Significance
The G protein-coupled receptor (GPCR) family is responsible for regulating much of human physiology, and GPCRs have become the most successful target class for drug development. Although many biologics have gained clinical approval, GPCR-targeted drugs remain almost exclusively small molecules, and developing GPCR-targeted antibodies remains challenging and often unsuccessful. We describe a methodology to isolate antibody fragment antagonists of a peptide-binding GPCR, and we show that one of these antibody fragments can be used to modulate blood pressure in vivo in mice. Antibody-based approaches may be useful in treating conditions where small-molecule drugs cannot be developed or where the unique pharmacological properties of antibodies are desirable.
Keywords: nanobodies, angiotensin, ligand, GPCR, hypertension
Abstract
There is considerable interest in developing antibodies as functional modulators of G protein-coupled receptor (GPCR) signaling for both therapeutic and research applications. However, there are few antibody ligands targeting GPCRs outside of the chemokine receptor group. GPCRs are challenging targets for conventional antibody discovery methods, as many are highly conserved across species, are biochemically unstable upon purification, and possess deeply buried ligand-binding sites. Here, we describe a selection methodology to enrich for functionally modulatory antibodies using a yeast-displayed library of synthetic camelid antibody fragments called “nanobodies.” Using this platform, we discovered multiple nanobodies that act as antagonists of the angiotensin II type 1 receptor (AT1R). Following angiotensin II infusion in mice, we found that an affinity matured nanobody antagonist has comparable antihypertensive activity to the angiotensin receptor blocker (ARB) losartan. The unique pharmacology and restricted biodistribution of nanobody antagonists may provide a path for treating hypertensive disorders when small-molecule drugs targeting the AT1R are contraindicated, for example, in pregnancy.
G protein-coupled receptors (GPCRs) are the largest family of transmembrane proteins in humans and constitute the single largest class of small-molecule drug targets (1). Still, many GPCR-targeted drugs lack selectivity for a single receptor subtype, and some therapeutically relevant GPCRs have not been successfully targeted by small-molecule drugs. Antibody-based therapeutics could provide a path to target otherwise undruggable GPCRs, as well as achieve high receptor subtype specificity, by recognizing extended epitopes outside the conserved ligand-binding pocket of GPCRs. Furthermore, antibodies are inherently endowed with potentially advantageous pharmacological properties distinct from small molecules, such as long half-lives, restricted distribution in vivo, and the ability to induce targeted cytotoxic effects by triggering immune responses via their Fc regions. Despite this potential, few functionally modulatory GPCR antibodies have been reported, and no broadly applicable approaches for GPCR antibody discovery have been established, although new screening methods are emerging (2).
To date, most examples of GPCR-targeted antibodies act on members of the chemokine receptor family, which bind small protein ligands (3–8). These antibodies likely function by occluding ligand recognition motifs on the receptors’ amino termini rather than by directly altering receptor signaling or competing for ligand binding to the seven-pass transmembrane (7TM) domain (7, 8). Similarly, the known antibody fragments targeting family B GPCRs such as the glucagon receptor and GLP-1 receptor (9, 10), parathyroid hormone receptor (11), and calcitonin gene-related peptide receptor (12) function by blocking receptor ectodomains rather than interacting with the 7TM core. However, the vast majority of GPCRs lack extracellular ligand recognition regions and instead bind small-molecule and peptide ligands entirely within their 7TM domain. Although functional antibody fragments were recently obtained via immunization for the peptide binding apelin receptor (13), there is a need for more broadly applicable methodologies to discover antibody fragments explicitly directed to the membrane-embedded domains with limited surface exposure.
The angiotensin II type 1 receptor (AT1R) is a GPCR that exemplifies the opportunities and the challenges surrounding antibody drug development. Both the endogenous peptide agonist of the AT1R (angiotensin II) and small-molecule inhibitors (angiotensin receptor blockers, ARBs) bind deep within the 7TM bundle (14–17). Since this receptor serves as one of the principal regulators of blood pressure and renal function, ARBs are frontline treatments for hypertension and kidney disease. While ARBs are safe and effective in most patients, they readily cross the placenta and exhibit severe on-target fetal toxicity that prevents their use during pregnancy (18). This toxicity restricts pharmacological treatments for hypertensive disorders during pregnancy, including preeclampsia, a leading cause of maternal and fetal death (19). The development of biologic-based AT1R antagonists may provide an alternative therapeutic approach for treating hypertensive disorders during pregnancy, as small proteins cannot cross the placental barrier (20).
We recently developed a fully in vitro camelid antibody fragment (“nanobody”) discovery platform (21), which allowed isolation of conformation-stabilizing nanobodies for the AT1R that bind the intracellular face of the receptor (14). We reasoned that similar approaches might equally apply to targeting the extracellular side of the receptor and yield antagonists and agonists. We designed a yeast selection scheme to enrich orthosteric ligands, resulting in multiple nanobodies targeting the extracellular side of AT1R. These nanobodies effectively bind the receptor in vitro, and a lead clone antagonizes AT1R signaling through both the G protein and β-arrestin pathways. Moreover, in mice this nanobody clone shows antihypertensive activity and blocks the pressor activity of angiotensin II infusion comparably to the small-molecule ARB losartan. These results demonstrate that synthetic yeast display library selections can effectively deliver receptor-modulating antibody fragments with activity both in vitro and in vivo, providing a path to drugging GPCRs.
Results
Discovery of Nanobody Ligands to the Angiotensin Receptor.
