Significance
Some cellulases exhibit “processivity”: the ability to degrade crystalline cellulose through successive hydrolytic catalytic reactions without the release of the enzyme from the substrate surface. We previously observed the movement of fungal processive cellulases by high-speed atomic force microscopy, and here, we use the same technique to directly observe the processive movement of bacterial cellobiohydrolases settling a long-standing controversy. Although fungal and bacterial processive cellulases have completely different protein folds, they have evolved to acquire processivity through the same strategy of adding subsites to extend the substrate-binding site and forming a tunnel-like active site by increasing the number of loops covering the active site. This represents an example of protein-level convergent evolution to acquire the same functions from different ancestors.
Keywords: cellobiohydrolase, processivity, crystalline cellulose, biomass utilization, cellulase
Abstract
Cellulose is the most abundant biomass on Earth, and many microorganisms depend on it as a source of energy. It consists mainly of crystalline and amorphous regions, and natural degradation of the crystalline part is highly dependent on the degree of processivity of the degrading enzymes (i.e., the extent of continuous hydrolysis without detachment from the substrate cellulose). Here, we report high-speed atomic force microscopic (HS-AFM) observations of the movement of four types of cellulases derived from the cellulolytic bacteria Cellulomonas fimi on various insoluble cellulose substrates. The HS-AFM images clearly demonstrated that two of them (CfCel6B and CfCel48A) slide on crystalline cellulose. The direction of processive movement of CfCel6B is from the nonreducing to the reducing end of the substrate, which is opposite that of processive cellulase Cel7A of the fungus Trichoderma reesei (TrCel7A), whose movement was first observed by this technique, while CfCel48A moves in the same direction as TrCel7A. When CfCel6B and TrCel7A were mixed on the same substrate, “traffic accidents” were observed, in which the two cellulases blocked each other’s progress. The processivity of CfCel6B was similar to those of fungal family 7 cellulases but considerably higher than those of fungal family 6 cellulases. The results indicate that bacteria utilize family 6 cellulases as high-processivity enzymes for efficient degradation of crystalline cellulose, whereas family 7 enzymes have the same function in fungi. This is consistent with the idea of convergent evolution of processive cellulases in fungi and bacteria to achieve similar functionality using different protein foldings.
Efficient production of biofuels and biobased products from biomass is one of the biggest challenges for the circular bioeconomy because of its potential to replace fossil-based fuels and products with biomass-based renewable fuels and products. In nature, biomass is broken down by the collaborative action of multiple functionally complementary enzymes that are secreted by biomass-degrading microorganisms. Cellulose, which is the most abundant biomass, is the main structural polysaccharide of plant cell walls and consists of linear polymers of β-1,4-linked d-glucose units. The chains are stabilized by intra- and intermolecular hydrogen bonds, forming an insoluble structure that is ∼70% crystalline (1). One of the most important factors influencing the efficiency of degradation of recalcitrant crystalline cellulose is the processivity of the cellobiohydrolases (CBHs) that degrade crystalline cellulose to soluble products. Processivity is defined as the ability of the cellulases to catalyze continuous degradation of a cellulose chain through successive hydrolytic catalytic reactions without dissociation from the chain. Recently, our research group succeeded in visualizing processive movements of fungal glycoside hydrolase (GH) family 7 CBHs by means of high-speed atomic force microscopy (HS-AFM). We found that Trichoderma reesei (TrCel7A) and Phanerochaete chrysosporium (PcCel7C and PcCel7D) cellulase molecules move unidirectionally from the reducing end to the nonreducing end of the chain on crystalline cellulose (2–4). Cellulose-degrading filamentous fungi also secrete other CBHs belonging to GH family 6, but their processive movement could not be visualized by HS-AFM under the same conditions, suggesting that family 6 and 7 CBHs differ in their ability to remain on the crystal surface (5). Comparison of their three-dimensional (3D) structures indicated that the catalytic domain of fungal family 6 CBHs is covered by a short loop, while that of family 7 CBHs has four loops covering the active site. These observations suggest that cellulolytic fungi secrete two types of CBHs, of which one binds strongly to the cellulose chain (like TrCel7A, PcCel7C, and PcCel7D) and shows processive degradation, while the other binds weakly to the cellulose chain (like TrCel6A), showing little or no processivity. However, less is known about bacterial cellulases, and the processivity of bacterial CBHs has never been confirmed by direct observation.
In the present study, we observed the processive movement of four bacterial cellulases, CfCel6A, CfCel6B, CfCel9A, and CfCel48A [formerly known as CenA (6), CbhA (7), CenB (8), and CbhB (9), respectively] from the cellulolytic actinobacteria Cellulomonas fimi. This microorganism is one of the best-known aerobic cellulolytic bacteria and expresses three endoglucanases, two exocellobiohydrolases and one xylanase in the supernatants of cultures grown on cellulose (10). CfCel6A and CfCel9A are thought to be endoglucanases (6, 8), while CfCel9A has a high amino acid sequence similarity to the possibly processive endoglucanase TfCel9A from Thermobifida fusca (11). CfCel6B and CfCel48A are CBHs (7, 9), and both have been predicted to have processivity. We also compare the findings for fungal and bacterial cellulases and discuss the requirements for processivity, as well as the likelihood of convergent evolution of processive cellulases in bacteria and fungi.
Results
General Characteristics of the Four C. fimi Cellulases.
The cellulolytic activities of the four cellulases were tested on several insoluble substrates, i.e., phosphoric acid–swollen cellulose (PASC), microcrystalline cellulose (Avicel), and crystalline celluloses Iα and IIII from green algae, and the amounts of reaction products were measured by high performance liquid chromatography (HPLC). Crystalline cellulose in nature has structures called Iα and Iβ (12, 13), which differ in interlayer chain stacking. When Iα and Iβ are treated with supercritical ammonia, the chain stacking arrangement changes to a completely different structure, called IIII (14). Cellulose IIII forms staggered layers with intra- and interlayer hydrogen-bonding interactions, unlike cellulose Iα, in which the chains pack into flat layers. Because of this change, cellulose IIII has a larger hydrophobic surface than cellulose Iα, making it more susceptible to attack by cellulase (15). Cellulose IIII is highly susceptible to degradation by cellulase while maintaining high crystallinity (16) and therefore is suitable for observing the degradation of crystalline cellulose by cellulase by means of HS-AFM. The hydrolytic progress curves are shown in SI Appendix, Fig. S1 A–D. The major product of CfCel6A, CfCel6B, and CfCel48A was cellobiose, whereas the major products of CfCel9A were glucose and cellotriose, in addition to cellobiose (SI Appendix, Fig. S1 E–H). CfCel6A degraded PASC and Avicel more efficiently than did the other three cellulases, while CfCel9A efficiently hydrolyzed crystalline cellulose Iα. CfCel6B degraded crystalline cellulose IIII more efficiently than did the other three enzymes. CfCel48A showed the lowest degradation activity toward insoluble cellulose substrates among the four cellulases.
