Significance
Posttranslational modifications (PTMs) play an important role in modulating α-syn structure, function, and pathology. Phosphorylation commonly applies to pathological α-syn aggregation in Lewy bodies and is a pathological hallmark of Parkinson’s disease (PD). Here, we obtain homogeneous α-syn fibrils formed by semisynthesized α-syn with site-specific phosphorylation at Y39. The synthetic fibril recapitulates the neuropathology and degradation resistance of endogenous pY39 α-syn aggregation in PD. Cryo-EM structure determination of the pY39 α-syn fibril reveals a fold of α-syn with the entire N terminus involved in the fibril core attracted by pY39. This structure illuminates the mechanism of pY39 pathology and highlights the importance of PTMs in defining the structure and pathology of amyloid aggregation in neurodegenerative diseases.
Keywords: amyloid fibril structure, α-synuclein, posttranslational modifications
Abstract
Posttranslational modifications (PTMs) of α-synuclein (α-syn), e.g., phosphorylation, play an important role in modulating α-syn pathology in Parkinson’s disease (PD) and α-synucleinopathies. Accumulation of phosphorylated α-syn fibrils in Lewy bodies and Lewy neurites is the histological hallmark of these diseases. However, it is unclear how phosphorylation relates to α-syn pathology. Here, by combining chemical synthesis and bacterial expression, we obtained homogeneous α-syn fibrils with site-specific phosphorylation at Y39, which exhibits enhanced neuronal pathology in rat primary cortical neurons. We determined the cryo-electron microscopy (cryo-EM) structure of the pY39 α-syn fibril, which reveals a fold of α-syn with pY39 in the center of the fibril core forming an electrostatic interaction network with eight charged residues in the N-terminal region of α-syn. This structure composed of residues 1 to 100 represents the largest α-syn fibril core determined so far. This work provides structural understanding on the pathology of the pY39 α-syn fibril and highlights the importance of PTMs in defining the polymorphism and pathology of amyloid fibrils in neurodegenerative diseases.
Abnormal aggregation of α-synuclein (α-syn) into amyloid fibrils is closely associated with Parkinson’s disease (PD) and other synucleinopathies, such as dementia with Lewy bodies (LBs) and multiple system atrophy (MSA) (1, 2). Accumulation of α-syn fibrils in LBs and Lewy neurites (LNs) is the histological hallmark of these diseases (3, 4). Moreover, the α-syn fibril is the key pathological entity for the cell-to-cell transmission of α-syn pathology in brain or from other organs, such as gut, to the brain (5–8). A variety of posttranslational modifications (PTMs) of α-syn, e.g., phosphorylation, acetylation, ubiquitination, and O-GlcNAcylation, have been identified in regulating α-syn biological function and pathological amyloid aggregation (9–12). Endogenous α-syn is generally acetylated at the N terminus (12). α-syn fibrils in LBs are commonly phosphorylated at S129 (pS129) and highly ubiquitinated in the N terminus (3, 13). O-GlcNAcylation in the non–amyloid-β component (NAC) region of α-syn (e.g., T72 and S87) can slow down or even eliminate α-syn fibrillation (9). Phosphorylation is a common PTM of α-syn. Besides pS129, phosphorylation has been reported on many other residues, including Y125, Y133, and Y135 in the C terminus, and S87 in the NAC region (10, 14, 15). Recently, Y39 in the N terminus has been identified to be phosphorylated by the c-Abl protein tyrosine kinase (16, 17). Phosphorylated Y39 (pY39) α-syn is elevated and accumulates in human brain tissues and LBs of sporadic PD patients (17). pY39 can decrease the lipid binding of α-syn and accelerate α-syn aggregation and neuropathology in the transgenic mouse PD model (16–19). α-syn phosphorylation by c-Abl protects α-syn against degradation via the autophagy and proteasome pathways in cortical neurons (16). However, the molecular mechanism of pY39 α-syn pathology is unknown.
In this work, we determined the near-atomic structure of full-length pY39 α-syn fibril by cryo-electron microscopy (cryo-EM). The structure comprises residues 1 to 100 covering the entire N terminus, the NAC region, and a short segment of the C terminus. Strikingly, the structure shows that pY39 attracts the lysine residues in the α-syn N terminus to form a hydrophilic channel in the center of the α-syn protofilament, which leads to complete rearrangement of the α-syn fibril structure. In addition, we show that the pY39 α-syn fibril exhibits enhanced propagation in primary cortical neurons and is more resistant to protease digestion, compared to the nonphosphorylated wild-type (WT) fibril. This work provides structural indication for the pathology of pY39 α-syn fibril and suggests that PTM may alter α-syn fibril structure and affect PD pathology.
Results
Phosphorylation at Y39 Alters α-Syn Fibril Morphology.