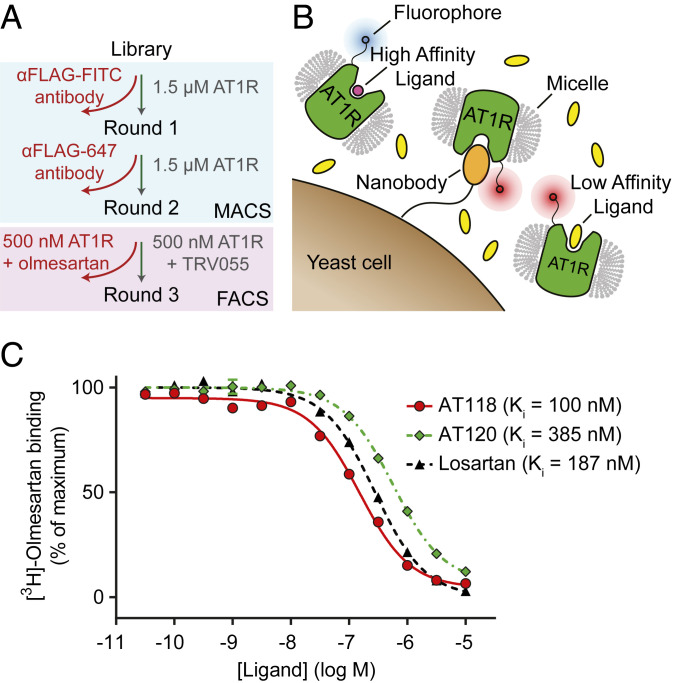
To discover nanobodies targeting the human AT1R, we employed a fully synthetic library of 5 × 108 unique nanobodies displayed on the surface of Saccharomyces cerevisiae yeast (21). Cells were incubated with purified AT1R protein in detergent, and receptor-binding nanobody clones were enriched using magnetic-activated cell sorting (Fig. 1A). The resulting population was used as a starting point to discover AT1R ligands. We reasoned that nanobodies that are orthosteric ligands would be able to displace low-affinity ligands but would be less effective at competing for binding with high-affinity ligands. Therefore, we stained the yeast simultaneously with two different populations of AT1R labeled with distinct fluorophores, one bound to a high-affinity ligand (olmesartan, population 1) and another bound to a low-affinity ligand (TRV055, population 2). The two ligands are mutually competitive with one another, both binding to the AT1R’s canonical orthosteric binding site (22). Additionally, the staining mixture contained 2.5 µM of free TRV055 (Fig. 1B) as an additional selective pressure to favor enrichment of high-affinity nanobodies capable of effectively competing with the low-affinity ligand. While ligand exchange is possible under these conditions, the slow off rate of olmesartan (T1/2 = 72 min) (23) minimizes mixing of the populations during the timescale of the experiment. We performed fluorescence-activated cell sorting (FACS) to isolate yeast cells whose nanobodies preferentially bound to the TRV055 population.
Fig. 1.
Discovery of synthetic nanobody ligands for the angiotensin receptor. (A) Flowchart of the selection method. Two rounds of MACS were performed on a yeast library displaying synthetic nanobodies to enrich for AT1R binders, as previously described (14). For the third round of selection, yeasts were simultaneously stained with two receptor populations, olmesartan-bound AT1R and TRV055-bound AT1R, and FACS was used to enrich for yeast displaying nanobodies that preferentially interact with the TRV055-bound AT1R population. (B) Cartoon representing FACS selection for yeast displaying nanobody ligands to AT1R. Yeasts were stained with two receptor populations, AT1R bound to a high-affinity (olmesartan, pink) or low-affinity (TRV055, yellow) ligand. Each of these receptor populations was labeled by a different fluorophore through dye-conjugated anti-FLAG antibody. Yeasts that preferentially interact with the low-affinity ligand-AT1R population were collected. (C) Radioligand competition binding of [3H]-olmesartan and nanobodies AT118 or AT120 to AT1R. AT118 (red), AT120 (green), and positive control losartan (black) compete with antagonist olmesartan for binding to detergent-solubilized AT1R.
After FACS, single clones were isolated and screened in on-yeast binding assays using flow cytometry. First, 48 clones were screened for preferential binding to TRV055-bound AT1R compared to AngII-bound AT1R. Nanobody ligands that compete with AngII and TRV055 would be expected to preferentially displace TRV055 due to its much lower AT1R-binding affinity than AngII. Two candidate clones from this screen were further evaluated for binding to olmesartan-bound AT1R. Both of these clones, AT117 and AT118, had weak binding to AT1R in the presence of high-affinity ligands (AngII and Olm) and stronger binding in the presence of a low-affinity ligand (TRV055). After isolating these two clones, we searched for additional antagonist nanobodies using a similar but less stringent approach by screening 48 clones for binding to either unliganded AT1R or olmesartan-bound AT1R. One candidate nanobody was chosen (AT120) for further evaluation from this screen. Clone AT117 showed very low receptor binding affinity, so we performed detailed characterization of only clones AT118 and AT120. Radioligand competition binding using purified AT1R in detergent showed that both nanobodies competed with the ARB [3H]-olmesartan with mid- to low-nanomolar affinities, confirming they bind to the receptor at the same site as a conventional AT1R antagonist (Fig. 1C). Sequences of selected nanobodies are summarized in SI Appendix, Table S1.
Affinity Maturation of Nanobody AT118.
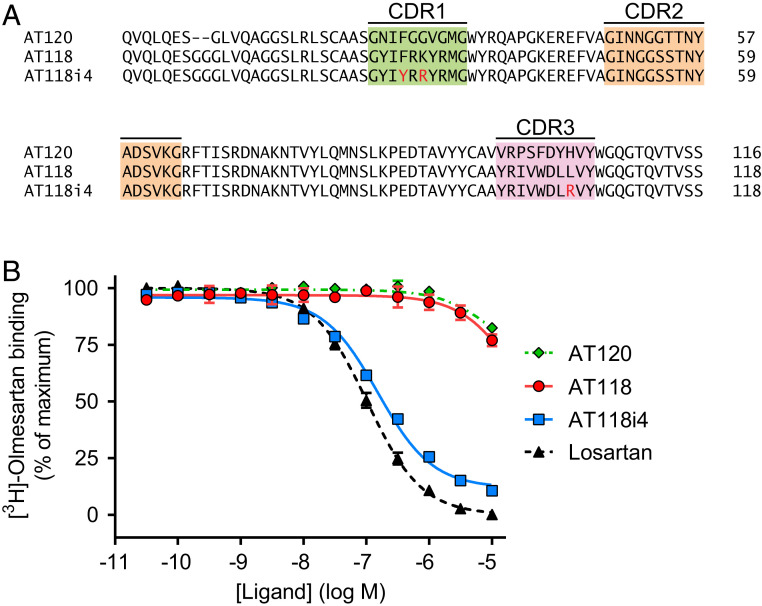
Both nanobody ligands showed a reduced ability to compete with radioligands in membranes (SI Appendix, Table S2). However, we reasoned that the reduced competition could be overcome by increasing the nanobody’s affinity for AT1R through affinity maturation. Using error-prone PCR to mutagenize AT118, we generated a new yeast-displayed library of 8.4 × 107 mutant AT118 clones. FACS selections were performed to enrich for higher-affinity AT118 mutants. Three mutations that were highly enriched after two sequential FACS selections were combined into a consensus clone called AT118i4 (Fig. 2A).
Fig. 2.
Affinity maturation of nanobody ligand AT118. (A) Sequence of affinity-matured AT118. The sequences of AT118, affinity-matured AT118i4, and AT120 were aligned. Residues mutated to generate AT118i4 are highlighted in red. (B) Radioligand competition binding of AT118i4 and [3H]-olmesartan to AT1R in cell membranes. AT118i4 (blue) competes with olmesartan for binding to AT1R in HEK cell membranes, demonstrating higher affinity than AT118 (red).
