ABSTRACT
Background
Different fatty acids (FAs) can vary in their obesogenic effect, and genetic makeup can contribute to fat deposition in response to dietary FA composition. However, the antiobesogenic effects of the interactions between dietary MUFAs and genetics have scarcely been tested in intervention studies.
Objective
We evaluated the overall (primary outcome) and genetically modulated (secondary outcome) response in body weight and fat mass to different levels of MUFA consumption.
Methods
In the Canola Oil Multicenter Intervention Trial II, a randomized, crossover, isocaloric, controlled-feeding multicenter trial, 44 men and 71 women with a mean age of 44 y and an increased waist circumference (men ∼108 cm and women ∼102 cm) consumed each of 3 oils for 6 wk, separated by four 12-wk washout periods. Oils included 2 high-MUFA oils—conventional canola and high-oleic canola (<7% SFAs, >65% MUFAs)—and 1 low-MUFA/high-SFA oil blend (40.2% SFAs, 22.0% MUFAs). Body fat was measured using DXA. Five candidate single-nucleotide polymorphisms (SNPs) were genotyped using qualitative PCR. Data were analyzed using a repeated measures mixed model.
Results
No significant differences were observed in adiposity measures following the consumption of either high-MUFA diet compared with the low-MUFA/high-SFA treatment. However, when stratified by genotype, 3 SNPs within lipoprotein lipase (LPL), adiponectin, and apoE genes influenced, separately, fat mass changes in response to treatment (n = 101). Mainly, the LPL rs13702-CC genotype was associated with lower visceral fat (high-MUFA: −216.2 ± 58.6 g; low-MUFA: 17.2 ± 81.1 g; P = 0.017) and android fat mass (high-MUFA: −267.3 ± 76.4 g; low-MUFA: −21.7 ± 102.2 g; P = 0.037) following average consumption of the 2 high-MUFA diets.
Conclusions
Common variants in LPL, adiponectin, and apoE genes modulated body fat mass response to dietary MUFAs in an isocaloric diet in adults with abdominal obesity. These findings might eventually help in developing personalized dietary recommendations for weight control. The trial was registered at clinicaltrials.gov as NCT02029833 (https://www.clinicaltrials.gov/ct2/show/NCT02029833?cond=NCT02029833&rank=1).
Keywords: dietary fatty acids, fat quality, genotype, gene–nutrient interaction, fatness, adiposity
Introduction
The composition of dietary fatty acids (FAs) has been recognized as a determinant of fat deposition and distribution (1–5). FAs can vary in their obesity-inducing effects by influencing energy expenditure, fat oxidation, and thermogenesis, and/or modulating appetite sensation (6–8). Increasing evidence has demonstrated that dietary MUFAs increase fat oxidation, diet-induced thermogenesis (8–10), and resting energy expenditure (11), and promote weight loss (12, 13) compared with SFAs. Our recent controlled feeding study showed that 2 test diets high in MUFAs—canola oil and a high-oleic canola oil—significantly reduced android fat mass compared with a high-PUFA flaxseed/safflower oil in participants with abdominal obesity (1). These favorable effects might be attributed partly to interactions between FAs and genetic polymorphisms.
The genetic contribution to obesity is well recognized, and heritability of obesity is estimated to be 40–70% (14, 15). The responses of individuals with obesity to weight-gain prevention and reduction strategies can also vary broadly based on their genetic makeup (16). Therefore, examining gene–nutrient interactions assists in estimating the role of qualitative intake of FAs on the onset/progression of obesity in a genotype-specific manner.
Evidence from controlled trials and observational studies suggests a contribution of the interactions between genetic polymorphisms and dietary FAs in modulating adiposity via several mechanisms. For instance, the consumption of a higher proportion of MUFAs (17–19) and PUFAs (20, 21) relative to SFAs was found to be associated with lower body weight in peroxisome proliferator activated receptor γ (PPARG) rs1801282-G allele carriers in different ethnic populations. Additionally, a meta-analysis of 14 studies in US and European Caucasians revealed a direct association between SFA consumption and BMI, as well as waist-to-hip ratio, in the rs2306692-TT genotype carriers of low-density lipoprotein receptor-related protein 1 (22).
Although current research provides emerging evidence for gene–MUFA interactions (17–19, 23–25), numerous polymorphisms in lipid metabolism–related genes have yet to be investigated. Moreover, to investigate fat deposition and distribution, surrogate biomarkers have been often used, potentially masking outcomes. This study aimed to assess associations of common genetic variants and changes of total and regional fat mass following 6-wk controlled isocaloric dietary interventions with different concentrations of dietary MUFAs. To meet our objective, we used whole-body DXA scanning, which provides a reliable identification of fat distribution and discrimination between different fat depots.
Methods
Study design and population
This study of gene–nutrient interactions was conducted within the framework of the Canola Oil Multicenter Intervention Trial (COMIT) II. COMIT II was a randomized, controlled, double-blind, crossover study designed to evaluate the response of body composition to 3 oils with different MUFA concentrations, including regular canola oil (RCO), high-oleic acid canola oil (HOCO), and a low-MUFA/high-SFA oil blend. This trial was conducted from 2014 to 2016 at 3 sites in Canada and 1 site in the United States: the Richardson Centre for Functional Foods and Nutraceuticals (RCFFN) at the University of Manitoba in Winnipeg; the Canadian Centre for Agri-Food Research in Health and Medicine at St Boniface Hospital Albrechtsen Research Centre in Winnipeg; the Institute of Nutrition and Functional Foods at Laval University in Quebec City; and the Departments of Nutritional Sciences and Biobehavioral Health at The Pennsylvania State University in University Park. The protocol was reviewed and approved by institutional ethics boards across the participating clinical sites. The trial was registered at clinicaltrials.gov as NCT02029833.
Participants aged 20–65 y were included if they had abdominal obesity according to the International Diabetes Federation cutoff point for waist circumference (94 cm in men and 80 cm in women) in addition to at least 1 of the following metabolic syndrome criteria: fasting concentrations of blood glucose ≥5.6 mmol/L, triglycerides (TGs) ≥1.7 mmol/L, and HDL-cholesterol <1 mmol/L (men) or <1.3 mmol/L (women); and blood pressure ≥130 mmHg (systolic) and/or ≥85 mmHg (diastolic). Individuals were excluded if they had unstable thyroid disease, kidney disease, diabetes mellitus, or liver disease. Current smokers, individuals consuming more than 14 alcoholic beverages per week, individuals taking medication known to affect lipid metabolism for at least the last 3 mo, or individuals who were unwilling to stop taking any supplement at least 2 wk before the study were not permitted to participate. Written informed consent was obtained from all participants upon enrollment. Participants were randomly assigned to 1 of 6 treatment sequences using a random number generator at randomization.com.
Study diets
This study consisted of 3 treatment periods during which the participants consumed a controlled isocaloric, full-feeding diet with a fixed macronutrient composition of 35% fat, 50% carbohydrate, and 15% protein of total energy, as well as ∼208 mg/3000 kcal/d cholesterol and ∼38 g/3000 kcal/d fiber. The macronutrient composition of the 3 experimental diets has been previously reported (26). Menus for the 3 phases were identical except for the type of treatment oil provided. Treatment phases extended for 6 wk and were separated by 6-wk washout periods (ranged from 4 to 12 wk for a few participants). During the washout periods, participants were instructed to consume their habitual diets. Participants were asked to maintain their usual level of physical activity throughout the entire study. Physical activity changes were monitored by a weekly checklist.
