Summary
The islets of Langerhans are dynamic structures that can change in size, number of cells, and molecular function in response to physiological and pathological stress. Molecular cues originating from the surrounding “peri-islet” acinar cells that could facilitate this plasticity have not been explored. Here, we combine single-molecule transcript imaging in the intact pancreas and transcriptomics to identify spatial heterogeneity of acinar cell gene expression. We find that peri-islet acinar cells exhibit a distinct molecular signature in db/db diabetic mice that includes upregulation of trypsin family genes and elevated mTOR activity. This zonated expression program seems to be induced by CCK that is secreted from islet cells. Elevated peri-islet trypsin secretion could facilitate the islet expansion observed in this model via modulation of the islet capsule matrix components. Our study highlights a molecular axis of communication between the pancreatic exocrine and endocrine compartments that may be relevant to islet expansion.
Keywords: pancreas, acinar cells, diabetes, single cell heterogeneity, db/db mice, smFISH, peri-islet acinar cells, zonation, single molecule imaging
Graphical Abstract
Highlights
-
•
Pancreatic acinar cells exhibit a zonated gene expression signature in db/db mice
-
•
Peri-islet acinar cells upregulate trypsin genes and mTOR activity
-
•
Islet-derived CCK induces the zonated acinar gene expression signature
-
•
Exocrine-endocrine crosstalk may be relevant to islet expansion
Egozi et al. identify a zonated gene expression signature of pancreatic acinar cells in diabetic mice. Peri-islet acinar cells elevate trypsin genes. This expression signature is regulated by islet-derived CCK and may be relevant for islet expansion.
Introduction
The endocrine pancreas, composed of the islets of Langerhans, is a dynamic structure that can change in size, number of cells, and function in response to physiological and pathological stress. In states of systemic insulin resistance, such as those observed in the metabolic syndrome or pregnancy, islets exhibit substantial changes that result in higher insulin secretion (Gunasekaran and Gannon, 2011). These changes include the induction of insulin by individual beta cells, an increase in beta cell size (hypertrophy), and an increase in beta cell numbers (hyperplasia). Understanding the molecular cues that facilitate such islet plasticity is important, since they may identify potential avenues to stimulate beta cell expansion. Different studies have highlighted systemic and intrinsic factors that facilitate beta cell expansion (Nielsen et al., 2001). Other studies have highlighted signals in the islet microenvironment, specifically from pericytes, that induce beta cell state and proliferative potential (Sakhneny et al., 2018). The potential involvement of acinar cells in islet expansion has not been explored.
The islets of Langerhans are embedded in the exocrine pancreas, which is predominantly composed of acinar cells. Acinar cells secrete digestive enzymes into pancreatic ducts that drain into the small intestine. Recent work has suggested that acinar cells are not identical, but rather show heterogeneous gene expression (Tosti et al., 2019). Acinar cells in close proximity to the islet, called “peri-islet” acinar cells, have been shown to be morphologically and functionally different from “tele-islet” acinar cells that are farther away (Adelson and Miller, 1989; Anzi et al., 2018; Hellman et al., 1962; Malaisse-Lagae et al., 1975; White and Swartz, 1980). Some of these differences, for example, cell size, have been attributed to the elevated levels of insulin, which could locally diffuse into the pancreas parenchyma (Hellman et al., 1962). However, technical limitations impeded the identification of differences in gene expression between peri-islet and tele-islet acinar cells. Since the quality of mRNA extracted from the pancreas is notoriously low, in situ approaches to image mRNA have largely been unsuccessful. Here, we apply a recently developed method for single-molecule fluorescence in situ hybridization (smFISH) in the intact pancreas (Farack et al., 2019; Farack, 2020) to explore whether acinar gene expression is zonated (Halpern et al., 2017; Moor et al., 2018), namely, whether the molecular identity of acinar cells changes as a function of their distance from the islets of Langerhans. We uncover a zonated gene expression signature of trypsin genes in diabetic mice. This signature may be induced by islet CCK and could facilitate islet expansion.
Results
smFISH in the Intact Pancreas Reveals Acinar Zonation in db/db Mice
To address potential zonation of acinar cells we chose to focus on adult db/db mice (9–17 weeks). These mice have been shown to be highly insulin resistant and to exhibit substantial islet expansion (Coleman, 1978; Hummel et al., 1966). These changes could result in higher levels of morphogens originating in the islet, which could in turn result in changes in the molecular identities of peri-islet acinar cells.
We examined pancreatic tissues from 9- to 17-week-old B6.BKS(D)-Leprdb/J db/db mice. These mice were shown to be hyperinsulinemic and hyperglycemic (Coleman, 1978; Table S1). We found that peri-islet acinar cells were significantly larger than tele-islet acinar cells, particularly in db/db mice (1.8-fold in db/db mice, p < 1.4e−12, 1.17-fold in control, p < 0.04; Figures 1A–1C). This was in line with previous studies that demonstrated morphological changes in peri-islet acinar cells in this mouse model (Hellman et al., 1962; White and Swartz, 1980).
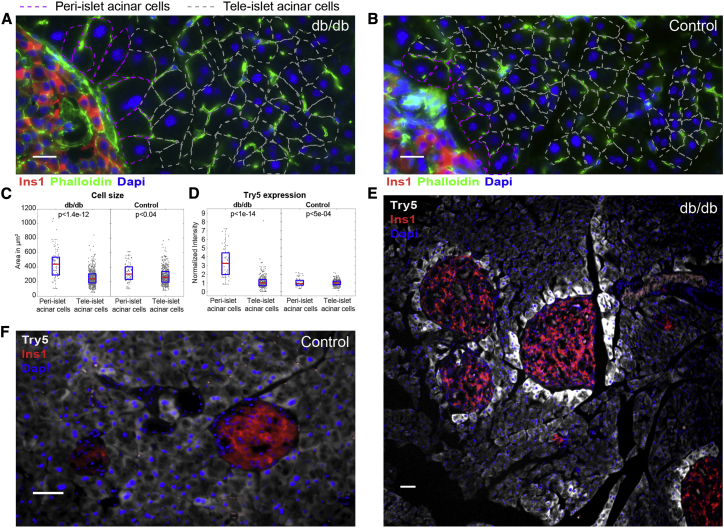
Figure 1.
Zonation of Acinar Cells in db/db mice
(A and B) Peri-islet acinar cells (purple dashed lines) are significantly larger than tele-islet acinar cells (gray dashed lines) in db/db mice (A) and to a lesser extent in control mice (B). Red is Ins1 mRNA, the membrane is stained by phalloidin (green), and nuclei are stained by DAPI (blue). Scale bars, 20 μm.
(C) Quantification of acinar cell size (10 islets from 5 mice, p < 1.4e-12 in db/db mice, p < 0.04 in control mice).
(D) Quantification of peri-islet acinar cell zonation of Try5 in db/db and control mice (10 islets from 5 mice, p < 1e-14 in db/db mice, p < 5e-04 in controls). Red lines are medians and blue boxes are 25th–75th percentiles. Of 358 cells, 5 are above the maximal shown scale.
(E) Try5 mRNA (gray) is significantly higher in peri-islet acinar cells compared to tele-islet acinar cells in db/db mice.
(F) Try5 mRNA (gray) is expressed in a non-zonated manner in control mice.
Islets in (E) and (F) marked by Ins1 mRNA (red), nuclei stained by DAPI (blue). Scale bars, 50 μm. (A), (B), (E), and (F) are examples of pancreatic tissue of 14- to 17-week-old db/db and control mice.
See also Figures S1, S2, and S3.