This consensus nanobody clone exhibited nearly 70-fold enhanced binding affinity when compared to the parent clone AT118 in a radioligand binding assay using membranes from HEK 293 cells overexpressing AT1R (Fig. 2B and SI Appendix, Table S2). Notably, the affinity of AT118i4 for AT1R on membranes was comparable to that of losartan, a commonly prescribed ARB.
Functional Characterization of Nanobody AT118i4.
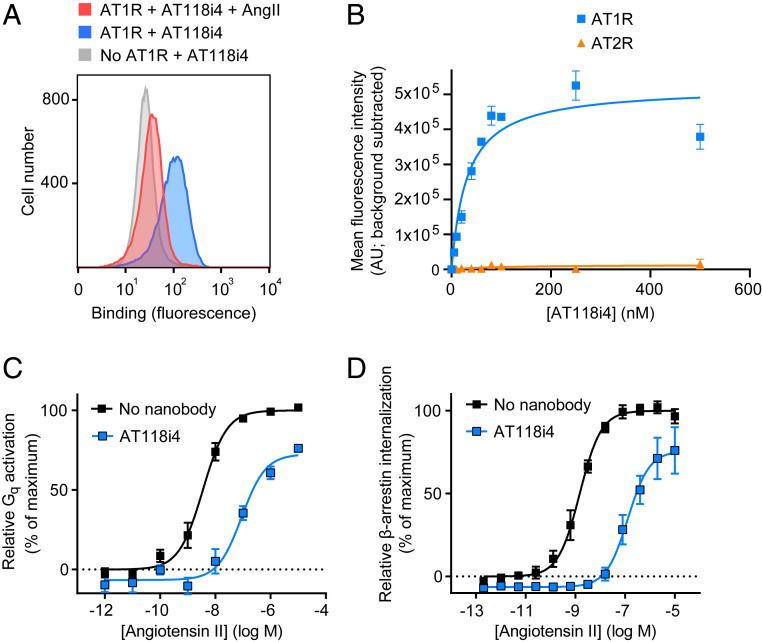
Competitive binding with orthosteric ligands suggested AT118 and AT118i4 likely bind the extracellular face of the receptor. However, this result could, in principle, also arise from an intracellular binder that has a high degree of negative cooperativity with orthosteric ligands. In order to definitively determine if AT118i4 binds to the extracellular side of the AT1R we stained nonpermeabilized HEK suspension cells overexpressing the AT1R with AT118i4. Strong binding was observed by flow cytometry in the absence of AT1R ligands, but this was attenuated in the presence of AngII, confirming that AT118i4 binds on the extracellular side of the AT1R (Fig. 3A). One potential benefit of antibody-based therapeutics that target GPCRs is the possibility of achieving very high receptor subtype specificity, which may reduce side effects in clinical applications. Indeed, when we stained cells expressing either AT1R or angiotensin II type 2 receptor (AT2R), we found that there was little or no binding of AT118i4 to AT2R expressing cells (Fig. 3B), while AT118i4 showed strong, saturable binding to AT1R-expressing cells (half-maximal effective concentration [EC50] = 31 nM).
Fig. 3.
Functional characterization of AT118i4. (A) Binding of AT118i4 to cells expressing AT1R demonstrated by flow cytometry. HEK suspension cells transiently expressing AT1R were treated with AT118i4-His (blue) or both AT118i4-His and AngII (red). Cells not expressing AT1R (transiently transfected with empty vector) were also treated with AT118i4-His (gray). Cells were stained with anti-His-488 before flow cytometry analysis. Data in A are representative of three independent experiments. (B) Subtype specificity of AT118i4 assessed by flow cytometry. HEK suspension cells transiently transfected with AT1R (blue) and AT2R (orange) were treated with AT118i4-V5-His and anti-V5 488, then analyzed by flow cytometry. Data are representative of three independent experiments. Error bars represent SEM from replicates. (C) Inhibition of Gq-mediated inositol monophosphate accumulation by AT118i4 in response to AngII. HEK cells were treated with 5 μM AT118i4 (blue) or no antibody (black). (D) AT118i4 treatment reduces translocation of β-arrestin2 to endosomes upon AngII stimulation.
Next, we sought to investigate the functional properties of AT118i4. In order to determine whether AT118i4 effectively antagonizes AT1R signaling, we treated HEK suspension cells stably overexpressing AT1R with AngII in the presence of AT118i4 (Fig. 3C and SI Appendix, Table S3). Gq signaling, assessed by inositol monophosphate accumulation, was significantly reduced in cells treated with AT118i4 (EC50 = 89 nM) when compared with vehicle (EC50 = 3.5 nM). We also observed a modest but reproducible decrease in basal activation of Gq signaling (Fig. 3C), indicating that AT118i4 acts as an inverse agonist. AT118i4 also reduced AT1R internalization (Fig. 3D and SI Appendix, Table S3), indicating that signaling through β-arrestin is suppressed. In addition, ERK phosphorylation, which is activated by both Gq and β-arrestin signaling, was profoundly reduced in cells treated with AT118i4 (SI Appendix, Fig. S1).
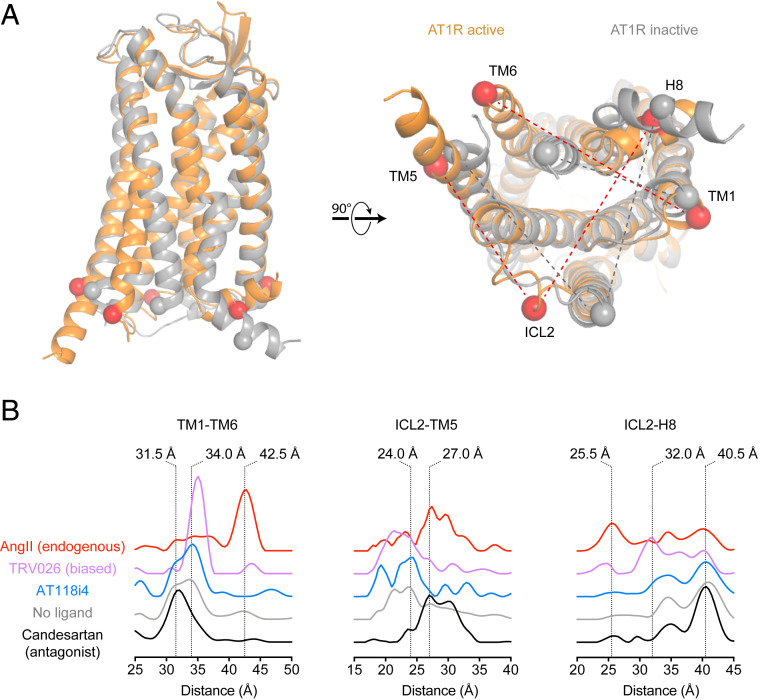
Reduced basal activation of the AT1R suggested that AT118i4 is likely stabilizing an inactive conformation of the receptor (i.e., it is an inverse agonist rather than a simple neutral antagonist). We used double electron–electron resonance (DEER) spectroscopy to monitor the effects of AT118i4 binding on the intracellular regions of the AT1R responsible for transducer activation. DEER measures the distribution of distances between two nitroxide spin labels, which are added to engineered cysteine residues on the AT1R. We previously validated pairs of AT1R sites that report on key conformational changes that occur upon binding to small-molecule inverse agonists, AngII, and agonists that are biased toward the activation of either Gq- or β-arrestin-mediated pathways. Analysis of AT118i4’s effect on several of these pairs shows that the nanobody stabilizes conformations sampled by the unliganded receptor and has a profile distinct from both ARBs and agonists (Fig. 4). In the TM1-TM6 pair, which reports on the ∼10 Å outward movement of TM6 that typically occurs upon GPCR activation (42.5 Å, AngII-stabilized peak in the AT1R), two conformations are present in the absence of ligand (31.5 and 34 Å peaks). Small-molecule ARBs shift the distribution toward the shorter 31.5 Å distance, but AT118i4 slightly stabilizes the 34 Å distance, although much more weakly than the β-arrestin-biased ligand TRV026. Like TRV026, AT118i4 depletes the population of distances at >25 Å in the ICL2-TM5 pair, which ARBs like candesartan strongly stabilize. However, like ARBs, AT118i4 fails to induce conformational changes in helix 8 (ICL2-H8 pair) that are characteristic of either TRV026 (25.5 Å) or AngII (32 Å).
