Abstract
The U antigen (MNS5) is one of 49 antigens belonging to the MNS blood group system (ISBT002) carried on glycophorins A (GPA) and B (GPB). U is present on the red blood cells in almost all Europeans and Asians but absent in approximately 1.0% of Black Africans. U negativity coincides with negativity for S (MNS3) and s (MNS4) on GPB, thus be called S–s–U–, and is thought to arise from homozygous deletion of GYPB. Little is known about the molecular background of these deletions. Bioinformatic analysis of the 1000 Genomes Project data revealed several candidate regions with apparent deletions in GYPB. Highly specific Gap-PCRs, only resulting in positive amplification from DNAs with deletions present, allowed for the exact genetic localization of 3 different breakpoints; 110.24- and 103.26-kb deletions were proven to be the most frequent in Black Americans and Africans. Among 157 CEPH DNAs, deletions in 6 out of 8 African ethnicities were present. Allele frequencies of the deletions within African ethnicities varied greatly and reached a cumulative 23.3% among the Mbuti Pygmy people from the Congo. Similar observations were made for U+<sup>var</sup> alleles, known to cause strongly reduced GPB expression. The 110- and 103-kb deletional GYPB haplotypes were found to represent the most prevalent hereditary factors causative of the MNS blood group phenotype S–s–U–. Respective GYPB deletions are now accessible by molecular detection of homo- and hemizygous transmission.
Keywords: GYPB deletion, MNS phenotypes, Glycophorin B
Introduction
Glycophorin A (GPA) and glycophorin B (GPB) are glycoproteins of the human erythrocyte membrane that carry the 49 antigens of the MNS blood group system [1], i.e., the second blood group system discovered in human history after ABO [2, 3, 4]. M/MNS1/N/MNS2 and S/MNS3/s/MNS4 are the two pairs of antithetical antigens with common prevalence found on the extracellular domains of GPA and GPB, respectively.
The antigen U/MNS5 was originally described in 1953 and was characterized as a high-frequency antigen that is absent in 1.2% of African Americans [5]. In Europeans, however, the phenotype S–s–U– is extremely rare [6]. In 1954, the association with the MNS blood group system and the concurrent S‒s‒ phenotype became evident [7]. In 1987, this phenotype was postulated to be caused by a homozygous deletion in GYPB [8, 9, 10]. Besides regular M and N expression, U– red blood cells (RBCs) are almost always S–s–. However, S–s– RBCs are often reacting weakly positive with anti-U antisera and are then referred to as S–s–U+var [6, 11, 12, 13]. Using adsorption/elution tests with particularly potent anti-U antisera, about 50% of apparent S–s–U– RBCs are in fact U+var, most frequently encoded by one of two distinct molecular variants of GYPB [13, 14].
The antigenic complexity of the blood group system MNS as well as levels of genetic diversity in African populations have led to speculation that this locus is under evolutionary selection due to its function as a ligand for the malaria parasite Plasmodium falciparum [15, 16, 17, 18]. Complete absence of GPA and GPB on the erythrocyte surface might therefore represent a genetic selection process, influencing invasion and consequent progression of malaria [19, 20, 21]. More recently, the MNS antigen Dantu was shown to represent one of the strongest protective genetic variants against infection with this parasite (OR 0.26–0.57) [22, 23, 24]. Leffler et al. [23]reported additional structural variation in the GYP locus, also including GYPB deletions, but differing from molecular variants reported earlier [9, 10, 11].
Still, accurate molecular detection of U negativity, e.g., displaying correct genotypes and distinguishing heterozygous (hemizygous) from S-s-U- homozygous individuals, remained extremely difficult, if not completely impossible until now. Therefore, 1000 Genomes Project (1000G) data were analyzed by us and revealed approximate locations for GYPB deletions [25]. Highly specific amplicons of Gap-PCRs, only positive in DNAs with deletions present, were then sequenced and delivered exact positions of 3 different deletional breakpoints. Using these novel insights into the GYP locus on chromosome 4 as diagnostic tools, comprehensive molecular detection of true S–s–U– genotypes in Black Africans was made possible.
Materials and Methods
Samples
Ethical approval for the study was obtained from the Cantonal Office of Public Health, Zurich, Switzerland (Swiss approval No. 2014-0408).
DNA samples were from various sources (Tables 1, 2). Table 1 DNA samples (116) were accumulated for S–s–U– blood group MNS pheno- and/or genotypes with and without S and s phenotypes available. They consisted of samples with proven S–s–U– (15) and proven S–s–U– or U+w phenotypes (12) from the Blood Research Institute, Versiti, Inc., Milwaukee, WI, USA. Three samples with S–s–U– phenotype came from the Austrian Red Cross, Vienna, the German Red Cross, Hagen, and the Clinical Immunology and Transfusion Medicine Department, Lund, Sweden. Another 27 samples were included due to their low GYPB:GYPE ratios, indicative of the lack of at least 1 parental GYPB gene; they were originally identified among 5,800 Swiss Caucasians and 50 Black Africans [26]. A panel of 55 DNA samples of known, or presumptive, Black African ancestry comprised donors or patients from the greater area of the Canton of Zurich, Switzerland: 33 with and 22 without S/s serology. The Coriell Cell Repository (Coriell Institute, Camden, NJ, USA) provided 2 DNA samples with expected GYPB deletions as identified during bioinformatic analysis of the 1000G data. Another 2 Coriell samples carried 1 GYPB*03N.02 allele [pers. commun., Mónica López-Martinez, Progenika-Grifols, Bilbao, Spain]. De-identified samples of EDTA-anticoagulated blood from Versiti, Inc., were provided under an approved material transfer agreement for DNA extraction (QIAamp 96 DNA blood kit; Qiagen, Valencia, CA, USA) to Zurich. DNA extraction of samples from the Blood Transfusion Service Zurich (Switzerland) was done using a QIAamp DNA blood kit (Qiagen, Hilden, Germany) or as described previously [27].
Table 1.
