories in China. Chin J Lab Med. 2015;38:637–41. [Google Scholar]
- 3.Krishnamurthy S, Korenblat KM, Scott MG. Persistent increase in aspartate aminotransferase in an asymptomatic patient. Clin Chem. 2009;55:1573–5. doi: 10.1373/clinchem.2008.120782. [DOI] [PubMed] [Google Scholar]
- 4.Miller WG, Myers GL. Commutability still matters. Clin Chem. 2013;59:1291–3. doi: 10.1373/clinchem.2013.208785. [DOI] [PubMed] [Google Scholar]
- 5.Miller WG, Jones GR, Horowitz GL, Weykamp C. Proficiency testing/external quality assessment: current challenges and future directions. Clin Chem. 2011;57:1670–80. doi: 10.1373/clinchem.2011.168641. [DOI] [PubMed] [Google Scholar]
- 6.Cobbaert C, Weykamp C, Franck P, de Jonge R, Kuypers A, Steigstra H, et al. Systematic monitoring of standardization and harmonization status with commutable EQA-samples—Five year experience from the Netherlands. Clin Chim Acta. 2012;414:234–40. doi: 10.1016/j.cca.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 7.Zhang T, Zeng J, Wang M, Zhang C, Zhang J, Zhao H, et al. The commutability of reference materials for serum glucose measurements. Chin J Lab Med. 2015;38:296–300. [Google Scholar]
- 8.Zhang S, Zeng J, Zhang C, Li Y, Zhao H, Cheng F, et al. Commutability of possible external quality assessment materials for cardiac troponin measurement. PLoS One. 2014;9:e102046. doi: 10.1371/journal.pone.0102046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge M, Zhao H, Yan Y, Zhang T, Zeng J, Wang Y, et al. Evaluation of the bias of serum magnesium measurements and the commutability of processed materials. Clin Lab. 2016;62:921–30. doi: 10.7754/clin.lab.2015.150944. [DOI] [PubMed] [Google Scholar]
- 10.Meng Q, Zhou W, Zhang C, Zeng J, Zhao H, Zhang T, et al. Serum triglyceride measurements: the commutability of reference materials and the accuracy of results. Clin Chem Lab Med. 2017;55:1284–90. doi: 10.1515/cclm-2016-0682. [DOI] [PubMed] [Google Scholar]
- 11.Zeng J, Qi T, Wang S, Zhang T, Zhou W, Zhao H, et al. Commutability of control materials for external quality assessment of serum apolipoprotein A-I measurement. Clin Chem Lab Med. 2018;56:789–95. doi: 10.1515/cclm-2017-0652. [DOI] [PubMed] [Google Scholar]
- 12.CLSI. Characterization and qualification of commutable reference materials for laboratory medicine; approved guideline. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. EP30-A. [Google Scholar]
- 13.Miller WG, Schimmel H, Rej R, Greenberg N, Ceriotti F, Burns C, et al. IFCC Working Group Recommendations for Assessing Commutability Part 1: General experimental design. Clin Chem. 2018;64:447–54. doi: 10.1373/clinchem.2017.277525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson G, Budd JR, Greenberg N, Delatour V, Rej R, Panteghini M, et al. IFCC Working Group Recommendations for Assessing Commutability Part 2: Using the difference in bias between a reference material and clinical samples. Clin Chem. 2018;64:455–64. doi: 10.1373/clinchem.2017.277541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Budd JR, Weykamp C, Rej R, MacKenzie F, Ceriotti F, Greenberg N, et al. IFCC Working Group Recommendations for Assessing Commutability Part 3: Using the calibration effectiveness of a reference material. Clin Chem. 2018;64:465–74. doi: 10.1373/clinchem.2017.277558. [DOI] [PubMed] [Google Scholar]
- 16.Schumann G, Bonora R, Ceriotti F, Férard G, Ferrero CA, Franck PF, et al. IFCC primary reference procedures for the measurement of catalytic activity concentrations of enzymes at 37 degrees C. International Federation of Clinical Chemistry and Laboratory Medicine. Part 4. Reference procedure for the measurement of catalytic concentration of alanine aminotransferase. Clin Chem Lab Med. 2002;40:718–24. doi: 10.1515/CCLM.2002.124. [DOI] [PubMed] [Google Scholar]