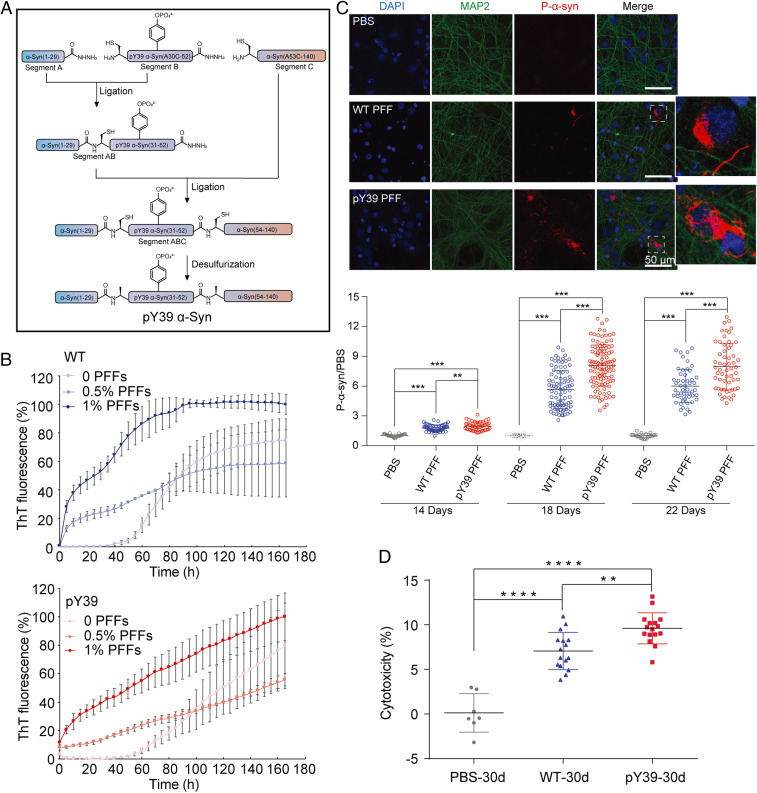
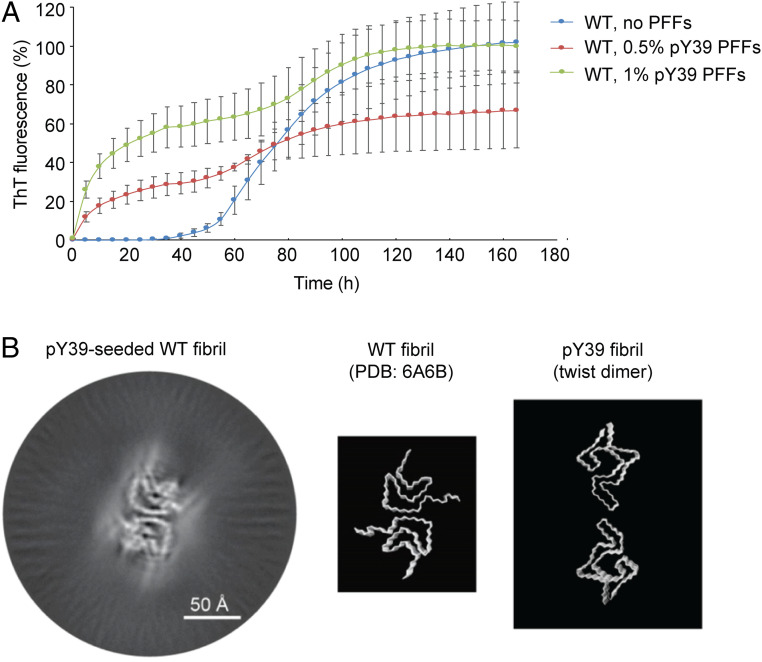
To investigate the influence of phosphorylation at Y39 on α-syn aggregation, we semisynthesized full-length α-syn with site-specific phosphorylation at Y39 (Fig. 1A and SI Appendix, Fig. S1). The synthetic α-syn was characterized for the correct sequence and phosphorylation at Y39 by analytical reversed-phase high-pressure liquid chromatography (RP-HPLC) and electrospray ionization mass spectrometry (ESI-MS) (SI Appendix, Fig. S2). The pY39 α-syn amyloid fibril was grown in the buffer containing 50 mM Tris, 150 mM KCl, pH 7.5 at 37 °C with agitation. The aggregation kinetics of pY39 α-syn was characterized and compared with that of nonphosphorylated WT α-syn by thioflavin T (ThT) kinetic assay. The result showed that the aggregation lag time of pY39 α-syn is ∼50 h, which is similar to that of WT α-syn (Fig. 1B), and consistent with the aggregation kinetics of pY39 α-syn previously characterized (19). Addition of preformed fibril seeds (PFFs) dramatically shortened the lag phase of both pY39 and WT α-syn aggregation (Fig. 1B). However, unlike the WT α-syn aggregation reaching plateau in ∼100 h, pY39 α-syn aggregation did not reach plateau during the monitored time (Fig. 1B). Consistently, negative-staining transmission electron microscopy (TEM) showed slower fibril formation of pY39 α-syn than that of WT α-syn (SI Appendix, Fig. S3A).
Fig. 1.
pY39 α-syn synthesis and pathology. (A) Workflow of the semisynthesis of human pY39 α-syn. (B) ThT kinetic assays of the WT and pY39 aggregation with or without the addition of PFFs. Mol% of added PFFs are indicated. Data shown are mean ± SD, n = 3. (C) α-syn PFFs induce endogenous α-syn aggregation in rat primary cortical neurons. Primary neurons were treated with 100 nM α-syn PFFs at DIV8 for 14 d. Fixed neurons were stained for DAPI (blue), MAP2 (green), and pS129 α-syn (red) and imaged by confocal microscopy. Measurement of the mean gray value of pS129 α-syn was shown Below. The images were analyzed by Image J. The mean gray values of pS129 α-syn were normalized to that of PBS; >10 images were randomly taken for each sample. More than five individual samples were analyzed for each time point. **P < 0.01; ***P < 0.001 for one-way ANOVA followed by Tukey honestly significant difference (HSD) post hoc test. The dashed line boxes stand for the region of zoom in images. (D) Cytotoxicity of WT and pY39 α-syn PFFs to primary neurons at DIV8 for 30 d measured by LDH assay. **P < 0.01; ****P < 0.0001 for one-way ANOVA followed by Tukey HSD post hoc test.
Furthermore, both negative-staining TEM and atomic force microscopy (AFM) revealed a mixture of three polymorphs of pY39 α-syn fibrils (SI Appendix, Fig. S3B). They include two types of left-handed twisted fibrils with a periodic spacing of ∼122 nm and ∼120 nm, respectively, and one type of straight fibril (SI Appendix, Fig. S3 C and D). AFM measurement showed that ∼80% of the total fibrils are twisted, with a nearly equal proportion of the two types of twisted fibrils, while only ∼20% are straight fibrils (SI Appendix, Fig. S3E). Notably, all of the pY39 fibril polymorphs showed different morphologies from the nonphosphorylated WT α-syn fibril (referred to as WT fibril) formed under the same condition (SI Appendix, Fig. S4). The distinct morphologies of pY39 fibrils indicate that phosphorylation at Y39 may alter the structure of the α-syn fibril.
The pY39 α-Syn Fibril Exhibits Enhanced Neuropathology in Rat Primary Cortical Neurons.