Overview and summarized genotyping results for 116 samples accumulated for GYPB deletions
| Source | N | Unknown S, s, serology | Known S, s, serology | Serology S–s–U– | Allele count |
GYPB* 03 |
GYPB* 03N.02 |
GYPB* 03N.03 | GYPB* 03N.04 | GYPB* 06.01 | GYPB* 06.02 | GYPB* 04 |
GYPB* 05N.01 (del 110 kb) |
GYPB* 05N.02 (del 103 kb) |
GYPB* 05N.03 (del 19 kb) |
GYPB* 05N unknown |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st Blood Center of Wisconsin Milwaukee, USA | 9 | 0 | 9 | 9 | 18 | − | − | − | − | − | − | − | 18 | − | − | − |
| 5 | 0 | 5 | 5 | 10 | − | − | − | − | − | − | − | 5 | 5 | − | − | |
| 1 | 0 | 1 | 1 | 2 | − | − | − | − | − | − | − | 1 | − | − | 1 | |
| 15 | 0 | 15 | 15 | 30 | 24 | 5 | 1 | |||||||||
| 2nd Blood Center of Wisconsin Milwaukee, USA | 4 | 0 | 4 | 4 | 8 | − | − | − | − | − | − | − | 8 | − | − | − |
| 1 | 0 | 1 | 1 | 2 | − | − | − | − | − | − | − | 1 | 1 | − | − | |
| 1 | 0 | 1 | 1 | 2 | − | − | − | − | − | − | − | 1 | − | − | 1 | |
| 2 | 0 | 2 | 0 | 4 | − | − | − | 2 | − | − | − | 2 | − | − | − | |
| 1 | 0 | 1 | 0 | 2 | − | − | − | − | − | − | 1 | − | − | − | ||
| 1 | 0 | 1 | 0 | 2 | − | − | − | 1 | − | − | 1 | − | − | − | − | |
| 1 | 0 | 1 | 0 | 2 | − | 1 | − | − | − | − | − | 1 | − | − | − | |
| 1 | 0 | 1 | 0 | 2 | − | 1 | − | − | − | − | 1 | − | − | − | − | |
| 12 | 0 | 12 | 6 | 24 | 1 | 2 | 3 | 2 | 14 | 1 | 1 | |||||
| Austrian Red Cross, Vienna* | 1 | 0 | 1 | 1 | 2 | − | n.t. | n.t. | n.t. | n.t. | n.t. | − | 1 | 1 | n.t. | − |
| DRK West, Hagen, Germany* | 1 | 0 | 1 | 1 | 2 | − | n.t. | n.t. | n.t. | n.t. | n.t. | − | 2 | − | n.t. | − |
| Lund Blood Bank, Sweden* | 1 | 0 | 1 | 1 | 2 | − | n.t. | n.t. | n.t. | n.t. | n.t. | − | 1 | 1 | n.t. | − |
| 3 | 0 | 3 | 3 | 6 | 4 | 2 | ||||||||||
| 18 of 20 with lowest B:E ratio Reference [26], Figure 4b | 7 | 0 | 7 | 0 | 14 | 7 | n.t. | − | − | − | − | 7 | − | − | n.t. | − |
| 8 | 0 | 8 | 0 | 16 | − | n.t. | − | − | − | − | 16 | − | − | n.t. | − | |
| 2 | 0 | 2 | 0 | 4 | − | n.t. | − | − | − | − | 2 | 2 | − | n.t. | − | |
| del GYPB 103 kb “Lorena”* |
1 | 0 | 1 | 0 | 2 | − | n.t. | − | − | − | − | 1 | − | 1 | n.t. | − |
| 18 | 0 | 18 | 0 | 36 | 7 | 26 | 2 | 1 | ||||||||
| 9 of 50 with lowest B:E ratio Reference [26], Figure 4c | 1 | 0 | 1 | 0 | 2 | − | n.t. | − | − | − | 1 | 1 | − | − | n.t. | − |
| 3 | 0 | 3 | 0 | 6 | − | n.t. | − | − | − | − | 3 | 3 | − | n.t. | − | |
| del GYPB 110 kb “Gerold”* |
1 | 0 | 1 | 0 | 2 | − | n.t. | − | − | − | − | 1 | 1 | − | n.t. | − |
| NY Blood Center, Uvar, “P876” | 1 | 0 | 1 | 0 | 2 | − | n.t. | − | 1 | − | − | − | 1 | − | n.t. | − |
| NY Blood Center, Uvar, “CW” | 1 | 0 | 1 | 0 | 2 | − | n.t. | − | 2 | − | − | − | − | − | n.t. | − |
| NY Blood Center, S–s–U–, “AO” | 1 | 0 | 1 | 1 | 2 | − | n.t. | − | − | − | − | − | 1 | − | n.t. | 1 |
| NY Blood Center, S–s–U–, “JB”* | 1 | 0 | 1 | 1 | 2 | − | n.t. | − | − | − | − | − | 2 | n.t. | ||
| 9 | 0 | 9 | 2 | 18 | 3 | 1 | 5 | 6 | 2 | 1 | ||||||
| BTS Zurich, Switzerland, “black” with S, s serology | 1 | 0 | 1 | 0 | 2 | 2 | n.t. | − | − | − | − | − | − | − | n.t. | − |
| 6 | 0 | 6 | 0 | 12 | 6 | n.t. | − | − | − | − | 6 | − | − | n.t. | − | |
| 18 | 0 | 18 | 0 | 36 | − | n.t. | − | − | − | − | 36 | − | − | n.t. | − | |
| 2 | 0 | 2 | 0 | 4 | 2 | n.t. | − | − | − | − | − | 2 | − | n.t. | − | |
| 4 | 0 | 4 | 0 | 8 | − | n.t. | − | − | − | − | 4 | 4 | − | n.t. | − | |
| 2 | 0 | 2 | 0 | 4 | − | n.t. | − | 2 | − | − | 2 | − | − | n.t. | − | |
| 33 | 0 | 33 | 0 | 66 | 10 | 2 | 48 | 6 | ||||||||
| BTS Zurich, Switzerland, “black” lacking S, s serology | 2 | 2 | 0 | 0 | 4 | 4 | n.t. | − | − | − | − | − | − | − | n.t. | − |
| 1 | 1 | 0 | 0 | 2 | 1 | n.t. | − | − | − | − | 1 | − | − | n.t. | − | |
| 7 | 7 | 0 | 0 | 14 | − | n.t. | − | − | − | − | 14 | − | − | n.t. | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | − | − | − | − | 2 | − | n.t. | − | |
| 8 | 8 | 0 | 0 | 16 | − | n.t. | − | − | − | − | 8 | 8 | − | n.t. | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | 1 | − | − | − | − | − | n.t. | 1 | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | 1 | − | 1 | − | − | − | n.t. | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | − | − | 1 | 1 | − | − | n.t. | − | |
| 22 | 22 | 0 | 0 | 44 | 5 | 2 | 2 | 24 | 10 | 1 | ||||||
| Coriell, HG02970*A1/”del 32 kb” | 1 | 1 | 0 | 0 | 2 | − | n.t. | n.t. | n.t. | n.t. | n.t. | − | 1 | 1 | − | − |
| del GYPB 19 kb, Coriell HG01880*C1* |
1 | 1 | 0 | 0 | 2 | 1 | n.t. | n.t. | n.t. | n.t. | n.t. | − | − | − | 1 | − |
| Coriell, NA19379*1 | 1 | 1 | 0 | 0 | 2 | 1 | 1 | − | − | − | − | − | − | − | n.t. | − |
| Coriell, NA19404*1 | 1 | 1 | 0 | 0 | 2 | − | 1 | − | − | − | − | 1 | − | − | n.t. | − |
| 4 | 4 | 0 | 0 | 8 | 2 | 2 | 1 | 1 | 1 | 1 | ||||||
| All numbers | 116 | 26 | 90 | 26 | 232 | 25 | 4 | 0 | 10 | 0 | 3 | 106 | 67 | 12 | 1 | 4 |
Samples indicated by “*” were the first 6 samples analyzed for GYPB deletions by Gap-PCRs. SEQ submission. Sequences of the deletional GYPB breakpoint regions of the samples marked in gray were submitted to GeneBank and were MN005664, MN005663, and MN005662 for the deletional GYPB haplotypes of 110-, 103-, and 19-kb types, respectively (online suppl. Table s5). §Though showing a Uvar phenotype, SNP-based genotyping did not deliver any of the known Uvar alleles. Sequencing of the respective sample was not performed.
