Significance
Parvoviruses (PVs) are ssDNA viruses, with T = 1 icosahedral symmetry, infecting deuterostome and protostome animals. Most PVs have a highly conserved phospholipase A2 domain (PLA2) in the N-terminal region of their minor capsid protein. Under acidic pH, during endosomal/lysosomal egress, the PLA2 domain is activated to disrupt vesicle membranes. However, certain PVs lack the PLA2 and thus must use a different escape mechanism. Our study offers insight into this enigma, showing how a recently discovered PV of marine crustacean has evolved a cation-dependent mechanism to accomplish this task. We also show how host-driven convergent evolution pushed two PVs, infecting the same host species, to adopt strikingly similar surface morphologies, despite distinct multimer interactions and lack of sequence similarity.
Keywords: Crustacea, capsid structure, Parvoviridae, densovirus, convergent evolution
Abstract
The giant tiger prawn (Penaeus monodon) is a decapod crustacean widely reared for human consumption. Currently, viruses of two distinct lineages of parvoviruses (PVs, family Parvoviridae; subfamily Hamaparvovirinae) infect penaeid shrimp. Here, a PV was isolated and cloned from Vietnamese P. monodon specimens, designated Penaeus monodon metallodensovirus (PmMDV). This is the first member of a third divergent lineage shown to infect penaeid decapods. PmMDV has a transcription strategy unique among invertebrate PVs, using extensive alternative splicing and incorporating transcription elements characteristic of vertebrate-infecting PVs. The PmMDV proteins have no significant sequence similarity with other PVs, except for an SF3 helicase domain in its nonstructural protein. Its capsid structure, determined by cryoelectron microscopy to 3-Å resolution, has a similar surface morphology to Penaeus stylirostris densovirus, despite the lack of significant capsid viral protein (VP) sequence similarity. Unlike other PVs, PmMDV folds its VP without incorporating a βA strand and displayed unique multimer interactions, including the incorporation of a Ca2+ cation, attaching the N termini under the icosahedral fivefold symmetry axis, and forming a basket-like pentamer helix bundle. While the PmMDV VP sequence lacks a canonical phospholipase A2 domain, the structure of an EDTA-treated capsid, determined to 2.8-Å resolution, suggests an alternative membrane-penetrating cation-dependent mechanism in its N-terminal region. PmMDV is an observed example of convergent evolution among invertebrate PVs with respect to host-driven capsid structure and unique as a PV showing a cation-sensitive/dependent basket structure for an alternative endosomal egress.
Densoviruses (DVs) are autonomous parvoviruses (PVs) of the family Parvoviridae infecting invertebrates. Until recently, all known PVs infecting invertebrate hosts were members of Densovirinae; however, they have recently been divided into two separate subfamilies: Densovirinae, composed of exclusively invertebrate-infecting PVs, and Hamaparvovirinae, which infect both invertebrates and vertebrates (1). The third Parvoviridae subfamily, Parvovirinae, contains exclusively vertebrate-infecting PVs. All PVs are nonenveloped, single-stranded DNA (ssDNA) viruses, with an approximate capsid diameter of 21.5 to 25 nm (2). They package relatively small genomes 3.9 to 6.3 kb, flanked by two inverted terminal repeat (ITR)-containing palindromic sequences forming various hairpin-shaped secondary structures. The genome organization is conserved and includes two major ORF expression cassettes. Conventionally these are referred to as rep, which encodes the nonstructural (NS) proteins, and cap, which encodes the capsid viral proteins (VPs), which may have different N-terminal extensions (2). Most Parvoviridae contain a phospholipase A2 (PLA2) domain in the N-terminal region of their VP1, which breach the endosomal membrane during cellular trafficking (3, 4). DVs are pathogenic for their hosts (5) and were isolated from both proto- and deuterostome invertebrates, mostly insects and other arthropods (6–18).
To date, parvovirus crustacean pathogens comprise three distinct lineages with divergent genome organizations and transcription patterns. Cherax quadricarinatus DV, isolated from the freshwater red-clawed crayfish (Cherax quadricarinatus), has an ambisense genome with a PLA2 domain and is now an assigned member of genus Aquambidensovirus of the Densovirinae (19). Genera Hepanhamaparvovirus and Penstylhamaparvovirus are members of subfamily Hamaparvovirinae, each with one species that lacks a PLA2 domain. They both infect penaeid shrimps, including Penaeus monodon and Litopenaeus stylirostris (20–26). Hepanhamaparvoviruses possess a larger genome ∼6.3 kb with 220-nt-long ITRs that form hairpins. The Penstylhamaparvovirus genome is only 3.9-kb long and instead of hairpins harbors direct terminal repeats (27), an exception among PVs.
In contrast to the ∼100 capsid structures determined for members of the Parvovirinae, there are only four high-resolution structures for invertebrate-infecting PVs (28). These include three members of the Densovirinae, Galleria mellonella DV (GmDV) of genus Protoambidensovirus at a resolution of 3.7 Å (29), Acheta domestica DV (AdDV) of genus Scindoambidensovirus at 3.5-Å resolution (30), and Bombyx mori DV 1 (BmDV1) of Iteradensovirus at 3.1-Å resolution (31), and one member of the Hamaparvovirinae, Penaeus stylirostris DV (PstDV) of genus Penstylhamaparvovirus, at a resolution of 2.5 Å (32). All PVs structures are T = 1 icosahedral capsids (point group operator 5.3.2), consisting of 60 VP subunits. The VP core is structurally conserved with a jellyroll fold (33) flanked by loops inserted between the β-strands of the jellyroll, and strands and helices, forming the surface morphology. In the case of the Parvoviridae, the BIDG sheet of the jellyroll is complemented with an additional β-strand, strand A (28). The PV fivefold axis assembly forms a channel-like opening reported to aid genome packaging and uncoating, and PLA2 domain externalization when required (34, 35).
This study reports the complete genome sequence, expression strategy, and near-atomic 3D structure of a DV, designated P. monodon metallodensovirus (PmMDV), isolated from P. monodon shrimp. Its relationship to other PVs by phylogenetic inference, transcription mapping, and expression analysis were also characterized. The PmMDV and PstDV capsids have convergently evolved similar morphologies. However, PmMDV incorporates unique strategies to stabilize its capsid and it has possibly evolved an alternative membrane-penetrating mechanism in the absence of the PLA2 that is dependent on divalent cations. Furthermore, PmMDV, as the third distinct lineage of PVs to infect penaeid shrimps, cannot be assigned to any of the current subfamilies and provides insights into PV capsid evolution.
Results
Virus Detection and Cloning.
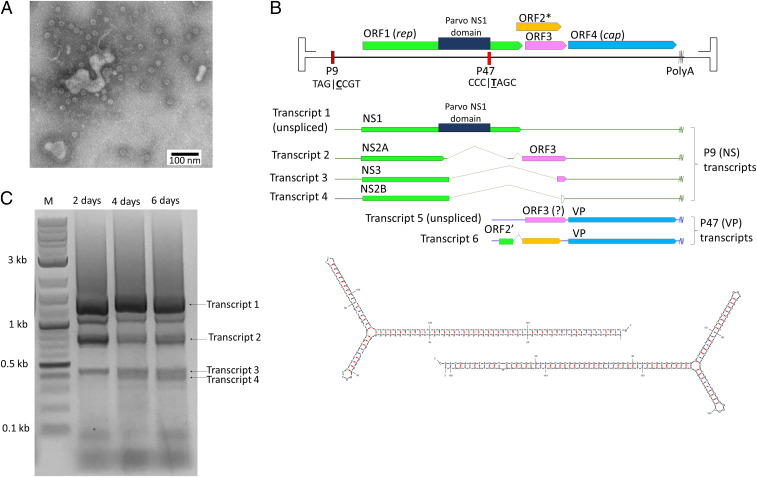
Deceased P. monodon specimens showing clinical signs of a red telson, uropodia, and pleopods, were acquired from a farm in the South Vietnam. Discoloration of the cephalothorax was observed, suggesting an underlying viral infection. Negative-staining electron microscopy (EM) from homogenized tissue revealed uniform ∼21-nm icosahedral particles (Fig. 1A). Consequently, extracted DNA was blunt-ended, cloned, and sequenced; it contained a previously unknown crustacean DV, designated PmMDV.
Fig. 1.
Discovery, genome organization, and transcription analysis of PmMDV. (A) Homogenized tissue of infected P. monodon specimens diluted and examined by negative-staining EM, showing icosahedral particles. (B) The complete genome and transcription pattern of PmMDV. The ITRs are shown by the thinner lines compared to the thick line indicating the coding region of the genome. The noncoding regions of the transcripts are marked as horizontal lines. ORFs, as well as the putative proteins expressed by them, are shown as arrows of various colors. The pattern of the ORF corresponds with the pattern of the protein-coding region of mRNA transcripts. Transcripts of the upstream promoter p9 (nonstructural) are highlighted in green, whereas transcripts of the downstream promoter p47 (structural) are in blue. The panel below displays the secondary structure of the terminal hairpins of PmMDV. (C) Agarose gel of the PCR products from rep transcripts using a forward primer at position 1,100 and a reverse primer at 2,564. RNA was extracted 2, 4, and 6 d posttransfection.
Complete Genome Characterization of the Crustacean DV.
