Significance
Fluorescence micrographs that capture the sizes, shapes, and distributions of liquid domains in model membranes have provided high standards of evidence to prove (and disprove) theories of how micron-scale domains form and grow. Some theories about smaller domains have remained untested, partly because experimental methods of identifying submicron domains in vitrified, hydrated vesicles have not been available. Here we introduce two such methods; both leverage cryo-electron tomography to observe membrane features far smaller than the diffraction limit of light. The first method is probe-free and identifies differences in thicknesses between liquid domains and their surrounding membranes. The second method identifies membrane regions labeled by an electron-dense, fluorescent protein, which enables direct comparison of fluorescence micrographs with cryo-electron tomograms.
Keywords: probe-free, height mismatch, phase separation, membrane
Abstract
Images of micrometer-scale domains in lipid bilayers have provided the gold standard of model-free evidence to understand the domains' shapes, sizes, and distributions. Corresponding techniques to directly and quantitatively assess smaller (nanoscale and submicron) liquid domains have been limited. Researchers commonly seek to correlate activities of membrane proteins with attributes of the domains in which they reside; doing so hinges on identification and characterization of membrane domains. Although some features of membrane domains can be probed by indirect methods, these methods are often constrained by the limitation that data must be analyzed in the context of models that require multiple assumptions or parameters. Here, we address this challenge by developing and testing two methods of identifying submicron domains in biomimetic membranes. Both methods leverage cryo-electron tomograms of ternary membranes under vitrified, hydrated conditions. The first method is optimized for probe-free applications: Domains are directly distinguished from the surrounding membrane by their thickness. This technique quantitatively and accurately measures area fractions of domains, in excellent agreement with known phase diagrams. The second method is optimized for applications in which a single label is deployed for imaging membranes by both high-resolution cryo-electron tomography and diffraction-limited optical microscopy. For this method, we test a panel of probes, find that a trimeric mCherry label performs best, and specify criteria for developing future high-performance, dual-use probes. These developments have led to direct and quantitative imaging of submicron membrane domains in vitrified, hydrated vesicles.
Seeing is believing, which makes images powerful. Current advances in microscopy have revolutionized our understanding of cellular components, macromolecular assemblies, protein structure, and membrane organization. For example, images of micrometer-scale synapses in stimulated immune cells have successfully led to the development of quantitative models of membrane protein interactions (1). Similarly, direct imaging has demonstrated that vacuole membranes in living yeast cells phase-separate (2, 3) and that model and cell-derived membranes exhibit critical phenomena (4, 5). However, in all of these examples, the membrane features span micrometer length scales. Challenges persist in observing membrane features that are far smaller than the diffraction limit of light, especially in model lipid vesicles under native solution conditions (a phrase that describes vitrified vesicles captured in a fully hydrated state). As a result, a wide range of quantitative questions has remained impossible to answer. For example, if a vesicle membrane contains nanodomains, what are the sizes and distributions of those domains across the vesicle surfaces? Similarly, do submicron domains fit quantitative predictions of modulated phases or of microemulsions (6, 7)?
New approaches are needed in order to overcome current limitations and to complement current methods. Transmission electron microscopy (TEM) can achieve near-atomic resolution, and freeze-fracture TEM has successfully been used to identify coexisting solid and liquid phases in simple lipid membranes (8–11). However, freeze-fracture is an unwieldy technique that images a metal-shadowed surface of a membrane. To date, freeze-fracture has achieved contrast between liquid domains and the rest of the membrane only when membranes contain large protein complexes (2). A more common way of identifying submicron liquid domains by TEM is gold labeling of proteins and lipids (e.g., refs. 12 and 13). This method results in overcounting (which can be misinterpreted as self-clustering proteins) if labeling uses both primary and secondary antibodies or if multiple labels are conjugated to a single antibody (14). Other methods of imaging submicron liquid domains have their own limitations. Atomic force microscopy (AFM) requires deposition of membranes on solid substrates (15–22). Near-field scanning optical microscopy (NSOM) places cantilevers in contact with membranes, which may alter membrane structures (23). Standard superresolution optical techniques cannot image small-enough features, and expansion microscopy relies on cross-linked proteins (24).
Here, we introduce and test two methods for identifying submicron domains in membranes under native solution conditions from direct images collected by cryo-electron tomography (cryo-ET) Of the two methods, the first is entirely label-free and leverages differences in thicknesses of the domains versus the rest of the membrane (Fig. 1). The companion paper by Heberle et al. (25) uses a similar approach with cryo electron microscopy projection images. We benchmark our label-free approach against known phase diagrams to demonstrate that it accurately quantifies the area fractions of coexisting liquid-ordered (Lo) and liquid-disordered (Ld) phases in membranes. The second method employs a probe that is fluorescent, electron-dense, and labels the membrane through a single binding site. Our goal is for the probe to enable direct, model-free comparisons for a single vesicle sample analyzed by both fluorescence microscopy and electron microscopy. We test a panel of probes and find that a trimeric mCherry label performs best in this role.
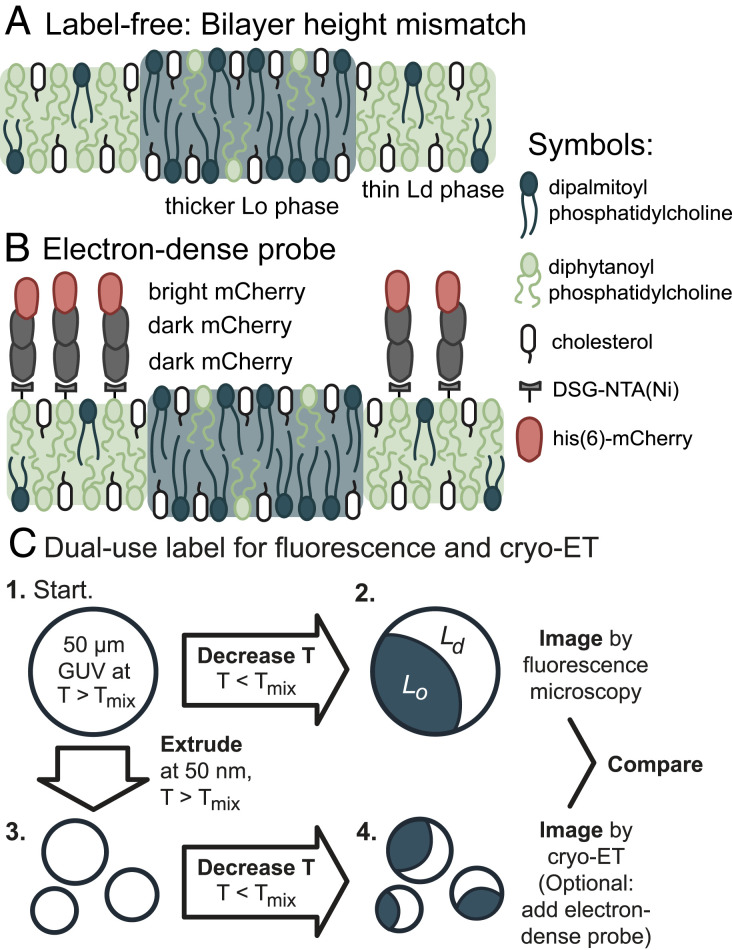
Fig. 1.
