Abstract
Lipids form an integral, structural, and functional part of all life forms. They play a significant role in various cellular processes such as membrane fusion, fission, endocytosis, protein trafficking, and protein functions. Interestingly, recent studies have revealed their more impactful and critical involvement in infectious diseases, starting with the manipulation of the host membrane to facilitate pathogenic entry. Thereafter, pathogens recruit specific host lipids for the maintenance of favorable intracellular niche to augment their survival and proliferation. In this review, we showcase the lipid-mediated host pathogen interplay in context of life-threatening viral and bacterial diseases including the recent SARS-CoV-2 infection. We evaluate the emergent lipid-centric approaches adopted by these pathogens, while delineating the alterations in the composition and organization of the cell membrane within the host, as well as the pathogen. Lastly, crucial nexus points in their interaction landscape for therapeutic interventions are identified.
Graphic Abstract
 Lipids act as critical determinants of bacterial and viral pathogenesis by altering the host cell membrane structure and functions.
Lipids act as critical determinants of bacterial and viral pathogenesis by altering the host cell membrane structure and functions.
Keywords: Host–pathogen interactions, Virulence-associated lipids, Lipid rafts, Lipid biosynthesis, Membrane organization, Drug development
Lipids in Bacterial Infections
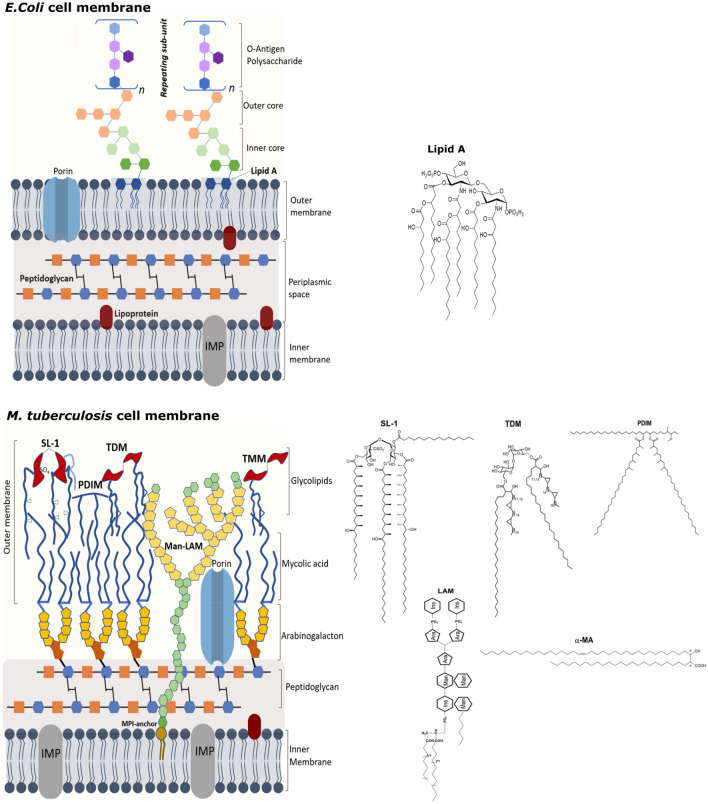
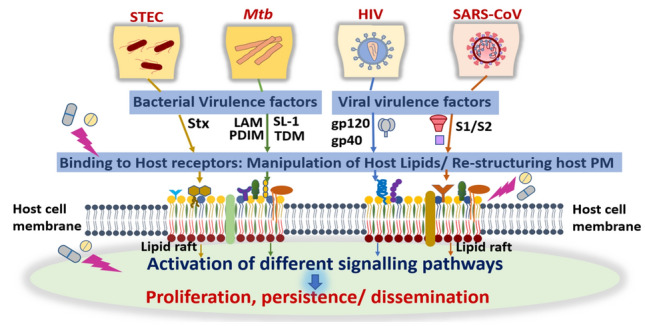
Bacterial cell membranes are specialized structures with specific spatiotemporal arrangement of complex lipids and proteins. They harbor all the components that enable bacterial survival and allows for selective bidirectional passage of nutrients and waste products. Numerous bacterial lipid-virulence factors are recognized by the human innate immune response, such as lipopolysaccharide (LPS) in Gram-negative bacteria, lipoteichoic acid in Gram-positive bacteria, and lipoglycans in mycobacteria (Larrouy-Maumus 2018). These underscore the role of lipids and their intricate interplay with the host cell machinery during bacterial infection. Below, we focus on lipids derived from two main disease-causing bacteria: (a) Escherichia coli (E.coli) and its pathogenic variant shiga toxin producing E.coli (STEC) responsible for bloody diarrhea and Hemolytic uremic syndrome (HUS), and (b) Mycobacterium tuberculosis (Mtb) that causes tuberculosis (TB). Both E.coli and Mtb differ vastly in their cell envelope composition and organization (Fig. 1), as well as their cell wall-associated lipid components, which serve as virulent factors causing disruption of host cellular signaling and manifestation of their respective diseases.
Fig. 1.
Schematic representation of the cell membrane architectures in E. coli and M. tuberculosis along with structures of key virulence-associated lipids
Structural and Functional Role of Lipids in E. Coli
Escherichia coli is a Gram-negative bacterium that is commonly found in the gut of humans and warm-blooded animals. It has been extensively studied and served as a role model for understanding of diverse fundamental biological processes.
The Bacterial Envelope
A typical Gram-negative bacterial cell wall consists of three principal layers: an outer membrane (OM), a peptidoglycan cell wall, and a cytoplasmic or inner membrane (IM) (Fig. 1). The aqueous cellular compartment delimited by the concentric IM and OM is called the periplasm. It is more viscous than the cytoplasm (Mullineaux et al. 2006) and is densely packed with proteins, essentially degradative enzymes like RNase and other periplasm-binding proteins that help in chemotaxis. The IM is essentially a phospholipid bilayer comprising of various membrane proteins that regulate energy production, lipid biosynthesis, protein secretion, and transport (Silhavy et al. 2010). The most abundant phospholipids in E. coli IM are phosphatidyl ethanolamine (PE) and phosphatidylglycerol (PG), and to a lesser extent, phosphatidylserine (PS) and cardiolipin (CL) (Raetz and Dowhan 1990). OM is a lipid bilayer that consists of phospholipids limited to the OM inner leaflet. The OM outer leaflet is composed of complex glycolipids, and LPS, (Fig. 1) which impart low fluidity and permeability. Overall, this asymmetry authors the impeccable barrier functions of OM (Shrivastava and Chng 2019). Rowlett et al. (2017) elucidated the precise roles played by membrane phospholipids in bacterial physiology and stress adaptation by means of genetically altered E. coli strains (i.e., having manipulated phospholipid contents). They inferred that membrane phospholipids have a global impact on both the membrane structure and the key metabolic pathways. For example, the cells lacking PE and CL result in changed LPS structure and affect the assembly and folding of OM proteins. Further, Rowlett et al. revealed alteration in phospholipids to impede global dehydrogenase activity, decreasing the levels of ATP, and increasing the intracellular oxidative stress. They observed metabolic changes in the upper glycolytic pathway, pentose phosphate pathway, and amino acid utilization, with altered membrane phospholipid synthesis and turnover. These results highlight the importance of proper membrane phospholipid composition in the maintenance of bacterial morphology, homeostasis, and adaptation to environmental factors. Through molecular genetics-based approaches, it is known that the absence of PE alters cell division and affects IM protein folding and assembly (Dowhan and Bogdanov 2009), thus emphasizing the role of PE in the localization, structure, and function of several IM bacterial proteins.
Lipopolysaccharides (LPS): The “Stageholder” Lipid Endotoxin
LPS is a notorious lipid macromolecule; it serves as a toxin inducing stimulation of the eukaryotic host cells, and also, protects the bacteria by providing essential permeability barrier against various antimicrobial peptides and clinical antibiotics (Di Lorenzo et al. 2019). LPS structure is highly conserved across bacterial species and consist of a covalently linked lipid component known as Lipid A, a core hydrophilic oligosaccharide and a O-antigen polysaccharide with repeating units of varying lengths (Fig. 1) (Wu et al. 2013). Lipid A, the key component of LPS, serves as the membrane anchor, with the polysaccharide component interacting with the external environment. The core oligosaccharide component is further divided into outer and inner core. The inner core, situated close to lipid A, is composed of rare sugars such as 2-keto-3-deoxyoctulosonate (Kdo) and l-glycero-d-manno-heptose and the outer core, that extends beyond lipid A, consists of common sugars such as hexoses and hexosamines. Attached to the outer core is the O-antigen that has either linear or branched arrangement of repeating saccharide units (Erridge et al. 2002). Wu et al. (2013) studied the structural dynamics of LPS through NMR and molecular dynamics simulation and inferred that water can penetrate through the inner core region and that the hydration is crucial for maintaining the bilayer structure. The core oligosaccharide and lipid A are negatively charged, resulting in strong affinity for divalent cations such as Ca2+. This strong electrostatic interaction is crucial for maintaining LPS structure–function integrity. The human-β-defensins and its analogues are cationic antimicrobial peptides that target LPS by electrostatic interactions and subsequently cause its disintegration leading to cell hemolysis (Krishnakumari et al. 2020). In fact, electrostatic interactions also play a role in communicating with the host cells. Through in silico studies and biophysical characterization, Bahl et al. (2011) showed that hemoglobin, which is a frontline host defense molecule, possesses evolutionarily conserved surface of cationic patches that function as potential LPS-binding sites. Through surface plasmon resonance (SPR), the high binding affinity regions in both the subunits of hemoglobin molecule responsible for LPS binding were revealed.
Lipid A is the endotoxin responsible for the toxic effects such as fever, diarrhea, and septic shock associated with the LPS-mediated bacterial infections (Erridge et al. 2002). Lipid A is recognized by different receptors on the host cell membranes. For instance, recognition by toll-like receptors (TLR)-4 and -2 leads to the stimulation of inflammatory responses by activation of caspase-4, caspase-5 as well as secretion of inflammatory cytokines. The process underlying the immune response signaling involves binding of LPS with TLR-4/MD-2 complex, with subsequent dimerization that initiates the signal transduction (Maeshima and Fernandez 2013). E. coli lipid A comprises of a bis-phosphorylated glucosamine disaccharide backbone decorated by six fatty acids with a 4 + 2 distribution, with 14:0 (3-OH) as the primary, 14:0 and 12:0 as the secondary acyl substituents (Fig. 1). X-ray crystallography studies of human TLR-4/MD-2 with E. coli hexa-acylated lipid A proved that five of the six acyl chains are buried within the MD-2 binding pocket, and the sixth acyl chain is partially exposed and interacts with TLR-4 (Park et al. 2009). This specific binding pattern facilitates the dimerization of LPS/TLR-4/MD-2 complex and suggests that the primary structures, as well as the conformation of LPS lipid, are important features that govern the immunopotency of LPS.
In addition to the immune-regulatory role, the lipid phase behavior and membrane organization of LPS modulates bacterial cell membrane properties toward drugs and host defense factors. In this regard, supramolecular assemblies of LPS such as spherical, lamellar, cubic and hexagonal or inverted hexagonal have been identified. These further depend on the LPS molecular geometry and environmental factors such as hydration, temperature, and presence of ions (Seydel et al. 1993; Wilkinson 1996). Studies examining the effect of structural and environmental factors on the phase transition behavior of LPS have revealed the following major observations: the main phase transition temperature (Tm) of LPS reduces upon removal of the polysaccharide unit and increases in the presence of divalent cations such as Mg2+ (Brandenburg and Seydel 1990; Nikaido 2003). In 2011, (Kubiak et al. 2011) Kubiak et al. examined the lateral organization of LPS in giant unilamellar vesicles (GUVs) reconstituted from E. coli polar lipid extracts. Rhodamine-DPPE—probe that preferentially partitions into fluid/disordered regions of the lipid membrane—labeled GUVs showed the existence of micrometer sized lipid domains at 10 mol % LPS concentration. Fluorescence correlation spectroscopy (using fluorescently labeled LPS) and Laurdan general polarization experiments corroborated the co-existence of fully disordered and gel-like lipid domains. These results indicated that LPS forms gel-like lipid clusters, with sizes depending on various LPS structural attributes.
Lipid-Dependent Pathogenic Manifestations in E. coli
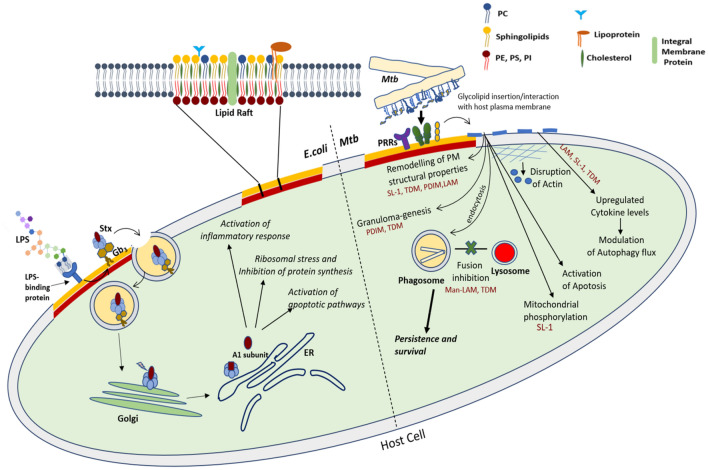
Escherichia coli is generally considered a harmless, friendly bacteria residing in the gastro-intestinal tract of humans and other warm-blooded animals, but the pathogenic strains cause diseases such as severe dysentery, urinary tract infections, meningitis and septicemia. The pathogenic strains differ from other commensal strain in their ability to express certain virulence factors, which disrupt the normal host physiology, and thus manifest diseased phenotypes (Donnenberg and Whittam 2001). Shiga toxin producing Escherichia coli (STEC) is one such strain among diarrheagenic E. coli that produces Shiga toxins (Stx) type 1 and type 2 virulence factors and is responsible for bloody diarrhea and HUS. Shiga toxins are ribosomal-inactivating proteins and are referred as “verotoxin-producing” or “verocytotoxin-producing” E. coli (VTEC), due to their ability to exert cytotoxic effect on the Vero monkey kidney cell line (Hunt 2010). The Stxs belong to a group of bacterial AB5 proteins (about 70 kDa) that inhibit protein synthesis in sensitive eukaryotic cells. Stxs preferentially target the microvascular endothelial cells of human kidneys and the brain (Melton-Celsa 2014; Bauwens et al. 2013). The pentamer of identical B subunits mediates toxin binding to the cellular receptor globotriaosylceramide (Gb3) (Melton-Celsa 2014), followed by endocytosis and retrograde trafficking of the toxin to the target organelle; the endoplasmic reticulum, ER (Fig. 2). Once at ER, the A1 fragment of the bacterial toxin (upon cleavage) exerts its ribotoxic effect resulting in the inhibition of protein biosynthesis followed by cell death (Legros et al. 2018). The precise molecular mechanism involves a complex interaction of the bacterial effector molecules and host lipids that manipulates the host signaling networks, such as apoptotic pathways (Karpman et al. 1998; Clements et al. 2012; Burlaka et al. 2013). The entry process of shiga toxins through the Gb3 receptors and the associated cellular effects are outlined in Fig. 2.
Fig. 2.