The complete genome sequence of PmMDV was deposited into GenBank under the accession number of MK028683 (Fig. 1B). Its length was 4,374 nt, flanked by ITRs of 416 nt, of which 161 nt fold into a regular, T-shaped hairpin (Fig. 1B) (36). There was a single nucleotide insertion in the stem of the right ITR, which was present in all three clones sequenced. The overall GC content of the genome was 45.6%, with 76.4% at the termini, presumably stabilizing the secondary structure. In silico analysis revealed three ORFs, with a length greater than 100 nt, as well as a fourth without a canonical start codon (summarized in SI Appendix, Table S1). The leftmost ORF, ORF1 (516 aa), displayed sequence identity with the major NS proteins of various PVs and bidensoviruses (family Bidnaviridae) with no greater coverage than 34% (B. mori bidensovirus 3; National Center for Biotechnology Information [NCBI] protein ID: ALJ76088) and identity less than 36% (Simian parvo-like virus 3; NCBI protein ID: APC23168), hence judged to encode the NS proteins. ORF2 and ORF3 (143 and 133 aa, respectively) also possessed no detectable homology. ORF4 (369 aa) did not have any amino acid similarity with proteins submitted to GenBank to date; however, due to its rightmost location, a major VP role was assumed. Screening the NCBI Whole-Genome Shotgun (WGS) and Transcriptome Shotgun Assembly (TSA) databases, using the tBLASTn algorithm, resulted in a significant hit for NS and ORF4, respectively. An endogenous sequence overlapping 74% of the PmMDV ORF1 with 42% identity was present in the draft genome sequence of an amphipod crustacean, Trinorchestia longiramus (WGS ID: VCRD01000839). A TSA transcript was derived from the transcriptome of a decapod, the marine Japanese blue crab (Portunus trituberculatus) (TSA ID: GFFJ01053568), encoding a C-terminal truncated protein of 30% amino acid identity with ORF4.
Transcription Strategy.
The transcriptome of PmMDV revealed six transcripts under the control of two promoters and coterminating at the same polyadenylation site of the genome (nucleotide 3829). The first promoter was identified at map unit nine, hence referred to as p9. Four of the six transcripts transcribed from this proximal promoter, of which three underwent splicing (Fig. 1B). The electrophoretrograms showing the intron–exon boundaries of these spliced transcripts are shown in SI Appendix, Fig. S1. The exact nucleotide position of each intron and exon is presented in SI Appendix, Table S2. RNA was isolated at days 2, 4, and 6 posttransfection. Transcript 4 was first detectable in the day 4 samples and remained expressed until day 6 posttransfection (Fig. 1C). The unspliced transcript 1 could express NS1 (ORF1) in its entire length. Transcript 2 was spliced twice and can express a truncated NS1 as a stop codon is incorporated at amino acid 271, creating the NS2A protein. The complex splicing pattern of this transcript also enabled the possible expression of the complete ORF3 (Fig. 1B). Transcript 3 was spliced once and can encode a 303-aa protein, including 270 aa of ORF1 and the C-terminal region of ORF3, designated NS3. Transcript 4 is also spliced once; however, this donor site adds a 2-aa extension to the truncated NS, forming a 2-aa longer version of NS2A, designated NS2B. The downstream p47 was responsible for transcribing two transcripts, assessed to encode the capsid VP. Transcript 5 was unspliced and could express ORF4 as well as ORF3. Transcript 6 was subjected to splicing, adding a short fragment at the C-terminal region of ORF1, translated from Met462 to Pro494, with ORF2, using the same donor site as transcript 2 for its second intron, possibly expressing ORF2′ (Fig. 1A). This spliced transcript could also express the complete ORF4.
Proteins.
All amino acid sequences were subjected to in silico analysis. NS1 contained an ATPase motif with an E-value of 1.4 (residues 285 to 426) and a Parvo_NS1 motif (Pfam ID: PF01057) of an E-value 2e-11, encompassing the tripartite helicase domain conserved throughout the Parvoviridae, corroborating its nonstructural function (SI Appendix, Fig. S2A). ORF3, encoded by one of the small auxillary ORFs, demonstrated a conserved domain of 0.01 E-value homologous to the catalytic domain of the precorrin-6x reductase enzyme, CbiJ. CibJ, together with CobK, are enzymes of the cobalamin-synthesis pathway, catalyzing the NADP-dependent reduction of precorrin-6x (37). ORF2′ had a Pfam domain with an E-value of 0.08, corresponding with the tetratricopeptide fold region of mitochondrial fission protein Fis1, responsible for scaffolding activity during mitochondrial fission (38) (SI Appendix, Fig. S2A). ORF4, similarly to other penaeid shrimp DVs, did not contain a PLA2 motif. Nevertheless, with the E-value of 0.02, an ADAM-TS metalloproteinase spacer 1 domain was identified, hence the name PmMDV (SI Appendix, Fig. S2A).
Structural similarity was detected between ORF4 and the VP2 of a birnavirus, infectious pancreatic necrosis virus (Birnaviridae, genus Aquabirnavirus) (P = 0.009) (PDB ID code 3IDE), while searching for potential homology modeling targets. To confirm its structural function and to determine the number of proteins comprising the capsid, expression studies were carried out with the Bac-to-Bac expression system. Briefly, Sf9 cultures were transfected with three recombinant bacmid constructs named PmMDV-Bac-complete, PmMDV-Bac-p47, and PmMDV-Bac-ORF4, containing the complete PmMDV genome, the p47 promoter, and downstream regions, as well as exclusively ORF4, respectively. All three supernatants contained icosahedral particles of ∼21 nm (SI Appendix, Fig. S2B). All purified samples have a single protein band at ∼41 kDa when analyzed by SDS-PAGE, suggesting the presence of only one VP or two with the same C-terminal core, from transcripts 5 and 6, assembling the PmMDV capsid (SI Appendix, Fig. S2C).
Structural Studies.
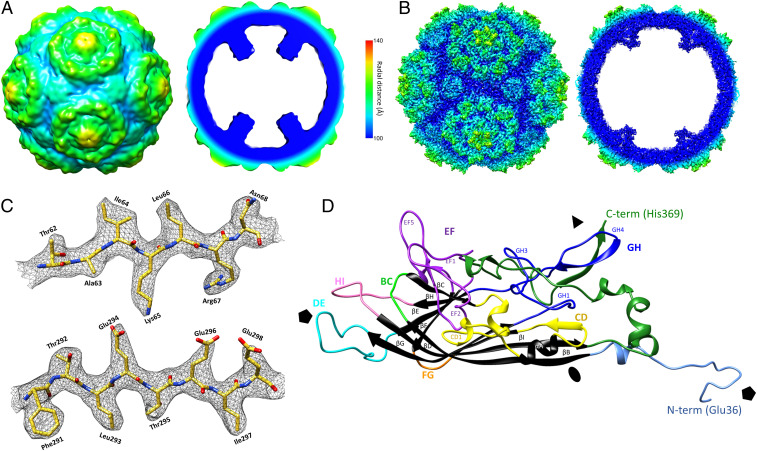
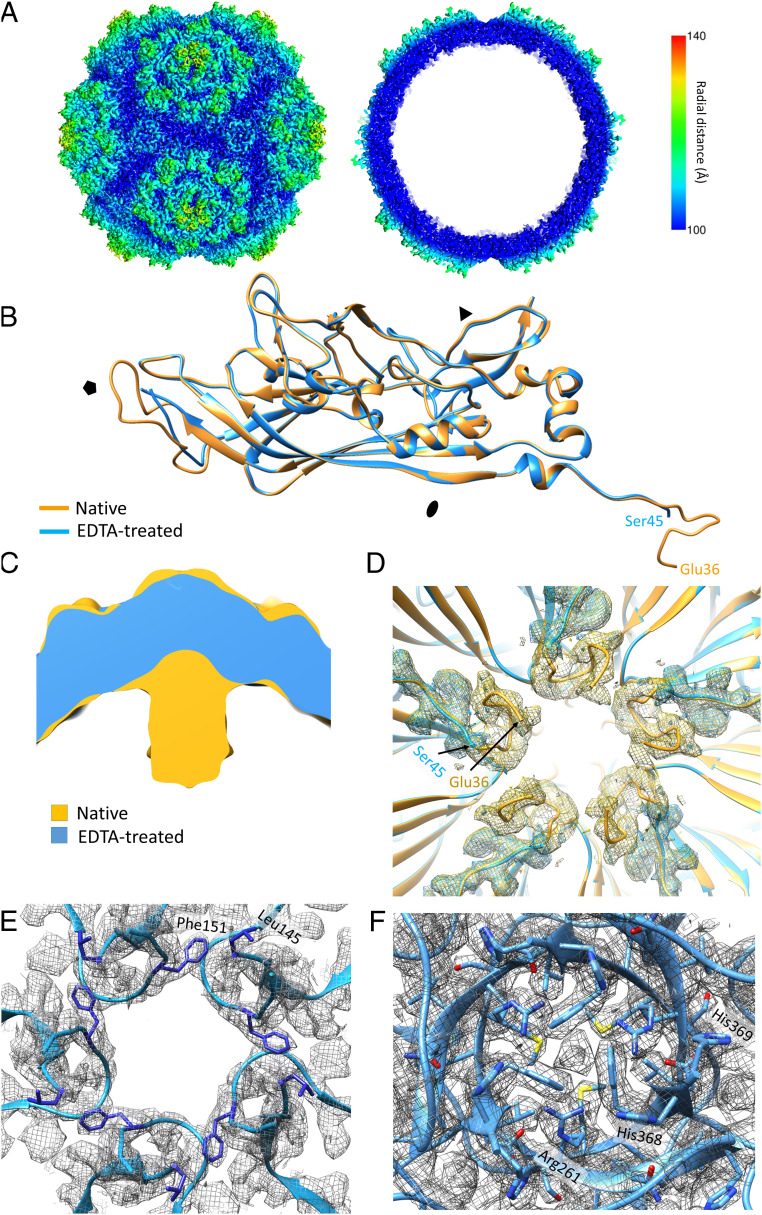
The PmMDV structure was determined using cryo-EM and image reconstruction. Particles purified from PmMDV-Bac-ORF4 expression were subjected to data collection by cryo-EM followed by single-particle image reconstruction (39). We obtained the capsid structure at high resolution of 3 Å (PDB ID code 6WH3) (40). As disordered and mobile protein regions, such as the parvoviral VP N terminus, are frequently absent from high-resolution protein structures but may be possible to visualize at a lower resolution (41), we determined the PmMDV capsid structure at the low resolution of 8.4 Å as well, from an independent data collection. Data collection parameters and refinement statistics are given in SI Appendix, Table S3. Both structures are similar and have T = 1 icosahedral symmetry (Fig. 2 A and B). PmMDV is the smallest parvoviral capsid isolated to date, ranging from 20 to 25 nm in diameter, with the smallest lumen size as shown in SI Appendix, Table S4. Overall, the surface morphology of PmMDV contains prominent protrusions surrounding the fivefold axes in two concentric rim-like circles, as well as small protruding threefold axes (Fig. 2 A and B). The interior of the capsid contains a “basket-like” structure at the base of each fivefold channel, attached to the wall by five “stalk-like” features (Fig. 2 A and B).
