Abstract
Background
MNS blood group system genes GYPA and GYPB share a high degree of sequence homology and gene structure. Homologous exchanges between GYPA and GYPB form hybrid genes encoding hybrid glycophorins GP(A-B-A) and GP(B-A-B). Over 20 hybrid glycophorins have been characterised. Each has a distinct phenotype defined by the profile of antigens expressed including Mi<sup>a</sup>. Seven hybrid glycophorins carry Mi<sup>a</sup> and have been reported in Caucasian and Asian population groups. In Australia, the population is diverse; however, the prevalence of hybrid glycophorins in the population has never been determined. The aims of this study were to determine the frequency of Mi<sup>a</sup> and to classify Mi<sup>a</sup>-positive hybrid glycophorins in an Australian blood donor population.
Method
Blood samples from 5,098 Australian blood donors were randomly selected and screened for Mi<sup>a</sup> using anti-Mi<sup>a</sup> monoclonal antibody (CBC-172) by standard haemagglutination technique. Mi<sup>a</sup>-positive red blood cells (RBCs) were further characterised using a panel of phenotyping reagents. Genotyping by high-resolution melting analysis and DNA sequencing were used to confirm serology.
Result
RBCs from 11/5,098 samples were Mi<sup>a</sup>-positive, representing a frequency of 0.22%. Serological and molecular typing identified four types of Mi<sup>a</sup>-positive hybrid glycophorins: GP.Hut (n = 2), GP.Vw (n = 3), GP.Mur (n = 5), and 1 GP.Bun (n = 1). GP.Mur was the most common.
Conclusion
This is the first comprehensive study on the frequency of Mi<sup>a</sup> and types of hybrid glycophorins present in an Australian blood donor population. The demographics of Australia are diverse and ever-changing. Knowing the blood group profile in a population is essential to manage transfusion needs.
Keywords: Mia (MNS7) antigen, MNS hybrid glycophorins, MNS blood group system, Miltenberger, Blood group antigen, Blood group genotyping
Introduction
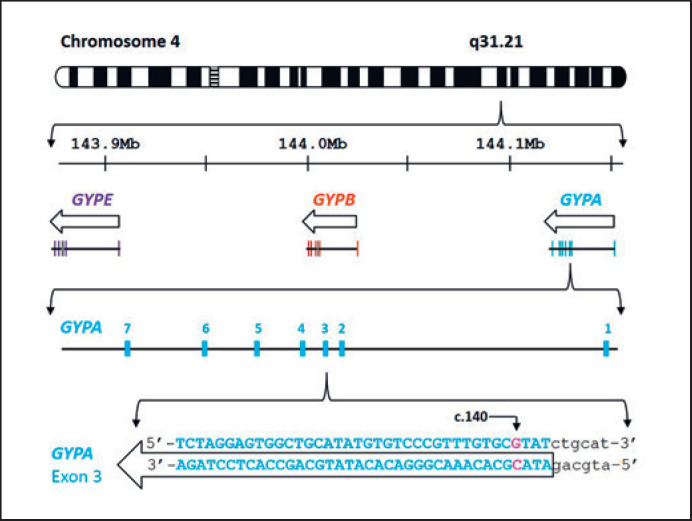
The glycophorin genes of the MNS blood group system are clustered in a 350-kb region on chromosome 4q28–q31 (Fig. 1) [1, 2, 3]. Glycophorin genes GYPA, GYPB, and GYPE encode GPA, GPB, and GPE, respectively, and share more than 95% sequence homology and gene structure [4, 5]. Homologous exchanges between GYPA and GYPB result in the formation of GYP(A-B-A) and GYP(B-A-B) hybrid genes and encode glycophorin molecules expressed on the red blood cell (RBC) surface [6, 7, 8]. These hybrid glycophorins display a distinct phenotype defined by a profile of antigens including Mia [6, 7, 8].
Fig. 1.
Schematic diagram of chromosome 4 showing the location and arrangement of GYPA, GYPB,and GYPE genes. Based on the GRCh38/hg38 assembly, the arrangement and location of the MNS blood group gene cluster is located on Chr4 q31.21 [2, 3]. The GenBank NG_007470 reference sequence was used as the basis for GYPA exon 3 sequence [37]. The nucleotide position c.140 is shown on GYPA exon 3. The molecular basis for GYP*Hut is c.140C>A (p.Thr47Lys) and that for GYP*Vw is c.140C>T (p.Thr47Met). Arrows above show the direction of transcription.
Mia (MNS7) is an antigen present in 7 hybrid glycophorins, namely: GP.Vw, GP.Hut, GP.HF, GP.Mur, GP.Hop, GP.Bun, and GP.Kip [7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17]. The distribution of these glycophorins varies between population groups. GP.Vw is found more commonly in Caucasians (up to 1.4% in south-east Switzerland) [18] while GP.Mur is more frequent in Asian populations – Malaysians (2.8%) [19], Indians (3.0%) [19], Chinese (6.5%) [20], Vietnamese (6.5%) [21], Filipinos (7.6%) [22], and Ami Taiwanese (88%) [23]. Over a quarter (26%) of Australia's population were born overseas, and several case studies have reported the presence of MNS hybrid glycophorins in Australian individuals [16, 24, 25, 26]. Antigens expressed on hybrid glycophorins are immunogenic and may stimulate an immune response when exposed to individuals who do not carry these antigens [6, 7].
