Significance
RNA polymerase II (Pol II) is stalled at the transcription starting site for a large number of genes in higher eukaryotes. We previously found that JMJD5/7 proteins act as both endopeptidases and exopeptidases to cleave arginine methylated histone tails including those of H2A, H3, and H4, thus generating relatively “tailless nucleosomes”, which can be overcome by Pol II. This report shows that knockout of JMJD5 in mice leads to accumulation of nucleosomes at +1 and loss of expression of a large number of genes. We find that the N-terminal domain of JMJD5 binds to CTD of Pol II with a unique combination of phosphorylation generated by CDK9. We propose that JMJD5 couples with CDK9 to release the paused Pol II.
Keywords: JMJD5 N-terminal, CTD interacting domain, CDK9, RNA polymerase II phosphorylation, pausing release
Abstract
More than 30% of genes in higher eukaryotes are regulated by RNA polymerase II (Pol II) promoter proximal pausing. Pausing is released by the positive transcription elongation factor complex (P-TEFb). However, the exact mechanism by which this occurs and whether phosphorylation of the carboxyl-terminal domain of Pol II is involved in the process remains unknown. We previously reported that JMJD5 could generate tailless nucleosomes at position +1 from transcription start sites (TSS), thus perhaps enable progression of Pol II. Here we find that knockout of JMJD5 leads to accumulation of nucleosomes at position +1. Absence of JMJD5 also results in loss of or lowered transcription of a large number of genes. Interestingly, we found that phosphorylation, by CDK9, of Ser2 within two neighboring heptad repeats in the carboxyl-terminal domain of Pol II, together with phosphorylation of Ser5 within the second repeat, HR-Ser2p (1, 2)-Ser5p (2) for short, allows Pol II to bind JMJD5 via engagement of the N-terminal domain of JMJD5. We suggest that these events bring JMJD5 near the nucleosome at position +1, thus allowing JMJD5 to clip histones on this nucleosome, a phenomenon that may contribute to release of Pol II pausing.
Pausing of RNA polymerase II (Pol II) at promoter regions is a unique transcription regulation mechanism seen in higher eukaryotes (1–7). It is well established that the positive transcription elongation factor P-TEFb, a protein complex that includes Cyclin T1 and CDK9, is essential for the release of paused Pol II (4, 8–10). P-TEFb is thought to act in this regard because the catalytic subunit of P-TEFb, CDK9, phosphorylates negative elongation factor (NELF) and 5,6-Dichloro-1-β-d-ribofuranosylbenzimidazole (DRB)-sensitivity-induced factor (DSIF), thus releasing Pol II from inhibition by these proteins (11–15). CDK9 has also been found to activate Pol II by phosphorylating Ser2 residues in the repeating heptad sequences in the C-terminal domain (CTD) of Pol II (16–24), but how this improves transcription progression by Pol II is controversial (25–28).
To search for mechanisms that might be involved in the release of Pol II from pausing, we investigated whether a member of the JmjC family of proteins; for example, JMJD5, might play a role. JMJD5 is a potential candidate because its knockout in mice leads to early embryonic death just before the onset of organogenesis (29, 30) and growth arrest of mouse fibroblasts (29, 31). JMJD5 is highly expressed in breast cancer cells and knockdown of JMJD5 leads to growth arrest of such cells (32). A role for JMJD5 in cell cycle regulation is also suggested by the fact that G1 is prolonged in human embryonic stem cells upon deletion of JMJD5 (29, 31).
In studies of JMJD5, we recently found that it has both endopeptidase and exopeptidase activities, with specificity for peptides containing methylated arginines in the amino terminal tails of histones H2A, H3, and H4. In line with this observation, we found that H3, H4, and their arginine methylated isoforms are greatly increased in cell lines with deficiency of JMJD5 in vivo (31). Binding studies showed that JMJD5 specifically recognizes peptides containing methylated arginine H3R2(me2s) with high affinity (∼110 nM) and has lower affinity for the nonmethylated form of, or the lysine-methylated form of, the same H3 peptide (∼7 μM or ∼4 μM, respectively) (31, 33). Others have shown that artificial tailless or mock acetylated nucleosomes are less of a mechanical barrier to Pol II (34). Thus, based on these discoveries, we hypothesized that JMJD5 could cleave the arginine methylated histone tails of nucleosomes at transcription start sites (TSSs), particularly at the +1 position (31, 33, 35), thereby reducing the barrier for Pol II progression past the +1 nucleosome (36, 37).
In support of the hypothesis, here we report that the promoters of many genes become less open in the absence of JMJD5. We show that the N-terminal domain of JMJD5 binds with highest affinity to the CTD of Pol II in which neighboring Ser2 residues and a Ser5 residue in the CTD have been phosphorylated and that this phosphorylation is catalyzed by CDK9. We therefore suggest that engagement of JMJD5 to the Pol II complex via its binding to the CTD may bring JMJD5 close to the +1 nucleosome, a location that would facilitate JMJD5 cleavage of the nucleosome’s histone tails, creation of relative tailless nucleosomes, and alleviation of the blockage to Pol II’s transcriptional progression.
Results
JMJD5 KO Impairs Promoter Accessibility Globally.