To identify the potential zonation of gene expression, we used an smFISH protocol optimized for pancreatic tissues (Farack et al., 2019; Farack, 2020). This protocol has been shown to be highly sensitive and quantitative, enabling the detection of individual mRNA molecules as diffraction-limited spots under a fluorescence microscope (Figures S1A and S1B). For highly abundant genes, where single transcripts cannot be resolved, the fluorescence signal intensity correlates with cellular mRNA levels (Farack et al., 2019; Little et al., 2013). We imaged the transcripts of acinar genes encoding key digestive enzymes, including Try5, encoding a family member of the serine protease cleaving enzymes (Figures 1D and 1E); Cel, encoding carboxyl ester lipase (Figure S1C); Cpb1, encoding carboxypeptidase B1 (Figure S1D); and Cela2a, encoding chymotrypsin-like elastase family member 2A (Figure S1E). Notably, Try5 exhibited significantly higher mRNA levels in peri-islet acinar cells compared to tele-islet acinar cells (Figures 1D, 1E, and S2). These elevated levels of Try5 in peri-islet acinar cells were not observed in age-matched control mice (Figures 1D, 1F, and S2). We observed a similar zonation of Try5 in peri-islet acinar cells in BKS.Cg-Dock7m+/+ Leprdb/J db/db mice, an additional insulin-resistant mouse model (Figures S3A–S3C). Our measurements thus indicate that peri-islet acinar cells carry a distinct zonated gene expression signature in db/db mice.
A Global Signature of Peri-islet Acinar Cells Obtained from Isolated Islets
We next sought to obtain a global view of acinar cell zonation in db/db mice. Islet isolation is a common technique for exploring the gene expression signatures of pancreatic endocrine cells (Jaafar et al., 2019; Neelankal John et al., 2018). The dissociation and extraction protocols used in this procedure could result in residual acinar cells. We hypothesized that such acinar cells would have remained attached to the islet capsule and would thus be enriched for peri-islet cells (Figure 2A). Therefore, exploring the differential gene expression between acinar transcripts in bulk-sequenced islets obtained from db/db mice and control mice could reveal additional zonated acinar genes.
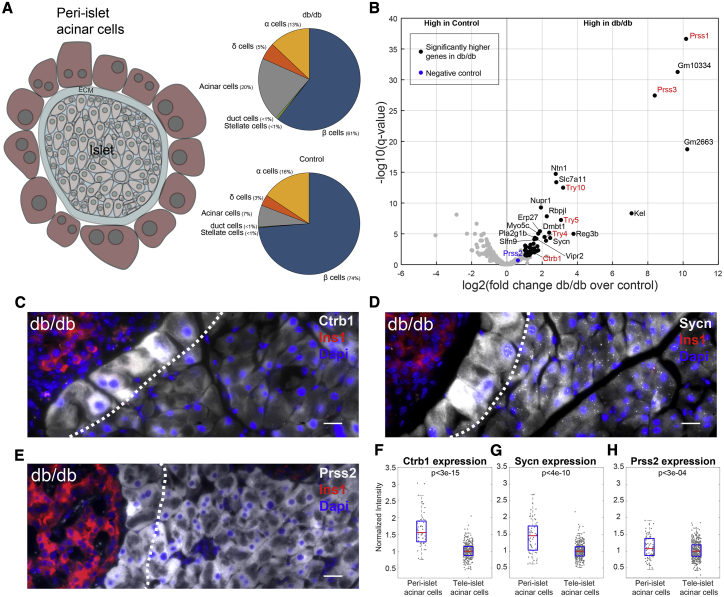
Figure 2.
Islet Isolation RNA-Seq Analysis Reveals a Zonated Acinar Cell Signature in db/db Mice
(A) Diagram demonstrating peri-islet acinar cells attached to an isolated islet. Pie charts show the relative fraction of cell-type-specific marker genes in Neelankal John et al. (2018) (see Method Details).
(B) Differential gene expression of acinar-specific genes reveals an upregulation of trypsin genes (red) in islets of db/db mice compared to control mice. The black dots mark genes with log2(fold change) > 1 and q < 0.05. Trypsin genes are in red, except Prss2 (blue), which is not significantly differentially expressed.
(C and D) smFISH images show higher levels of Ctrb1 (C) and Sycn (D) in peri-islet acinar cells compared to tele-islet acinar cells. Scale bars, 20 μm.
(E) smFISH image shows comparable levels of Prss2 in peri-islet acinar cells and tele-islet acinar cells.
(F–H) Quantification of smFISH images. Results are based on 10–16 islets from 5–6 mice. p < 3e-15 for Ctrb1, p < 4e-10 for Sycn, p < 3e-04 for Prss2.
Islets in (C)–(E) marked by Ins1 mRNA (red), nuclei stained by DAPI (blue), and the gene of interest in gray. Scale bars, 20 μm; dashed white lines demarcate peri-islet acinar cells. (C)–(E) are examples of the pancreatic tissue of 14- to 17-week-old db/db mice. The red lines in (F)–(H) are medians and the blue boxes are 25th–75th percentiles.
We analyzed datasets of bulk RNA sequencing (RNA-seq) extracted from islets isolated from db/db and control mice (Jaafar et al., 2019; Neelankal John et al., 2018). We used the Tabula Muris dataset (Tabula Muris Consortium, 2018) to assemble a panel of 50 cell-type-specific markers for the main pancreatic cell types—alpha cells, beta cells, delta cells, stellate cells, duct cells, and acinar cells (Table S2). We found that the extracted islets contained substantial representation of acinar transcripts (20% of the transcripts analyzed in db/db mice and 7% of the transcripts analyzed in control mice; Figure 2A).
We next performed differential gene expression analysis between db/db and control islets (Neelankal John et al., 2018) over acinar-specific genes and identified profound differences in acinar gene expression (Figure 2B; Table S3). Acinar genes induced in db/db mice included all of the trypsin gene family—Try5 (8.5-fold, q = 6e−8), Try4 (4.4-fold, q = 3.5e−5), Try10 (9.2-fold, q = 3.5e−13), Prss1 (1,162-fold, q = 2.5e−37), Prss3 (338-fold, q = 3.8e−28), and Ctrb1 (2.8-fold, q = 0.009)—except Prss2 (1.6-fold, q = 0.22). Other upregulated genes included the zymogen granule protein Sycn (5.5-fold, q = 5e−5). Analysis of a second RNA-seq dataset (Jaafar et al., 2019) revealed high overlap in the induced acinar genes (Figure S4A). Moreover, the peri-islet acinar expression program was highly similar when compared to the db/+ heterozygotes rather than the wild-type (WT) controls (Figure S4B). We validated and quantified the predicted upregulation of some of these genes in peri-islet acinar cells using smFISH, observing substantial upregulation of Ctrb1 and Sycn (>1.4-fold higher expression in peri-islet acinar cells; Figures 2C, 2D, 2F, and 2G). In contrast, Prss2 was expressed to only slightly (yet significant) higher levels in peri-acinar cells (1.07-fold; Figures 2E and 2H). Our analysis thus indicates that peri-islet acinar cells upregulate a global gene expression signature composed of trypsin genes.
Islet CCK Induces the Peri-islet Zonation Program
Which molecules could induce the peri-islet acinar zonation program we observed in db/db mice? To address this question, we sought to identify ligand molecules (Ramilowski et al., 2015) that are upregulated in db/db islets. We found that Cck, encoding the intestinal entero-endocrine hormone cholecystokinin, was the most highly induced ligand (653-fold induction in db/db mice, q = 1.3e−17; Figure 3A; Table S3). Similar increases in intra-islet Cck levels were previously shown in ob/ob mice (Lavine et al., 2010). Additional ligands that were significantly upregulated in the db/db mice islets included central matrisome components (Naba et al., 2016), such as collagens and Mmp genes (Figure 3A; Table S3). Using smFISH, we identified abundant expression of Cck in db/db islet cells (∼4% of islet cells positive for Cck in db/db mice and no detectable expression in control islets; Figures 3B and 3C). As expected, db/db islet cells also exhibited higher mRNA levels of the proliferation marker Mki67 compared to control islets (Figure 3D).