Fig. 4.
Conformations sampled by AT1R. (A) Labeling sites in the intracellular regions of the AT1R. Spheres indicate positions of Cα of labeled residues in the receptor in inactive (gray, Protein Data Bank [PDB] ID code 4YAY) and active (orange, PDB ID code 6DO1) conformations. (B) DEER distance probability distributions in the presence of AT118i4 and various classes of ligands (candesartan, ARB; TRV026, β-arrestin-biased ligand; AngII, endogenous agonist) for AT1R pairs monitoring the conformations of the cytoplasmic ends of the indicated transmembrane helices, intracellular loop 2, and helix 8.
Activity of AT118i4 In Vivo.
Given its potent inhibition of angiotensin II signaling in cells that is comparable to clinically used ARBs, we reasoned that AT118i4 could serve as an antihypertensive in vivo. To examine whether AT118i4 could be functional in a mouse model, we performed a radioligand competition binding assay with [3H]-olmesartan and AT118 using human AT1R or murine angiotensin II receptor type 1a (AT1Ra). Compared to human AT1R, murine AT1aR has substantially reduced (19-fold) affinity for AT118i4 but not [3H]-olmesartan (SI Appendix, Fig. S2). Since murine AT1Ra and human AT1R are very highly conserved and differ at only nine positions in their extracellular regions, we introduced these differences as mutations into human AT1R to determine if they influence AT118 binding. Extracellular mutations (I164V/F170Y and Q187R) significantly decreased AT118i4 affinity, while other mutations were tolerated.
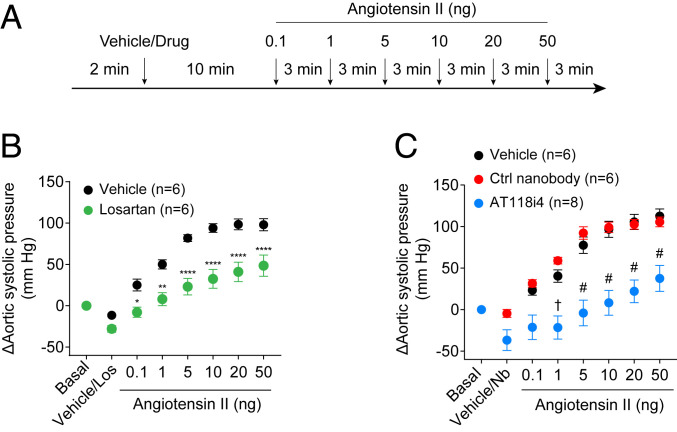
We then tested whether AT118i4 could suppress AngII activation of the AT1R in vivo. Anesthetized mice were pretreated with an intravenous (i.v.) bolus of the ARB losartan (10 mg/kg), AT118i4 (7 to 10 mg/kg), or the control nanobody AT110S (10 mg/kg, which binds the intracellular surface of the AT1R), followed by a graded dose of AngII (Fig. 5A and SI Appendix, Fig. S3). Continuous monitoring of aortic blood pressure with a high-fidelity micromanometer catheter showed that, like losartan (Fig. 5B), AT118i4 significantly attenuates the AngII-induced rise in blood pressure (Fig. 5C). In contrast, a control nanobody that binds the intracellular side of the AT1R did not block the physiological effects of AngII (Fig. 5C). Notably, AT118i4 was even more efficacious in blocking the effects of AngII in female mice than in male mice (SI Appendix, Table S3).
Fig. 5.
AT118i4 blocks hypertension caused by angiotensin II infusion in mice. (A) Treatment protocol for measuring changes in mouse blood pressure upon exposure to AngII. (B) Measurements of the effect of losartan on aortic systolic pressure in mice. The effects of the antihypertensive or ARB losartan (green) on systolic pressure in mice exposed to increasing concentrations of AngII were compared to untreated mice (black) in a positive control experiment. Statistical analysis was performed using two-way ANOVA with Sidak multiple comparison posttests; *P < 0.05, **P < 0.01, ****P < 0.0001, compared with the vehicle-treated mice. (C) Measurements of the effect of nanobody AT118i4 on aortic systolic pressure in mice. Systolic pressure in mice exposed to increasing concentrations of AngII was measured after pretreatment with nanobody AT118i4 (blue), negative control nanobody (red), or vehicle PBS (black). Statistical analysis was performed using two-way ANOVA with Sidak multiple comparison posttests; †P < 0.01, #P < 0.0001, compared with the vehicle-treated mice.
Another critical step in adapting antibody fragments for therapeutic applications is humanization to reduce immunoreactivity. Encouraged by the in vivo activity of AT118i4, we tested whether its camelid framework could be humanized without adversely affecting its functional properties. We created a panel of nanobody variants with varying numbers of humanizing mutations to better resemble the heavy chain of a human antibody. AT118i4h32, with 11 total humanizing amino acid changes, was the most aggressively mutagenized clone with unaltered affinity for the AT1R (SI Appendix, Fig. S4). AT118i4h32 further displays enhanced protein expression, solubility, and stability compared to AT118i4. These data demonstrate that AT118i4 and other nanobodies derived from our yeast display library are readily amenable to optimization for therapeutic applications.