In the exocellulase reaction, cellobiose is produced mainly through processive hydrolysis, whereas glucose and cellotriose are the primary products released at initial hydrolysis events. Therefore, processivity can be assessed in terms of the ratio of cellobiose/(glucose + cellotriose), as discussed previously (4, 17). Processivity ratios above 10 were achieved for all four insoluble substrates by CfCel6B and CfCel48A; these ratios are within the range measured for processive GH family 7 cellulases (4, 17). In contrast, ratios under 5 and under 0.7 were observed for CfCel6A and CfCel9A, respectively. These results suggest that CfCel6B and CfCel48A are highly processive cellulases, whereas CfCel6A and CfCel9A are less processive cellulases.
The random hydrolysis catalyzed by an endocellulase results in a greater decrease of viscosity compared with the restricted action of an exocellulase, and therefore, plots of viscosity-lowering versus incubation time during the hydrolysis of carboxymethyl cellulose (CMC) are commonly used to characterize exocellulase or endocellulase activity. The viscometric activities of the four cellulases are shown in SI Appendix, Fig. S2. CfCel6A and CfCel9A quickly reduced CMC viscosity, indicating that they are very active endocellulases. This is consistent with an earlier observation that CfCel6A acts randomly on cellulose, leading to particle fragmentation (18).
HS-AFM Observation of the Four C. fimi Cellulases.
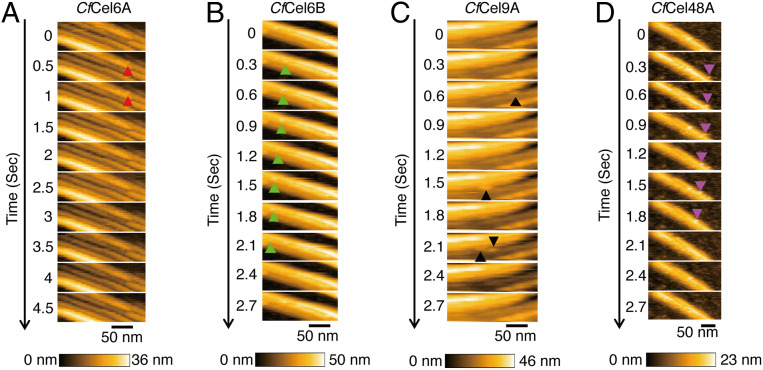
The processive movements of the four cellulases was observed by means of HS-AFM (Fig. 1). A suspension of crystalline cellulose IIII (average degree of polymerization [DP] of 880 ± 500; see SI Appendix, Fig. S3) was dropped on a highly oriented pyrographite (HOPG) disk for HS-AFM, and the cellulases were added to the observation buffer during the imaging. As shown in Fig. 1B and Movie S1, CfCel6B moved unidirectionally (from right to left in this image) on the surface of the cellulose IIII. CfCel48A also moved unidirectionally on the surface of cellulose IIII, as shown in Fig. 1D and Movie S3, but molecules moving on the substrate were rarely observed, compared to the case of CfCel6B. This is consistent with the fact that the cellulolytic activity of CfCel6B toward crystalline cellulose IIII was 170-fold higher than that of CfCel48A (SI Appendix, Fig. S1D). CfCel6B showed faster unidirectional movement and faster detachment from the cellulose IIII surface compared with TrCel7A, as described later. Furthermore, CfCel6A and CfCel9A molecules were bound to cellulose but did not move and were easily detached from the surface (Fig. 1 A and C and Movie S2 [CfCel9A]).
Fig. 1.
HS-AFM images of the four C. fimi cellulases. (A) Real-time observation of crystalline cellulose IIII incubated with 1 µM CfCel6A. The time interval between images is 0.5 s (200 × 70 nm2, 120 × 42 pixels). The positions of CfCel6A are marked by red arrowheads. (B) Real-time observation of crystalline cellulose IIII incubated with 1 µM CfCel6B. The time interval between images is 0.3 s (200 × 80 nm2, 120 × 48 pixels). These images are taken from Movie S1. The positions of CfCel6B are marked by green arrowheads. (C) Real-time observation of crystalline cellulose IIII incubated with 1 µM CfCel9A. The time interval between images is 0.3 s (200 × 80 nm2, 120 × 40 pixels). These images are taken from Movie S2. The positions of CfCel9A are marked by black arrowheads. (D) Real-time observation of crystalline cellulose IIII incubated with 1 µM CfCel48A. The time interval between images is 0.3 s (200 × 100 nm2, 120 × 50 pixels). These images are taken from Movie S3. The positions of CfCel48A are marked by purple arrowheads.
Different Orientations of Movement of CfCel6B and CfCel48A or of CfCel6B and TrCel7A.
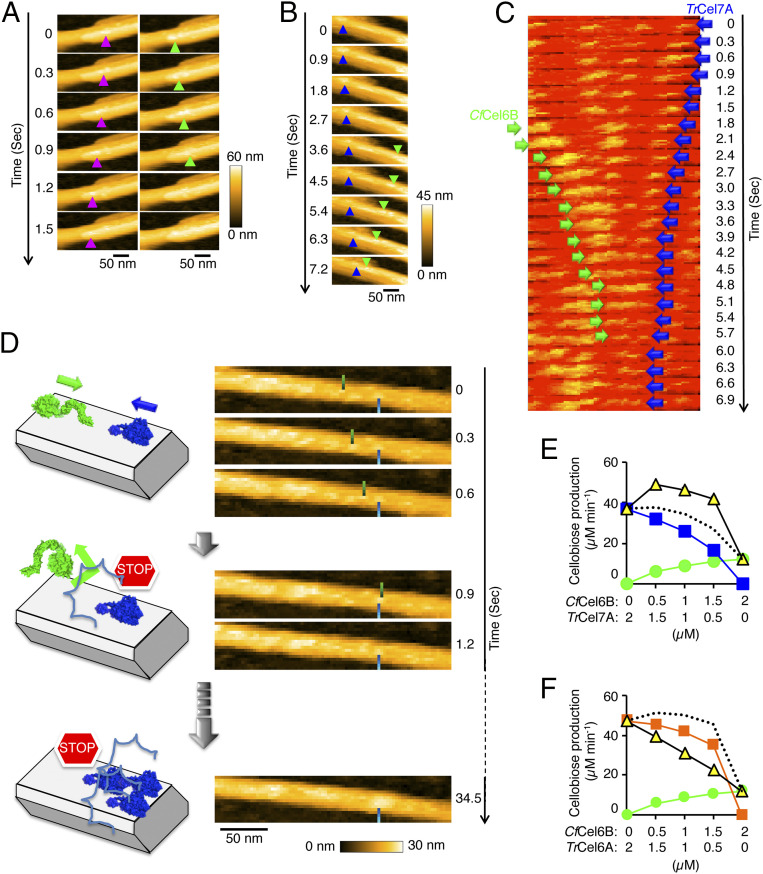
During the HS-AFM observation, enzymes can be added to the observation solution, and their real-time movements on the same microfibrils can be followed before and after the addition of another enzyme. In the present work, we directly confirmed that molecules of CfCel6B and CfCel48A or of CfCel6B and TrCel7A move in opposite directions using crystalline cellulose with parallel orientation of substrate chains (Fig. 2 A and B and Movies S4 and S5). When CfCel48A alone was initially observed, the enzyme molecules moved from the right to the left, as shown in Fig. 2A, Left, while movement from the left to the right was observed after CfCel6B was added (Fig. 2A, Right and Movie S4). Fig. 2B and Movie S5 show TrCel7A molecules moving from the left to the right, while CfCel6B molecules move in the opposite direction. The absolute direction of movement of cellulase molecules cannot be determined from HS-AFM observation, but reported kinetic data for labeled oligosaccharides (19, 20), morphological observations of crystalline cellulose after degradation by cellulases (21, 22), and the high-resolution molecular structure determination of the cellulases (23–31) all clearly indicate that TrCel7A and CfCel48A move from the reducing end to the nonreducing end, and CfCel6B moves from the nonreducing end to the reducing end.