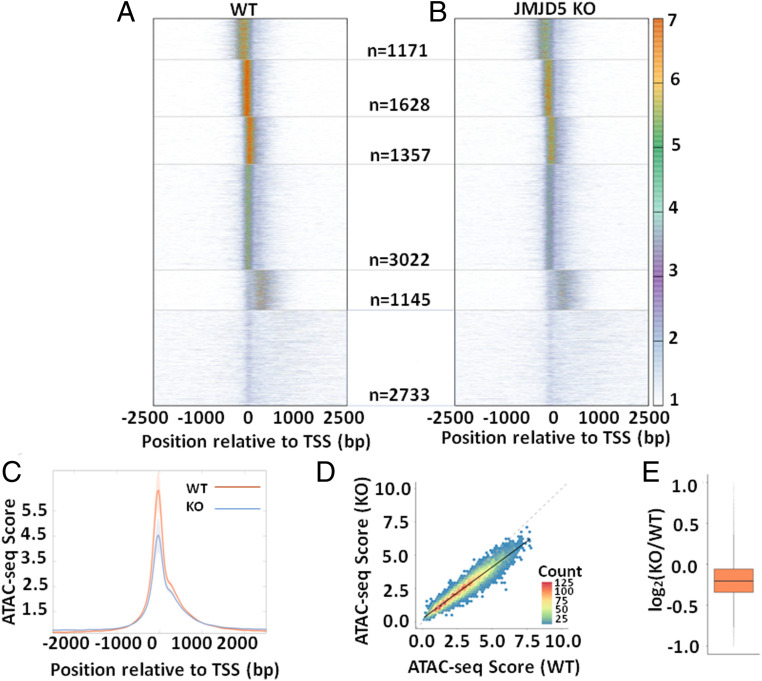
To ask if JMJD5 influenced the promoter accessibility of expressed genes, we performed ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) on mouse wildtype (WT) and JMJD5 knockout (KO) male mouse embryonic fibroblasts (male MEF). ATAC-seq measures chromatin accessibility based on the extent of transposition after treating nuclei with Tn5 transposase. Tn5 acts preferentially at regions of open chromatin like promoters and enhancers as they present naked DNA for transposition at a higher rate compared to the rest of the genome. The ATAC-seq for the wildtype and JMJD5 KO was analyzed as previously described (38). Briefly, the density of ATAC-seq reads over a given position was divided by the mean number of ATAC-seq reads over TSS ± 2,000 bp to yield an enrichment over mean signal at each position of the TSS ± 2,000 bp region. This normalization enabled comparison of the “openness” of promoters of different genes. We then clustered ATAC-seq enrichment patterns around TSS using k-means method in R (https://www.R-project.org) into six clusters and observed clusters with high ATAC-seq signal at the TSS and just upstream and downstream of the TSS in WT cells (Fig. 1A). When we plotted ATAC-seq enrichment for KO cells with the same order of genes as WT, we observed similar patterns of ATAC-seq signals in each cluster. However, we observed a significantly decreased enrichment of such signals in KO cells compared to WT cells (Fig. 1B). This is evident from the global weakened accessibility at TSS in JMJD5 KO cells by comparison with WT when the ATAC-seq enrichment averaged over all expressed genes relative to the TSS was plotted (Fig. 1C). Next, to determine the extent of loss of accessibility in JMJD5 KO at an individual gene level, we calculated the average ATAC-seq enrichment in the interval TSS ± 150 bp for each gene. We then plotted these scores from WT against similar scores from JMJD5 KO. We found significant correlation in the scores (r2 = 0.91). However, the slope was 0.77 (95% confidence interval: 0.766 to 0.775), indicating a significant decrease in accessibility globally in JMJD5 KO (Fig. 1D), with a median Log2 fold change of −0.2 (13% reduction). In summary, absence of JMJD5 results in a global loss of accessibility at promoters, indicating that JMJD5 might function in maintaining accessibility at expressed promoters genome-wide.
Fig. 1.
Loss of promoter accessibility in JMJD5 KO cells. (A) ATAC-seq enrichment of WT cells relative to TSS is plotted as a heatmap. Genes (n = 11,056) are ordered based on clusters generated using k-means method. (B) ATAC-seq enrichment of JMJD5 KO cells is plotted as a heatmap. The order of genes is the same as WT. The number of genes in each cluster is indicated in the middle between the two heatmaps. (C) Average of ATAC-seq enrichment relative to TSS for all expressed genes (n = 11,056) is plotted for WT and JMJD5 KO cells. SE of mean calculated from four replicates of the ATAC-seq data are plotted as the shaded region around each line. The difference in accessibility at the promoter is significant (paired t test P value = 0.025). (D) ATAC-seq enrichment score in the interval TSS ± 150 bp was calculated for each gene for WT and JMJD5 KO cells and then plotted as a two-dimensional (2D) histogram with hexagonal binning. The line of best fit (solid black line) has a slope (0.77) significantly lower than the x = y line (gray dashed line), indicating a global decrease in accessibility at promoters upon JMJD5 KO. (E) The ratio of promoter enrichment scores of JMJD5 KO to WT are plotted as a boxplot.
A drop in accessibility at promoter regions could cause lowered overall transcription of the relevant gene body. To find out if this was true, we combined four ATAC-seq data sets and performed ATAC-seq analyses alongside the RNA-seq data (transcription activities). We first found drastic changes of accessibility in a group of genes were coupled with a drastic drop of messenger RNA (mRNA). One example that stands out is the locus around Kdm5d with JMJD5 knockout on Y chromosome and some other genes (SI Appendix, Figs. S1–S3). This result is consistent with the early role of JMJD5, knockout of which leads to the severe lethal phenotype of embryo at E8 in mice (30).
JMJD5 Is Recruited by a Combinational Form of Ser2 Phosphorylated Pol II.