- 17.Schumann G, Bonora R, Ceriotti F, Férard G, Ferrero CA, Franck PF, et al. IFCC primary reference procedures for the measurement of catalytic activity concentrations of enzymes at 37 degrees C. International Federation of Clinical Chemistry and Laboratory Medicine. Part 5. Reference procedure for the measurement of catalytic concentration of aspartate aminotransferase. Clin Chem Lab Med. 2002;40:725–33. doi: 10.1515/CCLM.2002.125. [DOI] [PubMed] [Google Scholar]
- 18.Westerhuis LW, Hafkenscheid JC. Apoenzyme content of serum aminotransferases in relation to plasma pyridoxal-5′-phosphate concentration. Clin Chem. 1983;29:789–92. [PubMed] [Google Scholar]
- 19.National Center for Clinical Laboratories. https://www.nccl.org.cn/(Updated on Nov 2019)
- 20.Bio CRM. Product center/customized service. http://www.biotech-crm.com/ (Updated on Nov 2019)
- 21.Nishino T, Yachie-Kinoshita A, Hirayama A, Soga T, Suematsu M, Tomita M. In silico modeling and metabolome analysis of long-stored erythrocytes to improve blood storage methods. J Biotechnol. 2009;144:212–23. doi: 10.1016/j.jbiotec.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Yang P, Zhou J, Kang Y, Gong L, Zhang J, Yu J, et al. Mannitol-adenine-phosphate: a novel solution for intraoperative blood salvage. Transfusion. 2014;54:1146–52. doi: 10.1111/trf.12370. [DOI] [PubMed] [Google Scholar]
- 23.Zou L, Zhao H, Wang D, Wang M, Zhang C, Xiao F. Expression and purification of a functional recombinant aspartate aminotransferase (AST) from Escherichia coli. J Microbiol Biotechnol. 2014;24:998–1003. doi: 10.4014/jmb.1402.02018. [DOI] [PubMed] [Google Scholar]
- 24.Carobene A, R⊘raas T, S⊘lvik UØ, Sylte MS, Sandberg S, Guerra E, et al. Biological variation estimates obtained from 91 healthy study participants for 9 enzymes in serum. Clin Chem. 2017;63:1141–50. doi: 10.1373/clinchem.2016.269811. [DOI] [PubMed] [Google Scholar]
- 25.Deprez L, Toussaint B, Zegers I, Schimmel H, Grote-Koska D, Klauke R, et al. Commutability assessment of candidate reference materials for pancreatic alpha-amylase. Clin Chem. 2018;64:1193–202. doi: 10.1373/clinchem.2018.289744. [DOI] [PubMed] [Google Scholar]
- 26.Korzun WJ, Nilsson G, Bachmann LM, Myers GL, Sakurabayashi I, Nakajima K, et al. Difference in bias approach for commutability assessment: application to frozen pools of human serum measured by 8 direct methods for HDL and LDL cholesterol. Clin Chem. 2015;61:1107–13. doi: 10.1373/clinchem.2015.240861. [DOI] [PubMed] [Google Scholar]
- 27.CLSI. Preparation and validation of commutable frozen human serum pools as secondary reference materials for cholesterol measurement procedures; approved guideline. Wayne, PA: Clinical and Laboratory Standards Institute; 1999. C37-A. [Google Scholar]
- 28.Cobbaert C, Weykamp C, Baadenhuijsen H, Kuypers A, Lindemans J, Jansen R. Selection, preparation, and characterization of commutable frozen human serum pools as potential secondary reference materials for lipid and apolipoprotein measurements: study within the framework of the Dutch project “Calibration 2000”. Clin Chem. 2002;48:1526–38. [PubMed] [Google Scholar]
- 29.Eto A, Shiki A, Chikaura Y, Oka T, Nakano NI. Multienzyme control serum (Seraclear-HE) containing human enzymes from established cell lines and other sources. 1: Preparation and properties. Clin Chem. 1995;41:872–80. [PubMed] [Google Scholar]
- 30.Nakano NI, Eto A, Chikaura Y, Oishi T. Multienzyme control serum (Seraclear-HE) containing human enzymes from established cell lines and other sources. 2: Evaluation as candidate working enzyme reference material for alanine and aspartate aminotransferases. Clin Chem. 1995;41:881–91. [PubMed] [Google Scholar]
- 31.Rami L, Roura M, Canalias F. Evaluation of commutability of several materials for harmonization alkaline phosphatase catalytic concentration measurements. Clin Chim Acta. 2012;413:1249–54. doi: 10.1016/j.cca.2012.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data Table S1.