Fig. 2.
The PmMDV capsid structure. (A) The structure at low resolution (8.45 Å), showing surface (Left) and cross-section (Right) views radially colored from the center according to the color key. (B) The PmMDV capsid structure at high resolution (2.96 Å), showing surface (Left) and cross-section (Right), radially colored similar to A. (C) Example of map density, shown as gray mesh, with the PmMDV model (residues labeled): Carbon in yellow, oxygen in red, nitrogen in blue. (D) Ribbon diagram of PmMDV VP monomer. The conserved jellyroll core and the αA helix, present in all parvoviral structures to date, are shown in black. The loops linking the β-strands and the N- and C-terminal regions are in multiple colors and labeled according to the β-strands that they connect.
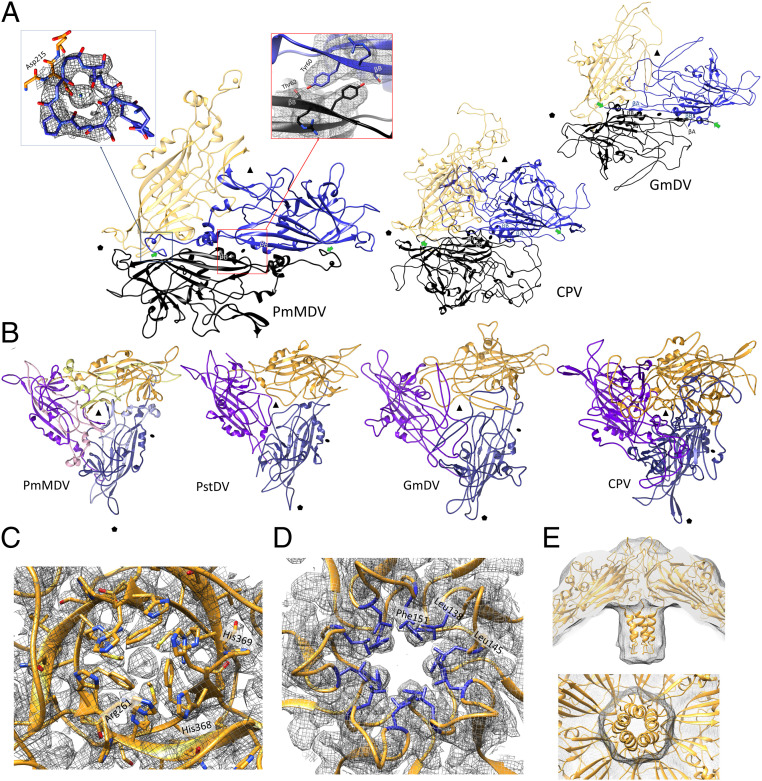
The cryo-EM electron density map of the VP could be built from Glu36 to the C-terminal residue, His369, for the high-resolution structure (Fig. 2 C and D). Each VP monomer contains an α helix (labeled A) and an eight-stranded β-barrel jellyroll core (βB–βI), but lack the analogous parvoviral βA strand. Unlike all DV structures determined so far, but similarly to PstDV, the VP contains a 12-aa-long insertion forming the CD1 subloop within the loop linking βC and βD (Fig. 2D). The same insertion is 10-aa-long in PstDV. Like all DV structures, the region located N-terminally from the βB was situated in a domain-swapped conformation. However, without the βA, there were no hydrogen bonds formed with the βB of the twofold-related monomer, a phenomenon shared by all DV structures to date (Fig. 3A). Instead, the N terminus formed a loop, within which carboxyl groups and the side-chain of Asp215, from a threefold-related VP monomer, coordinate unmodeled diffuse electron density (Fig. 3 A, Left Inset). Because of its coordination, the density was proposed to be a Ca2+ ion. Interestingly, in the absence of a βA, the βB strands of twofold-related VP monomers formed polar interactions between residues Tyr60 and Thr62 (Fig. 3 A, Right Inset). Unlike any PV structure to date, the C terminals of the PmMDV VP monomers are located on the surface at the threefold axes and create a β-annulus structure (Fig. 3B). Associated with this annulus, toward the capsid surface, is an additional β-strand, comprised of the C-terminal region of a predicted ADAM-TS spacer domain (Fig. 3B).
Fig. 3.
Multimer interactions of the PmMDV capsid. (A) Dimer interaction of two PmMDV subunits, shown as ribbon diagrams, compared to those of a vertebrate (canine parvovirus, CPV) and an invertebrate (GmDV) parvovirus. The hydrogen bonds linking the βB strands together are shown in the zoom to the top right of the PmMDV dimer. The interaction of the PmMDV subunit N terminus with the threefold neighboring subunit is shown to the left of the PmMDV dimer, indicating the extra density corresponding with the presence of a bivalent cation, predicted to be Ca2+. The twofold axes are indicated by the ellipsoids, threefold axes by the triangles, and fivefold axes by the pentagons. The N termini are indicated by green arrows. (B) Ribbon diagrams of trimer interactions of PmMDV, hamaparvovirus-PstDV, densovirus-GmDV, and vertebrate protoparvovirus, CPV. The location of the ADAM-TS metalloproteinase spacer domain is indicated by the lighter ribbon color compared to the colors of the subunit ribbons themselves. The location of the icosahedral symmetry axes are as in A. (C) Density and model at the threefold axis. The PmMDV threefold structure harbors a double β-annulus–like structure. His-369 has a double conformation. The pore opening of the annulus contains a piece of unassigned density. (D) Ribbon diagram and density map of the PmMDV fivefold channel, viewed from the top, as it appears from the capsid surface. The sidechains of the hydrophobic residues lining the fivefold channel are shown in blue. Example amino acid residues are labeled (in black) in (C) and (D). (E) Ribbon diagram of the model built into the low-resolution PmMDV density map at 8.45 Å. The basket-like density under the fivefold channel is modeled with the backbone of 20 additional N-terminal amino acids, arranged in an α-helical conformation, shown from a lateral view (Upper) and from the luminal surface of the capsid (Lower).
Both of the C-terminal–most residues were histidines, His368 and His369, of which His369 adopts a dual conformation, facing either toward or away from the annulus center (Fig. 3C). Interestingly, this annulus channel is wider than the fivefold pore (∼17 vs. ∼10 Å), and is filled by unassigned diffuse density, coordinated by positively charged His and Arg sidechains (Fig. 3C). In contrast, the two luminal layers were occupied by hydrophobic sidechains, such as Phe and Met (Fig. 3C). At the fivefold symmetry axes, the canonical pore is lined by the sidechains of bulky hydrophobic residues (Fig. 3D). The density of the basket below this pore could not be modeled at the sidechain level. However, the low-resolution structure and secondary structure predictions suggested a helical propensity, with five α-helices forming a pentamer helix bundle (Fig. 3E and SI Appendix, Fig. S3). We could model 20 more N-terminal residues in an α-helical arrangement into the basket density of the low-resolution structure. A stretch of basic residues (RKRRRH) is present in each α-helix.
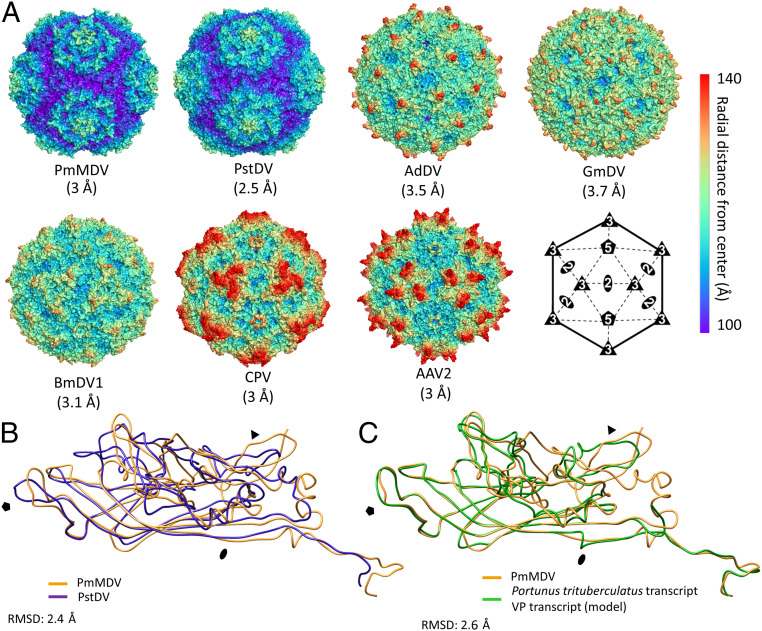
The surface morphology of PmMDV and its size were most similar to that of PstDV, when compared with available capsid structures for six Parvoviridae genera: Iteradensovirus, Protoambidensovirus, Scindoambidensovirus, Penstylhamaparvovirus, Dependoparvovirus, and Protoparvovirus (Fig. 4A). This observation was supported by a DALI search (z-score of 22.5) indicating that the PmMDV capsid is structurally most like PstDV, despite low sequence identity (13%). The two VP structures could be superposed with an RMSD of 2.4 Å for the Cα positions of 258 residues (Fig. 4B). With the exception of the threefold region, all loops superposed, although their conformation differed substantially. Interestingly, a homology model built from the P. trituberculatus transcript indicates that, if this transcript is of viral origin, the VP topology will be very similar to the PstDV and PmMDV, with variations in the surface loops (Fig. 4C).