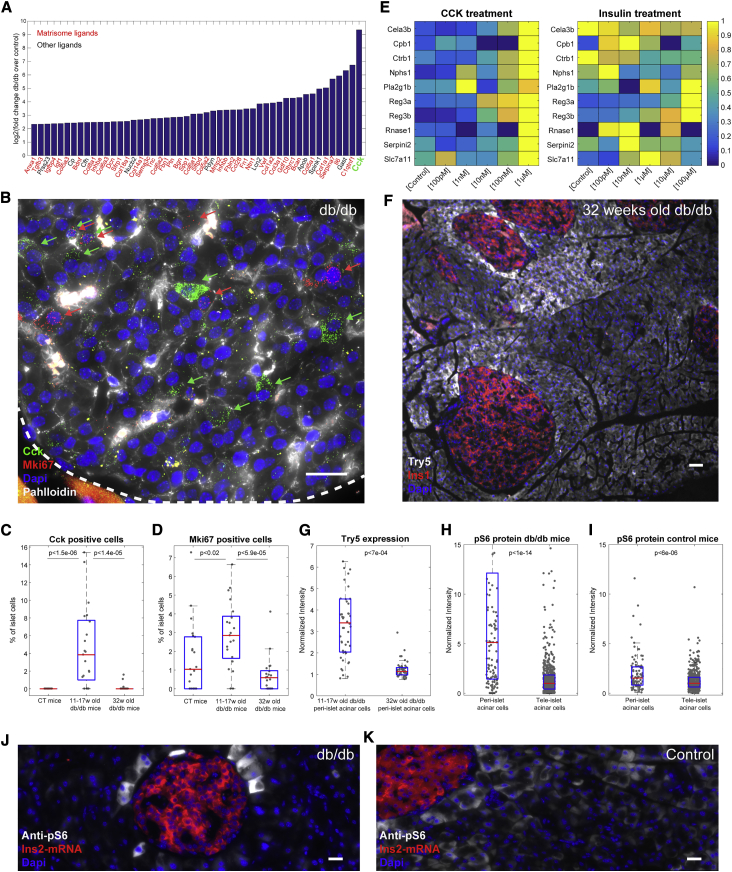
Figure 3.
Elevated Intra-islet Cck and Acinar Cell Zonation
(A) Bar plot of the fold change of the top 50 ligands between db/db islet and control islets. Cck is marked in green and the matrisome ligands are marked in red.
(B) smFISH image of Cck (green dots, positive cells marked by green arrows) and Mki67 (red dots, positive cells marked by red arrows) in db/db islets. The membrane is stained with phalloidin (gray) and the nuclei are stained by DAPI (blue). Scale bar, 20 μm.
(C and D) Quantification of the fractions of islet cells that are Cck positive (C, p < 1.5e-06 between control mice and young db/db mice, p < 1.4e-05 between young and old db/db mice) and Mki67 positive (D, p < 0.02 between control mice and young db/db mice, p < 5.9e-05 between young and old db/db mice). The data include 3 mice from each condition and 7 islets for each mouse.
(E) Heatmap of upregulated genes in CCK-exposed AR42J cells and insulin-exposed AR42J cells. Shown are all of the acinar genes that were induced by either CCK or insulin and also zonated in vivo. For each gene, the expression is normalized to the maximum across all of the hormone concentrations.
(F) smFISH staining of a 32-week-old db/db mouse, demonstrating the reduction in Try5 (gray) zonation. Islets are marked by Ins1 mRNA (red) and the nuclei are stained by DAPI (blue); scale bar, 50 μm.
(G) Peri-islet Try5 is significantly more zonated in young versus old db/db mice (p < 7e-04). Of 97 cells, 5 are above the maximal shown scale. The results are based on 9–10 islets from 3–5 mice.
(H and I) Quantification of peri-islet acinar cell zonation of anti-pS6 in db/db mice (H, p < 1e-14) and control mice (I, p < 6e-06). 39 of 683 cells (H) and 2 of 563 cells (I) are above the maximal shown scale. The results are based on 9–15 islets from 3–5 mice.
(J and K) Examples of islets in a db/db mouse (J) and a control mouse (K) stained with anti-pS6 (gray). Islets are marked by Ins2 mRNA (red) and the nuclei are stained by DAPI (blue); scale bar, 20 μm.
In (C), (D), and (G)–(I), the red lines are medians and the blue boxes are 25th–75th percentiles. (B), (J), and (K) are examples of the pancreatic tissue of 14- to 17-week-old db/db and control mice.
To mechanistically interrogate whether CCK can elicit the peri-islet acinar expression program, we incubated AR42J acinar rat cells (Rosewicz et al., 1992) with increasing levels of CCK or insulin in their medium and sequenced their transcriptomes. Systemic insulin concentrations are ∼174 pM (Garzilli and Itzkovitz, 2018) and are expected to be 100-fold higher locally within the peri-islet acinar cells (Unger and Cherrington, 2012). To mimic the high insulin levels that peri-islet acinar cells are exposed to, we therefore exposed cells to concentrations ranging from 100 pM to 100 μM (Figure 3E). These levels were much higher than the insulin levels in the growth medium (7 pM). We focused on acinar-specific genes that were also annotated in the rat genome. Of these 146 genes, 15 were induced by CCK, and 9 of the 15 were also elevated in peri-islet acinar cells in vivo (hypergeometric p = 0.0015; Figure 3E). This set included Ctrb1. In contrast, only 3 of the 11 acinar-specific genes that were induced by insulin in the AR42J cell line were elevated in vivo (hypergeometric p = 0.32; Figure 3E). Our experiments thus indicate that a significant subset of the db/db peri-islet acinar gene signature is transcriptionally induced by CCK.
We further examined acinar zonation and Cck islet expression in older db/db mice (32 weeks old). Unlike the younger db/db mice, Try5 zonation was significantly decreased in the older mice (p < 7e−4; Figures 3F, 3G, and S2). Notably, the older mice had significantly lower levels of both intra-islet Cck mRNA and Mki67 mRNA (p < 1.4e−5 for Cck and p < 5.9e−5 for Mki67; Figures 3C and 3D). The correlated decrease in islet Cck and peri-islet acinar zonation in older mice further suggest that Cck may be inductive for the peri-islet gene expression signature.
Our study identified increases in both peri-islet acinar cell size (Figures 1A and 1C) and the expression of genes encoding trypsin enzymes, which are massively translated and secreted. The mammalian target of rapamycin (mTOR) pathway has been shown to control both cell size (Fingar et al., 2002) and translation rates (Saxton and Sabatini, 2017). We therefore hypothesized that peri-islet acinar cells may exhibit elevated levels of mTOR signaling. We found that the levels of phosphorylated S6 ribosomal protein (pS6), a downstream target of mTOR complex 1 (mTORC1), were significantly higher in peri-islet acinar cells of 9- to 17-week-old db/db mice (5.1-fold, p < 1e−14; Figures 3H and 3J). This zonated mTORC1 activity was also apparent in control mice, although at a significantly less pronounced level (1.5-fold change, p < 6e−06; Figures 3I and 3K). Peri-islet acinar cells in db/db mice therefore exhibit a substantial increase in mTOR signaling.