It has been reported that phosphorylation at Y39 can exacerbate α-syn neuropathology (16, 17). Thus, we sought to assess the biological relevance of the in vitro prepared pY39 α-syn fibril by testing its neuropathology. We firstly used a well-documented neuronal propagation assay to monitor the aggregation of endogenous α-syn induced by exogenous pY39 α-syn PFFs (6, 20, 21). We added PFFs into the culture medium of rat primary cortical neurons and detected the formation of pathological α-syn aggregation by the antibody for pS129 α-syn. The result showed that pY39 PFFs induced a significant elevation of pathological pS129 α-syn than the WT PFFs (Fig. 1C and SI Appendix, Fig. S5). To validate the aggregation property of the induced pS129 α-syn, we further assessed the immunoreactivity of PFF-treated neurons for LB markers: ubiquitin and P62, respectively. The result showed colocalization of pS129 α-syn, ubiquitin, and P62, which confirms that the PFF-induced pS129 α-syn is a pathological aggregation in LBs (SI Appendix, Fig. S6). Next, we used the lactate dehydrogenase (LDH) assay to evaluate the toxicity of the pY39 fibril to neurons. The result showed that pY39 α-syn PFFs are more toxic to neurons than the WT PFFs (Fig. 1D). Together, these results indicate that the pY39 fibril prepared in this work exhibits consistent neuropathology to that of the pY39 α-syn aggregation in vivo (17).
Cryo-EM Structure Determination of the pY39 α-Syn Amyloid Fibril.
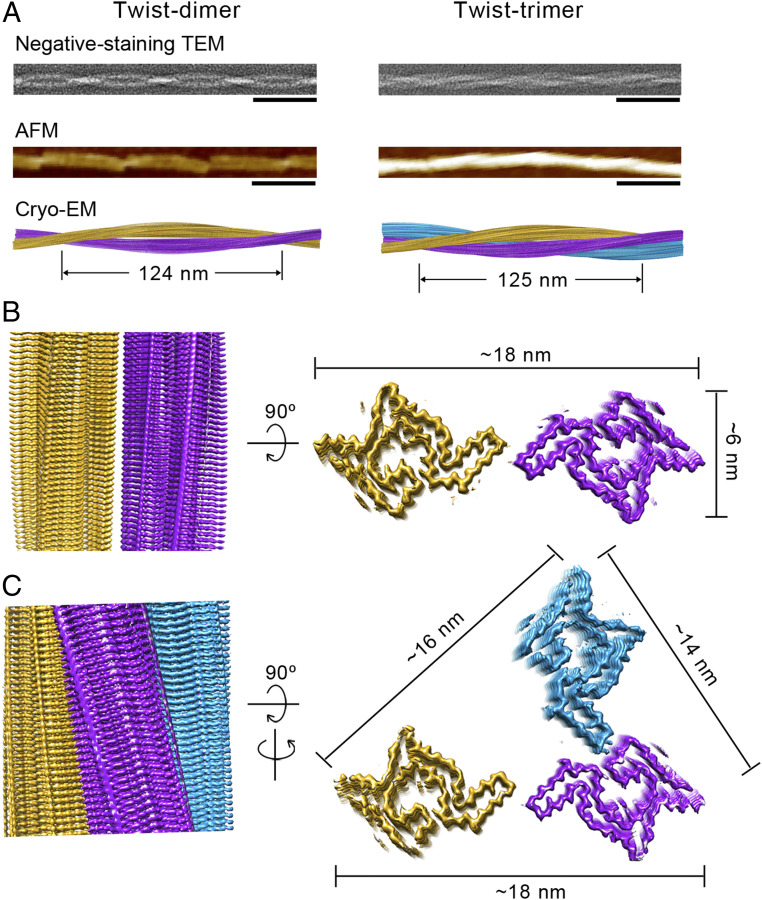
To understand how phosphorylation at Y39 alters the structure and pathology of the α-syn fibril, we set out to determine the atomic structure of pY39 α-syn fibrils by cryo-EM. We prepared cryo-EM grids with polydispersed pY39 fibrils and collected 3,177 micrographs. Consistent with the AFM data, two-dimensional (2D) classification from helical image processing of 10,460 fibril segments revealed two major polymorphs of twisted fibrils (>80% of the total): One consists of two protofilaments (termed twist-dimer); the other consists of three protofilaments (termed twist-trimer) (SI Appendix, Fig. S7) Besides, there was also a small species of straight fibril (<20%), which consists of two protofilaments aligned in parallel (SI Appendix, Fig. S8).
We were able to reconstruct the three-dimensional (3D) density maps for the two polymorphs of twisted fibrils (Fig. 2 and Table 1). The overall resolutions of the density maps are 3.22 Å for the twist-dimer fibril and 3.37 Å for the twist-trimer fibril (SI Appendix, Fig. S9). Both fibrils are reconstructed as left-handed helices according to AFM images (SI Appendix, Fig. S3D). The twist-dimer fibril spine features a width of ∼18 nm and a half pitch (a pitch is termed as the length of a 360° helical turn of the whole fibril) of 124 nm. The two protofilaments intertwine along an approximate twofold screw axis with a helical twist of 179.65° (in the presence of an approximate twofold symmetry, the helical twist represents the relative turning angle of the two protofilaments) and rise of 2.41 Å (Fig. 2 and Table 1). The twist-trimer fibril spine features a width of ∼18 nm and a half pitch of 125 nm. The helical twist of the twist-trimer fibril is −0.69° (in the absence of symmetry, the helical twist represents the turning angle of a protofilament) and rise of 4.80 Å (Fig. 2 and Table 1).
Fig. 2.
Structure determination of pY39 α-syn fibrils. (A) The negative-staining TEM images, AFM images, and cryo-EM reconstruction density maps of twist-dimer and twist-trimer fibrils. (Scale bars: 100 nm.) Half pitches of the fibrils are indicated. (B and C) Cryo-EM density maps of the pY39 α-syn twist-dimer fibril (B) and twist-trimer fibril (C). Fibril widths are indicated. Protofilaments are colored individually.
Table 1.