Fig. 2.
Opposite directions of processive movement, leading to traffic accidents. (A) HS-AFM images of crystalline cellulose IIII degradation by CfCel6B and CfCel48A. The time interval between images is 0.3 s (200 × 120 nm2, 120 × 60 pixels). These images are taken from Movie S4. Purple and green arrowheads show the positions of CfCel48A and CfCel6B, respectively. The reference processive direction was taken as that when CfCel48A alone was incubated with the substrate. (B) HS-AFM images of crystalline cellulose IIII degradation by CfCel6B and TrCel7A. The time interval between images is 0.3 s (200 × 80 nm2, 120 × 48 pixels). These images are taken from Movie S5. Blue and green arrowheads show the positions of TrCel7A and CfCel6B, respectively. The reference processive direction was taken as that when CfCel6B alone was incubated with the substrate. (C) Kymographic analysis of CfCel6B and TrCel7A in a traffic accident on crystalline cellulose IIII. These images are taken from Movie S6. The time interval between images 0.3 s (254 × 75 nm2, 153 × 45 pixels). (D) Actual HS-AFM images and schematic illustration of the traffic accident between CfCel6B and TrCel7A, followed by a traffic jam. The images are taken from Movie S6. Blue and green bars show the positions of TrCel7A and CfCel6B, respectively. (E and F) Synergy and negative synergy between CfCel6B and TrCel7A or CfCel6B and TrCel6A in cellobiose production from crystalline cellulose IIII. Green circles, blue squares, and orange squares show the rates of cellobiose production by CfCel6B, TrCel7A, and TrCel6A, respectively. The yellow triangles show the activity between the two cellulases. The dotted line indicates the sum of cellobiose production calculated from the green, blue, and orange plots. n = 3.
The HS-AFM observations revealed that CfCel6B molecules showed shorter and faster movement compared with TrCel7A molecules. Sometimes CfCel6B and TrCel7A collided head-on, and in all these cases, CfCel6B was knocked off the crystalline cellulose substrate (Movie S6 and Fig. 2 C and D). We also observed that these “traffic accidents” between CfCel6B and TrCel7A triggered a “traffic jam” of TrCel7A (Movie S6 and Fig. 2D). The halting of one TrCel7A molecule blocked the following TrCel7A molecules (moving from the right to the left in Movie S6), which then stacked up behind it. However, mixing CfCel6B and TrCel7A still resulted in synergistic degradation of crystalline cellulose (Fig. 2E), whereas a combination of enzymes from the same family, i.e., CfCel6B and TrCel6A, resulted in impaired cellulose degradation, possibly because of competition between the two enzymes (Fig. 2F). A competitive effect of TrCel6A and TfCel6B (formally Thermomonospora fusca cellulase E3, bacterial GH6) in the degradation of filter paper has been reported, in contrast to a synergistic effect of the combination of TrCel7A and TfCel6B (32).
Analysis of Moving Velocity and Processivity of CfCel6B.
The movements of CfCel6B molecules were further analyzed in order to estimate their kinetic characteristics. As shown in SI Appendix, Fig. S4A, the time courses of movement of CfCel6B showed that 24.0% of observed CfCel6B molecules (total of 500 molecules) stopped after moving on the cellulose for a while and then became detached from the substrate. SI Appendix, Fig. S4B shows a histogram of the moving velocities of CfCel6B (n = 500), which exhibited a Gaussian distribution with a mean ± SD of 12.7 ± 7.6 nm s−1 (Table 1). The average movement velocity of CfCel6B was twice that of TrCel7A (6.8 ± 3.5 nm s−1) and was nearly equal to that of PcCel7C (14.7 ± 9.1 nm s−1), which shows the fastest velocity we have ever measured (4). We made a histogram of the distribution of the duration of processive movement (SI Appendix, Fig. S4C) and estimated the half-life of processive movements of CfCel6B molecules to be 1.3 ± 0.1 s. From the velocity and half-life values, the processivity of CfCel6B was estimated to be 16.1 successive reactions, taking the length of the cellobiose repeating unit as 1.0 nm in crystalline cellulose (14).
Table 1.
Comparison of statistical analysis data of CfCel6B and GH family 7 cellulases
| Cellulase | Dissociation rate constant,* s−1 | Half-life of movement,* s | Average velocity,† nm s−1 | Calculated processivity, No. of reactions |
| CfCel6B | 0.55 ± 0.02 | 1.3 ± 0.1 | 12.7 ± 7.6 | 16.1 |
| PcCel7C‡ | 0.51 ± 0.01 | 1.4 ± 0.0 | 14.7 ± 9.1 | 19.9 |
| PcCel7D‡ | 0.32 ± 0.04 | 2.2 ± 0.1 | 9.4 ± 3.7 | 20.3 |
| TrCel7A‡ | 0.20 ± 0.01 | 3.5 ± 0.2 | 6.8 ± 3.5 | 23.6 |
| CfCel6B (with TrCel7A) | 0.47 ± 0.03 | 1.5 ± 0.1 | 19.4 ± 8.1 | 28.8 |
| TrCel7A (with CfCel6B) | 0.05 ± 0.02 | 13.5 ± 4.6 | 4.6 ± 1.8 | 62.2 |
The values are mean ± SE of the fitting with exponential decay.