Two methods for direct identification of lipid domains in vesicles using cryo-ET. (A) The label-free method identifies the difference in thickness between the Lo phase and Ld phase. (B) The labeling method deploys a linear trimer of mCherry that is both fluorescent and electron-dense. A his6-tag on the terminal mCherry protein binds to a nickel-chelated lipid that preferentially partitions to the Ld phase. (C) The mCherry label enables imaging a single starting solution of GUVs by both fluorescence microscopy and cryo-ET.
Results
Fig. 1 summarizes the two methods we developed for identifying coexisting Ld and Lo phases in vesicles by cryo-ET. The first method exploits the difference in bilayer thicknesses of the phases. The second method employs a label that preferentially partitions to the Ld phase. We tested these methods on systems that represent the broad class of membranes known to separate into macroscopic Ld and Lo phases in giant unilamellar vesicles (GUVs). Specifically, we imaged uncharged membranes composed of ternary mixtures, consisting of a lipid with a low melting temperature (diphytanoyl-phosphocholine, DiPhyPC), a lipid with a high melting temperature (dipalmitoyl-phosphocholine, DPPC), and a sterol (cholesterol) (26). Cryo-ET resolves submicron features of intact vesicles in aqueous environments. We imaged submicron domains in membranes with coexisting liquid phases under native solvent conditions.
To maximize differences between the Ld and Lo phases so they are distinguishable, we mixed the lipids in ratios that fall along an unusually long tie line (Fig. 2A and Table 1) (26). The endpoints of tie-lines represent the lipid compositions of the two phases. As a result, when we use cryo-ET to image submicron vesicles, we expect to observe Lo domains that are significantly (∼1 nm) thicker than membranes of the surrounding Ld phase (20). The same concept of employing a long tie line to maximize contrast applies when we use fluorescence microscopy. For example, when GUVs composed of the same lipid ratio are labeled with Texas Red dihexadecanoyl-PE (DHPE), Lo domains are significantly darker than the surrounding Ld phase (Fig. 2B and ref. 26).
Fig. 2.
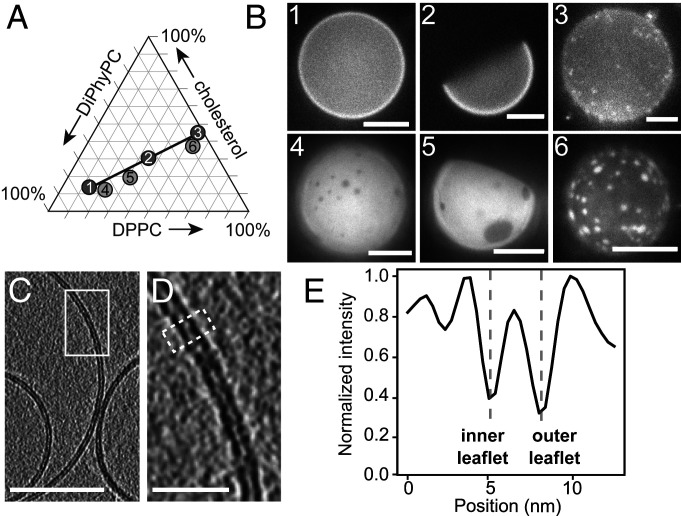
The fraction of membrane area in the Lo phase increases monotonically along tie-lines. (A) All possible ternary mixtures of DiPhyPC, DPPC, and cholesterol fall within the triangle. Ratios 1, 2, and 3 lie on a tie-line at 22 °C (26). Ratios 4, 5, and 6 lie on an extrapolated line parallel to the known tie-line. (B) Representative fluorescence micrographs of GUVs made from ratios 1 through 6. The GUVs contain 0.8 mol % of the dye Texas Red DHPE, which preferentially partitions to the Ld phase. To facilitate visualization of area fractions, images were captured shortly after domains nucleated, before all domains completely coalesced. (Scale bars, 20 µm.) (C) Slice at 0° through a cryo-ET tomogram of a field of vesicles made from ratio 4. Bilayer regions are resolvable as two distinct monolayer leaflets. (Scale bar, 50 nm.) The field of vesicles containing C is reproduced in SI Appendix, Fig. S2. (D) Enlarged image of the area in the white box in C. (Scale bar, 10 nm.) (E) A linescan reveals two troughs, which correspond to the clearly resolved dark bands of the inner and outer leaflets of the membrane in D. The linescan was 10 pixels wide and taken across the area in D outlined in the white dashed line.
Table 1.
Vesicles were produced from six lipid ratios
| Ratio | DiPhyPC, mol % | DPPC, mol % | Cholesterol, mol % | Lo phase, % |
| Ratio 1 | 68 | 17 | 15 | 0 |
| Ratio 2 | 35 | 25 | 30 | 50 |
| Ratio 3 | 6 | 52 | 42 | 100 |
| Ratio 4 | 66 | 21 | 13 | 10 |
| Ratio 5 | 48 | 32 | 20 | 20 |
| Ratio 6 | 5 | 58 | 37 | 90 |
Ratios 1, 2, and 3 fall on a tie-line measured at 22 °C from (26). Ratios 4, 5, and 6 fall on a parallel line. DGS-NTA(Ni) lipids were included in each mixture only when trimeric mCherry probes were used and only in ratios 2 and 5, replacing, 2 mol % of DiPhyPC. Values of the mole percent of all lipids that are expected to be in the Lo phase are derived from two methods: fluorescence microscopy of GUVs and NMR of multilamellar vesicles (26). Uncertainties in the mole percent of Lo phase are ±6%, propagated from uncertainties ≤3% for each tie-line endpoint, as measured by NMR (26).