Statistics of cryo-EM data collection and refinement
| Name | |||
| pY39 twist-dimer fibril | pY39 twist-trimer fibril | pY39-seeded WT fibril | |
| PDB ID | 6L1T | 6L1U | |
| EMDB ID | EMD-0801 | EMD-0803 | |
| Data collection | |||
| Magnification | 22,500 | 22,500 | 105,000 |
| Pixel size, Å | 0.6535 | 0.6535 | 0.678 |
| Defocus range, μm | −2.6 to −1.8 | −2.6 to −1.8 | −2.1 to −1.3 |
| Voltage, kV | 300 | 300 | 300 |
| Camera | K2 summit | K2 summit | K2 summit |
| Microscope | Titan Krios | Titan Krios | Titan Krios |
| Exposure time, s per frame | 0.25 | 0.25 | 0.25 |
| No. of frames | 32 | 32 | 32 |
| Total dose, e−/Å | 49 | 49 | 47 |
| Reconstruction | |||
| Micrographs | 3,177 | 3,177 | 1,918 |
| Manually picked fibrils | 10,460 | 10,460 | 26,128 |
| Box size, pixels | 320 | 320 | 320 |
| Interbox distance, Å | 41.8 | 41.8 | 43.4 |
| Segments extracted | 136,605 | 136,605 | 375,466 |
| Segments after class 2D | 51,716 | 125,661 | 350,265 |
| Segments after class 3D | 25,368 | 14,178 | 47,915 |
| Resolution, Å | 3.22 | 3.37 | 4.21 |
| Map sharpening B-factor, Å2 | −83.62 | −85.84 | −123.91 |
| Helical rise, Å | 2.41 | 4.80 | 4.79 |
| Helical twist, ° | 179.65 | −0.69 | −0.72 |
| Symmetry imposed | C1 | C1 | C1 |
| Atomic model | |||
| Map sharpening B-factor, Å | −164.17 | −139.74 | |
| Nonhydrogen atoms | 7,010 | 10,515 | |
| Protein residues | 1,000 | 1,500 | |
| Ligands | 0 | 0 | |
| rmsd bond lengths | 0.009 | 0.009 | |
| rmsd bond angles | 0.738 | 0.728 | |
| All-atom clashscore | 12.89 | 15.33 | |
| Rotamer outliers, % | 0.00 | 0.00 | |
| Ramachandran outliers, % | 0.00 | 0.00 | |
| Ramachandran allowed, % | 12.63 | 15.79 | |
| Ramachandran favored, % | 87.37 | 84.21 | |
No atomic model data available for pY39-seeded WT fibril.
Relation between the Different Polymorphs of pY39 α-Syn Fibrils.
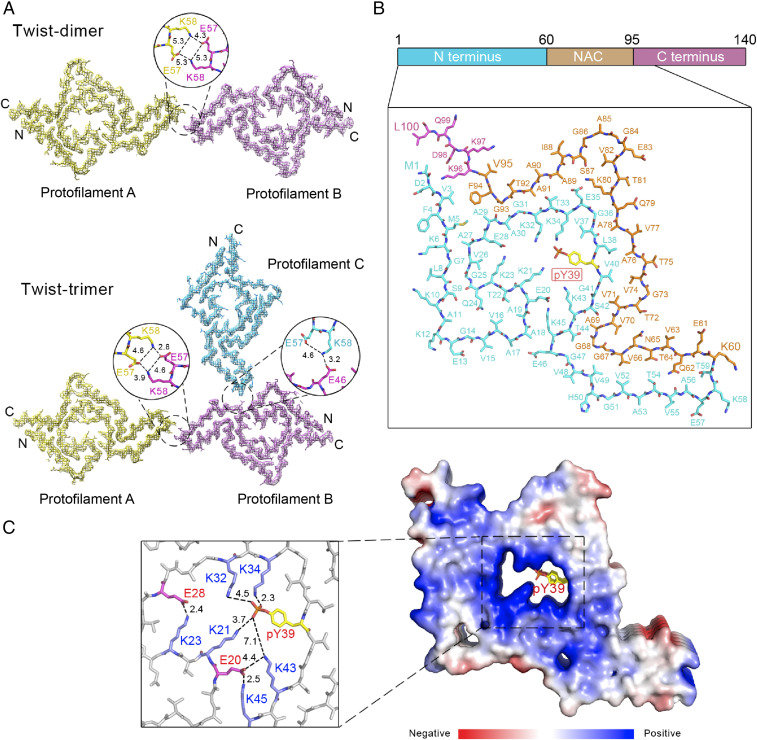
According to the density maps, we were able to unambiguously build the atomic model for the two twisted polymorphs (Fig. 3A). We aligned the α-syn monomers from protofilament A of the two polymorphs and found that they are nearly identical with an rmsd of 0.336 Å (93 out of 100 Cα atoms are aligned) (SI Appendix, Fig. S10A). Further, we aligned the α-syn dimers from protofilaments A and B of the two polymorphs and found that they are also nearly identical with an rmsd of 0.406 Å (194 out of 200 Cα atoms are aligned) (SI Appendix, Fig. S10B).
Fig. 3.
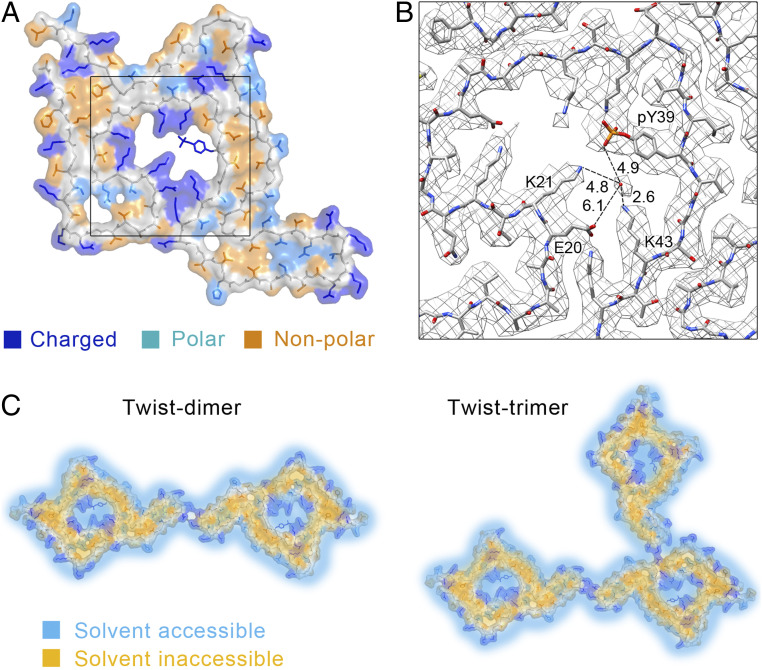
Cryo-EM structures of the pY39 α-syn fibrils. (A) Density maps with structural models for twist-dimer (Upper) and twist-trimer (Lower) fibrils. One dimer/trimer is shown and colored by molecules. Molecular interfaces are shown in the zoom-in views with involved residues and interactions labeled. Distances are shown in Å. (B) The structure of pY39 α-syn that is conserved in different twisted polymorphs is shown and colored by domains. The domain organization of α-syn is shown at the top. Each individual residue is labeled. (C) Electrostatic surface of five layers of the pY39 α-syn fibril. The central positive hole is shown in the zoom-in view with the charged residues and electrostatic interactions labeled. Glu is in red; Lys is in blue; pY39 is in yellow; the phosphate group of pY39 is in red. Distances are in Å.