The values are mean ± SD for Gaussian distribution.
Data are from ref. 4.
We also analyzed the molecular movements when CfCel6B and TrCel7A were mixed and worked together and estimated the kinetic characteristics. SI Appendix, Fig. S4D shows the time courses of movement of CfCel6B (total of 625 molecules) in the presence of TrCel7A. CfCel6B molecules moved for longer in the presence of TrCel7A than in its absence, and it was rare to observe CfCel6B molecules stopping and then becoming detached (1.8% of 625 total molecules) (SI Appendix, Fig. S4D). The movement of molecules showed a Gaussian distribution with a mean ± SD of 19.4 ± 8.1 nm s−1, i.e., 1.5 times faster than the movement of CfCel6B molecules alone (SI Appendix, Fig. S4E and Table 1). The half-life of processive movements of CfCel6B in the presence of TrCel7A was estimated to be 1.5 ± 0.1 s (SI Appendix, Fig. S4F and Table 1), corresponding to ∼28.8 reactions, nearly double that in the absence of TrCel7A (16.1). In contrast, the average velocity of TrCel7A molecules (n = 214) was 4.6 ± 1.8 nm s−1 in the presence of CfCel6B (SI Appendix, Fig. S4H), which is lower than that of TrCel7A alone (6.8 ± 3.5 nm s−1) (Table 1). However, the half-life of processive movements of TrCel7A in the presence of CfCel6B was estimated to be 13.5 ± 4.6 s (SI Appendix, Fig. S4I), which is 3.9 times longer than that in the absence of CfCel6B (3.5 ± 0.2 s) and corresponds to ∼62.2 reactions, i.e., 2.6 times higher than that in the absence of CfCel6B (23.6) (Table 1). These results can well explain the synergy detected by biochemical measurement (Fig. 2E).
Discussion
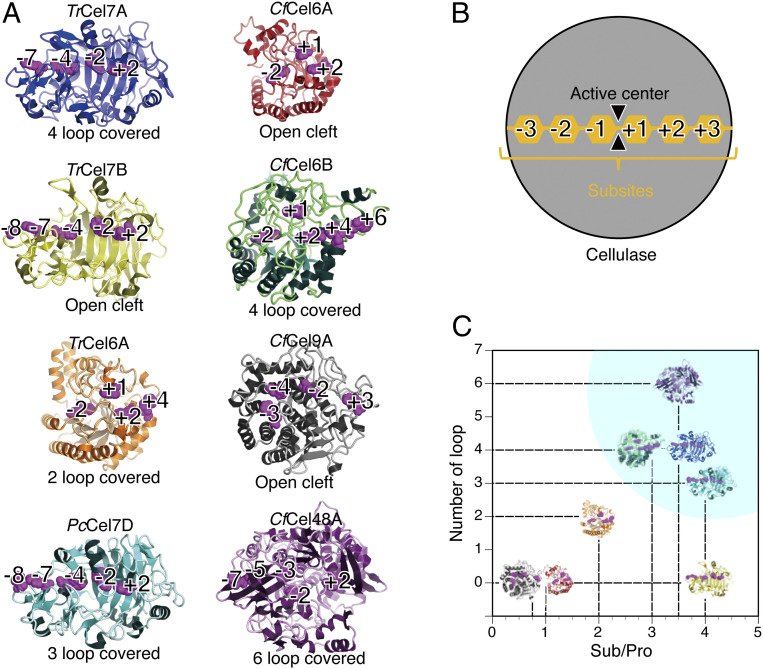
The processivity of glycanases is a critical property for effective degradation of crystalline polysaccharides. In this study, we employed HS-AFM to directly observe the movements of four bacterial cellulases from C. fimi on insoluble cellulose substrates. We observed unidirectional movement of CfCel6B (GH6) and CfCel48A (GH48) on crystalline cellulose IIII. The other two cellulases, CfCel6A (GH6) and CfCel9A (GH9), were bound to cellulose IIII but did not move on the cellulose surface. To understand these results, we compared the molecular structures of the four C. fimi cellulases with those of four previously studied fungal cellulases: TrCel7A (33), TrCel7B (34), and TrCel6A (24), derived from T. reesei, and PcCel7D (35), derived from P. chrysosporium (Fig. 3A). The structures of the four C. fimi cellulases were predicted using the Phyre2 server (36). Phyre2 is a web-based bioinformatics server for the prediction of protein structure based on the principles of homology-based modeling. Among these cellulases, processive movements of TrCel7A, PcCel7D, CfCel6B, and CfCel48A have been confirmed by HS-AFM, but processive movements of TrCel7B, TrCel6A, CfCel6A, and CfCel9A have not been detected.
Fig. 3.
(A) Structural comparison of the catalytic domains of the four fungal cellulases and four C. fimi cellulases. The fungal cellulases are T. reesei TrCel7A (PDB:1CEL), TrCel7B (PDB:1EG1), and TrCel6A (PDB:1QK2) and P. chrysosporium PcCel7D (PDB:1GPI). The catalytic domain structures of the C. fimi cellulases were predicted using Phyre2. Aromatic residues forming the subsites are shown in purple. (B) Definition of the Sub/Pro ratio. The active cleft of a cellulase consists of multiple binding sites for glucose units of cello-oligo sugars. These sites are defined as “subsites” and are designated as −3, −2, −1, active center, +1, +2, and +3 here. The subsites before the active center are defined as subsites for substrate (Sub) binding. The subsites after the active center are defined as subsites for product (Pro) binding. The Sub/Pro ratio was calculated. Cellulases which degrade substrates from the reducing end side to the nonreducing end of sugar chains are expressed as −3, −2, and −1 (Sub); active center; and +1, +2, and +3 (Pro). Cellulases that degrade substrates from the nonreducing end to the reducing end are expressed as +3, +2, and +1 (Sub); active center; and −1, −2, and −3 (Pro). (C) Structural classification of the eight cellulase catalytic domains. The vertical axis shows the number of loops covering the catalytic cleft. The horizontal axis shows the Sub/Pro ratio, which is the ratio of the number of binding subsites for Sub divided by the number of binding subsites for Pro. The Sub/Pro ratios are TrCel7A, 3.5; TrCel7B, 4.0; PcCel7D, 4.0; TrCel6A, 2.0; CfCel6A, 1.0; CfCel6B, 3.0; CfCel9A, 0.75; and CfCel48A, 3.5. Blue highlighting indicates the area observed by HS-AFM.