Table 2.
Overview and summarized genotyping results for 157 samples representing 8 African ethnicities and provided by CEPH (Centre d'Etude du Polymorphism Humain, Foundation Jean Dausset, Paris, France)
| Source (CEPH “African”) | n | Unknown S, s, serology | Known S, s, serology | Serology S–s–U– | Allele count | GYPB* 03 | GYPB* 03N.02 | GYPB* 03N.03 | GYPB* 03N.04 |
GYPB* 06.01 |
GYPB* 06.02 | GYPB* 04 |
GYPB* 05N.01 (del 110 kb) |
GYPB* 05N.02 (del 103 kb) |
GYPB* 05N.03 (del 19 kb) |
GYPB* 05N unknown |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biaka Pygmy Central African Republic | 6 | 6 | 0 | 0 | 12 | 6 | n.t. | − | − | − | − | 6 | − | − | − | − |
| 15 | 15 | 0 | 0 | 30 | − | n.t. | − | − | − | − | 30 | − | − | − | − | |
| 3 | 3 | 0 | 0 | 6 | − | n.t. | 3 | − | − | − | 3 | − | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | 1 | n.t. | − | 1 | − | − | − | − | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | 1 | − | − | 1 | − | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | 2 | − | − | − | − | − | − | − | |
| 8 | 8 | 0 | 0 | 16 | − | n.t. | − | − | − | − | 8 | − | 8 | − | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | − | − | − | − | − | 2 | − | − | |
| 36 | 36 | 0 | 0 | 72 | 7 | 3 | 4 | 48 | 10 | |||||||
| Haplotype frequency, % | 100.0 | 9.7 | 4.2 | 5.6 | 66.7 | 13.9 | ||||||||||
| Mbuti Pygmy Democratic Republic of Congo | 2 | 2 | 0 | 0 | 4 | − | n.t. | − | − | − | − | 4 | − | − | − | − |
| 3 | 3 | 0 | 0 | 6 | − | n.t. | − | 6 | − | − | − | − | − | − | − | |
| 3 | 3 | 0 | 0 | 6 | − | n.t. | − | 3 | − | − | 3 | − | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | − | − | − | − | 2 | − | − | − | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | − | − | − | 1 | 1 | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | 1 | − | − | − | 1 | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | − | − | − | − | 1 | 1 | − | − | |
| 3 | 3 | 0 | 0 | 6 | − | n.t. | − | 3 | − | − | − | − | 3 | − | − | |
| 15 | 15 | 0 | 0 | 30 | 13 | 2 | 8 | 3 | 4 | |||||||
| Haplotype frequency, % | 100.0 | 43.3 | 6.7 | 26.7 | 10.0 | 13.3 | ||||||||||
| Bantu, NE Kenya | 1 | 1 | 0 | 0 | 2 | 2 | n.t. | − | − | − | − | − | − | − | − | − |
| 3 | 3 | 0 | 0 | 6 | 3 | n.t. | − | − | − | − | 3 | − | − | − | − | |
| 6 | 6 | 0 | 0 | 12 | − | n.t. | − | − | − | − | 12 | − | − | − | − | |
| 2 | 2 | 0 | 0 | 4 | − | 2 | − | − | − | − | 2 | − | − | − | − | |
| 12 | 12 | 0 | 0 | 24 | 5 | 2 | 17 | |||||||||
| Bantu, SE and SW South Africa | 2 | 2 | 0 | 0 | 4 | 2 | n.t. | − | − | − | − | 2 | − | − | − | − |
| 5 | 5 | 0 | 0 | 10 | − | n.t. | − | − | − | − | 10 | − | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | − | − | − | 1 | 1 | − | − | − | |
| 8 | 8 | 0 | 0 | 16 | 2 | 13 | 1 | |||||||||
| Mandenka Senegal | 4 | 4 | 0 | 0 | 8 | 4 | n.t. | − | − | − | − | 4 | − | − | − | − |
| 14 | 14 | 0 | 0 | 28 | − | n.t. | − | − | − | − | 28 | − | − | − | − | |
| 1 | 1 | 0 | 0 | 4 | − | − | − | − | 3 | 1 | − | − | − | − | − | |
| 2 | 2 | 0 | 0 | 4 | − | n.t. | − | 2 | − | − | 2 | − | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | − | − | − | 1 | 1 | − | − | − | |
| 2 | 2 | 0 | 0 | 2 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | 2 | |
| 24 | 24 | 0 | 0 | 48 | 4 | 2 | 3 | 1 | 35 | 1 | 2 | |||||
| Mozabite Algeria (Mzab) | 1 | 1 | 0 | 0 | 2 | 2 | n.t. | − | − | − | − | − | − | − | − | − |
| 7 | 7 | 0 | 0 | 14 | 7 | n.t. | − | − | − | − | 7 | − | − | − | − | |
| 18 | 18 | 0 | 0 | 36 | − | n.t. | − | − | − | − | 36 | − | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | − | n.t. | − | 1 | − | − | 1 | − | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | 1 | n.t. | − | − | − | − | − | 1 | − | − | − | |
| 2 | 2 | 0 | 0 | 4 | − | n.t. | − | − | − | − | 2 | 2 | − | − | − | |
| 30 | 30 | 0 | 0 | 60 | 10 | 1 | 46 | 3 | ||||||||
| San Namibia | 1 | 1 | 0 | 0 | 2 | 1 | n.t. | − | − | − | − | 1 | − | − | − | − |
| 1 | 1 | 0 | 0 | 2 | − | − | − | − | − | 1 | 1 | − | − | − | − | |
| 5 | 5 | 0 | 0 | 10 | − | n.t. | − | − | − | − | 10 | − | − | − | − | |
| 7 | 7 | 0 | 0 | 14 | 1 | 1 | 12 | |||||||||
| Yoruba Nigeria | 3 | 3 | 0 | 0 | 6 | 3 | n.t. | − | − | − | − | 3 | − | − | − | − |
| 15 | 15 | 0 | 0 | 30 | − | n.t. | − | − | − | − | 30 | − | − | − | − | |
| 2 | 2 | 0 | 04 | − | n.t. | -2 | − | − | 2 | − | − | − | − | |||
| 2 | 2 | 0 | 0 | 4 | − | n.t. | − | − | − | − | 2 | 2 | − | − | − | |
| 1 | 1 | 0 | 0 | 2 | 1 | n.t. | − | − | − | − | − | − | 1 | − | − | |
| 2 | 2 | 0 | 0 | 4 | − | n.t. | − | − | − | − | 2 | − | 2 | − | − | |
| 25 | 25 | 0 | 0 | 50 | 4 | 2 | 39 | 2 | 3 | |||||||
|
CEPH all, n |
157 | 157 | 0 | 0 | 314 | 33 | 2 | 3 | 22 | 5 | 2 | 218 | 10 | 17 | 0 | 2 |
| Haplotype frequencies, | 100 | 10.51 | 0.64 | 0.96 | 7.01 | 1.59 | 0.64 | 69.43 | 3.18 | 5.41 | 0.00 | 0.64 | ||||
Table 2 DNA samples (157) came from different African ethnicities, were derived from lymphoblastoid cell lines, and had been provided by the Centre d'Etude du Polymorphism Humain (CEPH, Foundation Jean Dausset, Paris, France).