Discussion
Our study uncovered a zonated gene expression signature in db/db mice consisting of a specific induction of trypsin genes in the layer of acinar cells that surrounds the islets of Langerhans. Previous studies of peri-islet acinar cells hypothesized that insulin that locally diffuses from islets within the pancreatic parenchyma would be the stimulator of peri-islet acinar cell phenotypes such as increased size and protein secretion (Trimble et al., 1985). In contrast, our study supports the role of CCK, a central secretagogue of acinar cells, as a facilitator of acinar cell zonation via three lines of evidence: (1) Cck was the most highly induced ligand in the db/db islets (Figures 3A–3C); (2) elevated CCK levels in acinar cell cultures elicit transcriptional changes that overlap the peri-islet acinar gene expression signature that we observed in vivo, an overlap that was not observed when treating cells with insulin (Figure 3E); and (3) a decline in intra-islet Cck expression in older mice coincided with a decrease in acinar zonation (Figures 3C, 3F, and 3G). Moreover, CCK is a potent activator of the mTOR pathway, as previously shown in AR42J cells (Inushima et al., 2001), which could also account for our observed increase in mTOR activity in peri-islet acinar cells in db/db mice (Figures 3H and 3J). Beta cell-secreted CCK has been shown to accelerate the progression of pancreatic ductal cancer in obese mice, supporting the ability of CCK to diffuse and affect the exocrine parenchyma (Chung et al., 2020). Notably, peri-islet acinar cell sizes (Figures 1B and 1C) and pS6 activity (Figures 3I and 3K) were also elevated in control mice compared to tele-islet acinar cells. These zonated patterns, at a stage when Cck is not expressed, indicate that other islet-secreted factors such as insulin may also be important regulators of peri-islet acinar cell identity. Future studies could apply cell-type-specific ablation of Cck in islet cells or its receptor genes in acinar cells under the db/db background to further establish the inductive role of CCK in peri-islet acinar cell zonation.
What could be the functional role of the CCK-induced peri-islet acinar zonation that we observed? Under normal physiology, intestinal Cck that is expressed by entero-endocrine cells stimulates the apical secretion of trypsin and other pancreatic acinar enzymes. These enzymes flow through pancreatic ducts into the small intestine, where they participate in protein digestion. Elevated levels of CCK have been shown to lead to mistrafficking of digestive enzymes, inhibition of apical secretion, and abnormal exocytosis redirected to the basolateral plasma membrane (Gaisano et al., 2001; Liang et al., 2017; Scheele et al., 1987; Chen, 2018). Such basal secretion of trypsin has been implicated in cellular damage during pancreatitis (Gaisano et al., 2001). Islets in insulin-resistant animals often expand in size (Chen et al., 2017) to meet the demands of increased insulin production. This increase entails changes in the expression of many matrisome components in the db/db islets (Figure 3A; Table S3). Basal secretion of trypsin from peri-islet acinar cells could facilitate these changes by cleaving capsule extracellular matrix (ECM) proteins (Figure 4). In line with this hypothesis, TAT, a trypsin-like protein, has been shown to contribute to the cell-mediated degradation of ECM in tumors (Koivunen et al., 1991). Trypsin and other proteases have been shown to act synergistically with collagenase in separating islets from the surrounding exocrine tissue (Wolters et al., 1992). Trypsin is also highly potent in dissociating islets and is commonly used in islet digestion protocols (Lu et al., 2017). These effects of trypsin could stem from their breakdown of collagen-binding proteoglycans and glycoproteins (Brandhorst et al., 2009). The mere change in ECM composition could also lead to the observed changes in peri-islet acinar gene expression, as matrix properties affect the gene expression of cells (Lelièvre, 2009). In contrast to this hypothesis, our ex vivo experiment, which assessed acinar cells in culture without the associated ECM, supports the direct role of CCK as the potential inducing factor.
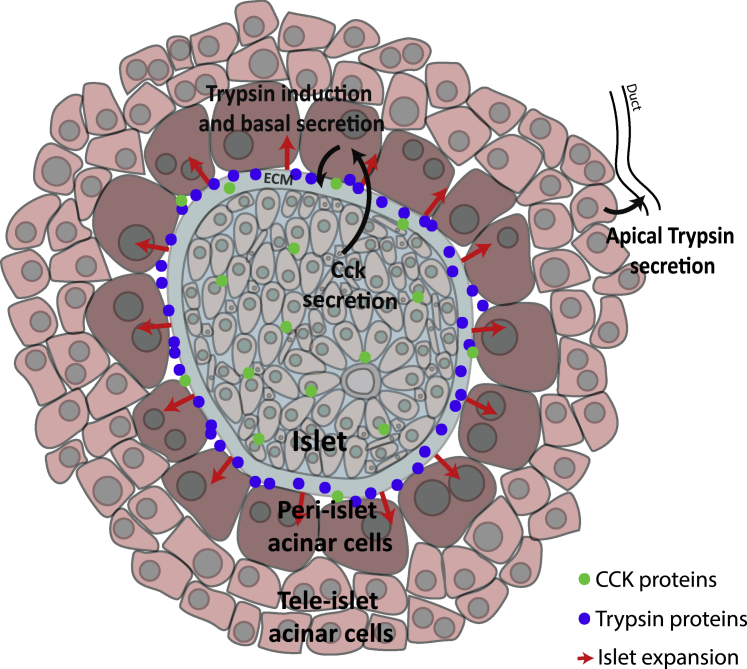
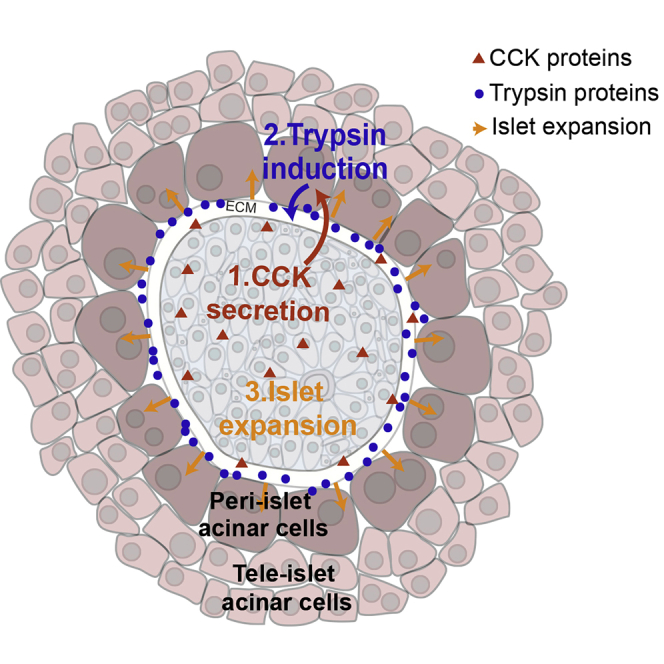
Figure 4.
Model for Acinar Cell Zonation in db/db Mice
Islet cells elevate CCK levels (green dots), which elicit a zonated increase in both the expression of trypsin genes in peri-islet acinar cells and their basal secretion (blue dots). Such induction could contribute to islet capsule ECM remodeling, potentially facilitating islet expansion (red arrows).
The decline in intra-islet Cck and acinar cell zonation in older db/db mice coincided with a decrease in intra-islet levels of the proliferation marker Mki67 (Figures 3C and 3D). Our study thus suggests an axis of communication whereby expanding islets induce Cck that locally diffuses and is sensed by peri-islet acinar cells. The CCK stimulation gives rise to both transcriptional induction of trypsin genes and, potentially, their basal secretion (Figure 4). It will be interesting to apply dynamic measurements of acinar cell secretion in pancreatic slices (Liang et al., 2017) to assess the ability of peri-islet acinar cells to secrete zymogen basally toward the islet capsule in the contexts of islet expansion studied here.