In Australia, a limited number of haemolytic transfusion reaction and haemolytic disease of the fetus and newborn cases have been reported due to antibodies against hybrid glycophorins likely stimulated by exposure to GP.Vw and GP.Mur RBCs [7]. However, there are no comprehensive data on the occurrence of Mia and the type of MNS hybrid glycophorins present in the current Australian population. The aims of the study were to determine the prevalence of Mia and to categorise Mia-positive hybrid glycophorin variants in an Australian blood donor population.
Methods
Blood Donor Samples
Blood samples from volunteer Australian blood donors in Queensland were randomly selected for this study between January 2011 and July 2013. A total of 5,098 blood samples were screened for Mia using monoclonal antibody anti-Mia CBC-172. For blood donors identified carrying the Mia antigen, an extra collection of 6-mL EDTA-whole blood sample was requested on their subsequent blood donation for DNA analysis.
RBC Preparation and Genomic DNA Isolation
RBCs from EDTA whole blood samples were washed with PBS and then suspended in PBS to a concentration of 3–5% for use in a standard haemagglutination test (tube method). For molecular biology testing, genomic DNA was extracted from EDTA whole blood samples using a DNA extraction kit (EZ1 DNA Blood 350µL kit; QIAGEN) in a robotic equipment (BioRobot EZ1 Workstation; QIAGEN) according to the manufacturer's instructions. Isolated DNA was quantitated and quality-checked on a spectrophotometer (NanoDrop 2000c; Thermo Fisher Scientific).
Phenotyping Reagents
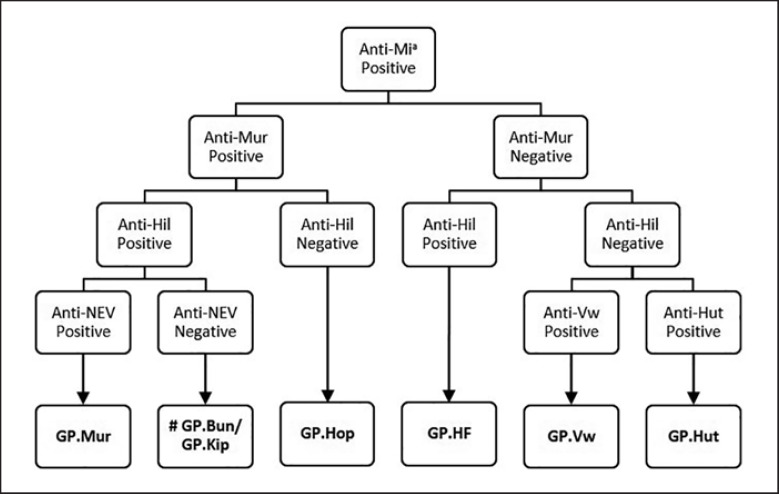
All Mia-positive RBCs were characterised by serology using a panel of typing antisera (monoclonal and polyclonal antibodies). The list of phenotyping reagents and the haemagglutination techniques applied are shown in Table 1. To further classify Mia-positive RBCs, an algorithm was developed to test RBCs sequentially for the presence of low-frequency antigens. The testing algorithm is presented in Figure 2.
Table 1.
Phenotyping reagents
| Antibody | Clone/ID | Technique | Source | |
|---|---|---|---|---|
| Anti-Mia | CBC-172 | Monoclonal, IgG | IAT | JRC |
| Anti-Mur | HIRO-138 (64-D6) | Monoclonal, IgM | SRT | JRC |
| Anti-Hil | Human | Polyclonal, IgG | IAT | JRC |
| Anti-NEV | CBC-181 (64-2A3) | Monoclonal, IgG | IAT | JRC |
| Anti-Vw | Human | Polyclonal, IgG | IAT | ARCBS |
| Anti-Hut | Human | Polyclonal, IgG | IAT | SCARF |
| Anti-Hop(+Nob) | Human | Polyclonal, IgG | IAT | JRC |
| Anti-Mia | GAMA210 | Monoclonal, IgG | SRT | Immucor |
Anti-Hop(+Nob), Anek anti-serum, is known to react with Mia-positive GP.Hop, GP.Bun, and GP.Kip RBCs. JRC, Japanese Red Cross; ARCBS, Australian Red Cross Blood Service; SCARF, Serum, Cells and Rare Fluids (Exchange program); IAT, indirect anti-globulin test; SRT, saline room temperature.
Fig. 2.
Phenotyping algorithm for classifying Mia-positive MNS hybrid glycophorins using a panel of typing sera. # Phenotyping reagents used above cannot differentiate GP.Bun from GP.Kip RBCs. However, anti-Hop antiserum reacts positive with GP.Bun and negative with GP.Kip RBCs [14, 16, 38]. Anti-Hop antiserum is rare and is only available in specialised or reference testing laboratories. Molecular typing by DNA sequencing or MALDI-TOF MS is a more practical approach for distinguishing GP.Bun from GP.Kip hybrid glycophorins [33].