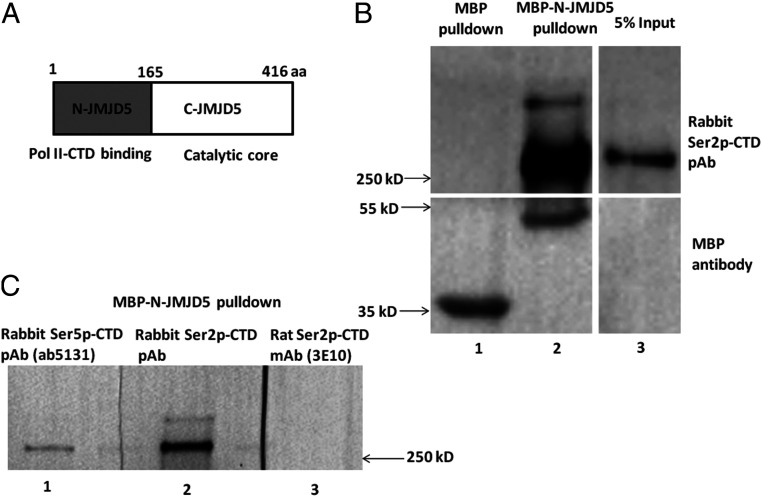
The data shown above suggest that JMJD5 affects progression of Pol II past the +1 nucleosome and it also leads to some questions. How is JMJD5 recruited to the transcription machine complex and how is this event coupled to release of the paused Pol II? Besides the catalytic C-terminal domain, JMJD5 contains a structurally and functionally unknown N-terminal domain (N-JMJD5) between residues 1 to 165 (Fig. 2A). Does N-JMJD5 make contacts with the transcription complex, thus promoting recruitment of JMJD5 to the Pol II complex? To answer this question, we expressed, in Escherichia coli, N-JMJD5 fused to maltose binding protein (MBP), purified the protein, and incubated it with nuclear extracts from HEK293 cells. MBP alone was used as control. After pulldown of MBP, the MBP or MBP-N-JMJD5 and their associated components were subjected to blotting analysis. The membrane was blotted with a polyclonal rabbit antibody against Ser2p-CTD with double Ser2p heptad repeats (39) or a mouse monoclonal anti-MBP antibody. Pull downs of MBP alone revealed MBP but no bands staining with antibody to Ser2p-CTD (Fig. 2B, lane 1). Pulldown of MBP-N-JMJD5 revealed a 55 kD band stained with anti-MBP, the molecular weight of MBP-N-JMJD5. This pull down also revealed a band of Pol II molecular weight (∼250 kD) stained with the antibody against Ser2p-CTD with double Ser2p heptad repeats (39) (a gift from Dr. David Bentley’s group) (Fig. 2B and SI Appendix, Fig. S3), the size of which is well consistent with the pol II in cell lysate as shown in the 5% input lane. Interestingly, a polyclonal antibody (ab5131, Abcam) against Ser5p-CTD also leads to the detection of a similar band (Lane 1, Fig. 2C). However, the pulldown component was barely recognized by a monoclonal antibody against Ser2p-CTD with a single Ser2p heptad. This antibody is widely used to detect the active elongating Pol II (3E10, ref. 40) (Lane 3, Fig. 2C and SI Appendix, Fig. 3). Hence, it is possible that the MBP-N-JMJD5 pulldown product contains both a site for Ser5p-CTD antibody recognition and a site for the double Ser2p-CTD antibody. Of note, a unique phosphorylation composition on CTD heptad repeats of this type has not been previously described.
Fig. 2.
N-JMJD5 pulls down a combinational specie of Ser2p-CTD of Pol II. (A) Domain structures of JMJD5, N-terminal Pol II binding and C-terminal catalytic core. (B) Upon incubation with human cell extract, MBP-N-JMJD5 pulls down a species of Ser2p-CTD of Pol II, recognized by a rabbit polyclonal antibody raised with Ser2p within each heptad repeat CTD of Pol II (Lane 2, Top). (C) The Ser2-CTD of Pol II pulled down by MBP-N-JMJD5 could be recognized by a rabbit Ser5p-CTD of Pol II polyclonal antibody (Lane 1) but not by a rat monoclonal antibody (3E10) raised by a single Ser2p-CTD peptide (lane 3).
N-JMJD5 Specifically Recognizes Dual Phosphorylated Ser2-CTD of Pol II.
No structural or functional studies of N-JMJD5 have been published. From secondary structural prediction, using Jpred (41) as well as three-dimensional (3D) prediction using Raptor X (42), we propose a tentative model for N-JMJD5 that contains seven alpha-helices and potentially has an overall structure that is similar to those of the Pol II CTD binding domains (CIDs) from proteins such as NRD1, PCF11, Ritt103, SCAF8, and RPRD1A/B. (SI Appendix, Fig. S4) (43–47). This similarity suggests that N-JMJD5 might bind to some forms of the CTD of Pol II.
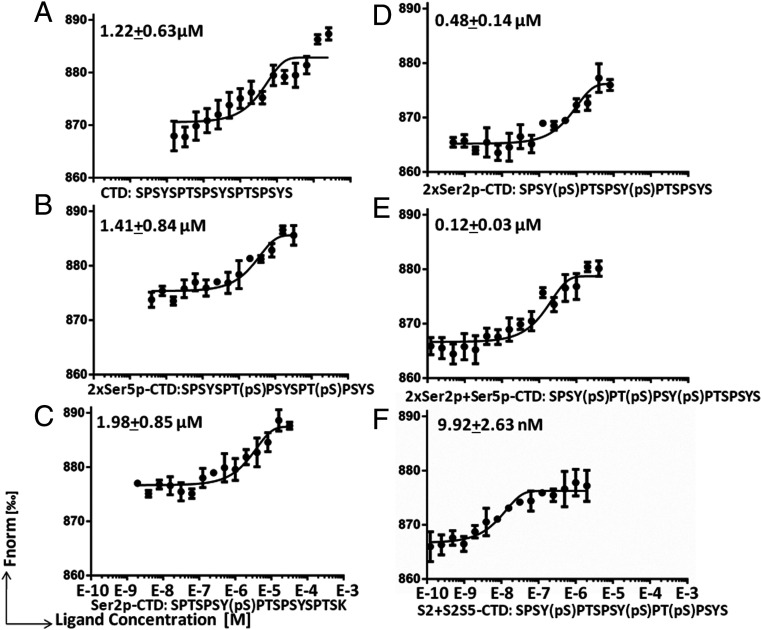
There are 52 heptad repeats (-YSPTSPS-) within the human or mouse Pol II CTD and countless patterns of posttranslational modifications of this region (25), so it is not feasible to screen all of the potential forms of the Pol II CTD. A report from Dirk Eick and his colleagues showed a drastic divergence in the patterns of Ser2p modifications of the Pol II-CTD of human versus that of yeast. For example, frequent double Ser2p signals from two neighbored heptad repeats occurred in human but not in yeast Pol II (22). Since Pol II pausing is absent in yeast, we hypothesized that the dual Ser2p-CTD of Pol II could be the target of N-JMJD5. To test this hypothesis, several forms of CTD heptad repeats were synthesized: native CTD peptide (-YSPTSPSYSPTSPS -); single Ser2p (-YSPTSPSYS(p)PTSPS-); dual Ser2p (-YS(p)PTSPSYS(p)PTSPS-); dual Ser5p (-YSPTS(p)PSYSPTS(p)PS-); dual Ser2p with a Ser5p in first repeat (-YS(p)PTS(p)PSYS(p)PTSPS-); and dual Ser2p with a Ser5p in the second repeat (-YS(p)PTSPSYS(p)PTS(p)PS-), HR-Ser2p (1, 2)-Ser5p (2) for short. These peptides were each subjected to binding analysis by MBP-N-JMJD5 using Microscale Thermophoresis (MST) (48, 49). MBP-N-JMJD5 showed some affinity for the native CTD peptide (∼1.22 μM) or the dual Ser5p-CTD peptide (∼1.41 μM) (Fig. 3 A and B) and had a similar binding affinity for the single Ser2p-CTD peptide (∼1.98 μM) (Fig. 3C). However, MBP-N-JMJD5 bound more strongly to the dual Ser2p CTD peptide (∼0.48 μM) (Fig. 3D) and had an even higher affinity for the dual Ser2p peptide containing a Ser5p in the first heptad repeat (∼0.12 μM) (Fig. 3E). The highest affinity binding was seen for MBP-N-JMJD5 binding to the dual Serp2 CTD in which the Ser5p was located in the second heptad repeat (∼9 nM) (Fig. 3F). This is ∼100 times stronger affinity than those of MBP-N-JMJD5 for native, Ser5p, or single Ser2p repeats of CTD and the highest binding affinity among CTD of Pol II and binding partners ever reported (43–47). This suggests that JMJD5 preferentially recognizes dual Ser2 phosphorylated CTD of Pol II with a Ser5p in vivo.
Fig. 3.
N-JMJD5 binds to dual Ser2p-CTD of Pol II with high affinity. (A) MicroScale Thermophoresis measurement of the binding of MBP-N-JMJD5 to native CTD peptide. (B) The binding of MBP-N-JMJD5 to dual Ser5p-CTD peptide. (C) The binding of MBP-N-JMJD5 to single Ser2p-CTD peptide. (D) The binding of MBP-N-JMJD5 to dual Ser2p-CTD peptide. (E) The binding of MBP-N-JMJD5 to dual Ser2p-CTD peptide with Ser5p within the first repeat. (F) The binding of MBP-N-JMJD5 to dual Ser2p-CTD peptide with Ser5p within the second repeat.
We did a control for the accuracy of our measurements by assessing, with the same methods, the affinity of RPRD1A for dual Serp2 CTD (SI Appendix, Fig. S5). We found the affinity is ∼7.2 μM, a value that is very close to the ∼8 μM affinity for this reaction measured previously by others (47).
The fact that N-JMJD5 binds to dual Ser2p plus Ser5p with the highest affinity of all of the peptides we tested may explain the results shown Fig. 2C in which Pol II CTD pulldown by JMJD5 was recognized by a rabbit polyclonal anti-CTD peptide with Ser2p in the neighboring heptad repeat (39) but not by a monoclonal antibody against a CTD peptide with a single Ser2p (40). Furthermore, the fact that the addition of Ser5p in the Ser2p repeat drastically increases the binding affinity is consistent with the positive staining of the pulldown of MBP-N-JMJD5 by the polyclonal Ser5p antibody (Lane 1, Fig. 2C).
CDK9 Phosphorylates the Ser2 of Pol II.
The results derived so far suggest that JMJD5 is recruited to the Pol II complex by reaction with the dual Ser2p plus Ser5p of the CTD of Pol II. This raises the issue of which kinase is responsible for generating the phosphorylated CTD target. Many reports by others suggest that CDK9 is the most likely enzyme to perform this function (16–23). However, there is some controversy over this idea (27). Firstly, CDK9 was reported to phosphorylate Ser5 instead of Ser2 (28), confusing the issue. Secondly, CDK9 has been suggested to be the homolog of BUR1 in yeast, an enzyme that is thought to play only a minor role in Pol II CTD Ser2 phosphorylation but instead phosphorylates the yeast Pol II rpb1 linker (50, 51). Thirdly, experiments have indicated that, in yeast, CTK1, a homolog of CDK12 and CDK13 in higher eukaryotes, produces Ser2p-CTD of Pol II (52). Fourthly, chromatin immunoprecipitation DNA sequencing (ChIP-seq) data indicate that the Ser2p-CTD of Pol II is found at high levels at the transcription end sites (TESs) of genes but is barely detected at TSSs either in yeast (53–56) or mammals (57, 58), suggesting loose connection to Pol II pausing regulation at promoter regions. Finally, Ser2p-CTD of modifications of Pol II exist both in yeast and higher eukaryotes but Pol II pausing exists only in higher eukaryotes (53, 55, 59). Because of this continuing controversy, we thought it was worthwhile to retest the idea that CDK9 is one of the major kinases that phosphorylates Pol II CTD Ser2 in mammals.