One of our central goals was to quantitatively benchmark cryo-ET results for 10- to 100-nm vesicles against fluorescence microscopy results for vesicles roughly a thousand times larger. To ensure that the lipid composition of vesicles did not vary with their size, we made careful choices about how we produced vesicles. Because different techniques incorporate different ratios of lipids into vesicles (27–29), we produced all vesicles by the same technique: electroformation (Fig. 1C). We maintained some of these vesicles as GUVs to image by fluorescence microscopy, and we extruded others through 50- or 100-nm pores to image by cryo-ET (Fig. 2 and SI Appendix, Figs. S1, S2, and S5). All vesicle solutions (whether GUVs or extruded vesicles) were diluted into buffer solutions. The dark areas in the representative images of Fig. 2B are consistent with the fraction of Lo phase in Table 1, which strongly suggests that the buffer does not shift the tie-line in Fig. 2A, within measurement uncertainty.
To identify Ld and Lo domains by the first cryo-ET method, we took a probabilistic approach, separating distributions of bilayer thicknesses into component parts associated with each phase. Critically, this approach sets no thickness cutoff and makes no assumptions about the size, spatial arrangement, or absolute thicknesses of domains. First, we collected cryo-electron tomograms for two types of control vesicles, which we made from lipid ratios that lay at the two ends of a tie-line (ratios 1 and 3). To identify the two leaflets of vesicle membranes, we performed a Canny edge filter to the central slice of each tomogram. We established an objective procedure to determine apparent bilayer thicknesses; specifically, we evaluated the minimum distance from every pixel on the inner leaflet of vesicles to all possible pixels on the outer leaflet (Figs. 3A and 4). Because this procedure identifies differences in thicknesses rather than absolute thicknesses, it is robust to subtle changes in how different research groups might defocus tomograms, apply contrast transfer functions, or create edge filter algorithms.
Fig. 3.
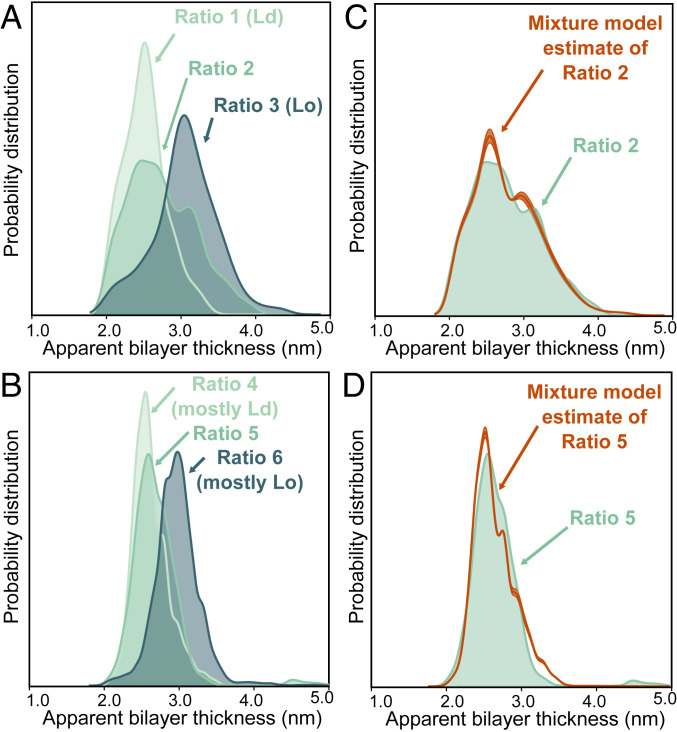
Quantitative agreement between ratios of Lo and Ld phases measured by cryo-ET and ratios expected from GUV phase diagrams. (A and B) Approximately 5,000 apparent bilayer thickness values are plotted for each lipid ratio (derived from measurements of the central slice of 20 to 30 vesicles, representing 10 tomograms of vesicle fields per lipid ratio, from one preparation session and one imaging session for ratios 1, 2, and 3 and another session for ratios 4, 5, and 6.). For all six ratios, Gaussian kernel density estimates (which plot the probability of measuring each distance, similar to a histogram) were calculated for all three ratios. (C and D) The ratio of the membrane area in the Lo phase vs. the Ld phase can be estimated directly from images using a mixture of kernel density estimates, calculated using ratios 1 and 3 for C and ratios 4 and 6 for D. For ratio 2 and ratio 5, this procedure yields area ratios of 43:57 ± 3 Ld:Lo and 83:17 ± 2 Ld:Lo, respectively.
Fig. 4.
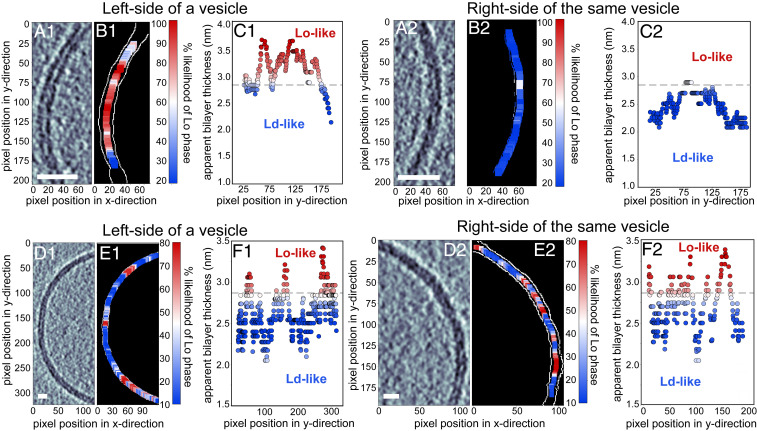
Apparent membrane thicknesses identify regions consistent with Lo vs. Ld phases. A–C are from a single vesicle made from the lipids in ratio 2. Similarly, D–F are from a single vesicle of ratio 4. (A1, A2, D1, and D2) Label-free tomogram slices showing bilayer regions resolved as two distinct monolayer leaflets. (Scale bars, 10 nm.) (B1, B2, E1, and E2) Each slice after detection of the two edges of the bilayer (white pixels). (C1, C2, F1, and F2) The location of each pixel on the inner edge of the bilayer vs. the minimum distance from that pixel to the outer edge. Colors represent the likelihood (from 0 to 100%) that each distance corresponds to the thicker, Lo, phase instead of the thinner, Ld, phase. A horizontal dashed line shows the apparent bilayer thickness for which there is a 50% likelihood of being in the Lo phase. More examples of colormap images are found in SI Appendix, Fig. S3.