The treatment oils, which comprised 20% of total energy, were incorporated into a smoothie beverage and were divided equally into 2 portions consumed at breakfast and supper. Treatment oils included: 1) RCO (Canola Harvest Canola Oil; Richardson International), which provided 6.6% SFAs, 65.3% MUFAs, 19.6% n–6 PUFAs, 8.5% n–3 PUFAs α-linolenic acid; 2) HOCO (Canola Harvest Canola Oil; Richardson International), which provided 6.7% SFAs, 75.9% MUFAs, 14.8% n–6 PUFAs, 2.6% n–3 PUFAs α-linolenic acid; and 3) a low-MUFA/high-SFA oil blend that provided 40.2% SFAs, 22.0% MUFAs, 29.6% n–6 PUFAs, 8.2% n–3 PUFAs α-linolenic acid. The low-MUFA/high-SFA oil blend was prepared using commercially available ghee/butter oil (36.0%, Verka), safflower oil (34.9%, eSutras), coconut oil (16.0%, eSutras), and flaxseed oil (13.1%, Shape Foods). Study food and treatment shakes were prepared based on a 7-d rotating menu cycle in the metabolic kitchen of the participating sites. Compliance was assessed by smoothie consumption where the participants were required to consume ≥90% of the smoothies provided at each phase. Participants signed a daily checklist to verify consumption of smoothies. To maximize compliance rate, participants were required to consume 1 smoothie at breakfast under the supervision of a clinical coordinator for 5 d/wk. During weekdays, participants were provided the rest of their meals and a second smoothie in a food cooler bag for consumption off-site. Weekend meals and treatment shakes were delivered to the participants’ residences or handed out to them, upon their request, at the clinical site on Fridays.
Measurement of fat mass
DXA scans were performed by a trained operator using Lunar Prodigy Advance DXA (GE Healthcare) with the default configurations. A DXA scan was performed for all participants at the initiation and termination of each dietary phase. Participants were asked to remove any metal items and heavy clothes before scanning. Regions of interests (ROIs) were manually adjusted using enCORE 2012 software (version 14.10.022) according to the manufacturer's instructions. Fat mass was analyzed as total fat mass, as well as 4 different districts including trunk, legs, android, and gynoid fat masses. The android and gynoid ROIs were identified as per the manufacturer's instructions. The android region has been defined as a portion of the abdomen that starts at the pelvis cut line and extends upward to include 20% of the distance between the pelvis and neck cut lines, with the outer arms’ cuts as the lateral boundaries. The gynoid region has been defined as a portion of the legs with upper boundary below the pelvis cut line by 1.5 times the height of the android region, extending downward to twice the height of the android ROI, with the outer legs’ cuts as the lateral boundaries. Further, visceral adipose tissue (VAT) was assessed by the CoreScan feature in enCORE 2012 software (version 14.10.022), and used to calculate the subcutaneous adipose tissue (SCAT) by subtracting VAT mass from android fat mass (27). VAT measurement using the CoreScan has been validated using computed tomography scanning (28). Criteria used to identify the anatomical ROIs were identical across all sites.
Genotyping
Twelve-hour fasting blood samples were collected and processed at the beginning of the trial, then stored at −80°C until being shipped to the RCFFN for analysis. Genomic DNA was extracted from the buffy coat samples of the first day of the first phase using a Qiagen DNeasy Blood and Tissue Kit according to the manufacturer's instructions (Qiagen Sciences, Inc.). A Thermo Scientific NanoDrop 2000 microvolume spectrophotometer was used to assess the concentration and purity of the extracted DNA (Thermo-Fisher Scientific, Inc.). TaqMan GTXpress Master Mix with allele-specific probes (Applied Biosystems, Life Technologies Inc) was used for genotyping of the single-nucleotide polymorphisms (SNPs) of interest. Amplification and detection of DNA were conducted with the 7500 Fast Real-Time PCR System (Applied Biosystems, Life Technologies Inc). Data were acquired by software StepOne 2.1 (Applied Biosystems, Life Technologies Inc). Polymorphisms located in lipogenesis/adipogenesis-related genes were selected for their various roles in obesity development, where each SNP chosen was either a functional SNP, had a minor allele frequency ≥5, and/or had been previously reported for gene–nutrient interactions. This study assessed possible gene–diet interactions in a total of 5 candidate variants (Supplemental Table 1), within 3 genes, namely adiponectin (ADIPOQ), apoE (APOE), and lipoprotein lipase (LPL). The role of LPL as the rate-limiting enzyme that catalyzes the hydrolysis of TGs underscores LPL as a candidate gene for obesity. The functional LPL rs13702 and rs3200218 are located in the 3′ untranslated region (UTR) region and they are involved in translational regulation (29, 30). LPL rs13702-C allele is suggested to disrupt the microRNA recognition elements seed site and abolish the microRNA-410–mediated repression of mRNA at the LPL 3′ UTR, therefore increasing the activity of LPL (31). Despite the master role of LPL in regulating the supply of FAs to adipose tissue, the effects of possible interactions between LPL polymorphism and dietary FA interactions on obesity have been scarcely studied. ADIPOQ rs266729 (−11,377 C/G) is located in the promoter region and has been shown to alter the circulating adiponectin concentration as well as the risk of obesity and insulin resistance (24, 32). Lastly, APOE4 isoform has been associated with abnormal lipid metabolism and increased risk for several health problems including obesity (33–35).
Statistical analyses and sample size
The primary aim of the COMIT II trial was to evaluate the effect of MUFA consumption on body composition, mainly VAT. Therefore, the sample size was calculated to detect a 55-g change in android fat mass using the variance parameter in android fat mass from our previous controlled trial (1). A total sample size of 140 was required to account for a dropout rate of 20%.
Statistical analyses were performed using SAS 9.4 (SAS Institute, Inc.) based on a per protocol approach. Normality was assessed using the Shapiro–Wilk test and the skewness values. Nonnormally distributed variables were log-transformed before analysis. The results are expressed as least-squares means ± SEMs unless otherwise specified, and statistical significance was set at P < 0.05. Multiple comparison was assessed using the Tukey–Kramer test. Changes in fat mass and body weight represent the difference over 6 wk between end point and baseline of each dietary phase. PROC MIXED (SAS Institute, Inc.) with repeated-measures procedure was used to assess the effect of the 3 dietary treatments on changes in body fat and body weight. Treatment, sex, age, and genotype were used as fixed effects, with participants as a repeated factor. Random effects were treatment sequence, clinical site, and participants. Prespecified potential confounders such as ethnicity, baseline body composition, baseline fasting concentrations of glucose, homeostatic model of insulin resistance, and cholesterol were investigated in all models. The Hardy–Weinberg equilibrium was assessed with a chi-square test.
For diet–gene interaction analysis, due to the considerable not exactly similar comparable concentrations of MUFAs in the 2 canola treatments compared with the low-MUFA/high-SFA treatment, the statistical analysis of diet-by-SNP interaction was conducted to compare the combined effect of the 2 high-MUFA diets (HOCO + RCO, averaged) with the low-MUFA/high-SFA diet on changes in body fat and body weight. This decision was also based on our inability to detect statistical differences between HOCO and RCO in body composition in the overall population in our previous trial (COMIT I) (1), and based on the predefined hypothesis that the small variation in the concentration of MUFAs between the 2 high-MUFA treatments will not significantly influence the effect of genes on obesity. Although our sample size was lower than the longitudinal, survey-based diet–gene interaction studies, the controlled, full-feeding, crossover design of this study reduced the need for a larger sample size because it eliminated a wide range of confounders associated with the former designs. However, we consider this analysis an exploratory study to identify SNPs that might influence the body fat response to dietary fat type.
