Abstract
Advancing knowledge of the transcriptome has revealed that circular RNAs (circRNAs) are widely expressed and evolutionarily conserved molecules that may serve relevant biological roles. More interesting is the accumulating evidence which demonstrates the implication of circRNAs in diseases, especially cancers. This revelation has helped to form the rationale for many studies exploring their utility as clinical biomarkers. CircRNAs are highly stable due to their unique structures, exhibit some tissue specificity, and are enriched in exosomes, which facilitate their detection in a range of body fluids. These properties make circRNAs ideal candidates for biomarker development in many diseases. This review will outline the discovery, biogenesis, and proposed functions of circRNAs.
Keywords: Circular RNA, RNA splicing, Prostate cancer, Biomarker, Cell-free RNA
Core tip: Circular RNAs are unusually stable RNA molecules that are tissue- and cell lineage-specific, abundantly expressed in cells, and enriched in exosomes. These properties facilitate their detection in different body fluids and probable utility as biomarkers. Herein, we review the literature that investigates their potential as biomarkers for prostate cancer.
INTRODUCTION
Whilst they are amongst the last addition to the RNA family, circular RNAs (circRNAs) are not new discoveries[1]. Circular transcripts were originally found to naturally exist in plant viroids in 1976[2] and in the hepatitis delta virus in 1986[3]. They were noted as endogenous molecules in eukaryotes by a study investigating splicing in the DCC gene[4]. In this study, splicing was observed to occur in a non-sequential fashion by means of “exon scrambling”; upstream exons moved downstream to bind exons and yielded circular transcripts[4]. Because their exons are inverted compared to the exonic arrangement on the genomic open reading frame, circRNAs were initially labeled as by-products of splicing error[5]. This narrative began to change upon discovery that the Syr gene in adult mice was only expressed as 1.23-kb circular transcripts[6]. Given the importance of this gene in sex determination during embryogenesis, it inferred possible pre-determined biological of circRNAs, albeit being grouped as non-coding RNAs (ncRNAs) at this time[7]. However, renewed interest in circRNAs occurred when Salzman et al[8] identified a myriad of circRNAs in a variety of normal, and malignant cell types. Additionally, the functional exploration of CDR1as revealed its ability to sponge miR-7 in neuronal tissue, inferring that miRNA sponging may be a function of other circRNAs as well[9]. Consequently, interest in the mechanistic machinery that drives the genesis of circRNAs, as well as their function has intensified over the last few years.
CIRCULAR RNA BIOGENESIS
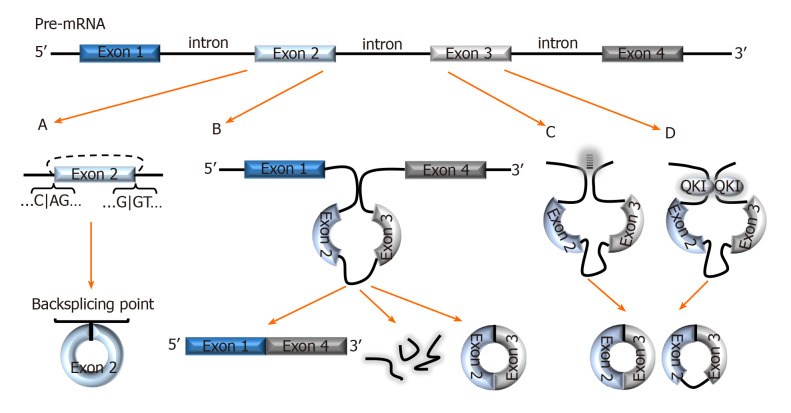
The combinatorial model best explains the alternative splicing (AS) mechanism that facilitates exon skipping. In this model, splicing regulatory factors coordinate the splicing order to determine which exons are included in the final mRNA transcript[10]. The outcome is multiple isoforms of a protein with different functions[11]. AS not only coordinates diversity amongst the linear transcriptome, it also facilitates a diverse group of circRNAs formed via backsplicing[12]. In the backsplicing process, circular transcripts are generated through covalently fusing the 5′ site of an upstream exon (acceptor) with the 3′ end of the same, or a downstream exon (donor)[5,13,14] (Figure 1A). The diversity amongst circRNAs was evidenced with multiple genes in a recent study- a salient example was the BIRC6 gene which was shown to generate over 500 circular isoforms[15]. Unsurprisingly, the study also highlighted that diversity amongst circular isoforms was directly proportional to exon counts in the gene[15].
Figure 1.
