Significance
Lung cancers are frequently driven by well-characterized mutations in proto-oncogenes, including ERBB2. Evidence from other cancers demonstrates that ERBB2 is also activated by alternative splicing that causes skipping of exon 16. Here, we find evidence for increased ERBB2 alternative splicing in a subset of lung cancers and a range of other solid tumors, including deletions and previously undescribed mutations promoting exon 16 skipping. We show that the ERBB2 isoform lacking exon 16 transforms lung epithelial cells in vitro and in vivo, arguing that alternative splicing of ERBB2 is a mechanism promoting lung carcinogenesis. These findings extend our knowledge of lung cancer biology and may lead to improved patient stratification to facilitate future use of ERBB2-targeted therapies.
Keywords: lung cancer, ErbB2 signaling, alternative splicing, oncogene activation, tumor progression
Abstract
Lung cancer causes more deaths annually than any other malignancy. A subset of non-small cell lung cancer (NSCLC) is driven by amplification and overexpression or activating mutation of the receptor tyrosine kinase (RTK) ERBB2. In some contexts, notably breast cancer, alternative splicing of ERBB2 causes skipping of exon 16, leading to the expression of an oncogenic ERBB2 isoform (ERBB2ΔEx16) that forms constitutively active homodimers. However, the broader implications of ERBB2 alternative splicing in human cancers have not been explored. Here, we have used genomic and transcriptomic analysis to identify elevated ERBB2ΔEx16 expression in a subset of NSCLC cases, as well as splicing site mutations facilitating exon 16 skipping and deletions of exon 16 in a subset of these lung tumors and in a number of other carcinomas. Supporting the potential of ERBB2ΔEx16 as a lung cancer driver, its expression transformed immortalized lung epithelial cells while a transgenic model featuring inducible ERBB2ΔEx16 specifically in the lung epithelium rapidly developed lung adenocarcinomas following transgene induction. Collectively, these observations indicate that ERBB2ΔEx16 is a lung cancer oncogene with potential clinical importance for a proportion of patients.
Over 50% of non-small cell lung cancers (NSCLCs) have one of several mutually exclusive mutations activating specific oncogenes, most commonly KRAS and EGFR (1, 2). Smaller but still significant proportions of the disease are driven by genomic events targeting other oncogenes, including MET, ALK, and RET (1, 2). Therapies targeting these oncogenes have demonstrated considerable clinical efficacy (3, 4), leading to concerted preclinical and clinical efforts to develop further personalized strategies for lung cancer patients based on the genomic profiles of their tumors. While EGFR is widely recognized as a major lung cancer driver, the closely related RTK ErbB2/HER2 (Human Epidermal Growth Factor Receptor 2) is also implicated in ∼3 to 7% of NSCLC cases, typically through activating mutations in its kinase and extracellular domains (5, 6) or through genomic amplification and overexpression (2, 7). Recent data from preclinical cancer models have identified other mechanisms of ERBB2 activation, including an alternative splicing event leading to the exclusion (or “skipping”) of exon 16, generating an ERBB2 variant with a 16-amino acid in-frame deletion in the juxta-transmembrane region, referred to as ErbB2ΔEx16. This variant constitutively forms disulfide-bonded homodimers that engage downstream pathways typically implicated in ErbB2-mediated transformation, including the Ras-MAP kinase and PI(3)-kinase/Akt pathways (8). While we and others have recently shown the significance of ERBB2 alternative splicing in breast cancer (9–11), its involvement in other tumors where ERBB2 activation has been implicated, including lung cancer, has not been fully explored. Here, we use genomic and transcriptomic profiles derived from patient samples, as well as in vitro and genetically engineered mouse models (GEMMs), to show that ERBB2 exon 16 skipping occurs in a subset of lung cancers, among other tumor types, and demonstrate its transforming potential. These observations support the contention that ERBB2ΔEx16 is an oncogenic driver in a subset of lung cancers and also suggest that ERBB2 alternative splicing may play a similar role in a range of other human malignancies.
Results
ERBB2ΔEX16 is Overexpressed in a Subset of Human Lung Cancers.
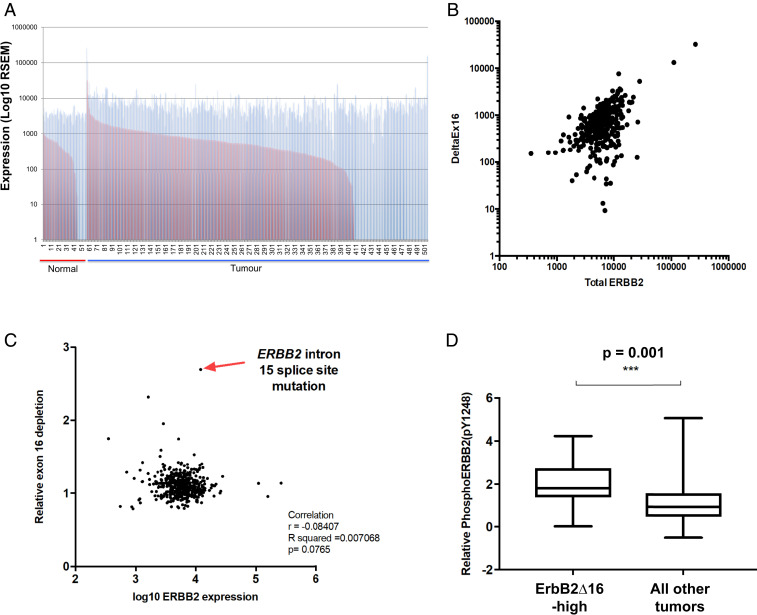
To investigate the prevalence of the ERBB2ΔEx16 transcript in lung cancers, we examined RNA-seq data from 57 normal lung tissue samples and 448 NSCLC (non-small cell lung carcinoma) cases available through The Cancer Genome Atlas (TCGA). We identified enrichment of ERBB2ΔEx16 in ∼4% (18/448) of samples by calculating the ratio between the number of reads representing the full-length ERBB2 transcript including exon 16 (ERBB2-FL) and those representing ERBB2ΔEx16 (Fig. 1A). While lung tumors expressing elevated overall levels of ERBB2 showed a corresponding increase in ERBB2ΔEx16 (Fig. 1B), we also observed that a high ERBB2ΔEx16:ERBB2-FL ratio did not always correlate with the overall expression level of ERBB2 (Fig. 1C), thereby indicating that expression of ERBB2ΔEx16 is not exclusively a function of elevated total ERBB2 gene expression. Given that ERBB2ΔEx16 is a constitutively active RTK that drives transformation (8), we correlated levels of tyrosine phosphorylated ERBB2, determined using reverse-phase protein array (RPPA) data, with ERBB2ΔEx16 expression. This revealed that lung tumors expressing the highest levels of ERBB2ΔEx16 exhibited significantly higher ERBB2 autophosphorylation (Fig. 1D), suggesting that elevation of ERBB2ΔEx16 expression enhances ERBB2 signaling in lung cancer.
Fig. 1.
A subset of human lung cancers with elevated expression of ErbB2ΔEx16 and tyrosine phosphorylation of ERBB2. (A) RNA-seq data from the TCGA lung adenocarcinoma dataset were analyzed to quantify full-length ERBB2 (blue bars) and ERBB2ΔEx16 (red bars) transcripts in samples of normal lung tissue and lung cancers. (B) The relationship between overall ERBB2 transcript level and ERBB2ΔEx16 level in the TCGA lung adenocarcinoma RNA-seq data was examined using Pearson’s correlation analysis. (C) The ratio of ERBB2ΔEx16 to full-length ERBB2 was plotted against the total level of ERBB2 transcript as determined from TCGA lung adenocarcinoma RNA-seq data, while identifying the tumor sample with highest ERBB2ΔEx16:ERBB2 ratio (arrow), which harbors a point mutation ablating the splice acceptor site in intron 15. (D) TCGA RPPA data for ERBB2 tyrosine 1248 phosphorylation in a subset of the same panel of lung adenocarcinoma samples was stratified by expression level of ERBB2ΔEx16 whereby the 5% of tumors with the highest ERBB2ΔEx16 transcript levels were considered ERBB2ΔEx16-high.