Each individual SNP was assessed separately using the aforementioned statistical model. All SNPs were analyzed in the additive model. Dominant and recessive models were analyzed only when the simple effect of heterozygous-by-MUFAs (in addition, to 1 homozygotes-by-MUFAs) showed a significant interaction. Only 4 APOE isoforms (encoded by rs429358 and rs7412) were obtained, and were analyzed and presented as non-E4 (ε2/ε3 and ε3/ε3 genotypes) and E4 (ε3/ε4 and ε4/ε4 genotypes).
Results
A total of 124 participants completed the counterbalanced trial and all of the required DXA scans. Three participants were excluded due to a high fasting blood glucose concentration (>7 mmol/L) and 6 participants were excluded due to large changes in body weight (weight change from baseline to endpoint >5%) at any dietary period (Supplemental Figure 1). Therefore, 115 participants (71 women and 44 men) were included in the analysis of the effect of dietary MUFAs on changes in body composition, as the primary outcome of the COMIT II trial. No significant differences were observed in changes in body weight or fat mass following the consumption of any of the 3 treatments (Supplemental Table 2).
The assessment of gene-by-diet interactions, the secondary outcome of the COMIT II trial, included 101 participants, because 14 participants did not consent for genetic analyses. Baseline characteristics are displayed in Table 1. Hardy–Weinberg equilibrium was not achieved for the ADIPOQ rs266729 and APOE. The effects of gene–diet interactions on changes in body weight, total fat mass, and selected regional fat mass were tested (Supplemental Tables 3 and 4). Diet was found to interact with common variants in LPL, ADIPOQ, and APOE to modify changes in body fatness in an isocaloric diet (Figures 1–3), as detailed below.
TABLE 1.
Characteristics of participants at the baseline of dietary intervention1
| Characteristic2 | Total (n = 101) | Female (n = 60) | Male (n = 41) |
|---|---|---|---|
| Age, y | 43.3 ± 1.29 | 45.7 ± 1.64a | 39.9 ± 1.98b |
| Ethnicity, n | |||
| Caucasian | 74 | 45 | 29 |
| African | 4 | 3 | 1 |
| Asian | 8 | 4 | 4 |
| Hispanic | 3 | 1 | 2 |
| Others | 12 | 7 | 5 |
| Waist circumference, cm | 104 ± 1.30 | 101 ± 1.60a | 108 ± 1.90b |
| Systolic BP, mmHg | 119 ± 1.30 | 118 ± 1.70 | 120 ± 2.00 |
| Diastolic BP, mmHg | 78.1 ± 1.08 | 77.5 ± 1.41 | 79.0 ± 1.70 |
| Total cholesterol, mmol/L | 5.19 ± 0.09 | 5.21 ± 0.12 | 5.15 ± 0.14 |
| TGs, mmol/L | 1.55 ± 0.07 | 1.46 ± 0.09 | 1.67 ± 0.11 |
| HDL-cholesterol, mmol/L | 1.35 ± 0.04 | 1.46 ± 0.04a | 1.20 ± 0.05b |
| LDL-cholesterol, mmol/L | 3.13 ± 0.08 | 3.09 ± 0.10 | 3.19 ± 0.12 |
| Glucose, mmol/L | 5.22 ± 0.04 | 5.21 ± 0.06 | 5.23 ± 0.07 |
| Insulin, pmol/L | 98.7 ± 6.10 | 94.3 ± 7.92 | 105.0 ± 9.58 |
| VAT mass, g | 1334 ± 84.0 | 1056 ± 99.0a | 1741 ± 120b |
| SCAT mass, g | 2213 ± 90.0 | 2293 ± 117 | 2097 ± 141 |
| Legs fat mass, g | 12,645 ± 430 | 13,790 ± 531a | 10,969 ± 642b |
| Trunk fat mass, g | 19,472 ± 721 | 18,584 ± 930 | 20,772 ± 1124 |
| Android fat mass, g | 3548 ± 145 | 3349 ± 187 | 3838 ± 226 |
| Gynoid fat mass, g | 6002 ± 206 | 6334 ± 264 | 5516 ± 319 |
| Total fat mass, g | 36,282 ± 1119 | 36,565 ± 1459 | 35,869 ± 1765 |
| Body weight, kg | 89.8 ± 1.88 | 83.0 ± 2.21a | 99.6 ± 2.67b |
| BMI, kg/m2 | 31.1 ± 0.53 | 31.0 ± 0.69 | 31.3 ± 0.84 |
Values are means ± SEMs unless otherwise specified. PROC MIXED (SAS Institute, Inc.) procedure was used to assess sex differences, P < 0.05 was considered significant. PROC MEANS (SAS Institute, Inc.) was used to determine the mean characteristics of the overall population. Labeled means within the same row without a common letter indicate sex-based statistical difference. BP, blood pressure; SCAT, subcutaneous adipose tissue; TG, triglyceride; VAT, visceral adipose tissue.
Lipid profiles and glucose concentrations were determined using Cobas enzymatic reagents on Roche/Hitachi c 501e automated clinical chemistry analyzers using serum samples. Serum insulin concentrations were measured with the Roche/Hitachi Cobas e immunoassay analyzer and electrochemiluminescence immunoassay kits.
FIGURE 1.
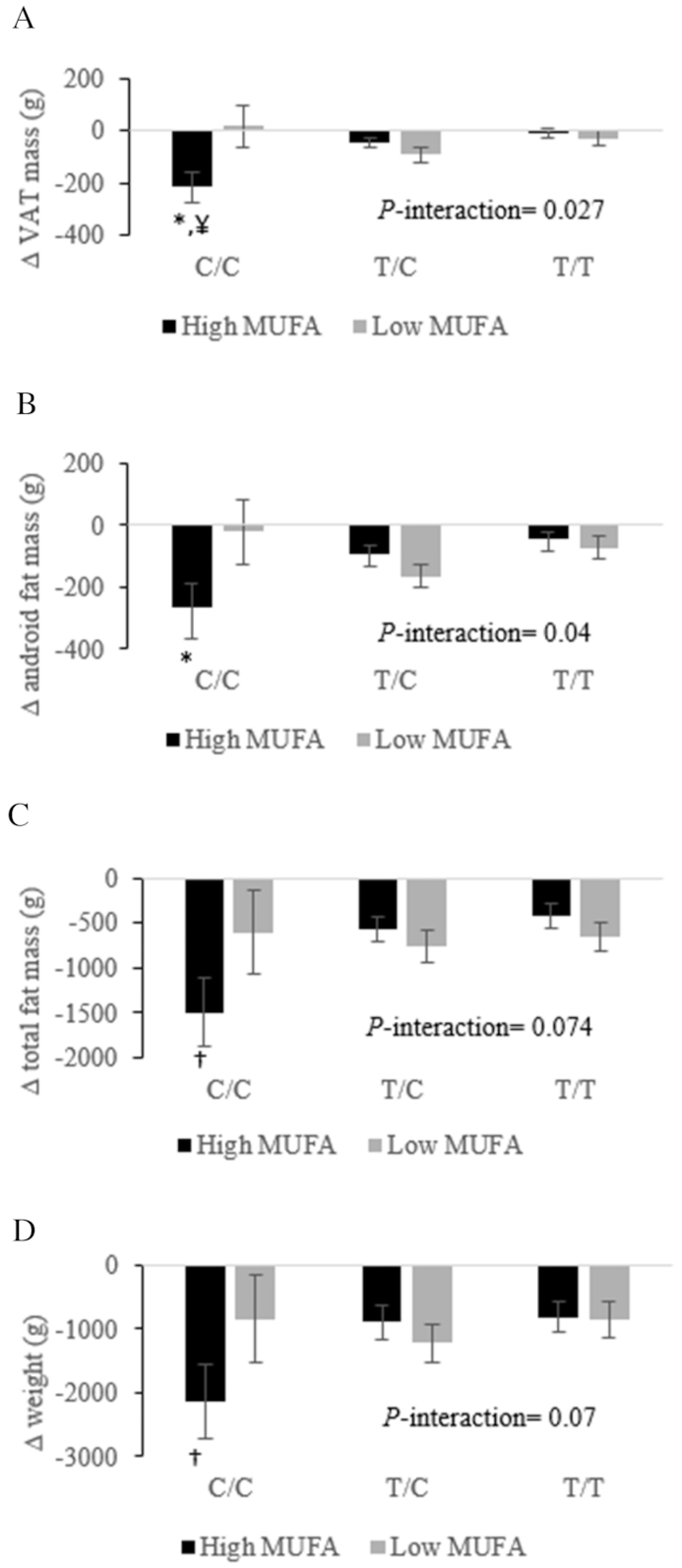