Biogenesis of circRNAs. A: In backsplicing, circRNAs are usually flanked by the canonical splicing motifs, AG-GT, and covalently fuse the 5′ site of an upstream exon (acceptor) with the 3′ end of a downstream exon (donor); B: In the exon skipping model, an unstable intermediate lariat consisting of introns and skipped exons are generated after splicing. The intermediate lariat is then spliced to produce circRNA; C: Flanking introns containing complementary sequences (Alu repeats) bind and increase the possibility of backsplicing; D: RNA-binding proteins, such as Quaking can bind to flanking introns and dimerize to create a closed RNA loop which facilitates backsplicing. QKI: Quaking.
Interestingly, backsplicing is flanked by the canonical splicing motif, AG-GT[15] and the circular RNAs and their relative linear RNAs share canonical splice sites suggesting that they are both generated by the same spliceosome machinery[16]. One study demonstrated that introducing mutations into the canonical splice sites significantly decreased circRNA production[16]. This study, as well as others[17] have also projected that circular and linear RNAs are competitively generated by the same spliceosome.
Liang et al[18] indicate that circRNAs are seldomly formed from the first or last exons as these exons lack splicing binding sites. Moreover, the number of exons in a single circRNA usually ranges between one and five exons, with several sources reporting that circRNAs with two to three exons are most prevalent[4,5,8,12]. Nonetheless, exons are not exclusive components of circRNAs; circularization of introns, long non-coding RNAs (lncRNAs), antisense transcripts, and intergenic regions is also possible[8,19]. Fascinatingly, there are multiple pieces of evidence of circRNAs consisting of both exonic and intronic regions[5,8,20,21], but exonic circRNAs are still most prevalent and studied[12,20]. Interestingly, Vo et al[15], mentioned a new subset of circRNAs generated from exons provided by adjacent genes on the same strand called read-through circRNAs (rt-circRNAs). The specific mechanisms of backsplicing are intricate and are still being investigated as bioinformatics of circRNA mapping improves. However, the following models are recurrently proposed to facilitate backsplicing: Exon skipping model (Lariat model), Intron-pairing, and the RNA-binding protein (RBP) models.
Exon skipping model (Lariat model)
In the exon skipping model, canonical splicing occurs first, producing the mRNA transcript, and an intermediate lariat consisting of introns and skipped exons[1,5] (Figure 1B). The intermediate lariat is unstable and undergoes further splicing (intra-lariat splicing) in which circRNA(s) are produced via backsplicing, and the intron lariat forms a separate RNA strand[1,5,20]. However, backsplicing via exon skipping can also occur independent of lariat formation by means of direct backsplicing[5].
Intron-pairing
A common feature amongst circularized exons is the presence of long flanking introns containing complementary sequences (Alu repeats)[20] (Figure 1C). This characteristic makes it possible to predict the backsplicing sites of circularization using bioinformatics. Hybridization of these complementary sequences increases the proximity of exonic backsplicing sites and facilitates backsplicing of said sites[18,20]. In this model, the circRNA generation is prioritized over linear transcripts, unlike in the exon skipping model[5,20]. Thus further suggesting that circRNAs are purposely produced, and according to Eger et al[5], explain the higher expression of certain circRNAs for some genes over linear transcripts. Interestingly, multiple studies propose that flanking intronic sequences represented in this model can be considered modulators in circularization efficiency[16,20,22]. Zhang et al[21] calls this model of backsplicing “alternative circularization”, and adds that alternative circularization in concert with alternative splicing, also enhances exonic circularization diversity from a single gene.
RBPs-mediated backsplicing
Multiple studies have demonstrated RBPs-mediated exon circularization with RBPs such as Quaking (QKI) and Muscleblind protein (MBL)[16,23]. In this model, RBPs bind to flanking introns (near to splicing sites) and dimerize to create a closed RNA loop that facilitates backsplicing[23,24] (Figure 1D). Conn et al[23] showed that inserting synthetic QKI into intron sites significantly induced circRNA formation and confirmed QKI-directed biosynthesis of circRNA. Similarly, in a prior study, circMbl formation was significantly increased after cells were transfected with MBL variants. This finding was accompanied by a reduction in linear Mbl generation[16]. Altogether, these results not only demonstrated RBP-regulated circRNA generation but also demonstrated the role of RBPs in competitive splicing to generate circular versus linear mRNAs.