Between-assay correlations of 21 assay pairs for clinical samples based on Deming regression
| Assay pairs (X–Y) | ALT | AST | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Outliers | r2 | DR slope (95% CI) | DR intercept (95% CI) | Outliers | r2 | DR slope (95% CI) | DR intercept (95% CI) | |
| Biosino-BSBE | 1/85 | 0.9994 | 1.00 (0.99–1.00) | 0.05 (0.04 to 0.07) | 1/85 | 0.998 7 | 0.99 (0.99–1.00) | −0.03 (−0.05 to 0.00) |
|
| ||||||||
| Biosino-DiaSys | 1/85 | 0.9989 | 0.98 (0.97–0.98) | 0.08 (0.05 to 0.10) | 3/85 | 0.999 2 | 1.00 (0.99–1.00) | 0.03 (0.01 to 0.05) |
|
| ||||||||
| Biosino-KHB | 0/85 | 0.9990 | 1.00 (1.00–1.01) | −0.11 (−0.13 to −0.09) | 3/85 | 0.999 4 | 1.00 (1.00–1.01) | −0.07 (−0.09 to −0.06) |
|
| ||||||||
| Biosino-LEADMAN | 2/85 | 0.9986 | 0.98 (0.98–0.99) | 0.02 (0.00 to 0.05) | 2/85 | 0.997 8 | 0.99 (0.98–1.00) | 0.10 (0.07 to 0.13) |
|
| ||||||||
| Biosino-Maccura | 0/85 | 0.9958 | 0.99 (0.98–0.99) | 0.03 (−0.01 to 0.07) | 1/85 | 0.999 0 | 1.00 (0.99–1.00) | 0.01 (−0.01 to 0.03) |
|
| ||||||||
| Biosino-Wako | 1/85 | 0.9991 | 0.98 (0.98–0.98) | 0.06 (0.04 to 0.08) | 2/85 | 0.998 8 | 1.01 (1.01–1.02) | −0.09 (−0.11 to −0.06) |
|
| ||||||||
| BSBE-DiaSys | 0/85 | 0.9991 | 0.98 (0.98–0.98) | 0.03 (0.01 to 0.05) | 1/85 | 0.998 8 | 1.01 (1.00–1.01) | 0.04 (0.02 to 0.07) |
|
| ||||||||
| BSBE-KHB | 2/85 | 0.9993 | 1.01 (1.00–1.01) | −0.15 (−0.17 to −0.14) | 2/85 | 0.999 1 | 1.01 (1.01–1.02) | −0.05 (−0.07 to −0.03) |
|
| ||||||||
| BSBE-LEADMAN | 3/85 | 0.9990 | 0.99 (0.98–0.99) | −0.03 (−0.05 to −0.01) | 0/85 | 0.996 7 | 1.00 (0.99–1.01) | 0.13 (0.09 to 0.16) |
|
| ||||||||
| BSBE-Maccura | 0/85 | 0.9984 | 0.98 (0.97–0.98) | 0.07 (0.04 to 0.09) | 0/85 | 0.999 2 | 1.00 (1.00–1.01) | 0.04 (0.02 to 0.05) |
|
| ||||||||
| BSBE-Wako | 1/85 | 0.9993 | 0.98 (0.98–0.99) | 0.01 (−0.01 to 0.03) | 1/85 | 0.998 5 | 1.02 (1.02–1.03) | −0.06 (−0.09 to −0.04) |
|
| ||||||||
| DiaSys-KHB | 2/85 | 0.9990 | 1.03 (1.02–1.03) | −0.18 (−0.21 to −0.16) | 2/85 | 0.999 3 | 1.01 (1.00–1.01) | −0.11 (−0.12 to −0.09) |
|
| ||||||||
| DiaSys-LEADMAN | 3/85 | 0.9989 | 1.01 (1.00–1.01) | −0.06 (−0.08 to −0.03) | 1/85 | 0.998 7 | 1.00 (0.99–1.00) | 0.07 (0.05 to 0.10) |
|
| ||||||||
| DiaSys-Maccura | 1/85 | 0.9990 | 0.99 (0.99–1.00) | 0.05 (0.03 to 0.07) | 1/85 | 0.999 1 | 1.00 (0.99–1.00) | −0.01 (−0.03 to 0.01) |
|
| ||||||||
| DiaSys-Wako | 0/85 | 0.9995 | 1.00 (1.00–1.01) | −0.02 (−0.03 to 0.00) | 2/85 | 0.999 3 | 1.02 (1.01–1.02) | −0.11 (−0.13 to −0.09) |
|
| ||||||||
| KHB-LEADMAN | 2/85 | 0.9985 | 0.98 (0.97–0.99) | 0.13 (0.10 to 0.15) | 2/85 | 0.997 8 | 0.99 (0.98–1.00) | 0.17 (0.14 to 0.20) |
|
| ||||||||