Zonation of gene expression has been demonstrated in several epithelial tissues such as the liver (Halpern et al., 2017), the intestine (Moor et al., 2018), and the kidney (Stewart et al., 2019). Our study demonstrates that the diabetic pancreas also exhibits zonated gene expression, possibly induced by islet-derived CCK. It will be important to explore whether similar zonation exists in other models of insulin resistance, such as high-fat diets and pregnancy, during pancreas development and in humans.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Phospho-S6 Ribosomal | Cell Signaling Technology | Cat. No. 4856; RRID: AB_2181037 |
| Goat anti-Rabbit IgG (Alexa Fluor 647) | Thermo Fisher | Cat. No. A-21245; RRID: AB_2535813 |
| Biological Samples | ||
| Fetal Bovine Serum | Biological Industries | Cat. No. 04-005-1A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| O.C.T. Compound Cryostat Embedding Medium | Scigen | Cat. No. 4586 |
| Poly-L-lysin solution | Sigma-Aldrich | Cat. No. P8920 |
| Formaldehyde, 37% (w/v) | J.T. Baker | Cat. No. JT2106 |
| PBS, pH 7.4, RNase-free, 10 × | Ambion | Cat. No. AM9625 |
| Water UltraPure Dnase/RNase-free Molecular Biology | Bio-Lab | Cat. No. 23217723 |
| Proteinase K | Ambion | Cat. No. AM2546 |
| Formamide, deionized, nuclease-free | Ambion | Cat. No. AM9342 |
| Dextran sulfate | Sigma-Aldrich | Cat. No. D8906 |
| E.coli tRNA, Roche | Sigma-Aldrich | Cat. No. R1753 |
| BSA | Ambion | Cat. No. AM2616 |
| Vanadyl- ribonucleoside complex | NEB | Cat. No. S1402S |
| Cy5 Mono-Reactive Dye Pack | Sigma-Aldrich | Cat. No. PA25001 |
| 6-TMRA, SE | Thermo Fisher | Cat. No. C6123 |
| Alexa Fluor 594 carboxylic acid, succinimidyl ester | Thermo Fisher | Cat. No. A20004 |
| DAPI | Sigma-Aldrich | Cat. No. D9542 |
| Alexa Fluor 488 Phalloidin | Thermo Fisher | Cat. No. A12379 |
| ProLong Gold Antifade Mountant | Molecular Probes | Cat. No. P36934 |
| CAS block | Thermo Fisher | Cat. No. 008120 |
| TRI reagent | Sigma-Aldrich | Cat. No. T9424 |
| (Tyr[SO3H]27) Cholecystokinin fragment 26-33 Amide | Sigma-Aldrich | Cat. No. C2175 |
| Insulin | Sigma-Aldrich | Cat. No. I1882 |
| Maxima H -RT | Thermo Fisher | Cat. No. EP0753 |
| dNTPs | Thermo Fisher | Cat. No. R0182 |
| PEG 8000 | Sigma-Aldrich | Cat. No. 89510 |
| Exonuclease I | Thermo Fisher | Cat. No. EN0582 |
| AM pure beads | Beckman Coulter | Cat. No. A63881 |
| Terra direct polymerase | Clontech | Cat. No. 639270 |
| Critical Commercial Assays | ||
| Direct-zol RNA Miniprep kit | Zymo Research | Cat. No. R2050 |
| Nextera XT DNA Library Prep Kit | Illumina | Cat. No. FC-131-1024 |
| NextSeq 500 Kits v2 (75 cycles) | Illumina | Cat. No. FC-404-2005 |
| Deposited Data | ||
| RNA sequencing of AR42J cells | This paper | GEO: GSE154533 |
| Islet isolation data | Neelankal John et al., 2018 | GEO: GSE107489 |
| Islet isolation data | Jaafar et al., 2019 | GEO: GSE132261 |
| Tabula Muris dataset | Tabula Muris Consortium, 2018 | https://tabula-muris.ds.czbiohub.org/; GEO: GSE109774 |
| Matrisome components data | Naba et al., 2016 | http://matrisomeproject.mit.edu/ |
| Ligand-receptor data | Ramilowski et al., 2015 | https://doi.org/10.1038/ncomms8866 |
| Experimental Models: Cell Lines | ||
| AR42J | James Dutton |
https://www.atcc.org/products/all/CRL-1492.aspx RRID:CVCL_0143 |
| Experimental Models: Organisms/Strains | ||
| B6.BKS(D)-Leprdb/J mice on C57BL/6J genetic background | The Jackson Laboratory | Strain: 000697 | B6 db |
| BKS.Cg-Dock7m +/+ Leprdb/J on C57BLKS/J genetic background | The Jackson Laboratory | Strain: 000642| BKS db |
| C57BL/6J mice | The Jackson Laboratory | Strain: 000664 | B6/J |
| C57BL/6 mice | Envigo | Strain: C57BL/6JOlaHsd |
| Oligonucleotides | ||
| smFISH probes, see Table S4 | This paper | N/A |
| SINGV6 primer: /5Biosg/ACA CTC TTT CCC TAC ACG ACG C | IDT (Standard Desalting) | N/A |
| barcoded RT primer | Bagnoli et al., 2018 (Standard Desalting) | N/A |
| TSO∗ E5V6NEXT: ACA CTC TTT CCC TAC ACG ACG CrGrG rG | IDT (RNA oligo RNase Free HPLC Purification) | N/A |
| P5NEXTPT5: AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG∗ A∗T∗C∗ T | IDT (HPLC purification) | N/A |
| Software and Algorithms | ||
| Stellaris FISH Probe Designer | Biosearch Technologies, Inc., Petaluma, CA | https://biosearchtech.com/products/rna-fish/ |
| R and RStudio v3.6.1 | R Consortium | https://rstudio.com/ |
| MATLAB R2016b | MathWorks | https://www.mathworks.com/ |
| ImageM | Lyubimova et al., 2013 | N/A |
| ImageJ 1.52d | Schindelin et al., 2012 | https://imagej.nih.gov/ij/; RRID:SCR_003070 |
| MetaMorph software | Molecular Devices, Downington, PA | https://www.biocompare.com/19333-Image-Analysis-Software-Image-Processing-Software/78845-MetaMorphreg-Microscopy-Automation-Image-Analysis-Software/ |
| NIS-Elements software | Nikon Instruments, Melville, NY | https://www.nikon.com/products/microscope-solutions/lineup/img_soft/nis-elements/ |
| edgeR version 3.26.8 | Robinson et al., 2010 | N/A |
| bcl2fastq | Illumina | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html RRID: SCR_015058 |
| zUMI package | Parekh et al., 2018 | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shalev Itzkovitz (shalev.itzkovitz@weizmann.ac.il).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The accession number for the data reported in this study is GEO: GSE154533.
Experimental Model and Subject Details
Animal experiments
All animal studies were approved by the Institutional Animal Care and Use committee of Weizmann Institute of Science. All C57BL/6 male mice were purchased from Envigo and Jackson Laboratory. Homozygous B6.BKS(D)-Leprdb/J (db/db) male and female mice were purchased from Jackson Laboratory. Homozygous BKS.Cg-Dock7m +/+ Leprdb/J (db/db) male mice were purchased from Jackson Laboratory and used for Figure S3 only. All mice were fed with normal chow ad libitum except the mouse in Figure S1A, which was fasted for 9 hours prior to sacrifice. Mice were sacrificed by cervical dislocation. All mice were allowed to acclimatize to the animal facility environment for at least 7 days before being used for experimentation.
B6.BKS(D)-Leprdb/J mice on C57BL/6J genetic background were sacrificed at ages 9-17 weeks and 32 weeks. C57BL/6J were used as controls. BKS.Cg-Dock7m +/+ Leprdb/J mice on C57BLKS/J genetic background were sacrificed at age 14 weeks. Body weight and blood glucose level for db/db and control mice are shown in Table S1.