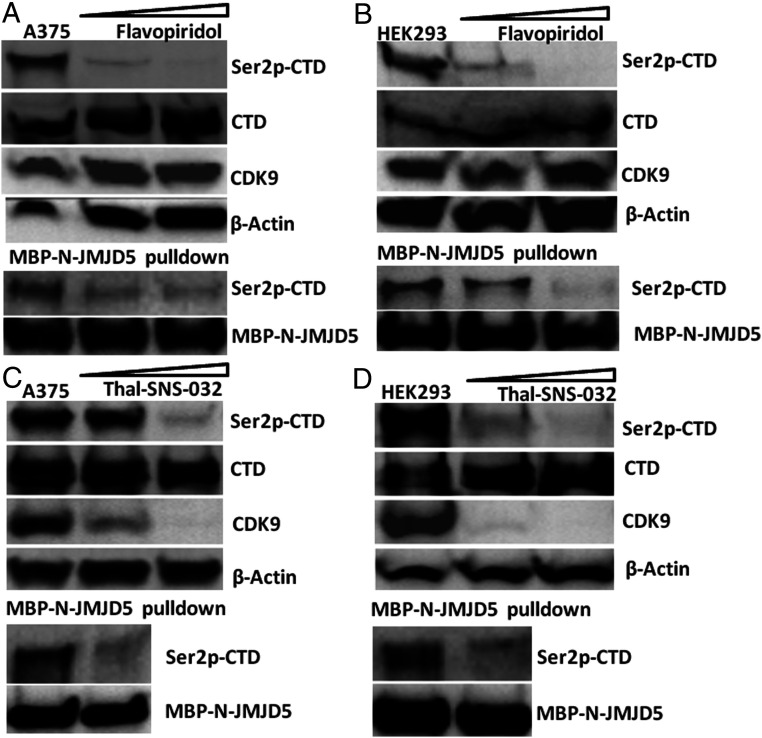
To address this question, the most straightforward experiment would be to examine changes in the level of dual Ser2p-CTD of Pol II in cells that do versus do not express CDK9. However, examination using cells which contain a complete knockout of CDK9 through CRISPR/CAS9 is not possible in our hands, cells without CDK9 are difficult to survive. Therefore, instead we treated human cells with an inhibitor of CDK9, flavopiridol, and evaluated changes in dual Ser2p-CTD of Pol II, which is recognized by JMJD5. The amount of dual Ser2p-CTD of Pol II detected (Fig. 4 A, Top) or pulled down (Fig. 4 A, Lower) by JMJD5 in the flavopiridol-treated cells was drastically reduced even though overall Pol II and CDK9 levels were unaffected by the inhibitor. This was true in two cell lines, A375 and 293. (Fig. 4B). In a different approach to the same subject, we treated A375 cells with THAL-SNS-032, a CDK9-binding inhibitor SNS-032 conjugated to a thalidomide derivative for E3 ubiquitin ligase (24). This drug binds CDK9 and causes degradation of the protein. Application of this ligand for 4 h to A375 cells led to a drastic drop of CDK9 and a concurrent drop of Ser2p-Ser5p-CTD of Pol II (Fig. 4 C, Top) as well as that pulled down by MBP-N-JMJD5 (Fig. 4 C, Lower). Again, the results could be repeated in HEK293 cells (Fig. 4D).
Fig. 4.
The dual Ser2p-CTD of Pol II is generated by CDK9. (A) A375 cells treated with Flavopiridol at 0.0 µM, 0.3 µM, and 1 µM for 24 h, (Top) shows cell extract blot. (Lower) Ser2p-CTD blot of MBP-N-JMJD5 pulldown from the above three groups. (B) HEK293 cells treated with Flavopiridol at 0.0 µM, 0.1 µM, and 0.3 µM for 24 h. (Top) shows cell extract blot. (Lower) Ser2p-CTD blot of MBP-N-JMJD5 pulldown from the above three groups. (C) A375 cells treated with Thal-SNS-032 at 0 nM, 100 nM, and 300 nM, respectively, for 4 h. (Top) shows cell extract blot. (Lower) Ser2p-CTD blot of MBP-N-JMJD5 pulldown from the 0 nM and 300 nM Thal-SNS-032 group. (D) HEK293 cells treated with Thal-SNS-032 at 0 nM, 30 nM, and 100 nM, respectively, for 4 h. (Top) shows cell extract blot. (Lower) Ser2p-CTD blot of MBP-N-JMJD5 pulldown from the 0 nM and 100 nM Thal-SNS-032 group.
These data suggest that, in mammals, CDK9 phosphorylates Ser2 of the CTD of Pol II and produces dual Ser2p of the CTD, which may be different from the Ser2p-CTD generated by CDK12/13 (or CTK1 in yeast), thereby generating a docking site for JMJD5 on Pol II and consequent release of paused Pol II. However, our data do not exclude the possibility that CDK9 might phosphorylate other components of the Pol II machinery, such as NELF and DSIF, which may also play a role in release of paused Pol II.
Discussion
Pol II pausing at promoter regions is a common mechanism of regulation in metazoans (3, 4, 60, 61). Such pausing does not happen in yeast or bacteria. In such organisms, Pol II activity is instead mostly regulated through recruitment of the polymerase to the promoter (62, 63). It is also well known that nucleosomes at +1 play dominant roles in the pause and release of Pol II (36, 37, 64, 65). However, the precise way in which Pol II pausing is relieved is not completely understood, although there may be a role for DSIF and NELF (66, 67). In the experiments here, we investigated a possible role for another mechanism, involving the proteolytic enzyme, JMJD5.
Our early discoveries showed that JMJD5 specifically recognizes argininemethylated histone tails and from these can generate, via its endopeptidase and exopeptidase activities, relative tailless nucleosomes. Since tailless nucleosomes are thought to present less of barrier to the progression of RNA polymerases through the gene body (34, 68), we proposed that JMJD5 may work at the +1 nucleosome to promote Pol II elongation of RNA (31, 33, 35). We therefore searched for a mechanism that would promote JMJD5 recruitment to paused Pol II, thus giving JMJD5 access to the nucleosome at +1 and reducing the nucleosomal barrier to Pol II.