Lipid-mediated cellular effects pertaining to Shiga toxin producing E. coli and Mtb infection
Gb3 have been shown to cluster in the host lipid rafts with host glycosphingolipid, GM1, on the apical surface of (Kovbasnjuk et al. 2001), rendered using fluorescence resonance energy transfer (FRET) microscopy studies. Lipid rafts are specialized domains in the host plasma membrane (PM) that are enriched in cholesterol, glycosphingolipids, sphingomyelin, and raftophilic proteins (Fig. 2). Fluctuations in local PM lipid composition leads to lipid raft domain formation, which act as major signaling hotspots, regulating cellular processes. In addition, lipid rafts also house actin anchoring (signaling) lipids such as phosphatidylinositol phosphates (PIPs) that regulate the structuring of the host cytoskeleton and hence are exploited by pathogens for their uptake. It has been shown that lipid rafts disruption significantly decreases the internalization of Stx 1 B-subunit. Although raft disruption by cholesterol depletion does not affect the amount of bound Stx 1 B-subunit, host lipid rafts are necessary for the toxin uptake across the apical host membrane. Consistently, functional hijacking of host lipid rafts and the associated cytoskeletal machinery have been implicated in infections by also other enterohemorrhagic E. coli strains. For instance, Riff et al. (2005) assessed the modulation of the host cytoskeleton during toxin invasion upon adherence of enterohemorrhagic E. coli (EHEC) O157:H7 strain CL56 and enteropathogenic E. coli (EPEC) O127:H6 strain E2348/69 to the epithelial cells. It was observed that upon cholesterol depletion from the PM, the cytoskeletal rearrangement was inhibited and the same was restored upon exogenous cholesterol addition. This indicates that host membrane cholesterol within lipid rafts is necessary for the cytoskeletal rearrangement following infection with attaching-effacing E. coli strains. Furthermore, the role of lipid rafts in invasion of colonic epithelial cells by Shiga toxigenic E. Coli O113:H21 is also documented (Rogers et al. 2012). Prior to this study, Duncan et al. (2004) elucidated the molecular basis for the invasion of uropathological E. coli (UPEC), responsible for urinary tract infection, into the bladder epithelial cells. They found that the signaling molecule required for E. coli invasion was located within the host lipid rafts and associated with caveolin-1, as the disruption of lipid rafts or lowering of caveolin expression inhibited the bacterial invasion.
Gb3 receptors belong to a diverse group of Glycosphingolipids (GSLs), which along with cholesterol and sphingomyelin, regulate the stabilization and spatial organization of PM microdomains. Recently, Legros et al. (2018) carried out a comprehensive study on the composition of Stx-binding glycosphingolipids (GSLs) in Madin-Darby canine kidney (MDCK) II epithelial cells. They demonstrated that the distribution of GSLs was limited to the detergent-resistant membranes (DRMs), while ascertaining the lipid composition of DRM and non-DRM preparations. Despite the high cellular content of the GSLs in MDCK cells, the cells showed a lack of Stx-binding GSLs in the apical PM, and, as a result, displayed high resistance toward the shiga toxins. The authors reasoned that this could be either a masking effect or structural modification such as a tilt of GSL polar head group by other membrane components like cholesterol, consistent with other studies (Lingwood 2011). Or could be a manifestation of the different routes of intercellular trafficking, though this hypothesis awaits verification. The authors finally concluded that the cellular content of GSLs and their biochemical detection in DRM preparations are not the only determinants of cellular sensitivity toward Stxs.
Lipid Microvesicle-Mediated Trafficking of Toxins
The bacterial virulence factors can direct their way to the target organs by gaining access to the bloodstream, causing widespread target organ damage, such as renal failure or brain damage (Villysson et al. 2017). One of the main mechanisms of toxin-induced systemic and targeted organ injury involves the transport of bacterial toxin through microvesicles (MV). In fact, Shiga toxin circulate within host MVs originating from neutrophils, monocytes, platelets and red blood cells, followed by uptake by glomerular endothelial cells leading to cell damage and thrombocytopenia (Ståhl et al. 2015). MVs are small lipid vesicles (100–1000 nm) that ubiquitously contribute to infection and immunity depending on their origin, i.e., the pathogen or host (Fig. 2). Host MVs expose PS on their outer surface, which are otherwise present in the inner leaflet of the membrane, and this flipping involves calcium-mediated activation of enzymes such as floppase (facilitating movement of lipids toward the outer leaflet) and scramblase (bidirectional activity). Inhibition of flippase (facilitating movement of lipids toward the inner leaflet) leads to host membrane remodeling, and perturbs the phospholipid asymmetry (Nagata et al. 2020). In acute cases of STEC-HUS infections, the MVs, mostly from platelets and monocytes bearing tissue factor, binds to annexin V through PS. The presence of tissue factor and PS on MVs contribute to the formation of microthrombi during the acute phases of infection (Villysson et al. 2017).
Apart from the transport of bacterial effectors through host-derived MVs, bacteria itself releases MVs transport proteins, lipids and genetic material directly into the host cells. For example, UPEC strain 536 produces MVs that contain the protein toxin hemolysin (Balsalobre et al. 2006), cytotoxic necrotising factor type 1 (Kouokam et al. 2006) and a range of RNA species (Ghosal et al. 2015; Blenkiron et al. 2016). Yaron et al. (2000) demonstrated the MVs isolated from the E. coli O157:H7 to mediate the transfer of virulent genes, which are then expressed by the recipient Salmonella enterica serovar Enteritidis or E. coli JM109. MVs transport molecular cargos and dispatch them to the target cells by either direct fusion with the cytoplasmic membrane and/or by endocytosis, modifying the host physiology (Fig. 2). Though understanding the molecular mechanisms of MV biogenesis is still at a nascent stage, modulation of lipid composition has been shown to enhance membrane curvature—a perquisite for the emergence of these extracellular vesicles (Gill et al. 2019). In fact, asymmetric distribution of phospholipids between the inner and outer leaflets of the OM enhances membrane curvature aiding in hypervesiculation. Thus, identification of the mechanism controlling the asymmetric distribution of lipids across the bacterial membrane is a guiding factor for the future research focused on MVs.
Pathogenic role-play of mycobacterial lipids
Mycobacteria belongs to the Actinobacteria genus that consists of over 170 different species. M. tuberculosis and M. leprae constitute the human pathogens, while others cause diseases in animals (M.bovis, M. avium) and some are generally non-pathogenic (M.kansasii, M. smegmatis). Tuberculosis (TB), caused by obligate human pathogen, Mycobacterium tuberculosis (Mtb), persists as one of the most fatal infectious disease involving multidrug resistance phenotype and therefore warrants development of novel therapies and preventive strategies. TB is a pulmonary disease and the infection initiates when Mtb is inhaled in the form of liquid aerosol droplets and infiltrate deeply into the lungs of a host. The lung alveolar macrophages are the host’s first line of defense that eradicates pathogens through a systematic receptor-mediated phagocytosis. In this regard, Mtb is peculiar as it evades the immune system by blocking the maturation of the phagosome (Fig. 2) and stays concealed within host cells or within the granulomatous caseous necrotic centers (Barry et al. 2009). This enables Mtb to resist degradation and eventually create a favorable niche in the phagosome for its replication. The most distinguishing feature of this bacillus is that it can persist during the long phase of latency.
Lipids that decorate the Mtb OM are complex non-covalently attached glycolipids (Fig. 1) and are intricately involved in regulating specific events of host–pathogen interaction like: penetration into the host cells, escaping the eradication machinery of the macrophages, proliferation, alteration of host cell membrane properties upon insertion and finally modulation of host signaling networks (Mishra et al. 2019; Dadhich et al. 2020), Fig. 2. These underline Mtb lipids as major virulence factors, in addition to much investigated Mtb proteins (Forrellad et al. 2013). Further, these outer membrane lipids are also responsible for the ineffectiveness of various drugs underscored by their limited passage across the Mtb OM. As an important fact, mycobacterium has about 250 genes encoding for lipid biosynthesis and metabolism, which further stresses upon the importance of lipids in the life cycle of Mtb (Smith 2003). This highlights the unmet need to investigate the various facets of Mtb lipids detailing their structural make-up, involvement in host interaction as well as their functional role in forming a robust permeability barrier against therapeutics.
Mycobacterial Lipid Cell Envelope: Avenues for Therapeutic Membrane Targeting
Mycobacterium tuberculosis has a complex cell membrane architecture (Fig. 1) with its IM consisting of conventional glycerophospholipids and phosphatidyl-dimannosides (Bansal-Mutalik and Nikaido 2014). Attached to IM is the cell wall core composed of peptidoglycan (PG) with covalent linkages to the heteropolysaccharide arabinogalactan (AG). AGs are further esterified at their non-reducing ends to α-alkyl, β-hydroxy long-chain (C60-C90) mycolic acids (MAs). The covalently bound MAs of the cell wall core form the inner leaflet of OM, while the outer leaflet of OM is constituted by non-covalently exposed lipids such as phthiocerol dimycocerosate (PDIM), Trehalose Dimycolate (TDM), sulfoglycolipids (SL), phosphatidylinositol mannosides (PIMs), lipoarabinomannan (man-LAM), phenolic glycolipid (PGL), and diacylglycerol (DAG) along with some phospholipids (Bansal-Mutalik and Nikaido 2014; Layre et al. 2011), Fig. 1. Asymmetric OM is also referred to as mycomembrane and imparts an exceptionally high impermeability barrier, and therefore, characteristic resistance to many therapeutic agents (Brennan and Nikaido 1995). Investigating the molecular organization of mycobacterial cell membrane has been the focus of numerous studies based on the structural characterization through Cryo-electron microscopy (Hoffmann et al. 2008; Sani et al. 2010; Zuber et al. 2008) and lipidomic determination (Layre et al. 2011; Sartain et al. 2011). It is extremely important to understand the lateral organization of Mtb’s long-chained lipids within the membrane plane to gain a clearer perspective of the associated structural and dynamic attributes that regulate mycobacterial membrane functions implicated in drug interactions and virulence.
Cell membrane organization controls cellular functions by regulating the lipid domain dynamics as well as biomolecular interactions within the membrane and thus depends on the structural properties of the constituent lipids. Lack of our knowledge on the physicochemical aspects of mycobacterial lipid greatly thwarts efforts directed toward therapeutic targeting of Mtb membranes. In this regard, recently, our group elucidated the lipid membrane properties of compositionally and spatially distinct mycobacterial IM and OM lipids as well as the functional significance of Mtb transmembrane bilayer arrangement (Adhyapak et al. 2020). In this work, the authors reconstituted the protein-free lipid extracts of IM and OM from M. smegmatis into model membranes in vitro and, characterized these using atomic force microscopy (AFM), fluorescence, infrared spectroscopy, and two-photon microscopy. These experiments revealed the existence of distinct lipid domains in mycobacterial IM and OM membranes with varying levels of membrane packing, order, hydration, and fluidity. The major observations were: (a) existence of loose acyl chain packing with high fluctuations and interfacial hydration of the branched and long-chained lipid in OM, (b) tighter packing at the hydrophobic lipid acyl chain region in IM, attributed to the saturated nature of IM lipid chains. Addition of peptidoglycan associated lipids (PAL) and lipoarabinomannan (LAM) to OM, fine-tuned the lipid-lipid interactions governed by the conformational heterogeneity in the meromycolate chains of MA within PALs. Further, the highly branched network of complex sugars in LAM negated the effect of MA, highlighting LAM’s involvement in attuning the morphological organization and lipid dynamics in the M. smegmatis mycomembrane. These findings could inspire designing of suitable therapeutic agents that can leverage the distinct biophysical properties of mycobacterial lipid membranes to enhance drug uptake. Design of selective small molecules exhibiting selective interactions with Mtb membrane over host membranes, thus obviating toxicity issues, is also foreseen from this study.
In line with above, a well-characterized mycobacterial membrane mimic can find openings in myriad technological applications, e.g., a cell-free drug screening platform and drug-delivery (Zhang et al. 2019). Aligning with the approach of lipid-guided drug screening in tuberculosis, Dadhich et al. developed membrane scaffolds specific to mycobacterial OM and demonstrated them as selective and novel platforms for investigating anti-tubercular drug interactions (Dadhich et al. 2019). Designed membrane scaffolds such as TDM:DAG:DPPG (9.1:18.2:72.7 mol %), TDM:DAG (33.3:66.6 mol %) and TDM were characterized for their interaction with antituberculotic drug, rifabutin. These scaffolds were also compared with DMPG, a pan-bacterial model lipid extensively used for examining drug interactions, and a negatively charged eukaryotic membrane mimic (DOPC:DOPG:DPPC:DPPG:Chol::20:5:45:5:25) to probe the specificity of drug interactions. Properties such as membrane order, lateral organization, lipid dynamics and fluidity were evaluated upon drug binding, followed by determination of the drug’s partitioning coefficients. It was revealed that the TDM-containing membrane systems show altered bilayer hydration governed by the MA chain dynamics/conformations, trehalose head group-water interactions, and molecular interactions induced by DAG and DPPG. Employing GUVs stained with n-Rh-DHPE (that selectively partitions into the fluid disordered lipid domains), authors elucidated the co-existence of ordered and disordered lipid domains within TDM-containing membrane mimics. Notably, the apparent phase transition (Tm) for TDM:DAG:DPPG agreed well with the thermodynamic behavior of mycobacterial cell wall, (Liu et al. 1995, 1996). This implies the suitability of the novel TDM-containing lipid systems to recapitulate the physical properties of native Mtb outer mycomembranes. Rifabutin was shown to remodel the domain architecture of Mtb model membranes by disrupting the ordered and disordered lipid domains. It was reasoned that the hydrophobic chain length mismatch governed by the line tension was the main driving force for the observed lateral phase segregation (García-Sáez et al. 2007). And in the presence of the drug, this interfacial energy or line tension at the periphery of the domains was either depleted or redistributed, leading to modulated lateral domain organization. Interestingly, attenuated interaction of rifabutin with DMPG and eukaryotic membranes highlighted the lipid composition specificity during drug-membrane interactions, and the same should be accounted for in drug discovery efforts tailored toward TB to increase specificity and reduce toxicity. Finally, deepened understanding of drug-Mtb membrane interaction would foster designing efficient liposomal drug-delivery systems, which have been therapeutically most successful till date.
Lipid Landscape in Mtb-Host Interactions
Mycobacterium tuberculosis synthesizes various atypical lipids (outlined in Fig. 1) with structural features that are unique to this species. These include trehalose head group, methyl branches, cyclopropane rings and long-chain lengths. Among all the Mtb lipids, the free-exposed lipids within the bacterial OM are of utmost interest as their location pre-disposes them to interact with the host cell membrane upon contact. These surface exposed bacterial lipids influence host functions either by binding to the host membrane receptors or upon insertion by altering the physicochemical properties of the host membranes (largely understudied). The latter modulates host membrane-associated cellular signaling and trafficking events. In the former mode of interaction, pathogen-associated molecular patterns (PAMP) on bacterial surface are recognized by the corresponding pattern recognition receptors (PRRs) on the host cells such as TLR, C-type lectin receptors (CLR), Fc receptors (FcR), scavenger receptors (SR), and cytosolic DNA sensors (Queval et al. 2017). As an outcome, various cellular processes are activated including apoptosis, antigen processing/presentation, inflammasome activation, phagosome maturation, and autophagy (Lugo-Villarino et al. 2011; Mortaz et al. 2015). Once phagocytosed inside the host, Mtb resides inside the macrophagal endosomes with its intracellular survival resting upon the ability to prevent phagosome-lysosome fusion, and related host processes.
Mycobacterium tuberculosis adopts a lipid-centric approach to maneuver its entry into the host followed by opportunistic alteration of various host cellular processes and is described below in a lipid specific fashion (Fig. 2).
LAM is a well-recognized Mtb effector molecule regulating the intracellular trafficking network, as well as immune responses in infected host cells (Mishra et al. 2011). The distinguishing immunological ramifications of mannose capped LAM, Man-LAM, in virulent Mtb appears to be linked to the distinct and well-defined structural characteristics of this molecule. These include degree of acylation, lengths of D-mannan, D-arabinan cores, and mannose caps, as well as the presence of other acidic constituents such as succinates. For example, existence of 5-methylthioxylosyl (MTX), a substituent present in the arabinan core of LAM is associated with its anti-oxidative effect (Turnbull et al. 2004). Man-LAM of the virulent Mtb strains interferes with the maturation of the phagosomes (Fig. 2). Recognition of man-LAM by mannose receptor, Dectin-2 (belonging to the CLR family), induces anti-inflammatory cytokine production leading to subdued oxidative response (Yonekawa et al. 2014), Fig. 2. Man-LAM interferes with the phosphatidylinositol 3-phosphate machinery—known to serve as a docking molecule by the lysosome peripheral proteins (Józefowski et al. 2008)—by blocking its formation and accumulation, and thus arresting phagosome maturation (Fratti et al. 2003; Vergne et al., 2003). Recently, Turner and Torrelles (2018) reviewed the subtle relationship between structure and biological functions of Man-LAM, dissecting its impact on the host–pathogen interactions. The structural motif in the tripartite structure of Man-LAM, which is composed of an MPI (Man-LAM phosphatidyl-myo-inositol) anchor, D-mannan and immunodominant D-arabinan cores, has been linked to its physical and biological properties (Fig. 1). The nature, number, and position of acyl chains in the MPI anchor add to the characteristic heterogeneity of Man-LAM. The unique biological functions involving host cell interference has been mainly derived from D-arabinan core motifs involving the mannose-cap units, succinate residue and the MTX residue.