Cell lines
AR42J male cells were used for culture experiment. Cells were thawed and let to recover in flasks at 37°C for a week before exposed to CCK or insulin. Growth medium used was low-glucose DMEM, 10% FBS, 1% Pen/strep, 1% NEAA, 1% Glutamine.
Method Details
SmFISH and imaging
smFISH was performed with a modified smFISH protocol that was optimized for pancreatic tissues as described in Farack et al. (2019), with an extended period of post fixation of 1 hour instead of 15 minutes. In brief, mice were sacrificed and their pancreas was extracted and rinsed with 1XPBS. Pancreas was fixed in 4% Formaldehyde (FA, J.T. Baker, JT2106) in PBS for 3 hours and subsequently agitated in 30% sucrose, 4% FA in PBS overnight at 4°C. Fixed tissues were embedded in OCT (Scigen, 4586). 5-7μm thick sections of fixed pancreas were sectioned onto poly L-lysine coated coverslips and fixed again in 4% FA in PBS for 1 hour followed by 70% Ethanol dehydration for 2h in 4°C. Tissues were treated for 10 min with proteinase K (10 μg/ml Ambion AM2546) and washed twice with 2 × SSC (Ambion AM9765). Tissues were incubated in wash buffer (30% Formamide Ambion AM9342, 2 × SSC) for 3-5 hours and mounted with the hybridization buffer (10% Dextran sulfate Sigma D8906, 30% Formamide, 1 mg/ml E.coli tRNA Sigma R1753, 2 × SSC, 0.02% BSA Ambion AM2616, 2 mM Vanadyl-ribonucleoside complex NEB S1402S) mixed with 1:3000 dilution of probes. Hybridization mix was incubated with tissues for overnight in a 30°C incubator. SmFISH probe libraries (Table S4) were coupled to Cy5, TMR or Alexa594. After the hybridization, tissues were washed with wash buffer containing 50 ng/ml DAPI (Sigma-Aldrich, D9542) for 15 min at 30 °C followed by GLOX buffer (0.4% Glucose, 1% Tris, 10% SSC) containing Phalloidin conjugated to Alexa Fluor 488 (1:500, Thermo Fisher, A12379) for 20 minutes for cell-membrane staining. Probe libraries were designed using the Stellaris FISH Probe Designer Software (Biosearch Technologies, Inc., Petaluma, CA), see Table S4.
Imaging was performed on either Nikon-Ti-E inverted fluorescence microscopes equipped with 100x and 60x oil-immersion objectives and a Photometrics Pixis 1024 CCD camera or Nikon eclipse Ti2 inverted fluorescence microscopes equipped with 100x and 60x oil-immersion objectives and a Photometrics Prime 95B 25MM EMCCD camera. Image stacks were collected with a z spacing of 0.3μm. Image stitching was performed with the fusion mode linear blending and default settings of the pairwise stitching plugin in Fiji (Preibisch et al., 2009; Schindelin et al., 2012). Quantification of smFISH was done using ImageM (Lyubimova et al., 2013).
Immunofluorescence (IF) combined with smFISH
smFISH was performed as described in the section above. At the second day of the protocol before DAPI staining, the tissues were washed twice with 30% WB for 15 minutes at room temperature, then washed 3 times with 1xPBS each for 5 minutes. Blocking using CAS-Block (Blocking solution, Thermo Fisher, 008120) was performed for 80 minutes at room temperature. Tissues were incubated with anti-Phospho-S6 antibody (1:100, Cell Signaling Technology 4856) in blocking solution for 2h at room temperature. Tissues were washed 3 times with 1xPBS each for 5 minutes and incubated with secondary antibody conjugated to Alexa Fluor 647 (1:200, Thermo Fisher, A-21245) for 45 minutes at room temperature. Tissues were next washed 3 times with 1xPBS each for 5 minutes. Cell borders were marked with Phalloidin conjugated to alexa fluor 488 (1:500) and nuclei were stained with 50 ng/ml DAPI before mounting. Imaging was carried out using the same setting as for the smFISH experiments.
AR42J cell line experiment
AR42J cells were obtained from James Dutton. Cells were thawed and let to recover for a week before exposed to CCK or insulin. Growth medium used was low-glucose DMEM, 10% FBS, 1% Pen/strep, 1% NEAA, 1% Glutamine. CCK (Sigma-Aldrich, C2175) was suspended to stock concentration (0.5mM) in NH4OH as specified in manufacturer instruction. Insulin (Sigma-Aldrich, I1882) was suspended to stock concentration (1.7mM) with 0.01 acetic acid in water as specified in manufacturer instruction. Stocks were diluted to working concentrations in growth medium. Cells were split into 6 well plates and growth medium with the different concentrations of CCK/insulin was added in triplicates after 24 hours. After additional 24 hours, cells were lysed in TRI reagent (Sigma-Aldrich, T9424) for RNA extraction. RNA was isolated by Direct-zol RNA MiniPrep kit (Zymo Research, R2050) according to the manufacturer instructions and processed with the mcSCRBseq protocol with minor modifications (Bagnoli et al., 2018). RT reaction was performed on 20ng of total RNA with a final volume of 10μl (1x Maxima H Buffer, 1mM dNTPs, 2μM TSO∗ E5V6NEXT, 7.5% PEG8000, 20U Maxima H enzyme, 1μl barcoded RT primer). Amplification step was 14 PCR cycles. Subsequent steps were applied as mentioned in the protocol. Library preparation was performed using Nextera XT kit (Illumina, FC-131-1024) on 0.6ng amplified cDNA. Library final concentration of 2.2pM was loaded on NextSeq 500 (Illumina, FC-404-2005) sequencing machine with the following cycle distribution: 16bp read1, 66bp read2 with a barcode (i7) length of 8bp. Raw files were converted to FASTQ files using bcl2fastq package (Illumina). To obtain the UMI counts, fastq reads were aligned to the rat reference genome (Rnor_6.0.96) using zUMI package (Parekh et al., 2018). Normalized expression tables were obtained by dividing the UMI counts for each gene by the summed UMI counts over all samples.
Quantification and Statistical Analysis
smFISH and IF quantification
In the imaging experiments, peri-islet cells were defined as the first layer of cells surrounding the islet. For each imaging field (islet), Wilcoxon rank-sum tests were used for comparing size / expression among peri-islet and tele-islet cells. Fisher method was used to combine p values among different islets. SmFISH expression values for each imaging field were divided by the median tele-islet cell expression in the field. smFISH size and intensity quantifications (Figures 1C, 1D, and 2F–2H) results were based on 10-16 islet stitches from 5-6 mice. pS6 intensity quantifications (Figures 3H and 3I) results were based on 9-15 islet stitches from 3-5 mice. Cck and Mki67 positive cells analysis was based on 21 islets from 3 mice for each condition (Figures 3C and 3D). Try5 quantifications on old versus young db/db mice (Figure 3G) were based on 9-10 islet stitches from 3-5 mice. Try5 quantifications on BKS.Cg-Dock7m +/+ Leprdb/J db/db mice (Figure S3C) were based on 12 islet stitches from 3 mice. Pixel intensities were quantified over 3 μm of the Z stacks, and divided by the segmented cell volume to obtain the mRNA concentration per cell. Intensities were background-subtracted using islet cells. Probe sequences used in this study are described in Table S4. Cck and Mki67 positive cell (Figures 3C and 3D) were defined as cells with more than two cytoplasmic mRNA molecules. For all analysis, arbitrary islets above 70μm in diameter were selected for control mice, and arbitrary islets above 120μm in diameter were selected for B6.BKS(D)-Leprdb/J db/db mice and islets above 100μm in diameter were picked for BKS.Cg-Dock7m +/+ Leprdb/J db/db mice (since islets are smaller in this strain).