During the course of this analysis, we observed that the sample with the highest proportion of ERBB2ΔEx16:ERBB2-FL ratio (TCGA-05-4384) had a point mutation located at the splice acceptor site in the 15th intron, adjacent to exon 16 (G to A, referred to as “X633_splice”) (Fig. 1C and SI Appendix, Fig. S1). Since the guanine residue at this position is essential for exon inclusion (12), this mutation would be highly likely to mediate constitutive skipping of exon 16. We confirmed that reads mapping to exon 16 were underrepresented compared to flanking exons in this sample, with increased presence of reads mapping to the junction of exons 15 and 17 compared to a sample with a low ERBB2ΔEx16:ERBB2-FL ratio (TCGA-73-7499) (SI Appendix, Fig. S1).
Identification of Genomic Events that Induce ERBB2 Exon 16 Skipping in Human Cancers.
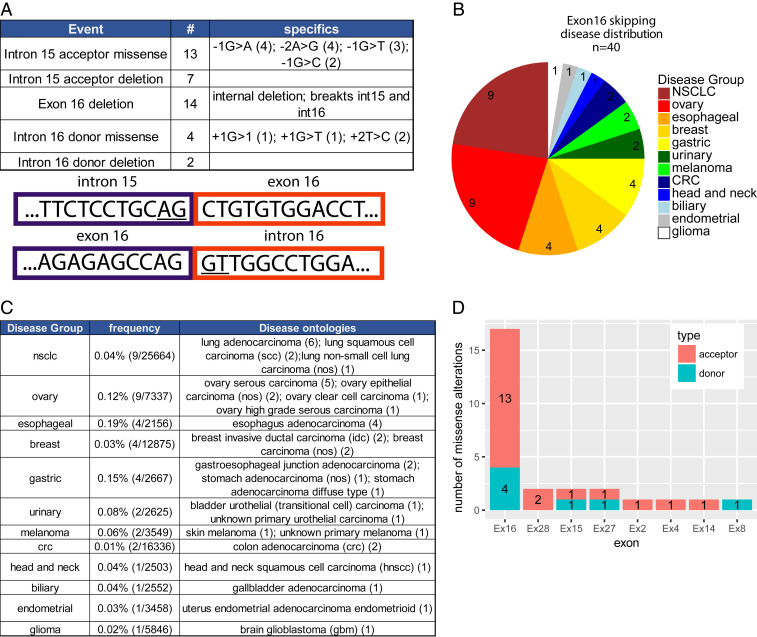
To explore the significance of these observations further, we mined a large database of tumor genome sequencing data derived from patients with a wide range of tumor types, including NSCLC, who underwent comprehensive genomic profiling (CGP) using a targeted exome sequencing assay (FoundationOne) run in a Clinical Laboratory Improvement Amendments (CLIA)-certified and College of American Pathologists (CAP)-accredited laboratory at Foundation Medicine (7, 13). An examination of ERBB2 in this dataset revealed 40 alterations that may impact exon 16 inclusion (Fig. 2A). These alterations spanned several classes, including 13 alterations of the intron 15 acceptor site, seven deletions of the intron 15 acceptor site, 14 complete deletions of exon 16 (each through an internal deletion with break points in introns 15 and 16), four missense alterations of the intron 16 donor site, and two deletions of the intron 16 donor site. Four of these patient samples had alterations that exactly matched the −1G>A splice acceptor mutation observed in the TCGA lung cancer study. The exon 16 splicing alterations were observed across 10 diverse disease groups and 18 individual disease ontologies, including NSCLC (n = 9), ovarian cancers (n = 9), esophageal cancers (n = 4), and breast carcinomas (n = 4) (Fig. 2B). Corrected for the number of samples, the frequency of ERBB2 splicing alterations did not markedly differ between disease groups (frequency range 0.03 to 0.19%) (Fig. 2C). Thus, while infrequent overall, mutations affecting the splicing sites that mediate inclusion of ERBB2 exon 16 occur in human cancers, including lung cancer. These findings are consistent with a recent study where deletion of ERBB2 exon 16 occurred in a single case of EGFR-mutated NSCLC that developed resistance to osimertinib, a tyrosine kinase inhibitor (TKI) targeting mutant EGFR (14).
Fig. 2.
ERBB2 splice site mutations in human cancers. (A) Table of genomic alterations potentially impacting exon 16 splicing (Top) in patients with a wide range of tumor types, including NSCLC, who underwent CGP using a targeted exome sequencing assay (FoundationOne). Graphic of the intron 15/exon 16 and exon 16/intron 16 junctions with the splice donor and acceptor sites underlined (Bottom). (B) Pie chart showing the disease group distribution of specimens from A. (C) Table of exon 16 splice site events broken down by disease group. Included are frequencies within the group (column 2) and specific disease ontologies as annotated in the Foundation Core database. (D) Count of core splice site (2 nucleotide) missense alterations in the donor and acceptor sites flanking each exon.
We found that missense alterations in splicing donor and acceptor sites across ERBB2 are almost exclusively confined to regions flanking exon 16 (n = 17). A smaller number of mutations impacted on exon 28 (n = 2), exon 15 (n = 2), and exon 27 (n = 2) while four additional exons had only a single splice site alteration present in the database (Fig. 2D). However, apart from exon 4, which was affected by a single mutation, exclusion of any of these exons would generate a frameshift in the final transcript, indicating that these are likely to be passenger mutations. Strikingly, most cancer cases harboring ERBB2 splicing mutations contained relatively few mutations in other known driving oncogenes. For the nine NSCLC specimens, only two had cooccurring alterations (one ALK, one KRAS) in canonical lung cancer drivers (KRAS, BRAF, EGFR, RET, ROS1, ALK, MET). Four tumors (three ovarian and one NSCLC) harbored only a splice site alteration in ERBB2 and no other known driver alterations. ERBB2 was amplified in 43% (17/40) of tumors with ERBB2 splice site alterations affecting exon 16. Interestingly, there is precedence for genomic amplification of activated, mutant forms of ERBB2 since human lung cancers with ERBB2 kinase domain mutations tend to also undergo ERBB2 genomic amplification (7), while mammary epithelial tumors initiated by expression of an activated ErbB2 point mutant under the transcriptional control of its endogenous promoter frequently amplify this transgene during progression (15).
Splice Acceptor Site Mutations Promote ERBB2 Exon 16 Skipping.