LPL rs13702 genotypes determine the effect of high- compared with low-MUFA consumption on 6-wk changes in VAT mass (A), android fat (B), total fat mass (C), and body weight (D) in adults with abdominal obesity. Changes were calculated by subtracting the baseline value of the selected fat mass from its corresponding 6-wk end-point value. Total participants = 101: n = 50 LPL rs13702-TT, n = 45 LPL rs13702-CT, and n = 6 LPL rs13702-CC. Values are least-squares means ± SEMs. PROC MIXED (SAS Institute, Inc.) with repeated-measures procedure was used to assess the effect of gene–MUFA interactions on fat mass changes, using participants’ identification code as a repeated factor. P < 0.05 was considered significant. *Statistical significance in the response of a specific fat mass to different concentrations of dietary MUFAs within the same genotype. †Trend toward statistical significance (0.06 > P > 0.05) in the response of a specific fat mass to different concentrations of dietary MUFAs within the same genotype. ¥Statistically significant (P = 0.017) greater reduction in VAT mass following high-MUFA consumption in CC carriers compared with TT carriers of LPL rs13702. LPL, lipoprotein lipase gene; VAT, visceral adipose tissue.
FIGURE 3.
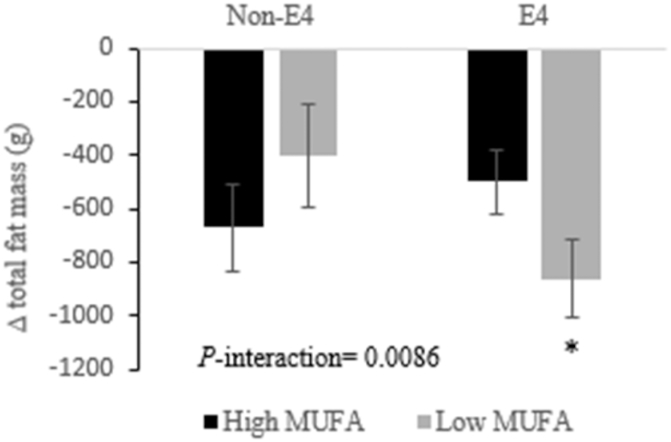
APOE genotypes determine the effect of high- compared with low-MUFA consumption on 6-wk changes in total fat mass in adults with abdominal obesity. Changes were calculated by subtracting the baseline value of the total fat mass from its corresponding 6-wk end-point value. Total participants = 101: n = 35 non-E4 and n = 66 E4. Values are least-squares means ± SEMs. PROC MIXED (SAS Institute, Inc.) with repeated-measures procedure was used to assess the effect of gene–MUFA interactions on fat mass changes, using participants’ identification code as a repeated factor. P < 0.05 was considered significant. *Statistical significance in the response of a specific fat mass to different concentrations of dietary MUFAs within the same genotype. APOE, apoE gene.
The LPL rs13702-CC genotype (Figure 1) was found to be associated with lower VAT and android fat mass following high-MUFA consumption compared with the low-MUFA diet. Likewise, carriers of the LPL rs13702-CC genotype showed trends toward less total fat mass and body weight following high-MUFA consumption compared with low-MUFA intake.
The consumption of high-MUFA diets protected the ADIPOQ rs266729-GG homozygotes from the increase in SCAT that was observed following consumption of the low-MUFA diet (Figure 2A). Further, in response to the low-MUFA diet, the carriers of ADIPOQ rs266729-GG homozygotes showed higher SCAT and android fat mass compared with C allele carriers (Figure 2). Lastly, E4 carriers had greater reductions in total fat mass following consumption of the low-MUFA diet compared with high-MUFA diets (Figure 3).
FIGURE 2.
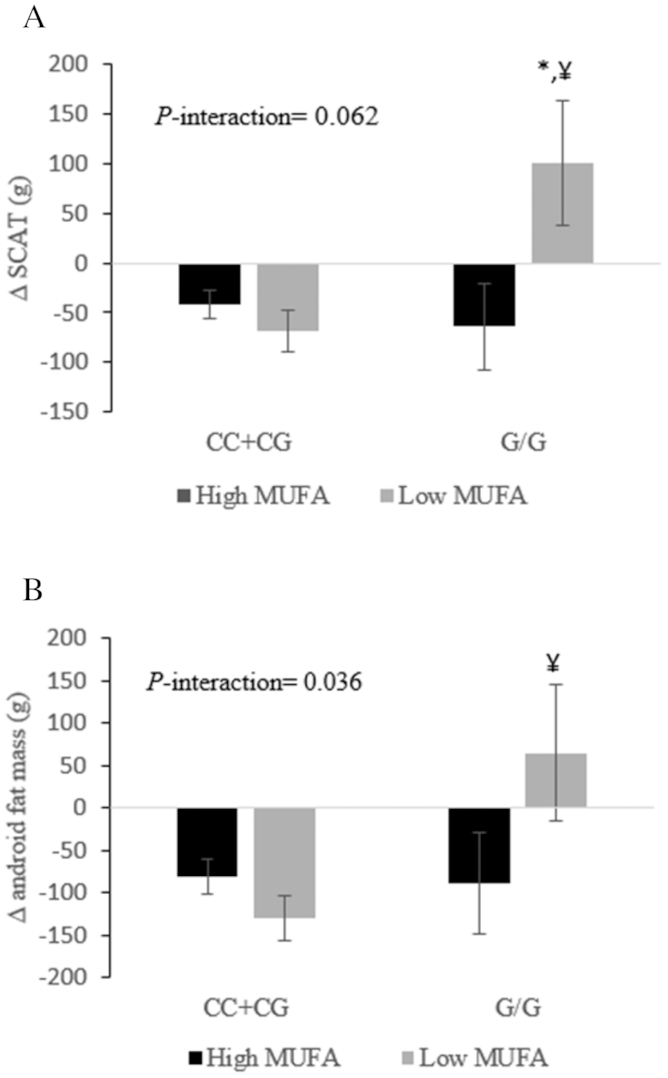
ADIPOQ rs266729 genotypes determine the effect of high- compared with low-MUFA consumption on 6-wk changes in SCAT (A) and android fat mass (B) in adults with abdominal obesity. Changes were calculated by subtracting the baseline value of the selected fat mass from its corresponding 6-wk end-point value. Total participants = 101: n = 91 ADIPOQ rs266729-CC + CG and n = 10 ADIPOQ rs266729-GG. Values are least-squares means ± SEMs. PROC MIXED (SAS Institute, Inc.) with repeated-measures procedure was used to assess the effect of gene–MUFA interactions on fat mass changes, using participants’ identification code as a repeated factor. P < 0.05 was considered significant. *Statistical significance in the response of a specific fat mass to different concentrations of dietary MUFAs within the same genotype. ¥Statistical significance of the greater reduction in SCAT mass and android fat mass following low-MUFA consumption in C carriers (P = 0.012 and 0.022, respectively) compared with GG carriers of ADIPOQ rs266729. Recessive model (CC + CG compared with GG) was analyzed because the simple effect of heterozygous-by-MUFA showed a significant interaction on ≥1 compartmental fat masses. ADIPOQ, adiponectin gene; SCAT, subcutaneous adipose tissue.