CIRCULAR RNA FUNCTIONS
Though there are several pieces of evidence supporting functions such as miRNA sponging in molecules like CDR1as, substantial investigation of general functionality have only been demonstrated in a handful of circRNAs. Herein, we highlight three proposed functions of circRNAs that have been investigated: MiRNA sponging, protein binding, and cap-independent translation. However, whether these functions are generally exhibited by all or most circRNAs is not known.
CircRNAs are miRNA sponges and intermediate miRNA reservoirs
Perhaps the most examined function of circRNAs is their ability to sponge miRNAs. Some circRNAs harbor microRNA response elements (MREs) which facilitate the competitive binding of miRNAs[25,26]. The sequestration of miRNAs by circRNAs modifies their activity in regards to mRNA target gene expression[1,25]. In essence, circRNAs are indirectly involved in mRNA gene expression through miRNA sponging. For example, CDR1as contains over 70 conserved binding sites for miR-7[9,25,27], and the binding capacity is 10 times higher than that of any other transcript or mRNA target[27]. Hansen and colleagues further add that the competition between miR-7 targets and CDR1as creates a buffer effect that prevents transient fluctuations in miR-7 expression[28]. Furthermore, cleavage of CDR1as-miR-7 by argonaute 2 (AGO2) results in the release of miR-7 and the subsequent inhibition of miR-7 targets[25,28,29]. As such, CDR1as functions not only as a miRNA sponge but also as an intermediate reservoir for miR-7[29].
Protein binding
Some circRNAs can competitively bind RBPs as well as store, sort, and sequester proteins in the cytoplasm to limit nuclear entry, regulate their function, and act as scaffolds for protein-protein interactions[30,31]. For example, CircFOXO3 binds and prevents the interaction of p21 and CKD1 to suppress cell cycle progression at the G1 stage in a non-tumor cell line[32], and scaffolds p53 and Mdm2 in breast cancer cell lines to promote Mdm2-induced p53 degradation[33]. The interaction between circMbl and MBL is interesting as MBL can prioritize the generation of circMbl over linear forms, which in turn regulates MBL levels by sponging[16].
CircRNAs mediated protein translation in a cap-independent manner
The predominant opinion on circRNAs is that they are ncRNAs that do not translate proteins. However, the advent of engineered circRNAs that translate protein[20] fostered questions as to whether protein-coding endogenous eukaryotic circRNAs exist. Whilst the predominant stance still aligns with the former view, it has since come to light that there is a minute proportion (< 1%) of circRNAs that contain the start AUG codon, and are able to associate with ribosomes. Amongst them is circZNF609, which consists of a start and stop codon similar to those in the linear transcript. In their study, Legnini et al[34] were able to identify circ-ZNF609 as eukaryotic circRNAs that associate with polysomes, and are protein-coding. In circular transcripts like circ-ZNF609, the 5’untranslated regions (5’UTR) are included in the circular sequence during circularization. The 5’UTRs undergo folding to form internal ribosomal entry sites (IRES) which facilitate ribosomal association[34]. Some circRNAs such as circ-FBXW7 are also able to translate protein by other mechanisms such as N6- adenosine methylation[12,29]. Considering that most circRNAs are less abundant than their linear counterparts, it is unsurprising that the aforementioned examples of protein-coding circRNAs are less efficient in this activity than linear transcripts. Accumulating evidence also suggests that cap-independent translation is a cellular stress response to generate immediate and selective changes in protein levels[34].
THE POTENTIAL OF CIRCULAR RNA AS BIOMARKERS
Abundance
CircRNAs represent approximately 10% of the total RNA content in cells[35], with some being more abundantly expressed than their linear isoforms[8,36]. Their global expression and abundance can be stage-or-age dependent[37] as evidenced by several studies demonstrating variation in circRNA expressions at different developmental stages. Two studies reported the induction of circRNA expression during embryonic development in humans and flies across a range of tissues[38,39]. For example, the circular RNA generated from the NCX1 gene (primarily expressed in cardiomyocytes) was most highly expressed during fetal development according to Szabo et al[38]. In the mouse brain, one study demonstrated that certain circRNAs were more expressed in aged mice versus mice half their age[40] suggesting a function in neuronal maturity; another study described circRNA abundance at different stages of hippocampus development in the brain[41]. Interestingly, circRNA abundance can be independent of linear RNA expression[42] indicating splicing preference for generating certain circRNAs at different biological stages and suggesting an overall function in development.