For the two control samples, distributions of thicknesses formed two distinguishable peaks corresponding to a thinner Ld membrane (ratio 1) and a thicker Lo membrane (ratio 3). We repeated this procedure for vesicles made from ratio 2, which lies between ratios 1 and 3. This intermediate composition falls in a region of the phase diagram in which membranes exhibit coexisting Ld and Lo membrane phases. Because this composition is far from a miscibility critical point, domains of Ld and Lo always coarsen into micrometer-scale regions in taut GUVs (30). Therefore, the area fraction of Ld and Lo phases that has been previously measured in taut GUVs (26) should be equivalent to the area fraction in submicron domains imaged by cryo-ET.
In Fig. 3A, it is clear that cryo-ET of submicron vesicles of ratio 2 indeed yields a distribution of bilayer thicknesses that corresponds to a mixture of thin and thick membranes. In Fig. 4, it is clear that this probe-free method can resolve submicron domains. Domains in Fig. 4 are constrained to submicron sizes for the obvious reason that the vesicle themselves are submicron, and perhaps also for the more subtle reason that excess area in nonspherical membranes allows submicron domain configurations (6). Next, we quantitatively evaluated the probability that each thickness corresponds to the Ld versus the Lo phase, using the mixture model described in Materials and Methods. This analysis led to the conclusion that vesicles made from ratio 2 contain domains and that 43 ± 3% of the membrane area is in the Ld phase.
This area fraction of 43 ± 3% Ld phase, measured by cryo-ET in submicron vesicles, is in statistical agreement with values measured in vesicles that are hundreds to thousands of times larger. Quantitative tie-lines have been previously measured by NMR of multilamellar vesicles of the same lipid composition (35/35/30 DiPhyPC/DPPC/cholesterol), and edges of liquid–liquid coexistence regions have been previously measured by fluorescence microscopy of GUVs (26). These previous experiments have firmly established that micrometer-scale GUVs made from lipids mixed in ratio 2 contain 50 ± 6 mol % Ld phase, which agrees with our cryo-ET values within the experimental uncertainty of the two methods.
Next, we tuned the lipid composition (and area fraction) of vesicles to show that thickness mismatches quantitatively identify domains in membranes that do not lie exactly on tie-lines, with controls that do not lie exactly at endpoints. For example, ratios 4, 5, and 6 lie on a line that is parallel and near the tie-line of ratios 1, 2, and 3. The two new controls (ratios 4 and 6) are near endpoints but are not purely Ld or Lo phases. Applying the cryo-ET imaging and analysis above leads to the conclusion that for vesicles made from ratio 5, 83 ± 2% of the area is Ld phase (Fig. 3D). This value is in excellent agreement with two independent measurements for micrometer-scale vesicles. A value of 80 ± 6 mol % of Ld phase is expected for intact vesicles (26), and 76 ± 6 area % of Ld phase was determined by AFM of GUVs ruptured on mica surfaces (20). In summary, a label-free approach of identifying domains by membrane thicknesses accurately quantifies the amount of Ld and Lo phases.
Switching our focus to identify Ld or Lo domains by the second method, namely by partitioning of a probe, we surveyed labels that are both fluorescent and electron-dense. Our goal was to find a single probe to serve two purposes: to image micrometer-scale GUVs by fluorescence microscopy and to image submicron domains in ∼100-nm vesicles by cryo-ET. This requirement of a dual use imposed several challenging criteria. The probe 1) must be highly electron-dense so that it is visible by cryo-ET, 2) must be fluorescent, and 3) must partition strongly with membrane domains. Moreover, for the probe to be nonperturbing, it must meet the following additional criteria. 4) Any fluorescent or electron-dense moiety must be attached to the probe through a single binding site in order to avoid overcounting (14) or cross-linking (31). 5) The probe must partition strongly to the membrane so that it can be used at low concentrations (32). 6) The probe must not severely perturb the membrane’s shape [as, for example, a BAR domain protein would (33)]. 7) The probe must not aggregate or induce membranes to stick to each other.
We tested a panel of seven probes (SI Appendix, Table S1 and Fig. S6) and found that our criteria were best met by an mCherry trimer that binds through a single site to DGS-NTA(Ni) lipids incorporated into electroformed vesicles. Other probes fell short by aggregating (A206K GFP, green fluorescent protein), causing vesicles to aggregate (A206K GFP), and/or producing no discernible contrast between membrane phases [14:0 PE-DTPA(Gd), GM1 lipids with Cholera Toxin B, monomeric mCherry, and 18:1 DGS-NTA(Ni) without mCherry].
In Fig. 5, we establish proof of principle that membranes labeled by a single probe can be imaged by both fluorescence microscopy and cryo-ET. To image micrometer-scale GUVs with our dual-use mCherry trimer, we added the probe directly to GUV solutions. Fluorescence micrographs in Fig. 5 A and B show that the probe strongly preferentially partitions to the Ld phase. To image smaller vesicles with our dual-use probe, we extruded unlabeled GUVs and then added mCherry trimer to the resulting solution (Fig. 1C). In many cryo-ET tomograms, clusters of mCherry trimers appear in a single layer on membrane surfaces (Fig. 5 C–F), consistent with the bright labeling of domains we observed in GUVs. Trimers in the clusters are evenly spaced ∼3 nm apart (SI Appendix, Fig. S7), consistent with monovalent binding to DGS-NTA(Ni) lipids in Ld domains.
Fig. 5.