Fig. 4.
Comparison of the PmMDV capsid structure with selected PVs. (A) The PmMDV capsid surface morphology is compared to those of vertebrate PVs of Parvovirinae: That is, CPV of genus Protoparvovirus and Adeno-associated virus serotype 2 (AAV2) of Dependoparvovirus; and invertebrate-infecting PVs of Densovirinae, GmDV of Protoambidensovirus, BmDV1 of Iteradensovirus, AdDV of Scindoambidensovirus, and PstDV of Penstylhamaparvovirus, subfamily Hamaparvovirinae. The capsids are radially colored, according to the scale to the right and are orientated according to the schematic icosahedron diagram. (B) The PmMDV VP monomer structure superimposed on the VP monomer structure of PstDV, shown as coil diagrams. (C) Coil diagram of the PmMDV VP monomer structure superimposed on the model constructed from the derived protein sequence of a transcript derived from the Japanese blue crab, P. trituberculatus (TSA ID: GFFJ01053568). In both B and C the RMSD values between the Cαs are shown.
The Effect of EDTA Treatment.
The potential contribution of divalent cations to additional unmodeled density in the PmMDV map and structure stability of the capsid was assessed by pretreating purified virus-like particles (VLPs) with 100 nmol EDTA, a divalent metal ion chelator, and determining the structure. As with the untreated capsid, the capsid-EDTA–treated structure was first determined at low resolution, 10.37 Å, then at high resolution of 2.82 Å (Fig. 5 A and B) (PDB ID code 6WH7) (42). The removal of the divalent cation (probably Ca2+ ion) from the N-terminal–threefold interaction resulted in the complete absence (disordering) of the fivefold basket and stalk as well as in the disorder of the loop involved in the intra–VP-cation coordination, making Ser45 the first ordered N-terminal residue rather than Glu36 (Fig. 5 C and D). The apex of the DE loop, from residues Leu138 to Thr147, forming the channel at the fivefold axes, was disordered. The hydrophobic ring forming the interior of the channel, including Phe151, was altered due to a change in the orientation of the sidechains (Fig. 5E). The structural change on the inside and outside of the capsid at the fivefold axes resulted in the opening of the channel in the EDTA-treated structure, with the pore expanding to 16.8 Å in diameter (Fig. 5E). The unmodeled density covering the threefold annulus was still present, albeit less coordinated, as the orientation of His369 changes to favor the conformation facing away from the density (Fig. 5F).
Fig. 5.
The EDTA-treated PmMDV capsid structure. (A) Surface morphology and cross-section of the high-resolution EDTA-treated structure at 2.78-Å resolution. (B) Ribbon diagram superposition of the native PmMDV and EDTA-treated VP monomer structures. The N termini are indicated by the first ordered residue in each structure. (C) Superposition of the low-resolution native and EDTA-treated PmMDV maps. The absence of the basket-like structure in the EDTA-treated structure is evident. (D) Superimposed density maps and ribbon diagrams of the native (yellow ribbon and mesh) and EDTA-treated (blue mesh and ribbon) N-terminal regions of the PmMDV high resolution structures located under the fivefold symmetry axis and viewed from the capsid lumen. The arrows indicate the first ordered residue in both structures. Density is contoured at 2ơ. (E) EDTA treatment affects the DE loop structure and expands the fivefold pore diameter to 16.8 Å. The model is shown in blue ribbon inside the mesh-like density of the map. The hydrophobic residues, previously facing toward the lumen of the channel, are highlighted in blue. (F) The threefold symmetry axis of the EDTA-treated structure. The central unassigned density is still present in the annulus center. The terminal histidine, His369, now adopts a single conformation (rather than the dual in the native structure) facing away from the annulus center.
Phylogenetic Reconstructions.
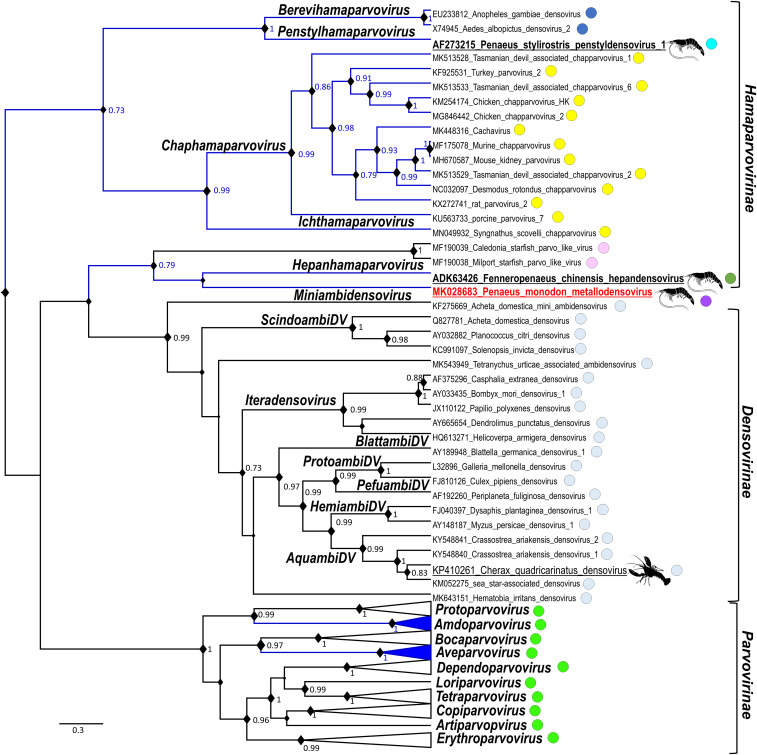
The only region of the deduced protein sequences of each ORF, which could be aligned reliably with other PV and DV sequences retrieved from GenBank, was the SF3 helicase domain of ORF1 (NS1). Fig. 6 presents the results of the Bayesian phylogeny inference. According to these criteria, the Parvoviridae is composed of four distantly related clades, where two correspond with subfamilies Parvovirinae and Densovirinae. As the fourth lineage, PmMDV pulled out genus Hepanhamaparvovirus and two hitherto unclassified DVs isolated from echinoderms, from the Hamaparvovirinae. The clustering of PmMDV with hepanhamaparvoviruses was not significantly supported, unlike the clade itself. Based on the results of a BlastP search, capsid VP shares the same origin within the Parvovirinae and Densovirinae, as well as within genera Chaphamaparvovirus and Ichthamaparvovirus of Hamaparvovirinae (Fig. 6).
Fig. 6.
The Bayesian phylogenetic inference based on the NS1 Pfam domain of family Parvoviridae (1U0J). Alignment was constructed incorporating structural data, retrieved automatically from the RCSB Protein Data Bank. The substitution model LG+I+G proved to be the most suitable based on both Akaike and Bayesian information criteria. Posterior probability values are indicated on the nodes if significant (0.7<). The area of node shapes is proportional with the posterior probability value of each node. Crustacean PVs are shown underlined and in larger font, whereas penaeid shrimp infecting viruses are in bold and underlined. The PmMDV is highlighted in red. The branches of lineages not including the PLA2 in their VP proteins are presented in blue. Capsid protein homology is indicated as circles of the same color.
Discussion
PmMDV is a newly discovered pathogen of the penaeid shrimp, P. monodon, with a significant amino acid sequence identity only within its SF3 helicase domain with other members of Parvoviridae. Its short ssDNA genome, flanked by the T-shaped secondary structures, and its T = 1 icosahedral capsid symmetry are all characteristics of members of the family Parvoviridae. Although members of two PV genera had been previously isolated from the same host so far, neither penstyl- nor hepanhamaparvoviruses were closely related to PmMDV. Consequently, PmMDV thus represents a virus of a third parvoviral lineage to infect penaeid shrimps to be assigned to a novel genus.
The subfamily Hamaparvovirinae was created to include all PVs that could not be assigned to either Densovirinae or Parvovirinae reliably (43). While the clade of genera Penstyl-, Brevi-, Chap-, and Ichthamaparvovirus appear to be monophyletic, the introduction of PmMDV splits Hamaparvovirinae into two clades. This suggests that PmMDV should not be assigned to the established subfamilies and its classification requires a new subfamily to be proposed to the International Committee on Taxonomy of Viruses Parvovirus Study Group (1). Although all members of the Parvovirinae and Densovirinae subfamilies possess homologous structural proteins, the high number of lineages lacking VP sequence similarity in Hamaparvovirinae and in the fourth clade could suggest the existence of several, only distantly related subfamily-level lineages. Since most of these PVs are of aquatic origin, this environment might facilitate the acquisition of VP-encoding genes from other viral sources. Chimerism between CRESS DNA viruses and ssRNA(+) viruses has been observed multiple times (44, 45). The finding of a homologous transcript of the PmMDV VP in another decapod crustacean, P. trituberculatus, and an endogenous element in an amphipod, a stemgroup of Malacostraca, suggested that the lineage, of which PmMDV is the first representative, might hold a wide host spectrum, involving the most diverse crustacean class, Malacostraca.