Antisera were supplied by the Japanese Red Cross, SCARF, and Immucor as gifts to the Australian Red Cross Blood Service. Epitopes recognised by anti-Mia CBC-172, anti-Mur HIRO-138 (64-D6), anti-NEV CBC-181 (64-2A3), and anti-Mia GAMA210 have been described in a previous publication report on the Fourth International Workshop on Monoclonal Antibodies [27].
Haemagglutination Techniques
Standard haemagglutination techniques were used in this study. Briefly, one drop of antiserum was added into a test tube followed by one drop of 3‒5% RBC suspension. Tubes were mixed and then incubated at room temperature for the saline room temperature (SRT) technique or at 37°C for the indirect antiglobulin test (IAT).
For the SRT technique using anti-Mia GAMA210 monoclonal antibody (Novaclone; Immucor), tubes were incubated for 10 min at room temperature and spun for 15 s at 1,000 g. For the SRT technique using anti-Mur antibody, tubes were incubated for 30 min at room temperature and spun for 815 g. Agglutination reactions were assessed by visual examination.
For the IAT technique, tubes were incubated at 37°C for 30 min. After incubation, RBCs were washed four times at 3,500 rpm using a cell washer (Rotolavit; Hettich) and then added with 2 drops of anti-human globulin (Anti-IgG reagent; CSL). Tubes were mixed and then spun at 2,000 rpm (Rotolavit, Hettich) for 15–20 s. Agglutination reactions were examined by visual inspection.
All agglutination reactions for SRT and IAT were scored on a scale of zero (negative) to 4+ (positive).
Murine Anti-Mia Monoclonal Reagents: CBC-172 and GAMA210
Monoclonal antibodies recognise different epitopes on Mia. Anti-Mia CBC-172 recognises amino acid sequence 29HKRDTYAA35 [27] while GAMA210 targets 24QTNDMHKR31 or 25TNDKHKRD32 [28]. The reactivity profile for these two monoclonal antibodies were compared using a panel of well-characterised RBCs expressing hybrid glycophorins: GP.Vw, GP.Hut, GP.Mur, GP.Hop, GP.Hil, GP.Bun, and GPB control cells. The IAT technique was used for anti-Mia CBC-172 and SRT for anti-Mia GAMA210 (Table 1).
Massively Parallel Sequencing for GYPA and GYP(B-A-B) Hybrids GYP*Vw and GYP*Hut
DNA was sequenced on a high-throughput, massively parallel sequencing (MPS) platform. MPS was conducted using a DNA sequencing kit (TruSight One Sequencing Panel [TSO]; Illumina). The TSO panel was used for library preparation according to the manufacturer's instructions. TSO amplifies exons and untranslated regions of over 4,800 genes associated with known clinical phenotypes which include the GYPA and GYPB blood group genes. Targeted sequencing was performed on an MPS sequencing equipment (MiSeq; Illumina) to generate paired-end, 150-bp reads. FASTQ files, MPS data generated by the MiSeq system software, were exported to a bioinformatics software (CLC Genomics Workbench software version 8.5; QIAGEN) for MPS data analysis [29].
High-Resolution Melting Analysis Assay for GYPB and GYP(B-A-B) Hybrids GYP*Mur and GYP*Bun
Genomic DNA was isolated from EDTA whole blood using a DNA extraction kit (EZ1 DNA Blood kit; QIAGEN) on a robotic instrument (EZ1 Advanced; QIAGEN). A polymerase chain reaction (PCR) and high-resolution melting (HRM) genotyping assay was used to genotype DNA from Mia serology-positive RBCs. One pair of primers, forward primer: P7-F22TT, 5′-ACGCAGTCACCTCATTCTTGTT-3′, and reverse primer: P9-R23GG, 5′-GGCTTTGGAGTAAAAGAGTTGGG-3′, was designed to amplify GYPB pseudoexon 3 and the GYP(B-A-B) exon 3 hybrid gene [30]. A 270-bp PCR product is expected for GYPB, GYP*Mur, and GYP*Bun. A DNA typing kit (Type-it HRM PCR Kit; QIAGEN) was used to perform the PCR HRM assay. In a PCR reaction mix, 12.5 µL of 2X HRM PCR Master Mix reagent, 5 µL of genomic DNA (10 ng/µL concentration), and 0.9 µL (10 µM) each of forward and reverse primers are combined with water (5.7 µL) to make a final volume of 25 µL. PCR reaction tubes were placed on a real-time PCR cycler (QIAGEN Rotor-Gene Q 5plex HRM). The PCR step was performed under the following conditions: activation step (5 min at 95°C) followed by 40 cycles of denaturation (95°C for 10 s) and annealing/extension (65°C for 30 s). At the end of the PCR step, the temperature was gradually increased by 0.1°C every 2 s from 73°C to 83°C [30].