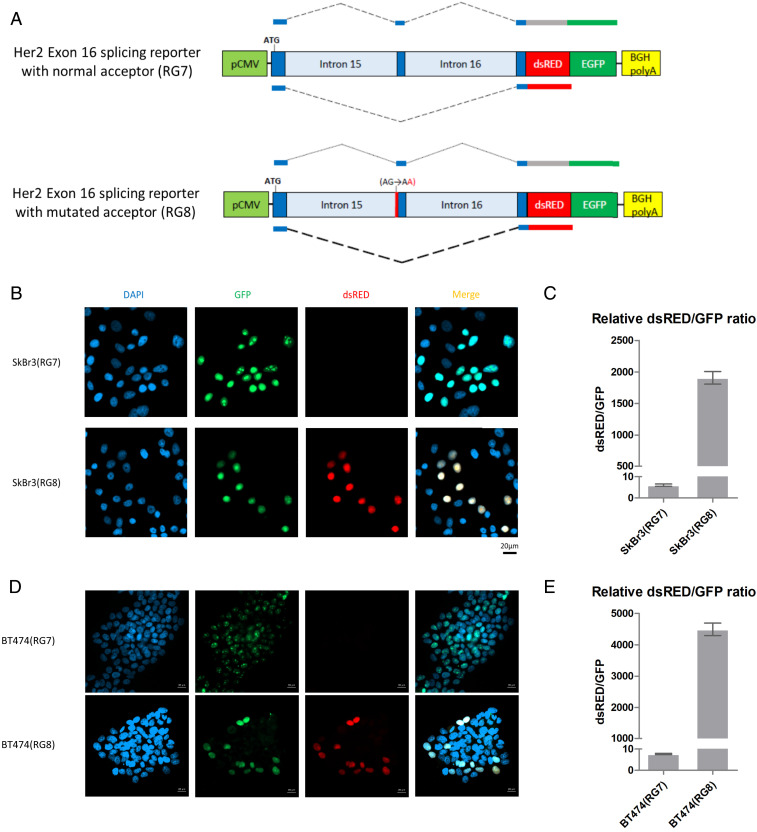
To confirm the functional role of the observed ERBB2 mutations in altering the alternative splicing patterns of ERBB2, we created a splicing reporter cassette (16) containing a 49-base pair sequence derived from ERBB2 exon 16 flanked by portions of ERBB2 introns 15 and 16 (Fig. 3A). Exon 16 skipping in the transcript generated from this reporter induces a frameshift, resulting in a switch from a GFP (exon 16 inclusion) to a dsRed (exon 16 skipping) fusion protein. We introduced two of the most frequently identified alterations from our genomic dataset (−1G>A and −1G>C) into the intron 15 splicing acceptor site and transfected wild-type and mutant reporter constructs into SkBr3 and BT474 breast cancer cells, where the endogenous level of ERBB2ΔEx16 comprises ∼10% of the total ERBB2 messenger RNA (mRNA) (SI Appendix, Fig. S2 A and B) (9). The splice acceptor site mutations prominently increased the ratio of the dsRed:GFP fusion protein compared to the wild-type reporter, indicating a significant elevation in exon 16 skipping (Fig. 3 B–E and SI Appendix, Fig. S2 C and D). Taken together with the RNA-seq data (Fig. 1 and SI Appendix, Fig. S1), these observations argue that ERBB2 splicing mutations found in human cancer facilitate exon 16 skipping and thereby promote overexpression of the highly oncogenic ERBB2ΔEx16 isoform.
Fig. 3.
Mutation of the splice acceptor site at the ERBB2 intron15/exon16 junction leads to skipping of exon 16. (A) Schematic diagram illustrating the ERBB2 splicing reporter construct. In this system, inclusion of exon 16 will result in expression of GFP whereas skipping of exon 16 will result in expression of dsRed. Reporters were constructed with a wild-type splice acceptor site between intron 15 and exon 16 (RG7, Top) and containing the G-A mutation in the splice acceptor site (RG8, Bottom) that was identified in five human cancers (Fig. 2B). (B–E) SkBr3 (B and C) or BT474 (D and E) cells were transfected with the splicing reporter constructs, GFP and dsRed fluorescence was observed using confocal microscopy, and corresponding transcripts were quantified using qRT-PCR.
ERBB2ΔEx16, but Not Full-Length ERBB2, Transforms Immortalized Epithelial Cells.
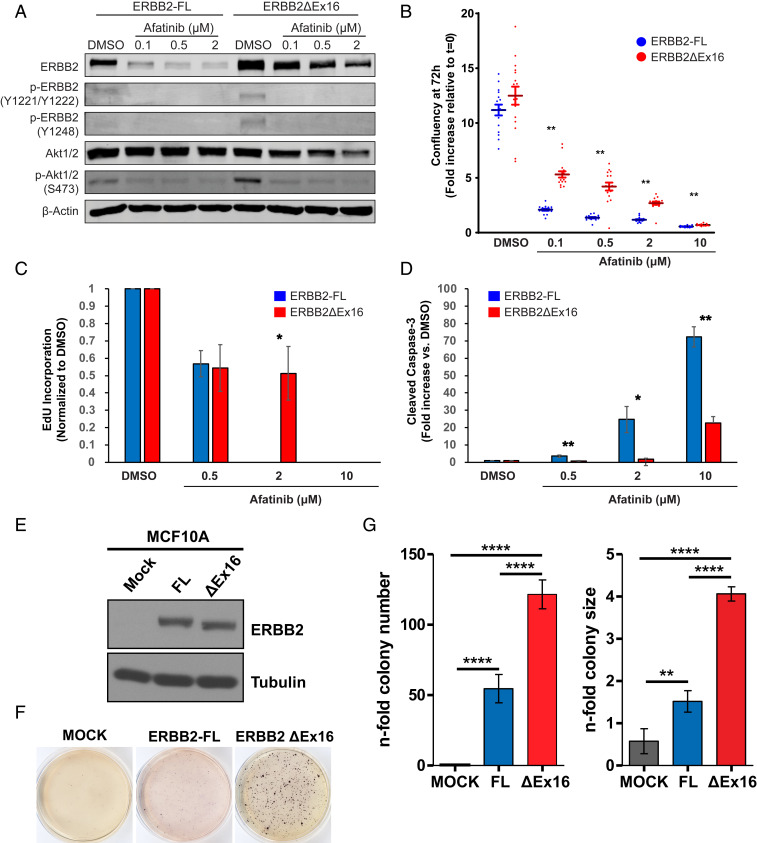
To assay the transforming capacity of ERBB2ΔEx16 in epithelial cells, we first transduced RLE-6TN, an immortalized but nontumorigenic line derived from rat lung alveolar type-II cells (17), with lentiviral vectors to stably express complementary DNA (cDNA) encoding either ERBB2-FL or ERBB2ΔEx16. While the amount of ERBB2 mRNA expressed was comparable between RLE-6TN cells expressing each ERBB2 isoform, the level of ERBB2ΔEx16 protein was higher than that of ERBB2-FL (SI Appendix, Fig. S3A). Cells expressing ERBB2ΔEx16 also exhibited elevated ERBB2 signaling, assessed by tyrosine phosphorylation of ERBB2 and downstream activation of the PI3K/Akt pathway (Fig. 4A). Treatment with afatinib, an irreversible, ATP-competitive pan-ERBB family TKI, inhibited ERBB2 signaling and cell growth in a dose-dependent manner in cells expressing either ERBB2 isoform (Fig. 4 A and B and SI Appendix, Fig. S3B), corresponding with diminished expression of markers of proliferation (Ki67) and elevated expression of markers of cell death (cleaved caspase-3) (Fig. 4 C and D and SI Appendix, Fig. S3C). However, despite similar inhibition of ERBB2 and Akt phosphorylation in both cell lines, effects on growth, proliferation, and death were significantly greater in cells expressing ERBB2-FL compared to those expressing ERBB2ΔEx16 at each concentration of afatinib tested. Interestingly, while in vitro proliferation assays indicated no significant difference in growth between lung epithelial cells expressing either ERBB2 isoform, in vivo tumorigenesis assays revealed that RLE-6TN cells expressing ERBB2ΔEx16 readily established ERBB2-positive tumors in immunocompromised mice whereas RLE-6TN cells expressing ERBB2-FL were not tumorigenic (SI Appendix, Fig. S3 D and E). Collectively, these data suggest that ERBB2ΔEx16 has a greater capacity to transform lung epithelial cells than ERBB2-FL, while also inducing a degree of resistance to ERBB2-targeting TKIs.
Fig. 4.