Homology modeling target selection suggested structural similarity between the VP2 of infectious pancreatic necrosis virus, a double-stranded RNA (dsRNA) virus, and ORF4 of PmMDV. Earlier structural studies revealed another birnavirus, infectious bursal disease virus, to provide a missing evolutionary link between the structure of the mature dsRNA virus capsid (families Reoviridae and Birnaviridae) and certain families of icosahedral positive-stranded ssRNA viruses, such as Nodaviridae and Tetraviridae (46). The VP2 of these families, which contain a jellyroll fold as its core element, self-assembles into an immature procapsid of T = 1 icosahedral symmetry, while the N-terminal helices come together into a pentamer helix bundle at the fivefold axis. In the case of family Tetraviridae, the bundle has been shown to undergo externalization and disrupt the plasma membrane during viral entry (47). Considering its fold-related position, the PmMDV basket, which also appears to be a pentamer helix bundle, might hold a similar function. Additionally, helical membrane-penetrating peptides often include similar arginine-rich basic motifs, similarly to the PmMDV VP N terminus (48, 49). It was shown that divalent cation removal triggered the externalization of the PmMDV fivefold basket from each threefold neighboring subunit. As PVs traffic through the endosomal–lysosomal network, following endocytosis, their environment becomes increasingly acidified, reaching a pH as low as 4.0 (50, 51). This could trigger the dissociation of a cation, suggesting that the PmMDV capsid might undergo the observed conformational changes during its natural trafficking pathway. Structural reorganization at the fivefold axis, manifesting as the expansion of the fivefold pore and loss of both the DE loop and the basket, could be potentially associated with the externalization of the N-terminal helical bundle. This mechanism is analogous to that used by members of the Tetraviridae to breach the endolysosome (47) and not the phospholipase activity, after PLA2 externalization, exploited by almost all PVs (3). However, basket-like structures have been reported for Parvovirinae genus Bocaparvovirus, although the Gly-rich flexible extension linking the VP2 N terminus to the VP1 PLA2 domain is likely involved in externalization and a role for membrane-penetrating helices has not been deduced (52, 53).
Despite the homology modeling target selection results, the PmMDV capsid has a remarkably similar surface morphology to another shrimp-infecting DV, PstDV, yet their VPs are low in sequence identity. The structural similarity, however, is limited because the PmMDV capsid possesses multimer interactions that differ from all other PVs. The N-termini of VPs of invertebrate-infecting PVs display a domain-swapped conformation; that is, the βA interacts with the βB of the twofold-neighboring subunit and not with its own βB as in case of the Parvovirinae (29–32). In PmMDV the VP N terminus interacts with the threefold neighboring subunit through a similar swapped-like conformation. Due to the lack of the βA strand, the PmMDV capsid luminal surface is assembled by the four-stranded βBIDG sheet, stabilized by βB–βB interactions instead of the canonical βB–βA observed for all Parvovoiridae with known structures (28). As a result, the capsid lumen is smaller, yet packages an average-sized PV genome of 4.3 kb. Finally, PmMDV and PstDV are only distantly related based on NS1 phylogeny, and possess different genome organizations and transcription strategies (27, 54). Consequently, the structural similarity between these two PVs is likely due to convergent evolution, attributed to infecting the same host and perhaps sharing similar aspects of receptor attachment and trafficking to the nucleus for packaged genome replication.
All four available DV structures possess a β-annulus–like opening at their threefold axes, similarly to T = 3 ssRNA viruses, such as Tombusviridae. This is a portal-like opening in GmDV and PmMDV, with a diameter of ∼18 and ∼17 Å, respectively (29–32, 55). The GmDV β-annulus is lined with large, positively charged sidechains similar to the upper layer of the PmMDV double β-annulus, and was speculated to be a flexible channel for packaging or uncoating the ssDNA genome (29). The belief is that the PmMDV fivefold axes portal may be the location of membrane penetration while the PmMDV threefold axes portal performs genome-associated functions, such as genome release or entry into the empty particle during uncoating or packaging, respectively. The shape of the unmodeled diffuse density occupying the annulus resembles a metal ion; however, it was not chelated with EDTA. This suggests that it is well coordinated by the three histidines, similarly to the zinc ions in a variety of metalloproteins (56). If indeed this is a zinc ion, it may have a role in catalyzing the formation of trimers during assembly (57). The role for the predicted ADAM-TS metalloproteinase spacer-like domain, surrounding the annulus and surface-exposed at the outer rim surrounding the fivefold axes, is unknown. These domains are widespread in eukaryotic proteins and have been associated with extracellular matrix attachment and signaling (58–60). Therefore, this region might play a role in virus–host interactions of PmMDV and may possibly be associated with attachment. This remains to be tested and confirmed.
The PmMDV genome, despite its 4.3-kb size, contains four predicted ORFs. Among PVs the presence of multiple, small ORFs, which frequently overlap or are encompassed by the NS or the VP cassette, are well-known (10, 13, 61–67). ORF2 and ORF3 were not related to any other PV ORF, suggesting their independent acquisition by horizontal gene transfer. This was supported by the presence of the Fis1 Pfam domain and the CobJ motif in the amino acid sequence of ORF2′ and ORF3, respectively, implying host or prokaryotic origin. The PmMDV genome encoded two major groups of NS proteins from an upstream promoter. The significance of expressing shorter C-terminal NS protein variants along a full-length NS1 is unclear but is common among PVs. Transcript 4, expressing NS2B, showed up later in the transcriptome, suggesting that each variant harbors different roles during replication. This has also been described in dependoparvoviruses, where each Rep possess two slightly different C-terminal–extended versions (68). Transcript 2 is spliced twice and may express ORF3 in addition to NS2B. ORF3 could potentially also be expressed from the unspliced p47 transcript. As this putative protein might possess enzymatic activity, the complex splicing pattern may have evolved to regulate its quantity as needed. Regulation of protein quantity via splicing has been observed in case of structural protein expression of several Parvovirinae genera, including Proto-, Amdo-, and Dependoparvovirus (2). In Densovirinae, leaky scanning acts as the main expression regulatory mechanism, although alternative splicing has evolved sporadically in various genera of Densovirinae, such as Proto-, Scindo-, and Pefuambidensovirus (5, 7, 10, 13, 54). The alternative splicing in PmMDV, however, probably evolved independently from these. ORF2′, expressed by a spliced transcript of p47, is a chimera of two ORFs acquiring its N terminus from the nonstructural protein gene, ORF1. As the tetratricopeptide region of the Fis1 protein has a scaffolding role during mitochondrial fission, it cannot be excluded that ORF2′ provides a similar function during PmMDV assembly. However, as the VLPs expressed by the PmMDV-Bac-ORF4 were capable of self-assembly, without such a function, it does not appear to be essential for capsid assembly.
Materials and Methods
Virus Detection, DNA Isolation, and Cloning.
Virus particles were purified by cesium chloride gradient to obtain viral DNA for cloning (detailed later) from the pooled, ∼500-g P. monodon-infected tissue, obtained from Vietnam. After homogenization in PBS and initial extraction with chloroform/butanol (1:1 volume), a clear supernatant containing viral particles was obtained by low-speed centrifugation. Virus stock was concentrated from the supernatant by ultracentrifugation at 40,000 rpm in a type 60Ti rotor for 2 h at 4 °C. Pellets were resuspended in small volume of PBS for DNA extraction or EM analysis. Viral DNA was extracted by the High Pure Viral Nucleic Acid Kit (Roche) and eluted in 40 μM of distilled water. The isolated DNA was blunt-ended utilizing T4 DNA polymerase and Large Klenow fragment of DNA polymerase I (New England Biolabs) in the presence of 33 μM of each dNTP and cloned into the EcoRV restriction site of a pBluescript KS+ vector. Starting with the M13 primer sites of the vector, the sequence of the PmMDV genome could be determined by primer walking. To obtain the sequences of the termini, several GC-rich cutter restriction enzymes were used to release secondary structures and the fragments subcloned and sequenced. Two GC cutters proved to be sufficient, namely ApaI and HaeIII, enabling cloning into the ApaI and EcoRV sites of the pBluescript KS− and KS+ vectors, respectively. We used the Sure Escherichia coli strain (Stratagene), and incubation at 30 °C, for both infectious clone construction and for subcloning of viral ITRs.
Cell Lines, Transfection, and Culturing Conditions.
Sf9 (ATCC CRL-1711), C6/36 (ATCC CRL-1660), and Schneider’s Drosophila Line 2 (ATCC CRL-1963) were tested for PmMDV susceptibility; however, none of these could sustain PmMDV replication. To perform transcription and expression studies, the Bac-to-Bac expression system was used (Invitrogen, Thermo Fisher Scientific), involving the transfection of Sf9 cells. Sf9 cultures were maintained in SF900 II medium (Gibco) in a serum-free system at 28 °C. Cellfectin II Reagent (Invitrogen) was used for DNA transfection with 8 × 105 cells per well, previously seeded on a six-well culturing dish and incubated overnight at 28 °C. The culturing medium was aspired and replaced by seeding medium of Grace’s complete insect medium supplemented with 5% FBS (Gibco) and Graces’s unsupplemented insect medium, mixed at a ratio of 1:6, respectively. After adding the transfection reagent–DNA mixture to the wells, cells were incubated for 5 h. The aspired transfection medium was replaced with SF900 II medium supplemented with 2% FBS. Cells were checked daily for signs of cytopathic effects (CPE) and the whole culture was collected when 70% of the cells detached from the dish or showed granulation. This was followed by three cycles of freeze–thaws on dry ice and 200 μL of this passage 1 (P1) stock was transferred to 25 mL of fresh Sf9 suspended cell culture in polycarbonate Erlenmeyer flasks (Corning) at the density of 2.5 × 106 cells/mL, to create the P2 stock, cultured in serum-free SF900 II medium without antibiotics.