HRM analysis is a post-PCR DNA analysis method used to characterise the dissociation profile of double-stranded DNA. HRM uses intercalating dyes that produce fluorescence only when bound to double-stranded DNA. When the temperature is increased, double-stranded DNA dissociates into single-stranded DNA, losing fluorescence. Total fluorescence and the rate of change in fluorescence was monitored and data acquired on the HRM channel. Rotor-Gene Q Series Software was used to analyse the HRM data. Melt profiles for unknown samples were compared to melt profiles of DNA control samples. A confidence threshold of 90% was set for HRM genotyping call. Genotype assignments for samples with a confidence percentage value below 90% were classified as “Variation” [20, 31, 32].
DNA
Controls Used in the HRM Assay
GYPB homozygote, GYP*Mur homozygote, and GYP*Mur/GYPB DNA controls used in the HRM assay were genotyped by matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry (MALDI-TOF MS) and DNA was fully sequenced [33]. These DNA controls have been used in previous publications [20, 30].
Results
Mia Antigen Screening
RBCs from 11 out of 5,098 blood samples tested with anti-Mia CBC-172 gave a positive agglutination reaction (Table 2). Extended phenotyping of these 11 samples revealed four types of hybrid glycophorins: GP.Hut (n = 2), GP.Vw (n = 3), GP.Mur (n = 5), and GP.Bun (n = 1). The reactivity pattern displayed by each of the 11 blood samples were consistent with previously reported serological profiles for Mia-positive hybrid glycophorins.
Table 2.
Serological profile of Mia-positive RBCs
| Donor RBCs | Phenotype | Anti-Mia | Anti-Mur | Anti-Hil | Anti-NEV | Anti-Vw | Anti-Hut |
|---|---|---|---|---|---|---|---|
| Donor 1 | GP.Hut | + | 0 | 0 | 0 | 0 | + |
| Donor 2 | GP.Hut | + | 0 | 0 | 0 | 0 | + |
| Donor 3 | GP.Vw | + | 0 | 0 | 0 | + | NT |
| Donor 4 | GP.Mur | + | + | + | + | 0 | NT |
| Donor 5 | GP.Bun | + | + | + | 0 | NT | NT |
| Donor 6 | GP.Vw | + | 0 | NT | NT | + | NT |
| Donor 7 | GP.Mur | + | + | + | + | NT | NT |
| Donor 8 | GP.Vw | + | 0 | NT | NT | + | NT |
| Donor 9 | GP.Mur | + | + | + | + | NT | NT |
| Donor 10 | GP.Mur | + | + | + | + | NT | NT |
| Donor 11 | GP.Mur | + | + | + | + | NT | NT |
Comparison of Two Anti-Mia Monoclonal Reagents
The reactivity profile for anti-Mia CBC-172 against a panel of phenotyped RBCs was compared with a commercially available reagent anti-Mia GAMA210. Both showed a negative agglutination reaction with GPB and GP.Hil RBCs and a strongly positive reaction (3+ to 4+) with GP.Vw, GP.Hut, GP.Mur, GP.Hop, and GP.Bun RBCs (Table 3).
Table 3.
Haemagglutination reaction comparison between two anti-Mia monoclonal antibodies against a panel of RBC-expressing MNS hybrid glycophorins
| RBC panel | CBC-172 | GAMA210 |
|---|---|---|
| GPB | 0 | 0 |
| GP.Vw | 4+ | 3+ |
| GP.Hut | 4+ | 4+ |
| GP.Mur | 4+ | 4+ |
| GP.Hop | 4+ | 4+ |
| GP.Hil | 0 | 0 |
| GP.Bun | 4+ | 4+ |
GPB RBCs were used as a negative control. GP.Hil, a GP(A-B) MNS hybrid glycophorin, does not express Mia antigen and was used as an Mia−negative hybrid glycophorin negative control.
Genotyping for Hybrid Glycophorins
DNA samples from 11 Mia-positive blood donors were segregated into two groups. Group 1 were the GP.Vw and GP.Hut blood donors (donors 1, 2, 3, 6, and 8) and were genotyped by MPS (Fig. 3). Group 2 were the GP.Mur and GP.Bun blood donors (donor, 4, 5, and 7) and were genotyped by the HRM assay (Fig. 4).
Fig. 3.