ErbB2ΔEx16 transforms immortalized epithelial cells and confers resistance to ErbB2 kinase inhibition compared to ERBB2-FL. (A) RLE-6TN cells stably expressing ERBB2 isoforms were treated with Afatinib at the indicated concentrations or DMSO (vehicle). After 24 h, cell lysates were immunoblotted with the indicated antibodies. (B) RLE-6TN stable cell lines as in A were treated with the indicated concentrations of Afatinib or with DMSO, and their proliferation was monitored by image-based analysis of cell confluency. Data shown are mean fold increase in confluency at 72 h posttreatment relative to the beginning of the assay (t = 0), +/− SEM, and are representative of two independent experiments, each with six replicates per condition. **P < 0.01, unpaired Student’s t test of RLE-6TN/ERBB2-FL vs. RLE-6TN/ERBB2ΔEx16 for each condition. For both cell lines, proliferation at each concentration of Afatinib was significantly (P < 0.01) lower than the DMSO control. (C and D) RLE-6TN stable cell lines were treated with the indicated concentrations of Afatinib or DMSO for 24 h, and immunofluorescence was used to quantify cell cycle progression (C, EdU incorporation) and apoptosis (D, cleaved Caspase-3 immunostaining) with normalization to the DMSO control. Data are the mean +/− SEM of six replicates. *P < 0.05, **P < 0.01, unpaired Student’s t test of RLE-6TN/ERBB2-FL vs. RLE-6TN/ERBB2ΔEx16 for each condition. (E) Cell lysates from MCF10-A cells stably expressing ERBB2 isoforms were immunoblotted with the indicated antibodies. (F and G) Anchorage-independent growth potential was assessed by soft agar assay of MCF10A cells expressing ERBB2 isoforms. Cells were stained with MTT and photographed after 2 wk (F), and colony number and size were quantified (G). Data are represented as mean ± SD. **P < 0.01, ****P < 0.0001, unpaired Student’s t test.
To confirm the oncogenic capacity of the ERBB2ΔEx16 splice variant in human epithelial cells, we used the nontumorigenic mammary epithelial cell line MCF10A (18), a well-characterized system that has been used extensively to study mutant forms of ERBB2 (19, 20). Despite comparable levels of expression of the ERBB2 isoforms in this system (Fig. 4E), anchorage-independent growth assays demonstrated that ERBB2ΔEx16 expression supported the growth of significantly more colonies of larger size compared to ERBB2-FL (Fig. 4 F and G). This indicates that, as in murine mammary (9) and rat lung epithelial cells (SI Appendix, Fig. S3 D and E), and consistent with previously published data (20), the transforming capacity of ERBB2ΔEx16 in this human-derived system is substantially greater than that of ERBB2-FL.
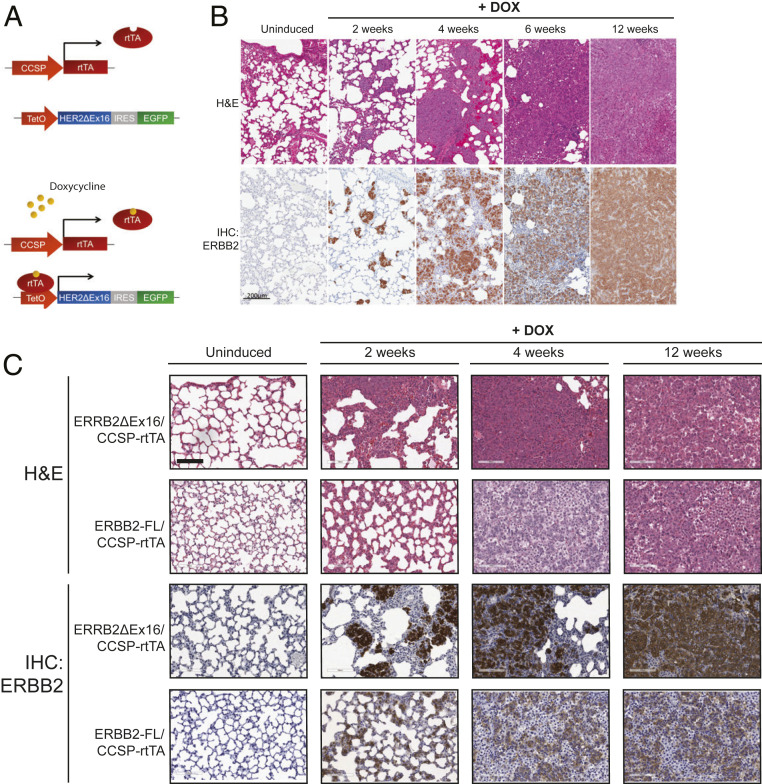
Inducible Expression of ERBB2ΔEx16 in a GEMM Drives Lung Carcinoma with an Immunosuppressive Tumor Microenvironment.
GEMMs have been used to confirm the transforming potential of human lung cancer oncogenes including KRASG12D (21) and mutant EGFR (22), as well as the “YVMA” kinase domain mutation of HER2, yielding models histologically similar to the human disease (23). These studies have frequently relied on the Club Cell Secretory Protein (CCSP) promoter, which drives gene expression in Club cells and subsets of Type-II pneumocytes (24, 25). To directly test whether elevated expression of ERBB2ΔEx16 can drive lung tumorigenesis, we constructed a GEMM combining TET-operator-driven ERBB2ΔEx16-IRES-GFP (9) and CCSP-rtTA (reverse tetracycline-dependent transactivator) (25) transgenes, resulting in ERBB2ΔEx16 and GFP expression in bitransgenic mice upon doxycycline (DOX) administration (Fig. 5A). Histological examination, immunohistochemical analysis of ERBB2 expression, and fluorescence microscopy of whole-mounted lung tissue to detect GFP expression were performed at defined time points following transgene induction with DOX, revealing the presence of ERBB2-expressing hyperplasias as early as 2 wk postinduction, with progression to large, invasive ERBB2/GFP-positive adenocarcinomas by 12 wk of DOX induction (Fig. 5B and SI Appendix, Fig. S4A). These observations were supported by quantitation of lung mass and the percentage of HER2-positive lung epithelial cells, both of which increased steadily during DOX induction (SI Appendix, Fig. S4B). To investigate the cell lineage that gives rise to ERBB2ΔEx16/CCSP-rtTA-driven tumors, we examined coexpression of ErbB2 with Surfactant Protein C (SP-C), a marker of type II pneumocytes, and CCSP, a marker of club cells. As observed in other CCSP oncogene-driven models (21) and in human NSCLC, lung lesions in the ERBB2ΔEx16/CCSP-rtTA model stained positively for SP-C, as well for Pro-SP-C (SP-C precursor) (SI Appendix, Fig. 4C). ERBB2ΔEx16/CCSP-rtTA tumor cells also expressed CC10, which occurs infrequently in lung cancer models (26) and may indicate that these tumors arise from an early epithelial progenitor population such as bronchioalveolar stem cells (BASCs), which coexpress these markers (27). Interestingly, mammary tumors induced by ERBB2ΔEx16 coexpress markers of basal and luminal epithelial cells, indicating an analogous state of poor or alternative differentiation (9).
Fig. 5.
Inducible expression of ErbB2ΔEx16 drives lung adenocarcinoma. (A) Schematic diagram illustrating the ERBB2ΔEx16-IRES-GFP/CCSP-rtTA cross. (B) H&E staining (Top) and ERBB2 immunohistochemistry (Bottom) were performed on lung tissue sections from ERBB2ΔEx16/CCSP-rtTA mice at the indicated time points following DOX administration. (Scale bar: 200 μm.) (C) H&E staining (Top Panels) and ERBB2 immunohistochemistry (Bottom) were performed on lung tissue sections from ERBB2-FL/CCSP-rtTA and ERBB2ΔEx16/CCSP-rtTA mice at the indicated time points following DOX administration. (Scale bar: 100 μm.)
To compare the in vivo transforming capacity of ERBB2 isoforms, we generated additional GEMMs using the strategy described above to inducibly express either ERBB2ΔEx16 or ERBB2-FL in the lung epithelium. Strikingly, whereas expression of ERBB2ΔEx16 induced the formation of focal lung lesions with rapid kinetics as described above, ERBB2-FL was expressed in a diffuse pattern, with disruption of normal lung architecture and histological evidence of inflammation but an absence of focal ERBB2-positive lesions (Fig. 5C and SI Appendix, Fig. S4 D and E). Immunohistochemical (IHC) staining of ERBB2 in these models was consistent with in vitro findings showing higher protein expression of ERBB2ΔEx16 compared to ERBB2-FL. The lung pathology in mice expressing ERBB2-FL correlated with deteriorating body condition and significant weight loss following DOX administration, phenotypes not observed in mice expressing ERBB2ΔEx16 (SI Appendix, Fig. S4F). To respect ethical guidelines, ERBB2/CCSP-rtTA mice were therefore not induced for periods longer than 12 wk, and mice exhibiting symptoms were euthanized. Overall, these data indicate that ERBB2ΔEx16 expression drives lung tumorigenesis while expression of full-length ERBB2 primarily induces an inflammatory state without the formation of discrete premalignant lesions or carcinomas in this model.