Tissue- and cell lineage-specificity
The expression of some circRNAs is cell and tissue-dependent[17,42,43] which suggests they can be used as molecular markers for different diseases. For example, the expression levels of circular isoforms of the DCC gene varied across human tissues and did not correlate with their linear counterparts[4]. Similarly, certain circRNAs are concentrated in different parts of mammalian brains, and also had varying ratios of circRNAs versus linear RNAs[17]. In mice, the circular forms of Rmst and Khl12 were highly expressed in the brains versus the liver and lungs[41]. These studies suggest that circRNA generation and subsequent expression is a widely regulated process. Furthermore, this regulation appears to be evolutionarily conserved across mammals, having had several studies document the conservation between mouse, pigs, flies, and humans in brain tissues[1,17,20,42].
Stability
Unlike linear transcripts, circRNAs are covalently closed loops that lack polyadenylated tails[8,20]. Hence, circRNAs are relatively more stable, and have increased protection from exonuclease degradation[8,20]. Considering that exonucleases, and not endonucleases are the predominant nucleases in host RNA cells[44], it is inferred that the accumulation and detection of circRNAs is favored over the linear transcripts. Though RNA circularization generally increases stability of RNA molecules, hepatitis delta virus (HDV) circular RNAs become more susceptible to degradation by nucleases as they increase in molecular size. However, there is evidence suggesting that these larger HDV circles can be stabilized by their interactions with RBPs such as Ag-S[45].
Unsurprisingly, most circRNAs also have a half-life that is approximately 2.5 times longer than their linear counterparts in mammalian cells[20,25]. Due to their relative stability, circRNAs can also be detected at higher levels (approximately 6.3 folds higher) in exosomes than in cells[46]. This is an important property which contributes to their detection in body fluids.
Exosome enrichment and detection in body fluids
CircRNAs are more enriched in exosomes compared to intracellular levels[30,46]. Exosomes are vesicles that facilitate cell-to-cell communication between parent and recipient cells[27]. CircRNAs are sorted into exosomes potentially as a response to stimuli or physiological needs[27]. Though the precise mechanism is largely unclear, the sorting of circRNAs into exosomes is considered to be a regulated and selective process and can be guided by different factors such as RBPs and miRNA abundance[30,46]. Because of their enrichment and stability in exosomes, circRNAs are detectable in a range of body fluids including saliva[47], plasma[48], urine[49], gastric fluid[50], and supports their consideration as minimally-invasive biomarkers. One study shows that a group of exosomal-circRNAs (exo-circRNAs) in serum could distinguish between colon cancer patients and healthy controls[46]. Another study demonstrated that circRNA-IARS in exosomes could be a potential early diagnostic and prognostic predictor of pancreatic ductal adenocarcinoma (PDAC)[51]. These two studies demonstrate the translational potential of exo-circRNAs as circulating clinical biomarkers.
Genomic information
Unlike protein biomarkers, circRNAs are transcriptomic molecules that entail nucleic acid sequences. These sequences could potentially convey genomic information pertaining to germline mutations, as well as therapy-related somatic mutations which may inform disease prognosis and facilitate therapy decision[52]. Although cell-free tumor DNA can also provide similar information, it reflects the tumor cell genome and is passively released from dead tumor cells. In contrast, circRNAs are gene transcripts and can be both passively and selectively released from tumor cells in exosomes. Therefore, circRNAs could be more effective early indicators of disease.
CIRCULAR RNA IN PROSTATE CANCER
Current biomarkers in prostate cancer
Prostate cancer (PCa) is one of the most common cancers amongst men worldwide[53,54]. Like many other cancers, PCa management is plagued with the possibility of metastasis, therapy resistance, and poor diagnostic and prognostic biomarkers for screening[54]. Despite the emergence of a plethora of potential prostate cancer biomarkers, the prostate-specific antigen (PSA) still remains the best tool to general screening, and monitoring post-treatment[54]. Still, PSA testing is not without its shortcomings and controversies. Whilst it is prostate-specific, the PSA is not PCa specific, and its level in the blood can be affected by other factors such as age, trauma, inflammation, benign prostatic hypertrophy (BPH), etc[55]. Moreover, the established normal range of PSA (< 4.0 ng/mL) insufficiently captures PCa cases and often lead to under-diagnoses and false-positives[56,57]. Reports show that only 25%-30% of elevated PSA within the grey zone (4.0-9.9 ng/mL) cases are confirmed with PCa when biopsied[57,58]. From their study, Thompson et al[57] showed that normal PSA is also possible in men with PCa and high Gleason grade- this was observed in 15% of their study participants with normal PSAs.