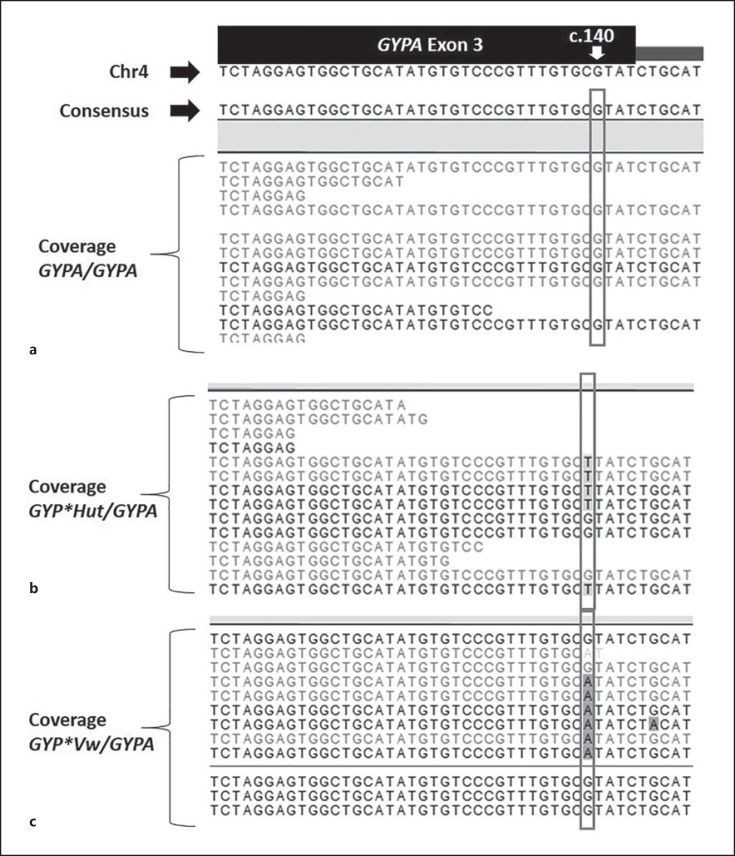
Sequence alignment for group 1 samples. a Representative sequences for GYPA/GYPA DNA control aligned to the GYPA exon 3 reference sequences. b Representative sequences for GYP*Hut/GYPA heterozygote – donors 1 and 2. c Representative sequences for GYP*Vw/GYPA heterozygote – donors 3, 6, and 8. Refer to diagram and legend of Figure 1 for GYPA exon 3 sequence. Nucleotide detected at c.140 predicts the base on the complementary strand. Grey box indicates nucleotides in position c.140 for group 1 DNA sequences.
Fig. 4.
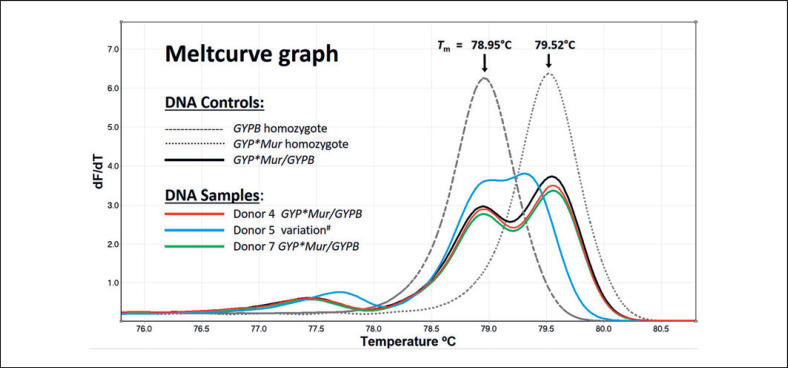
Meltcurve graph for group 2 samples. Meltcurve graph plots the rate of change in fluorescence (y axis) against temperature (x axis). The meltcurve profile for DNA controls GYPB homozygote (single peak, continuous grey dashes, Tm = 78.95°C), GYP*Mur homozygote (single peak, grey dots, Tm = 79.52°C), and GYP*Mur/GYPB (solid black line, double peaks, Tm = 78.96 and 79.55°C). High-resolution melting analysis showed that the meltcurve profiles for donor 4 (red line) and donor 7 (green line) are similar to DNA control GYP*Mur/GYPB while the profile for donor 5 (blue line) is distinct from all three DNA controls. # DNA from donor 5 had been used in a previous study and had been genotyped as GYP*Bun/GYPB [33]. Tm, melting temperature.
In group 1, sequences were aligned to a reference sequence (Fig. 3a). MPS analysis of the GYPA/GYPA DNA control at position c.140 showed G/G predicting a C/C on the complementary strand (Fig. 3a). Analysis of sequences at c.140 detected T/G (predicting A/C on the complementary strand) for donors 1 and 2 interpreted as heterozygous for GYP*Hut/GYPA(Fig. 3b). For donors 3, 6, and 8, A/G were detected at c.140 (predicting T/C on the complementary strand) interpreted as heterozygous for GYP*Vw/GYPA (Fig. 3c).
In group 2, HRM analysis showed that the DNA meltcurve profiles for donors 4 and 7 matched the DNA control GYP*Mur/GYPB with 99.53 and 99.23% confidence, respectively (Fig. 4). The meltcurve pattern for donor 5 gave a result of 32.5% confidence when compared to GYP*Mur/GYPB DNA control. Since 32.5% is below the 90% threshold, the genotype call assigned for donor 5 was “Variation.” DNA analysis for donor 5 on the HRM meltcurve graph revealed two peaks showing melting temperature at 78.99 and 79.32°C. Donor 5 was originally reported as heterozygous for GYP*Bun/GYPB in a study assessing the viability of MALDI-TOF MS as a genotyping platform for hybrid glycophorins [33].