Discussion
Results of the current study indicate that changes in total and compartmental fat mass and body weight in response to dietary fat substitutions are modified by common variants within lipid metabolism–related genes. Minor allele homozygotes of either LPL or ADIPOQ had lower body fat indices following consumption of high-MUFA diets compared with a low-MUFA diet, whereas APOE4 carriers had lower body fat indices upon the consumption of a low-MUFA diet compared with a high-MUFA intake. These results highlight the genetic contribution to the responsiveness of body fatness to dietary MUFAs and could explain our inability to detect significant effects of MUFA consumption on body weight and fat mass compared with the low-MUFA/high-SFA diet in spite of the existing evidence (12, 13). Identifying the contribution of genetic architecture to the body's response to dietary modifications can direct the pathway toward the era of personalized nutrition. From our dietary intervention trial, we cannot conclude on the exact mechanisms of the observed phenomena and we need to refer to future biochemical studies. However, some existing knowledge might help to illuminate the biochemistry underlying the present findings.
LPL is the rate-limiting enzyme that catalyzes the hydrolysis of TGs in the core of TG-rich lipoprotein constituents as well as facilitating the uptake of FAs by adipocytes (36, 37). These functions highlight LPL as a candidate gene for obesity. Ma et al. (38) reported no influence of the SNP LPL rs13702 under different dietary FA interventions on BMI or waist circumference in 2 independent populations. The C allele in the functional LPL rs13702 is suggested to increase the hydrolytic activity of LPL (30, 31); however, to our knowledge, no interactions between LPL polymorphisms and dietary FAs to modulate regional fat masses have been reported. The consistent decrease of 4 distinct regions of fat mass in LPL rs13702-CC homozygotes following the high-MUFA diets (∆ high-MUFA compared with low-MUFA greater than −200 g per site) in an isocaloric condition provides validity to this interaction. Given the previously proposed LPL rs13702-C allele–induced elevation in LPL activity, MUFA-rich diets might, therefore, protect the LPL rs13702-CC carriers from an increment in fat mass by the activation of obesity-opposing pathways, such as increasing the activity of hormone-sensitive lipase (39) or elevating the ratio of skeletal muscle to adipose tissue LPL activity (40), which would reduce the propensity for fat deposition.
The ADIPOQ gene encodes the peptide hormone adiponectin, which modulates a number of metabolic processes including lipid oxidation in muscle and liver (41). The ADIPOQ rs266729-G allele has been identified as a risk factor for obesity in several studies (32, 42), and has been associated with a lower risk for obesity following the consumption of a higher percentage of energy derived from fat (43). The present study shows, despite the controlled isocaloric diet, that compared with the low-MUFA diet a higher MUFA intake significantly reduced SCAT in the android region (∆ high-MUFA compared with low-MUFA approximately −160 g) among the ADIPOQ rs266729-GG homozygotes. However, the C-allele carriers had greater benefits following low-MUFA consumption (∆ low-MUFA compared with high-MUFA approximately −160 g and −190 g in SCAT and android fat mass, respectively) compared with high-MUFA intake. This finding constitutes further refinement of existing obesity associations, specifically because a previous study found no effect of dietary MUFAs on the association between the ADIPOQ rs266729 and obesity (24). However, we did not assess depot-specific concentrations of adiponectin or its receptors, leaving the validation of the underlying mechanism to future studies.
The APOE gene encodes apoE, which mediates the catabolism of the TG-rich lipoprotein particles in an isoform-dependent manner (44, 45). The E4/E4 genotype has been associated with abnormal lipid metabolism and increased risk for several health problems including obesity (33–35, 46–49) and has previously shown responsiveness to dietary interventions (45, 46, 50). Mice carrying the human non-E4 allele were heavier when fed low- and high-fat diets compared with E4/E4 mice (51). The same study found that FA mobilization was lower in non-E4 than in E4/E4 mice, whereas E4 mice overexpressed proteins involved in FA oxidation in skeletal muscle. Our results add another dimension to the evidence that APOE isoforms differentially influence fat mass, through demonstrating that the concentration of dietary MUFAs modulated fat loss in E4 carriers (∆ low-MUFA compared with high-MUFA approximately −360 g) in an isocaloric condition. The E4/E4 genotype was found to be associated with increased basal mitochondrial uncoupling and FA oxidation in mice (52), and this mechanism might be modulated by the quantity of dietary MUFAs, especially given that dietary MUFAs could increase fat oxidation and thermogenesis (8).
Assessing adiposity using DXA scanning provided a comprehensive assessment of the effect of these SNPs on total and regional adiposity. Another strength of this study was the crossover design with a controlled, isocaloric dietary intervention, which eliminated a range of confounders that might be inherent with free-living and/or parallel study designs. However, an important limitation of the current study was that we did not apply stringent control for multiple testing, which could lead to a potential overstatement of our findings. Thus, large-scale studies are highly encouraged to evaluate these associations between the quality of dietary fat and polymorphisms within lipid metabolism–related genes. Additionally, Kien et al. (11) reported an attractive effect of dietary MUFA in which a high-MUFA consumption elevated physical activity levels compared with a high-SFA intake. The fact that participants of this study were instructed to maintain the same level of physical activity throughout the trial, and the lack of objective evaluation of physical activity, could have hindered the effect of different dietary FAs on physical activity; and consequently adiposity. The mixed ethnicity of this study population could also be perceived as a limitation, but might also provide generalizability of the current findings.
In summary, we report the contribution of common variants in LPL, ADIPOQ, and APOE genes to changes in body fatness in response to dietary MUFAs. These changes in body fat were observed regardless of the controlled isocaloric scenario. Although the observed changes in total and compartmental fat mass in response to gene–diet interactions were small over 6 wk, their statistical significance might indicate a potentially substantial clinical effect in weight reduction/maintenance regimens over prolonged periods.
Supplementary Material
Acknowledgments
We thank Stephanie Jew for overseeing site–site study coordination, and Julia Rempel, clinical coordinator at RCFFN. The authors’ responsibilities were as follows—SSH, JS, XC, PC, VG, JM-G, DP, AW, SC, PZ, KJB: were involved in the clinical trial conduction and data collection; SSH: performed the laboratory work, data interpretation, and wrote the manuscript; PE: contributed expertise for genotyping studies; PWC, BL, DJAJ, CT, SGW, PMK-E, PJHJ: designed the project and supervised the study; and all authors: critically reviewed and read and approved the final manuscript.
Notes
This study was supported by Agriculture and Agri-Food Canada, the Canola Council of Canada, Alberta Canola, SaskCanola, Dow Agro Sciences, and the Manitoba Canola Growers. SSH was a recipient of The University of Jordan Scholarship for Graduate Studies. DJAJ was funded by the government of Canada through the Canada Research Chair Endowment.