The difference in processivity between the two bacterial GH6 cellulases CfCel6A and CfCel6B is presumably due to their structural difference (Fig. 3A). As proposed by Varrot et al., GH family 6 cellulases can be clustered in four subfamilies based on amino acid sequence similarity (37), and CfCel6A belongs to subfamily 2, while CfCel6B belongs to subfamily 4. Among subfamily 2 cellulases, the structures of MtCel6A (amino acid identity with the catalytic domain of CfCel6A of 47%) and TfCel6A (identity of 43%) have been solved (37, 38), and these two molecules possess shortened loops or lack two loops that cover the active cleft in exotype TrCel6A (Fig. 3A); consequently, they exhibit open-cleft active sites that are typical of GH6 endoglucanases (37, 38). Moreover, GH6 cellulases in subfamily 2 lack the +4 tryptophan subsite that plays an essential role in crystalline cellulose degradation by the fungal family 6 cellulase TrCel6A (39). In contrast, subfamily 4 cellulase CfCel6B, which has 53% amino acid identity with the catalytic domain of TfCel6B, has the +4 subsite, and there are four loops covering the active sites, producing a tunnel-shaped active-site structure (29, 30). There is also a +6 subsite. These differences between CfCel6A and CfCel6B seem consistent with the observed difference in processivity (Fig. 3A). Although our previous HS-AFM study of TrCel6A demonstrated that the processivity of the fungal family 6 enzyme is less than that of family 7 cellulase (3), the present work clearly demonstrates a strong processive movement of CfCel6B, suggesting that the presence of four loops that cover the active-site cleft and construct a longer tunnel than in TrCel6A could be important for enabling GH6 subfamily 4 cellulases to grip the cellulose chain well and thus to achieve increased processivity.
Our previous study of the processivity of three fungal GH7 CBHs revealed that the processivity is strongly influenced by the presence and nature of the loop regions covering the active-site cleft (4). The high processivity observable with HS-AFM seems to be associated with the presence of three or more loops covering the active cleft and a substrate-binding/product-binding subsite (Sub/Pro) ratio of 3 or more (Fig. 3C). Cellulases without a loop covering the active cleft, such as CfCel9A, CfCel6A, and TrCel7B, can all be classified biochemically as endotype cellulases. For two GH7 cellulases, PcCel7D and TrCel7B, which have the same Sub/Pro ratio, the presence or absence of the loop covering the active cleft is considered to lead to the presence or absence of processivity, respectively. Interestingly, there is a clear difference between these processive cellulases and GH18 chitinases, which exhibit processivity, as observed by HS-AFM, even though the chitinases have no loop covering the active cleft (40). The Sub/Pro ratio in the catalytic domain of GH18 chitinases is only about 2, indicating that the catalytic domain itself cannot show processivity, at least according to our Sub/Pro ratio vs. number of loops analysis. However, they are connected to a chitin-binding module via a short linker peptide, and the connected two-domain structure extends the subsites and affords a Sub/Pro ratio of more than 5, which is consistent with higher processivity. In fact, the higher Sub/Pro ratio of chitinase A (ChiA) than chitinase B (ChiB) is reflected in higher processivity of ChiA than ChiB. Thus, in cellulases, the processivity arises from the combination of loops covering the active cleft and the Sub/Pro ratio of the catalytic domain, whereas chitinases acquire processivity only via the Sub/Pro ratio. It should be noted that chitin is a polymer of acetylglucosamine, which has an acetoamide modification at the C2 position of glucose and thus is bulkier than cellulose, a simple glucose polymer. This could be the reason why the active cleft of chitinase is not covered with loops.
Next, we employed HS-AFM observation to see what kinds of phenomena occur at the single-molecule level to produce the synergistic effect of the combination of GH6 and GH7 cellulases. Statistical analysis revealed that in the presence of both CfCel6B and TrCel7A, the moving distances of the two cellulases on the substrate became longer, and the processivity of both enzymes was increased. This may imply that the use of two complementary types of cellulases is effective for removing obstacles on the cellulose crystal surface. This, in turn, might increase the area of the cellulose surface accessible to the degradative enzymes, as well as reducing the frequency of their detachment from the cellulose surface, thus promoting decomposition of the cellulose chain (Fig. 2E) and increasing the processivity (Table 1). An alternative explanation could be that once a chain has been removed from the surface by the action of one enzyme, the neighboring chains are less tightly held together and thus are more accessible to the other enzyme. In addition, it was observed that CfCel6B sometimes collided with TrCel7A, causing a traffic jam (Fig. 2D and Movie S6). T. reesei is thought to degrade crystalline cellulose by employing the combination of TrCel7A and TrCel6A, both of which are thought to be processive CBHs. However, we could not observe processivity of TrCel6A by means of HS-AFM, which may mean that the retaining force on the cellulose surface is weaker than that of TrCel7A or CfCel6B. This situation could have evolved to prevent the occurrence of immovable traffic jams because TrCel6A would be knocked off the substrate when a collision occurs. The combination of CfCel6B and TrCel7A did show collisions resulting in congestion, which may be the reason why the synergistic effect between CfCel6B and TrCel7A (1.4 times, Fig. 2E) is less than that between TrCel6A and TrCel7A [2.3 times (3)]. Similarly, C. fimi secretes CfCel9A, which is considered to be the major endoprocessive enzyme, but no processive action was observed with HS-AFM, suggesting that the relationship between CfCel9A and CfCel6B may be similar to that between TrCel7A and TrCel6A from T. reesei. Although T. reesei and C. fimi are completely different types of microorganisms (eukaryote and prokaryote, respectively), they both share a combination of cellulases with high and low processivity, clearly indicating that such a combination is favorable for the deconstruction of cellulose. However, it is noteworthy that in C. fimi, the highly processive CfCel6B moves from the nonreducing to the reducing end, while in T. reesei, the highly processive TrCel7A moves in the opposite direction, from the reducing to the nonreducing end (Fig. 2B).