Genotyping calls for 8 out of 11 Mia-positive blood donors were consistent with the observed phenotypes (Table 4). Three blood donors (Donors 9, 10, and 11) did not present back to the Blood Service for their subsequent blood donation since their RBCs were initially tested for Mia between 2011 and 2013 and, therefore, DNA samples from these blood donors were unavailable for genotyping.
Table 4.
Phenotyping and genotyping comparison of Mia-positive blood donors
| Blood donor | Phenotype | Genotype |
|---|---|---|
| Donor 1 | GP.Hut | GYP*Hut/GYPA |
| Donor 2 | GP.Hut | GYP*Hut/GYPA |
| Donor 3 | GP.Vw | GYP*Vw/GYPA |
| Donor 4 | GP.Mur | GYP*Mur/GYPB |
| Donor 5 | GP.Bun | GYP*Bun/GYPB |
| Donor 6 | GP.Vw | GYP*Vw/GYPA |
| Donor 7 | GP.Mur | GYP*Mur/GYPB |
| Donor 8 | GP.Vw | GYP*Vw/GYPA |
| Donor 9 | GP.Mur | Not tested |
| Donor 10 | GP.Mur | Not tested |
| Donor 11 | GP.Mur | Not tested |
DNA from donors 9, 10, and 11 was not isolated from their primary blood donation. These donors did not return to donate blood since the preliminary screening. DNA analysis for donor 5 was initially reported in a previous study and had been genotyped as GYP*Bun/GYPB [33].
Discussion
In this study, 11 out of 5,098 blood samples were Mia-positive which represents a frequency 0.22% in the Australian blood donor population. Of the 11 Mia-positive MNS hybrid glycophorin blood donors identified, GP.Mur was the most common type followed closely by GP.Vw. Although Mia-positive GP.Hop and GP.Kip were not detected in this study, each was reportedly found in an individual in Australia described in two independent case studies [16, 24]. Collectively, data from this study and two case studies indicate that the MNS hybrid glycophorin blood group profile in the Australia population is diverse.
The discovery of these hybrid glycophorin types in the blood donor pool is instructive and practical. RBCs from these blood donors have become a valuable resource of reagent RBCs for the identification of antibodies against MNS hybrid glycophorins, particularly anti-Mia. Anti-Mia antibody is clinically significant and reported to cause haemolytic transfusion reactions and haemolytic disease of the fetus and newborn [7]. In an Australian study, peptide ELISA assay detected anti-Mia antibody in the serum/plasma of 3.8% blood donors, 4% antenatal patients (5/124), and 1.4% multi-transfused patients (3/215) [34]. Recently, we identified an anti-Mia antibody in a patient that reacted positive to a particular packed RBC unit during routine pre-transfusion cross-match. Further serological and molecular studies typed the RBCs as GP.Mur homozygote, S– s+/–, predicting a JENU-negative phenotype [35].
Mia is considered a low-frequency antigen (0.23%, 9/3,844) in the Caucasian population [36] but generally appears at a higher frequency in several Asian population groups [11, 20, 23]. Although the Australian demographics are diverse and ever-changing, the frequency of Mia at 0.22% suggests that the Australian blood donor population resembles a Caucasian profile. However, for GP.Mur, the frequency reported in this study is 1 in 1,020 in contrast to the historically reported frequency of 1 in 10,020 in a Caucasian population [18].
This is the first comprehensive study to report on the frequency of Mia and the type of MNS hybrid glycophorins found in an Australian blood donor population. Knowing the blood group profile in a population is essential to effectively manage transfusion needs.
Statement of Ethics
Consent was given for the investigation of blood groups at the time of collection of the samples. This study has approval from the Australian Red Cross Blood Service Human Research Ethics Committee (Application No. 2010#07).
Disclosure Statement
The authors have no conflicts of interest to disclose.
Acknowledgements
The Australian government funds the Australian Red Cross Blood Service for the provision of blood, blood products, and services to the Australian community. We thank Dr. Makoto Uchikawa (Japanese Red Cross) and Immucor (through Carol G. Judy, Asia-Pacific Regional Director, Immucor) for the gift of monoclonal antibodies used in this study.