Mammary-epithelial expression of ERBB2ΔEx16 induces a highly inflammatory tumor microenvironment characterized by infiltration of immune cells including macrophages and T cells (9). Using immunofluorescent staining to detect immune cell markers, we found that ERBB2-positive lesions in the lungs of DOX-induced ERBB2ΔEx16/CCSP-rtTA and ERBB2-FL/CCSP-rtTA mice were infiltrated extensively by macrophages, many of which expressed CD206, suggestive of their polarization toward an “M2” phenotype associated with tumor progression in many cancers (28) (SI Appendix, Fig. S5A). However, consistent with an antitumor immune response, ERBB2-positive regions of lung tissue from DOX-induced ERBB2-FL/CCSP-rtTA were infiltrated by CD8+ cytotoxic T cells that were interspersed among the ERBB2+ lung epithelial cells. In contrast, CD8+ cells were largely restricted to the stroma surrounding ERBB2+ lesions in DOX-induced ERBB2ΔEx16/CCSP-rtTA mice (SI Appendix, Fig. S5B) where a prominent population of immunosuppressive CD4/FOXP3-positive regulatory T cells (T-regs) (29) was also observed (SI Appendix, Fig. 5C). T-regs are associated with lung cancer progression in mouse models and human patients (30–32), particularly when their abundance is increased relative to Th17 cells, another CD4+ population (30). Interestingly, analysis of systemic cytokine levels revealed that cytokines strongly associated with T-regs, such as IL-10 (33), were more abundant in DOX-induced ERBB2ΔEx16/CCSP-rtTA mice whereas IL-17, the signature cytokine of Th17 cells (34), was more abundant in ERBB2-FL/CCSP-rtTA mice (SI Appendix, Fig. 5D). This is consistent with alterations in the balance of CD4+ T cell populations that may favor tumor progression in the lungs of DOX-induced ERBB2ΔEx16/CCSP-rtTA mice. Overall, cytokines involved in antitumor immune responses [e.g., IL-12 (35) and IL-15 (36)] were elevated in the blood of ERBB2-FL/CCSP-rtTA tumor-bearing mice relative to ERBB2ΔEx16/CCSP-rtTA mice, whereas cytokines associated with tumor progression, including IL-10, IL-16 (37), and CCL21 (38), were more prevalent in the blood of ERBB2ΔEx16/CCSP-rtTA tumor-bearing mice (SI Appendix, Fig. S5D). These observations argue that lung epithelial expression of ERBB2-FL or ERBB2ΔEx16 leads to distinct immune microenvironments that may, at least in part, underlie the different phenotypes of these GEMMs.
Discussion
In this study we report that a subset of NSCLC overexpresses a constitutively active isoform of ERBB2, referred to as ERBB2ΔEx16, which is associated in some cases with either somatic mutation of splicing sites that mediate inclusion of exon 16 or deletion of exon 16. We show that ERBB2ΔEx16 transforms immortalized murine (9), rat, and human (20) epithelial cells and drives the formation of rapidly progressing lung adenocarcinomas when expressed in GEMMs. Interestingly, the full-length ERBB2 isoform (ERBB2-FL) is unable to transform immortalized epithelial cells (Fig. 4 and SI Appendix, Fig. S3) (9) while a GEMM expressing ERBB2-FL did not form discrete neoplastic lung lesions. These results suggest that, consistent with earlier studies of breast cancer models (9, 20), ERBB2ΔEx16 is a more potent oncogene than ERBB2-FL in lung epithelial cells. While overexpression of ERBB2-FL in a GEMM was sufficient to induce lung adenocarcinoma in other studies (39), these differences may be due to variation in the genetic backgrounds of the mice used, which is a major factor affecting phenotypes observed in GEMMs of cancer (40), or to experimental conditions differing between studies. Importantly, the poor condition of these mice did not permit us to evaluate whether sustained expression of ERBB2-FL would eventually result in tumorigenesis in this setting. Future studies incorporating other genomic modifications into these GEMMs, guided by patient genomic data, may also improve our understanding of the oncogenic properties of ERBB2 isoforms.
The immune/inflammatory responses in the lungs of mice expressing ERBB2-FL and those expressing ERBB2ΔEx16 are also consistent with findings from mammary cancer models, where tumor cells expressing ERBB2ΔEx16 suppress the immune response against the antigenic human ERBB2 protein. Our histopathological and cytokine profiling studies are consistent with a role for the immune system in the distinct phenotypes of GEMMs expressing either ERBB2-FL or ERBB2ΔEx16. For example, regions of ERBB2-FL–expressing lung epithelial cells were extensively infiltrated by cytotoxic T cells, whereas these were excluded from ERBB2ΔEx16-expressing lung lesions, which were surrounded by a stroma rich in immunosuppressive regulatory T cells. Based on these findings, further studies are warranted to examine the functions of the immune system in directing the initiation and progression of ERBB2-driven NSCLC in these models and to identify potential strategies for immunotherapy.
While genomic events affecting ERBB2 exon 16 are rare in individual tumor types (Fig. 2 A and B), they occur in a broad range of prevalent solid tumors beyond NSCLC, including those of the lung, ovary, stomach, colon, and breast, increasing their potential clinical signficance. Interestingly, tumors with ERBB2 splicing mutations tended to have few other alterations in known oncogenes. Indeed, in this dataset, they were accompanied mainly by mutations in tumor suppressor genes, most notably TP53, as well as by amplification of the ERBB2 locus. Overall, our data suggest the importance of investigating ERBB2ΔEx16 as a potential driver in additional tumor types beyond breast and lung cancer. Importantly, genomic events triggering ERBB2 exon 16 skipping can be detected using next-generation sequencing (NGS)-based CGP assays that are currently being implemented in the clinic (41–43). In NSCLC, elevated expression of ERBB2ΔEx16 also occurs in the absence of genomic events affecting ERBB2 mRNA splicing (Fig. 1A). These results are consistent with reports showing that specific mRNA splicing factors overexpressed in some cancers, such as SRSF3 and hnRNP H1, can induce ERBB2 exon 16 skipping (44). It is conceivable that alterations in specific splicing regulators could be used as biomarkers to identify additional tumors exhibiting elevated ERBB2ΔEx16 expression (Fig. 1A). The development of a quantitative assay for ERBB2 isoforms that could be applied to clinical samples would also facilitate the identification of cancer cases where ERBB2ΔEx16 may be a driver.
Stratifying patients according to overexpression of ERBB2ΔEx16 has important therapeutic implications. Clinical trial results have suggested efficacy of anti-ERBB2 antibody-drug conjugates in lung cancers with ERBB2 mutations (45, 46), while TKIs are efficacious in preclinical models of ERBB2-driven NSCLC (23, 39), although their performance in the clinic has been variable (47, 48). Heterogeneous responses to ERBB2 targeted therapies in lung cancer are thought to be at least partially due to properties unique to specific molecular alterations in ERBB2 (49, 50). Our work provides a basis for further studies examining the available suite of ERBB2 targeted therapies and other modalities, such as immunotherapies, to identify effective strategies for cancers driven by ERBB2ΔEx16. The role of ERBB2 alternative splicing in resistance to targeted therapy also merits further study, given our observations of TKI resistance in ERBB2ΔEx16-expressing cells (Fig. 4), and the published observation of ERBB2 exon 16 deletion in acquired resistance to Osimertinib in EGFR-mutant NSCLC (14). Such studies, which may be facilitated by the models established here, could then inform the design of clinical trials where molecular profiling is used to match patients with elevated ERBB2ΔEx16 levels with appropriate therapies.
Methods
Lung Cancer Patient Data Analysis.