Islet isolation bulk RNA-Seq analysis
Bulk islet RNaseq data was taken from Neelankal John et al. (2018) (Figures 2A, 2B, 3A, and S4B) and from Jaafar et al. (2019) (Figure S4A). The tabula Muris dataset (Tabula Muris Consortium, 2018) was used to define genes that are specific to acinar cells. Acinar specific genes were defined as genes with 3-fold higher expression in acinar cells compared to the maximum expression in all other pancreatic cell types (beta, alpha, delta, stellate and ductal cells). Differential gene expression between db/db and control islet transcriptomes were performed with edgeR version 3.26.8 (Robinson et al., 2010). Genes with less than 10 counts were removed. Q-values were computed using the Benjamini-Hochberg false discovery rate correction (Benjamini and Hochberg, 1995). This analysis was performed on all genes (Table S3) as well as on the acinar-specific genes (Figure 2B; Table S3). Ligand and matrisome gene data (Figure 3A; Table S3) were extracted from Table S3 of all genes. To estimate the degree of representation of non-islet transcripts we examined the islet RNaseq expression of a concise subset of 50 cell-type specific genes for each of the 6 cell types considered (Figure 2A; Table S2). To this end, we identified the genes in Tabula Muris with mean expression above 10−4 of cellular transcriptome, ranked the ratio between the mean expression in the cell type of interest and the maximal mean expression in any other cell type and maintained the top 50 genes. We computed the relative fraction of the summed expression of each cell-type specific gene set. Tabula Muris dataset was normalized to the sum of UMIs.
AR42J cell line sequencing analysis
In the AR42J experiment, genes induced by CCK or insulin were defined as genes with maximal expression level observed in the highest hormone dose, that was above 10−6 of all transcripts and at least 2-fold higher than the expression with no stimulation. Hypergeometric tests were used to compute the overlap between the genes induced by either hormone and the genes induced in peri-islet cells, defined as acinar genes with levels in db/db islets that were at least 1.5-fold higher than in controls, with q-values < 0.1.
Acknowledgments
We thank Yuval Dor and Michael Walker for valuable discussions. S.I. is supported by the Wolfson Family Charitable Trust, the Edmond de Rothschild Foundations, the Fannie Sherr Fund, the Helen and Martin Kimmel Institute for Stem Cell Research grant, Israel Science Foundation grant no. 1486/16, Broad Institute-Israel Science Foundation grant no. 2615/18, European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program grant no. 768956, Chan Zuckerberg Initiative grant no. CZF2019-002434, the Bert L. and N. Kuggie Vallee Foundation, and the Howard Hughes Medical Institute (HHMI) international research scholar award.
Author Contributions
S.I. and A.E. conceived the study. A.E. designed and performed most of the experiments. A.E., K.B.H., and L.F. performed the smFISH experiments and image analysis. A.E. and H.R. performed the cell line experiment. A.E. and S.I. performed the data analysis. S.I. and A.E. wrote the manuscript. All of the authors discussed the results and commented on the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: August 18, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108043.
Supplemental Information
Shown are the 50 genes with the highest fold-change in expression for each cell type. Related to Figure 2.
Table contains the ratio between db/db and control islets, q-value, p value, and counts for all samples (3 controls and 3 db/db). Data taken from Neelankal John et al. (2018). Related to Figures 2 and 3.
References
- Adelson J.W., Miller P.E. Heterogeneity of the exocrine pancreas. Am. J. Physiol. 1989;256:G817–G825. doi: 10.1152/ajpgi.1989.256.5.G817. [DOI] [PubMed] [Google Scholar]
- Anzi S., Stolovich-Rain M., Klochendler A., Fridlich O., Helman A., Paz-Sonnenfeld A., Avni-Magen N., Kaufman E., Ginzberg M.B., Snider D. Postnatal Exocrine Pancreas Growth by Cellular Hypertrophy Correlates with a Shorter Lifespan in Mammals. Dev. Cell. 2018;45:726–737.e3. doi: 10.1016/j.devcel.2018.05.024. [DOI] [PubMed] [Google Scholar]
- Bagnoli J.W., Ziegenhain C., Janjic A., Wange L.E., Vieth B., Parekh S., Geuder J., Hellmann I., Enard W. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat. Commun. 2018;9:2937. doi: 10.1038/s41467-018-05347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- Brandhorst H., Friberg A., Andersson H.H., Felldin M., Foss A., Salmela K., Lundgren T., Tibell A., Tufveson G., Korsgren O., Brandhorst D. The importance of tryptic-like activity in purified enzyme blends for efficient islet isolation. Transplantation. 2009;87:370–375. doi: 10.1097/TP.0b013e31819499f0. [DOI] [PubMed] [Google Scholar]
- Chen X. 2018. Protein Composition and Biogenesis of the Pancreatic Zymogen Granules. American Pancreatic Association’s Pancreapedia.https://www.pancreapedia.org/reviews/protein-composition-and-biogenesis-of-pancreatic-zymogen-granules [Google Scholar]
- Chen C., Cohrs C.M., Stertmann J., Bozsak R., Speier S. Human beta cell mass and function in diabetes: recent advances in knowledge and technologies to understand disease pathogenesis. Mol. Metab. 2017;6:943–957. doi: 10.1016/j.molmet.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K.M., Singh J., Lawres L., Dorans K.J., Garcia C., Burkhardt D.B., Robbins R., Bhutkar A., Cardone R., Zhao X. Endocrine-Exocrine Signaling Drives Obesity-Associated Pancreatic Ductal Adenocarcinoma. Cell. 2020;181:832–847.e18. doi: 10.1016/j.cell.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman D.L. Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia. 1978;14:141–148. doi: 10.1007/BF00429772. [DOI] [PubMed] [Google Scholar]
- Farack L. Protocol for Single-Molecule Fluorescence In Situ Hybridization for Intact Pancreatic Tissue. STAR Protocols. 2020;1 doi: 10.1016/j.xpro.2019.100007. https://www.sciencedirect.com/science/article/pii/S2666166719300073?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farack L., Golan M., Egozi A., Dezorella N., Bahar Halpern K., Ben-Moshe S., Garzilli I., Tóth B., Roitman L., Krizhanovsky V., Itzkovitz S. Transcriptional Heterogeneity of Beta Cells in the Intact Pancreas. Dev. Cell. 2019;48:115–125.e4. doi: 10.1016/j.devcel.2018.11.001. [DOI] [PubMed] [Google Scholar]
- Fingar D.C., Salama S., Tsou C., Harlow E., Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaisano H.Y., Lutz M.P., Leser J., Sheu L., Lynch G., Tang L., Tamori Y., Trimble W.S., Salapatek A.M. Supramaximal cholecystokinin displaces Munc18c from the pancreatic acinar basal surface, redirecting apical exocytosis to the basal membrane. J. Clin. Invest. 2001;108:1597–1611. doi: 10.1172/JCI9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzilli I., Itzkovitz S. Design principles of the paradoxical feedback between pancreatic alpha and beta cells. Sci. Rep. 2018;8:10694. doi: 10.1038/s41598-018-29084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran U., Gannon M. Type 2 diabetes and the aging pancreatic beta cell. Aging (Albany NY) 2011;3:565–575. doi: 10.18632/aging.100350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern K.B., Shenhav R., Matcovitch-Natan O., Tóth B., Lemze D., Golan M., Massasa E.E., Baydatch S., Landen S., Moor A.E. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman B., Wallgren A., Petersson B. Cytological characteristics of the exocrine pancreatic cells with regard to their position in relation to the islets of Langerhans. A study in normal and obese-hyperglycaemic mice. Acta Endocrinol. (Copenh.) 1962;39:465–473. doi: 10.1530/acta.0.0390465. [DOI] [PubMed] [Google Scholar]