References
- 1.Cook PJ, Lindenbaum RH, Salonen R, de la Chapelle A, Daker MG, Buckton KE, et al. The MNSs blood groups of families with chromosome 4 rearrangements. Ann Hum Genet. 1981 Feb;45((1)):39–47. doi: 10.1111/j.1469-1809.1981.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013 Jan;41((Database issue)):D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genome Browser UC UCSC Genome Browser on Human. 2013 Dec; (GRCh38/hg38) Assembly. http://genome.ucsc.edu/ [Google Scholar]
- 4.Kudo S, Fukuda M. Structural organization of glycophorin A and B genes: glycophorin B gene evolved by homologous recombination at Alu repeat sequences. Proc Natl Acad Sci USA. 1989 Jun;86((12)):4619–23. doi: 10.1073/pnas.86.12.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vignal A, Rahuel C, London J, Cherif Zahar B, Schaff S, Hattab C, et al. A novel gene member of the human glycophorin A and B gene family. Molecular cloning and expression. Eur J Biochem. 1990 Aug;191((3)):619–25. doi: 10.1111/j.1432-1033.1990.tb19166.x. [DOI] [PubMed] [Google Scholar]
- 6.Reid ME. MNS blood group system: a review. Immunohematology. 2009;25((3)):95–101. [PubMed] [Google Scholar]
- 7.Heathcote DJ, Carroll TE, Flower RL. Sixty years of antibodies to MNS system hybrid glycophorins: what have we learned? Transfus Med Rev. 2011 Apr;25((2)):111–24. doi: 10.1016/j.tmrv.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Lomas-Francis C. Miltenberger phenotypes are glycophorin variants: a review. ISBT Sci Ser. 2011;6((2)):296–301. [Google Scholar]
- 9.Graydon JJ. A rare iso-haemagglutinogen. Med J Aust. 1946 Jul;2((1)):9–10. [PubMed] [Google Scholar]
- 10.Simmons RT, Albrey JA, McCULLOCH WJ. The duplication of the Gr (Graydon) blood group by Vw (Verweyst) Vox Sang. 1959 Apr;4((2)):132–7. doi: 10.1111/j.1423-0410.1959.tb04026.x. [DOI] [PubMed] [Google Scholar]
- 11.Chandanyingyong D, Pejrachandra S. Studies on the Miltenberger complex frequency in Thailand and family studies. Vox Sang. 1975;28((2)):152–5. doi: 10.1111/j.1423-0410.1975.tb02753.x. [DOI] [PubMed] [Google Scholar]
- 12.Chandanayingyong D, Pejrachandra S, Poole J. Three antibodies of the MNSs system and their association with the Miltenberger Complex of antigens. I. Anek serum. Vox Sang. 1977;32((5)):272–3. doi: 10.1111/j.1423-0410.1977.tb00643.x. [DOI] [PubMed] [Google Scholar]
- 13.Chandanayingyong D, Pejrachandra S. Separable anti-Hut which is specific for class II of the Miltenberger complex. Vox Sang. 1975;28((2)):149–51. doi: 10.1111/j.1423-0410.1975.tb02752.x. [DOI] [PubMed] [Google Scholar]
- 14.Giles CM. Serological activity of low frequency antigens of the MNSs system and reappraisal of the Miltenberger complex. Vox Sang. 1982;42((5)):256–61. doi: 10.1111/j.1423-0410.1982.tb00754.x. [DOI] [PubMed] [Google Scholar]
- 15.Flower RL, Wei L, Ji YL, Luo GP, Lopez GH, Hyland CA. GP.Kipp and GP.Yak BAB hybrid glycophorins: no difference in sequence or serology. Vox Sang. 2013;105(Suppl 2):10. [Google Scholar]
- 16.Green C, Poole J, Ford D, Glameyer T. A postulated glycophorin B-A-B hybrid demonstrating heterogeneity of anti-Hop and anti-Nob sera. [Abstract P64] Transfus Med. 1992;2((Suppl. 1)):67. [Google Scholar]
- 17.Uchikawa M, Ogasawara K, Suzuki Y, Saito M, Tsuneyama H, Morimoto K, et al. A new GP(B-A-B) hybrid molecule (GP.Yak) with Miltenberger Phenotype. Vox Sang. 2012;103(Suppl. 1):214. [Google Scholar]
- 18.Race RR, Sanger R. Blood Groups in Man. 6th ed. Oxford: Blackwell Scientific Publications; 1975. [Google Scholar]
- 19.Prathiba R, Lopez CG, Usin FM. The prevalence of GP Mur and anti-“Mia” in a tertiary hospital in Peninsula Malaysia. Malays J Pathol. 2002 Dec;24((2)):95–8. [PubMed] [Google Scholar]
- 20.Wei L, Lopez GH, Zhang Y, Wen J, Wang Z, Fu Y, et al. Genotyping analysis of MNS blood group GP(B-A-B) hybrid glycophorins in the Chinese Southern Han population using a high-resolution melting assay. Transfusion. 2018 Jul;58((7)):1763–71. doi: 10.1111/trf.14641. [DOI] [PubMed] [Google Scholar]
- 21.Huynh NT, Ford DS, Duyen TT, Huong MT. Jk and Mi.III phenotype frequencies in North Vietnam. Immunohematology. 2003;19((2)):57–8. [PubMed] [Google Scholar]