Blood donor reference samples “Verena”, “Bruno”, “Helmut,” and “Urs” were from the Canton of Zurich (Switzerland) and had MMSS, MMss, NNSS, and NNss phenotypes, respectively.
Phenotyping Procedures
Samples from Switzerland had M/N and S/s serology performed as described previously [28]. Samples originating from Versiti, Inc., were S/s phenotyped from EDTA blood using a test tube method following the manufacturers' recommendations (Bio-Rad Laboratories, Hercules, CA, USA; Immucor, Norcross, GA, USA; Ortho Clinical Diagnostics, Raritan, NJ, USA). Serological phenotype U testing was performed using an unlicensed human source reagent. S–s–U– blood donors were phenotyped prior to confirmatory genotyping.
Analytical and Diagnostic Gap-PCRs, Diagnostic PCRs using Sequence-Specific Priming, and Sequencing
PCRs and sequencing were done as described previously [29]. Primers, their concentrations, and combinations are provided in the supplementary Table s1 (for all online suppl. Material, see www.karger.com/doi/10.1159/000504946). Confirmatory genotyping for GYPB c.143C>T, c.230C>T, and c.270 + 5G>T was performed on all samples originating from the USA using a nanofluidic open-array system (OpenArray real-time PCR system; Life Technologies Corporation) [30]. DNA that failed to amplify all 3 targets [21] were deemed to have a GYPB homozygous deletion and were confirmed by S/s phenotyping with licensed reagents and an unlicensed anti-U. Due to DNA depletion of some samples, further analysis of all Table 1 and 2 samples were preterminated whenever unambiguous genotypes, as defined by the unequivocal identification of 2 different parental alleles/haplotypes, occurred (online suppl. Material).
Briefly, methods for bioinformatic analysis of the 1000G phase 3 data included the following core steps: Potentially true large deletion candidates in the region of interest were confirmed if (a) paired-end reads were found with an extremely large separation distance after alignment, especially if a soft-clipped part of one end could be mapped in the region of the other end, or (b) distinct coverage drops were found, or (c) complete coverage gaps were found, or (d) a combination of several of these criteria was true. Details of the bioinformatic 1000G phase 3 data, homology analysis of the GYP locus, and allele frequency (AF) calculations are given in the online supplementary material.
Results
Homology Analysis of the GYP Locus
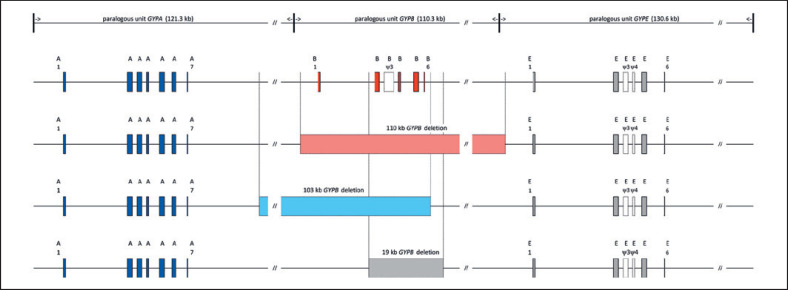
Using GenBank accession No. NC_00004.12 (GRCh38.p12) as a reference, multiple sequence alignment of the paralogous GYP locus on chromosome 4 highlighted the 3 tandemly organized units (Fig. 1; online suppl. Fig. s1). Each segmental duplication consisted of the gene and continued with a large intergenic sequence, summing up to their individual unit lengths of 121.3, 110.3, and 130.6 kb, for GYPA, GYPB, and GYPE, respectively. Sequence similarity was mainly interrupted by unit-specific insertions, the longest with approximately 16.0 kb, carrying exon 6 and 7 of GYPA and being absent from GYPB and GYPE.
Fig. 1.
GYP locus on chromosome 4 and GYPB deletions identified in Black Africans. Paralog units of 121.3-kb GYPA, 110.3-kb GYPB, and 130.6-kb GYPE are indicated in the upper most line. The GYPA homologous unit started approximately 7.3 kb, 5′ of its start codon before ending 7.3 kb 5′ of GYPB. Homology of the 3rd unit started 10.8 kb 5′ of GYPE. The drawing is in scale with exons magnified 10 times in relation and 60.0 kb not shown for every unit as indicated by //. In the second line, exons of the 3 genes as found in a wild-type GYP locus and given as vertical boxes colored in blue, red, and gray for GYPA, GYPB, and GYPE, respectively. Pseudoexons (ψ) present in GYPB and GYPE are uncolored and numbered so that homologous exons maintain the same number in all 3 genes. Lines 3–5 show the relative positions and extent of the 110-kb (light red), 103-kb (light blue), and 19-kb (grey) deletions of GYPB observed in this study. Sizes of the 110- and 103-kb GYPB deletions approximately corresponded to the length of 1 paralog unit.
Fine Mapping of Breakpoint Candidate Regions
Bioinformatic analysis of the 1000G phase 3 data revealed multiple hits for 2 distinct GYPB deletions, ∼110 and ∼110 kb in length, with corresponding haplotype frequencies of approximately 0.05 and 0.02, respectively, and 1 hit each for a ∼19- and ∼32-kb GYPB deletion. All deletions were identified among Black Africans (AFR of the 1000G data set). Both ∼100-kb GYPB deletions affected all GYPB, the ∼19- and ∼32-kb deletions were located within GYPB. Due to the high paralogy of the GYP locus, sequences derived from GenBank accession No. NT_016354.20 were reviewed for accuracy by a comparison to sequences derived from 4 wild-type DNAs of the MMSS, NNSS, MMss, and NNss phenotype, respectively (Fig. 2).
Fig. 2.