Discussion
Drugs targeting GPCRs represent roughly 35% of marketed therapeutics (1), illustrating the profound physiological importance of this class of receptors. There has been growing interest in targeting GPCRs with antibody-based therapeutics, which offer advantages over small-molecule drugs for certain applications. Importantly, antibodies have the potential for exquisite subtype specificity by virtue of their large and extensive interaction surfaces with target antigens. The ability to target specific GPCRs within closely related families could reduce side effects and enable therapeutic development for previously inaccessible GPCRs. Likewise, antibodies that recognize naturally occurring GPCR variants could be useful for treating rare diseases (24). Notably, as genetically encoded entities, additional attributes can be engineered readily into antibody ligands. For example, tissue- or cell-type-specific activity can be achieved through engineering bispecific antibodies, and Fc-dependent immune effector functions can be used to achieve antibody-dependent cell-mediated cytotoxicity.
Despite the therapeutic potential of GPCR-targeted antibody ligands, there are a number of challenges associated with their discovery. Many GPCRs are highly conserved across species, creating barriers to generating antibody therapeutics from animal-derived libraries or immunization approaches. Moreover, most GPCRs are biochemically unstable upon purification, resulting in rapid denaturation following injection into an animal. Our in vitro selection approach addresses both challenges. Since our library is fully synthetic, it has not been negatively selected for tolerance of self. In addition, selections can be performed on short timescales under optimized conditions (e.g., low temperature, defined buffer composition, no endogenous proteases or receptor ligands), ensuring that receptors remain folded during the selection process. In addition, counterselections can be carried out simultaneously, increasing the probability of finding a candidate molecule possessing the desired properties.
The small convex paratope and long CDR3 loops of nanobodies may make these antibody fragments particularly suited to targeting the buried pockets typically responsible for ligand binding. Indeed, nanobodies are frequently used in place of prototypical antibodies to access the cytosolic transducer binding pocket of GPCRs and to stabilize active conformations as “G protein mimetics” for crystallography. Recently, a camelid-derived nanobody was reported to antagonize the human apelin receptor (APJ) through extensive interactions between the nanobody and extracellular loops of APJ and insertion of CDR3 into the activating peptide binding site (13). The APJ nanobody antagonist was rationally engineered into an APJ agonist, demonstrating the potential for rapid modification of antibody ligand pharmacology through protein engineering. Nanobody attachments to GPCR ligands have also been developed in order to impart additional functionality to the ligands (25, 26).
We discovered two nanobody antagonists to the AT1R from a synthetic yeast-displayed nanobody library. Our selection methodology took advantage of the range of affinities of small-molecule AT1R ligands, which facilitates a selection methodology that enriches for nanobodies that are competitive with orthosteric ligands. The high-affinity AT1R nanobody antagonist, AT118i4, antagonizes AT1R signaling comparably to clinically used antagonists and exhibits exceptional subtype specificity, with negligible binding to the AT2R receptor subtype. Despite reduced affinity for murine AT1R, AT118i4 significantly reduced the rise in blood pressure in mice in response to infusion of the endogenous agonist angiotensin II. In both in vitro and in vivo experiments, we observed a reduction in basal AT1R signaling, suggesting that AT118i4 acts as an inverse agonist. DEER spectroscopy indicates that AT118i4 stabilizes an inactive state of the AT1R that is distinct from small-molecule AT1R antagonists.
AT1R-targeted nanobody ligands may have therapeutic potential for treating hypertensive and kidney diseases during pregnancy, as ARBs and ACE inhibitors both exhibit severe fetal toxicity, making pharmacologic modulation of the renin–angiotensin–aldosterone system largely impossible with existing therapeutics. Unlike traditional small-molecule drugs, a protein would not pass through the placental barrier without the capacity to bind to the FcRn receptor (27, 28). Our approach for discovering an orthosteric nanobody may serve as a blueprint for future efforts aimed at generating other GPCR-targeted antibody ligands.
Materials and Methods
Nanobody Discovery.
A yeast-displayed library of nanobody AT1R binders was generated using magnetic-activated cell sorting (MACS), which has been previously described (14). In summary, 1 × 1010 cells displaying a synthetic library of nanobodies were stained for 40 min at 4 °C with anti-fluorescein isothiocyanate (FITC) microbeads (Miltenyi) and FITC-labeled anti-FLAG antibody. After centrifugation and resuspension in buffer (20 mM Hepes [pH 7.5], 150 mM NaCl, 2.8 mM CaCl2, 0.05% maltose neopentyl glycol [MNG], 0.005% cholesterol hemisuccinate [CHS], 0.1% bovine serum albumin [BSA], 0.2% maltose), cells were passed through an LD column (Miltenyi) to remove yeast displaying nanobodies that bound to FLAG antibody or microbeads. Collected yeast was then stained again, with 1.5 µM of FLAG-tagged AT1R and 1 µM of anti-FLAG FITC antibody for 1 h at 4 °C, followed by staining with anti-FITC microbeads for 15 min at 4 °C, and then passed into an LS column (Miltenyi). Bound yeast was eluted and grown for a second round of MACS. The second round was performed similarly to the first, but with 4 × 108 cells and anti-FLAG antibody labeled with Alexa Fluor 647 and anti-647 microbeads. Yeast enriched for binding to AT1R after these two rounds of MACS was simultaneously stained with 500 nM AT1R prebound to the high-affinity ligand olmesartan and 500 nM AT1R bound to a lower-affinity ligand, TRV055, and the receptors were fluorescently labeled using 150 nM of M1 anti-FLAG 647 antibody and M1 anti-FLAG 488, respectively, before staining. The staining buffer was composed of 20 mM Hepes (pH 7.5), 150 mM NaCl, 3 mM CaCl2, 2.5 µM TRV055, 0.1% MNG, 0.01% CHS, 0.1% BSA, 0.2% maltose. FACS was performed on the stained yeast, and yeast cells displaying nanobodies that preferentially bound to the AT1R-TRV stained population were collected and grown. From this FACS library, AT118 was isolated by screening with a BD Accuri C6 Plus flow cytometer for yeast-displayed nanobody clones that interact more robustly with TRV055-bound AT1R versus olmesartan- or angiotensin II–bound AT1R, and AT120 was isolated by screening for clones that interact more strongly with unliganded AT1R versus olmesartan-bound AT1R.
Affinity Maturation.