Transcription Studies.
To study the transcriptome of PmMDV, we cloned the entire viral genome, including the ITRs, into a pFB dual vector, where both the p10 and the PH (polyhedrin) promoters had been knocked out previously; hence, transcription of the whole PmMDV genome could be analyzed in the recombinant Autographa californica nuclear polyhedrosis virus (NPV) genome without interference of the NPV promoters. This construct was designated PmMDV-Bac-complete. The yield of DNA, however, of bacmid minipreps from the DH10Bac (Invitrogen) cells was below 500 μg/mL; hence, the large ITRs were removed. This yielded over 2 mg/mL bacmid DNA of each preparation. Total RNA was extracted using the Direct-zol RNA MiniPrep Kit (Zymo Research), where the denaturation step was executed by adding TRIzol Reagent (Thermo Fisher Scientific). RNA was treated by digestion with the TURBO DNA-free Kit (Ambion) to get rid of residual DNA contamination, as well as subjected to a control PCR for the remaining DNA fragments. Reverse transcription was performed only on entirely DNA-negative preparations using the SuperScript IV or the SuperScript III enzymes (Thermo Fisher Scientific), supplemented with random nonamers (Sigma-Aldrich). To avoid false-detection of splicing, isolated and Dnase-treated RNA was dephosphorylated by adding Antarctic phosphatase (New England Biolabs) and incubated for 30 min at 37 °C. Primers were designed at the following positions of the PmMDV genome: 1,100 nt (reverse and forward), 2,300 nt (reverse and forward), 2,564 nt (reverse and forward), 3,038 nt (reverse), and 3,511 (reverse and forward).
Anchored oligo(dT) primers were used together with the 2,300 forward and 3,511 forward primers for 3′ RACE (rapid amplification of cDNA ends). To perform 5′ RACE to map transcription start sites, we designed adaptors with the sequence of ATCCACAACAACTCTCCTCCTC’3. Dnase-treated RNA was subjected to dephosphorylation. by alkaline calf intestinal phosphatase (New England Biolabs) and after phenol-chloroform extraction to dephosphorization by tobacco acid pyrophosphatase (Ambion) to remove 5′ RNA caps. After the ligation of adaptors using T4 RNA ligase (New England Biolabs), reverse transcription was executed as described above. PCR was performed with the readaptor primers together with oligos 1,100 reverse and 2,564 reverse. All PCRs were performed using Phusion Hot Start Flex DNA Polymerase (New England Biolabs) in a 25-μL final reaction volume, including 2 μL of purified cDNA target, 0.5 μL of both primers in 50-pmol concentration, 0.5 μL dNTP mix with 8 μmol of each nucleotide, 0.75 μL of 50 mM MgCl2 solution, and 0.25 μL of enzyme. PCR reactions were executed under a program of 5-min denaturation at 95 °C followed by 35 cycles of 30-s denaturation at 95 °C, 30-s annealing at 48 °C, and 1 or 2 min of elongation at 72 °C. The final elongation step was 8-min long at 72 °C. In case of the 5′RACE reactions, 0.5 μL of enzyme was used and the number of cycles was reduced to 25. For the 3′RACE, the reaction was supplemented with 1 μL of 50 mM MgCl2 and the annealing step was left out.
Protein Expression and Purification of VLPs.
The plasmids PmMDV-bac-complete and PmMDV-Bac-p47 were constructed by using a pFB dual vector (Invitrogen), from which both the polyhedrin (PH) and the p10 promoters had been removed, while PmMDV-Bac-ORF4 was of pFB1 backbone, driven by the PH promoter (Invitrogen). For the expression studies, the P2 baculovirus stock was used in the case of all three constructs detailed above. The P2 stocks were incubated for at least 7 d and monitored for CPE every third day. When at least 70% of the cells showed signs of CPE, the culture was collected, centrifuged at 3,000 × g, and the pelleted cells disrupted by three cycles of freeze–thaws on dry ice. This lysed cell pellet was then resuspended in 1 mL of 1× TNTM pH8 (50 mM Tris pH8, 100 mM NaCl, 0.2% Triton X-100, 2 mM MgCl2) and centrifuged again. Supernatant was mixed back together with the cell culture supernatant and was subjected to treatment with 250 units of Benzonase Nuclease (Sigma-Aldrich) per every 10 mL. The liquid was mixed with 1× TNET pH8 (50 mM Tris pH8, 100 mM NaCl, 0.2% Triton X-100, 1 mM EDTA) in a 1:1 ratio and concentrated on a cushion of 20% sucrose in TNET, using a type 60 Ti rotor for 3 h at 4 °C at 45,000 rpm on a Beckman Coulter S class ultracentrifuge. The pellet was resuspended in 1 mL of 1× TNTM pH8 and after overnight incubation purified on a 5 to 40% sucrose step gradient for 3 h at 4 °C at 35,000 rpm on the same instrument in a SW 41 Ti swinging bucket preparative ultracentrifuge rotor. The visible single band that formed at the 15 to 20% sucrose interface was then collected by needle puncture and a 10-mL volume syringe. In the case of constructs PmMDV-Bac-complete and PmMDV-Bac-p47, both expressed by the own promoters of PmMDV, sucrose gradient purification did not result in a visible band; hence, the cushion-concentrated pellet after TNTM resuspension was purified in cesium chloride instead, dissolved in TNTM at a density of 1.38 g/cm3. After 24-h ultracentrifugation at 40,000 rpm at 16 °C in a SW 55 Ti rotor, the VLPs could be aspired using a needle and a 1-mL syringe. The aspirate was dialyzed into 1× HCB buffer (50 mM Hepes, 4.3 mM MgCl2 × 6H2O, 0.15 M NaCl) at pH 7.4 to remove the sucrose or the cesium chloride. As PmMDV demonstrated better stability at the higher pH of its natural extracellular environment, particles purified were dialyzed into 1× universal buffer (20 mM Hepes, 20 mM MES, 20 mM sodium acetate, 0.15 M NaCl, 3.7 mM CaCl2) at pH 8.2, which is equivalent with the pH of tropical marine water.
In Silico Analyses.
After obtaining the sequence of the ITRs and genome clones, the complete genome sequence of PmMDV was assembled using Staden package v4.11.2 (69). The assembled genome was annotated, as well as the transcripts assembled and splice sites investigated in Artemis Genome Browser by the Sanger Institute (70). Homology searching at amino acid levels was carried out two ways: To determine sequential similarity, the Blast algorithms were applied with different expectation value levels (71), whereas structural similarity was predicted by the genThreader, pDomThreader, and pGenThreader algorithms of the PSIpred web server (http://bioinf.cs.ucl.ac.uk/psipred/) (72). To investigate the conserved motifs and domains with known homologs in the derived aa sequences, the DomPred algorithm of the PSIpred server as well as the SMART web application was used (73). Structural similarity of the resolved capsid structures with those available in the RCSB Protein Data Bank (PDB) was investigated using the DALI server (74). The tBLASTn algorithm was utilized to screen the RefSeq, Whole Genome Shotgun Contigs, High-Throughput Genomic Sequences, and Transcript Shotgun Assembly databases, targeting 5,000 hits with an expectation value of 10 (71).
For phylogenic inference, alignments, incorporating the outputs of pairwise, multiple, and structural aligners, were constructed using the Expresso algorithm of T-Coffee (75). Structural data were obtained using the PDB mode of this algorithm. The constructed alignment was further edited using Unipro Ugene (76). Model selection was executed by ProtTest v2.4, suggesting the LG + I + G + F substitution model based on both the Bayesian and Akaike information criteria (77). The distance matrix to the starting trees were constructed using the Prodist program of Phylip v3.695 with a Johns–Thorton–Taylor method and the starting tree was constructed from this using the Fitch–Margoliash method of the Fitch program with global rearrangements (78). Bayesian inference was executed by the BEAST v1.10.4 package, incorporating the predicted LG + I + G + F model, using a log-normal relaxed clock with a Yule speciation prior, throughout 50,000,000 generations (79). Convergence diagnostics were carried out using Tracer v1.7.1, which indicated the Markov-chain Monte Carlo runs to have converged (80). Phylograms were edited and displayed in the FigTree 1.4.1 program of the Beast package.
To investigate possible homologous VPs throughout the family, we used the Blast P and PSI Blast NCBI algorithms with an expect threshold of 1,000 targeting the maximum number of 5,000 sequences (81). As query, one VP-derived amino acid sequence was submitted for each recognized parvovirus species as well as one for each complete, unclassified entry. In the case of the PLA2-containing VPs, the N-terminal sequence, containing this conserved domain, was removed to avoid false hits. Screening was performed using the substitution matrices Blosum62 and PAM250. Two sequences were marked as homologs in case the search resulted in a hit. The search was limited to family Parvoviridae in order to filter out false positives.
To construct the model of the eight stranded β-barrel jellyroll core, which could be docked into the PmMDV high-resolution structure density, the I-TASSER Standalone Package v5.1 was used (82). As a template search failed to detect structural similarity with any PV capsid structure, threading was restricted to the PstDV VP monomer (PDB ID code 3N7X) and incorporated secondary structure predictions, obtained by the PSIpred server (72). To achieve the best possible fit of the core, the N- and C-terminal regions up to and following the first and last β-sheets were removed from the amino acid sequence.