The interfaces between protofilaments A and B in twist-dimer and twist-trimer are the same, composed of salt bridges formed by E57 and K58 (Fig. 3A). If not considering the pseudo twofold symmetry, the helical twist of the twist-dimer fibril would be −0.70° and the helical rise would be 4.81 Å, which are nearly identical to the parameters of the twist-trimer fibril (−0.69° and 4.80 Å) (Table 1). These results indicate that the twist-trimer fibril is formed by an additional protofilament (protofilament C) twining around the twist-dimer fibril. Indeed, we observed a successive appearance of twist-dimer and twist-trimer during the fibril growth by TEM (SI Appendix, Fig. S11A). Moreover, we observed the coexistence of twist-dimer, twist-trimer, and straight polymorphs in a single fibril (SI Appendix, Fig. S11B). In addition, the width of the protofilament of straight fibril is ∼6.1 nm measured from the 2D class averages (SI Appendix, Fig. S8), which is very close to the width of the conserved protofilament of twisted fibrils. These results indicate that the polymorphism of the pY39 α-syn fibril may derive from different packing of a conserved protofilament in the process of fibril maturation.
pY39 Drives the Formation of a Distinctively Large Fibril Core.
In the cryo-EM fibril structure, pY39 α-syn folds into a hook-like architecture composed of resides 1 to 100 with the entire N terminus, NAC domain, and a short segment of the C terminus involved (Fig. 3B), which presents the largest fibril core of α-syn reported so far. Impressively, in the center of the hook head, lysine residues (K21, K23, K32, K34, K43, and K45) of the N terminus form a positive hole, which is neutralized by E20, E28, and the phosphate group of the pY39 (Fig. 3C). A network of electrostatic interactions is formed between E28/K23, E20/K43/K45, and pY39/K21/K32/K34 (Fig. 3C). Residues V52 to V66 form a hetero-steric zipper and reach out to mediate interprotofilamental interactions for the further assembly of mature fibrils (Fig. 3 A and B).
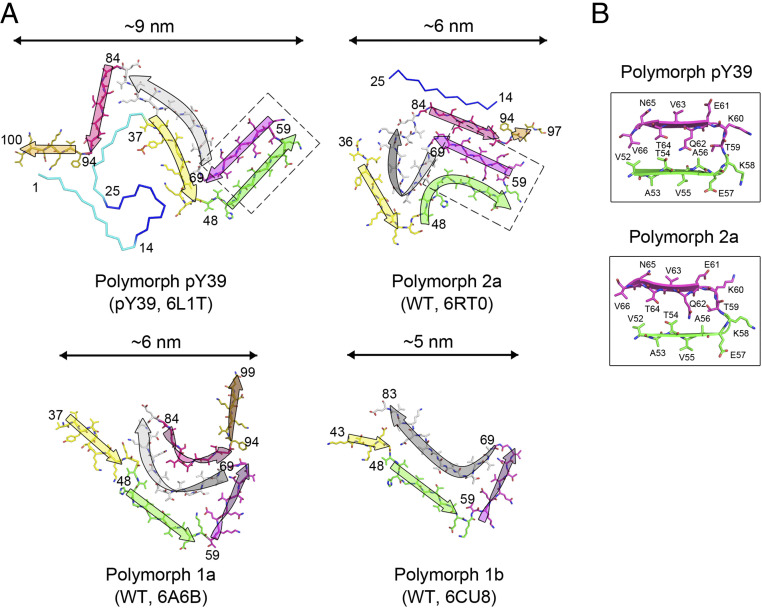
The overall topology of the pY39 structure is distinct from any known structures of α-syn fibrils formed by full-length α-syn (22–26) (Fig. 4A) or C terminus truncated WT α-syn (27), or mutant α-syn (28–31), except that the out-reaching steric zipper of the pY39 fibril also exists in the structure of the polymorph 2a WT fibril reported lately (26) (Fig. 4B). Different polymorphs of the α-syn fibril cores reported previously are composed by a common segment covering residues approximately from 37 to 99 (polymorph 1b is even smaller). This segment is able to fold into various compact structures for the polymorphic formation of α-syn amyloid fibrils (Fig. 4A). In contrast, this segment is relatively stretched in the fold of the pY39 fibril and, together with the rest of the N-terminal residue (residues 1 to 36), forms a distinctly large fibril core (Fig. 4A).
Fig. 4.
Comparison of polymorphic α-syn fibril structures. (A) Topology of α-syn in different polymorphs of amyloid fibrils. α-Syn variants and PDB IDs are shown in parentheses. The sequence involved in the WT fibril cores is shown in sticks and colored regionally. The N terminus that is usually flexible in the WT fibril but involved in the core of the pY39 fibril is shown in ribbons and colored in cyan and blue. The arrows indicate the topology of the structures. Flip of the arrow indicates flip of the amino acid chain. Selected residue numbers are shown. The name of WT fibril polymorphs follows ref. 26. Widths of the fibril cores are indicated. Dashed boxes highlight the conserved hetero-steric zipper in pY39 and polymorph 2a. (B) A zoom-in view of the conserved steric zipper highlighted in A, shown in schematic and sticks. Residue numbers are labeled.
Hydrophilic Channel in the Center of the pY39 α-Syn Fibril Core.
The involvement of the entire N terminus not only enlarges the fibril core but also significantly increases the proportion of charged residues, including K, D, and E (SI Appendix, Fig. S12). Note that α-syn does not contain arginine. Besides, phosphorylation further modifies Y39 into a strong negatively charged residue. With such a high amount of charged residues, pY39 α-syn remarkably folds with eight charged residues (six lysine and two glutamate) plus pY39 to form a hydrophilic channel in the center of the fibril core, which might be accessible to the solvent (Fig. 5A). Actually, we observed isolated round density in the channel at the local resolution of 2.9 Å (Fig. 5B and SI Appendix, Fig. S9A), which may represent water molecules hydrogen-bonding with the side chains of pY39, K21, and K43. Thus, the pY39 α-syn fibril may contain an inner and an outer surface to contact with the environment (Fig. 5C).
Fig. 5.