| KHB-Maccura | 1/85 | 0.9980 | 0.97 (0.96–0.98) | 0.21 (0.19 to 0.24) | 2/85 | 0.999 5 | 0.99 (0.99–1.00) | 0.09 (0.07 to 0.10) |
|
| ||||||||
| KHB-Wako | 2/85 | 0.9989 | 0.98 (0.97–0.98) | 0.16 (0.14 to 0.19) | 2/85 | 0.999 1 | 1.01 (1.01–1.01) | −0.01 (−0.02 to 0.01) |
|
| ||||||||
| LEADMAN-Maccura | 2/85 | 0.9985 | 0.99 (0.98–0.99) | 0.11 (0.08 to 0.13) | 1/85 | 0.997 3 | 1.00 (0.99–1.01) | −0.08 (−0.12 to −0.05) |
|
| ||||||||
| LEADMAN-Wako | 3/85 | 0.9992 | 1.00 (0.99–1.00) | 0.05 (0.03 to 0.07) | 1/85 | 0.998 1 | 1.02 (1.02–1.03) | −0.18 (−0.21 to −0.15) |
|
| ||||||||
| Maccura-Wako | 1/85 | 0.9994 | 1.01 (1.00–1.01) | −0.05 (−0.06 to −0.03) | 0/85 | 0.998 9 | 1.02 (1.01–1.02) | −0.10 (−0.12 to −0.08) |
Abbreviations: DR, Deming regression; r2, determination of coefficient; CI, confidence interval.
Supplemental Data Table S2.
Commutability for ALT activity measurements in candidate EQA materials according to the CLSI approach
| Assay pairs (X–Y) | A01 | A02 | A03 | A04 | A05 | A06 | B01 | B02 | B03 | C01 | C02 | C03 | D01 | D02 | D03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biosino-BSBE | C | C | C | C | C | C | C | C | C | NC | C | C | C | C | C |
| Biosino-DiaSys | C | C | C | C | C | C | C | C | C | C | C | C | NC | C | C |
| Biosino-KHB | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-LEADMAN | C | C | C | NC | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-Maccura | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| BSBE-DiaSys | C | C | C | C | C | C | C | C | C | C | C | C | NC | C | C |
| BSBE-KHB | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| BSBE-LEADMAN | C | C | C | NC | C | C | C | C | C | NC | C | C | C | C | C |
| BSBE-Maccura | C | C | C | C | C | C | C | C | C | NC | C | C | C | C | C |
| BSBE-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-KHB | C | C | C | C | C | C | C | C | C | C | C | C | NC | C | C |
| DiaSys-LEADMAN | C | C | C | NC | C | C | C | C | C | C | C | C | NC | C | C |
| DiaSys-Maccura | C | C | C | NC | C | C | C | C | C | NC | C | C | C | C | C |
| DiaSys-Wako | C | C | C | C | C | C | C | C | C | C | C | C | NC | C | C |
| KHB-LEADMAN | C | C | C | NC | C | NC | C | C | C | C | C | C | C | C | C |
| KHB-Maccura | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| KHB-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| LEADMAN-Maccura | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| LEADMAN-Wako | C | C | C | NC | C | C | C | C | C | C | C | C | C | C | C |
| Maccura-Wako | C | C | C | C | C | C | NC | C | C | NC | C | C | C | C | C |
Abbreviations: C, commutable; NC, noncommutable; EQA, external quality assessment.
Supplemental Data Table S3.