Structural Studies.
The statistics for each reconstruction and refinement are given in SI Appendix, Table S3. Three-microliter aliquots of the PmMDV VLPs without/with EDTA (∼1 mg/mL) were applied to glow-discharged C-flat holey carbon grids with a thin layer of carbon (Protochips) and vitrified using a Vitrobot Mark IV (FEI) at 95% humidity and 4 °C. The quality and suitability of the grids for cryo-EM data collection was determined by screening with a 16-megapixel charge-coupled device camera (Gatan) in a Tecnai G2 F20-TWIN transmission electron microscope operated at 200 kV under low-dose exposure (∼20 e−/Å2) prior to data collection. For collecting the low-resolution native and EDTA-treated PmMDV datasets, the same microscope was used at 50 frames per 10 s using a K2 direct electron detector (DED) at the University of Florida Interdisciplinary Center for Biotechnology Research electron microscopy core.
High-resolution data collection was carried out at two locations: the Florida State University (FSU) for the EDTA-treated PmMDV dataset, and the University of California, Los Angeles (UCLA) for the native dataset. At both locations, a Titan Krios electron microscope (FEI) was used, operating at 300 kV, equipped with a DE64 DED (Direct Electron Detector) at FSU and Gatan K2 DED at UCLA. At UCLA, the scope also contained a Gatan postcolumn imaging filter (Gatan) and a free-path slit width of 20 eV. Movie frames were recorded using the Leginon semiautomated application at both sites (83). At FSU, the frame rate was 50 per 10 s with ∼60 e−/Å2 electron dosage. At UCLA, images were collected at 50 frames per 10 s with an ∼75 e−/Å2 electron dosage. Movie frames collected at both locations, as well as for the low-resolution data set collected at the University of Florida, were aligned using the MotionCor2 application with dose weighting (84).
To reconstruct the 3D structure from the micrographs, particles were extracted using EMAN2 interactive boxing (85) and the AUTO3DEM software suite (86). Individual particle image normalization and apodization was performed using the AUTOPP subroutine of AUTO3DEM, with options F and O. Estimations of the defocus values for the micrographs used the ctffind4 subroutine (87) in AUTO3DEM (option 3X) to enable correction of the microscope-related contrast transfer functions. Initial models at low resolution (30 Å) were generated from the images of 100 particles by an ab initio random-model method, applying icosahedral symmetry. Orientations and origins of each particle were determined based on the initial model and the final map was obtained after a number of refinement iterations (15–31) in AUTO3DEM, until the resolution could not be further improved. The resolution of each cryoreconstructed map was calculated based on a Fourier shell correlation (FSC) of 0.143. The number of particles used to compute the final density maps are in SI Appendix, Table S3. A noise-suppression factor was applied in AUTO3DEM to avoid amplification of noise in the density maps. The homology model of the VP core structure used to generate a 60 mer, was fitted into the density map using the University of California, San Francisco Chimera visualization system (88). Each map was resized to the voxel size determined in Chimera (by maximizing correlation coefficient) using the e2proc3D.py subroutine in EMAN2 and then converted to the CCP4 format using the program MAPMAN (89). Once the βB strand could be confidently built, using the Coot application (90), the entire ill-fitting docked model was deleted and the density remodeled residue by residue. The EDTA-treated virion structure was built by docking the native structure model into the density, followed by refinement and rebuilding where necessary. Then, each model was refined against the map utilizing the rigid body, real space, and B-factor refinement subroutines in Phenix (91).
Supplementary Material
Acknowledgments
The authors thank Dr. Hanh T. Van and Chien P. Le from The Institute of Tropical Biology in Hochiminh City for helping with the collection of shrimp specimens; Micheline Letarte at Institut national de la recherche scientifique-Institut Armand-Frappier microscopy and the University of Florida Interdisciplinary Center for Biotechnology Research electron microscopy core for access to electron microscopes utilized for negative-stain electron microscopy and cryoelectron micrograph screening. The Spirit and TF20 cryoelectron microscopes were provided by the University of Florida College Of Medicine and Division of Sponsored Programs. We thank Dr. Hong Zhou (University of California, Los Angeles) and the NIH “West/Midwest Consortium for High-Resolution Cryo Electron Microscopy” project for access to the Electron Imaging Center for Nanomachines’s Titan Krios and K2 Direct Electron Detector utilized for high-resolution data collection (Multi-PI: Hong Zhou, M.A.-M., and others). Data collection at Florida State University was made possible by NIH Grants S10 OD018142-01 for purchase of a direct electron camera for the Titan-Krios at Florida State University (Principal Investigator Taylor), S10 RR025080-01 for purchase of a Field Electron and Ion (company) Titan Krios for three-dimensional electron microscope (Principal Investigator Taylor), and U24 GM116788 The Southeastern Consortium for Microscopy of MacroMolecular Machines (Principal Investigator Taylor). The University of Florida College of Medicine and NIH Grants R01 GM109524 and GM082946 (to M.A.-M. and R.M.) provided funds for the research efforts at the University of Florida. H.T.P. and J.J.P. acknowledge the financial support during their doctoral and postdoctoral studies, respectively. Part of this work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant (to P.T.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008191117/-/DCSupplemental.
Data Availability.
The complete genome sequence of PmMDV, accompanied by its annotation, has been deposited in the GenBank database (accession no. MK028683). The structure of the PmMDV capsid has been deposited to the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank, https://www.rcsb.org/ (PDB ID code 6WH3 for the native PmMDV capsid; PDB ID code 6WH7 for the EDTA-treated PmMDV capsid).
References
- 1.Pénzes J. J. et al., Reorganizing the family Parvoviridae: Proposal for a revised taxonomy independent from the canonical approach based on host affiliation. Arch. Virol., 10.1007/s00705-020-04632-4 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Cotmore S. F. et al.; Ictv Report Consortium , ICTV virus taxonomy profile: Parvoviridae. J. Gen. Virol. 100, 367–368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zádori Z. et al., A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1, 291–302 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Farr G. A., Zhang L. G., Tattersall P., Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. U.S.A. 102, 17148–17153 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tijssen P., Bergoin M., . “Densoviruses: A highly diverse group of arthropodparvoviruses” in Insect Virology, Asgari S., Johnson K. N., Eds. (Caister Academic Press, Norwich, UK, 2010), pp. 57–90. [Google Scholar]
- 6.Li Y. et al., Genome organization of the densovirus from Bombyx mori (BmDNV-1) and enzyme activity of its capsid. J. Gen. Virol. 82, 2821–2825 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Tijssen P. et al., Organization and expression strategy of the ambisense genome of densonucleosis virus of Galleria mellonella. J. Virol. 77, 10357–10365 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H., Zhang J., Hu Y., Complete sequence and organization of Periplaneta fuliginosa densovirus genome. Acta Virol. 44, 315–322 (2000). [PubMed] [Google Scholar]
- 9.Kapelinskaya T. V., Martynova E. U., Schal C., Mukha D. V., Expression strategy of densonucleosis virus from the German cockroach, Blattella germanica. J. Virol. 85, 11855–11870 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu K. et al., The Acheta domesticus densovirus, isolated from the European house cricket, has evolved an expression strategy unique among parvoviruses. J. Virol. 85, 10069–10078 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pham H. T., Yu Q., Bergoin M., Tijssen P., A novel ambisense densovirus, Acheta domesticus mini ambidensovirus, from crickets. Genome Announc. 1, e00914-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boublik Y., Jousset F. X., Bergoin M., Complete nucleotide sequence and genomic organization of the Aedes albopictus parvovirus (AaPV) pathogenic for Aedes aegypti larvae. Virology 200, 752–763 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Baquerizo-Audiot E. et al., Structure and expression strategy of the genome of Culex pipiens densovirus, a mosquito densovirus with an ambisense organization. J. Virol. 83, 6863–6873 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivaram A. et al., Isolation and characterization of densonucleosis virus from Aedes aegypti mosquitoes and its distribution in India. Intervirology 52, 1–7 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Thao M. L., Wineriter S., Buckingham G., Baumann P., Genetic characterization of a putative Densovirus from the mealybug Planococcus citri. Curr. Microbiol. 43, 457–458 (2001). [DOI] [PubMed] [Google Scholar]
- 16.François S. et al., A new prevalent densovirus discovered in Acari. Insight from metagenomics in viral communities associated with two-spotted mite (Tetranychus urticae) populations. Viruses 11, 233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudenkauf B. M., Eaglesham J. B., Aragundi W. M., Hewson I., Discovery of urchin-associated densoviruses (family Parvoviridae) in coastal waters of the Big Island, Hawaii. J. Gen. Virol. 95, 652–658 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Hewson I. et al., Densovirus associated with sea-star wasting disease and mass mortality. Proc. Natl. Acad. Sci. U.S.A. 111, 17278–17283 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bochow S., Condon K., Elliman J., Owens L., First complete genome of an Ambidensovirus; Cherax quadricarinatus densovirus, from freshwater crayfish Cherax quadricarinatus. Mar. Genomics 24, 305–312 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Sukhumsirichart W., Attasart P., Boonsaeng V., Panyim S., Complete nucleotide sequence and genomic organization of hepatopancreatic parvovirus (HPV) of Penaeus monodon. Virology 346, 266–277 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Tang K. F., Pantoja C. R., Lightner D. V., Nucleotide sequence of a Madagascar hepatopancreatic parvovirus (HPV) and comparison of genetic variation among geographic isolates. Dis. Aquat. Organ. 80, 105–112 (2008). [DOI] [PubMed] [Google Scholar]
- 22.La Fauce K. A., Elliman J., Owens L., Molecular characterisation of hepatopancreatic parvovirus (PmergDNV) from Australian Penaeus merguiensis. Virology 362, 397–403 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Bonami J. R., Mari J., Poulos B. T., Lightner D. V., Characterization of hepatopancreatic parvo-like virus, a second unusual parvovirus pathogenic for penaeid shrimps. J. Gen. Virol. 76, 813–817 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Jeeva S. et al., Complete nucleotide sequence analysis of a Korean strain of hepatopancreatic parvovirus (HPV) from Fenneropenaeus chinensis. Virus Genes 44, 89–97 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Shike H. et al., Infectious hypodermal and hematopoietic necrosis virus of shrimp is related to mosquito brevidensoviruses. Virology 277, 167–177 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Tang K. F. et al., Geographic variations among infectious hypodermal and hematopoietic necrosis virus (IHHNV) isolates and characteristics of their infection. Dis. Aquat. Organ. 53, 91–99 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Pham H. T., Molecular Biology of Single-Stranded DNA Viruses in Shrimps and Crickets. PhD thesis, Université duQuébec, Quebec City, QC, Canada (2015).