Hydrophilic central channel of the pY39 α-syn fibril. (A) The surface of the pY39 α-syn is shown. The side chains of charged residues are colored in blue; polar residues are in light blue; nonpolar residues are in orange. The main chain is colored in gray. A density map of the framed region is shown in B. (B) Density map of the central hole shows the density of water. Distances are in Å. (C) The potential solvent-accessible (light blue) and inaccessible (orange) surfaces of the twist-dimer and twist trimer fibrils are shown.
pY39 α-Syn Fibrils Are More Resistant to Protease Digestion.
We used trypsin to digest the WT and pY39 α-syn fibrils, respectively. Of note, since trypsin cleaves proteins at K or R, accordingly to the pY39 structure, only one cleavage site (K102) is outside of the fibril core and most available for trypsin digestion. Indeed, upon trypsin digestion, the pY39 fibril generated only one major band of cleaving product, which covers residues 1 to 102 analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (SI Appendix, Fig. S13A). In contrast, the WT fibril was digested into several fragments. Slower degradation of pY39 fibril was also observed by protease K (PK) digestion (SI Appendix, Fig. S13B). In addition, pY39 and WT α-syn monomers were used to confirm the activity of the proteases (SI Appendix, Fig. S13C). These results indicate that phosphorylation at Y39 may cause α-syn fibrils to be more difficult for clearance in cells.
pY39 PFFs Cross-Seed WT α-Syn to Form Amyloid Fibrils Featuring the WT Fibril Structure.
To understand the potential for the spread of pY39 fibrils, we sought to see if the pY39 fibrils can cross-seed the fibril formation of WT α-syn. We monitored the aggregation kinetics of WT α-syn in the presence of pY39 PFFs by the ThT assay. Intriguingly, the result showed that, similar to WT PFFs, pY39 PFFs effectively seeded the amyloid aggregation of WT α-syn (Figs. 1B and 6A). However, it is difficult to imagine that pY39 PFFs could possibly template nonphosphorylated α-syn into the same fibril structure.
Fig. 6.
pY39 PFFs cross-seed WT α-syn fibril formation. (A) ThT kinetic assay of the pY39-seeded WT α-syn aggregation. Mol% of added PFFs are indicated. Data shown are mean ± SD, n = 3. (B) Cryo-EM 3D reconstruction of the pY39-seeded WT α-syn fibril (Left). Structural models of WT α-syn fibril (PDB ID: 6A6B) and pY39 twist-dimer are shown on the Right.
To understand whether and how WT α-syn lacking the phosphate group at Y39 could fold into the structure as that in the pY39 fibril, we performed cryo-EM to determine the structure of the pY39-seeded WT α-syn fibril. Although the image processing did not allow sufficiently high resolution for model building, the result of 3D reconstruction showed that the cross-seeded fibril features the structure of the WT fibril (polymorph 1a) rather than the pY39 fibril (Fig. 6B). This result confirms the key role of Y39 phosphorylation in the formation of the pY39 fibril. Without pY39, WT α-syn can hardly form the structure as that in the pY39 fibril.
While the question remains how pY39 cross-seeds WT α-syn fibril formation, we are reminded that there is a subpopulation of ∼20% straight fibrils in pY39 (SI Appendix, Figs. S3E and S11A), whose structure has not been able to be determined yet due to technical limitation. It is possible that these straight fibrils also contribute to the ability of pY39 to cross-seed WT fibrils. Another possibility is that there is a trace amount of the WT fibril structure formed by pY39 α-syn. This possibility is less likely though. As we seeded WT α-syn aggregation with a gradient of low percentages of WT PFFs, the ThT result showed that <0.05% WT PFFs, which is equivalent to <5% WT fibril structure in 1% pY39 PFFs, showed apparently weaker seeding effect than that of 1% pY39 PFFs (Fig. 6A and SI Appendix, Fig. S14). Actually, we did not find any WT fibril morphology in the pY39 fibrils during cryo-EM data processing (SI Appendix, Fig. S3E). Note that the WT fibrils are also polymorphic, and thus comparison of the relative seeding efficiency between pY39 and WT fibrils is actually unquantifiable. Finally, the observed cross-seeding effect could also be derived from surface-mediated secondary nucleation (32), which can be less dependent on fibril structures. Although the mechanism of pY39 cross-seeding WT α-syn fibril formation remains unknown, the phenomenon is not surprising since the cross-seeding effect has been widely found between different amyloid proteins, such as between α-syn and Aβ (33), between Aβ and IAPP (34, 35), and between Aβ and Tau (36).
Discussion
In this work, we reveal that Y39 phosphorylation leads to the formation of polymorphs of the α-syn amyloid fibril, which features a distinctly large fibril core. Structural polymorphism is a common feature of amyloid fibrils formed by different peptides/proteins, such as α-syn, Tau, and Aβ (22, 24, 37–41). Different fibril polymorphs may exhibit different pathologies under disease conditions (6, 42). For instance, α-syn fibrils derived from PD and MSA patients exhibit distinctive morphologies and pathologies (6). Thus, structural understanding on different fibril polymorphs is important for the understanding of the complicated pathology of the disease. In recent years, technological advance in cryo-EM have enabled the structure determination of various full-length amyloid fibrils at the near-atomic resolution (43). Polymorphic core structures have been determined in the WT and mutant α-syn fibrils (22–31), which are commonly composed of segment ∼40 to 100 or smaller. Both the C terminus and the majority of the N terminus remain flexible and thus are invisible in the cryo-EM structures. The C terminus is highly enriched in acidic residues, and the N terminus is enriched in alkaline residues. The high proportion of charged residues precludes the engagement of the two termini in the fibril core structure. Intriguingly, a recent structural study on the human brain-derived α-syn fibril showed that in vivo cofactors can participate into α-syn fibril structure and introduce more N-terminal region into the fibril core (44), which recalls the observation in the pY39 fibril. Phosphorylation at Y39 involves the entire N terminus into the fibril core and rearranges the fibril core structure. pY39 changes the electrostatic property of the N terminus and stabilizes multiple lysine residues to form a hydrophilic channel in the center of the fibril core. Thus, the patient-derived and pY39 α-syn fibril structures together suggest the important role of the N terminus in the pathological fibril formation of α-syn. In addition, pY39 fibril structure highlights the importance of PTM in defining the structure and pathology of amyloid fibrils.