To test whether JMJD5 might play a role in release of Pol II pausing, we first analyzed the consequences for cells absent of JMJD5. We found that nucleosomes accumulated at the +1 position in cells lacking JMJD5 and that this was coupled with down-regulation of transcription of a large number of genes (Fig. 1) including, interestingly, all encoded by the Y chromosome, a phenomenon that we will follow up in future studies (SI Appendix, Figs. S1–S3). Searching for a mechanism that might concentrate JMJD5 on the +1 nucleosome, we focused on the previously uncharacterized N-terminal domain of JMJD5 (N-JMJD5). Sequence comparisons predicted that this domain might be structurally similar to domains in other proteins that bind the CTD of Pol II (43–47). To follow up this idea, we performed coprecipitation studies with N-JMJD5. These revealed that the N-terminal domain of JMJD5 pulled down a particular species of Pol II, a species that was recognized by a rabbit polyclonal antibody raised against a dual Ser2p CTD of Pol II, but not recognized by a monoclonal antibody specific for CTD peptide that contained only one Ser2p. We used affinity measurements to show that N-JMJD5 bound with highest affinity to CTD with dual Ser2p within neighboring heptad repeats plus an additional phosphorylation at Ser5-CTD within the second heptad repeat. Furthermore, we found that the dual Ser2p species of Pol II is specifically generated by CDK9 since inhibition or depletion of CDK9 in A375 or HEK293 cells resulted in a drastic loss of the dual Ser2p-CTD derivative of Pol II and lack of engagement of N-JMJD5. These data suggest not only that CDK9 works directly upstream of JMJD5, but also demonstrate that CDK9 is responsible for the generation of this unique Ser2p-CTD of Pol II. As for the additional modification, Ser5p, of Pol II’s CTD that increases further its affinity for N-JMJD5, we hypothesize that this modification is performed by another, quite likely CDK7 or CDK8, kinase and generated at initiation before action of CDK9. This speculation needs further investigation.
As demonstrated recently by us, the release of CDK9 from 7SK snRNP is controlled by JMJD6, which is unique in higher eukaryotes (69). Interestingly, along this line, the regulation of Pol II pausing, the 7SK small nuclear ribonucleoprotein (snRNP) complex (which includes CDK9), and JMJD5 only exist in higher eukaryotes, suggesting the possibility that these three have a related function, in agreement with the results discussed above. However, as mentioned in the Results section of this manuscript, there is preexisting evidence that CDK9 does not phosphorylate Pol II’s CTD. For example, CDK9 is thought to be the analog of yeast Bur1, an enzyme that, in yeast phosphorylates the linker region of Rbp of Pol II, a modification that recruits Spt6 (50). We speculate, therefore, that some other kinase is the analog in higher eukaryotes, of Bur1. A question raised here is how to discriminate Ser2p generated by CDK9 from Ser2p generated by CDK12/13, CTK1 in yeast, which are required to modify Pol IIs CTD to stimulate functions such as transcription termination, polyadenylation, and recruitment of spliceosomes (70). We speculate that there are two forms of Ser2p coexisting in higher eukaryotes; this idea is consistent with the report showing that the patterns of Ser2p-CTD Pol II are different between humans and yeast (22).
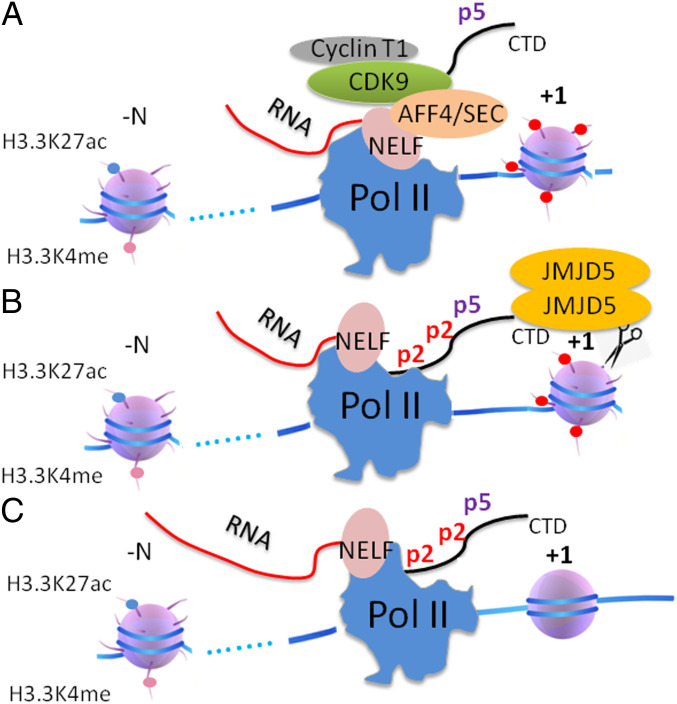
Finally, the above data and data published earlier by our group (31, 33) suggest that the phosphorylation of Ser2-CTD of Pol II by CDK9, Pol II pausing regulation, the high turnover rate of histone in nonproliferating cells, and the generation of tailless nucleosomes by JMJD5, are closely coupled. Based on these accumulating results, a simple transcription elongation model for paused Pol II could be derived (Fig. 5). Upon stimulating signals, with the help of BRD4 and JMJD6 (35, 69), CDK9 is recruited to paused Pol II coupled with super elongation complex and phosphorylates neighboring Ser2-CTD of Pol II. This, together with Ser5p-CTD of Pol II generated at initiation, creates a high affinity ligand for JMJD5. Once the complex of Pol II and JMJD5 is formed, JMJD5 is now ideally placed to cleave the histone tails of the barrier nucleosomes at +1 of genes, thus generating relatively tailless nucleosomes and promoting progression of Pol II down the gene body. However, questions remain. For example, how does Pol II overcome the barriers of other nucleosomes further down the gene body? How is Pol II transcription of housekeeping genes in higher eukaryotes or all genes in yeast controlled? These are puzzles that remain to be addressed.
Fig. 5.
The transcription elongation model of paused Pol II in higher eukaryotes. (A) The phosphorylation of CTD of Pol II on Top of Ser5p-CTD by CDK9 with the help of super elongation complex. (B) Recruitment of JMJD5 by Ser2 phosphorylated CTD. (C) JMJD5 generates tailless nucleosome by cleaving arginine methylated histone tails.
Materials and Methods
Antibodies.
A full list of antibodies can be found in the SI Appendix, Table S1.
Cells.
The JMJD5 knockout cell line was generated by Dr. Ralf Janknecht’s laboratory. The representative image of wildtype and JMJD5 knockout MEF cell can be found in SI Appendix, Fig. S6.
ATAC-Seq.