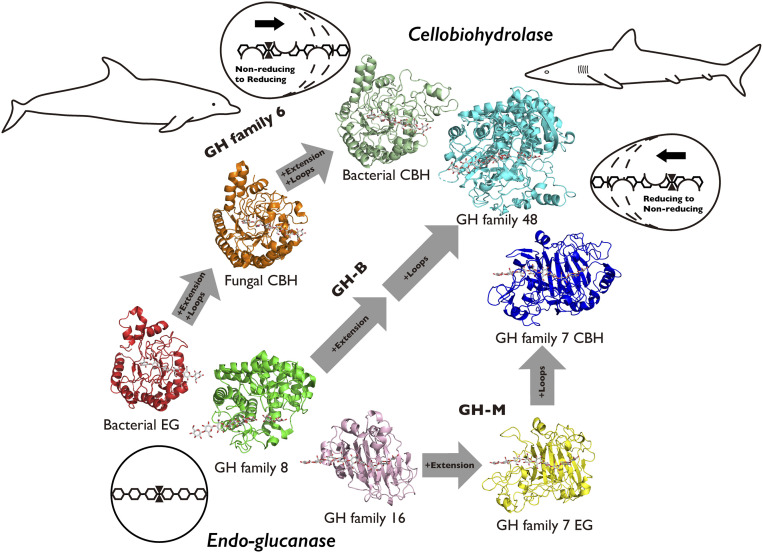
A comparison of GH6 cellulases with different processivities clearly indicated that increased processivity is favored by 1) an increase in the number of subsites available for substrate binding (41), 2) the presence of an asymmetric active site with a decreased number of subsites for the product (39), and 3) an increase in the number of loop structures to hold the substrate more tightly in the active site (29), as shown in Fig. 4. The importance of these features is supported by the results of a molecular phylogenetic analysis of GH6 cellulases (SI Appendix, Fig. S5). The 3D structures of two GH7 cellulases and one GH16 endo-β-1,3-1,4-glucanase are compared in Fig. 4. The two GH families are classified into clan GH-B, sharing the β-jellyroll fold (42). PtLic16A, a GH16 β-1,3-1,4-glucanase from Paecilomyces thermophila, has an almost symmetrical subsite (Sub/Pro ratio of 5/4) with no loop covering the active center (43). However, in GH7 endocellulase TrCel7B the subsite is asymmetric (Sub/Pro ratio of 8/2), while in TrCel7A the active center is covered with loops (23). Molecular phylogenetic analysis of GH7 cellulases and GH16 β-1,3-1,4-glucanases also supports the significance of these features (SI Appendix, Fig. S6). The structures of GH8 cellulase and GH48 cellulase are compared in Fig. 4. The two GH family enzymes are both classified in clan GH-M, sharing the (α/α)6 fold. GH8 from Clostridium thermocellum (HtCel8A) has a symmetrical subsite (Sub/Pro ratio of 3/3) with no loop covering the active center (44), while GH48 cellulase from Clostridium cellulolyticum (RcCel48F) has an asymmetric subsite (Sub/Pro ratio of 7/3), and the active center is covered with loops (26). The results of molecular phylogenetic analysis of GH8 β-1,3-1,4-glucanases and cellulases and GH48 cellulases also support the relationship of higher processivity with increasing numbers of subsites in the active site and loop structures over the active site (SI Appendix, Fig. S7). For example, the structure of CfCel6B with four loops and a Sub/Pro ratio of 6/2 is quite similar to that of TrCel7A, which has four loops and a Sub/Pro ratio of 8/2, even though their degradation directions are opposite to each other. Bacteria do not have GH7 cellulases, but they have evolved GH6 cellulases, which have different protein foldings, into highly processive cellulases that can degrade recalcitrant crystalline cellulose.
Fig. 4.
Schematic representation of the putative convergent evolution of bacterial and fungal cellulases, employing different protein foldings. Modification of the symmetric architecture at positive and negative subsites in endoglucanases to generate asymmetric architecture by blocking part of the product binding region and/or by extending the substrate binding subsites increases the processivity (lower left to middle part). The presence of loops covering the active site increases the processivity (upper right). A combination of additional loops and asymmetricity results in high processivity for cellobiohydrolases.
The well-known concept of “convergent evolution,” which is often applied to explain the shape of animals, such as sharks and dolphins having a long nose and streamlined body for effective movement in water even though they were originally different animals (fish and mammalian), also seems applicable to the changes in the shape of endoglucanase to generate a processive cellulase. Thus, the GH6, GH48, and GH7 processive cellulases all utilize common strategies: extending the protein “nose” in the substrate direction to form asymmetric subsite architecture and covering the subsite with loops. Degradation of crystalline cellulose is essential for both fungi and bacteria, and processive CBHs are necessary for this purpose. However, since fungi and bacteria produce glycosyl hydrolases with different folding types, they have evolved processive CBHs using different protein folds as templates. In other words, completely different evolutionary strategies of cellulolytic enzymes in bacteria and fungi have yielded functionally similar and highly processive cellulases, providing an interesting example of “protein-level convergent evolution” in cellulolytic microorganisms.
Methods
Materials.
Crystalline celluloses Iα and IIII were prepared from green algae Cladophora spp. according to a previous report (14). PASC was prepared from Avicel (Funacel II; Funakoshi Chemical) according to a previous report (4). CMC was purchased from Hercules. Cellooligosaccharides were acquired from Seikagaku Kogyo. Cellulomonas fimi ATCC 484 was used as a source of CfCel6A, CfCel6B, CfCel9A, and CfCel48A genes. Trichoderma reesei ATCC 66589 was used as a source of the TrCel6A gene. Escherichia coli JM109 (Takara) was used as the subcloning host. E. coli Rosetta2 (DE3), Tuner (DE3) (Novagen), and Brevibacillus choshinensis HPD31-SP3 (Takara) were used for heterologous production of the recombinant proteins.
Cloning of Four Bacterial Cellulases from C. fimi.
Four C. fimi cellulase genes were amplified by PCR. The CfCel6A gene and CfCel6B gene were cloned into the E. coli expression vector. The CfCel9A gene and CfCel48A gene were cloned into the B. choshinensis expression vector. For details, see SI Appendix.
Production and Purification of Four Bacterial Cellulases from C. fimi.
Four recombinant C. fimi cellulases were purified (SI Appendix, Fig. S8) according to the following protocol, as described in detail in SI Appendix.
The concentrations of the four cellulases were determined using the following values of molecular absorption coefficient at 280 nm: 107,745 M−1 cm−1 for CfCel6A, 134,105 M−1 cm−1 for CfCel6B, 217,875 M−1 cm−1 for CfCel9A, and 280,325 M−1 cm−1 for CfCel48A. Calculations were performed using the ProtParam tool (45).
Enzyme Assay.
Enzyme activity toward insoluble celluloses was determined in a 150 µL reaction mixture containing 100 mM sodium acetate buffer (pH 5.0), 1.0 mg mL−1 substrate, and 1 µM enzyme at 37 °C. The reaction was stopped by heating the sample at 98 °C for 5 min. Soluble sugars were recovered by passing the reaction mixture through a 0.22 µm Durapore polyvinylidene difluoride (PVDF) membrane filter (MultiScreen HTS-GV; Merck Millipore) and were analyzed by HPLC (LC-2000 series; Jasco) using a Corona charged aerosol detector (Corona CAD; ESA Biosciences) and a prepacked Shodex Asahipak NH2P-50 column (4.6 × 250 mm; Showa Denko K. K.). The eluent was water with a linear gradient of CH3CN from 60 to 40% over 12.5 min. The column was kept at 40 °C and eluted at a flow rate of 1.0 mL min−1. The amounts of glucose oligomers up to cellohexaose were identified and quantitated by comparing the retention times and peak areas with those of authentic standards. Concentrations of soluble sugars in the reaction mixtures were fitted to the equations previously described (4) to calculate the production velocity of soluble sugars. The ratio of cellobiose/glucose + cellotriose production was estimated according to a previous report (17).