Phosphorylation widely exists in different amyloid proteins, such as α-syn, Tau, Aβ, and FUS (10, 13, 45, 46). However, whether and how phosphorylation influences the fibril formation is poorly understood. Recently, a fibril structure of phosphorylated Ser8 (pS8) Aβ40 has been determined by solid-state NMR while the segment containing pS8 is flexible due to limited NMR constraints (46). A recent cryo-EM study on α-syn reconstructed a low-resolution density map for the pS129 α-syn fibril, which at that resolution is similar to that of the WT fibril (24). Tau is hyperphosphorylated at multiple sites (e.g., S202, S262, S356, and S396) in the progress of amyloid aggregation (10) whereas phosphorylation cannot be determined in the cryo-EM structures of Tau fibrils derived from different tauopathies, although the fibrils are indeed phosphorylated probed by antibodies (37, 38, 41). A possible explanation is that phosphorylation of native fibrils is heterogeneous, and thus invisible in cryo-EM structures. As well, the phosphorylation sites that are not involved in the fibril core are unable to be determined by cryo-EM. In this work, we use semisynthetic α-syn with site-specific phosphorylation at Y39 to form amyloid fibrils in vitro. The pathology and protease resistance of the fibril are consistent to that of native pY39 α-syn aggregates (16, 17), which indicates the biological relevance of the synthetic fibril. Moreover, the unique structure of the pY39 α-syn fibril, in particular the key role of pY39 in stabilizing the fibril structure via electrostatic interactions, may limit the possibility of alternative structures. Indeed, although pY39 PFFs can seed WT fibril formation, the seeded fibrils still feature WT rather than pY39 fibril structure. Thus, although prepared in vitro, the unique pY39 fibril structure presented in this work is informative for the structural understanding of native pY39 fibril.
Previous studies suggest that pY39 phosphorylation can enhance α-syn pathology and protect α-syn against degradation via the autophagy and proteasome pathways in mouse cortical neurons (16). Consistently, we found that the pY39 fibril exhibits enhanced propagation and cytotoxicity in the neuronal model and tends to be more resistant to protease cleavage than the WT fibril. The structures of pY39 fibrils may provide a structural explanation for these phenomena. The involvement of the entire N terminus into the fibril core enlarges the folded structure of α-syn, which may protect α-syn from protease digestion; on the other hand, chaperones that recognize the N terminus of α-syn (47) may be inefficient to prevent α-syn aggregation or fail to mediate its clearance. Besides, other factors, such as the internalization efficiency, may also influence the pathology of pY39 fibrils. The enhanced pathology of pY39 fibrils may play a role in the progression of PD and α-synucleinopathies.
Materials and Methods
Semisynthesis of pY39 α-Syn.
We referred to the synthesis strategy of Lashuel and coworkers (19) and used the peptide hydrazide strategy of Liu and coworkers (48) to improve the synthesis strategy for pY39 α-syn. To obtain the full-length pY39 α-syn, we performed N-to-C sequential native chemical ligation using three segments, including segment A (α-Syn 1-29NHNH2), segment B (pY39 α-syn A30C-52NHNH2), and segment C (α-Syn A53C-140) (SI Appendix, Fig. S1) (19, 48, 49). The segments A and B were manually synthesized by Fmoc-based solid phase peptide synthesis (Fmoc-SPPS). The sequence of segment A is MDVFMKGLSKAKEGVVAAAEKTKQGVAEA-NHNH2. The sequence of segment B is CGKTKEGVL[pY]VGSKTKEGVVHGV-NHNH2. The segment A and segment B were purified with RP-HPLC and characterized by analytical RP-HPLC and ESI-MS (SI Appendix, Fig. S2).
To obtain segment C (α-Syn A53C-140), we fused the SUMO-tag to the N terminus of α-syn 54-140 and overexpressed the SUMO-fused α-syn 54-140 in BL21 (DE3) Escherichia coli cells (49). Protein expression was induced by addition of 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) and incubation of cells for 20 h at 16 °C. Then, the cells were harvested in buffer (50 mM NaH2PO4, 500 mM NaCl, pH 7.4) and lysed by ultrasonication (twice on ice, each for 20 min). The fusion protein was purified by nickel-nitrilotriacetic acid (Ni-NTA). To remove the SUMO tag, the SUMO-C-α-syn 54-140 was treated with protease ULP1 in cleavage buffer (50 mM NaH2PO4, 500 mM NaCl, pH 8.0) at 16 °C overnight. Then, the SUMO tag and ULP1 were removed using Ni-NTA. The segment C was purified by preparative RP-HPLC with Proteonavi column. Characterization of segment C was achieved by using analytical HPLC and ESI-MS (SI Appendix, Fig. S2).
Then, the three segments were N-to-C sequentially linked by native chemical ligation (NCL) for producing the full-length pY39 α-syn. Firstly, segment A and segment B were ligated to produce segment AB (pY39 α-syn 1-52NHNH2, A30C). Then, 2 mM segment A was dissolved in 400 μL of 0.2 M sodium phosphate buffer containing 6 M GuHCl (PS buffer) at pH 3.0 and put into an ice–salt bath to precool. In order to oxidize the peptide hydrazide to the corresponding azide, 0.05 M NaNO2 was added in the reaction for 15 min. To convert the peptide azide to the thioester, 0.2 M 4-mercaptophenylacetic acid (MPAA) in 400 μL of PS buffer was added, and then the pH was adjusted to 6.8. To complete the ligation reaction, 1.2 equivalent (Eq.) segment B was added and reacted at 16 °C overnight. The segment AB was further purified with RP-HPLC and characterized by analytical RP-HPLC and ESI-MS (SI Appendix, Fig. S2). Secondly, we ligated segment AB and segment C to obtain segment ABC (pY39 α-syn 1-140, A30C, A53C). The ligation of segments AB and C was similar to that of A and B. The segment ABC was further purified with RP-HPLC and characterized by analytical RP-HPLC and ESI-MS (SI Appendix, Fig. S2). Finally, we changed Cys to Ala in segment ABC by chemical desulfurization to obtain the full-length pY39 α-syn protein. Under free radical conditions [0.4 M Tris(2-carboxyethyl)phosphine (TCEP), 0.5 mM 2,2′-Azobis[2-(2-imidazolin-2-yl)propane]dihydrochloride (VA-044), and 10% (vol/vol) 2-methylpropane-2-thiol in 0.2 M PS buffer, pH 7.0], the ligation product was desulfurized at 37 °C overnight. pY39 was synthesized in milligram amounts with high purity as assessed by RP-HPLC and ESI-MS (SI Appendix, Fig. S2). Note that the N terminus of the synthesized pY39 α-syn is not acetylated.