Whole transcriptome sequencing data from lung tumors were downloaded from the TCGA Data Portal (level 3 data). ErbB2 gene-level and junction-level read counts were extracted for analysis. The relative abundance of ErbB2ΔEx16 was determined using the average read count of the E15:E16 (exon 15:exon 16) and E16:E17 junctions compared to the average of the E14:E15 and E17:E18 junctions. ERBB2 expression, obtained using RNA-Seq by Expectation-Maximization (51), was extracted from the cBIO Portal (http://www.cbioportal.org), and total abundance of ErbB2ΔEx16 was determined by adjusting this value based on the ratio of the junction counts containing exon 16 for each sample. For visualization of sequencing data, aligned reads (bam format) for samples TCGA-05-4384 (containing the ×633_splice mutation) and TCGA-73-7499 (nonmutated) were obtained from the National Cancer Institute Genomics Data Commons (GDC) portal (dbgap Project ID: 6208). Sliced bam files containing reads mapped to the ERBB2 genes (hg 38 built) were generated using the available GDC toll and used for the visualization analysis. Coverage and junction information for both samples was visualized using the Integrative Genomics Viewer (IGV) (version 2.4, Broad Institute). The large memory version (10 GB, 64-bit Java) was used. The scale for all tracks was equalized in order to make histogram height and junction sashimi plot comparable. Reverse-phase protein array data (Replicates-Based Normalized values) for the TCGA lung adenocarcinoma tumor set were downloaded from the University of California Santa Cruz Cancer Browser (http://xena.ucsc.edu/). Samples were separated into groups based on ERBB2ΔEX16 expression, and ERBB2-pY1248 was compared between the high ERBB2ΔEX16-expressing samples (top 5%) and all other tumors using the Mann–Whitney U test.
Targeted NGS.
A series of 121,509 clinical cases of various disease origins were analyzed using CGP in a Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited laboratory (FoundationOne, Foundation Medicine Inc., Cambridge, MA). The pathologic diagnosis of each case was confirmed by routine hematoxylin and eosin (H&E) staining, and all samples forwarded for DNA and/or RNA extraction contained a minimum of 20% tumor nuclei. Extensive technical descriptions and validation of the genomic profiling assays used to analyze these samples in the course of clinical care have been published previously (13). In summary, 50 ng of DNA was extracted from 40 μm of tumor sample from slides or formalin-fixed, paraffin-embedded tissue blocks. Targeted NGS was performed on hybridization-captured, adaptor ligation-based libraries for all coding exons of at least 236 cancer-related genes, including ERBB2, plus select introns from at least 19 genes frequently rearranged in cancer. Sequencing of captured libraries was performed using the Illumina HiSeq 2000 or HiSeq 2500 platforms to an average exon coverage depth of 710×, and resultant sequences were analyzed for base substitutions, insertions, deletions, copy number alterations (focal amplifications and homozygous deletions), and select gene fusions. All patients were consented for research.
Construction of ErbB2 Splicing Reporters.
RG7 (wild-type splicing sites) was derived from an original splicing reporter RG6 (16). Primer sets ctgctctagaggtgagtccaacggtcttttctgc with ataagaatgcggcgcccttgtcatccaggtccac, and ataagaatgcggccgccgccgagcagagagccagg with gctcagcaaccggtcagagatgatggacgtccgaggg were used to amplify genomic sequences of ErbB2 intron 15 and intron 16. These sequences were then cloned into RG6, replacing the fragment between the XbaI and AgeI sites. RG8 and RG9 were derived from RG7, using the Q5 site-directed mutagenesis kit (Qiagen) according to the manufacturer’s instructions to introduce the A-G (RG8) and A-C (RG9) mutations in the intron 15 splice acceptor site.
Cell Culture and Retroviral Transduction.
The following cell lines (all human, female) were obtained from American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% (vol/vol) fetal bovine serum (FBS): HEK293T (CRL-3216), SkBr3 (HTB-30), and BT474 (HTB-20). RLE-6TN cells (rat, male) were obtained from ATCC (CRL-2300) and cultured in DMEM/F12 media with 2 mM l-glutamine supplemented with 0.01 mg/mL bovine pituitary extract, 0.005 mg/mL insulin, 2.5 ng/mL insulin-like growth factor, 0.00125 mg/mL transferrin, and 2.5 ng/mL EGF. MCF10-A cells (human, female) were kindly provided by G. Charras, University College London, London, United Kingdom and were cultured in DMEM/F12 medium with 2 mM l-glutamine, supplemented with 5% (vol/vol) horse serum, 10 μg/mL insulin, 0.5 μg/mL hydrocortisone, and 20 ng/mL EGF (Invitrogen), 100 IU/mL penicillin, and 100 μg/mL streptomycin. For retroviral transduction, ERBB2-FL and ERBB2ΔEx16 in the pMSCV-Puro retroviral vector (9) were each transfected into 293T cells (ATCC) with cotransfection of pMD2.G and psPAX2, gifts from Didier Trono, École polytechnique fédérale de Lausanne, Lausanne, Switzerland (plasmids 12259 and 12260; Addgene). Lipofectamine 3000 (L3000075; Invitrogen) was used for transfection according to the manufacturer’s instructions, and virus-containing media were harvested and filtered through a 0.45-µm filter at 24 and 48 h posttransfection. RLE-6TN cells were transduced in the presence of 10 µg/mL polybrene (107689; Sigma). Transduced cell lines were selected and maintained in Complete Media with 2 μg/mL puromycin (PUR333; BioShop). For lentiviral infection of MCF10A cells, GFP, ERBB2-FL-6Myc, and ERBB2ΔEx16-6Myc in the pLenti-puro lentiviral vector were each transfected using the calcium/phosphate method into HEK293T cells (ATCC) with cotransfection of pMDLg/pRRE, pRSV-Rev, and pMD2-VSVG. Virus-containing media were harvested and filtered through a 0.45-μm filter 48 h posttransfection. MCF10-A cells were infected in the presence of 4 μg/mL polybrene. Three days after viral infection, cells were treated and selected with 3 μg/mL puromycin. Cells were frequently PCR-tested to exclude mycoplasma contamination and used within 1 to 2 mo after thawing.
qRT-PCR.
Total RNAs were extracted using an RNeasy kit (Qiagen) and converted to cDNA using M-MuLV reverse transcriptase (New England Biolabs [NEB]). qPCR was performed with LightCycler 480 (Roche) using primers specific for full-length ErbB2 (GTGGACCTGGATGACAAGGG; TGCTGCCGTCGCTTGATGAG), ErbB2Δ16 (CAGCGGTGTGAAACCTGACC; TGGACGTCAGAGGGGAGTGG), GFP (GTGGACCTGGATGACAAGGG; CTCCATGCGCACCTTGAAGC), dsRED (GGATCTAGAGCCCTCGGACG; CCTCGATCTCGAACTCGTGG), GAPDH (human) (GTGGTCTCCTCTGACTTCAAC; GTTGCTGTAGCCAAATTCGTTG), and Gapdh (rat) (GAGAATGGGAAGCTGGTCAT; GAAGACGCCAGTAGACTCCA).
Immunoblotting.
Cells were lysed on ice in ice-cold RIPA buffer (50 mM Tris⋅HCl, pH 7.4, 150 mM sodium chloride, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 2 mM ethylenediaminetetraacetic acid, 0.5 mM 4-benzenesulfonyl fluoride hydrochloride [Santa Cruz, sc-202041], 25 mM β-glycerophosphate [G5422; Sigma], 1 mM sodium orthovanadate [SOV664; BioShop], and 10 mM sodium fluoride [S7920; Sigma]). Protein concentrations were determined by Bradford assay (5000006; Bio-Rad), and 40 μg of total protein was analyzed by immunoblot, using a Li-COR Odyssey system (Li-COR Biosciences) for fluorescent detection. The following primary antibodies were used for immunoblotting: ErbB2 (sc-284, Santa Cruz; and 06-562, Millipore); Phospho-Her2/ErbB2 (Tyr1221/1222) (2243; Cell Signaling); Phospho-Her2/ErbB2 (Tyr1248) (2247; Cell Signaling); Akt (pan) (58295; Cell Signaling); Phospho-Akt Ser473 (4060; Cell Signaling); Beta-actin (4700; Cell Signaling); and Tubulin (T5168; Sigma).