Error-prone PCR was performed on AT118 using the GeneMorph II Random Mutagenesis Kit (Agilent). The error-prone PCR product was then amplified with Q5 High-Fidelity DNA Polymerase (New England Biolabs) and transformed into BJ5465 yeast using an ECM 830 Electroporator (BTX-Harvard Apparatus). The resulting yeast-displayed library comprised 8.4 × 107 transformed clones with an average mutation rate of 1.8 amino acid changes per clone. The nanobody expression in the error-prone yeast library was induced, and cells were stained with 250 nM of AT1R and then subsequently with 2 µg/mL anti-HA-488 (Cell Signaling Technologies) and 400 nM of M1 anti-FLAG 647 antibody. Thirty-nine million cells were sorted, and 17,000 cells were collected using a gate drawn for cells displaying higher-affinity nanobodies. A second FACS was performed using a lower concentration of AT1R to further enrich for cells displaying higher-affinity nanobodies. To generate high-affinity clones, 50 colonies were Sanger sequenced, and enriched mutations were identified and added to a consensus clone, AT118i4.
Protein Expression and Purification.
DNA encoding AT118 and AT120 was cloned into pET26b, which contains an N-terminal pelB signal sequence for periplasmic expression and C-terminal His-tag for purification. A C-terminal V5-epitope (GKPIPNPLLGLDST) was inserted prior to the His-tag for flow cytometry experiments. All constructs were verified by Sanger sequencing.
Escherichia coli strain BL21(DE3) was transformed with nanobody encoding plasmids. Transformed cells were grown in Terrific Broth media at 37 °C with 50 μg mL−1 kanamycin to an optical density at 600 nm = 0.7 to 1.5, cooled to 17 °C, and induced with 200 μM isopropyl β-D-1-thiogalactopyranoside overnight. Cell pellets were resuspended in 50 mL sucrose, ethylenediaminetetraacetic acid (EDTA), Tris buffer (20 mM Tris [pH 8], 500 mM sucrose, 500 µM EDTA) per liter of culture; 100 mL cold H2O and 100 U of benzonase nuclease per liter of culture supplemented with 5 mM MgCl2 were added to resuspended pellets and stirred for 1 h at room temperature. Resuspended cells were cleared by centrifugation (14,000 × g, 30 min). Supernatant was supplemented with 100 mM NaCl and stirred at room temperature for 15 min. Supernatant was passed through a glass microfiber filter and applied to protein A agarose resin (GoldBio) or a HiTrap FF rProtein A column (GE Healthcare). The resin was washed with 10 column volumes of wash buffer (10 mM NaH2PO4 [pH 7.5], 100 mM NaCl). Protein was eluted with 100 mM NaH2PO4 (pH 2.5) and 100 mM NaCl directly into 2 M Hepes (pH 8; 0.2 mL/mL elution). Purified nanobody was buffer exchanged into 20 mM Hepes (pH 7.5), 150 mM NaCl, and 10% glycerol through size exclusion chromatography (Superdex S75 10 × 300 mm) or dialysis.
To remove endotoxin for animal experiments, AT118i4 was immobilized on Nickel NTA resin (GoldBio) and washed with 50 column volumes of sterile endotoxin-free phosphate-buffered saline (PBS) and 0.1% Triton X-114 at a flow rate of 1 mL/min at 4 °C. Triton X-114 was removed by washing the resin with five column volumes of sterile PBS four times at a flow rate of 1 mL/min at 4 °C. AT118i4 was eluted in sterile PBS containing 200 mM imidazole. Imidazole was subsequently removed using a PD-10 column, and AT118i4 was concentrated to 1 mg/mL, filter sterilized (0.2 µm), snap frozen in liquid nitrogen, and stored at −80 °C. Endotoxin levels were measured using the limulus amebocyte assay (Thermo Fisher), and all AT118i4 preparations contained less than five endotoxin units per milligram of protein.
Radioligand Binding.
In competition radioligand binding experiments with detergent-solubilized Flag-AT1R wild type,∼60 ng of purified receptor were added to serial dilutions of nanobodies and 2.5 nM [3H]-olmesartan (American Radiolabeled Chemicals) in 20 mM Hepes (pH 7.4), 100 mM NaCl, 0.01% maltose lauryl neopentyl glycol, and 0.1% BSA in 200 μL reactions. After 90 min at room temperature, reactions were harvested on GF/B filters using a 96-well Brandel harvester and quickly washed three times with cold 20 mM Hepes (pH 7.4) and 100 mM NaCl. Crude cell membranes for radioligand binding experiments were prepared as previously described (22) from Expi293F Inducible cells (Thermo Fisher) transfected with the indicated AT1R construct cloned in empty pcDNA-Zeo-tetO (29) according to the manufacturer’s instructions. Membranes were incubated with serially diluted nanobodies and 2.5 nM [3H]-olmesartan in 50 mM Tris (pH 7.4), 150 mM NaCl, 12.5 mM MgCl2, 0.2% BSA, leupeptin, and benzamidine. After 90 min at room temperature, reactions were harvested on GF/B filters using a 96-well Brandel harvester and quickly washed three times with cold 50 mM Tris (pH 7.4). Data points were fit to a one-site competition binding model in GraphPad Prism. To determine inhibitory constant (Ki) values, radioligand affinity was determined by saturation binding using a serial dilution of [3H]-olmesartan in the presence or absence of 10 μM candesartan. Data were fit to a one-site saturation-binding model in GraphPad Prism. Data shown represent the mean and SE of three independent experiments.
Flow Cytometry Assays.
Expi293F Inducible cells were transiently transfected with empty pcDNA-Zeo-tetO (29), pcDNA-Zeo-tetO-FLAG-AT1R wild type (human), or pcDNA-Zeo-tetO-FLAG-AT2R wild type (human) according to the manufacturer’s protocol and induced with doxycycline (4 μg/mL) and sodium butyrate (5 mM) 2 d later. Three days posttransfection, cells were harvested and resuspended in cold assay buffer (Hank’s Balanced Salt Solution, 10 mM Hepes [pH 7.4], 3 mM CaCl2, 0.1% BSA) at a density of 8 × 106 live cells/mL and kept at 4 °C for the duration of the experiment. Cells were incubated with or without 250 μM AngII for 30 min before the addition of His-tagged AT118i4 (160 nM). Cells were incubated with the nanobody for 30 min, washed twice with assay buffer, and then incubated with a 1:250 dilution of anti-6X His Dylight 488 (abcam) and 15 nM Dylight 650–labeled M1 anti-FLAG for 30 min. Cells were washed twice and then resuspended in assay buffer for flow cytometry (BioRad S3e cell sorter). Data were analyzed with FlowJo 10 software. AT1R- and AT2R-transfected cells were gated for M1-positive singlets. For saturation-binding experiments cells were harvested, washed, and resuspended in flow cytometry buffer (20 mM Hepes [pH 7.5], 150 mM NaCl, 0.1% BSA) 3 d posttransfection; 1 × 105 cells were stained with AT118i4 containing a C-terminal V5 epitope tag for 1 h at 4 °C. Cells were then washed twice with flow cytometry buffer, supplemented with 1 mM CaCl2, and incubated with a 1:500 dilution of Alexa Fluor 657-labeled anti-V5 antibody (Thermo Fisher) and 100 nM Alexa Fluor 488-labeled M1 anti-FLAG for 20 min. Cells were washed, resuspended in flow cytometry buffer with 1 mM CaCl2, and analyzed with a BD Accuri C6 flow cytometer. AT1R- and AT2R-transfected cells were gated for M1-positive singlets. Data were analyzed with BD Accuri C6 Plus software.