ATAC-seq was performed according to Buenrostro et al., (38) with 50,000 cultured WT MEFs and JMJD5 KO MEF cells. Cells were lysed to extract nuclei. Nuclei were resuspended in 50 μL 1 × TD (Tagment DNA) buffer containing 2.5 μL transposase (Nextera, Illumina). The transposase reaction was conducted for 40 min at 37 °C. Library amplification and barcoding were performed using Illumina-compatible index primers purchased from IDTdna. PCR was conducted for 12 to 15 cycles. Library purification was performed with the MinElute PCR Purification Kit (Qiagen) and size selection was using AMPureXP beads (Beckman Coulter). Libraries were quantified and size distribution was assessed using the Bioanalyzer High Sensitivity DNA Kit (Agilent). Paired-end sequencing was performed on a HiSEq 2500 (Illumina) with 50 cycles for each read. Raw data from the sequencer was demultiplexed and FASTQ files were generated using bcl2fastq Conversion Software (Illumina). Paired-end reads were aligned to mm10 version of the mouse genome using bowtie2 with following parameters:–local–very-sensitive-local–no-unal–no-mixed–no-discordant -I 10 -X 700. The fraction of reads mapped at each nucleotide was multiplied by the total number of nucleotides mapped genome-wide, which was arbitrarily chosen as approximate size of the genome (2.8 × 109), to give a normalized count at that position. The reads were then aggregated to 25 base pair windows, and only paired-end reads of length 40 to 120 bp were used in the analysis as they usually represent areas of transcription factor binding.
RNA-Seq.
Total RNA from MEF and JMJD5 knockout cell was extracted with a TRIzol kit (ThermoFisher Scientific). It was then sent to Quick Biology (Quick Biology, Pasadena) for further mRNA purification and analysis. Mouse genome mm10 was used as reference.
MBP-N-JMJD5 and Ser2p-CTD of Pol II Pulldown.
N-terminal human JMJD5 (17-148) was inputted after MBP in pMAL vector. The fused protein was induced for expression at 18 °C overnight after the optical density (O.D.) of the transformed bacteria reached 0.8. The fused protein was purified using Amylose beads and further purified with size exclusion column. Approximately 2 million human embryonic kidney cell line 293 cells were lysed with Pierce immunoprecipitation (IP) lysis buffer. For the pulldown assay, equal molar amounts of MBP protein and MBP-N-JMJD5 were incubated with the same amount of cell lysate overnight at 4 °C in Hepes buffer (10 mM Hepes, pH 7.5, 150 mM NaCl) with 5% glycerol. Amylose beads were added and incubated for another hour. The beads were then spun down and washed extensively with Hepes buffer. The bound protein was eluted with Hepes buffer with 10 mM maltose and used for blotting with phosphorylated-serine2 –Pol II CTD antibody provided by Dr. Bentley and MBP antibody produced in our laboratory (71).
MST MBP-N-JMJD5 Binding Assays.
MBP- N-JMJD5 (17-148) was cloned into pET-15b vector in order to add a 6*Histidine tag at the N-terminal of MBP protein. The His-MBP-N-JMJD5 protein was purified in the same way as MBP-N-JMJD5. The 6*Histidine tag was labeled with a His-tag labeling kit from NanoTemper technology (NanoTemper, South San Francisco). Labeled His-MBP-N-JMJD5 (10 nM) was incubated with serial dilution of synthesized Pol II CTD peptides with different modifications. The affinity was measured with Monolith NT.115 pico from NanoTemper technology. For each peptide, three repeats of measurement were done to obtain SD for each data point. The binding curve and affinity were modeled and analyzed in the MO. (Monolith) Control software. The original data were exported and the binding curve was drawn with GraphPad Prism software.
CDK9 Inhibition and Depletion.
CDK9-specific cyclin-dependent kinase inhibitor SNS-032 with conjugated cereblon E3 ligase ligand, thalidomide (Thal-SNS-032), was purchased from R&D systems. HEK293 cell or A375 cell were treated with different concentrations of Thal-SNS-032 (30 nM or 100 nM for HEK293 cell, 100 nM or 300 nM for A375 cell) for 4 h. Cells were then lysed and an equal amount of cell lysate was used for MBP-N-JMJD5 pulldown. The primary antibody for CDK9 detection was from Santa Cruz Biotechnology (sc-13130). Flavopiridol treatment was carried out in the same way except that the drug concentration and incubation time are different. HEK293 cell was incubated with 0.1 µM or 0.3 µM flavopiridol overnight. A375 cell was incubated with 0.3 µM or 1 µM flavopiridol overnight.
Supplementary Material
Acknowledgments
We thank James Crapo and other researchers at National Jewish Health (NJH) for their kind support and the David Bentley laboratory for Pol II ChIP-Seq, related antibodies, and R35GM118051 (to David Bentley). We thank Jack Greenblatt for complementary DNA (cDNA) of RPRD1A/B; Xin Bi for yeast genomic DNA; and Joshua Loomis for help with cell imaging. The binding data were obtained from the structural biology core facility at Anschutz Medical Center, University of Colorado Denver. We also thank Dr. Yongmei Jiang, Shi-Ning Xu, Cheng-Yuan Zhang, Dr. Peng Sun, and Yun-Xia Jiang for private funding. H.L. is supported by New Buddies (NB) Life Laboratory LLC, and S.L. is supported by NIH Training Grant T32AI007405-28 (to P.M.). This work is supported by NIH grant R01GM135421 (to G.Z.).
Footnotes
Competing interest statement: During the middle of the project, H.L. became an employee of NB Life Laboratory, LLC. P.P. was a scientist from Nano Temper Technologies, Inc. G.Z. holds equity in NB Life Laboratory LLC.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005745117/-/DCSupplemental.
Data Availability.
All data sets of RNA-seq and ATAC-seq have been deposited to Gene Expression Omnibus with the accession number GSE153322.