Viscometric Assay.
Enzymatic hydrolysis of CMC was measured on an Automated Micro Viscometer (AMVn; Anton Paar) based on the rolling time of a ball through viscous liquids according to the Höppler principle. The AMVn measuring system employed a 1.6 mm diameter glass capillary and a 1.5 mm diameter steel ball (density; 7.67 g/cm3). Calibration parameters were 0.797 mPa s for dynamic viscosity, 0.99336 g/cm3 for buffer density, 20° for minimum angle, and 80° for maximum angle. The reaction mixture contained 20 mM sodium acetate buffer (pH 5.0), 5.0 mg mL−1 CMC, and 10 nM enzyme, and the enzyme reaction was performed at 37 °C.
High-Speed AFM Observation.
The movements of molecules of the four bacterial cellulases and TrCel7A were observed by high-speed atomic force microscopy as described in previous reports (2–4, 40, 46). A suspension (1 mg mL−1) of highly crystalline cellulose IIII was dropped on a stage of HOPG and incubated at 25 °C for 10 min. Unbound cellulose IIII was washed off with 100 mM sodium acetate buffer (pH 5.0). Then the HOPG substrate was immersed in the observation pool filled with the same buffer (volume of 76 µL), and 4 µL of 20 µM cellulase solution were added to the observation pool. We observed molecules whose movement started and ended within the observation area. More than five independent cellulose microfibrils with a length greater than 0.8 µm were observed to estimate the velocity and processivity of cellulases.
For statistical analysis of the moving velocities of individual cellulase molecules, semiautomatic tracking of each molecule was carried out with a kymograph (40). The moving velocities of cellulase molecules were analyzed by IGOR Pro-6 (WaveMetrics) and fitted to a Gaussian distribution. Histogram data were tested for normality using the Shapiro–Wilk test (Prism7, GraphPad). Values satisfied the Shapiro–Wilk test, ensuring a normal distribution (P ≥ 0.05). Adsorption times of molecules were calculated from the number of frames in which the molecules were observed, and plots of the number of molecules in each range of adsorption time were fitted to the previously reported equation (4, 40).
Determination of the Degree of Polymerization of Cellulose IIII.
The DP of cellulose IIII was estimated from the fiber lengths measured in HS-AFM scanning images (scanning area: 0.64 to 9.00 µm2). A histogram of fiber lengths (n = 363) was created from the results. Histogram data were tested for normality using the Shapiro–Wilk test (Prism7, GraphPad). Values satisfied the Shapiro–Wilk test, ensuring a normal distribution (P ≥ 0.05). The DP was calculated by taking the length of the cellobiose repeating unit in crystalline cellulose IIII to be 1.0 nm (14).
Phylogenetic Tree Analysis.
The amino acid sequences of the enzymes belonging to each GH family whose function is known were collected from the Carbohydrate-Active enZymes (CAZy) database (47). The obtained amino acid sequences were subjected to BlastP analysis, and homologous sequences [conserved protein domain family (48)] representing each GH family were excised. These sequences were subjected to phylogenetic analysis. Sequence alignment and phylogenetic analysis were performed using the Phylogeny.fr web server (49). The “one-click” analysis mode was applied to each set of sequences, using MUSCLE (version 3.5) (50) for multiple sequence alignment, PhyML (version 3.0) (51) and aLRT (52) for phylogenetic analysis, and TreeDyn (53) for tree rendering. Branches with aLRT branch support values under 50% were collapsed.
Supplementary Material
Acknowledgments
This research was supported by Grants-in-Aid for Innovative Areas (Grant 18H05494 to K.I. and Grant 18H04512 and 19H05389 to T. Uchihashi) from the Japanese Ministry of Education, Culture, Sports and Technology and by Grants-in-Aid for Scientific Research (B: Grant 18H02252 to K.I. and Grant 18H01837 to T. Uchihashi; C: Grant 15K07383 to T. Uchiyama) from the Japan Society for the Promotion of Science. K.I. thanks the Finnish Funding Agency for Innovation for the support of the Finland Distinguished Professor Program “Advanced approaches for enzymatic biomass utilisation and modification (BioAD).”
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011366117/-/DCSupplemental.
Data Availability.
All data obtained and analyzed in the present work are available in SI Appendix and Movies S1 to S6.