The limitation of PSA also lies in deciding which cases move forward with biopsy for pathological diagnosis of PCa, which has been the blame for hundreds of thousands of unnecessary prostate biopsies in the United States yearly[59]. Serum levels of other PSA isoforms (e.g. p2PSA) show improved specificity to the PSA blood test[55]. Other potential biomarkers such as the prostate cancer antigen 3 (PCA3) score has shown utility in PCa diagnosis and monitoring[60]. PCA3 is a long non-coding RNA that is highly expressed in PCa (primary and metastatic cases)[60]. Whilst possessing a higher specificity than serum PSA, PCA3 score has variable sensitivity and requires a digital rectal examination to collect the specimen, which limits its clinical usage[61]. As evidenced by one study, using PCa-specific circRNAs (circ_0057558 and circ_0062019) from tissues and PSA levels together could offer a diagnostic advantage over just the PSA test[62]. In this study, the combination increased the AUC, specificity, and sensitivity for distinguishing between BPH and PCa[62]. However, reliable, and minimally-invasive PCa clinical biomarkers that can provide diagnostic and prognostic information solely, or in supplementation to the PSA test is still lacking.
CircRNAs as potential biomarkers of prostate cancer
The advancement of transcriptomic profiling has revealed a plethora of circRNAs worthy of further investigations for PCa biomarker development[15,36,63,64]. Chen and colleagues identified a group of circRNAs that are able to distinguish between localized PCa and normal prostate[36]. This study also proposed that circRNA abundance may not only be tissue-dependent but also based on functional roles in the tumor such as cell proliferation[36]. The functional analyses conducted in this study have strengthened the consideration of circRNAs as PCa biomarkers.
Along with establishing the MiOncoCirc catalog of circRNAs, Vo and colleagues identified a subset of circRNAs able to distinguish between PCa subtypes using tissue biopsies[36]. From this subset, circAMACR was upregulated and associated with androgen receptor (AR) amplification in castration-resistant prostate cancer. Additionally, circAURKA was upregulated in the suggestion of neuroendocrine prostate cancer (NEPC)[36]. These are promising markers for therapy-resistant PCa progression and warrant further investigations in clinical settings in different patient cohorts.
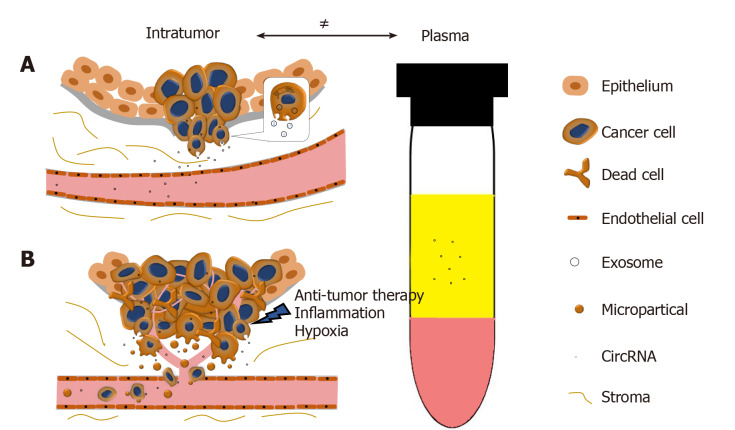
In collaboration with Yan Dong’s Lab, we reported and validated that multiple circRNAs are encoded by the AR gene, and are widespread in PCa cells and xenograft models[65]. We have further demonstrated that one of the AR circRNAs, namely circAR3, is abundantly expressed in prostate tissues and detectable in patient plasma in prostate- and prostate cancer-specific manners[52]. It is worth to be noticed that the levels of intratumoral circAR3 reduced in high Gleason tumors, while plasma circAR3 is positively associated with high Gleason scores and positive lymph node metastasis, making it suitable for biomarker development in PCa[52]. This disproportional expression of circRNAs in tissue and blood may likely be explained by the release rates of circRNAs from tissue to bloodstream that can be affected by multiple factors (Figure 2): (1) CircRNAs can be selectively packaged into exosomes and actively released from the tumor into the circulatory system where they are detectable in plasma; (2) With PCa development, the prostate architecture is disrupted leading to faster release of circRNAs from the tissues into the stromal space. They can circumvent the endothelial cells of the blood vessels and enter the bloodstream. Similar to PSA, the plasma concentration of PCa-specific circRNAs can be increased in this way; (3) Cell death induced by stresses such as hypoxia, inflammation, and anti-tumor therapies can increase the release of circRNAs into the bloodstream; and (4) As tumor invasion and metastasis occur, microparticles containing circRNAs are shed from tumor cells, subsequently increasing the circRNA concentration in plasma. As indicated with circAR3, plasma levels were higher in lymph node metastasis than without[52]. Altogether, these form a complex network that constitutes the disproportion between circRNA levels in tumors versus plasma.