References
- 1.Rougvie A. E., Lis J. T., The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54, 795–804 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Nechaev S. et al., Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327, 335–338 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adelman K., Lis J. T., Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonkers I., Lis J. T., Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeitlinger J. et al., RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39, 1512–1516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley D. L., Groudine M., A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature 321, 702–706 (1986). [DOI] [PubMed] [Google Scholar]
- 7.Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M., Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330, 489–493 (1987). [DOI] [PubMed] [Google Scholar]
- 8.Marshall N. F., Price D. H., Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270, 12335–12338 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Lis J. T., Mason P., Peng J., Price D. H., Werner J., P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14, 792–803 (2000). [PMC free article] [PubMed] [Google Scholar]
- 10.Peterlin B. M., Price D. H., Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Kwak H., Lis J. T., Control of transcriptional elongation. Annu. Rev. Genet. 47, 483–508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi Y., Shibata H., Handa H., Transcription elongation factors DSIF and NELF: Promoter-proximal pausing and beyond. Biochim. Biophys. Acta 1829, 98–104 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Guo J., Price D. H., RNA polymerase II transcription elongation control. Chem. Rev. 113, 8583–8603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J. B., Sharp P. A., Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276, 12317–12323 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Fujinaga K. et al., Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 24, 787–795 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz J. C. et al., FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 26, 2690–2695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon I. et al., Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou M. et al., Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20, 5077–5086 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall N. F., Peng J., Xie Z., Price D. H., Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271, 27176–27183 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Hsin J. P., Manley J. L., The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26, 2119–2137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni Z., Schwartz B. E., Werner J., Suarez J. R., Lis J. T., Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13, 55–65 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Schüller R. et al., Heptad-specific phosphorylation of RNA polymerase II CTD. Mol. Cell 61, 305–314 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Chen F. X., Smith E. R., Shilatifard A., Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 19, 464–478 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Olson C. M. et al., Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat. Chem. Biol. 14, 163–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eick D., Geyer M., The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 113, 8456–8490 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Egloff S., Dienstbier M., Murphy S., Updating the RNA polymerase CTD code: Adding gene-specific layers. Trends Genet. 28, 333–341 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Paparidis N. F., Durvale M. C., Canduri F., The emerging picture of CDK9/P-TEFb: More than 20 years of advances since PITALRE. Mol. Biosyst. 13, 246–276 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Czudnochowski N., Bösken C. A., Geyer M., Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat. Commun. 3, 842 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Ishimura A. et al., Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression. Development 139, 749–759 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Oh S., Janknecht R., Histone demethylase JMJD5 is essential for embryonic development. Biochem. Biophys. Res. Commun. 420, 61–65 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Liu H. et al., Clipping of arginine-methylated histone tails by JMJD5 and JMJD7. Proc. Natl. Acad. Sci. U.S.A. 114, E7717–E7726 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsia D. A. et al., KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 107, 9671–9676 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H. et al., Specific recognition of arginine methylated histone tails by JMJD5 and JMJD7. Sci. Rep. 8, 3275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bintu L. et al., Nucleosomal elements that control the topography of the barrier to transcription. Cell 151, 738–749 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H., Lee S., Zhang Q., Chen Z., Zhang G., The potential underlying mechanism of the leukemia caused by MLL-fusion and potential treatments. Mol. Carcinog. 59, 839–851 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Weber C. M., Ramachandran S., Henikoff S., Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell 53, 819–830 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Voong LN et al., Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell 167, 1555–1570.e15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glover-Cutter K., Kim S., Espinosa J., Bentley D. L., RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 15, 71–78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapman R. D. et al., Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318, 1780–1782 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Drozdetskiy A., Cole C., Procter J., Barton G. J., JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Källberg M., Margaryan G., Wang S., Ma J., Xu J., RaptorX server: A resource for template-based protein structure modeling. Methods Mol. Biol. 1137, 17–27 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Meinhart A., Cramer P., Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430, 223–226 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Noble C. G. et al., Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat. Struct. Mol. Biol. 12, 144–151 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Lunde B. M. et al., Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 17, 1195–1201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker R., Loll B., Meinhart A., Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 283, 22659–22669 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Ni Z. et al., RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat. Struct. Mol. Biol. 21, 686–695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duhr S., Braun D., Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. U.S.A. 103, 19678–19682 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seidel S. A. et al., Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–315 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chun Y. et al., Selective kinase inhibition shows that Bur1 (Cdk9) phosphorylates the Rpb1 linker in vivo. Mol. Cell. Biol. 39, e00602-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keogh M. C., Podolny V., Buratowski S., Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 23, 7005–7018 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartkowiak B. et al., CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 24, 2303–2316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim H. et al., Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 17, 1279–1286 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bataille A. R. et al., A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol. Cell 45, 158–170 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Mayer A. et al., Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17, 1272–1278 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Tietjen J. R. et al., Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 17, 1154–1161 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koch F. et al., Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 18, 956–963 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Brookes E. et al., Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10, 157–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhee H. S., Pugh B. F., Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Q., Li T., Price D. H., RNA polymerase II elongation control. Annu. Rev. Biochem. 81, 119–143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shao W., Zeitlinger J., Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 49, 1045–1051 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Ptashne M., Gann A., Transcriptional activation by recruitment. Nature 386, 569–577 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Zhang G. et al., Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98, 811–824 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Bondarenko V. A. et al., Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24, 469–479 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Mavrich T. N. et al., Nucleosome organization in the Drosophila genome. Nature 453, 358–362 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vos S. M. et al., Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560, 607–612 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Vos S. M., Farnung L., Urlaub H., Cramer P., Structure of paused transcription complex Pol II-DSIF-NELF. Nature 560, 601–606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koster M. J., Snel B., Timmers H. T., Genesis of chromatin and transcription dynamics in the origin of species. Cell 161, 724–736 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Lee S. et al., JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes. eLife 9, e53930 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsin J. P., Xiang K., Manley J. L., Function and control of RNA polymerase II C-terminal domain phosphorylation in vertebrate transcription and RNA processing. Mol. Cell. Biol. 34, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu H. et al., A rapid method to characterize mouse IgG antibodies and isolate native antigen binding IgG B cell hybridomas. PLoS One 10, e0136613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data sets of RNA-seq and ATAC-seq have been deposited to Gene Expression Omnibus with the accession number GSE153322.