Sequence alignments of breakpoint regions corresponding to the 110-kb (a), 103-kb (b), and 19-kb GYPB deletions (c). Unit-specific nucleotides of the homologous breakpoint regions corresponding to the 110-, 103-, and 19-kb GYPB deletions are given in blue for unit GYPA, in red for unit GYPB, and in gray for unit GYPE. For each gene, reference sequence NT_016354.20 and wild-type sequences of 4 blood donors, dubbed Verena, Bruno, Helmut, and Urs of MMSS, MMss, NNSS, and NNss blood group MNS phenotype, respectively, are given (DNAs c). With the exemption of 2 unit GYPE-specific wild-type sequences, all other and 1 breakpoint sequence of every deletional type each, e.g. the 110, 103, and 19 kb, presented in a–c were submitted to GenBank (accession Nos. MN005662–MN005698; online suppl. Table s5). Breakpoints of the 3 deletions are indicated by a vertical black line. Position numbers given in the header line indicate nucleotide positions in relation to the deletional breakpoint (negative counts for positions downstream of the breakpoint and positive counts for positions upstream of the breakpoint). Unavailable sequence information is indicated by “*”, and deletion of nucleotides is indicated by “–”. Framed nucleotides show linkage to either the M/N, or S/s causative SNPs in exon 2 of GYPA (e.g., rs7682260) and exon 4 of GYPB (e.g., rs7683365). a Breakpoint region corresponding to the 110-kb GYPB deletion (GYPB*05N.01). The 110.24-kb deletion completely removed GYPB and extended from genomic coordinates 144025065–143914828 (GRCh38.p12, NC_000004.12). A hybrid sequence between unit GYPBand unit GYPE was created. Four resultant hybrid sequences are shown in the lower part of the alignment. Three were from samples of our study, 1 submitted to GenBank (MN005664) and 1 (DEL1) was found by Leffler et al. [23]. Nucleotides shown in green (“privates”), i.e., −800A, −217, and +36A, were identified as SNPs with potential specificity for the 110-kb deletional GYPB haplotype. Subsequent genotyping of 2 SNPs (−800A and +36A) in all 157 African CEPH samples, however, did not confirm suspected full linkage of these polymorphisms to the 110-kb GYPB deletion (online suppl. Table s2). b Breakpoint region corresponding to the 103-kb GYPB deletion (GYPB*05N.02). The 103.26-kb deletion completely removed GYPB and extended from genomic coordinates 144094973–143991719 (GRCh38.p12, NC_000004.12). A hybrid sequence between unit GYPA and unit GYPB was created. Three resultant hybrid sequences are shown in the lower part of the alignment. All were from samples within our study, 1 was submitted to GenBank (MN005663). At first, nucleotides shown in green, “privates,” were specifically identified on hybrid sequences only. Subsequent typing of nucleotide −402T and +70C among all 157 African CEPH samples resulted in full linkage of these polymorphisms to the 103-kb GYPB deletion (online suppl. Table s2). c Breakpoint region corresponding to the 19-kb GYPB deletion (GYPB*05N.03). The 18.61-kb deletion removed all DNA from 1.48 kb 5′ of the donor splice site of exon 2 until 12.63 kb downstream of the stop codon of GYPB and extended from genomic coordinates 144002272–143990949 (GRCh38.p12, NC_000004.12). The hybrid sequence was created within GYPB and could only be identified in sample HG01880*C1 of the Coriell Human Genetic Cell Repository. Breakpoint region sequence of HG01880*C1 was submitted to GenBank (MN005662). In this alignment, continuation of wild-type sequences into the area of the deletion indicated by 2 vertical black lines is shown. The gray vertical bar represents approximately 18.61 kb of sequence not shown. Whereas the hybrid sequence 3′ of the breakpoint clearly shows specificity for GYPB. Green nucleotides up to position −207, 5′ of the breakpoint, however, lack homology to either of the 3 genes.
Gap-PCRs to Track GYPB Deletions
Combining the working principle of Gap-PCRs [31] with PCR amplification using sequence-specific priming [32, 33] allowed for bridging the GYPB deletions by exclusively deletion-specific amplicons (Gap-PCRs, suppl. table s1A). Gap-PCRs were tested on an original 4 DNAs of known S–s– individuals and 2 samples with low GYPB:GYPE gene ratios (Table 1), indicative of the lack of 1 parental GYPB gene [26]. Gap-PCRs for the 2 expected ∼100-kb GYPB deletions resulted in positive amplification in 4 of the 6 samples each and were reproduced on additional samples with low GYPB:GYPE gene ratios. Amplicons were sequenced (suppl. table s1A, B) from 3 independent samples each and showed identical breakpoint nucleotide sequences for both distinct ∼100-kb GYPB deletions (Fig. 2a, b). The breakpoint of the expected ∼19-kb GYPB deletion could only be identified and sequenced from the 1000G Coriell sample HG01880*C1 (Fig. 2c). Linkage exploration of the 110- and 103-kb GYPB deletions is detailed in the supplementary Material (online suppl. Results and online suppl. Tables s2, s3).
Appearance of 3 GYPB Deletions
One 110.24-kb deletion stretched from 5.78 kb upstream of the GYPB start codon until 9.32 kb upstream of the GYPE start codon (genomic coordinates NC_000004.12, GRCh38.p12, 144025065–143914828). The 103.26-kb deletion started 16.47 kb downstream of the GYPA stop codon and ended 4.58 kb downstream of the GYPB stop codon (coordinates 144094973–143991719). Both deletions encompassed the whole GYPB gene and involved flanking paralogous sequences, separated in undeleted haplotypes by the approximate length of a single GYP unit. The ∼19-kb GYPB deletion was 18.61 kb in length and started 1.48 kb 5′ of the donor splice site of exon 2 and reached until 12.63 kb downstream of the stop codon (coordinates 144002272–143990949) (ALl shown in Figure 1, 2; online suppl. Fig. s1.)
Diagnostic Typing for the 110-, 103-, and 19-kb GYPB Deletions
Diagnostic Gap-PCRs different from the analysis described above specifically detected the presence of any of the 3 GYPB deletions. Additional detection of undeleted (wild-type) sequences at the homologous positions of the 5′ and 3′ deletional endpoints (“breakpoints”) enabled the discrimination of homo- versus heterozygous 110-, 103-, and 19-kb GYPB deletions, respectively (Fig. 3; online suppl. Table s1C).
Fig. 3.
Diagnostic PCRs for the 110- and 103-kb GYPB deletions. Primers used and reaction numbers of diagnostic Gap-PCRs and PCR sequence-specific priming are given in the supplementary Table s1C. A positive amplification control is run in every lane (480 bp). Lane 1 corresponds to 103-1.3 and detects the presence of the 103-kb deletion of GYPB (GYPB*05N.02). Lane 4 corresponds to 110-1.3 and detects the presence of the 110-kb GYPB deletion (GYPB*05N.01). Reactions 2, 3, 4, and 6 are detecting wild-type sequences corresponding to the 5′ and 3′ breakpoint ends of both deletional haplotypes, respectively. Typing for 4 DNA samples is shown, and their pheno- and genotype is given. The uppermost sample represents a wild-type DNA of either SS, Ss, or ss blood group MNS phenotype, with all reactions positive except for Nos. 1 and 4. GYPB*0x denotes any GYPB allele other than the deletional haplotypes GYPB*05N. The lowermost sample is compound heterozygous for the 110- and 103-kb GYPB deletion, e.g., has the compound heterozygous genotype GYPB*05N.01/GYPB*05N.02, with reactions 1, 2, 4, and 6 positive. Wild-type reactions 2 and 6 are also positive, since reaction 2 detects the corresponding wild-type sequence present on GYPB*05N.01, and reaction 6 vice versa.
Among all 26 samples (Table 1) of S–s–U– phenotype with proven serology, 14 were homozygous for the 110-kb GYPB deletion, 8 were heterozygous for 110- and 103-kb deletions, 1 was homozygous for the 103-kb deletion, and 3 were heterozygous for the 110-kb deletion and a second GYPB deletion not yet further analyzed. Among these 26 samples, 39 (75.0%), 10 (19.2%), and 3 (5.8%) deletional GYPB haplotypes were observed for the 110-kb, the 103-kb, and 3 GYPB deletions of yet unknown specificity, respectively. With respect to genotypes, 23 of the 26 (88.4%) samples with proven S–s–U– phenotype were exclusively defined by the 110- and 103-kb GYPB deletions. Among all other 90 samples of Table 1, a further 28 samples carried the 110-kb and 2 samples carried the 103-kb GYPB deletional haplotypes. The 1000G Coriell sample originally identified as representing a putative ∼32-kb GYPB deletion (Coriell HG02970*A1) turned out to be heterozygous for the 2 deletions of 110 kb and 103 kb, causing a compound overlap void of any GYPB sequences with a calculated length of 33.3 kb. Another 8 samples of Table 1 and 1 sample of Table 2 were also shown to share this heterozygous 110-/103-kb GYPB deletion genotype (Tables 1, 2).