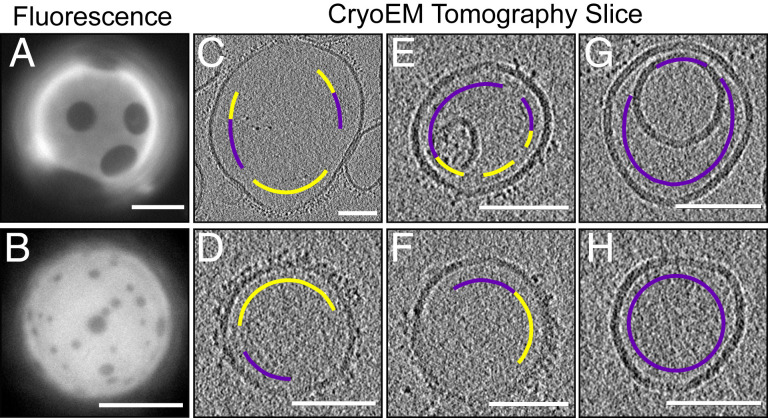
Trimeric his6-mCherry as a dual-use probe to image GUVs by fluorescence microscopy and to image submicron vesicles by cryo-ET. Lipids were mixed in ratios 2 (top row) and 4 (bottom row). (A and B) Fluorescence micrographs in which trimeric his6-mCherry labels the Ld phase of GUVs. (Scale bars, 20 µm.) (C–H) Cryo-ET tomographic central slices of extruded vesicles. On the exterior of vesicles, regions that are densely covered by a brush of trimeric his6-mCherry (yellow arc) are clearly distinguishable from areas that are devoid of his6-mCherry (magenta arc). (Scale bars, 100 nm.) A larger version of this figure appears in SI Appendix, Figs. S7 and S8.
The utility of any probe, including the mCherry trimer, to image domains by cryo-ET or to determine areas of Lo vs. Ld phases is mitigated by four observations, all of which are illustrated in Fig. 5. 1) When vesicles touch, it is unclear if an absence of probe denotes an Lo domain or simply inaccessibility of the probe to the membrane surface. For applications in which the addition of charged lipids is acceptable, their presence can help maintain separation between membranes. 2) When membrane regions are sparsely labeled, it is unclear whether they should be assigned to the Lo phase or to the Ld phase. 3) Uniform mixing of the probe is challenging to achieve—some vesicles appear unlabeled (Fig. 5 G and H). 4) Two regions of the sample are unusable: the interior vesicles of multilamellar structures, which are inaccessible to the probe, and the air–water interfaces, which trap unbound probes (Movie S1). An advantage of tomography is that the air–water interface can be computationally sliced away.
Discussion and Conclusion
Here, we leveraged cryo-ET imaging to develop two methods of identifying liquid domains in membranes. Both methods present strengths and limitations.
The main advantage of using membrane thickness to identify liquid domains in unstained vesicles is that the technique is probe-free. The method is nonspecific to the type of lipids, as long as there is a measureable thickness mismatch between phases. For example, membranes containing sphingomyelins (which more closely mimic eukaryotic plasma membranes) have significant thickness mismatches between Lo and Ld phases (16).
In addition, using membrane thickness to identify liquid domains avoids all concern that labels may shift transition temperatures (32) or lead to oxidation (34). Because the analysis is statistical, it works well when distributions are built from a large number of thickness measurements. For example, the distributions of ratios 1 and 3 in Fig. 3A reflect ∼5,000 points. Conversely, the method will fail if images are not representative. For example, in isolation, the micrograph in Fig. 4 A2 could be misinterpreted as signifying that all membranes in ratio 2 are nearly entirely in the Ld phase. Another feature of the technique is that it does not require vesicles to be unilamellar or spherical, so it can be applied to uncharged vesicles extruded through 100-nm pores, which are typically neither unilamellar (35) nor spherical (Figs. 2 and 5). Membrane thickness differences can also be used to qualitatively identify domains in membranes derived from cells, as in the companion article by Heberle et al. (25).
However, a quantitative version of this approach is limited to membranes in which the difference in thicknesses between the Ld and Lo phases is resolvable. Luckily, many membranes fulfill this criterion (16–21). Thickness differences can be maximized through savvy choices for the types of lipids in the system and the ratios at which they are mixed. For example, mixing long, saturated lipids with short, unsaturated or methylated lipids typically results in thick Ld phases and thin Lo phases (16–20). Existing phase diagrams and tie-lines, which are reviewed in refs. 36 and 37 and discussed further in SI Appendix, can be leveraged to quantitatively and accurately measure the relative amounts of Ld and Lo phases.
The main advantage of using an mCherry probe to identify liquid domains is that the label is both fluorescent and electron-dense, enabling similarly prepared vesicles (made from the same batch of lipids, on the same day, using the same methods up to the final extrusion step) to be directly imaged by both fluorescence microscopy and cryo-ET. A potential biological application of dual-use probes is in identifying lipid domains in yeast vacuole membranes. Briefly, vacuole membranes of Saccharomyces cerevisiae demix into coexisting liquid phases after the cells experience nutrient depletion (3). A probe could be used to facilitate correlation of fluorescence images with cryogenic electron micrographs in order to identify when liquid domains first appear in the membranes.
Dual-use probes of this type are also necessary for some types of controls. For example, domain sizes in some types of membranes are reported to be different in GUVs and in ∼60-nm vesicles (38); a dual-purpose probe could be used to distinguish if the discrepancy is due to different vesicle sizes or merely due to sample-to-sample variations in lipid composition. This point is powerful because lipid ratios in electroformed GUVs can differ from ratios in hydrated, multilamellar vesicles (the basis of most protocols for imaging submicron domains) (39). An additional advantage of the mCherry probe is that it attaches to a lipid through a single binding site, which avoids overcounting.
With all probes, it can be difficult to achieve uniform labeling, especially when vesicles are near an air–water interface or are in contact with each other, as is common when vesicles are composed of only zwitterionic lipids. Likewise, for all probes, it is difficult to determine whether clusters of probes originally nucleated on the membrane or in solution. An advantage of the mCherry probe is that it has been shown to aggregate only at relatively high concentrations [>25 µM (40)]. A remaining challenge is that the mCherry probe appears to strongly partition to the air–water interface of cryo-ET grids, and any protein at this location has the potential to denature. Looking to the future, productive approaches could include synthesis of quantum dots that label membranes via a single linker.