Cellular Assays.
Gq activation.
Expi293F cells stably expressing wild-type FLAG-AT1R (human) were diluted to 2 × 106 cells/mL the day before the experiment. On the day of the experiment, 2 × 104 cells/well were plated in a low-volume 96-well plate, pretreated ±5 μM AT118i4, and then stimulated with the indicated concentration of AngII for 1 h at 37 °C. IP1 was quantified using the IP-One Gq Kit (Cisbio) on a CLARIOstar plate reader (BMG Labtech).
β-arrestin2 internalization.
U2OS cells stably expressing β-arrestin2 with an enzyme acceptor tag and endosome-localized ProLink were transiently transfected with wild-type FLAG-AT1R (human). One day posttransfection, cells were plated at a density of 35,000 cells per well. Two days posttransfection, cells were treated with ±5 μM AT118i4 for 30 min and then with the indicated concentration of AngII for 3 h at 37 °C. Chemiluminescence resulting from the complementation of the enzyme acceptor/ProLink β-galactosidase fragments due to β-arrestin endocytosis was detected with a PathHunter kit (Eurofins, DiscoverX) using a CLARIOstar plate reader (BMG Labtech).
PhosphoERK stimulation.
Expi293F cells stably expressing wild-type FLAG-AT1R (human) were diluted to 2 × 106 cells/mL the day before the experiment. On the day of the experiment, 500 μL of cells were pretreated with ±5 μM AT118i4 and then with 10 nM AngII for the indicated time at 37 °C. Cells were solubilized by the addition of 4× Laemmli sample buffer and sonicated. Total and phosphorylated ERK were detected by Western blotting using anti-MAPK 1/2 (1:10,000; EMD Millipore) and anti-p44/42 MAPK (1:4,000; Cell Signaling) and quantified by densitometry (ImageJ, NIH). Densitometry values for pERK were normalized to their respective total ERK values, and relative pERK values in each experiment were normalized to the 5 min time point in the absence of a nanobody. Data shown represent the mean and SE of three independent experiments.
DEER Spectroscopy.
AT1R constructs were purified and spin labeled with bis-(2,2,5,5-tetramethyl-3-imidazoline-1-oxyl-4-yl)disulfide (Enzo) to install the V1 spin label as previously described (30). Briefly, minimal cysteine versions of N-terminal FLAG-tagged human AT1R with engineered cysteines (F55C D236C, TM1-TM6; R139C K220C, ICL2-TM5; R139C R311C, ICL2-H8) in pACMV-tetO were transiently transfected into Expi293F cells stably expressing the tetracycline repressor (29) using an ExpiFectamine transfection kit (Thermo Fisher) according to the manufacturer’s instructions. Two days after transfection, doxycycline (4 μg/mL), sodium butyrate (5 mM), and losartan (5 μM) were added to the cultures, and cells were harvested and flash frozen with liquid nitrogen ∼30 h thereafter. Following hypotonic lysis (10 mM Tris [pH 7.4], 2 mM EDTA, 10 mM MgCl2, 1 μM losartan, 2.5 U/mL benzonase [Sigma], protease inhibitors benzamidine and leupeptin), cell pellets were solubilized with lauryl MNG (0.5%, 0.05% CHS, 20 mM Hepes [pH 7.4], 500 mM NaCl, 10 mM MgCl2, 1 μM losartan, 2.5 U/mL benzonase, benzamidine, leupeptin) for 2h at 4 °C. After insoluble material was cleared by centrifugation (18,000 × g for 30 min at 4 °C), 2 mM CaCl2 was added to the solubilized material, which was loaded onto M1 anti-FLAG resin at 4 °C. After washing with the same buffer containing a 50-fold lower concentration of MNG and CHS and Hepes at pH 6.8, the receptor was eluted with 5 mM EDTA and 0.2 mg/mL FLAG peptide (in wash buffer without CaCl2). A 10-fold molar excess of bis-(2,2,5,5-tetramethyl-3-imidazoline-1-oxyl-4-yl)disulfide was added to the receptor and incubated at ambient temperature for 3 h. Spin-labeled receptor was purified and buffer exchanged (40 mM Hepes [pH 7.4], 100 mM NaCl, 0.01% MNG, 0.001% CHS, D2O) by size exclusion chromatography. After monomeric fractions were concentrated, ligands (10-fold excess) or AT118i4 (twofold molar excess) were added to the receptor, along with dimethyl sulfoxide and d8-glycerol to final concentrations of 2% and 20%, respectively, so that all samples had the same buffer composition. Samples were incubated at room temperature for 1 h and then flash frozen in 1.4/1.7 mm or (inner diameter/outer diameter) borosilicate capillaries (VitroCom).
Q-band four-pulse DEER measurements were conducted on an Elexsys E580 pulsed electron paramagnetic resonance spectrometer equipped with an ER5106QT-2 cavity resonator (Bruker). During experiments a recirculating helium cryocooling system (ColdEdge Technologies) was used to maintain sample temperature at 50 K. Observed pulse lengths were 18 to 22 ns (π/2) and 36 to 44 ns (π), as determined by an echo nutation experiment. A linear frequency-swept (chirp) pulse of 50 MHz half-width, generated by an arbitrary waveform generator (Bruker) and applied 70 MHz below the observer frequency, served as the pump pulse. All pulses were further amplified via an external 150 W TWT amplifier (Applied Engineering Systems). LongDistances (version 590 or newer, available at http://www.biochemistry.ucla.edu/Faculty/Hubbell/software.html) was used for background correction and model-free analysis of dipolar evolution data. The l-curve criterion was used to select a single smoothness parameter for each spin pair that was used for all ligand conditions.
Blood Pressure Measurements in Mice.
For this study 10- to 14-wk-old C57BL/6J wild-type mice of both sexes were used. Animal experiments carried out for this study were handled according to approved protocols and animal welfare regulations mandated by the Institutional Animal Care and Use Committee of Duke University Medical Center.