- 28.Mietzsch M., Pénzes J. J., Agbandje-McKenna M., Twenty-five years of structural parvovirology. Viruses 11, 362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson A. A., Chipman P. R., Baker T. S., Tijssen P., Rossmann M. G., The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 A resolution. Structure 6, 1355–1367 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng G. et al., The structure and host entry of an invertebrate parvovirus. J. Virol. 87, 12523–12530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufmann B. et al., Structure of Bombyx mori densovirus 1, a silkworm pathogen. J. Virol. 85, 4691–4697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufmann B. et al., Structure of Penaeus stylirostris densovirus, a shrimp pathogen. J. Virol. 84, 11289–11296 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossmann M. G. et al., Structural comparisons of some small spherical plant viruses. J. Mol. Biol. 165, 711–736 (1983). [DOI] [PubMed] [Google Scholar]
- 34.Plevka P. et al., Structure of a packaging-defective mutant of minute virus of mice indicates that the genome is packaged via a pore at a 5-fold axis. J. Virol. 85, 4822–4827 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkatakrishnan B. et al., Structure and dynamics of adeno-associated virus serotype 1 VP1-unique N-terminal domain and its role in capsid trafficking. J. Virol. 87, 4974–4984 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penzes J. J., Pham H. T., Agbandje-McKenna M., Tijssen P., Penaeus monodon metallodensovirus strain 2014CDN, complete genome. NCBI GenBank. https://www.ncbi.nlm.nih.gov/nuccore/1784324175. Deposited 9 October 2018.
- 37.Blanche F. et al., Precorrin-6x reductase from Pseudomonas denitrificans: Purification and characterization of the enzyme and identification of the structural gene. J. Bacteriol. 174, 1036–1042 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dohm J. A., Lee S. J., Hardwick J. M., Hill R. B., Gittis A. G., Cytosolic domain of the human mitochondrial fission protein fis1 adopts a TPR fold. Proteins 54, 153–156 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo F., Jiang W., Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 1117, 401–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Penzes J. J., et al. , Capsid structure of Penaeus monodon metallodensovirus at pH 8.2. RCSB Protein Data Bank. https://www.rcsb.org/structure/6wh3. Deposited 7 April 2020.
- 41.Kaufmann B., Chipman P. R., Kostyuchenko V. A., Modrow S., Rossmann M. G., Visualization of the externalized VP2 N termini of infectious human parvovirus B19. J. Virol. 82, 7306–7312 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penzes J. J., et al. , P. Capsid structure of Penaeus monodon metallodensovirus following EDTA treatment. RCSB Protein Data Bank. https://www.rcsb.org/structure/6WH7. Deposited 7 April 2020.
- 43.Pénzes J. J. et al., “Re-organize the family Parvoviridae” in Taxonomy Proposal, EC Approved, (International Committee on Taxonomy of Viruses, 2019). [Google Scholar]
- 44.Diemer G. S., Stedman K. M., A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol. Direct 7, 13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krupovic M. et al., Multiple layers of chimerism in a single-stranded DNA virus discovered by deep sequencing. Genome Biol. Evol. 7, 993–1001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coulibaly F. et al., The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120, 761–772 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Domitrovic T., Matsui T., Johnson J. E., Dissecting quasi-equivalence in nonenveloped viruses: Membrane disruption is promoted by lytic peptides released from subunit pentamers, not hexamers. J. Virol. 86, 9976–9982 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kämper N. et al., A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J. Virol. 80, 759–768 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang P., Monteiro da Silva G., Deatherage C., Burd C., DiMaio D., Cell-penetrating peptide mediates intracellular membrane passage of human papillomavirus L2 Protein to trigger retrograde trafficking. Cell 174, 1465–1476.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mani B. et al., Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J. Virol. 80, 1015–1024 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nam H. J. et al., Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking. J. Virol. 85, 11791–11799 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kailasan S. et al., Structure of an enteric pathogen, bovine parvovirus. J. Virol. 89, 2603–2614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mietzsch M. et al., Structural insights into human bocaparvoviruses. J. Virol. 91, e00261-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tijssen P., Pénzes J. J., Yu Q., Pham H. T., Bergoin M., Diversity of small, single-stranded DNA viruses of invertebrates and their chaotic evolutionary past. J. Invertebr. Pathol. 140, 83–96 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Sherman M. B. et al., Near atomic resolution cryo-electron microscopy structures of cucumber leaf spot virus and red clover necrotic mosaic virus; evolutionary divergence at the icosahedral three-fold axes. J. Virol. 94, e01439-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou L. et al., Interaction between histidine and Zn(II) metal ions over a wide pH as revealed by solid-state NMR spectroscopy and DFT calculations. J. Phys. Chem. B 117, 8954–8965 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Laitaoja M., Valjakka J., Jänis J., Zinc coordination spheres in protein structures. Inorg. Chem. 52, 10983–10991 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Luken B. M. et al., The spacer domain of ADAMTS13 contains a major binding site for antibodies in patients with thrombotic thrombocytopenic purpura. Thromb. Haemost. 93, 267–274 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Tortorella M. et al., The thrombospondin motif of aggrecanase-1 (ADAMTS-4) is critical for aggrecan substrate recognition and cleavage. J. Biol. Chem. 275, 25791–25797 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Gao G. et al., Activation of the proteolytic activity of ADAMTS4 (aggrecanase-1) by C-terminal truncation. J. Biol. Chem. 277, 11034–11041 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Fédière G., Li Y., Zádori Z., Szelei J., Tijssen P., Genome organization of Casphalia extranea densovirus, a new iteravirus. Virology 292, 299–308 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Pham H. T. et al., Expression strategy of Aedes albopictus densovirus. J. Virol. 87, 9928–9932 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pénzes J. J., de Souza W. M., Agbandje-McKenna M., Gifford R. J., An ancient lineage of highly divergent parvoviruses infects both vertebrate and invertebrate hosts. Viruses 11, 525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Day J. M., Zsak L., Determination and analysis of the full-length chicken parvovirus genome. Virology 399, 59–64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z. et al., Human bocavirus NP1 inhibits IFN-β production by blocking association of IFN regulatory factor 3 with IFNB promoter. J. Immunol. 189, 1144–1153 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Tse H. et al., Discovery and genomic characterization of a novel ovine partetravirus and a new genotype of bovine partetravirus. PLoS One 6, e25619 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zádori Z., Szelei J., Tijssen P., SAT: A late NS protein of porcine parvovirus. J. Virol. 79, 13129–13138 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Im D. S., Muzyczka N., Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J. Virol. 66, 1119–1128 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staden R., Beal K. F., Bonfield J. K., The Staden package, 1998. Methods Mol. Biol. 132, 115–130 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Carver T., Harris S. R., Berriman M., Parkhill J., McQuillan J. A., Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 72.Lobley A., Sadowski M. I., Jones D. T., pGenTHREADER and pDomTHREADER: New methods for improved protein fold recognition and superfamily discrimination. Bioinformatics 25, 1761–1767 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Letunic I., Bork P., 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holm L., DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Armougom F. et al., Expresso: Automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 34, W604–W608 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okonechnikov K., Golosova O., Fursov M.; UGENE team , Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 28, 1166–1167 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Darriba D., Taboada G. L., Doallo R., Posada D., ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Felsenstein J., PHYLIP (Phylogeny Inference Package) (version 3.6, Department of Genome Sciences, University of Washington, Seattle).
- 79.Suchard M. A. et al., Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A., Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Altschul S. F. et al., Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang J. et al., The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 12, 7–8 ((2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suloway C. et al., Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 84.Zheng S. Q. et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang G. et al., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Spear J. M. et al., The influence of frame alignment with dose compensation on the quality of single particle reconstructions. J. Struct. Biol. 192, 196–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pettersen E. F. et al., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Kleywegt G. J., Jones T. A., xdlMAPMAN and xdlDATAMAN—Programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr. D Biol. Crystallogr. 52, 826–828 (1996). [DOI] [PubMed] [Google Scholar]
- 90.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Adams P. D. et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genome sequence of PmMDV, accompanied by its annotation, has been deposited in the GenBank database (accession no. MK028683). The structure of the PmMDV capsid has been deposited to the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank, https://www.rcsb.org/ (PDB ID code 6WH3 for the native PmMDV capsid; PDB ID code 6WH7 for the EDTA-treated PmMDV capsid).