Among the 157 CEPH-DNAs (100.00%) representing 8 different African ethnicities (Table 2), 9 samples (5.73%) carried one 110-kb deletional haplotype, 14 (8.91%) carried one 103-kb haplotype, 1 (0.64%) showed compound heterozygosity for 110/103 kb, and 1 (0.64%) homozygosity for the 103-kb deletional GYPB haplotypes, respectively. Pygmy people of the Democratic Republic of the Congo (DR Congo) and the Central African Republic (CAR) showed the highest frequencies of the 2 GYPB deletions, summing up to a combined 23.33% in Mbuti and 13.89% for the solely present 103-kb deletion in the Biaka. Overall, African (CEPH) frequencies were 3.18 and 5.41% for the 110- and 103-kb deletional GYPB haplotypes, respectively (Table 2). Despite the low number of individuals investigated, differences in the geographic distribution of the 2 different GYPB deletions were observed. The 110-kb GYPB deletion seemed to be more widely spread throughout the African continent, whereas the 103-kb deletional haplotype was observed exclusively in Equatorial Africa (Fig. 4).
Fig. 4.
Distribution of relative allele and haplotype frequencies for the various GYPB variants in CEPH samples of African cohorts. Frequencies shown are according to Table2. Red, GYPB*05N.01 (del GYPB 110 kb); blue, GYPB*05N.02 (del GYPB 103 kb); green, GYPB*03N.04; purple, GYPB*03N.03; gray, all other GYPB alleles. Black dots represent the approximate geographical location of the various populations investigated and were: Mozabite (Algeria, Mzab, n = 30), Mandenka (Senegal, n = 24), Biaka Pygmy (Central African Republic, n = 36), Mbuti Pygmy (Democratic Republic of the Congo, n = 15), Yoruba (Nigeria, n = 25), Bantu (NE, Kenya, n = 12), San (Namibia, n = 7), and Bantu (SE and SW, South Africa, n = 8).
Comprehensive Molecular Typing of All Samples
Following the described typing hierarchy (online suppl. Materials), all samples were tested for SNPs GYPA c.59C>T (M/N, rs7682260) and GYPB c.143C>T (S/s, rs7683365), for deletions of GYPB of the 110-, 103-, or 19-kb type plus 5′ and 3′ breakpoint positions only present in wild-type haplotypes, and GYPB SNPs c.59T>G (He) and U+w alleles as defined by GYPB SNPs c.230C>T and c.270 + 5g>t. All 157 CEPH-DNAs of African origin (Table 2) were additionally tested for “private” SNPs (Fig. 2a, b) −800A (144025051, rs143076335 T) and +36A (143913980, no rs) of the 110-kb GYPB deletion and −402T (genomic coordinate 144095375, rs186872886, A) and +70C (143991649, rs142534144, G) of the 103-kb GYPB deletion for further linkage analysis (online suppl. Materials). Detailed results of the CEPH-DNA are given in online supplementary Table s4.
AFs among Biaka and Mbuti Pygmy People
The investigated 36 Biaka Pygmy people from the CAR were the only carriers of the U+var, GYPB*03N.03 alleles with a local AF of 4.2%. Among the 15 Mbuti Pygmy from the DR Congo 13 U+var, GYPB*03N.04 alleles were observed, resulting in a local AF of 43.3%. Beside the combined local haplotype frequency for the 110- and 103-kb deletional GYPB haplotypes in Mbuti of 23.3%, only 8 regular (defined by SNP-based typing only) GYPB*04 alleles were found, representing 26.7% local AF (Table 2).
Data Submission and Allele/Haplotype Names
All novel sequences were submitted to the GenBank under accession Nos. MN005662–MN005698 (online suppl. table s5). Reference sequences of the breakpoint regions of the characterized GYPB deletions are: BP_GYPBdel19kb_HG01880*C1, accession Nos. MN005662, BP_GYPB-del103kb_Lorena, MN005663, and BP_GYPB-del110kb_Gerold, accession No. MN005664. Due to their overall frequency, GYPB*05N.01 was granted as allele name for the GYPB deletional haplotype of 110-kb (spanning from 5′ across GYPB to GYPE) and GYPB*05N.02 for the 103-kb (spanning from 3′ of GYPA across GYPB) variant from the Terminology Working Party of the ISBT [1].
Discussion
GYPB is not essential for RBC development or survival in humans. Hence, the precise functional role of GYPB in humans remains unclear. Its polymorphism, however, is essential for the survival of patients in the context of blood transfusion. Fatal hemolytic transfusion reactions or hemolytic disease of the fetus and newborn have been reported in GPB-negative individuals with antibodies against S, s, or U antigens, normally expressed on GPB [6, 34]. Bioinformatic data analysis of phase 3 1000G whole genomes delivered approximate positions for GYPB deletions on chromosome 4 among Black Africans. Positions of the approximate breakpoints were analyzed in detail and resulted in the identification of 110- and 103-kb deletions, both including the complete GYPB gene. In 26 Black Africans, most of them from the US and all of them with a serologically proven negativity for S, s, and U, 23 (88.4%) were homo- or compound heterozygous for these 2 deletions. Only 3 individuals carried a second, yet unidentified, deletional haplotype involving parts of GYPB aside a 110-kb deletional haplotype. Among the 26 samples, the 110- and 103-kb GYPB deletions had haplotype frequencies of 75.0 and 19.2%, respectively, representing 94.2% of all deletional haplotypes causative of the MNS S–s–U– phenotype.
In 157 CEPH samples without serological data for S, s, or U, the 110-kb GYPB deletion was found in 5 of 8 different African ethnicities. Presence of the 103-kb GYPB deletion appeared to be limited to Equatorial Africa, e.g., the Yoruba people from Nigeria, and Biaka and Mbuti pygmy people from the CAR and DR Congo, respectively. Recently, Leffler et al. [23] reported on structural variants, e.g., duplications and deletions affecting the GYP locus. Though lacking phenotypically derived proof for U negativity, they identified different deletions among the 1000G phase 3 reference sample set, 5 of them also affecting GYPB. Breakpoint sequences of their DEL1 and our 110-kb GYPB deletion seem to be identical with respect to the overlapping sequence information. For DEL2 and DEL8, no sequences were given, but the first seems to correspond to our 103-kb GYPB deletion, and the latter to our 19-kb deletion, where all coding sequences except GYPB exon 1 is absent. Other deletional GYPB variants have been reported earlier but did not correspond to the ones reported here or previously [8, 9, 10, 23].