Commutability for AST activity measurements in candidate EQA materials according to the CLSI approach
| Assay pairs (X–Y) | A01 | A02 | A03 | A04 | A05 | A06 | B01 | B02 | B03 | C01 | C02 | C03 | D01 | D02 | D03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biosino-BSBE | C | C | C | C | NC | C | C | C | C | C | C | C | C | C | C |
| Biosino-DiaSys | C | C | NC | NC | NC | C | C | C | C | C | C | C | C | C | C |
| Biosino-KHB | C | NC | NC | C | NC | NC | C | C | C | C | C | C | C | C | C |
| Biosino-LEADMAN* | NC | NC | NC | C | C | C | C | C | C | C | NC | NC | NC | NC | NC |
| Biosino-Maccura | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-Wako | C | C | C | C | NC | NC | C | C | C | C | C | C | C | C | C |
| BSBE-DiaSys | NC | NC | NC | C | C | C | C | C | C | C | C | C | NC | C | C |
| BSBE-KHB | C | C | C | C | C | NC | C | C | C | C | C | C | C | C | C |
| BSBE-LEADMAN* | NC | NC | NC | C | NC | C | C | C | C | C | NC | NC | NC | NC | NC |
| BSBE-Maccura | C | C | C | C | NC | C | C | C | C | C | C | C | NC | C | C |
| BSBE-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-KHB | NC | NC | NC | C | C | NC | C | C | C | C | C | C | C | C | C |
| DiaSys-LEADMAN* | C | NC | NC | C | NC | NC | C | C | C | NC | NC | NC | C | NC | NC |
| DiaSys-Maccura | C | NC | NC | NC | NC | C | C | C | C | C | C | C | C | C | C |
| DiaSys-Wako | NC | NC | NC | C | C | NC | C | C | C | C | C | C | C | C | C |
| KHB-LEADMAN* | NC | NC | NC | C | C | NC | C | C | C | NC | NC | NC | NC | NC | NC |
| KHB-Maccura | C | NC | C | NC | NC | NC | C | C | C | C | C | C | C | C | C |
| KHB-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| LEADMAN-Maccura* | NC | NC | NC | NC | C | NC | C | C | C | C | NC | NC | C | NC | NC |
| LEADMAN-Wako* | NC | NC | NC | C | NC | NC | C | C | C | NC | NC | NC | NC | NC | NC |
| Maccura-Wako | C | C | C | NC | NC | C | C | C | C | C | C | C | C | C | C |
Routine assay pairs, including LEADMAN.
Abbreviations: C, commutable; NC, noncommutable; EQA, external quality assessment.
Supplemental Data Table S4.
Commutability for ALT activity measurements in candidate EQA materials according to the IFCC approach
| Assay pairs (X–Y) | A01 | A02 | A03 | A04 | A05 | A06 | B01 | B02 | B03 | C01 | C02 | C03 | D01 | D02 | D03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biosino-BSBE | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-DiaSys | C | C | C | C | C | C | C | C | C | C | C | C | C | C | I |
| Biosino-KHB | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-LEADMAN | C | C | C | NC | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-Maccura | C | C | C | I | C | C | I | C | C | I | C | C | C | C | C |
| Biosino-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| BSBE-DiaSys | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| BSBE-KHB | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| BSBE-LEADMAN | C | C | C | I | C | C | C | C | C | I | C | C | C | C | C |
| BSBE-Maccura | C | C | C | C | C | C | I | C | C | NC | C | C | C | C | C |
| BSBE-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-KHB | C | C | C | C | C | C | I | C | C | C | C | C | C | C | I |
| DiaSys-LEADMAN | C | C | C | I | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-Maccura | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| KHB-LEADMAN | C | C | C | I | C | I | C | C | C | C | C | C | C | C | C |
| KHB-Maccura | C | C | C | I | C | C | I | C | C | I | C | C | C | C | C |
| KHB-Wako | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| LEADMAN-Maccura | C | C | C | C | C | C | I | C | C | C | C | C | C | C | C |
| LEADMAN-Wako | C | C | C | I | C | C | C | C | C | C | C | C | C | C | C |
| Maccura-Wako | C | C | C | C | C | C | I | C | C | I | C | C | C | C | C |
Abbreviations: C, commutable; NC, noncommutable; I, inconclusive; EQA, external quality assessment.
Supplemental Data Table S5.