Immunofluorescence.
RLE-6TN stable cell lines were plated on glass coverslips in 24-well plates at a density of 4 × 104 cells per well, treated overnight with dimethyl sulfoxide (DMSO) or Afatinib at indicated concentrations and fixed for 20 min with 2% paraformaldehyde (PFA). Coverslips were rinsed three times with phosphate-buffered saline (PBS), permeabilized with PBS containing 0.2% Triton X-100 for 10 min, blocked at room temperature in PBS with 0.2% Triton X-100, 0.05% Tween-20, and 2% BSA for 30 min, and incubated with primary antibodies for 1 h at room temperature or overnight at 4 °C. Following three rinses in PBS with 0.2% Triton X-100 and 0.05% Tween 20, coverslips were incubated with Alexa Fluor-conjugated antibodies (all at 1/1,000 dilution; Invitrogen) for 40 min. After a further three rinses as above, nuclei were counterstained with 0.5 ng/mL DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride) for 10 min and rinsed with ddH2O twice. Coverslips were mounted on glass slides using ImmuMount (Thermo Scientific), allowed to dry overnight and imaged using a Zeiss LSM 800 confocal microscope running ZEN software (Zeiss). The primary antibody used was anti-Cleaved Caspase-3 (9661; Cell Signaling) at a 1/100 dilution. For EdU incorporation, the Click-IT EdU Incorporation kit (MP 10338) was used according to the manufacturer’s instructions with a final EdU concentration of 10 μM.
Cell Proliferation Assays.
Cell proliferation was analyzed through phase contrast-based image analysis of cell confluency using the Incucyte Zoom instrument (Essen Biosciences), with images taken at defined time points using a 10× objective and analysis using the associated software according to the manufacturer’s protocols.
Anchorage-Independent Growth Assays.
MCF10-A cells (1 × 104) were detached and resuspended in a top layer of growth media (DMEM/F12 with 2 mM l-glutamine, supplemented with 5% (vol/vol) horse serum, 10 μg/mL insulin, 0.5 μg/mL hydrocortisone, and 20 ng/mL EGF, 100 IU/mL penicillin, and 100 μg/mL streptomycin) containing 0.3% Noble agar and seeded in a 35-mm non-TC treated culture dish over a bottom layer of growth media containing 0.6% Noble Agar. Cells were fed twice a week with growth media. Colonies were stained after 15 d with a 0.5-mg/mL solution of Thiazolyl Blue Tetrazolium Bromide (MTT) reagent (Sigma-Aldrich). The number of colonies (normalized to the initial number of cells seeded) and colony size were quantified with ImageJ software.
Animal Studies.
All procedures involving mice were conducted under an approved protocol in accordance with national (Canadian Council for Animal Care) and institutional (McGill Facility Animal Care Committee) guidelines. CCSP-rtTA and TetO-ERBB2D16-IRES-GFP mice were described previously (9, 25). TetO-ERBB2D16-IRES-Cre and TetO-ERBB2FL-IRES-Cre constructs were generated by first subcloning the Cre recombinase sequence into the pTE-ERBB2D16-IRES-GFP vector, replacing the GFP, and then inserting the 48 nucleotide sequence derived from exon 16 using the Q5 Site-Directed Mutagenesis Kit (E0554S; NEB). Linearized plasmids were prepared by digestion with FspI and XhoI (both NEB) followed by agarose gel electrophoresis and purification using a QIAquick gel extraction kit (28704; Qiagen). Transgenic mouse lines were established by the McGill Integrated Core for Animal Modelling by pronuclear injection of the linearized constructs into FVB/N single-cell embryos, according to standard facility protocols. Founder lines were screened for germline transmission of the transgene using PCR genotyping. For studies involving GEMMs, DOX (2 mg/mL) was administered to bitransgenic TetO-ERBB2Δ16-IRES-GFP/CCSP1-rTTA, TetO-ERBB2Δ16-IRES-Cre/CCSP1-rTTA, or TetO-ERBB2-FL-IRES-Cre/CCSP1-rTTA mice at 6 wk of age via drinking water. Mice lacking either the ERBB2 or rTTA transgenes or mice not induced with DOX were used as negative controls. Mice were monitored daily and weighed weekly and were euthanized at appropriate stages as indicated, with tissues collected for further analysis. For xenograft studies, athymic Nude (NCr) mice (Taconic) were injected with 1 × 106 RLE-6TN cells expressing either full-length ERBB2 or ERBB2Δ16, generated as described above, or pMSCV-Puro Vector control, subcutaneously into the left flank or into the left inguinal mammary fat pad. Mice were monitored for tumor formation by daily manual palpation, and tumors were measured twice weekly. Tumors were allowed to reach a maximum size of 1.5 cm3, at which point the mice were euthanized.
Immunohistochemistry and Immunofluorescence on Formalin-Fixed, Paraffin-Embedded Tissue.
Harvested tissues were fixed in 10% neutral buffered formalin (3800600; Leica) for 24 h. Tissues were embedded in paraffin and sections cut at 4 μm by the Goodman Cancer Research Centre Histology Innovation Platform. Sections were deparaffinized in xylene and antigen retrieval was performed with 10 mM sodium citrate (pH 6) using a pressure cooker. Sections were then blocked with 10% Power Block (HK083; BioGenex) in PBS for 10 min at room temperature and endogenous peroxidase activity was quenched by incubation in 3% hydrogen peroxide for 20 min (for IHC only). Sections were incubated with primary antibody at 4 °C overnight and washed three times in PBS. For IHC, sections were incubated with ImmPRESS HRP polymer reagents to detect rabbit and mouse antibodies (Vector Elite, MP-7401/MP-7402; Vector Elite) according to the manufacturer’s instructions. After three further washes in PBS, IHC staining was visualized using the SignalStain DAB Substrate kit (8059S; Cell Signaling) according to the manufacturer’s instructions. Sections were then counterstained with hematoxylin, dehydrated, and mounted with Clearmount (10058832; Invitrogen). Images were acquired using an Aperio-XT slide scanner. For immunofluorescence, sections were incubated with secondary antibodies for one hour at room temperature, followed by DAPI (ThermoFisher, D1306) for 15 min, washed three times in PBS and mounted in ImmuMount (Thermo Scientific, 9990412). Immunostained samples were imaged using a Zeiss LSM800 confocal microscope and analyzed with ZEN software. Primary antibodies used were anti-ErbB2 (A0485; DAKO), SP-C (sc-7706; Santa Cruz), CCSP (07-623; Millipore), Pro-SP-C (AB3786; Millipore), F4/80 (7076; Cell Signaling), CD206 (AF2535; R&D Systems), CD3 (99940; Cell Signaling), CD8 (98941; Cell Signaling), CD4 (25229; Cell Signaling), and FoxP3 (12653; Cell Signaling). Slides were imaged using a Zeiss LSM800 confocal microscope running ZEN software (Zeiss).
Cytokine Arrays.
Cytokine levels were analyzed in serum harvested from cohorts of four ERBB2Δ16/CCSP-rRTA and four ERBB2-FL/CCSP-rRTA mice following 4 wk of transgene induction with DOX. Serum from two uninduced mice of each strain was harvested as a control for baseline cytokine expression. A multiplexed, fluorescent bead-based assay was used by Eve Technologies (https://www.evetechnologies.com/, Calgary, AB, Canada) for simultaneous detection of 44 murine cytokines according to their standard procedures.
Statistical Analysis.
Pearson’s correlation analysis was used to examine relationships between ERBB2 isoforms in RNA-seq data. ERBB2 phosphorylation data from RPPA were compared using a Mann–Whitney U test. All other analysis of significance was performed using a two-tailed, unpaired Student’s t test unless otherwise indicated.
Study Approval.