Mycobacterial cord factor or trehalose-6,6-dimycolate (TDM) is another Mtb glycolipid involved in the biogenesis of phagolysosome. The Mincle receptor, belonging to another sub-family of CLRs specifically recognizes TDM (Ishikawa et al. 2017) and this interaction induces several responses such as production of pro-inflammatory cytokines, generation of Th1/Th17 immune responses and induction of granuloma-genesis (Ishikawa et al. 2009; Mishra et al. 2017), Fig. 2. In an earlier study by Spargo et al. (1991), TDM was shown to inhibit calcium-induced vesicle fusion, suggesting a role of TDM in the inhibition of fusion between the phagosome and lysosomes. A detailed description of macrophage receptors for Mtb has been reviewed by Ernst (1998).
Phthiocerol dimycocerosate (PDIM) is a widely investigated Mtb virulent lipid (Camacho et al. 2001). The mode of action of DIM/PDIM includes modulation of phagocytosis and is correlated to cholesterol-linked CR (Compliment Receptor)-3 receptors (Astarie-Dequeker et al. 2009). Of most interest is the ability of PDIM to directly impact biophysical properties of the host membrane upon interaction—a field rapidly gaining attention (Astarie-Dequeker et al. 2009; Augenstreich et al. 2019; Mishra et al. 2019). DIM incorporates into the host membranes and modulates its organization by increasing the membrane rigidity. More recently, Augenstreich et al. (2019) corroborated the incorporation of DIM into the host THP-1 cell membrane by MALDI-TOF mass spectroscopy. The proposed mechanism involves either direct insertion of free DIM into the host membrane, or vesicle-mediated transfer and/or endocytosis. Further, using multi-scale molecular modeling and 31P-NMR, they revealed that DIM adopts a conical shape in host membranes. DIM also aggregates in the stalks formed between two opposing lipid bilayers, leading to formation of non-bilayer (inverted hexagonal) structures that act as fusion intermediates. Conical shape of PDIM helps the vesicle to attain the required curvature for the formation of fusion intermediates and finally fusion with the host membrane (Chernomordik and Kozlov 2008). In fact, formation of the non-bilayer structures is associated with the ease of the fusion process (Zick et al. 2014). In the work by Augenstriech et al., infection of macrophages pre-treated with lipids of various shapes uncovered a general role for conical lipids in promoting phagocytosis. Thus, the conical shape of DIM and its effect on disorganizing the membrane may play a role in the induction of phagosomal membrane rupture and apoptosis (Augenstreich et al. 2017). Many Mtb glycolipids extend their inhibitory effect via interaction of their sugar moiety with the target receptors (Astarie-Dequeker et al. 2010). However, as DIM lacks such a polysaccharide unit (Fig. 1), the molecular mechanism involving DIM may be affiliated to a wide ranging effect on the physical properties of the host cell membrane, such as fluidity, membrane order and hydration (Fig. 2). In addition, PDIM aids the escape of Mtb from its intracellular vacuole into the cytosol, leading to host cell necrosis and macro-autophagy (Quigley et al. 2017). In a very recent report, Augenstreich et al. (2020) investigated the membrane perturbation by DIM using polarity sensitive fluorophore, C-Laurdan, and multi-photon microscopy. A decrease in the membrane polarity at the site of contact between the bacilli and host membranes was observed and this decrease extended over distance of 1–1.5 μm around the bacterium. This is in corroboration to their earlier MD simulation findings that revealed diffusion of DIM across the host membrane (Augenstreich et al. 2019). Since DIM induces membrane curvature; its effect on the activity of membrane-associated proteins like CR-3 (receptor for Mtb phagocytosis) and EsxA (virulence factor) was evaluated. The authors observed that DIM caused an enhancement in activity of both CR-3 and EsxA. All-together, DIM disperses and remodels lipid organization within the host cell membrane influencing the activity of host cell receptors and bacterial effectors and consequently alters the cell signaling pathways. These outcomes reveal the molecular mechanisms by which Mtb exploit DIM to rewire the host cells (Augenstreich et al. 2020).
Sulfoglycolipid-1 (SL-1), a tetra-acylated glycolipid is the most abundant sulfatide in Mtb OM (Fig. 1). It features a sulfated trehalose headgroup with four acyl chains with CH3 branches of distinct stereochemistry. It is uniquely expressed in the virulent strains of Mtb and is present as a non-covalently bound free lipid on the OM surface (Domenech et al. 2004; Blanc et al. 2017). SL-1 mediated effects in the host cell include disruption of the phagosome-lysosome fusion, disarray of the mitochondrial phosphorylation and activation as well as suppression of cytokine levels in human leukocytes (Gilmore et al. 2012), Fig. 2. Mishra et al. (2019) specifically addressed the ability of SL-1 to alter host cell membrane properties, akin to PDIM, to validate the host cell membrane insertion as a “generic” mode of action of Mtb virulent lipids. The authors characterized the biophysical, nanomechanical and cell biological properties of live THP-1 macrophage cell membranes upon insertion of SL-1 (Mishra et al. 2019). SL-1 was demonstrated to remodel the host cell membrane organization with higher effects on the ordered lipid raft regions. Moreover, SL-1 increased the membrane fluidity in a spatiotemporal fashion. Further, SL-1 decreased the elastic modulus of host cell membrane by approximately twofold. Interestingly, the authors demonstrated the SL-1 effect to percolate to the underlying actin cytoskeleton leading to autophagy activation (Fig. 2). Inspired by these results, the same group adopted a chemical biology approach, wherein they synthesized various SL-1 analogues to explore the functional role of specific chemical moieties of SL-1 lipid structure during host membrane interactions. Biophysics, cell biology and molecular dynamics simulations furnished insights into the structure–function relationship of SL-1 to elucidate a direct correlation between host membrane structure modification and modulation of membrane-associated autophagy and cytokine signaling (Dadhich et al. 2020). It was revealed that the fatty acid acyl chains at 6,6’ position on SL-1 headgroup are crucial for membrane ordering during interaction with host membranes, leading to host lipid raft modulation. The sulfate group was shown to have a role in blocking autophagosome maturation and modulating host autophagy flux during signaling. SL-1 exhibited distinct conformational states of its acyl chains, which markedly differed from TDM, thus underlining the observed differences in biological output with distinct Mtb lipids. These findings highlight a mechanism whereby Mtb uses specific chemical moieties on its lipids to fine-tune host lipid interactions and regulate the downstream host signaling by modifying cell membrane structure and function. These results are expected to foster development of chemotherapeutics against Mtb by counteracting the effects of Mtb lipids on host cell membrane. The authors also reported a hitherto unknown existence of phase heterogeneity in single component SL-1 lipid GUVs. The phase co-existence was corroborated with the fluorescence lifetime measurements, infrared spectroscopy and AFM imaging. Finally, MD simulation attributed the observed phase co-existence to the distinct conformational states of Mtb SL-1 acyl chains.
Apart from Mtb lipids, the host lipid components such as cholesterol are also involved in facilitating the optimum lipid interface for the entry of the mycobacterium through lipid rafts (Gatfield and Pieters 2000). For example, cholesterol-dependent CR-3 has not been observed in entry mechanism of other bacteria. Entry of M. kansasii into the host neutrophil cells is through the cholesterol-rich raft domains with the involvement of CR-3 receptors, associated with GPI-anchored proteins. Viswanathan et al. (2015) reported the inhibition of M. smegmatis entry into the host macrophages upon depletion of cholesterol and the effect was reversed upon replenishment of membrane cholesterol. More recently, the same group (Viswanathan et al. 2018) also assessed the specific role-play of macrophage sphingolipids in the internalization of M. smegmatis. The authors divulged that metabolic depletion of sphingolipids in host macrophages lead to a significant reduction in the entry of M. smegmatis, where, on the other hand, the entry of Escherichia coli into host macrophages under similar conditions remains unaffected. These imply the specificity for the requirement of sphingolipids in mycobacterial entry.
Lipids in Viral Infections
Lipids are critically involved in the life cycle of viruses, right from the membrane fusion for entry, to the envelopment of the replicated viral genetic material for release. In more recent times, the role of lipids in viral infections has gained considerable recognition. Viruses not only manipulate cellular lipids and membranes inside host cells but also induce global lipidic metabolic changes within infected cells, facilitating viral multiplication (Heaton and Randall 2011; Martín-Acebes et al. 2013; Ketter and Randall 2019). A viral life cycle essentially involves events like entry of the viral genome, its integration with the host cell genome, translation of viral proteins and finally, assembly and budding of new virus particles (Fig. 3). Below, we focus on the effects of host and viral lipids on some of the above steps involved in pathogenesis of two deadly infectious viruses, human immunodeficiency virus (HIV-1) and Human Coronavirus (HCoV). Both these viruses are enveloped with double membranes and have RNA as their genome. Enveloped viruses have lipid bilayers as integral part of their structure, and evoke fusion of the viral membrane with the membrane of the target cell (Harrison 2015). In this way, both the viruses share an analogous fusion mechanism for inserting into the host membrane. It has also been reported that structural similarities exist between coronavirus fusion,4 protein (S2) and HIV envelope fusion protein (gp 41) (Kliger and Levanon 2003; Zhang and Yap 2004). The viral assembly for multiplication and viral proliferation in both the cases are distinct though; a common feature is the intricate membrane rearrangement leading to recruitment of respective replicating factors.
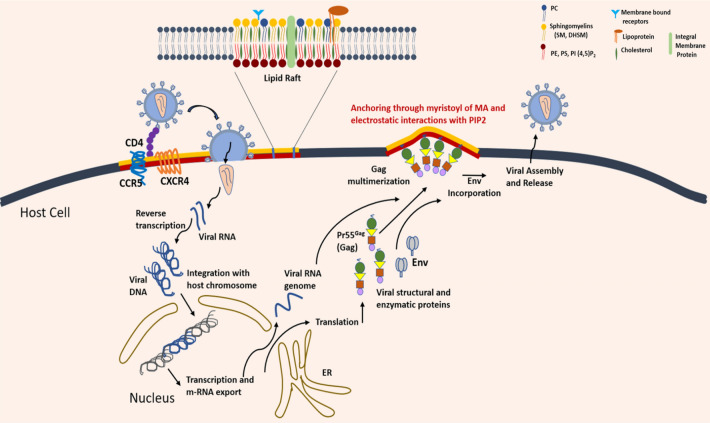
Fig. 3.
Schematic depicting HIV life cycle within the host cells. The entry process begins with the binding of viral spike protein gp120 to CD4 receptors and CCR5 and CXCR4 co-receptors situated within the host lipid raft domains, followed by a conformational change that exposes the fusion peptide of gp41. Upon fusion, the virus releases its single-stranded RNA-genome along with its reverse transcriptases for the formation of viral DNA, followed by integration with the host DNA and replication leading to the release of m-RNA. In the cytosol, the structural and enzymatic viral proteins are expressed including Pr55Gag, which are critical for the viral assembly and budding. HIV RNA assembles at the inner leaflet of the cell membrane and form an immature HIV virus. The subdomains of Gag protein are involved at different stages of the budding process. The MA domain is anchored to the cell membrane via a myristate along with basic amino acids that preferentially interact with acidic lipids of the host raft domains. The capsid domain (CA) contains amino acids that promote Gag-Gag interactions (multimerization) that invaginate the membrane, initiating the budding process. The nucleocapsid domain (NC) binds to viral RNA that are packaged in the virus particle. Before release, viral proteases cleave Gag to form a mature virus. Upon maturation, the new virus buds out from the infected host cell along with a part of the host cell membrane as its own viral membrane
Lipid Involvement in HIV Pathogenesis
HIV is a complex RNA virus belonging to retroviridae family with its nucleocapsid harboring two identical copies of the 9.8 kb single-stranded positive polarity RNA gene. The lipid bilayer enveloping the virus is decorated with external spikes, formed by two major envelope (Env) glycoproteins: gp120 and gp41.
Viral Binding and Membrane Fusion: The Grand Entry
The primary events during HIV infection (outlined in Fig. 3) involves binding to the target cell receptors, followed by membrane fusion with the cell membrane of macrophages and T-lymphocytes leading to the viral capsid entry (Dumas and Haanappel 2017). Membrane fusion itself is an intricate process that broadly involves (a) bringing the two membranes together in a way that they are closely apposed to each other; a highly energy demanding process, (b) mixing of apposing outer membranes to form a hemi-fusion or stalk-like intermediate, followed by (c) reorganization of inner leaflet lipids leading to pore formation and eventual content mixing (Martens and McMahon 2008; Chernomordik and Kozlov 2008). The fusion proteins facilitate binding of the viral envelope to the target host cell and catalyze the fusion process. In case of HIV, the viral envelope glycoprotein gp120, is the docking protein that binds to CD4 receptors and G-protein coupled receptors CXCR4 or CCR5 (co-receptors) on the host PM (Fig. 3). This binding event induces a conformational change in the viral envelope fusion protein gp41 resulting in the fusion of the two membranes (Lai et al. 2012). gp41 has a cholesterol binding domain (Vincent et al. 2002) enabling anchorage to membrane and hence ascertains that cholesterol plays a dominating role in the viral entry process (Veiga and Castanho 2007; Lai et al. 2012; Yang et al. 2015). The conformational change in gp41 exposes its N-terminal hydrophobic fusion peptide (HIV-FP), whose insertion into the target cell membrane causes rearrangement of lipids leading to membrane softening (Agrawal et al. 2016; Sáez-Cirión et al. 2002). The sequence of these events highlights that the lateral organization of involved receptors and co-receptors on the host membranes depends on their lipid environment. The CD4 receptor are, in fact, known to colocalize with the membrane raft domains (Popik et al. 2002). Many studies have revealed that the preferred sites of viral fusion are liquid-ordered, lo domains (raft domains) rich in cholesterol and sphingolipids (Waheed and Freed 2009). But, thermodynamically, it would be energetically challenging due to the stiff and efficiently packed nature of lo domains owing to the fact that fusion mechanism involves processes like membrane bending and non-bilayer lipid intermediates requiring substantial flexibility of membrane structures (Chernomordik and Kozlov 2008). Thus, Yang et al. (2015) proposed the role of the edges of lo domains, rather than the bulk region, to be the preferred sites for fusion. Later-on, they verified the mechanism of fusion to be driven by the effect of hydrophobic mismatch at the edges of lo−ld (liquid-ordered–liquid-disordered) domains. Thus, gp41-mediated fusion is driven by the line tension energy at the lipid domain boundaries (Yang et al. 2016). These findings bring to the forefront the potential application of Linactins and related compounds against HIV. Linactins or line-active compounds such as α-tocopherol or vitamin E lower the domain line tension in heterogenous membranes (i.e., lo−ld phase segregated) impeding membrane fusion, and hence may be evaluated as natural inhibitors against HIV.
The fusogenic property of the host membrane during pathogen entry is controlled by the membrane composition and the resulting curvature it imparts. The role of lipid composition in altering the conformation of class-I fusion peptide in case of HIV, Influenza virus and SARS-CoV been recently reviewed by Meher and Chakraborty (Meher and Chakraborty 2019). Negatively charged lipids such as phosphatidylethanolamine (PE) and phosphatidic acid (PA) modulate the membrane curvature, which impacts the structure and function of viral fusion peptides, and thus strongly controls the fusion efficiency. Owing to the intrinsic inverted cone-shaped structure, PE confers spontaneous negative curvature to the membrane and helps in stabilizing the early fusion intermediates, specifically the stalk-like intermediates. The stress generated in the bilayer leaflet due to the negative curvature promotes formation of membrane defects which serves as the source of energy for the fusion of the two membranes (Meher and Chakraborty 2019; Kreutzberger et al. 2017). Additionally, PE is also known to stabilize inverted hexagonal (HII) state and transition from lamellar (L) to HII at the point of contact is an essential step that governs efficient outer membrane mixing (Siegel and Epand 1997).