Haplotype frequencies of the 110- and 103-kb deletions were 3.18 and 5.14% among all 157 African (CEPH) samples investigated. African Americans, in contrast, showed the 110-kb deletional haplotype almost 6 times more frequently than the 103-kb type. In Africa, Leffler et al. [23] found the 110-kb deletional haplotype, e.g., their DEL1 (our 110-kb deletion), at a frequency of 4.10% and their DEL2 (our 103-kb deletion) at a frequency of 0.75%. Taking all data into account, the 110-kb GYPB deletion seems to be the most frequent hereditary factor causative of the S–s–U– blood group phenotype, followed by the 103-kb GYPB deletion. Therefore, GYPB*05N.01 and GYPB*05N.02 were assigned as allele names for the 110- and 103-kb deletional GYPB haplotypes by the ISBT Working Party Red Cell Immunogenetics and Blood Group Terminology, respectively [1].
With the elevated frequencies observed as the only indicator, it could be speculated that both deletions originated in CAR Pygmy people and that the 110-kb GYPB deletion represents the more ancestral mutation, since it is more widely spread throughout the African continent (Fig. 4). Additional interest for the MNS blood group genetics of Pygmy people arose from the U+var, GYPB*03N.03 allele, exclusively found among the Biaka people of the CAR. Also, only Biaka people showed the 103-kb GYPB deletional haplotype but had no 110-kb deletions. Mbuti Pygmy people from the DR Congo additionally exhibited high frequencies for variants of GYPBs, 74.3% of all chromosomes either carried alleles of the He (GYPB*06.01), U+var(GYPB*03N.04), or 1 of the 2 GYPB deletions, all of which are known to express lowered, trace amounts, or no GPB at all, respectively [13, 35]. As a consequence of these high AFs, only 6 out of 15 Mbuti Pygmy people were shown to express an apparently normal wild-type S–s+ phenotype, whereas 9 out of 15 were predicted to have an altered, remnant, or lacking expression of GPB on their erythrocyte surface (Table 2; online suppl. Table s5). Though only analyzed within a small sample collection, these frequencies are highly remarkable.
Investigating structural variation in the GYP locus, Leffler et al. [23] were not able to prove a protective effect against malaria of altered GPB expression other than the presence of the MNS antigen Dantu [23, 24]. However, a potentially cumulative effect of all allelic/haplotypic variation in GYPB causative of variant, remnant, or full absence of GPB expression, as shown among Pygmy people, has not been ruled out in the course of protection against malaria until now [19, 20, 21]. It might be of interest to specifically address such cumulative blood group MNS antigen variation and its molecular background on the protective effect against malaria and other diseases.
Our work allowed for the design of diagnostic Gap-PCRs, capable of positively detecting the presence of either of the 2 GYPB deletional haplotypes and additionally included testing for the presence of wild-type sequences at their 5′ and 3′ junctions. Comprehensive molecular genotyping, e.g., calling both parental haplotypes in all their zygosities, is now possible for U and U+var. Primarily in Black Africans and admixed populations, this will allow for the discrimination of true phenotype S–s–U– individuals from carriers of GYPB genes still expressing trace amounts of GPB, as expressed in U+var phenotypes [13]. DNA-based diagnosis of U is now also possible in prenatal settings.
As stated by Daniels [6], MNS was the second blood group system to be discovered, but probably second only to Rh in its complexity. The complexity of MNS has again increased. In any case, access to the 103- and 110-kb GYPB deletions, accounting for the most frequent hereditary factors causative of the MNS blood group phenotype S–s–U–, is now diagnostically available.
Statement of Ethics
Ethical approval for the study was obtained from the Cantonal Office of Public Health, Zurich, Switzerland (Swiss approval No. 2014-0408).
Disclosure Statement
C.G. acts as a consultant to inno-train Diagnostik GmbH, Kronberg, Germany. Diagnostic procedures for the molecular detection of GYPB deletions of the 110- and 103-kb type have been submitted as a patent application (pending). G.D. is on the speaker bureau for Grifols S.A., Barcelona, Spain. All other authors declare no conflicts of interest.
Funding Sources
The present study was financed by the Blood Transfusion Service Zurich, Swiss Red Cross, Zürich-Schlieren, Switzerland.
Author Contributions
Contribution: C.G., C.P., S.M., N.T., M.P.M.-G., Y.-L.S., C.E., K.M.B., and M.F. performed experiments and analyzed data; G.D., C.J., B.J., J.R.S., and A.F. contributed important material; C.G., C.P., M.P.M.-G., M.F., A.F., and B.M.F. discussed the results and commented on the manuscript; C.G. designed and supervised the study, made the figures, and wrote the manuscript; and all authors edited the manuscript.
Supplementary Material
Supplementary data
References
- 1.International Society of Blood Transfusion (ISBT) Working Party Red Cell Immunogenetics and Blood Group Terminology. http://www.isbtweb.org/working-parties/red-cell-immunogenetics-and-blood-group-terminology/ [Google Scholar]
- 2.Landsteiner K, Levine P. A new agglutinable factor differentiating individual human bloods. Proc Soc Exp Biol NY. 1927;24((6)):600–2. [Google Scholar]
- 3.Walsh RJ, Montgomery CM. A new human iso-agglutinin subdividing the MN blood groups. Nature. 1947 Oct;160((4067)):505. [PubMed] [Google Scholar]
- 4.Levine P, Kuhmichel AB, Wigod M, Koch E. A new blood factor, s, allelic to S. Proc Soc Exp Biol Med. 1951 Oct;78((1)):218–20. doi: 10.3181/00379727-78-19025. [DOI] [PubMed] [Google Scholar]
- 5.Wiener AS, Unger LJ, Gordon EB. Fatal hemolytic transfusion reaction caused by sensitization to a new blood factor U: report of a case. J Am Med Assoc. 1953 Dec;153((16)):1444–6. doi: 10.1001/jama.1953.02940330028010. [DOI] [PubMed] [Google Scholar]
- 6.Daniels G. Human Blood Groups. 3rd ed. Oxford, UK: Wiley-Blackwell; 2013. [Google Scholar]
- 7.Greenwalt TJ, Sasaki T, Sanger R, Sneath J, Race RR. An Allele of the S(s) Blood Group Genes. Proc Natl Acad Sci USA. 1954 Dec;40((12)):1126–9. doi: 10.1073/pnas.40.12.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CH, Johe K, Moulds JJ, Siebert PD, Fukuda M, Blumenfeld OO. Delta glycophorin (glycophorin B) gene deletion in two individuals homozygous for the S—s—U— blood group phenotype. Blood. 1987 Dec;70((6)):1830–5. [PubMed] [Google Scholar]
- 9.Tate CG, Tanner MJ, Judson PA, Anstee DJ. Studies on human red-cell membrane glycophorin A and glycophorin B genes in glycophorin-deficient individuals. Biochem J. 1989 Nov;263((3)):993–6. doi: 10.1042/bj2630993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahuel C, London J, Vignal A, Ballas SK, Cartron JP. Erythrocyte glycophorin B deficiency may occur by two distinct gene alterations. Am J Hematol. 1991 May;37((1)):57–8. doi: 10.1002/ajh.2830370115. [DOI] [PubMed] [Google Scholar]