Commutability for AST activity measurements in candidate EQA materials according to the IFCC approach
| Assay pairs (X–Y) | A01 | A02 | A03 | A04 | A05 | A06 | B01 | B02 | B03 | C01 | C02 | C03 | D01 | D02 | D03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biosino-BSBE | C | I | C | C | NC | C | I | C | C | C | C | I | C | I | I |
| Biosino-DiaSys | C | C | I | I | NC | C | C | C | C | C | C | C | I | C | C |
| Biosino-KHB | I | I | I | I | I | I | C | C | C | C | C | C | C | C | C |
| Biosino-LEADMAN* | NC | NC | NC | I | C | I | C | I | C | I | NC | NC | NC | I | NC |
| Biosino-Maccura | C | C | C | I | C | C | C | C | C | C | C | C | C | C | C |
| Biosino-Wako | I | I | I | I | NC | I | C | I | C | I | C | C | C | C | C |
| BSBE-DiaSys | I | I | I | I | C | C | I | C | C | C | C | C | I | C | C |
| BSBE-KHB | C | C | C | I | I | I | I | C | C | C | C | I | C | C | I |
| BSBE-LEADMAN* | NC | NC | NC | I | NC | I | I | I | C | I | NC | NC | NC | NC | NC |
| BSBE-Maccura | C | C | C | I | NC | C | I | C | C | C | C | C | I | C | C |
| BSBE-Wako | C | C | C | I | C | I | I | C | I | I | C | I | C | I | I |
| DiaSys-KHB | I | NC | NC | C | C | C | C | C | C | C | C | C | C | C | C |
| DiaSys-LEADMAN* | I | I | NC | C | I | NC | C | C | C | I | I | NC | I | NC | NC |
| DiaSys-Maccura | C | I | I | NC | NC | C | C | C | C | C | C | C | C | C | C |
| DiaSys-Wako | I | I | I | C | I | I | C | C | C | I | C | I | I | C | I |
| KHB-LEADMAN* | NC | NC | NC | C | I | NC | C | C | C | I | NC | NC | NC | NC | NC |
| KHB-Maccura | C | I | C | NC | NC | C | C | C | C | C | C | C | C | C | C |
| KHB-Wako | C | C | I | C | I | C | I | C | C | I | C | C | C | C | C |
| LEADMAN-Maccura* | NC | NC | NC | NC | C | NC | C | C | C | I | NC | NC | I | NC | NC |
| LEADMAN-Wako* | NC | NC | NC | C | NC | NC | C | C | C | NC | NC | I | NC | I | I |
| Maccura-Wako | I | I | C | NC | NC | I | C | C | I | I | C | I | I | C | I |
Routine assay pairs, including LEADMAN.
Abbreviations: C, commutable; NC, noncommutable; I, inconclusive; EQA, external quality assessment.
Supplemental Data Table S6.
Correlation analysis between seven routine assays and the means of all assays for 85 clinical samples
| Assay | ALT | AST | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| r2 | Slope (95% CI) | Intercept (95% CI) | Sy.x | r2 | Slope (95% CI) | Intercept (95% CI) | Sy.x | |
| Biosino | 0.9999 | 1.04 (1.04–1.05) | −0.88 (−1.11 to −0.66) | 0.63 | 0.9999 | 1.02 (1.02–1.02) | −0.30 (−0.52 to −0.08) | 0.69 |
|
| ||||||||
| BSBE | 0.9999 | 1.07 (1.07–1.08) | −0.58 (−0.87 to −0.28) | 0.83 | 0.9998 | 0.94 (0.94–0.95) | 0.40 (0.06 to 0.74) | 1.06 |
|
| ||||||||
| Diasys | 0.9999 | 0.98 (0.98–0.98) | 1.01 (0.76 to 1.26) | 0.71 | 0.9999 | 1.02 (1.01–1.02) | 0.39 (0.14 to 0.63) | 0.76 |
|
| ||||||||
| KHB | 0.9999 | 0.95 (0.94–0.95) | −0.95 (−1.22 to −0.69) | 0.75 | 1.0000 | 0.96 (0.96–0.96) | −0.34 (−0.51 to −0.17) | 0.52 |
|
| ||||||||
| LEADMAN* | 0.9998 | 0.96 (0.96–0.97) | 0.14 (−0.22 to 0.50) | 1.02 | 0.9997 | 1.06 (1.06–1.07) | 0.75 (0.26 to 1.24) | 1.51 |
|
| ||||||||
| Maccura | 0.9998 | 1.00 (1.00–1.00) | 0.80 (0.43 to 1.17) | 1.04 | 0.9999 | 1.00 (1.00–1.01) | 0.09 (−0.21 to 0.40) | 0.93 |
LEADMAN routine assay.
Abbreviations: r2, determination of coefficient; CI, confidence interval; Sy.x, residual standard deviation.