Mice were anesthetized with ketamine (100 mg/kg) and xylazine (2.5 mg/kg). After bilateral vagotomy, a 1.4 French (0.46 mm) high-fidelity micromanometer catheter (ADInstruments) connected to a pressure transducer (ADInstruments) was inserted into the right carotid artery just above the aortic valve to monitor aortic blood pressure. Drugs were administered by i.v. injection through a jugular vein. Recording of basal blood pressure was performed at steady state after the catheter insertion (2 to 3 min after insertion). Animals then received vehicle (100 µL PBS), the AT1R nanobody AT118i4 (7 to 10 mg/kg, in 100 µL PBS), the control nanobody AT110s (10 mg/kg, in 100 µl PBS), or the angiotensin II receptor blocker, losartan (10 mg/kg, in 100 µL PBS). Ten minutes after mice received nanobody or vehicle solutions blood pressure was recorded, and then mice were injected with a graded dose of AngII (0.1 to 50 ng, each in 100 µL water) at 3 min intervals. The blood pressure was continuously monitored and recorded as described. Data analysis was performed using LabChart 8 software (ADInstruments).
Data are expressed as mean ± SEM. Statistical significance was determined with two-way repeated-measures ANOVA with Sidak correction for multiple comparison in GraphPad Prism. Differences were considered statistically significant at P < 0.05.
Supplementary Material
Acknowledgments
Funding to support this research was provided by NIH Grants R01HL056687 (H.A.R.), R01HL075443 (H.A.R.), DP5OD021345 (A.C.K.), R21HD101596 (A.C.K.), and R01HL16037 (R.J.L.); the Vallee Foundation (A.C.K.); the Smith Family Foundation (A.C.K.); the Helen Hay Whitney Foundation (M.A.S.); and the Mandel Center for Hypertension and Atherosclerosis at Duke (R.J.L.). We thank the Duke Cardiovascular Research Center Small Animal Physiology Core for performing the in vivo mouse hemodynamic experiments.
Footnotes
Competing interest statement: C.M., D.P.S., L.M.W., M.A.S., A.C.K., and R.J.L. are coinventors on a patent application for AT1R blocking nanobodies. A.C.K. is a cofounder and consultant for Tectonic Therapeutic Inc. and for the Institute for Protein Innovation, a nonprofit research institute.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009029117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Santos R. et al., A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren H. et al., Function-based high-throughput screening for antibody antagonists and agonists against G protein-coupled receptors. Commun. Biol. 3, 146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jo M., Jung S. T., Engineering therapeutic antibodies targeting G-protein-coupled receptors. Exp. Mol. Med. 48, e207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchings C. J., Koglin M., Marshall F. H., Therapeutic antibodies directed at G protein-coupled receptors. mAbs 2, 594–606 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maussang D. et al., Llama-derived single variable domains (nanobodies) directed against chemokine receptor CXCR7 reduce head and neck cancer cell growth in vivo. J. Biol. Chem. 288, 29562–29572 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jähnichen S. et al., CXCR4 nanobodies (VHH-based single variable domains) potently inhibit chemotaxis and HIV-1 replication and mobilize stem cells. Proc. Natl. Acad. Sci. U.S.A. 107, 20565–20570 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boshuizen R. S. et al., A combination of in vitro techniques for efficient discovery of functional monoclonal antibodies against human CXC chemokine receptor-2 (CXCR2). mAbs 6, 1415–1424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng L., Damschroder M. M., Cook K. E., Wu H., Dall’Acqua W. F., Molecular basis for the antagonistic activity of an anti-CXCR4 antibody. mAbs 8, 163–175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koth C. M. et al., Molecular basis for negative regulation of the glucagon receptor. Proc. Natl. Acad. Sci. U.S.A. 109, 14393–14398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennen S. et al., Structural insight into antibody-mediated antagonism of the glucagon-like peptide-1 receptor. Sci. Rep. 6, 26236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarkar K. et al., Modulation of PTH1R signaling by an ECD binding antibody results in inhibition of β-arrestin 2 coupling. Sci. Rep. 9, 14432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garces F. et al., Molecular insight into recognition of the CGRPR complex by migraine prevention therapy aimovig (Erenumab). Cell Rep. 30, 1714–1723.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Ma Y. et al., Structure-guided discovery of a single-domain antibody agonist against human apelin receptor. Sci. Adv. 6, eaax7379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wingler L. M., McMahon C., Staus D. P., Lefkowitz R. J., Kruse A. C., Distinctive activation mechanism for angiotensin receptor revealed by a synthetic nanobody. Cell 176, 479–490.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wingler L. M. et al., Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR. Science 367, 888–892 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H. et al., Structural basis for ligand recognition and functional selectivity at angiotensin receptor. J. Biol. Chem. 290, 29127–29139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H. et al., Structure of the Angiotensin receptor revealed by serial femtosecond crystallography. Cell 161, 833–844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullo M., Tschumi S., Bucher B. S., Bianchetti M. G., Simonetti G. D., Pregnancy outcome following exposure to angiotensin-converting enzyme inhibitors or angiotensin receptor antagonists: A systematic review. Hypertension 60, 444–450 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Mol B. W. J. et al., Pre-eclampsia. Lancet 387, 999–1011 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Buse M. G., Roberts W. J., Buse J., The role of the human placenta in the transfer and metabolism of insulin. J. Clin. Invest. 41, 29–41 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon C. et al., Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 25, 289–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strachan R. T. et al., Divergent transducer-specific molecular efficacies generate biased agonism at a G protein-coupled receptor (GPCR). J. Biol. Chem. 289, 14211–14224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le M. T., Pugsley M. K., Vauquelin G., Van Liefde I., Molecular characterisation of the interactions between olmesartan and telmisartan and the human angiotensin II AT1 receptor. Br. J. Pharmacol. 151, 952–962 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauser A. S. et al., Pharmacogenomics of GPCR drug targets. Cell 172, 41–54.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheloha R. W. et al., Improved GPCR ligands from nanobody tethering. Nat. Commun. 11, 2087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrants H. et al., SNAP-tagged nanobodies enable reversible optical control of a G protein-coupled receptor via a remotely tethered photoswitchable ligand. ACS Chem. Biol. 13, 2682–2688 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simister N. E., Placental transport of immunoglobulin G. Vaccine 21, 3365–3369 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Roy S. et al., M281, an anti-FcRn antibody, inhibits IgG transfer in a human ex vivo placental perfusion model. Am. J. Obstet. Gynecol. 220, 498.e1-498.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Staus D. P. et al., Sortase ligation enables homogeneous GPCR phosphorylation to reveal diversity in β-arrestin coupling. Proc. Natl. Acad. Sci. U.S.A. 115, 3834–3839 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wingler L. M. et al., Angiotensin analogs with divergent bias stabilize distinct receptor conformations. Cell 176, 468–478.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.