HIV membranes are enriched in phosphoinositides (PI), and if present on the inner leaflet, may provide positive curvature required for fusion pore formation (Chan et al. 2008). Despite the cholesterol and sphingomyelin rich composition, HIV lipid envelope does not exist as a laterally homogeneous lo-like membrane. Huarte et al. (2016) experimentally demonstrated the existence of lateral discontinuities in the highly ordered viral membranes, while addressing the molecular basis underlying the packing and lateral heterogeneity in HIV model membranes. They identified the role of individual viral lipid components for the maintenance of high order and lateral demixing. Deleterious effects of the membrane-active compounds on the viral entry rendered by fluidification of the disordered lipid regions along with aggregation of ordered nanodomains were documented. These finding highlight that perturbing the functional organization of the viral membrane envelope may represent a lucrative strategy for antiviral drug design. Importantly, membrane-targeted antiviral approaches are less likely to select for antiviral resistance due to the non-biogenic nature of viral lipid membranes and hence their inability to correct for membrane perturbations.
Dendritic cells (DCs) generate the initial immune response and are critical for protection against pathogen invasion. HIV virus, however, evades immune response by proliferating within DCs without causing active infection. Eventually the infection disseminates into the more susceptible CD4+ T cells. HIV entry through DC involves gp120-independent mechanism of viral binding and a non-infectious endocytic mechanism (Izquierdo-Useros et al. 2007). It is known that sialyllactose on HIV-1 membrane gangliosides serve as a novel recognition pattern that mediates viral binding and internalization of HIV into mature DCs (Izquierdo-useros et al. 2012). Gangliosides are acidic glycosphingolipids carrying one or more terminal sialic acid and are abundantly present in the PM raft domains, where the HIV budding and assembly takes place (Fig. 3). Gangliosides GM1, GM2 and GM3 are the key molecules that mediate liposome uptake in mature DCs and hence serve as viral attachment factors, in addition to their role as cellular receptors for the pathogen entry.
Emerging Contributions of Lipids in Viral Assembly and Budding
The assembly and release/ budding of viral particles involves recruitment of Gag (Group-specific antigen) polyproteins (encoded by the HIV genome) to the PM at the site of release (Ono et al. 2004). Post translation, Gag recruits the dimeric positive RNA viral genome in the cytoplasm and then assembles them at the PM (Fig. 3). They are known to multimerize at the PM to form nucleation sites composed of Gag-RNA complexes (Finzi et al. 2007; Jouvenet et al. 2009; Kutluay and Bieniasz 2010). Gag polyprotein Pr55Gag, comprises of four subdomains: the matrix (MA), capsid (CA), nucleocapsid (NC), and two small spacer peptides (SP1 and SP2)). The N-terminal myristoylated portion of the matrix along with a highly basic peptide region enables Gag-PM binding afforded by hydrophobic insertion of the lipid anchor as well electrostatic interactions (Fig. 3); CA and NC motifs facilitate Gag multimerization (Li et al. 2007). Lipidomic characterization of HIV-1 viral membranes revealed cholesterol and sphingomyelin (SM) to be enriched in the virus at concentrations similar to detergent-resistant ordered membranes (Brügger et al. 2006; Lorizate et al. 2013). Further, as compared to the bulk PM, the viral membrane contains specific lipids, including aminophospholipids, dihydrosphingomyelin (DHSM), plasmenyl-phosphoethanolamine (pl-PE), PI (Chan et al. 2008; Huarte et al. 2016). Most importantly, acidic lipids such as PI, phosphatidylinositol-4,5-biphosphate [PI(4,5)P2] and PS play key roles in the specific binding of Gag to the inner PM leaflet (Ono et al. 2004). Apart from cholesterol, there are several reports that identify PI(4,5)P2 as the most important determinant in targeting Gag to PM, leading to Gag multimerization. This stresses the importance of PM lateral domain architecture for the viral exit, as Gag recognizes specific PM microdomains leading to the exit events. On the contrary, there is another hypothesis that argue that Gag leads to de novo lipid clustering instead of binding to the already existing heterogenous PM domains. The authors propose selective lipid recruitment by the budding viruses to form new domains. HIV membranes exhibit high level of consistency—in their lipid composition despite different origins (Chan et al. 2008). A Coarse-Grained simulation study (Charlier et al. 2014) proposed that the matrix domain of Gag proteins segregates and selectively clusters PIP2 due to electrostatic interactions, leading to acidic lipid-enriched domains (Fig. 3). Another study further reinforced that Gag self-assembly is responsible for the formation of PIP2 lipid nanoclusters and these nanodomains are enriched in cholesterol, but not in SM (Yandrapalli et al. 2016). Through FRET, it was established that Gag partitions into ld domains of the model lipid membranes. These studies show that instead of targeting pre-existing PM lipid domains, Gag generates new cholesterol-PIP2 enriched lipid nanodomains at the inner PM leaflet during early events of virus assembly.
Budding of new viral particles implicate tetraspanins-enriched microdomains (TEMs), especially CD9 and CD81 (Dahmane et al. 2019). Tetraspanins are proteins, which among other functions, act as molecular organizers within host PM to form an efficient dynamic network of protein–protein interactions inclusive of transmembrane proteins. TEM co-localizes with Gag (Jolly and Sattentau 2007; Grigorov et al. 2009) where tetraspanin components incorporate into retrovirus particles. In a very recent study by means of correlative dSTORM/AFM (Dahmane et al. 2019), the authors revealed that CD9 is specifically trapped within the nascent viral particles, especially at buds tips. This suggests that Gag mediates release of CD9 and CD81 from the PM leading to a differential membrane composition in the viral particles compared with the host PM. They proved that CD9 is organized within small membrane assemblies, which coalesce upon Gag expression. This disclosure, along with the earlier studies based on co-localization and clustering analysis by FRET, suggests that Gag induces integration of clustered rafts and TEMs. In addition, the active involvement of cytoskeleton to modulate Gag oligomerization and subsequent coalescence of lipid domains to form a “single” growing assembly has been documented (Rahman et al. 2014; Thomas et al. 2015).
Lipid Exosomes in HIV Proliferation
Exosomes are membrane-bound, extracellular vesicles (EVs) produced in the endosomal compartment of eukaryotic cells, followed by secretion into the extracellular environment. Formation of multivesicular bodies (MVBs) involves inward folding of the endosomal compartments through ESCRT pathway (endosomal sorting complex required for transport), and upon fusion with the PM, the MVBs are released as exosomes. Interestingly, recent studies elucidate the role of exosomes derived from infected cells in HIV pathogenesis, and characterize their mechanisms underpinning cell-to-cell transmission (Madison and Okeoma 2015; Chahar et al. 2015; Kulkarni and Prasad 2017). For instance, Chen et al. (2018) observed that exosomes released from HIV-infected T cells stimulate proliferation, migration, and invasion of oral/oropharyngeal and lung cancer cells. Kulkarni and Prasad 2017 demonstrated that the exosomes derived from HIV-infected T cells and DCs differ in their efficiency to trans-infect other healthy cells. They revealed that exosomes derived from HIV-1 infected DCs are fourfold more virulent than those from T cells or cell-free HIV-1. They also observed upregulation of pro-inflammatory cytokine (IFN-γ, TNF-α, IL-1β) highlighting their potential as possible antiviral therapeutic targets.
Lipidic Crown of Coronavirus Infections
Coronaviruses (CoVs) belong to the family Coronaviridae, which constitutes a group of enveloped, positive, single-stranded RNA viruses (having largest genome; 26–32 kb). Owing to the crown like appearance on their surface (Fig. 4), as observed through electron microscopy, they are termed as “CoV” (Su et al. 2016; Ye et al. 2020). Among the seven CoVs which infect humans, HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, are responsible for mild conditions like common cold, diarrhea, etc. (Yan et al. 2019). As a result, the HCoVs were considered harmless until 2003 when the severe acute respiratory syndrome coronavirus (SARS-CoV) outbreak surfaced as the first well-documented HCoV-caused pandemic in human history. This was followed by the persistent Middle-East respiratory syndrome coronavirus (MERS-CoV) epidemic in 2012. Currently, we are witnessing the upsurge of SARS-CoV-2 (COVID-19) causing global devastation and bringing CoVs to the center stage again. SARS-CoV, MERS-CoV, and SARS-CoV-2 are highly pathogenic viruses, causing severe lower respiratory tract infection and are associated with extrapulmonary manifestations (Ye et al. 2020).
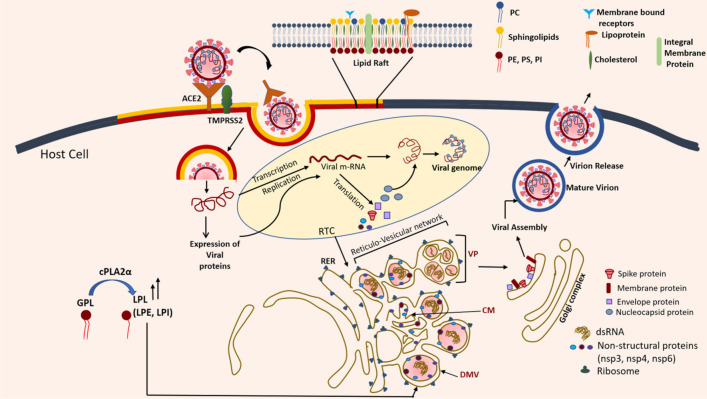
Fig. 4.
Schematic representing major events in the SARS-CoV life cycle in the host cell. Entry of SARS-CoV occurs through the binding of spike protein with the host ACE receptors facilitated by transmembrane serine protease 2 (TMPRSS2), embedded in the host cell raft domains. Once the viral genome is released, transcription-replication complex (RTC) carries out the expression of viral structural and non-structural proteins. The entire RTC is known to be situated in the viral-induced compartments formed by the rearrangement of cellular membranes. The schematic shows formation of DMVs (Double-membrane vesicles), CMs (convoluted membranes) and VPs (vesicle pockets) through rough endoplasmic reticulum (RER) in conjunction with a complex interplay of viral non-structural proteins, nsp3-4–6. The presence of double-stranded RNA (dsRNA) as observed by Knoops et al. is also shown within the compartments of the reticulo-vesicular network
Viral Invasion: Receptor-Binding and Fusion
Lipids play indispensable roles in the coronavirus life cycle by influencing steps such as insertion; assembly of the replication machinery and budding of newly enveloped viral particles (Fig. 4). Viral entry involves binding to the target cell’s cellular receptors, followed by fusion of the viral envelope with the host PM (Millet and Whittaker 2018). Viral entry is also facilitated through the pH- and receptor-dependent endocytic pathways, where it involves fusion with the late endosomes. Previous reports claim these pathways to be clathrin- and caveolae-independent (Wang et al. 2008), which are otherwise commonly exploited by animal viruses. For both SARS-CoV and SARS-CoV-2, glycoproteins S (spike proteins) binds to the angiotensin-converting enzyme 2 (ACE2) receptor on the human respiratory epithelial cells (Baglivo et al. 2020), Fig. 4. This binding is activated by another enzyme, TMPRSS2 (transmembrane serine protease 2), which is also a key player in the entry of the virus, and presently considered as a potential drug target for SARS-CoV2 (Hoffmann et al. 2020), Fig. 4. S-protein is a class-I viral fusion protein (Bosch et al. 2003) that plays a critical role in the viral entry by alleviating the kinetic barrier of lipid fusion between the pathogen and host membranes (Harrison 2015). Fusion of two membranes is a thermodynamically favorable event, however, the “repulsive hydration force” creates a kinetic impediment, which increases steeply as the distance between the surfaces of the two bilayers fall below 20 Å. Here, the fusion proteins act as catalysts to overcome the strong hydration forces, allowing the apposed membrane surfaces to fuse. The HIV envelope proteins (gp120 and gp40) and influenza virus hemagglutinins (HA1 and HA2) also appertain to the class-I fusion proteins (Millet and Whittaker 2018). The S-protein is further divided into the S1 receptor-binding subunit and S2 fusion domain, usually separated by a cleavage site (S1/S2). S1 contains the receptor-binding domain by which the coronavirus binds to the peptidase domain of the ACE2 receptor. This binding induces a conformational change in the S2 fusion domain. The S2 fusion peptide is, thereafter, directly involved in mediating the membrane fusion reaction by disrupting and connecting the inner leaflets of the two bilayers.
As with HIV, lipid rafts are also associated with the SARS-CoV entry. More specifically, cholesterol-rich host microdomains provide a platform facilitating the productive interaction of the S-protein with the cellular receptor ACE2 (Li et al. 2007; Lu et al. 2008), Fig. 4. Though discrepancies exist regarding the membrane location of ACE2 (raft vs non-raft regions) (Lu et al. 2008; Warner et al. 2005; Li et al. 2007), modulation of host cholesterol has been shown to alter the fusion process. For instance, PM cholesterol depletion by methyl-β-cyclodextrin (mβCD) attenuates the crosstalk of fusion proteins with the membrane receptors for several coronaviruses (Glende et al. 2008; Nomura et al. 2004; Choi et al. 2005). Lu et al., through confocal bioimaging, demonstrated co-localization of the ectodomain of the spike protein with the raft markers such as GM1, thus supporting the raft localization of ACE2. Natural small molecules such as phytosterols and cyclodextrins, with cholesterol depletive activity, represent effective antiviral agents as they inhibit the spike protein-ACE2 interactions via disruption of the host membrane lipid rafts (Baglivo et al. 2020).
Role of calcium (Ca2+) during the fusogenic event in SARS-CoV and MERS-CoV has been explored in recent years (Straus et al. 2020; Lai et al. 2017). Through electron spin resonance (ESR), SARS-CoV fusion peptide was shown to induce Ca2+-dependent membrane ordering upon interaction with the lipid acyl chains. Straus et al. (2020) showed that SARS-CoV fusion peptide binds to two Ca2+ ions as opposed to MERS-CoV fusion peptide which binds to only one Ca2+; even though the role of Ca2+ is more critical for MERS-CoV entry. Increased ordering in the headgroup region enhances dipolar interactions and lowers electrostatic energy, thus providing the energy source for membrane fusion (Ge and Freed 2009). Moreover, membrane ordering also aids in enhancing the negative curvature of the bilayer during the fusion process, thus depleting the repulsive energy between two opposing membranes while bending. Tang et al. (2020) reasoned that the greater membrane ordering induced by the fusion peptide in the presence of Ca2+ can be attributed to Ca2+-induced stabilization of the fusion peptide conformation that re-organizes lipids in a manner that promotes merging of the two membranes.
Reticulo-Vesicular Network: Exclusive Sites for Virion Assembly and Budding
CoV replication assembly is associated with the induction of multiple distinct cellular membrane alterations. The ultrastructure characterization of SARS-CoV in vitro revealed, for the first-time, existence of three distinct structural features: double-membrane vesicle, nucleocapsid inclusion, and large granular areas of cytoplasm (Goldsmith et al. 2004). These features were also corroborated in vivo using bronchiolar lavage specimens from SARS patients. It is now well known that CoVs expropriate intracellular host cell membranes to generate new compartments such as double-membrane vesicles (DMVs) and convoluted membranes (CMs) for the amplification of the viral genome (Hagemeijer et al. 2012), Fig. 4. These rearranged cellular membranes of the host cell provide a structural scaffold for the viral replication/transcription complexes (RTCs) and also help evading the cellular factors generated through antiviral host response (Martín-Acebes et al. 2013; Heaton and Randall 2011; Miller and Krijnse-Locker 2008). DMVs are double-layered vesicular structures that harbor viral proteins and a specific array of imbibed host factors, thus integrating a unique lipid environment facilitating viral replication (Fig. 4). Through electron tomography of the cryofixed SARS-CoV infected Vero E6 cells, Knoops et al. (2008) identified a unique reticulo-vesicular network of modified endoplasmic reticulum (ER) that conjoined: convoluted membranes, numerous interconnected DMVs (diameter 200–300 nm), and “vesicle pockets” (VPs) arising from the DMV merger (Fig. 4). Formation of the reticulo-vesicular network during the viral life cycle of SARS-CoV within the infected host cell is depicted schematically in Fig. 4.