In conclusion, we have developed two methods to identify submicron domains in membranes. One method employs thickness differences, and the other employs dual-use probes. We generate direct cryo-ET images of <100 nm liquid domains in protein-free, model membranes under native solvent conditions. We use these images to quantitatively correlate the area fractions of Ld (and Lo) phase in small vesicles on length scales smaller than 100 nm and in giant vesicles on length scales greater than micrometers.
Our approach complements existing methods for imaging <100-nm liquid domains in membranes, including freeze-fracture TEM (8–11), TEM of gold-labeled membranes (12, 13), AFM (15–21), and NSOM (23). All of these methods suffer from low throughput. Nevertheless, they are valuable because they circumvent limitations of spectroscopic methods (e.g., NMR, electron paramagnetic resonance, and fluorescence resonance energy transfer) and scattering methods (e.g., X-ray and neutron scattering) (38, 41–44). These indirect methods typically measure only average properties of domains and typically generate data that must be analyzed in the context of models. Direct imaging of 10- to 100-nm liquid domains is valuable because it can potentially allay concerns that submicron domains are artifacts (45). Direct imaging is vital for testing domain nucleation theories (46), evaluating microemulsion mechanisms (7), assessing simulations (47), and probing discrepancies between results in GUV and in ∼60-nm vesicles (38).
Materials and Methods
Lipids.
Phosphocholine (PC) lipids (Avanti Polar Lipids), cholesterol (Sigma-Aldrich), and Texas Red DHPE (Life Technologies) were used as purchased without further purification. Lipid stock solutions in chloroform contained, at minimum, a ternary mixture of DiPhyPC (4 ME 16:0 PC), DPPC (16:0 PC), and cholesterol. DiPhyPC and DPPC are zwitterionic; cholesterol is uncharged. Features of this ternary mixture is that its miscibility phase diagram has been mapped in detail (26), and the saturated carbon chains of DiPhyPC resist oxidation (5). Stock solutions for cryo-ET experiments with mCherry labels, which were limited to lipid ratios 2 and 5, also contained 2 mol % of a nickel-chelating lipid of dioleoylglycerosuccinylimino-diacetic acid [18:1 DGS-NTA(Ni), which replaced 2 mol % DiPhyPC], whereas stocks for fluorescence microscopy controls of GUVs contained 0.8 mol % Texas Red DHPE.
GUV Electroformation.
Solutions containing 2 × 10−6 mol (∼0.76 mg) of lipids were spread evenly on slides coated with indium tin oxide. The slides were placed under vacuum for >30 min to evaporate the chloroform. A capacitor was created by sandwiching 0.3-mm Teflon spaces between two lipid-coated slides. The gap was filled with 335 mM sucrose, and the edges were sealed with vacuum grease. Sucrose has been previously shown to not shift the miscibility transition temperature of electroformed vesicles of DiPhyPC/DPPC/cholesterol (48). GUVs 10 to 100 µm in diameter were electroformed (39) by application of an AC voltage of 1.5 V at 10 Hz across the capacitor for 1 h at 60 °C.
Extrusion.
Two solutions were produced: a “thickness mismatch buffer” of 150 mM NaCl and 25 mM Hepes and an “mCherry buffer” of 150 mM NaCl, 25 mM Hepes, and 1 mM Tris(2-carboxyethyl)phosphine. GUVs were diluted fivefold in one of the buffers and then concentrated by centrifugation at ∼10,000 rcf for 10 min. Supernatant was removed. Approximately 10 to 20 µL of centrifuged and concentrated GUVs were rediluted in 100 µL of buffer and stored at 60 °C for <15 min before extrusion in order to ensure that vesicles were well above their mixing temperatures, which ranged from 25 °C to 48 °C. A miniextruder (Avanti Polar Lipids) with a heat block and two 1-mL gas-tight syringes were maintained in an oven at 75 °C before use. Most GUVs were extruded 29 times at 75 °C in polycarbonate membranes with 50-nm pores [to maximize consistency with the companion paper by Heberle et al. (25), which used 50-nm pores]. The only exception was that GUVs of lipid ratios 4, 5, and 6 for thickness mismatch experiments were extruded through membranes with 100-nm pores. The resulting small vesicles were stored at room temperature for <1 h before vitrification.
Despite initial electroformation, extruded vesicles were often not unilamellar. This is surprising because electroformation typically yields unilamellar vesicles. We found that vesicles electroformed and extruded in pure water were always multilamellar, with at least two lamellae per vesicle. When we increased the ionic strength of the buffer with NaCl, we produced more unilamellar vesicles. Interestingly this result is the opposite to that reported in ref. 35.
Introduction of Trimeric mCherry.
Extruded vesicles containing 2 mol % DGS-NTA(Ni) lipids were diluted in “mCherry buffer,” lightly vortexed with 2 mM trimeric mCherry (49), and allowed to incubate for >25 min at room temperature. The DSG-NTA(Ni) concentration that we used is an order of magnitude lower than the concentration of DPIDA determined by Scheve et al. (50) to shift a membrane's miscibility transition temperature. Specifically, to induce steric crowding, Scheve et al. (50) used 25 mol % DPIDA, a lipid that is similar to DSG-NTA(Ni) and that binds a histidine-tagged protein roughly the same size as mCherry, his-GFP. To ensure that the mCherry trimer was not aggregated in solution, we performed dynamic light scattering and found an average particle radius of 3.04 ± 0.3 nm, which is close to the measured radius for GFP (∼2 nm; Protein Data Bank ID code 1GFL), a protein almost identical to mCherry (SI Appendix, Fig. S9 and Text S4). The mCherry trimer presents a larger volume of electron-dense material than a single mCherry molecule. The first mCherry in the trimer is fluorescent, and the last mCherry binds monovalently through a his6-tag to the nickel atom on a DGS-NTA(Ni) lipid (Fig. 1). At pH 7.4 (the pH of our “mCherry buffer”), the free carboxyl group on DGS-NTA(Ni) is negatively charged. By cryo-ET we did not observe a difference in shape or lamellarity of vesicles with or without DGS-NTA(Ni).
Fluorescence Imaging.