Author disclosures: SSH, PE, JS, XC, PWC, PC, VG, JM-G, PMK-E, KJB, DP, AW, and SC, no conflicts of interest. BL is Chair of Nutrition at Université Laval, which is supported by private endowments from Pfizer, La Banque Royale du Canada, and Provigo-Loblaws. BL has received funding in the last 5 y from the Canadian Institutes for Health Research, the Natural Sciences and Engineering Research Council of Canada, Agriculture and Agri-Food Canada [Growing Forward program supported by the Dairy Farmers of Canada (DFC), Canola Council of Canada, Flax Council of Canada, Dow Agrosciences], Dairy Research Institute, Dairy Australia, Merck Frosst, Pfizer, and Atrium Innovations. BL served as the Chair of the peer review Expert Scientific Advisory Council of DFC. BL is also an Advisory Board member of the Canadian Nutrition Society and has received honoraria from the International Chair on Cardiometabolic risk, DFC, and the World Dairy Platform as invited speaker in various conferences. SGW has received consulting funds, travel funds, and research funding from the Canola Council of Canada and the McCormick Science Institute. She has received research and travel funds from Flax Canada, the California Walnut Commission, American Pistachio Growers, and Hershey's. She has received research funding from the Almond Board of California, the Hass Avocado Board, the National Fisheries Institute, Dairy Management Incorporated, General Mills, Reliant Pharmaceuticals, Unilever, and the National Cattleman's Beef Fund. DJAJ has received research grants from Saskatchewan Pulse Growers, the Agricultural Bioproducts Innovation Program through the Pulse Research Network, the Advanced Foods and Material Network, Loblaw Companies Ltd, Unilever, Barilla, the Almond Board of California, Agriculture and Agri-Food Canada, Pulse Canada, Kellogg's Company, Canada, Quaker Oats, Canada, Procter & Gamble Technical Centre Ltd, Bayer Consumer Care, Springfield, NJ, Pepsi/Quaker, International Nut & Dried Fruit Council (INC), Soy Foods Association of North America, the Coca-Cola Company (investigator initiated, unrestricted grant), Solae, Haine Celestial, the Sanitarium Company, Orafti, the International Tree Nut Council Nutrition Research and Education Foundation, the Peanut Institute, the Canola and Flax Councils of Canada, the Calorie Control Council (CCC), the Canadian Institutes of Health Research (CIHR), the Canada Foundation for Innovation, and the Ontario Research Fund. DJAJ has received in-kind supplies for trials as research support from the Almond Board of California, Walnut Council of California, American Peanut Council, Barilla, Unilever, Unico, Primo, Loblaw Companies, Quaker (Pepsico), Pristine Gourmet, Bunge Limited, Kellogg Canada, and WhiteWave Foods. DJAJ has been on the speakers’ panel, served on the scientific advisory board, and/or received travel support and/or honoraria from the Almond Board of California, Canadian Agriculture Policy Institute, Loblaw Companies Ltd, the Griffin Hospital (for the development of the NuVal scoring system), the Coca-Cola Company, EPICURE, Danone, Diet Quality Photo Navigation (DQPN), FareWell, Verywell, True Health Initiative, Saskatchewan Pulse Growers, Sanitarium Company, Orafti, the Almond Board of California, the American Peanut Council, the International Tree Nut Council Nutrition Research and Education Foundation, the Peanut Institute, Herbalife International, Pacific Health Laboratories, Nutritional Fundamental for Health, Barilla, Metagenics, Bayer Consumer Care, Unilever Canada and Netherlands, Solae, Kellogg, Quaker Oats, Procter & Gamble, the Coca-Cola Company, the Griffin Hospital, Abbott Laboratories, the Canola Council of Canada, Dean Foods, the California Strawberry Commission, Haine Celestial, PepsiCo, the Alpro Foundation, Pioneer Hi-Bred International, DuPont Nutrition and Health, Spherix Consulting and WhiteWave Foods, the Advanced Foods and Material Network, the Canola and Flax Councils of Canada, the Nutritional Fundamentals for Health, Agri-Culture and Agri-Food Canada, the Canadian Agri-Food Policy Institute, Pulse Canada, the Saskatchewan Pulse Growers, the Soy Foods Association of North America, the Nutrition Foundation of Italy, Nutra-Source Diagnostics, the McDougall Program, the Toronto Knowledge Translation Group (St Michael's Hospital), the Canadian College of Naturopathic Medicine, The Hospital for Sick Children, the Canadian Nutrition Society (CNS), the ASN, Arizona State University, Paolo Sorbini Foundation, and the Institute of Nutrition, Metabolism and Diabetes. He received an honorarium from the United States Department of Agriculture to present the 2013 W.O. Atwater Memorial Lecture. DJAJ received the 2013 Award for Excellence in Research from the INC. He received funding and travel support from the Canadian Society of Endocrinology and Metabolism to produce mini cases for the Canadian Diabetes Association. DJAJ is a member of the International Carbohydrate Quality Consortium. His wife is a director and partner of Glycemic Index Laboratories, Inc., and his sister received funding through a grant from the St Michael's Hospital Foundation to develop a cookbook for one of his studies. CGT and PZ have received funding in the past 5 y from CIHR, the Natural Sciences and Engineering Research Council of Canada (NSERC), Agriculture and Agri-Food Canada (Growing Forward and the Canola/Flax Agri-Science Cluster, Pulse Agri-Science Cluster, Canada-Manitoba Agri-Food Research Development Initiative), Research Manitoba, Manitoba Energy Science and Technology, Manitoba Agri-Health Research Network, Alberta Innovates, Alberta Crop Industry Development Fund, Alberta Canola Producers Commission, Alberta Pulse, Saskatchewan Pulse Growers, Canadian Diabetes Association, and the Mathematics of Information Technology and Complex Systems (MITACS). PJHJ's research related to a variety of oils and fats has been supported by grants and contracts from both industry and nonindustry sources, including the Canola Council of Canada, Dairy Farmers of Canada, Canadian Institutes for Health Research, Natural Sciences and Engineering Research Council of Canada, Heart and Stroke Foundation of Canada, and National Institutes of Health Rare Diseases Network. PJHJ also serves as a committee member for the Soy Nutrition Institute, and the North American International Life Sciences Institute's Lipids Committee. PJHJ is President of Nutritional Fundamentals for Health Inc., which markets functional foods and nutraceuticals. The funding agencies played no role in defining the study design; in the collection, analysis, and interpretation of data; in the writing of the manuscript; or in the decision to submit the manuscript for publication.
Supplemental Tables 1–4 and Supplemental Figure 1 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: ADIPOQ, adiponectin; COMIT, Canola Oil Multicenter Intervention Trial; FA, fatty acid; HOCO, high-oleic canola oil; LPL, lipoprotein lipase; RCFFN, Richardson Centre for Functional Foods and Nutraceuticals; RCO, regular canola oil; ROI, region of interest; SCAT, subcutaneous adipose tissue; SNP, single nucleotide polymorphism; TG, triglyceride; UTR, untranslated region; VAT, visceral adipose tissue.