CoV non-structural membrane spanning proteins (nsps) 3, 4 and 6, play critical roles in membrane rearrangement and anchoring of RTCs (Hagemeijer et al. 2014, 2010; Angelini et al. 2013). Previously, it was observed that the interaction between nsp3 and nsp4 and their co-expression results in their re-localization from ER into distinct perinuclear foci (Hagemeijer et al. 2011). This correlates well with the observation by Knoops et al. showing that with the progress of infection, DMVs largely concentrate in the perinuclear region, often having mitochondria lying in between, Fig. 4. Apparently, nsps regulate alterations in the double-layered ER membranes leading to the formation of DMVs. The most intriguing observation by both Goldsmith et al. and Knoops et al. was the localization of double-stranded RNA in the DMV interior, presumably revealing the site of viral RNA synthesis. However, the authors could not discern a connection between DMV interior and cytosol, and hence some pieces of the puzzle-regarding the actual site of SARS-CoV RNA replication-remain missing. In this aspect, Moriel-Carretero (2020) recently hypothesized the role of lipid droplets (LD) for the above concerns while explaining the mechanism of DMV formation.
LD are complex, dynamic, membrane-enclosed organelles formed within the hydrophobic core of the ER bilayer and are composed of non-polar lipids such as triacylglycerols and steryl esters. LDs are regularly exploited by pathogens, for example for virion assembly and immune modulation (McLauchlan 2009; Cheung et al. 2010; Samsa et al. 2009). At the initial stages of the infection, some of these vesicles bear a single lipid layer and seem to be embedded in the ER, thus resembling the nascent LD (Moriel-Carretero 2020). Later during the infection, the interior of the SARS-CoV-induced vesicles become dense, filled with spider web-like contents. Certain lipids such as phosphatidic acid (PA) have a propensity to form cubic phase lipid arrangement, where the bilayer bends and splits to form curves, owing to its conical shape. This results in negative curvature (Kooijman et al. 2003). Enhanced PA concentration at ER appears to drive the lamellar-to-cubic bilayer transition facilitating LD formation. This process is a coordinated effect of altered host lipid metabolism by the viral effectors (as discussed ahead) and is well orchestrated by the nsp3-4-6 viral proteins. CoVs nsps interact with each other through their cytosolic domains and manipulate their transmembrane segments to tether together and appose ER membranes. Therefore, the structure of DMVs can be described as thin, closed inner membranes suggestive of a LD monolayer; the outer membrane is a bilayer common to all vesicles and corresponds to the ER membrane itself. This architecture can explain how the inner space of the DMVs is well shielded from cytoplasm, providing a protected niche for viral amplification and assembly, but still cytoplasmic (by definition), i.e., in the form of LDs entangled within the cubic ER membranes (Moriel-Carretero 2020), also depicted in Fig. 4. Though pending validation, this is the first report showing involvement of LDs in SARS-CoV infection, but unlike in HCV or Dengue, they are not colonized or hijacked by the invading virus (Filipe and McLauchlan 2015; Samsa et al. 2009). Instead there is a subversion of the LD birth environment, providing a protective niche for SARS-CoV replication. Chloroquine, which is considered as an effective drug treatment against SARS-CoV2, is a specific inhibitor of phospholipase-D that reduces PA levels; LDs exhibit high levels of PA. This indicates that the virus indeed exploits this lipid precursor for replication through LDs.
CM clusters (size 0.2–2 μm) are continuous with the outer membrane of DMVs and ER-cisternae suggesting a link to RTC. During the later stages of viral insertion, DMVs merge to form a large, single membraned cytoplasmic vacuole, referred to as vesicle pocket (VP), (diameters 1–5 μm), that facilitates the budding of new SARS-CoV particles, Fig. 4. These distinct membrane reformations are endowed with large curvatures and are attributed to the effect of lipid-modifying enzymes. Such enzymes may cause dynamic changes in the lipid composition, thus presenting a suitable microenvironment, which recruits viral and cellular membrane shaping proteins. In future, lipidomic analysis of viral DMVs, and CMs will enrich our understanding of the involvement of host lipids to coronavirus assembly and budding. Such studies are also expected to shed light on the specific viral lipid repertoire necessary for the aforementioned events. By targeting the synthesis of the implicated lipids, distinct steps in the coronavirus replication assembly may be inhibited. Inhibition of LDs also appears as an attractive target candidate for therapeutic interventions against COVID-19.
HCoVs Evoke Reprogramming of Lipid Metabolism
Virion morphogenesis hugely depends on the enzymes involved in lipid synthesis and is associated with significant alterations in the cellular lipid metabolism. Müller et al. (2017) conducted lipidome analysis on HCoV-229E infected cells to gain insights into the roles of lipids in coronavirus replication and DMV formation. They monitored the abundances of 359 lipids of 14 classes. The lipid profile analysis revealed no significant change in the total abundance of membrane lipids and neutral lipids (triacylglycerols, diacylglycerols, and cholesteryl esters); however, a decrease in PA levels and increase in Cer were observed. Most significant increase was observed with lysophospholipids (LPL), prominent ones being lysophosphatidylethanolamine (LPE) and lysophosphatidylinositols (LPI). Next, the role of cytosolic phospholipase A2α (cPLA2α), which cleaves the sn-2 position of glycerophospholipids giving rise to the lyso-lipid species (LPLs) was investigated, Fig. 4. The impeding effect of cPLA2 inhibitor, pyrrolidine-2 (Py-2), on coronavirus replication cycle was also studied. Transmission electron microscopy divulged that the DMV formation in infected cells was severely impaired in the presence of the inhibitor. Immunofluorescence microscopy analysis of coronavirus RTCs exhibited typical punctate perinuclear staining pattern pertaining to the detection of the double-stranded RNA (dsRNA) and viral proteins. This pattern was altered upon treatment of HCoV-229E infected cells with Py-2. Their major observations were: (a) viral RTCs co-localizes with LPL-containing membranes (b) cellular LPL concentrations increase in coronavirus-infected cells, and (c) LPL increase was diminished in the presence of cPLA2α inhibitor, Py-2. Overall, their findings indicate that cPLA2α is intricately involved in the viral life cycle, most likely by generating LPL that form specialized membrane compartments crucial for viral RNA amplification. Aptly, the authors propose cPLA2α as a potential target for the antiviral drug therapy. As lyso-lipid species induce membrane curvature, it appears that the virus utilizes the phospholipase A2α machinery, to its advantage, for production of highly curved membranous structures. In another recent UPLC-MS based lipidomic approach on HCoV-229E coronavirus by Yan et al. (2019), a significant amount of remodeling of human host lipid metabolism was revealed. Up regulation of lysophospholipids (largely lyso-PCs and lyso-PEs) and fatty acids such as linoleic acid, arachidonic acid, palmitic acid, etc., was observed. Through detailed pathway analysis, it was revealed that linoleic acid and arachidonic acid metabolism axis were the most perturbed upon HCoV-229E infection in VeroE6 cells.
These recent studies clearly indicate critical and functional involvement of lipids in coronavirus infections and warrant more studies to configure the lipidic interaction landscape of CoVs for identifying suitable lipid-centric therapeutic intervention points.
Lipid-Inspired Therapeutics in Infectious Diseases
Emergence of drug resistance has created an unmet need to identify novel drug targets for designing effective antivirals and antibiotics. One key aspect underlining this effort is to gain deepened understanding of the intricate web of host–pathogen interactions, thus identifying therapeutically actionable (pathogenic and host) targets. For many decades, therapeutic efforts were centered on host and/or pathogenic proteins; however, recent times have elucidated lipid molecules as critical regulators of host–pathogen interactions. Hence, development of therapeutic strategies targeting lipid-mediated cellular process, specifically lipid synthesis, metabolism, localization and transport, implicated in infectious diseases are gaining speed.
PS anionic lipids are present in many pathogenic viral and bacterial membranes, wherein recognition by corresponding PS receptors on host cells foster their uptake, also known as “apoptosis mimicry” (Amara and Mercer 2015). Thus, targeting exposed PS is a viable strategy. Bavituximab, a PS targeting monoclonal antibody effectively blocks the infectivity of enveloped viruses, both in vitro and in vivo (Corbin-Lickfett et al. 2010; Soares et al. 2008). Further, inhibition of phospholipid scramblase-1 with small molecule R5421 attenuates viral-induced exposure of PS on cell surface, thus reducing viral infection (Cheshenko et al. 2018). As PS flipping is also implicated in bacterial infections such as STEC-HUS, particularly on multivesicles (MVs), extending the PS targeting approaches may prove beneficial here.
A class of rigid amphipathic fusion inhibitors (RAFIs) with inverted cone molecular geometry, block infectivity of enveloped viruses by conferring a positive curvature to their lipid membranes (Colpitts et al. 2013; St Vincent et al. 2010). Lipids with large polar head groups such as lyso-lipid species confer a positive curvature by bending the membranes away from polar heads. The positive curvature hinders the formation of fusion intermediates (hemi-fusion state) prior to the formation of fusion pore. This is by virtue of impeding the transition from a lamellar to an inverted hexagonal phase, thereby raising the energy barrier for the hemi-fusion stalk formation. RAFIs, by inhibiting fusion of the viral membrane with the host PM, have an antagonizing effect on the well-orchestrated action of viral fusion proteins (Melikyan 2010). This renders an exciting proof-of-concept for developing broad-spectrum (pan-CoV) inhibitors that could block the fusion of most enveloped viruses. In addition, this therapeutic approach possesses a high barrier to resistance by targeting the lipid rearrangements in the “virustatic or non-biogenic” viral membranes. The leading RAFI, aUY11, has an ethynyl-perylene hydrophobic and an uracil-arabinose polar moiety. aUY11 intercalates in viral envelope and inhibits virion-to-cell fusion in a broad spectrum of otherwise unrelated enveloped viruses (Speerstra et al. 2018). Other antiviral compounds intercalate within the viral membrane and modulate the membrane fluidity. Glycyrrhizin decreases bilayer fluidity and is weakly active against several enveloped viruses, including HIV (Harada 2005), and SARS (Cinatl et al. 2003).
Another broad-spectrum antiviral drug that selectively targets viral membrane properties is LJ001. It is a photosensitizer and a membrane-binding compound (Wojcechowskyj and Doms 2010; Wolf et al. 2010). Its mechanism of action involves enhanced unsaturated fatty acid hydroxylation that primes the formation of a hydroxyl group in the middle of the hydrophobic lipid bilayer (Vigant et al. 2013). This leads to viral membrane disruption and hence attenuated infection. LJ001 remarkably inhibits infection from a wide range of enveloped viruses including HIV, influenza, and Ebola. Interestingly LJ001 does not affect host cellular membranes due to active processes that protect against phospholipid hydroperoxides. Newer membrane-intercalating photosensitizing compounds such as oxazolidine-2,4-dithiones have also been designed (Vigant et al. 2013), clearly highlighting the potential of the viral membrane as antiviral targets.
Activation of acid sphingomyelinase (Asm), a hydrolase that generates Cer from SM, produces Cer-enriched domains (akin to lipid rafts) in the PM and is used by bacteria for entering host cells. Chemical inhibition of Asm with imipramine restricts the growth of pathogenic mycobacterial species (Godbole et al. 2015). More directly, inhibition of SM synthase with SPK-601, MS-209 and D609 or inhibition of uridine diphosphate-glucose Cer synthase by N-butyldeoxynojirimycin reduces viral loads in infection models (Khan et al. 2014; Martín-Acebes et al. 2016). Pathogens exploit cholesterol within the raft and non-raft region of host PM to trigger their internalization into host cells. Thus, pharmacological disruption of cholesterol flows (e.g., by use of statins) and synthesis not only impairs the assembly of lipid rafts but also provides efficient strategies against bacteria by reducing their entry and replication. Consistently cholesterol depletion studies (in vitro and in vivo), have shown restricted entry of various pathogens such as mycobacteria, salmonella, Staphylococcus pneumoniae, aureus and E. coli (Kaul et al. 2004; Parihar et al. 2014; Catron et al. 2004; Huang 2011; Bergman et al. 2011; Jerwood and Cohen 2008; Al-Kuraishy et al. 2018). The same approach has been proven efficient against viral infections (at least in laboratory), where statins are used to inhibit HMGCR (HMG-CoA reductase). Statins hamper the rate by which HMGCR produces mevalonate, the next molecule in the cascade that generates cholesterol (Stancu and Sima 2001).
PI-signaling is a major host signaling network tapped by pathogens for modulating various host cell processes such as membrane dynamics, cytoskeletal arrangements, etc. Secreted virulent proteins alter host PI lipid levels by rewiring the activity of kinases and phosphatases that maintain an optimum PI abundance. In this regard, PI3K inhibitors such as wortmannin and LY294002 curb bacterial infections (Ireton et al. 1996). Non-vesicular modes of lipid transfer are encountered during host–pathogen interactions and are faster than vesicular trafficking in producing local lipid enrichment with membrane asymmetry. The major mode of non-vesicular lipid transfer involves the use of proteins that shield the hydrophobic acyl anchor of lipids from water-rich cytoplasm. One such protein is oxysterol-binding protein (OSBP) that exchanges PI4P and cholesterol between golgi and ER, wherein transport of cholesterol is dependent on PI4KIII activity and PI4P levels at Golgi membranes. Itraconazole, OSW-1 and TTP-8307 are small molecules that deter binding of cholesterol to OSBP and thus are active against various viruses, such as enteroviruses, HCV, etc. (Albulescu et al. 2015, 2017; Strating et al. 2015).
Targeting enzymes involved in biosynthesis of pathogenic lipids is another promising anti-infective approach, though the field is still in infancy. Particularly for mycobacterial infections, proof-of-concept studies have focussed on elucidating the functional role of specific lipids rendered by genetic depletion studies. For instance, depletion of sulfotransferase, Stf0 (that initiates Mtb SL-1 biosynthesis), augments Mtb survival in human macrophages (Gilmore et al. 2012), suggesting SL-1 to negatively regulate the intracellular growth of Mtb. Furthermore, it has been shown that removal of fatty acid chains in Mtb SL-1 at 6,6′ position attenuates autophagy induction (Dadhich et al. 2020). As such, targeting of enzymes involved in biosynthesis of SL-1, specifically enzymes that add fatty acyl chains to diacylated SL-1 intermediate (AC2SL) such as Chp1 and Chp2 (Seeliger et al. 2012), might represent possible therapeutic avenues to revamp autophagy and hence restrict pathogen growth within the host. This is supported by the knowledge that autophagy is a promising strategy adopted by invading mycobacterial pathogens to create favorable intracellular niches within the host to persist and survive with abundant energy and food.
Finally, identification of the mechanism of host cell membrane modification by virulent lipids is an emergent research field. It has a high potential to furnish additional cues to protect host cell membrane via selective incorporation of membrane-active agents that may attenuate the membrane perturbing effect of pathogenic lipids. More intensive work is required to achieve actionable targets in this regard.
It is worth mentioning here that apart from conventional anti-infection strategies, nanotechnological advancements have furnished cell membrane mimicking decoys such as liposomes, reconstituted lipoproteins and cell membrane-derived nanostructures that can trap and detail pathogens (Rao et al. 2020). These lipid-based nanostructure-pathogen complexes are being exploited as vaccine candidates. For example, Hu et al. (2013) demonstrated spontaneous interactions between red blood cell coated lipid nanodecoys and pore-forming toxins, safely delivery the latter for immune processing. Moreover, the nanoparticle-detained toxin show better protective immunity compared with heat-inactivated toxin.
Conclusions
Gaining a deepened mechanistic understanding of virulence factors is imperative for developing innovative therapeutic strategies against infections. For many decades, the host–pathogen interactions were focused on proteins and nucleic acids. However, advances in lipidomics, lipid chemical synthesis and analytical techniques have brought to the forefront, the indispensable roles played by lipids in the host–pathogen interactions. Lipids actively intervene in cellular signaling, trafficking, membrane fusion, and protein function, which form the “cornerstones” of the host–pathogen interaction landscape. The compositional variation in the lipidome of host and/or the pathogen upon interaction has been linked with de novo lateral reorganization of lipid domains. This subsequently regulates lipid and protein clustering that fine-tunes protein functions to impact membrane-associated cellular processes in favor of the pathogen. Moreover, host cell membrane insertion and modification by pathogenic lipids is now a well-accepted mode of action. This event induces changes in the host cell membrane biophysical properties such as fluidity, order, lateral organization or dynamics. An altered membrane property in turn affects protein localization, lipid/protein diffusion, lateral membrane organization, and lipid-protein interactions, and eventually their activity. Optimal membrane properties critically control the signaling efficiency in regulating various cellular processes, thus it is harnessed by pathogens for their survival and infection.