All patients providing samples to the TCGA or for CGP using the FoundationOne assay gave informed consent for research. All procedures involving mice were conducted under an approved protocol in accordance with national (Canadian Council for Animal Care) and institutional (McGill Facility Animal Care Committee) guidelines.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Cancer Research Society (21068), a Canada Research Councils Chair in Molecular Oncology (950-2310-33), and a Foundation award from the Canadian Institutes of Health Research (CIHR-FDN-148373) (all to W.J.M.).
Footnotes
Competing interest statement: E.S.S., D.C.P., G.M.F., and J.C. are current or former employees of Foundation Medicine, the manufacturer of the Comprehensive Genomic Profiling assay used for analysis of clinical samples in this study.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007474117/-/DCSupplemental.
Data Availability.
RNA-seq data analyzed in this study are available from the TCGA Data Portal. Raw data derived from exome sequencing of patient samples, as part of the clinical management of these patients, were analyzed by Foundation Medicine, and only the results of the analysis were provided. Requests for data should be addressed directly to Foundation Medicine. All other data and protocols supporting the findings of this study are available within the main text and SI Appendix.
References
- 1.Pao W., Girard N., New driver mutations in non-small-cell lung cancer. Lancet Oncol. 12, 175–180 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network , Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). Correction in: Nature514, 262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch T. J. et al., Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Chen Z., Fillmore C. M., Hammerman P. S., Kim C. F., Wong K.-K., Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 14, 535–546 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C. et al., Lung adenocarcinomas with HER2-activating mutations are associated with distinct clinical features and HER2/EGFR copy number gains. J. Thorac. Oncol. 7, 85–89 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Greulich H. et al., Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc. Natl. Acad. Sci. U.S.A. 109, 14476–14481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalmers Z. R. et al., Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 9, 34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel P. M., Ryan E. D., Cardiff R. D., Muller W. J., Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: Implications for human breast cancer. EMBO J. 18, 2149–2164 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turpin J. et al., The ErbB2ΔEx16 splice variant is a major oncogenic driver in breast cancer that promotes a pro-metastatic tumor microenvironment. Oncogene 35, 6053–6064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castiglioni F. et al., Role of exon-16-deleted HER2 in breast carcinomas. Endocr. Relat. Cancer 13, 221–232 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Castagnoli L. et al., Activated d16HER2 homodimers and SRC kinase mediate optimal efficacy for trastuzumab. Cancer Res. 74, 6248–6259 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Chen M., Manley J. L., Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frampton G. M. et al., Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu C. C. et al., Exon 16-skipping HER2 as a novel mechanism of osimertinib resistance in EGFR L858R/T790M-positive non-small cell lung cancer. J. Thorac. Oncol. 15, 50–61 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Andrechek E. R. et al., Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 97, 3444–3449 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orengo J. P., Bundman D., Cooper T. A., A bichromatic fluorescent reporter for cell-based screens of alternative splicing. Nucleic Acids Res. 34, e148 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driscoll K. E. et al., Establishment of immortalized alveolar type II epithelial cell lines from adult rats. In Vitro Cell. Dev. Biol. Anim. 31, 516–527 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Soule H. D. et al., Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50, 6075–6086 (1990). [PubMed] [Google Scholar]
- 19.Muthuswamy S. K., Li D., Lelievre S., Bissell M. J., Brugge J. S., ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat. Cell Biol. 3, 785–792 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alajati A. et al., Mammary tumor formation and metastasis evoked by a HER2 splice variant. Cancer Res. 73, 5320–5327 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Fisher G. H. et al., Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 15, 3249–3262 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji H. et al., The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell 9, 485–495 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Perera S. A. et al., HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc. Natl. Acad. Sci. U.S.A. 106, 474–479 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha M., Lowell C. A., Efficiency and specificity of gene deletion in lung epithelial doxycycline-inducible cre mice. Am. J. Respir. Cell Mol. Biol. 57, 248–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tichelaar J. W., Lu W., Whitsett J. A., Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J. Biol. Chem. 275, 11858–11864 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Xu X. et al., The cell of origin and subtype of K-Ras-induced lung tumors are modified by Notch and Sox2. Genes Dev. 28, 1929–1939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim C. F. B. et al., Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P., Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bos P. D., Plitas G., Rudra D., Lee S. Y., Rudensky A. Y., Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. J. Exp. Med. 210, 2435–2466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall E. A. et al., Emerging roles of T helper 17 and regulatory T cells in lung cancer progression and metastasis. Mol. Cancer 15, 67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganesan A.-P., et al. , Tumor-infiltrating regulatory T cells inhibit endogenous cytotoxic T cell responses to lung adenocarcinoma. J. Immunol. 191, 2009–2017 (2013). Corrected in: J. Immunol.191, 5319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips J. D. et al., Preferential expansion of pro-inflammatory tregs in human non-small cell lung cancer. Cancer Immunol. Immunother. 64, 1185–1191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asseman C., Mauze S., Leach M. W., Coffman R. L., Powrie F., An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J. Exp. Med. 190, 995–1004 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korn T., Bettelli E., Oukka M., Kuchroo V. K., IL-17 and Th17 cells. Annu. Rev. Immunol. 27, 485–517 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Trinchieri G., Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3, 133–146 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Guo Y., Luan L., Patil N. K., Sherwood E. R., Immunobiology of the IL-15/IL-15Rα complex as an antitumor and antiviral agent. Cytokine Growth Factor Rev. 38, 10–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richmond J., Tuzova M., Cruikshank W., Center D., Regulation of cellular processes by interleukin-16 in homeostasis and cancer. J. Cell. Physiol. 229, 139–147 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Shields J. D., Kourtis I. C., Tomei A. A., Roberts J. M., Swartz M. A., Induction of lymphoidlike stroma and immune escape by tumors that express the chemokine CCL21. Science 328, 749–752 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Liu S. et al., Targeting HER2 aberrations in non-small cell lung cancer with osimertinib. Clin. Cancer Res. 24, 2594–2604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taft R. A., Davisson M., Wiles M. V., Know thy mouse. Trends Genet. 22, 649–653 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Goodman A. M. et al., Next generation sequencing reveals potentially actionable alterations in the majority of patients with lymphoid malignancies. JCO Precis. Oncol. 1, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodman A. M. et al., Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 16, 2598–2608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blumenthal D. T. et al., Clinical utility and treatment outcome of comprehensive genomic profiling in high grade glioma patients. J. Neurooncol. 130, 211–219 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Gautrey H. et al., SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol. 12, 1139–1151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modi S. et al., Antitumor activity and safety of trastuzumab deruxtecan in patients with HER2-low-expressing advanced breast cancer: Results from a phase Ib study. J. Clin. Oncol. 38, 1887–1896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B. T. et al., Ado-trastuzumab emtansine for patients with HER2-mutant lung cancers: Results from a phase II basket trial. J. Clin. Oncol. 36, 2532–2537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai W. V. et al., Afatinib in patients with metastatic or recurrent HER2-mutant lung cancers: a retrospective international multicentre study. Eur. J. Cancer 109, 28–35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peters S. et al., Activity of afatinib in heavily pretreated patients with ERBB2 mutation-positive advanced NSCLC: Findings from a global named patient use program. J. Thorac. Oncol. 13, 1897–1905 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Robichaux J. P. et al., Pan-cancer landscape and analysis of ERBB2 mutations identifies poziotinib as a clinically active inhibitor and enhancer of T-DM1 activity. Cancer Cell 36, 444–457.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang W. et al., Mutation variants and co-mutations as genomic modifiers of response to afatinib in HER2-mutant lung adenocarcinoma. Oncologist 25, e545–e554 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B., Dewey C. N, RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data analyzed in this study are available from the TCGA Data Portal. Raw data derived from exome sequencing of patient samples, as part of the clinical management of these patients, were analyzed by Foundation Medicine, and only the results of the analysis were provided. Requests for data should be addressed directly to Foundation Medicine. All other data and protocols supporting the findings of this study are available within the main text and SI Appendix.