Figure 2.
The disproportion of circRNAs between tumor and plasma. A: CircRNAs can be selectively enriched in exosomes and actively released into plasma as exosomes. During PCa progression, the integrity of normal prostatic tissues will be interrupted; this facilitates the release of circRNAs into the bloodstream; B: Stresses such as hypoxia, inflammation, and anti-tumor therapies will cause cell death and increase the release of circRNAs. Microparticles containing circRNAs shed from the metastasizing tumor will subsequently increase the circRNA concentration in plasma.
The functional characterization of circRNAs in PCa cells further advocates that certain circRNAs could be developed into PCa biomarkers. CircRNA-miRNA mapping has revealed that studying the interaction between circRNAs and miRNAs may further help to characterize the role of certain circRNAs in PCa development. In vitro investigations of interactions such as CDR1as-miR-7[66], circRNA-MYLK- miR-29a[64], and circBAGE2-miR-103a[66] have implicated tumor suppressive and oncogenic roles of circRNAs, which could imply their utility as biomarkers as well as therapeutic targets[64]. Other studies have shown that some circRNAs may play roles in contributing to therapy-resistance PCa. For example, downregulated circFOXO3 promotes PCa progression to be resistance to docetaxel[67], while hsa_circ_0004870 downregulation is correlated with enzalutamide resistance[11].
CONCLUSION
The surmounting evidence linking circRNA expression to the development of PCa is promising. Their presence and stability in body fluids such as plasma and urine allow their expressions to be analyzed in regards to a range of urologic diseases. Moreover, their detectability in said body fluids is a key pro in regards to convenient, minimally invasive sample collection which is an important feature for ideal biomarkers. Most exciting is the validation of a circRNA that is prostate and prostate-cancer specific, and detectable in the plasma of patients. Overall, further investigations are needed to truly label circRNAs as biomarkers. Firstly, it might be useful to focus on functionally characterizing specific circRNAs in pathogenesis and or tumorigenesis.
Molecular pathological epidemiology (MPE) research focuses on the etiology and pathogenesis of diseases. The inclusion of MPE studies in the future could provide clearer correlations between circRNAs, tumor characteristics/molecular changes, risk factors (environmental, lifestyle, microbiome, genetic mutations, etc.), and disease outcome (including tumor subtypes) in PCa patients. It would also be interesting to see whether the findings of such studies could expand on the potential clinical applications of circRNAs in cancer management; specifically as it relates to constructing predictive models that could improve screening and personalized medicine. But, the success of MPE research is hindered by challenges such as the need for trans-disciplinary experts, and poorer success rates with funding applications[68]. Nonetheless, MPE research generally have strong impact[68], thus it is a promising direction for elevating prostate cancer research with circRNAs.
Furthermore, considering the wide expression of circRNAs, perhaps closer attention should be on defining disease-specific circRNA panels which could be used in addition to traditional diagnostic markers. Additionally, for clinical validation, sample processing, detection method, and interpretation (cut-off) values need to be standardized across studies prior to truly establishing their clinical capacity as biomarkers. Nonetheless, with the growing capacity of next-generation sequencing and bioinformatics, the knowledge of circRNAs and their biomarker potential will undoubtedly continue to expand.
Footnotes
Conflict-of-interest statement: The authors have no conflict-of-interest related to this manuscript.
Manuscript source: Invited manuscript
Peer-review started: December 31, 2019
First decision: March 15, 2020
Article in press: July 19, 2020
Specialty type: Oncology
Country/Territory of origin: Canada
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ogino S, Taylor J, Yu B S-Editor: Zhang H L-Editor: A P-Editor: Li JH
Contributor Information
Dwayne Tucker, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6H 3Z6, Canada.
Wei Zheng, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6H 3Z6, Canada; Department of Urology, Zhejiang Provincial People's Hospital, Hangzhou 310024, Zhejiang Province, China.
Da-Hong Zhang, Department of Urology, Zhejiang Provincial People's Hospital, Hangzhou 310024, Zhejiang Province, China.
Xuesen Dong, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6H 3Z6, Canada. xdong@prostatecentre.com.