References
- 1. Liu X, Kris-Etherton PM, West SG, Lamarche B, Jenkins DJ, Fleming JA, McCrea CE, Pu S, Couture P, Connelly PW et al.. Effects of canola and high-oleic-acid canola oils on abdominal fat mass in individuals with central obesity. Obesity (Silver Spring). 2016;24:2261–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alves RD, Moreira AP, Macedo VS, de Cassia Goncalves Alfenas R, Bressan J, Mattes R, Costa NM. Regular intake of high-oleic peanuts improves fat oxidation and body composition in overweight/obese men pursuing a energy-restricted diet. Obesity (Silver Spring). 2014;22:1422–9. [DOI] [PubMed] [Google Scholar]
- 3. Summers LK, Fielding BA, Bradshaw HA, Ilic V, Beysen C, Clark ML, Moore NR, Frayn KN. Substituting dietary saturated fat with polyunsaturated fat changes abdominal fat distribution and improves insulin sensitivity. Diabetologia. 2002;45:369–77. [DOI] [PubMed] [Google Scholar]
- 4. Mumme K, Stonehouse W.. Effects of medium-chain triglycerides on weight loss and body composition: a meta-analysis of randomized controlled trials. J Acad Nutr Diet. 2015;115:249–63. [DOI] [PubMed] [Google Scholar]
- 5. Hammad SS, Jones PJ.. Dietary fatty acid composition modulates obesity and interacts with obesity-related genes. Lipids. 2017;52:803–22. [DOI] [PubMed] [Google Scholar]
- 6. Mennella I, Savarese M, Ferracane R, Sacchi R, Vitaglione P. Oleic acid content of a meal promotes oleoylethanolamide response and reduces subsequent energy intake in humans. Food Funct. 2015;6:204–10. [DOI] [PubMed] [Google Scholar]
- 7. Stevenson JL, Clevenger HC, Cooper JA. Hunger and satiety responses to high-fat meals of varying fatty acid composition in women with obesity. Obesity (Silver Spring). 2015;23:1980–6. [DOI] [PubMed] [Google Scholar]
- 8. Krishnan S, Cooper JA. Effect of dietary fatty acid composition on substrate utilization and body weight maintenance in humans. Eur J Nutr. 2014;53:691–710. [DOI] [PubMed] [Google Scholar]
- 9. Kien CL, Bunn JY. Gender alters the effects of palmitate and oleate on fat oxidation and energy expenditure. Obesity (Silver Spring). 2008;16:29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piers LS, Walker KZ, Stoney RM, Soares MJ, O'Dea K. The influence of the type of dietary fat on postprandial fat oxidation rates: monounsaturated (olive oil) vs saturated fat (cream). Int J Obes Relat Metab Disord. 2002;26:814–21. [DOI] [PubMed] [Google Scholar]
- 11. Kien CL, Bunn JY, Tompkins CL, Dumas JA, Crain KI, Ebenstein DB, Koves TR, Muoio DM. Substituting dietary monounsaturated fat for saturated fat is associated with increased daily physical activity and resting energy expenditure and with changes in mood. Am J Clin Nutr. 2013;97:689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Piers LS, Walker KZ, Stoney RM, Soares MJ, O'Dea K. Substitution of saturated with monounsaturated fat in a 4-week diet affects body weight and composition of overweight and obese men. Br J Nutr. 2003;90:717–27. [DOI] [PubMed] [Google Scholar]
- 13. Gillingham LG, Robinson KS, Jones PJ. Effect of high-oleic canola and flaxseed oils on energy expenditure and body composition in hypercholesterolemic subjects. Metabolism. 2012;61:1598–605. [DOI] [PubMed] [Google Scholar]
- 14. Andersen MK, Sandholt CH. Recent progress in the understanding of obesity: contributions of genome-wide association studies. Curr Obes Rep. 2015;4:401–10. [DOI] [PubMed] [Google Scholar]
- 15. Fall T, Ingelsson E. Genome-wide association studies of obesity and metabolic syndrome. Mol Cell Endocrinol. 2014;382:740–57. [DOI] [PubMed] [Google Scholar]
- 16. Moreno-Aliaga MJ, Santos JL, Marti A, Martinez JA. Does weight loss prognosis depend on genetic make-up?. Obes Rev. 2005;6:155–68. [DOI] [PubMed] [Google Scholar]
- 17. Garaulet M, Smith CE, Hernandez-Gonzalez T, Lee YC, Ordovas JM. PPARgamma Pro12Ala interacts with fat intake for obesity and weight loss in a behavioural treatment based on the Mediterranean diet. Mol Nutr Food Res. 2011;55:1771–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Memisoglu A, Hu FB, Hankinson SE, Manson JE, De Vivo I, Willett WC, Hunter DJ. Interaction between a peroxisome proliferator-activated receptor gamma gene polymorphism and dietary fat intake in relation to body mass. Hum Mol Genet. 2003;12:2923–9. [DOI] [PubMed] [Google Scholar]
- 19. Rosado EL, Bressan J, Martinez JA, Marques-Lopes I. Interactions of the PPARgamma2 polymorphism with fat intake affecting energy metabolism and nutritional outcomes in obese women. Ann Nutr Metab. 2010;57:242–50. [DOI] [PubMed] [Google Scholar]
- 20. Franks PW, Jablonski KA, Delahanty L, Hanson RL, Kahn SE, Altshuler D, Knowler WC, Florez JC. The Pro12Ala variant at the peroxisome proliferator-activated receptor gamma gene and change in obesity-related traits in the Diabetes Prevention Program. Diabetologia. 2007;50:2451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luan J, Browne PO, Harding AH, Halsall DJ, O'Rahilly S, Chatterjee VK, Wareham NJ. Evidence for gene-nutrient interaction at the PPARgamma locus. Diabetes. 2001;50:686–9. [DOI] [PubMed] [Google Scholar]
- 22. Smith CE, Ngwa J, Tanaka T, Qi Q, Wojczynski MK, Lemaitre RN, Anderson JS, Manichaikul A, Mikkila V, van Rooij FJ et al.. Lipoprotein receptor-related protein 1 variants and dietary fatty acids: meta-analysis of European origin and African American studies. Int J Obes (Lond). 2013;37:1211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosado EL, Bressan J, Hernandez JA, Martins MF, Cecon PR. Effect of diet and PPARgamma2 and beta2-adrenergic receptor genes on energy metabolism and body composition in obese women. Nutr Hosp. 2006;21:317–31. [PubMed] [Google Scholar]
- 24. Warodomwichit D, Shen J, Arnett DK, Tsai MY, Kabagambe EK, Peacock JM, Hixson JE, Straka RJ, Province MA, An P et al.. ADIPOQ polymorphisms, monounsaturated fatty acids, and obesity risk: the GOLDN study. Obesity (Silver Spring). 2009;17:510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garaulet M, Smith CE, Gomez-Abellan P, Ordovas-Montanes M, Lee YC, Parnell LD, Arnett DK, Ordovas JM. REV-ERB-ALPHA circadian gene variant associates with obesity in two independent populations: Mediterranean and North American. Mol Nutr Food Res. 2014;58:821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bowen KJ, Kris-Etherton PM, West SG, Fleming JA, Connelly PW, Lamarche B, Couture P, Jenkins DJA, Taylor CG, Zahradka P et al.. Diets enriched with conventional or high-oleic acid canola oils lower atherogenic lipids and lipoproteins compared to a diet with a Western fatty acid profile in adults with central adiposity. J Nutr. 2019;149:471–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miazgowski T, Krzyzanowska-Swiniarska B, Dziwura-Ogonowska J, Widecka K. The associations between cardiometabolic risk factors and visceral fat measured by a new dual-energy X-ray absorptiometry-derived method in lean healthy Caucasian women. Endocrine. 2014;47:500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaul S, Rothney MP, Peters DM, Wacker WK, Davis CE, Shapiro MD, Ergun DL. Dual-energy X-ray absorptiometry for quantification of visceral fat. Obesity (Silver Spring). 2012;20:1313–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cho YS, Go MJ, Han HR, Cha SH, Kim HT, Min H, Shin HD, Park C, Han BG, Cho NH et al.. Association of lipoprotein lipase (LPL) single nucleotide polymorphisms with type 2 diabetes mellitus. Exp Mol Med. 2008;40:523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goodarzi MO, Wong H, Quinones MJ, Taylor KD, Guo X, Castellani LW, Antoine HJ, Yang H, Hsueh WA, Rotter JI. The 3' untranslated region of the lipoprotein lipase gene: haplotype structure and association with post-heparin plasma lipase activity. J Clin Endocrinol Metab. 2005;90:4816–23. [DOI] [PubMed] [Google Scholar]