- Hummel K.P., Dickie M.M., Coleman D.L. Diabetes, a new mutation in the mouse. Science. 1966;153:1127–1128. doi: 10.1126/science.153.3740.1127. [DOI] [PubMed] [Google Scholar]
- Inushima K., Okabayashi Y., Sakaguchi K., Matsumura Y., Kimura S., Inoue Y., Kasuga M. Cholecystokinin activation of 70-kDa S6 kinase in exocrine pancreas. Dig. Dis. Sci. 2001;46:1437–1443. doi: 10.1023/a:1010683703232. [DOI] [PubMed] [Google Scholar]
- Jaafar R., Tran S., Shah A.N., Sun G., Valdearcos M., Marchetti P., Masini M., Swisa A., Giacometti S., Bernal-Mizrachi E. mTORC1 to AMPK switching underlies β-cell metabolic plasticity during maturation and diabetes. J. Clin. Invest. 2019;129:4124–4137. doi: 10.1172/JCI127021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen E., Ristimäki A., Itkonen O., Osman S., Vuento M., Stenman U.H. Tumor-associated trypsin participates in cancer cell-mediated degradation of extracellular matrix. Cancer Res. 1991;51:2107–2112. [PubMed] [Google Scholar]
- Lavine J.A., Raess P.W., Stapleton D.S., Rabaglia M.E., Suhonen J.I., Schueler K.L., Koltes J.E., Dawson J.A., Yandell B.S., Samuelson L.C. Cholecystokinin is up-regulated in obese mouse islets and expands beta-cell mass by increasing beta-cell survival. Endocrinology. 2010;151:3577–3588. doi: 10.1210/en.2010-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelièvre S.A. Contributions of extracellular matrix signaling and tissue architecture to nuclear mechanisms and spatial organization of gene expression control. Biochim. Biophys. Acta. 2009;1790:925–935. doi: 10.1016/j.bbagen.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T., Dolai S., Xie L., Winter E., Orabi A.I., Karimian N., Cosen-Binker L.I., Huang Y.-C., Thorn P., Cattral M.S., Gaisano H.Y. Ex vivo human pancreatic slice preparations offer a valuable model for studying pancreatic exocrine biology. J. Biol. Chem. 2017;292:5957–5969. doi: 10.1074/jbc.M117.777433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S.C., Tikhonov M., Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 2013;154:789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Yu Y., Ungrin M. Enhancing the efficiency of human pancreatic islet dissociation. J. Undergrad. Res. Alberta. 2017;6:25–28. [Google Scholar]
- Lyubimova A., Itzkovitz S., Junker J.P., Fan Z.P., Wu X., van Oudenaarden A. Single-molecule mRNA detection and counting in mammalian tissue. Nat. Protoc. 2013;8:1743–1758. doi: 10.1038/nprot.2013.109. [DOI] [PubMed] [Google Scholar]
- Malaisse-Lagae F., Ravazzola M., Robberecht P., Vandermeers A., Malaisse W.J., Orci L. Exocrine pancreas: evidence for topographic partition of secretory function. Science. 1975;190:795–797. doi: 10.1126/science.1105788. [DOI] [PubMed] [Google Scholar]
- Moor A.E., Harnik Y., Ben-Moshe S., Massasa E.E., Rozenberg M., Eilam R., Bahar Halpern K., Itzkovitz S. Spatial Reconstruction of Single Enterocytes Uncovers Broad Zonation along the Intestinal Villus Axis. Cell. 2018;175:1156–1167.e15. doi: 10.1016/j.cell.2018.08.063. [DOI] [PubMed] [Google Scholar]
- Naba A., Clauser K.R., Ding H., Whittaker C.A., Carr S.A., Hynes R.O. The extracellular matrix: tools and insights for the “omics” era. Matrix Biol. 2016;49:10–24. doi: 10.1016/j.matbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelankal John A., Ram R., Jiang F.-X. RNA-Seq Analysis of Islets to Characterise the Dedifferentiation in Type 2 Diabetes Model Mice db/db. Endocr. Pathol. 2018;29:207–221. doi: 10.1007/s12022-018-9523-x. [DOI] [PubMed] [Google Scholar]
- Nielsen J.H., Galsgaard E.D., Møldrup A., Friedrichsen B.N., Billestrup N., Hansen J.A., Lee Y.C., Carlsson C. Regulation of beta-cell mass by hormones and growth factors. Diabetes. 2001;50(Suppl 1):S25–S29. doi: 10.2337/diabetes.50.2007.s25. [DOI] [PubMed] [Google Scholar]
- Parekh S., Ziegenhain C., Vieth B., Enard W., Hellmann I. zUMIs - a fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience. 2018;7:giy059. doi: 10.1093/gigascience/giy059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S., Saalfeld S., Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B., Forrest A.R. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosewicz S., Riecken E.O., Wiedenmann B. The amphicrine pancreatic cell line AR42J: a model system for combined studies on exocrine and endocrine secretion. Clin. Investig. 1992;70:205–209. doi: 10.1007/BF00184652. [DOI] [PubMed] [Google Scholar]
- Sakhneny L., Rachi E., Epshtein A., Guez H.C., Wald-Altman S., Lisnyansky M., Khalifa-Malka L., Hazan A., Baer D., Priel A. Pancreatic Pericytes Support β-Cell Function in a Tcf7l2-Dependent Manner. Diabetes. 2018;67:437–447. doi: 10.2337/db17-0697. [DOI] [PubMed] [Google Scholar]
- Saxton R.A., Sabatini D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele G., Adler G., Kern H. Exocytosis occurs at the lateral plasma membrane of the pancreatic acinar cell during supramaximal secretagogue stimulation. Gastroenterology. 1987;92:345–353. doi: 10.1016/0016-5085(87)90127-2. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart B.J., Ferdinand J.R., Young M.D., Mitchell T.J., Loudon K.W., Riding A.M., Richoz N., Frazer G.L., Staniforth J.U.L., Vieira Braga F.A. Spatiotemporal immune zonation of the human kidney. Science. 2019;365:1461–1466. doi: 10.1126/science.aat5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris Consortium Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosti L., Hang Y., Trefzer T., Steiger K., Ten F.W., Lukassen S., Ballke S., Kuehl A.A., Spieckermann S., Bottino R. Single nucleus RNA sequencing maps acinar cell states in a human pancreas cell atlas. bioRxiv. 2019 doi: 10.1101/733964. [DOI] [PubMed] [Google Scholar]
- Trimble E.R., Bruzzone R., Gjinovci A., Renold A.E. Activity of the insulo-acinar axis in the isolated perfused rat pancreas. Endocrinology. 1985;117:1246–1252. doi: 10.1210/endo-117-3-1246. [DOI] [PubMed] [Google Scholar]
- Unger R.H., Cherrington A.D. Glucagonocentric restructuring of diabetes: a pathophysiologic and therapeutic makeover. J. Clin. Invest. 2012;122:4–12. doi: 10.1172/JCI60016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.W., Swartz F.J. Changes in polyploidization of exocrine pancreas in db/db diabetic and normal mice. Acta Endocrinol. (Copenh.) 1980;94:523–528. doi: 10.1530/acta.0.0940523. [DOI] [PubMed] [Google Scholar]
- Wolters G.H., Vos-Scheperkeuter G.H., van Deijnen J.H., van Schilfgaarde R. An analysis of the role of collagenase and protease in the enzymatic dissociation of the rat pancreas for islet isolation. Diabetologia. 1992;35:735–742. doi: 10.1007/BF00429093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shown are the 50 genes with the highest fold-change in expression for each cell type. Related to Figure 2.
Table contains the ratio between db/db and control islets, q-value, p value, and counts for all samples (3 controls and 3 db/db). Data taken from Neelankal John et al. (2018). Related to Figures 2 and 3.
Data Availability Statement
The accession number for the data reported in this study is GEO: GSE154533.