References
- 1.Dragomir M, Calin GA. Circular RNAs in Cancer - Lessons Learned From microRNAs. Front Oncol. 2018;8:179. doi: 10.3389/fonc.2018.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 4.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 5.Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN. Circular RNA Splicing. Adv Exp Med Biol. 2018;1087:41–52. doi: 10.1007/978-981-13-1426-1_4. [DOI] [PubMed] [Google Scholar]
- 6.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 7.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 10.Mabon SA, Misteli T. Differential recruitment of pre-mRNA splicing factors to alternatively spliced transcripts in vivo. PLoS Biol. 2005;3:e374. doi: 10.1371/journal.pbio.0030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greene J, Baird AM, Casey O, Brady L, Blackshields G, Lim M, O'Brien O, Gray SG, McDermott R, Finn SP. Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci Rep. 2019;9:10739. doi: 10.1038/s41598-019-47189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holdt LM, Kohlmaier A, Teupser D. Circular RNAs as Therapeutic Agents and Targets. Front Physiol. 2018;9:1262. doi: 10.3389/fphys.2018.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 14.Carrara M, Fuschi P, Ivan C, Martelli F. Circular RNAs: Methodological challenges and perspectives in cardiovascular diseases. J Cell Mol Med. 2018;22:5176–5187. doi: 10.1111/jcmm.13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 18.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan PH, Liu ZH, Pei YJ, Wu ZG, Yu Y, Yang YF, Liu X, Che L, Ma CJ, Xie YK, Hu QJ, Wan ZY, Wang HQ. Landscape of RNAs in human lumbar disc degeneration. Oncotarget. 2016;7:63166–63176. doi: 10.18632/oncotarget.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA. RNA Biol. 2017;14:1000–1006. doi: 10.1080/15476286.2016.1227904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: an emerging key player in RNA world. Brief Bioinform. 2017;18:547–557. doi: 10.1093/bib/bbw045. [DOI] [PubMed] [Google Scholar]
- 26.Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, Tang W, Cao H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Liang W, Zhang P, Chen J, Qian H, Zhang X, Xu W. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36:152. doi: 10.1186/s13046-017-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 29.Haddad G, Lorenzen JM. Biogenesis and Function of Circular RNAs in Health and in Disease. Front Pharmacol. 2019;10:428. doi: 10.3389/fphar.2019.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 31.Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and Characterizing circRNA-Protein Interaction. Theranostics. 2017;7:4183–4191. doi: 10.7150/thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, Zeng Y, Hua JT, Petricca J, Guo H, Wang M, Yousif F, Zhang Y, Donmez N, Ahmed M, Volik S, Lapuk A, Chua MLK, Heisler LE, Foucal A, Fox NS, Fraser M, Bhandari V, Shiah YJ, Guan J, Li J, Orain M, Picard V, Hovington H, Bergeron A, Lacombe L, Fradet Y, Têtu B, Liu S, Feng F, Wu X, Shao YW, Komor MA, Sahinalp C, Collins C, Hoogstrate Y, de Jong M, Fijneman RJA, Fei T, Jenster G, van der Kwast T, Bristow RG, Boutros PC, He HH. Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell. 2019;176:831–843.e22. doi: 10.1016/j.cell.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 37.Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC, Rajewsky N. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) 2017;95:1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gruner H, Cortés-López M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, Xiang Y, Liu L, Zhong S, Han L, He C. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984–992. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 44.Taylor JM. Structure and replication of hepatitis delta virus RNA. Curr Top Microbiol Immunol. 2006;307:1–23. doi: 10.1007/3-540-29802-9_1. [DOI] [PubMed] [Google Scholar]
- 45.Lazinski DW, Taylor JM. Expression of hepatitis delta virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J Virol. 1994;68:2879–2888. doi: 10.1128/jvi.68.5.2879-2888.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kölling M, Haddad G, Wegmann U, Kistler A, Bosakova A, Seeger H, Hübel K, Haller H, Mueller T, Wüthrich RP, Lorenzen JM. Circular RNAs in Urine of Kidney Transplant Patients with Acute T Cell-Mediated Allograft Rejection. Clin Chem. 2019;65:1287–1294. doi: 10.1373/clinchem.2019.305854. [DOI] [PubMed] [Google Scholar]