Immediately before imaging, GUV solutions were further diluted 10-fold in one of the buffers and sandwiched between two coverslips. The edges of the coverslips were sealed with vacuum grease. Both DGS-NTA(Ni) and Texas Red DHPE preferentially partition to the Ld phase, which appears bright by fluorescence microscopy; the Lo phase appears dark. GUV images were viewed through an air objective on a Nikon Y-FL upright epifluorescence microscope, captured on a Photometrics CoolSnapFX camera, and manipulated using ImageJ (https://imagej.nih.gov/ij/). To preserve the fidelity of the data, image manipulation was limited to adjusting overall brightness or implementing linear (γ = 1) contrast enhancements.
Cryo-ET.
Solutions of extruded vesicles were mixed with 6 nm colloidal gold fiducial markers (Aurion) and applied to glow-discharged C-flat holey carbon grids (Electron Microscopy Science) or QUANTIFOIL R 2/2 holey carbon grids (Quantifoil Micro Tools GmbH) and plunge-frozen into liquid ethane using a Vitrobot Mark IV (FEI) at 100% humidity and temperatures of either 25 °C (for height mismatch) or 4 °C (for trimeric mCherry).
The thinness of the water film on the grid can perturb larger vesicles by flattening them and by introducing interactions with the air–water interface. We estimated the thickness of the water film as follows. The average thickness of each tomogram is 126.7 nm (499 unbinned pixels). The thinnest region of vitreous ice containing a vesicle is 244 pixels in the reconstructed tomogram. Due to resolution anisotropy of tomographic reconstructions from limited angular sampling, the z-dimension is stretched relative to the x- and y-dimensions. To account for this, the gold fiducial markers were measured perpendicular to the tilt axis and were found to be an average diameter of 28.2 pixels (7.1 nm). Measurement of the gold markers in the z-dimension indicates that the data are elongated by factors ranging from 1.2 to 1.7. Incorporating the most extreme elongation factor, the minimum observed ice thickness is ∼36 nm. Given that membrane domains were observed in vesicles over the entire range of sizes, with expected area fractions, this perturbation is minor at the 0° plane where vesicles were evaluated.
Additional details are given in SI Appendix.
Analysis of Thickness Mismatches.
Cryo-electron tomograms were analyzed to yield the fraction of bilayer corresponding to the Lo and Ld phases. A detailed schematic of the analysis process can be found in SI Appendix, Fig. S1. We cropped fields of tomograms to retain only the areas in which the membrane was resolvable in the central tomographic slice as two bands of lipid headgroups with high electron density (Fig. 2C).
Additional details are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank Eva Schmid and Dan Fletcher for their kind gift of trimeric mCherry. We thank Jeanne Stachowiak for her kind gift of monomeric mCherry and A206K GFP, as well as for expert advice on the development of probes for electron microscopy. We thank Adam Nguyen for dynamic light scattering experiments. We thank Avanti Polar Lipids for the gift of Aurora-DSG nanoparticles. We thank Ilya Levental and Fred Heberle for outstanding collegiality; they saw us present the results in this manuscript at the 2019 Biophysical Society Meeting, informed us of their complementary project, and simultaneously submitted with us. We hope our feedback influenced their manuscript as positively as their feedback influenced ours. Seed funding for this project was provided by UW Royalty Research Grant A122781 to S.L.K. Lipid research in the S.L.K. laboratory is supported by National Science Foundation grants MCB-1402059 and MCB-1925731. K.K.L. was supported by NIH grant R01-GM099989. C.E.C. and A.M. were funded by the National Institutes of General Medical Sciences of the National Institutes of Health under award T32GM008268 (to C.E.C.) and T32-GM007750 (to A.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002245117/-/DCSupplemental.
Data Availability.
Some study data available.
References
- 1.Grakoui A. et al., The immunological synapse: A molecular machine controlling T cell activation. Science 285, 221–227 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Moor H., Mühlethaler K., Fine structure in frozen-etched yeast cells. J. Cell Biol. 17, 609–628 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rayermann S. P., Rayermann G. E., Cornell C. E., Merz A. J., Keller S. L., Hallmarks of reversible separation of living, unperturbed cell membranes into two liquid phases. Biophys. J. 113, 2425–2432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honerkamp-Smith A. R., Machta B. B., Keller S. L., Experimental observations of dynamic critical phenomena in a lipid membrane. Phys. Rev. Lett. 108, 265702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honerkamp-Smith A. R. et al., Line tensions, correlation lengths, and critical exponents in lipid membranes near critical points. Biophys. J. 95, 236–246 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornell C. E. et al., Tuning length scales of small domains in cell-derived membranes and synthetic model membranes. Biophys. J. 115, 690–701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schick M., Strongly correlated rafts in both leaves of an asymmetric bilayer. J. Phys. Chem. B 122, 3251–3258 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Grant C. W., Wu S. H., McConnell H. M., Lateral phase separations in binary lipid mixtures: Correlation between spin label and freeze-fracture electron microscopic studies. Biochim. Biophys. Acta 363, 151–158 (1974). [DOI] [PubMed] [Google Scholar]
- 9.Luna E. J., McConnell H. M., Multiple phase equilibria in binary mixtures of phospholipids. Biochim. Biophys. Acta 509, 462–473 (1978). [DOI] [PubMed] [Google Scholar]
- 10.Stewart T. P., Hui S. W., Portis A. R. Jr., Papahadjopoulos D., Complex phase mixing of phosphatidylcholine and phosphatidylserine in multilamellar membrane vesicles. Biochim. Biophys. Acta 556, 1–16 (1979). [DOI] [PubMed] [Google Scholar]
- 11.Lentz B. R., Alford D. R., Hoechli M., Dombrose F. A., Phase behavior of mixed phosphatidylglycerol/phosphatidylcholine multilamellar and unilamellar vesicles. Biochemistry 21, 4212–4219 (1982). [DOI] [PubMed] [Google Scholar]