References
- 1.Park S., Baker J. O., Himmel M. E., Parilla P. A., Johnson D. K., Cellulose crystallinity index: Measurement techniques and their impact on interpreting cellulase performance. Biotechnol. Biofuels 3, 10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Igarashi K. et al., High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J. Biol. Chem. 284, 36186–36190 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Igarashi K. et al., Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333, 1279–1282 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Nakamura A. et al., Trade-off between processivity and hydrolytic velocity of cellobiohydrolases at the surface of crystalline cellulose. J. Am. Chem. Soc. 136, 4584–4592 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Nakamura A. et al., Single-molecule imaging analysis of binding, processive movement, and dissociation of cellobiohydrolase Trichoderma reesei Cel6A and its domains on crystalline cellulose. J. Biol. Chem. 291, 22404–22413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong W. K. R. et al., Characterization and structure of an endoglucanase gene cenA of Cellulomonas fimi. Gene 44, 315–324 (1986). [DOI] [PubMed] [Google Scholar]
- 7.Meinke A., et al. , Cellobiohydrolase A (CbhA) from the cellulolytic bacterium Cellulomonas fimi is a β-1,4-exocellobiohydrolase analogous to Trichoderma reesei CBH II. Mol. Microbiol. 12, 413–422 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Owolabi J. B., Beguin P., Kilburn D. G., Miller R. C. Jr., Expression in Escherichia coli of the Cellulomonas fimi structural gene for endoglucanase B. Appl. Environ. Microbiol. 54, 518–523 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen H., Gilkes N. R., Kilburn D. G., Miller R. C. Jr., Warren R. A. J., Cellobiohydrolase B, a second exo-cellobiohydrolase from the cellulolytic bacterium Cellulomonas fimi. Biochem. J. 311, 67–74 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandercock L. E., Meinke A., Gilkes N. R., Kilburn D. G., Warren R. A. J., Degradation of cellulases in cultures of Cellulomonas fimi. FEMS Microbiol. Lett. 143, 7–12 (1996). [Google Scholar]
- 11.Sakon J., Irwin D., Wilson D. B., Karplus P. A., Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 4, 810–818 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Atalla R. H., Vanderhart D. L., Native cellulose: A composite of two distinct crystalline forms. Science 223, 283–285 (1984). [DOI] [PubMed] [Google Scholar]
- 13.VanderHart D. L., Atalla R. H., Studies of microstructure in native cellulose using solid-state 13 C NMR. Macromolecules 17, 1465–1472 (1984). [Google Scholar]
- 14.Wada M., Chanzy H., Nishiyama Y., Langan P., Cellulose IIII crystal structure and hydrogen bonding by synchrotron X-ray and neutron fiber diffraction. Macromolecules 37, 8548–8555 (2004). [Google Scholar]
- 15.Igarashi K., Wada M., Samejima M., Activation of crystalline cellulose to cellulose IIII results in efficient hydrolysis by cellobiohydrolase. FEBS J. 274, 1785–1792 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Wada M., Heux L., Sugiyama J., Polymorphism of cellulose I family: Reinvestigation of cellulose IVI. Biomacromolecules 5, 1385–1391 (2004). [DOI] [PubMed] [Google Scholar]
- 17.von Ossowski I. et al., Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A. A comparison with Phanerochaete chrysosporium Cel7D. J. Mol. Biol. 333, 817–829 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Stålbrand H. et al., Analysis of molecular size distributions of cellulose molecules during hydrolysis of cellulose by recombinant Cellulomonas fimi β-1,4-glucanases. Appl. Environ. Microbiol. 64, 2374–2379 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vrsanská M., Biely P., The cellobiohydrolase I from Trichoderma reesei QM9414: Action on cello-oligosaccharides. Carbohydr. Res. 227, 19–27 (1992). [Google Scholar]
- 20.Barr B. K., Hsieh Y.-L., Ganem B., Wilson D. B., Identification of two functionally different classes of exocellulases. Biochemistry 35, 586–592 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Chanzy H., Henrissat B., Unidirectional degradation of Valonia cellulose microcrystals subjected to cellulase action. FEBS Lett. 184, 285–288 (1985). [Google Scholar]
- 22.Imai T., Boisset C., Samejima M., Igarashi K., Sugiyama J., Unidirectional processive action of cellobiohydrolase Cel7A on Valonia cellulose microcrystals. FEBS Lett. 432, 113–116 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Divne C., Ståhlberg J., Teeri T. T., Jones T. A., High-resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J. Mol. Biol. 275, 309–325 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Zou J. Y., et al. , Crystallographic evidence for substrate ring distortion and protein conformational changes during catalysis in cellobiohydrolase Cel6A from Trichoderma reesei. Structure 7, 1035–1045 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Varrot A., Schülein M., Davies G. J., Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase, Cel6A, upon oligosaccharide binding. Biochemistry 38, 8884–8891 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Parsiegla G. et al., Crystal structures of the cellulase Cel48F in complex with inhibitors and substrates give insights into its processive action. Biochemistry 39, 11238–11246 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Guimarães B. G., Souchon H., Lytle B. L., David Wu J. H., Alzari P. M., The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome. J. Mol. Biol. 320, 587–596 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Ubhayasekera W., Muñoz I. G., Vasella A., Ståhlberg J., Mowbray S. L., Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors. FEBS J. 272, 1952–1964 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Wu M. et al., Loop motions important to product expulsion in the Thermobifida fusca glycoside hydrolase family 6 cellobiohydrolase from structural and computational studies. J. Biol. Chem. 288, 33107–33117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandgren M. et al., The structure of a bacterial cellobiohydrolase: The catalytic core of the Thermobifida fusca family GH6 cellobiohydrolase Cel6B. J. Mol. Biol. 425, 622–635 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Kostylev M. et al., Cel48A from Thermobifida fusca: Structure and site directed mutagenesis of key residues. Biotechnol. Bioeng. 111, 664–673 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Irwin D. C., Spezio M., Walker L. P., Wilson D. B., Activity studies of eight purified cellulases: Specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42, 1002–1013 (1993). [DOI] [PubMed] [Google Scholar]
- 33.Divne C. et al., The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science 265, 524–528 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Kleywegt G. J. et al., The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 A resolution, and a comparison with related enzymes. J. Mol. Biol. 272, 383–397 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Muñoz I. G., et al. , Family 7 cellobiohydrolases from Phanerochaete chrysosporium: Crystal structure of the catalytic module of Cel7D (CBH58) at 1.32 Å resolution and homology models of the isozymes. J. Mol. Biol. 314, 1097–1111 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varrot A. et al., Mycobacterium tuberculosis strains possess functional cellulases. J. Biol. Chem. 280, 20181–20184 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Spezio M., Wilson D. B., Karplus P. A., Crystal structure of the catalytic domain of a thermophilic endocellulase. Biochemistry 32, 9906–9916 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Koivula A. et al., Tryptophan 272: An essential determinant of crystalline cellulose degradation by Trichoderma reesei cellobiohydrolase Cel6A. FEBS Lett. 429, 341–346 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Igarashi K. et al., Two-way traffic of glycoside hydrolase family 18 processive chitinases on crystalline chitin. Nat. Commun. 5, 3975 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Larsson A. M. et al., Crystal structure of Thermobifida fusca endoglucanase Cel6A in complex with substrate and inhibitor: The role of tyrosine Y73 in substrate ring distortion. Biochemistry 44, 12915–12922 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Henrissat B., Bairoch A., Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316, 695–696 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng Y.-S., et al. , Structural and mutagenetic analyses of a 1,3-1,4-β-glucanase from Paecilomyces thermophila. Biochm. Biophys. Acta 1844, 366–373 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Guérin D. M. A. et al., Atomic (0.94 A) resolution structure of an inverting glycosidase in complex with substrate. J. Mol. Biol. 316, 1061–1069 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Gasteiger E. et al., . “Protein identification and analysis tools on the ExPASy server” in The Proteomics Protocols Handbook, Walker J. M., Ed. (Springer, Totowa, NJ, 2005), pp. 571–607. [Google Scholar]
- 46.Igarashi K. et al., Visualization of cellobiohydrolase I from Trichoderma reesei moving on crystalline cellulose using high-speed atomic force microscopy. Methods Enzymol. 510, 169–182 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B., The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu S. et al., CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dereeper A. et al., Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guindon S. et al., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Anisimova M., Gascuel O., Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Chevenet F., Brun C., Bañuls A. L., Jacq B., Christen R., TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7, 439 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data obtained and analyzed in the present work are available in SI Appendix and Movies S1 to S6.