- 50.Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, Guo J. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173–1180. doi: 10.1002/cam4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo J, Li Y, Zheng W, Xie N, Shi Y, Long Z, Xie L, Fazli L, Zhang D, Gleave M, Dong X. Characterization of a Prostate- and Prostate Cancer-Specific Circular RNA Encoded by the Androgen Receptor Gene. Mol Ther Nucleic Acids. 2019;18:916–926. doi: 10.1016/j.omtn.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neuhaus J, Yang B. Liquid Biopsy Potential Biomarkers in Prostate Cancer. Diagnostics (Basel) 2018;8:68. doi: 10.3390/diagnostics8040068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim MCJ, Baird AM, Aird J, Greene J, Kapoor D, Gray SG, McDermott R, Finn SP. RNAs as Candidate Diagnostic and Prognostic Markers of Prostate Cancer-From Cell Line Models to Liquid Biopsies. Diagnostics (Basel) 2018;8:60. doi: 10.3390/diagnostics8030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alford AV, Brito JM, Yadav KK, Yadav SS, Tewari AK, Renzulli J. The Use of Biomarkers in Prostate Cancer Screening and Treatment. Rev Urol. 2017;19:221–234. doi: 10.3909/riu0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barry MJ. Clinical practice. Prostate-specific-antigen testing for early diagnosis of prostate cancer. N Engl J Med. 2001;344:1373–1377. doi: 10.1056/NEJM200105033441806. [DOI] [PubMed] [Google Scholar]
- 57.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, Minasian LM, Ford LG, Lippman SM, Crawford ED, Crowley JJ, Coltman CA., Jr Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 58.Parekh DJ, Ankerst DP, Troyer D, Srivastava S, Thompson IM. Biomarkers for prostate cancer detection. J Urol. 2007;178:2252–2259. doi: 10.1016/j.juro.2007.08.055. [DOI] [PubMed] [Google Scholar]
- 59.Vickers A, Cronin A, Roobol M, Savage C, Peltola M, Pettersson K, Scardino PT, Schröder F, Lilja H. Reducing unnecessary biopsy during prostate cancer screening using a four-kallikrein panel: an independent replication. J Clin Oncol. 2010;28:2493–2498. doi: 10.1200/JCO.2009.24.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N, Isaacs WB. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 61.Saini S. PSA and beyond: alternative prostate cancer biomarkers. Cell Oncol (Dordr) 2016;39:97–106. doi: 10.1007/s13402-016-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia Q, Ding T, Zhang G, Li Z, Zeng L, Zhu Y, Guo J, Hou J, Zhu T, Zheng J, Wang J. Circular RNA Expression Profiling Identifies Prostate Cancer- Specific circRNAs in Prostate Cancer. Cell Physiol Biochem. 2018;50:1903–1915. doi: 10.1159/000494870. [DOI] [PubMed] [Google Scholar]
- 63.Kong Z, Wan X, Zhang Y, Zhang P, Zhang Y, Zhang X, Qi X, Wu H, Huang J, Li Y. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;493:1217–1223. doi: 10.1016/j.bbrc.2017.07.162. [DOI] [PubMed] [Google Scholar]
- 64.Dai Y, Li D, Chen X, Tan X, Gu J, Chen M, Zhang X. Circular RNA Myosin Light Chain Kinase (MYLK) Promotes Prostate Cancer Progression through Modulating Mir-29a Expression. Med Sci Monit. 2018;24:3462–3471. doi: 10.12659/MSM.908009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao S, Ma T, Ungerleider N, Roberts C, Kobelski M, Jin L, Concha M, Wang X, Baddoo M, Nguyen HM, Corey E, Fazli L, Ledet E, Zhang R, Silberstein JL, Zhang W, Zhang K, Sartor O, Dong X, Flemington EK, Dong Y. Circular RNAs add diversity to androgen receptor isoform repertoire in castration-resistant prostate cancer. Oncogene. 2019;38:7060–7072. doi: 10.1038/s41388-019-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang C, Xiong J, Yang Q, Wang Y, Shi H, Tian Q, Huang H, Kong D, Lv J, Liu D, Gao X, Zi X, Sun Y. Profiling and bioinformatics analyses of differential circular RNA expression in prostate cancer cells. Future Sci OA. 2018;4:FSOA340. doi: 10.4155/fsoa-2018-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen Z, Zhou L, Zhang C, Xu J. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020;468:88–101. doi: 10.1016/j.canlet.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 68.Ogino S, Nowak JA, Hamada T, Milner DA, Jr, Nishihara R. Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu Rev Pathol. 2019;14:83–103. doi: 10.1146/annurev-pathmechdis-012418-012818. [DOI] [PMC free article] [PubMed] [Google Scholar]