- 22.Hsu K, Lin YC, Chao HP, Lee TY, Lin M, Chan YS. Assessing the frequencies of GP.Mur (Mi.III) in several Southeast Asian populations by PCR typing. Transfus Apheresis Sci. 2013 Oct;49((2)):370–1. doi: 10.1016/j.transci.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Broadberry RE, Lin M. The distribution of the MiIII (Gp.Mur) phenotype among the population of Taiwan. Transfus Med. 1996 Jun;6((2)):145–8. doi: 10.1046/j.1365-3148.1996.d01-64.x. [DOI] [PubMed] [Google Scholar]
- 24.Storry JR, Poole J, Condon J, Reid ME. Identification of a novel hybrid glycophorin gene encoding GP.Hop. Transfusion. 2000 May;40((5)):560–5. doi: 10.1046/j.1537-2995.2000.40050560.x. [DOI] [PubMed] [Google Scholar]
- 25.Lopez GH, Wei L, Ji Y, Condon JA, Luo G, Hyland CA, et al. GYP*Kip, a novel GYP(B-A-B) hybrid allele, encoding the MNS48 (KIPP) antigen. Transfusion. 2016 Feb;56((2)):539–41. doi: 10.1111/trf.13450. [DOI] [PubMed] [Google Scholar]
- 26.Australian Bureau of Statistics Cultural Diversity in Australia: 2016 Census Article. 2016 https://www.abs.gov.au. [Google Scholar]
- 27.Reid ME, Lisowska E, Blanchard D. Section 3: epitope determination of monoclonal antibodies to glycophorin A and glycophorin B. Coordinator's report. Antibodies to antigens located on glycophorins and band 3. Transfus Clin Biol. 2002 Jan;9((1)):63–72. doi: 10.1016/s1246-7820(01)00219-1. [DOI] [PubMed] [Google Scholar]
- 28.Chen V, Halverson G, Wasniowska K, Lisowska E, Chen J, Moulds M, et al. Direct evidence for the existence of Miltenberger antigen. Vox Sang. 2001 May;80((4)):230–3. doi: 10.1046/j.1423-0410.2001.00042.x. [DOI] [PubMed] [Google Scholar]
- 29.Schoeman EM, Lopez GH, McGowan EC, Millard GM, O'Brien H, Roulis EV, et al. Evaluation of targeted exome sequencing for 28 protein-based blood group systems, including the homologous gene systems, for blood group genotyping. Transfusion. 2017 Apr;57((4)):1078–88. doi: 10.1111/trf.14054. [DOI] [PubMed] [Google Scholar]
- 30.Lopez GH, Wilson B, Liew YW, Kupatawintu P, Emthip M, Hyland CA, et al. An alloantibody in a homozygous GYP*Mur individual defines JENU (MNS49), a new high-frequency antigen on glycophorin B. Transfusion. 2017 Mar;57((3)):716–7. doi: 10.1111/trf.13952. [DOI] [PubMed] [Google Scholar]
- 31.Lopez GH, Morrison J, Condon JA, Wilson B, Martin JR, Liew YW, et al. Duffy blood group phenotype-genotype correlations using high-resolution melting analysis PCR and microarray reveal complex cases including a new null FY*A allele: the role for sequencing in genotyping algorithms. Vox Sang. 2015 Oct;109((3)):296–303. doi: 10.1111/vox.12273. [DOI] [PubMed] [Google Scholar]
- 32.Lopez GH, Mcbean RS, Wilson B, Irwin DL, Liew YW, Hyland CA, et al. Molecular typing for the Indian blood group associated 252G[{GT}]C single nucleotide polymorphism in a selected cohort of Australian blood donors. Blood Transfus. 2015 Jan;13((1)):78–85. doi: 10.2450/2014.0336-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei L, Lopez GH, Ji Y, Condon JA, Irwin DL, Luo G, et al. Genotyping for Glycophorin GYP(B-A-B) Hybrid Genes Using a Single Nucleotide Polymorphism-Based Algorithm by Matrix-Assisted Laser Desorption/Ionisation, Time-of-Flight Mass Spectrometry. Mol Biotechnol. 2016 Oct;58((10)):665–71. doi: 10.1007/s12033-016-9966-6. [DOI] [PubMed] [Google Scholar]
- 34.Flower RL, Lin M, Kamhieh S, Chen Q, Morel-Kopp MC, Sztynda T, et al. Antibodies to variant MNS (Miltenberger) antigens detected in Australia and Asia: are some cases of ‘anaemia of prematurity’ undiagnosed haemolytic disease of the newborn? Transfus Med. 2002;12:159. [Google Scholar]
- 35.Powley T, Lopez GH, Wilson B, Morrison J, Hyland CA, Flower RL, et al. Rare JENU negative donor identified through incompatible crossmatch. Vox Sang. 2019;114:176. [Google Scholar]
- 36.Mohn JF, Lambert RM, Rosamilia HG, Wallace J, Milne GR, Moores P, et al. On the relationship of the blood group antigens Mia and Vw to the MNSs system. Am J Hum Genet. 1958 Sep;10((3)):276–86. [PMC free article] [PubMed] [Google Scholar]
- 37.Blumenfeld OO, Huang CH. GenBank NG_007483, Homo sapiens glycophorin B (MNS blood group) (GYPB), RefSeqGene on chromosome 4. https://www.ncbi.nlm.nih.gov/nuccore/NG_007483. [Google Scholar]
- 38.Giles CM, Chandanayingong D, Webb AJ. Three antibodies of the MNSs system and their association with the Miltenberger complex of antigens. III. Anek, Raddon and Lane antisera in relation to each other and the Miltenberger complex. Vox Sang. 1977;32((5)):277–9. doi: 10.1111/j.1423-0410.1977.tb00645.x. [DOI] [PubMed] [Google Scholar]