Given the above multifaceted role of lipids in infectious diseases, agents disrupting the biosynthesis of lipid (such as cholesterol, ceramide, and phospholipids), or small molecules inhibiting lipid-transporting proteins (e.g., OSBP) have been shown to interfere with pathogenic growth. Agents that alter lipid raft domains, such as statins, are efficient in restricting lipid raft-associated process during infections. Moreover, disruption of pathogenic membranes by incorporation of wedge-shaped molecules impedes fusion processes during infection. Importantly, strategies that target lipid membranes and their biophysical properties (instead of a specific lipid/protein) are less likely to suffer from resistance issues.
Finally, structural, metabolic and signaling functions of lipids often arise from interactions with proteins, which are validated nodes for drug action. The emerging functions of lipids in mammalian biology have already furnished number of drug targets (lipid-binding proteins) for diseases such as sclerosis, inflammation, cancer, and immunology. However, our understanding of the pathogenic lipid-host protein interactions and its accessibility to pharmacological perturbation, remains limited. In this regard, development of lipid-inspired chemical probes to inventory the landscape of lipid-binding host proteins to identify novel drug targets in viral and bacterial infection holds great potential.
Taken together, lipid-centric therapeutic approaches based on specific aspects of host–pathogen interactions are promising avenues against infectious diseases caused by viruses and bacteria.
Acknowledgements
RD acknowledges DST-Woman Scientist Scheme WOS-A/CS-16/2018, India for research funding. Work in our lab is supported by DST-SERB Grant EMR/2016/005414, BRNS Grant 201805BRE01RP04922, Wadhwani Research Centre for Bioengineering (WRCB), DBT Ramalingaswami Fellowship, and DST-Inspire Faculty Award. We acknowledge BEI Resources, NIAID, NIH for purified mycobacterial lipids and antibodies supporting our work.
Abbreviations
- LPS
Lipopolysaccharide
- E. coli
Escherichia coli
- STEC
Shiga toxin producing Escherichia coli
- HUS
Hemolytic uremic syndrome
- Mtb
Mycobacterium tuberculosis
- TB
Tuberculosis
- OM
Outer membrane
- IM
Inner membrane
- PE
Phosphatidyl ethanolamine
- PG
Phosphatidylglycerol
- PS
Phosphatidylserine
- Cer
Ceramides
- SM
Sphingomyelin
- CL
Cardiolipin
- TLR
Toll-like receptors
- MD
Myeloid differentiation factor
- GUV
Giant unilamellar Vesicles
- Stx
Shiga toxin
- Gb3
Globotriaosylceramide
- FRET
Fluorescence resonance energy transfer
- GSL
Glycosphingolipids
- DRM
Detergent-resistant membranes
- MV
Microvesicles
- EHEC
Enterohemorrhagic Escherichia coli
- EPEC
Enteropathogenic Escherichia coli
- UPEC
Uropathogenic Escherichia coli
- PDIM
Phthiocerol dimycocerosate
- TDM
Trehalose Dimycolate
- SL
Sulfoglycolipid
- PIM
Phosphatidylinositol mannoside
- Man-LAM
Mannose capped lipoarabinomannan
- PGL
Phenolic glycolipid
- DAG
Diacylglycerol
- DPPG
Dipalmitoyl phosphatidylglycerol
- DMPG
Dimyristoyl phosphatidylglycerol
- PAL
Peptidoglycan associated lipids
- CLR
C-type lectin receptor
- CR
Compliment receptor
- MALDI-TOF
Matrix-assisted laser desorption ionization-time of flight
- AFM
Atomic force microscopy
- HCoV
Human corona virus
- HIV
Human immunodeficiency virus
- lo
Liquid-ordered domains
- ld
Liquid-disordered domains
- CD
Cluster of differentiation
- PM
Plasma membrane
- PIP2
Phosphatidylinositol-4,5-biphosphate
- gp
Glycoprotein
- Gag
Group-specific antigen
- dSTORM
Direct stochastic optical reconstruction microscopy
- MVB
Multivesicular bodies
- TEM
Tetraspanins-enriched microdomains
- SARS
Severe acute respiratory syndrome
- MERS
Middle-East respiratory syndrome
- ACE 2
Angiotensin-converting enzyme 2
- nsps
Non-structural membrane spanning proteins
- RTC
Replication/transcription complexes
- DMV
Double-membrane vesicles
- CM
Convoluted membrane
- VP
Vesicle pockets
- LPS
Lysophospholipids
- UPLC-MS
Ultra-performance liquid chromatography-Mass Spectrometry
- cPLA2α
Cytosolic phospholipase A2α
- LD
Lipid droplets
- HMGCR
3-Hydroxy-3-methylglutaryl-CoA reductase
- OSBP
Oxysterol-binding protein
Compliance with ethical standards
Conflict of interests
The authors declare that they have no conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ruchika Dadhich, Email: ruchikadadhich@iitb.ac.in.
Shobhna Kapoor, Email: shobhnakapoor@chem.iitb.ac.in.
References
- Adhyapak P, Srivatsav AT, Mishra M, Singh A, Narayan R, Kapoor S. Dynamical organization of compositionally distinct inner and outer membrane lipids of mycobacteria. Biophys J. 2020;118(6):1279–1291. doi: 10.1016/j.bpj.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal H, Zelisko M, Liu L, Sharma P. Rigid proteins and softening of biological membranes-with application to HIV-induced cell membrane softening. Sci Rep. 2016;6:1–12. doi: 10.1038/srep25412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Kuraishy HM, Al-Gareeb AI, Al-Buhadily AK. Rosuvastatin as forthcoming antibiotic or as adjuvant additive agent: in vitro novel antibacterial study. J Lab Phys. 2018;10(3):271–275. doi: 10.4103/jlp.jlp_170_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albulescu L, Bigay J, Biswas B, Weber-Boyvat M, Dorobantu CM, Delang L, van der Schaar HM, Jung YS, Neyts J, Olkkonen VM, van Kuppeveld FJM, Strating JRPM. Uncovering oxysterol-binding protein (OSBP) as a target of the anti-enteroviral compound TTP-8307. Antivir Res. 2017;140:37–44. doi: 10.1016/j.antiviral.2017.01.008. [DOI] [PubMed] [Google Scholar]
- Albulescu L, Strating JRPM, Thibaut HJ, van Der Linden L, Shair MD, Neyts J, van Kuppeveld FJM. Broad-range inhibition of enterovirus replication by OSW-1, a natural compound targeting OSBP. Antivir Res. 2015;117:110–114. doi: 10.1016/j.antiviral.2015.02.013. [DOI] [PubMed] [Google Scholar]
- Amara A, Mercer J. Viral apoptotic mimicry. Nat Rev Microbiol. 2015;13:461–469. doi: 10.1038/nrmicro3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4(4):e00524–e13. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astarie-Dequeker C, Le Guyader L, Malaga W, Seaphanh FK, Chalut C, Lopez A, Guilhot C. Phthiocerol dimycocerosates of M. tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog. 2009;5(2):e1000289. doi: 10.1371/journal.ppat.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astarie-Dequeker C, Nigou J, Passemar C, Guilhot C. The role of mycobacterial lipids in host pathogenesis. Drug Disc Today. 2010;7(1):e33–41. doi: 10.1016/j.ddmec.2010.09.003. [DOI] [Google Scholar]
- Augenstreich J, Arbues A, Simeone R, Haanappel E, Wegener A, Sayes F, Le Chevalier F, Chalut C, Malaga W, Guilhot C, Brosch R, Astarie-Dequeker C. ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell Microbiol. 2017;19(7):e12726. doi: 10.1111/cmi.12726. [DOI] [PubMed] [Google Scholar]
- Augenstreich J, Haanappel E, Ferré G, Czaplicki G, Jolibois F, Destainville N, Guilhot C, Milon A, Astarie-Dequeker C, Chavent M. The conical shape of DIM lipids promotes Mycobacterium tuberculosis infection of macrophages. Proc Natl Acad Sci USA. 2019;116(51):25649–25658. doi: 10.1073/pnas.1910368116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augenstreich J, Haanappel E, Sayes F, Simeone R, Guillet V, Mazeres S, Chalut C, Mourey L, Brosch R, Guilhot C, Catherine A. Phthiocerol dimycocerosates from Mycobacterium tuberculosis increase the membrane activity of bacterial effectors and host receptors. BioRxiv. 2020 doi: 10.1101/2020.05.13.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglivo M, Baronio M, Natalini G, Beccari T, Chiurazzi P, Fulcheri E, Petralia P, Michelini S, Fiorentini G, Miggiano GA, Morresi A, Tonini G, Bertelli M. Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: a possible strategy for reducing SARS-COV-2 infectivity? Acta Biomedica. 2020;91(1):161–164. doi: 10.23750/abm.v91i1.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl N, Du R, Winarsih I, Ho B, Tucker-Kellogg L, Tidor B, Ding JL. Delineation of lipopolysaccharide (LPS)-binding sites on hemoglobin: from in silico predictions to biophysical characterization. J Biol Chem. 2011;286(43):37793–37803. doi: 10.1074/jbc.M111.245472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre C, Silván JM, Berglund S, Mizunoe Y, Uhlin BE, Wai SN. Release of the type I secreted α-haemolysin via outer membrane vesicles from Escherichia coli. Mol Microbiol. 2006;59(1):99–112. doi: 10.1111/j.1365-2958.2005.04938.x. [DOI] [PubMed] [Google Scholar]
- Bansal-Mutalik R, Nikaido H. Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc Natl Acad Sci USA. 2014;111(13):4958–4963. doi: 10.1073/pnas.1403078111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CE, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn JA, Schnappinger D, Wilkinson RJ, Young D. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7(11):845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwens A, Betz J, Meisen I, Kemper B, Karch H, Müthing J. Facing glycosphingolipid-Shiga toxin interaction: dire straits for endothelial cells of the human vasculature. Cell Mol Life Sci. 2013;70(3):425–457. doi: 10.1007/s00018-012-1060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman P, Linde C, Pütsep K, Pohanka A, Normark S, Henriques-Normark B, Andersson J, Björkhem-Bergman L. Studies on the antibacterial effects of statins - in vitro and in vivo. PLoS One. 2011;6(8):e24394. doi: 10.1371/journal.pone.0024394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc L, Gilleron M, Prandi J, Song OR, Jang MS, Gicquel B, Drocourt D, Neyrolles O, Brodin P, Tiraby G, Vercellone A, Nigou J. Mycobacterium tuberculosis inhibits human innate immune responses via the production of TLR2 antagonist glycolipids. Proc Natl Acad Sci USA. 2017;114(42):11205–11210. doi: 10.1073/pnas.1707840114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron C, Simonov D, Muthukaruppan A, Tsai P, Dauros P, Green S, Hong J, Print CG, Swift S, Phillips AR. Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. PLoS One. 2016;11(8):e0160440. doi: 10.1371/journal.pone.0160440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–8811. doi: 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg K, Seydel U. Investigation into the fluidity of lipopolysaccharide and free lipid a membrane systems by Fourier-transform infrared spectroscopy and differential scanning calorimetry. Euro J Biochem. 1990;191(1):229–236. doi: 10.1111/j.1432-1033.1990.tb19114.x. [DOI] [PubMed] [Google Scholar]
- Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;4(10):a021105. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- Brügger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Kräusslich HG. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci USA. 2006;103(8):2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlaka I, Liu XL, Rebetz J, Arvidsson I, Yang L, Brismar H, Karpman D, Aperia A. Ouabain protects against shiga toxin-triggered apoptosis by reversing the imbalance between Bax and Bcl-xL. J Am Soc Nephrol. 2013;24(9):1413–1423. doi: 10.1681/ASN.2012101044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho LR, Constant P, Raynaud C, Lanéelle MA, Triccas JA, Gicquel B, Daffé M, Guilhot C. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem. 2001;276:19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- Catron DM, Lange Y, Borensztajn J, Sylvester MD, Jones BD, Haldar K. Salmonella enterica serovar typhimurium requires nonsterol precursors of the cholesterol biosynthetic pathway for intracellular proliferation. Infect Immun. 2004;72(2):1036–1042. doi: 10.1128/IAI.72.2.1036-1042.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahar HS, Bao X, Casola A. Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses. 2015;7(6):3204–3225. doi: 10.3390/v7062770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R, Uchil PD, Jin J, Shui G, Ott DE, Mothes W, Wenk MR. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J Virol. 2008;82(22):11228–11238. doi: 10.1128/jvi.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier L, Louet M, Chaloin L, Fuchs P, Martinez J, Muriaux D, Favard C, Floquet N. Coarse-grained simulations of the HIV-1 matrix protein anchoring: revisiting its assembly on membrane domains. Biophys J. 2014;106(3):577–585. doi: 10.1016/j.bpj.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Feng Z, Yue H, Bazdar D, Mbonye U, Zender C, Harding CV, Bruggeman L, Karn J, Sieg SF, Wang B, Jin G. Exosomes derived from HIV-1-infected cells promote growth and progression of cancer via HIV TAR RNA. Nat Commun. 2018;9(1):4585. doi: 10.1038/s41467-018-07006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15(7):675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheshenko N, Pierce C, Herold BC. Herpes simplex viruses activate phospholipid scramblase to redistribute phosphatidylserines and Akt to the outer leaflet of the plasma membrane and promote viral entry. PLoS Pathog. 2018;14(1):e1006766. doi: 10.1371/journal.ppat.1006766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W, Gill M, Esposito A, Kaminski CF, Courousse N, Chwetzoff S, Trugnan G, Keshavan N, Lever A, Desselberger U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J Virol. 2010;84(13):6782–6798. doi: 10.1128/jvi.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KS, Aizaki H, Lai MMC. Murine coronavirus requires lipid rafts for virus entry and cell-cell fusion but not for virus release. J Virol. 2005;79(15):9862–9871. doi: 10.1128/jvi.79.15.9862-9871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361(9374):2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements A, Young JC, Constantinou N, Frankel G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes. 2012;3(2):71–87. doi: 10.4161/gmic.19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpitts CC, Ustinov AV, Epand RF, Epand RM, Korshun VA, Schang LM. 5-(Perylen-3-yl)ethynyl-arabino-uridine (aUY11), an arabino-based rigid amphipathic fusion inhibitor, targets virion envelope lipids to inhibit fusion of influenza virus, hepatitis C virus, and other enveloped viruses. J Virol. 2013;87(7):3640–3654. doi: 10.1128/jvi.02882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin-Lickfett K, Gowan B, Sefing E, Reiben K, Peters CJ, Grant A, Thorpe P, Soares M, Carrion R, Wakabayashi M, Schlunegger K, Freimark B, Empig C (2010) Targeting of anionic phospholipids exposed on infected cells and virions: potential broad-spectrum antiviral therapy. Chemical & Biological Defense Science & Technology Conference.