- 31. Richardson K, Nettleton JA, Rotllan N, Tanaka T, Smith CE, Lai CQ, Parnell LD, Lee YC, Lahti J, Lemaitre RN et al.. Gain-of-function lipoprotein lipase variant rs13702 modulates lipid traits through disruption of a microRNA-410 seed site. Am J Hum Genet. 2013;92:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karmelic I, Lovric J, Bozina T, Ljubic H, Vogrinc Z, Bozina N, Sertic J. Adiponectin level and gene variability are obesity and metabolic syndrome markers in a young population. Arch Med Res. 2012;43:145–53. [DOI] [PubMed] [Google Scholar]
- 33. Tabatabaei-Malazy O, Fakhrzadeh H, Qorbani M, Amiri P, Larijani B, Tavakkoly-Bazzaz J, Amoli MM. Apolipoprotein E gene polymorphism and its effect on anthropometric measures in normoglycemic subjects and type 2 diabetes. J Diabetes Metab Disord. 2012;11:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zarkesh M, Daneshpour MS, Faam B, Hedayati M, Azizi F. Is there any association of apolipoprotein E gene polymorphism with obesity status and lipid profiles? Tehran Lipid and Glucose Study (TLGS). Gene. 2012;509:282–5. [DOI] [PubMed] [Google Scholar]
- 35. Alharbi KK, Syed R, Alharbi FK, Khan IA. Association of apolipoprotein E polymorphism with impact on overweight university pupils. Genet Test Mol Biomarkers. 2017;21:53–7. [DOI] [PubMed] [Google Scholar]
- 36. Wang H, Eckel RH. Lipoprotein lipase: from gene to obesity. Am J Physiol Endocrinol Metab. 2009;297:E271–88. [DOI] [PubMed] [Google Scholar]
- 37. Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: structure, function, regulation, and role in disease. J Mol Med (Berl). 2002;80:753–69. [DOI] [PubMed] [Google Scholar]
- 38. Ma Y, Tucker KL, Smith CE, Lee YC, Huang T, Richardson K, Parnell LD, Lai CQ, Young KL, Justice AE et al.. Lipoprotein lipase variants interact with polyunsaturated fatty acids for obesity traits in women: replication in two populations. Nutr Metab Cardiovasc Dis. 2014;24:1323–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rivellese AA, Giacco R, Annuzzi G, De Natale C, Patti L, Di Marino L, Minerva V, Costabile G, Santangelo C, Masella R et al.. Effects of monounsaturated vs. saturated fat on postprandial lipemia and adipose tissue lipases in type 2 diabetes. Clin Nutr. 2008;27:133–41. [DOI] [PubMed] [Google Scholar]
- 40. Simsolo RB, Ong JM, Kern PA. The regulation of adipose tissue and muscle lipoprotein lipase in runners by detraining. J Clin Invest. 1993;92:2124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nigro E, Scudiero O, Monaco ML, Palmieri A, Mazzarella G, Costagliola C, Bianco A, Daniele A. New insight into adiponectin role in obesity and obesity-related diseases. Biomed Res Int. 2014;2014:658913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park JW, Park J, Jee SH. ADIPOQ gene variants associated with susceptibility to obesity and low serum adiponectin levels in healthy Koreans. Epidemiol Health. 2011;33:e2011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Santos JL, Boutin P, Verdich C, Holst C, Larsen LH, Toubro S, Dina C, Saris WH, Blaak EE, Hoffstedt J et al.. Genotype-by-nutrient interactions assessed in European obese women. A case-only study. Eur J Nutr. 2006;45:454–62. [DOI] [PubMed] [Google Scholar]
- 44. Moreno JA, Perez-Jimenez F, Marin C, Gomez P, Perez-Martinez P, Moreno R, Bellido C, Fuentes F, Lopez-Miranda J. The effect of dietary fat on LDL size is influenced by apolipoprotein E genotype in healthy subjects. J Nutr. 2004;134:2517–22. [DOI] [PubMed] [Google Scholar]
- 45. Saito M, Eto M, Nitta H, Kanda Y, Shigeto M, Nakayama K, Tawaramoto K, Kawasaki F, Kamei S, Kohara K et al.. Effect of apolipoprotein E 4 allele on plasma LDL cholesterol response to diet therapy in type 2 diabetic patients. Diabetes Care. 2004;27:1276–80. [DOI] [PubMed] [Google Scholar]
- 46. Paula RS, Souza VC, Benedet AL, Souza ER, Toledo JO, Moraes CF, Gomes L, Alho CS, Cordova C, Nobrega OT. Dietary fat and apolipoprotein genotypes modulate plasma lipoprotein levels in Brazilian elderly women. Mol Cell Biochem. 2010;337:307–15. [DOI] [PubMed] [Google Scholar]
- 47. Chouinard-Watkins R, Plourde M.. Fatty acid metabolism in carriers of apolipoprotein E epsilon 4 allele: is it contributing to higher risk of cognitive decline and coronary heart disease?. Nutrients. 2014;6:4452–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kolovou GD, Anagnostopoulou KK, Kostakou P, Giannakopoulou V, Mihas C, Hatzigeorgiou G, Vasiliadis IK, Mikhailidis DP, Cokkinos DV. Apolipoprotein E gene polymorphism and obesity status in middle-aged men with coronary heart disease. In Vivo. 2009;23:33–9. [PubMed] [Google Scholar]
- 49. Rodriguez-Carmona Y, Perez-Rodriguez M, Gamez-Valdez E, Lopez-Alavez FJ, Hernandez-Armenta CI, Vega-Monter N, Leyva-Garcia G, Monge-Cazares T, Barrera Valencia D, Balderas Monroy M et al.. Association between apolipoprotein E variants and obesity-related traits in Mexican school children. J Nutrigenet Nutrigenomics. 2014;7:243–51. [DOI] [PubMed] [Google Scholar]
- 50. Chouinard-Watkins R, Conway V, Minihane AM, Jackson KG, Lovegrove JA, Plourde M. Interaction between BMI and APOE genotype is associated with changes in the plasma long-chain-PUFA response to a fish-oil supplement in healthy participants. Am J Clin Nutr. 2015;102:505–13. [DOI] [PubMed] [Google Scholar]
- 51. Huebbe P, Dose J, Schloesser A, Campbell G, Gluer CC, Gupta Y, Ibrahim S, Minihane AM, Baines JF, Nebel A et al.. Apolipoprotein E (APOE) genotype regulates body weight and fatty acid utilization—studies in gene-targeted replacement mice. Mol Nutr Food Res. 2015;59:334–43. [DOI] [PubMed] [Google Scholar]
- 52. Slim KE, Vauzour D, Tejera N, Voshol PJ, Cassidy A, Minihane AM. The effect of dietary fish oil on weight gain and insulin sensitivity is dependent on APOE genotype in humanized targeted replacement mice. FASEB J. 2017;31:989–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