- 11.Francis BJ, Hatcher DE. MN blood types. The S-s-U+ and the M1 phenotypes. Vox Sang. 1966 Mar-Apr;11((2)):213–6. doi: 10.1111/j.1423-0410.1966.tb04224.x. [DOI] [PubMed] [Google Scholar]
- 12.Storry JR, Reid ME. Characterization of antibodies produced by S-s- individuals. Transfusion. 1996 Jun;36((6)):512–6. doi: 10.1046/j.1537-2995.1996.36696269509.x. [DOI] [PubMed] [Google Scholar]
- 13.Storry JR, Reid ME, Fetics S, Huang CH. Mutations in GYPB exon 5 drive the S-s-U+(var) phenotype in persons of African descent: implications for transfusion. Transfusion. 2003 Dec;43((12)):1738–47. doi: 10.1046/j.0041-1132.2003.00585.x. [DOI] [PubMed] [Google Scholar]
- 14.Reid ME, Storry JR, Maurer J, Nance ST. Practical method for determination of the U status of S-s- erythrocytes. Immunohematology. 1997;13((4)):111–4. [PubMed] [Google Scholar]
- 15.Pasvol G, Jungery M, Weatherall DJ, Parsons SF, Anstee DJ, Tanner MJ. Glycophorin as a possible receptor for Plasmodium falciparum. Lancet. 1982 Oct;2((8305)):947–50. doi: 10.1016/s0140-6736(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 16.Baum J, Thomas AW, Conway DJ. Evidence for diversifying selection on erythrocyte-binding antigens of Plasmodium falciparum and P. vivax. Genetics. 2003 Apr;163((4)):1327–36. doi: 10.1093/genetics/163.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko WY, Kaercher KA, Giombini E, Marcatili P, Froment A, Ibrahim M, et al. Effects of natural selection and gene conversion on the evolution of human glycophorins coding for MNS blood polymorphisms in malaria-endemic African populations. Am J Hum Genet. 2011 Jun;88((6)):741–54. doi: 10.1016/j.ajhg.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HY, Tang H, Shen CK, Wu CI. Rapidly evolving genes in human. I. The glycophorins and their possible role in evading malaria parasites. Mol Biol Evol. 2003 Nov;20((11)):1795–804. doi: 10.1093/molbev/msg185. [DOI] [PubMed] [Google Scholar]
- 19.Dolan SA, Proctor JL, Alling DW, Okubo Y, Wellems TE, Miller LH. Glycophorin B as an EBA-175 independent Plasmodium falciparum receptor of human erythrocytes. Mol Biochem Parasitol. 1994 Mar;64((1)):55–63. doi: 10.1016/0166-6851(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 20.Gaur D, Storry JR, Reid ME, Barnwell JW, Miller LH. Plasmodium falciparum is able to invade erythrocytes through a trypsin-resistant pathway independent of glycophorin B. Infect Immun. 2003 Dec;71((12)):6742–6. doi: 10.1128/IAI.71.12.6742-6746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dankwa S, Chaand M, Kanjee U, Jiang RH, Nobre LV, Goldberg JM, et al. Genetic Evidence for Erythrocyte Receptor Glycophorin B Expression Levels Defining a Dominant Plasmodium falciparum Invasion Pathway into Human Erythrocytes. Infect Immun. 2017 Sep;85((10)):e00074–17. doi: 10.1128/IAI.00074-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang CH, Blumenfeld OO. Characterization of a genomic hybrid specifying the human erythrocyte antigen Dantu: dantu gene is duplicated and linked to a delta glycophorin gene deletion. Proc Natl Acad Sci USA. 1988 Dec;85((24)):9640–4. doi: 10.1073/pnas.85.24.9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leffler EM, Band G, Busby GB, Kivinen K, Le QS, Clarke GM, et al. Malaria Genomic Epidemiology Network Resistance to malaria through structural variation of red blood cell invasion receptors. Science. 2017 Jun;356((6343)):eaam6393. doi: 10.1126/science.aam6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ndila CM, Uyoga S, Macharia AW, Nyutu G, Peshu N, Ojal J, et al. MalariaGEN Consortium Human candidate gene polymorphisms and risk of severe malaria in children in Kilifi, Kenya: a case-control association study. Lancet Haematol. 2018 Aug;5((8)):e333–45. doi: 10.1016/S2352-3026(18)30107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The International Genome Sample Resource (IGSR) Providing ongoing support for the 1000 Genomes Project data. doi: 10.1093/nar/gkw829. https://www.internationalgenome.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer S, Vollmert C, Trost N, Sigurdardottir S, Portmann C, Gottschalk J, et al. MNSs genotyping by MALDI-TOF MS shows high concordance with serology, allows gene copy number testing and reveals new St(a) alleles. Br J Haematol. 2016 Aug;174((4)):624–36. doi: 10.1111/bjh.14095. [DOI] [PubMed] [Google Scholar]
- 27.Gassner C, Meyer S, Frey BM, Vollmert C. Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry-based blood group genotyping—the alternative approach. Transfus Med Rev. 2013 Jan;27((1)):2–9. doi: 10.1016/j.tmrv.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Meyer S, Vollmert C, Trost N, Brönnimann C, Gottschalk J, Buser A, et al. High-throughput Kell, Kidd, and Duffy matrix-assisted laser desorption/ionization, time-of-flight mass spectrometry-based blood group genotyping of 4000 donors shows close to full concordance with serotyping and detects new alleles. Transfusion. 2014 Dec;54((12)):3198–207. doi: 10.1111/trf.12715. [DOI] [PubMed] [Google Scholar]
- 29.Gourri E, Denomme GA, Merki Y, Scharberg EA, Vrignaud C, Frey BM, et al. Genetic background of the rare Yus and Gerbich blood group phenotypes: homologous regions of the GYPC gene contribute to deletion alleles. Br J Haematol. 2017 May;177((4)):630–40. doi: 10.1111/bjh.14578. [DOI] [PubMed] [Google Scholar]
- 30.Hopp K, Weber K, Bellissimo D, Johnson ST, Pietz B. High-throughput red blood cell antigen genotyping using a nanofluidic real-time polymerase chain reaction platform. Transfusion. 2010 Jan;50((1)):40–6. doi: 10.1111/j.1537-2995.2009.02377.x. [DOI] [PubMed] [Google Scholar]
- 31.Harteveld CL. State of the art and new developments in molecular diagnostics for hemoglobinopathies in multiethnic societies. Int J Lab Hematol. 2014 Feb;36((1)):1–12. doi: 10.1111/ijlh.12108. [DOI] [PubMed] [Google Scholar]
- 32.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989 Apr;17((7)):2503–16. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu DY, Ugozzoli L, Pal BK, Wallace RB. Allele-specific enzymatic amplification of beta-globin genomic DNA for diagnosis of sickle cell anemia. Proc Natl Acad Sci USA. 1989 Apr;86((8)):2757–60. doi: 10.1073/pnas.86.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piliay GS, Womack B, Sandler SG. Immune-mediated hemolysis in a postoperative patient Case report: anti-U and differential diagnosis. Immunohematology. 1993;9((2)):41–6. [PubMed] [Google Scholar]
- 35.Reid ME, Lomas-Francis C, Olsson ML. The Blood Group Antigen FactsBook. 3rd ed. London, UK: Elsevier; 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data