- Dadhich R, Mishra M, Ning S, Jana S, Sarpe VA, Mahato J, Duan M, Kulkarni SS, Kapoor S. A virulence-associated glycolipid with distinct conformational attributes: impact on lateral organization of host plasma membrane, autophagy, and signaling. ACS Chem Biol. 2020;15(3):740–750. doi: 10.1021/acschembio.9b00991. [DOI] [PubMed] [Google Scholar]
- Dadhich R, Singh A, Menon AP, Mishra M, Athul CD, Kapoor S. Biophysical characterization of mycobacterial model membranes and their interaction with rifabutin: towards lipid-guided drug screening in tuberculosis. Biochim Biophys Acta- Biomembranes. 2019;1861(6):1213–1227. doi: 10.1016/j.bbamem.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Dahmane S, Doucet C, Le Gall A, Chamontin C, Dosset P, Murcy F, Fernandez L, Salas D, Rubinstein E, Mougel M, Nollmann M, Milhiet PE. Nanoscale organization of tetraspanins during HIV-1 budding by correlative dSTORM/AFM. Nanoscale. 2019;11(13):6036–6044. doi: 10.1039/C8NR07269H. [DOI] [PubMed] [Google Scholar]
- Di Lorenzo F, De Castro C, Silipo A, Molinaro A. Lipopolysaccharide structures of gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol Rev. 2019;43(3):257–272. doi: 10.1093/femsre/fuz002. [DOI] [PubMed] [Google Scholar]
- Domenech P, Reed MB, Dowd CS, Manca C, Kaplan G, Barry CE. The role of MmpL8 in sulfatide biogenesis and virulence of mycobacterium tuberculosis. J Biol Chem. 2004;279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- Donnenberg MS, Whittam TS. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J Clin Invest. 2001;107(5):539–548. doi: 10.1172/JCI12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan W, Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu Rev Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas F, Haanappel E. Lipids in infectious diseases—the case of AIDS and tuberculosis. Biochim Biophys Acta Biomembr. 2017;1859(9):1636–1647. doi: 10.1016/j.bbamem.2017.05.007. [DOI] [PubMed] [Google Scholar]
- Duncan MJ, Li G, Shin JS, Carson JL, Abraham SN. Bacterial penetration of bladder epithelium through lipid rafts. J Biol Chem. 2004;279:18944–18951. doi: 10.1074/jbc.M400769200. [DOI] [PubMed] [Google Scholar]
- Ernst JD. Macrophage receptors for mycobacterium tuberculosis. Infect Immun. 1998;66(4):1277–1281. doi: 10.1128/iai.66.4.1277-1281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erridge C, Bennett-Guerrero E, Poxton IR. Structure and function of lipopolysaccharides. Microbes Infect. 2002;4(8):837–851. doi: 10.1016/S1286-4579(02)01604-0. [DOI] [PubMed] [Google Scholar]
- Filipe A, McLauchlan J. Hepatitis C virus and lipid droplets: finding a niche. Trends Mol Med. 2015;21(1):34–42. doi: 10.1016/j.molmed.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Finzi A, Orthwein A, Mercier J, Cohen ÉA. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J Virol. 2007;81(14):7476–7490. doi: 10.1128/jvi.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrellad MA, Klepp LI, Gioffré A, García JS, Morbidoni HR, de la Paz SM, Cataldi AA, Bigi F. Virulence factors of the mycobacterium tuberculosis complex. Virulence. 2013;4(1):3–66. doi: 10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti RA, Chua J, Vergne I, Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci USA. 2003;100(9):5437–5442. doi: 10.1073/pnas.0737613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sáez AJ, Chiantia S, Schwille P. Effect of line tension on the lateral organization of lipid membranes. J Biol Chem. 2007;282:33537–33544. doi: 10.1074/jbc.M706162200. [DOI] [PubMed] [Google Scholar]
- Gatfield J, Pieters J. Essential role for cholesterol in entry of mycobacteria into macrophages. Science. 2000;288(5471):1647–1650. doi: 10.1126/science.288.5471.1647. [DOI] [PubMed] [Google Scholar]
- Ge M, Freed JH. Fusion peptide from influenza hemagglutinin increases membrane surface order: an electron-spin resonance study. Biophys J. 2009;96(12):4925–4934. doi: 10.1016/j.bpj.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal A, Upadhyaya BB, Fritz JV, Heintz-Buschart A, Desai MS, Yusuf D, Huang D, Baumuratov A, Wang K, Galas D, Wilmes P. The extracellular RNA complement of Escherichia coli. MicrobiologyOpen. 2015;4(2):252–266. doi: 10.1002/mbo3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, Catchpole R, Forterre P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol Rev. 2019;43(3):273–303. doi: 10.1093/femsre/fuy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore SA, Schelle MW, Holsclaw CM, Leigh CD, Jain M, Cox JS, Leary JA, Bertozzi CR. Sulfolipid-1 biosynthesis restricts mycobacterium tuberculosis growth in human macrophages. ACS Chem Biol. 2012;7(5):863–870. doi: 10.1021/cb200311s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glende J, Schwegmann-Wessels C, Al-Falah M, Pfefferle S, Qu X, Deng H, Drosten C, Naim HY, Herrler G. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology. 2008;381(2):215–221. doi: 10.1016/j.virol.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbole AA, Ahmed W, Bhat RS, Bradley EK, Ekins S, Nagaraja V. Targeting mycobacterium tuberculosis topoisomerase I by small-molecule inhibitors. Antimicrob Agents Chemother. 2015;59(3):1549–1557. doi: 10.1128/AAC.04516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith CS, Tatti KM, Ksiazek TG, Rollin PE, Comer JA, Lee WW, Rota PA, Bankamp B, Bellini WJ, Zaki SR. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10(2):320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorov B, Attuil-Audenis V, Perugi F, Nedelec M, Watson S, Pique C, Darlix JL, Conjeaud H, Muriaux D. A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology. 2009;6:28. doi: 10.1186/1742-4690-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer MC, Ulasli M, Vonk AM, Reggiori F, Rottier PJM, de Haan CAM. Mobility and interactions of coronavirus nonstructural protein 4. J Virol. 2011;85(9):4572–4577. doi: 10.1128/jvi.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer MC, Monastyrska I, Griffith J, van der Sluijs P, Voortman J, van Bergen en Henegouwen PM, Vonk AM, Rottier PJM, Reggiori F, De Haan CAM. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459(1):125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer MC, Rottier PJM, de Haan CAM. Biogenesis and dynamics of the coronavirus replicative structures. Viruses. 2012;4(11):3245–3269. doi: 10.3390/v4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer MC, Verheije MH, Ulasli M, Shaltiël IA, de Vries LA, Reggiori F, Rottier PJM, de Haan CAM. Dynamics of coronavirus replication-transcription complexes. J Virol. 2010;84(4):2134–2149. doi: 10.1128/jvi.01716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S. The broad anti-viral agent glycyrrhizin directly modulates the fluidity of plasma membrane and HIV-1 envelope. Biochem J. 2005;392(1):191–199. doi: 10.1042/BJ20051069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Virology. 2015;479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19(7):368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Pro Natl Acad Sci USA. 2008;105(10):3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CMJ, Fang RH, Luk BT, Zhang L. Nanoparticle-detained toxins for safe and effective vaccination. Nature Nanotech. 2013;8:933–938. doi: 10.1038/nnano.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FC. Plasma membrane cholesterol plays a critical role in the Salmonella-induced anti-inflammatory response in intestinal epithelial cells. Cell Immunol. 2011;271(2):480–487. doi: 10.1016/j.cellimm.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Huarte N, Carravilla P, Cruz A, Lorizate M, Nieto-Garai JA, Kräusslich HG, Pérez-Gil J, Requejo-Isidro J, Nieva JL. Functional organization of the HIV lipid envelope. Sci Rep. 2016;6:1–14. doi: 10.1038/srep34190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt JM. Shiga toxin-producing escherichia coli (STEC) Clin Lab Med. 2010;31(1):21–45. doi: 10.1016/j.cll.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Payrastre B, Chap H, Ogawa W, Sakaue H, Kasuga M, Cossart P. A role for phosphoinositide 3-kinase in bacterial invasion. Science. 1996;274(5288):780–782. doi: 10.1126/science.274.5288.780. [DOI] [PubMed] [Google Scholar]
- Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206(13):2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa E, Mori D, Yamasaki S. Recognition of mycobacterial lipids by immune receptors. Trends Immunol. 2017;30(1):66–76. doi: 10.1016/j.it.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Izquierdo-Useros N, Blanco J, Erkizia I, Fernández-Figueras MT, Borràs FE, Naranjo-Gómez M, Bofill M, Ruiz L, Clotet B, Martinez-Picado J. Maturation of blood-derived dendritic cells enhances human immunodeficiency virus type 1 capture and transmission. J Virol. 2007;81(14):7559–7570. doi: 10.1128/jvi.02572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-useros N, Lorizate M, Contreras F, Rodriguez-plata MT, Martinez-picado J, Kra G. Sialyllactose in viral membrane gangliosides is a novel molecular recognition pattern for mature dendritic cell capture of HIV-1. Plos Biol. 2012;10(4):e1001315. doi: 10.1371/journal.pbio.1001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerwood S, Cohen J. Unexpected antimicrobial effect of statins. J Antimicrob Chemother. 2008;61(2):362–364. doi: 10.1093/jac/dkm496. [DOI] [PubMed] [Google Scholar]
- Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol. 2007;81(15):7873–7884. doi: 10.1128/jvi.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci USA. 2009;106(45):19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Józefowski S, Sobota A, Kwiatkowska K. How mycobacterium tuberculosis subverts host immune responses. BioEssays. 2008;30(10):943–954. doi: 10.1002/bies.20815. [DOI] [PubMed] [Google Scholar]
- Karpman D, Håkansson A, Ferez MTR, Isaksson C, Carlemalm E, Caprioli A, Svanborg C. Apoptosis of renal cortical cells in the hemolytic-uremic syndrome: In vivo and in vitro studies. Infect Immun. 1998;66(2):636–644. doi: 10.1128/IAI.66.2.636-644.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul D, Anand PK, Verma I. Cholesterol-sensor initiates M. tuberculosis entry into human macrophages. Mol Cell Biochem. 2004;258:219–222. doi: 10.1023/B:MCBI.0000012851.42642.be. [DOI] [PubMed] [Google Scholar]
- Ketter E, Randall G. Virus impact on lipids and membranes. Annu Rev Virol. 2019;6(1):319–340. doi: 10.1146/annurev-virology-092818-015748. [DOI] [PubMed] [Google Scholar]
- Khan I, Katikaneni DS, Han Q, Sanchez-Felipe L, Hanada K, Ambrose RL, Mackenzie JM, Konan KV. Modulation of hepatitis C virus genome replication by glycosphingolipids and four-phosphate adaptor protein 2. J Virol. 2014;88(21):12276–12295. doi: 10.1128/jvi.00970-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliger Y, Levanon EY. Cloaked similarity between HIV-1 and SARS-CoV suggests an anti-SARS strategy. BMC Microbiol. 2003;3:20. doi: 10.1186/1471-2180-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K, Kikkert M, Van Den Worm SHE, Zevenhoven-Dobbe JC, Van Der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):1957–1974. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman EE, Chupin V, de Kruijff B, Burger KNJ. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 2003;4(3):162–174. doi: 10.1034/j.1600-0854.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- Kouokam JC, Wai SN, Fällman M, Dobrindt U, Hacker J, Uhlin BE. Active cytotoxic necrotizing factor 1 associated with outer membrane vesicles from uropathogenic Escherichia coli. Infect Immun. 2006;74(4):2022–2030. doi: 10.1128/IAI.74.4.2022-2030.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovbasnjuk O, Edidin M, Donowitz M. Role of lipid rafts in Shiga toxin 1 interaction with the apical surface of Caco-2 cells. J Cell Sci. 2001;114(22):4025–4031. doi: 10.1242/jcs.114.22.4025. [DOI] [PubMed] [Google Scholar]
- Krishnakumari V, Binny TM, Adicherla H, Nagaraj R. Escherichia coli Lipopolysaccharide modulates biological activities of human-β-defensin analogues but not non-ribosomally synthesized peptides. ACS Omega. 2020;5(12):6366–6375. doi: 10.1021/acsomega.9b03770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutzberger AJB, et al. Assymteric phosphatidylethanolamine distribution controls fusion pore lifetime and probability. Biophys J. 2017;113:1912–1915. doi: 10.1016/j.bpj.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiak J, Brewer J, Hansen S, Bagatolli LA. Lipid lateral organization on giant unilamellar vesicles containing lipopolysaccharides. Biophys J. 2011;100(4):978–986. doi: 10.1016/j.bpj.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni R, Prasad A. Exosomes derived from HIV-1 infected DCs mediate viral trans-infection via fibronectin and galectin-3. Sci Rep. 2017;7:14787. doi: 10.1038/s41598-017-14817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay SB, Bieniasz PD. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6(11):e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AL, Millet JK, Daniel S, Freed JH, Whittaker GR. The SARS-CoV fusion peptide forms an extended bipartite fusion platform that perturbs membrane order in a calcium-dependent manner. J Mol Biol. 2017;429(24):3875–3892. doi: 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AL, Moorthy AE, Li Y, Tamm LK. Fusion activity of HIV gp41 fusion domain is related to its secondary structure and depth of membrane insertion in a cholesterol-dependent fashion. J Mol Biol. 2012;418(1–2):3–15. doi: 10.1016/j.jmb.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrouy-Maumus G. Lipids as biomarkers of cancer and bacterial infections. Curr Med Chem. 2018;26(11):1924–1932. doi: 10.2174/0929867325666180904120029. [DOI] [PubMed] [Google Scholar]
- Layre E, Sweet L, Hong S, Madigan CA, Desjardins D, Young DC, Cheng TY, Annand JW, Kim K, Shamputa IC, McConnell MJ, Debono CA, Behar SM, Minnaard AJ, Murray M, Barry CE, Matsunaga I, Moody DB. A comparative lipidomics platform for chemotaxonomic analysis of mycobacterium tuberculosis. Chem Biol. 2011;18(12):1537–1549. doi: 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros N, Pohlentz G, Steil D, Kouzel IU, Liashkovich I, Mellmann A, Karch H, Müthing J. Membrane assembly of Shiga toxin glycosphingolipid receptors and toxin refractiveness of MDCK II epithelial cells. J Lipid Res. 2018;59(8):1383–2140. doi: 10.1194/jlr.M083048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM, Li YG, Yamate M, Li SM, Ikuta K. Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle. Microbes Infect. 2007;9(1):96–102. doi: 10.1016/j.micinf.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood CA. Glycosphingolipid functions. Cold Spring Harb Perspect Biol. 2011;3(7):a004788. doi: 10.1101/cshperspect.a004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Barry CE, Besra GS, Nikaido H. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J Biol Chem. 1996;271:29545–29551. doi: 10.1074/jbc.271.47.29545. [DOI] [PubMed] [Google Scholar]
- Liu J, Rosenberg EY, Nikaido H. Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc Natl Acad Sci USA. 1995;92(24):11254–11258. doi: 10.1073/pnas.92.24.11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorizate M, Sachsenheimer T, Glass B, Habermann A, Gerl MJ, Kräusslich HG, Brügger B. Comparative lipidomics analysis of HIV-1 particles and their producer cell membrane in different cell lines. Cell Microbiol. 2013;15(2):292–304. doi: 10.1111/cmi.12101. [DOI] [PubMed] [Google Scholar]