- 12.Wilson B. S. et al., Markers for detergent-resistant lipid rafts occupy distinct and dynamic domains in native membranes. Mol. Biol. Cell 15, 2580–2592 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaRocca T. J. et al., Proving lipid rafts exist: Membrane domains in the prokaryote Borrelia burgdorferi have the same properties as eukaryotic lipid rafts. PLoS Pathog. 9, e1003353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veatch S. L. et al., Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS One 7, e31457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stottrup B. L., Veatch S. L., Keller S. L., Nonequilibrium behavior in supported lipid membranes containing cholesterol. Biophys. J. 86, 2942–2950 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Sáez A. J., Chiantia S., Schwille P., Effect of line tension on the lateral organization of lipid membranes. J. Biol. Chem. 282, 33537–33544 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Connell S. D., Heath G., Olmsted P. D., Kisil A., Critical point fluctuations in supported lipid membranes. Faraday Discuss. 161, 91–111, discussion 113–150 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Bhatia T., Husen P., Ipsen J. H., Bagatolli L. A., Simonsen A. C., Fluid domain patterns in free-standing membranes captured on a solid support. Biochim. Biophys. Acta 1838, 2503–2510 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Khadka N. K., Ho C. S., Pan J., Macroscopic and nanoscopic heterogeneous structures in a three-component lipid bilayer mixtures determined by atomic force microscopy. Langmuir 31, 12417–12425 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Bleecker J. V., Cox P. A., Keller S. L., Mixing temperatures of bilayers not simply related to thickness differences between Lo and Ld phases. Biophys. J. 110, 2305–2308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bleecker J. V. et al., Thickness mismatch of coexisting liquid phases in noncanonical lipid bilayers. J. Phys. Chem. B 120, 2761–2770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sibold J., Tewaag V. E., Vagedes T., Mey I., Steinem C., Phase separation in pore-spanning membranes induced by differences in surface adhesion. Phys. Chem. Chem. Phys. 22, 9308–9315 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Sezgin E., Super-resolution optical microscopy for studying membrane structure and dynamics. J. Phys. Condens. Matter 29, 273001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen F., Tillberg P. W., Boyden E. S., Optical imaging. Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heberle F. A. et al., Direct label-free imaging of nanodomains in biomimetic and biological membranes by cryogenic electron microscopy. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.2002200117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veatch S. L., Gawrisch K., Keller S. L., Closed-loop miscibility gap and quantitative tie-lines in ternary membranes containing diphytanoyl PC. Biophys. J. 90, 4428–4436 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angelova M. I., “Lipid swelling and liposome formation in electric fields,” PhD thesis, Institute of Biophysics, Bulgarian Academy of Sciences, Sofia, Bulgaria (1998).
- 28.Blosser M. C., Horst B. G., Keller S. L., cDICE method produces giant lipid vesicles under physiological conditions of charged lipids and ionic solutions. Soft Matter 12, 7364–7371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dürre K., Bausch A. R., Formation of phase separated vesicles by double layer cDICE. Soft Matter 15, 9676–9681 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Stanich C. A. et al., Coarsening dynamics of domains in lipid membranes. Biophys. J. 105, 444–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammond A. T. et al., Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 102, 6320–6325 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veatch S. L., Leung S. S. W., Hancock R. E. W., Thewalt J. L., Fluorescent probes alter miscibility phase boundaries in ternary vesicles. J. Phys. Chem. B 111, 502–504 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Simunovic M., Prévost C., Callan-Jones A., Bassereau P., Physical basis of some membrane shaping mechanisms. Philos. Trans. A Math. Phys. Eng. Sci. 374, 20160034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayuyan A. G., Cohen F. S., Lipid peroxides promote large rafts: Effects of excitation of probes in fluorescence microscopy and electrochemical reactions during vesicle formation. Biophys. J. 91, 2172–2183 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott H. L. et al., On the mechanism of bilayer separation by extrusion, or why your LUVs are not really unilamellar. Biophys. J. 117, 1381–1386 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marsh D., Cholesterol-induced fluid membrane domains: A compendium of lipid-raft ternary phase diagrams. Biochim. Biophys. Acta 1788, 2114–2123 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Blosser M. C., Cornell C. E., Rayermann S. P., Keller S. L., “Phase diagrams and tie-lines in GUVs” in The Giant Vesicle Book, Dimova R., Marques C., Eds. (Taylor & Francis, Milton Park, 2019). [Google Scholar]
- 38.Heberle F. A. et al., Bilayer thickness mismatch controls domain size in model membranes. J. Am. Chem. Soc. 135, 6853–6859 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Angelova M. I. et al., Preparation of giant vesicles by external AC electric fields. Kinetics and applications. Prog. Colloid Polym. Sci. 89, 127–131 (1992). [Google Scholar]
- 40.Wu B., Chen Y., Müller J. D., Fluorescence fluctuation spectroscopy of mCherry in living cells. Biophys. J. 96, 2391–2404 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veatch S. L., Polozov I. V., Gawrisch K., Keller S. L., Liquid domains in vesicles investigated by NMR and fluorescence microscopy. Biophys. J. 86, 2910–2922 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ionova I. V., Livshits V. A., Marsh D., Phase diagram of ternary cholesterol/palmitoylsphingomyelin/palmitoyloleoyl-phosphatidylcholine mixtures: Spin-label EPR study of lipid-raft formation. Biophys. J. 102, 1856–1865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J. et al., Phase studies of model biomembranes: Complex behavior of DSPC/DOPC/cholesterol. Biochim. Biophys. Acta 1768, 2764–2776 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heftberger P., Kollmitzer B., Rieder A. A., Amenitsch H., Pabst G., In situ determination of structure and fluctuations of coexisting fluid membrane domains. Biophys. J. 108, 854–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heerklotz H., Triton promotes domain formation in lipid raft mixtures. Biophys. J. 83, 2693–2701 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williamson J. J., Olmsted P. D., Nucleation of symmetric domains in the coupled leaflets of a bilayer. Soft Matter 11, 8948–8959 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Fowler P. W., Williamson J. J., Sansom M. S. P., Olmsted P. D., Roles of interleaflet coupling and hydrophobic mismatch in lipid membrane phase-separation kinetics. J. Am. Chem. Soc. 138, 11633–11642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blosser M. C., Starr J. B., Turtle C. W., Ashcraft J., Keller S. L., Minimal effect of lipid charge on membrane miscibility phase behavior in three ternary systems. Biophys. J. 104, 2629–2638 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmid E. M. et al., Size-dependent protein segregation at membrane interfaces. Nat. Phys. 12, 704–711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheve C. S., Gonzales P. A., Momin N., Stachowiak J. C., Steric pressure between membrane-bound proteins opposes lipid phase separation. J. Am. Chem. Soc. 135, 1185–1188 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Some study data available.