- Lu Y, Liu DX, Tam JP. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem Biophys Res Commun. 2008;369(2):344–349. doi: 10.1016/j.bbrc.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo-Villarino G, Hudrisier D, Tanne A, Neyrolles O. C-type lectins with a sweet spot for Mycobacterium tuberculosis. Eur J Microbiol Immun. 2011;1(1):25–40. doi: 10.1556/eujmi.1.2011.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison MN, Okeoma CM. Exosomes: implications in HIV-1 pathogenesis. Viruses. 2015;7(7):4093–4118. doi: 10.3390/v7072810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima N, Fernandez RC. Recognition of lipid A variants by the TLR4-MD-2 receptor complex. Front Cell Infect Microbiol. 2013;3:3. doi: 10.3389/fcimb.2013.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- Martín-Acebes MA, Blázquez AB, de Oya NJ, Escribano-Romero E, Shi PY, Saiz JC. A single amino acid substitution in the core protein of west nile virus increases resistance to acidotropic compounds. PLoS One. 2013;8(7):e69479. doi: 10.1371/journal.pone.0069479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Acebes MA, Gabandé-Rodríguez E, García-Cabrero AM, Sánchez MP, Ledesma MD, Sobrino F, Saiz JC. Host sphingomyelin increases West Nile virus infection in vivo. J Lipid Res. 2016;57(3):422–432. doi: 10.1194/jlr.M064212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J. Lipid droplets and hepatitis C virus infection. Biochim Biophys Acta—molecular and cell biology of lipids. 2009;1796(1):552–559. doi: 10.1016/j.bbalip.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Meher G, Chakraborty H. Membrane composition modulates fusion by altering membrane properties and fusion peptide structure. J Membr Biol. 2019;252:261–272. doi: 10.1007/s00232-019-00064-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan GB. Driving a wedge between viral lipids blocks infection. Proc Natl Acad Sci USA. 2010;107(40):17069–17070. doi: 10.1073/pnas.1012748107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton-Celsa AR. Shiga Toxin (Stx) classification, structure, and function. Microbiol Spectr. 2014;2(4):EHEC-0024-2013. doi: 10.1128/microbiolspec.ehec-0024-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet JK, Whittaker GR. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3–8. doi: 10.1016/j.virol.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Akhtar S, Jagannath C, Khan A. Pattern recognition receptors and coordinated cellular pathways involved in tuberculosis immunopathogenesis: emerging concepts and perspectives. Mol Immunol. 2017;87:240–248. doi: 10.1016/j.molimm.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Mishra AK, Driessen NN, Appelmelk BJ, Besra GS. Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol Rev. 2011;35(6):1126–1157. doi: 10.1111/j.1574-6976.2011.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Adhyapak P, Dadhich R, Kapoor S. Dynamic remodeling of the host cell membrane by virulent mycobacterial sulfoglycolipid-1. Sci Rep. 2019;9:12844. doi: 10.1038/s41598-019-49343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriel-Carretero M. The hypothetical role of phosphatidic acid in subverting ER membranes during SARS-CoV infection. Traffic. 2020;21(8):545–551. doi: 10.1111/tra.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortaz E, Adcock IM, Tabarsi P, Masjedi MR, Mansouri D, Velayati AA, Casanova JL, Barnes PJ. Interaction of pattern recognition receptors with mycobacterium tuberculosis. J Clin Immunol. 2015;35:1–10. doi: 10.1007/s10875-014-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C, Hardt M, Schwudke D, Neuman BW, Pleschka S, Ziebuhr J. Inhibition of cytosolic phospholipase A2α impairs an early step of coronavirus replication in cell culture. J Virol. 2017;92(4):e01463–e1517. doi: 10.1128/jvi.01463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux CW, Nenninger A, Ray N, Robinson C. Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J Bacteriol. 2006;188(10):3442–3448. doi: 10.1128/JB.188.10.3442-3448.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Sakuragi T, Segawa K. Flippase and scramblase for phosphatidylserine exposure. Curr Opinion Immunol. 2020;62:31–38. doi: 10.1016/j.coi.2019.11.009. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003 doi: 10.1128/mmbr.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura R, Kiyota A, Suzaki E, Kataoka K, Ohe Y, Miyamoto K, Senda T, Fujimoto T. Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J Virol. 2004;78(16):8701–8708. doi: 10.1128/jvi.78.16.8701-8708.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101(41):14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parihar SP, Guler R, Khutlang R, Lang DM, Hurdayal R, Mhlanga MM, Suzuki H, Marais AD, Brombacher F. Statin therapy reduces the mycobacterium tuberculosis burden in human macrophages and in mice by enhancing autophagy and phagosome maturation. J Infect Dis. 2014;209(5):754–763. doi: 10.1093/infdis/jit550. [DOI] [PubMed] [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458(7242):1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- Popik W, Alce TM, Au WC. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4+ T Cells. J Virol. 2002;76(10):4709–4722. doi: 10.1128/jvi.76.10.4709-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval CJ, Brosch R, Simeone R. The macrophage: a disputed fortress in the battle against Mycobacterium tuberculosis. Front Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.02284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley J, Hughitt VK, Velikovsky CA, Mariuzza RA, El-Sayed NM, Briken V. The cell wall lipid PDIM contributes to phagosomal escape and host cell exit of Mycobacterium tuberculosis. Mbio. 2017;8(2):e00148–e17. doi: 10.1128/mBio.00148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, Dowhan W. Biosynthesis and function of phospholipids in Escherichia coli. J Biol Chem. 1990;265(3):1235–1238. [PubMed] [Google Scholar]
- Rahman SA, Koch P, Weichsel J, Godinez WJ, Schwarz U, Rohr K, Lamb DC, Kräusslich HG, Müller B. Investigating the role of F-actin in human immunodeficiency virus assembly by live-cell microscopy. J Virol. 2014;88(14):7904–7914. doi: 10.1128/jvi.00431-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L, Tian R, Chen X. Cell-Membrane-Mimicking Nanodecoys against Infectious Diseases. ACS Nano. 2020;14(3):2569–2574. doi: 10.1021/acsnano.0c01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riff JD, Callahan JW, Sherman PM. Cholesterol-enriched membrane microdomains are required for inducing host cell cytoskeleton rearrangements in response to attaching-effacing Escherichia coli. Infect Immun. 2005;73(11):7113–7125. doi: 10.1128/IAI.73.11.7113-7125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers TJ, Thorpe CM, Paton AW, Paton JC. Role of lipid rafts and flagellin in invasion of colonic epithelial cells by Shiga-toxigenic Escherichia coli O113:H21. Infect Immun. 2012;80(8):2858–2867. doi: 10.1128/IAI.00336-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett VW, Mallampalli VKPS, Karlstaedt A, Dowhan W, Taegtmeyer H, Margolin W, Vitrac H. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J Bacteriol. 2017;199(13):e00849–e916. doi: 10.1128/JB.00849-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Cirión A, Nir S, Lorizate M, Agirre A, Cruz A, Pérez-Gil J, Nieva JL. Sphingomyelin and cholesterol promote HIV-1 gp41 pretransmembrane sequence surface aggregation and membrane restructuring. J Biol Chem. 2002;277:21776–21785. doi: 10.1074/jbc.M202255200. [DOI] [PubMed] [Google Scholar]
- Samsa MM, Mondotte JA, Iglesias NG, Assunção-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5(10):e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani M, Houben ENG, Geurtsen J, Pierson J, De Punder K, Van Zon M, Wever B, Piersma SR, Jiménez CR, Daffé M, Appelmelk BJ, Bitter W, Van der Wel N, Peters PJ. Direct visualization by Cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010;6(3):e1000794. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartain MJ, Dick DL, Rithner CD, Crick DC, Belisle JT. Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel “Mtb LipidDB”. J Lipid Res. 2011;52(5):861–872. doi: 10.1194/jlr.M010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger JC, Holsclaw CM, Schelle MW, Botyanszki Z, Gilmore SA, Tully SE, Niederweis M, Cravatt BF, Leary JA, Bertozzi CR. Elucidation and chemical modulation of sulfolipid-1 biosynthesis in Mycobacterium tuberculosis. J Biol Chem. 2012;287(11):7990–8000. doi: 10.1074/jbc.M111.315473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydel U, Koch MHJ, Brandenburg K. Structural polymorphisms of rough mutant lipopolysaccharides Rd to Ra from Salmonella minnesota. J Struct Biol. 1993;110(3):232–243. doi: 10.1006/jsbi.1993.1026. [DOI] [PubMed] [Google Scholar]
- Shrivastava R, Chng SS. Lipid trafficking across the Gram-negative cell envelope. J Struct Biol. 2019;294(39):14175–14184. doi: 10.1074/jbc.AW119.008139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel DP, Epand RM. The mechanism of lamellar-to-inverted hexagonal phase transitions in phosphatidylethanolamine: implications for membrane fusion mechanisms. Biophys J. 1997;73(6):3089–3111. doi: 10.1016/S0006-3495(97)78336-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb perspect Biol. 2010;2(5):a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I. Mycobacterium tuberculosis pathogenesis and molecular. Clin Microbiol Rev. 2003;16(3):463–496. doi: 10.1128/CMR.16.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MM, King SW, Thorpe PE. Targeting inside-out phosphatidylserine as a therapeutic strategy for viral diseases. Nat Med. 2008;14(12):1357–1362. doi: 10.1038/nm.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spargo BJ, Crowe LM, Ioneda T, Beaman BL, Crowe JH. Cord factor (α, α-trehalose 6,6’-dimycolate) inhibits fusion between phospholipid vesicles. Proc Natl Acad Sci USA. 1991;88(3):737–740. doi: 10.1073/pnas.88.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speerstra S, Chistov AA, Proskurin GV, Aralov AV, Ulashchik EA, Streshnev PP, Shmanai VV, Korshun VA, Schang LM. Antivirals acting on viral envelopes via biophysical mechanisms of action. Antiviral Res. 2018;149:164–173. doi: 10.1016/j.antiviral.2017.11.018. [DOI] [PubMed] [Google Scholar]
- St Vincent MR, Colpitts CC, Ustinov AV, Muqadas M, Joyce MA, Barsby NL, Epand RF, Epand RM, Khramyshev SA, Valueva OA, Korshun VA, Tyrrell DLJ, Schang LM. Rigid amphipathic fusion inhibitors, small molecule antiviral compounds against enveloped viruses. Proc Natl Acad Sci USA. 2010;107(40):17339–17344. doi: 10.1073/pnas.1010026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl AL, Arvidsson I, Johansson KE, Chromek M, Rebetz J, Loos S, Kristoffersson AC, Békássy ZD, Mörgelin M, Karpman D. A novel mechanism of bacterial toxin transfer within host blood cell-derived microvesicles. PLoS Pathog. 2015;11(2):e1004619. doi: 10.1371/journal.ppat.1004619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancu C, Sima A. Statins: Mechanism of action and effects. J Cell Mol Med. 2001;5(4):378–387. doi: 10.1111/j.1582-4934.2001.tb00172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strating JRPM, van der Linden L, Albulescu L, Bigay J, Arita M, Delang L, Leyssen P, van der Schaar HM, Lanke KHW, Thibaut HJ, Ulferts R, Drin G, Schlinck N, Wubbolts RW, Sever N, Head SA, Liu JO, Beachy PA, DeMatteis MA, Shair MD, Olkkonen VM, Neyts J, van Kuppeveld FJM. Itraconazole inhibits enterovirus replication by targeting the oxysterol-binding protein. Cell Rep. 2015;10(4):600–615. doi: 10.1016/j.celrep.2014.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus MR, Tang T, Lai AL, Flegel A, Bidon M, Freed JH, Daniel S, Whittaker GR. Ca2+ ions promote fusion of Middle East respiratory syndrome coronavirus with host cells and increase infectivity. J Virol. 2020;94(13):e00426–e520. doi: 10.1128/jvi.00426-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Mariani-Floderer C, López-Huertas MR, Gros N, Hamard-Péron E, Favard C, Ohlmann T, Alcamí J, Muriaux D. Involvement of the Rac1-IRSp53-wave2-Arp2/3 signaling pathway in HIV-1 gag particle release in CD4 T cells. J Virol. 2015;89(16):8162–8181. doi: 10.1128/jvi.00469-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull WB, Shimizu KH, Chatterjee D, Homans SW, Treumann A. Identification of the 5-methylthiopentosyl substituent in Mycobacterium tuberculosis lipoarabinomannan. Angewandte Chemie—Int Ed. 2004;43(30):3918–3922. doi: 10.1002/anie.200454119. [DOI] [PubMed] [Google Scholar]
- Turner J, Torrelles JB. Mannose-capped lipoarabinomannan in Mycobacterium tuberculosis pathogenesis. Pathog Dis. 2018;76(4):fty026. doi: 10.1093/femspd/fty026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga AS, Castanho MARB. The influence of cholesterol on the interaction of HIV gp41 membrane proximal region-derived peptides with lipid bilayers. FEBS J. 2007;274(19):5096–5104. doi: 10.1111/j.1742-4658.2007.06029.x. [DOI] [PubMed] [Google Scholar]
- Vergne I, Chua J, Deretic V. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J Exp Med. 2003;198(4):653–659. doi: 10.1084/jem.20030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigant F, Lee J, Hollmann A, Tanner LB, Akyol AZ, Yun T, Shui G, Aguilar HC, Zhang D, Meriwether D, Roman-Sosa G, Robinson LR, Juelich TL, Buczkowski H, Chou S, Castanho MARB, Wolf MC, Smith JK, Banyard A, Kielian M, Reddy S, Wenk MR, Selke M, Santos NC, Freiberg AN, Jung ME, Lee B. A mechanistic paradigm for broad-spectrum antivirals that target virus-cell fusion. PLoS Pathog. 2013;9(4):e1003297. doi: 10.1371/journal.ppat.1003297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villysson A, Tontanahal A, Karpman D. Microvesicle involvement in Shiga toxin-associated infection. Toxins. 2017;9(11):376. doi: 10.3390/toxins9110376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent N, Genin C, Malvoisin E. Identification of a conserved domain of the HIV-1 transmembrane protein gp41 which interacts with cholesteryl groups. Biochim Biophys Acta—Biomembr. 2002;1567:157–164. doi: 10.1016/S0005-2736(02)00611-9. [DOI] [PubMed] [Google Scholar]
- Viswanathan G, Jafurulla M, Kumar GA, Raghunand TR, Chattopadhyay A. Dissecting the membrane cholesterol requirement for mycobacterial entry into host cells. Chem Phys Lipids. 2015;189:19–27. doi: 10.1016/j.chemphyslip.2015.05.006. [DOI] [PubMed] [Google Scholar]
- Viswanathan G, Jafurulla M, Kumar GA, Raghunand TR, Chattopadhyay A. Macrophage sphingolipids are essential for the entry of mycobacteria. Chem Phys Lipids. 2018;213:25–31. doi: 10.1016/j.chemphyslip.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143(2):162–176. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18(2):290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner FJ, Lew RA, Smith AI, Lambert DW, Hooper NM, Turner AJ. Angiotensin-converting enzyme 2 (ACE2), but not ACE, is preferentially localized to the apical surface of polarized kidney cells. J Biol Chem. 2005;280(47):39353–39362. doi: 10.1074/jbc.M508914200. [DOI] [PubMed] [Google Scholar]
- Wilkinson SG. Bacterial lipopolysaccharides—themes and variations. Prog Lipid Res. 1996;35(3):283–343. doi: 10.1016/S0163-7827(96)00004-5. [DOI] [PubMed] [Google Scholar]
- Wojcechowskyj JA, Doms RW. A potent, broad-spectrum antiviral agent that targets viralmembranes. Viruses. 2010;2(5):1106–1109. doi: 10.3390/v2051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MC, Freiberg AN, Zhang T, Akyol-Ataman Z, Grock A, Hong PW, Li J, Watson NF, Fang AQ, Aguilar HC, Porotto M, Honko AN, Damoiseaux R, Miller JP, Woodson SE, Chantasirivisal S, Fontanes V, Negrete OA, Krogstad P, Dasgupta A, Moscona A, Hensley LE, Whelan SE, Faull KF, Holbrook MR, Jung ME, Lee B. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci USA. 2010;107(7):3157–3162. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu EL, Engström O, Jo S, Stuhlsatz D, Yeom MS, Klauda JB, Widmalm G, Im W. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys J. 2013;105(6):1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Chu H, Yang D, Sze KH, Lai PM, Yuan S, Shuai H, Wang Y, Kao RYT, Chan JFW, Yuen KY. Characterization of the lipidomic profile of human coronavirus-infected cells: implications for lipid metabolism remodeling upon coronavirus replication. Viruses. 2019;11(1):73. doi: 10.3390/v11010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandrapalli N, Lubart Q, Tanwar HS, Picart C, Mak J, Muriaux D, Favard C. Self assembly of HIV-1 Gag protein on lipid membranes generates PI(4,5)P2/Cholesterol nanoclusters. Sci Rep. 2016;6:1–13. doi: 10.1038/srep39332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ST, Kiessling V, Simmons JA, White JM, Tamm LK. HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains. Nat Chem Biol. 2015;11(6):424–431. doi: 10.1038/nchembio.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ST, Kiessling V, Tamm LK. Line tension at lipid phase boundaries as driving force for HIV fusion peptide-mediated fusion. Nat Commun. 2016;7:11401. doi: 10.1038/ncomms11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron S, Kolling GL, Simon L, Matthews KR. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol. 2000;66(10):4414–4420. doi: 10.1128/AEM.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020;16(10):1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa A, Saijo S, Hoshino Y, Miyake Y, Ishikawa E, Suzukawa M, Inoue H, Tanaka M, Yoneyama M, Oh-hora M, Akashi K, Yamasaki S. Dectin-2 is a direct receptor for mannose-capped lipoarabinomannan of mycobacteria. Immunity. 2014;41(3):402–413. doi: 10.1016/j.immuni.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Zhang W, Wang X, Han X. Multilayer giant unilamellar vesicles as a model of artificial tissue for drug screen. Chem Phys Lett. 2019;717(16):34–37. doi: 10.1016/j.cplett.2018.12.041. [DOI] [Google Scholar]
- Zhang XW, Yap YL. Structural similarity between HIV-1 gp41 and SARS-CoV S2 proteins suggests an analogous membrane fusion mechanism. J Mol Struct (Thoechem) 2004;677:73–76. doi: 10.1016/j.theochem.2004.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M, Stroupe C, Orr A, Douville D, Wickner WT. Membranes linked by trans-SNARE complexes require lipids prone to non-bilayer structure for progression to fusion. eLife. 2014;3:e01879. doi: 10.7554/eLife.01879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffé M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol. 2008;190(16):5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]