Fibril Preparation of WT and pY39 α-Syn.
Fibrillation was conducted by diluting recombinant nonmodified WT α-syn and synthetic pY39 α-syn to 100 μM in buffer (50 mM Tris, pH 7.5, 150 mM KCl, 0.05% NaN3). WT and pY39 α-syn were incubated at 37 °C with constant agitation at 900 rpm in ThermoMixer (Eppendorf) for a week, respectively. The fibrils were sonicated for 30 s (1 s on, 1 s off) on ice at 20% power by JY92-IIN sonicator (Xinyi Sonication Equipment Company, Ningbo, China). The sonicated fibrils (0.5%, vol/vol) were mixed with 100 μM α-syn monomer and further incubated at 37 °C with agitation (900 rpm) for 2 wk to obtain fibrils for cryo-EM and AFM experiments. To prepare α-syn PFFs for the primary neuron transmission and protease digestion experiments, the fibrils used for cryo-EM were concentrated by centrifugation (14,000 rpm, 25 °C, 45 min), washed with phosphate-buffered saline (PBS) for three times, and sonicated again using the same sonication condition.
ThT Kinetic Assay.
pY39 and WT α-syn PFFs were prepared by sonication at 20% power on ice for 15 times (1 s per time, 1-s interval) by JY92-IIN sonicator (Xinyi Sonication Equipment Company, Ningbo, China). Then, 50 μM pY39 or WT α-syn monomer in buffer (50 mM Tris, pH 7.5, 150 mM KCl, 0.05% NaN3) was incubated with or without PFFs in clear 384-well plates (Thermo Scientific Nunc). Final concentration of 10 μM ThT was added to the reaction mixture. The plate was detected by a FLUOstar Omega Microplate Reader (BMG LABTECH). Fluorescence was measured at 440-nm (excitation) and 485-nm (emission) wavelengths, with a bottom read. Three replicates were performed for each sample.
Negative-Staining TEM.
An aliquot of 5 μL of fibril solution was loaded onto a glow-discharged grid with carbon support film (Beijing Zhongjingkeyi Technology Co., Ltd.). After 45 s, the grid was washed with 5 μL of double-distilled water and 5 μL of 3% wt/vol uranyl acetate. Then 5 μL of 3% wt/vol uranyl acetate was used to stain the sample for 45 s. Filter paper was used to remove the excess solution. The samples were imaged using a Tecnai T12 microscope (FEI Company) operated at 120 kV.
AFM.
Samples were diluted to a final concentration of 50 μM in the fibril growth buffer (50 mM Tris, 150 mM KCl, pH 7.5). Then, 8 μL of the sample were deposited on the surface of freshly cleaved mica at room temperature for 3 min and washed with Milli-Q water for AFM imaging. The samples were recorded in air by using Nanoscope V Multimode 8 (Bruker) in ScanAsyst mode using probe SNL-10. Continuous curves were recorded with a frequency of 65 kHz. Images were acquired at 1,024 × 1,024 pixels at a line rate of 1.5 Hz. The height of samples was processed on the NanoScope Analysis software (version 1.5).
Primary Neuronal Cultures.
Primary cortical neurons were prepared from the cortex of embryonic day (E) 16 to E18 Sprague–Dawley rat (Shanghai SIPPR BK Laboratory Animals Ltd.) embryos as previously described (20). In brief, dissociated cortical neurons were plated onto coverslips in a 24-well plate at a density of 150,000 cells per well, coated with poly-d-lysine. Neurons were then treated with PBS and WT or pY39 α-syn PFFs at 8 d in vitro (DIV) and collected for immunocytochemistry at 14/18/22 d posttreatment. All rat experiments were performed according to the protocols approved by the Animal Care Committee of the Interdisciplinary Research Center on Biology and Chemistry (IRCBC), Chinese Academy of Sciences (CAS). Each experiment was repeated for more than three times.
Immunofluorescence Imaging.
Primary cortical neurons were fixed with 4% paraformaldehyde and 4% sucrose in PBS and then permeabilized with 0.15% Triton X-100. Fixed coverslips were blocked with 5% bovine serum albumin (BSA) in PBS for 1 h at room temperature. Neurons were incubated with anti–α-syn (2642, 1:1,000 dilution; Cell Signaling Technology), anti–p129-α-syn (ab51253, 1:1,000 dilution; Abcam), anti-MAP2 (ab5392, 1:1,000 dilution; Abcam), anti-Ub (sc-8017, 1:200; Santa Cruz Biotechnology), anti-P62 (sc-28359, 1:200; Santa Cruz Biotechnology) antibodies, respectively. After washing with PBS for three times, neurons were stained with Alexa Fluor 488-, Alexa Fluor 568-, or Alexa Fluor 633-conjugated secondary antibodies (A11039, A11036, and A21070, 1:1,000 dilution, respectively; Life Technologies) for 1 h at room temperature. After washing with PBS for an additional three times, DAPI was stained with ProLong Gold antifade reagent when coverslips were mounted onto glass slides. The fluorescent images were acquired by a confocal microscope. The mean gray values of the pS129 α-syn signal for primary cortical neurons were quantified by using ImageJ software.
LDH Assay.
Primary neurons were treated with 100 nM WT and pY39 α-syn PFFs or PBS at DIV8. The neurons were harvested 30 d after treatment. The LDH cytotoxicity assay was performed following the manufacturer’s instructions. The absorbance was recorded at 490 nm and 680 nm (background) with BioTek Synergy 2 Multi-Mode Microplate Readers (Thermo Scientific). The experiment was independently repeated for more than three times. Statistical significance was calculated by one-way ANOVA using GraphPad Prism.
Cryo-EM Imaging and Data Processing.
The pY39 α-syn fibril was diluted to 5 μM. An aliquot of 4 μL of pY39 α-syn fibril solution was applied to a glow-discharged carbon electron microscope grid (Quantifoil R1.2/1.3, 300 mesh). After 10 s of incubation, the grid was blotted for 6.5 s, and plunge-frozen into liquid ethane using an FEI Vitrobot Mark IV. Cryo-EM data were collected on an FEI Titan Krios microscope operated at 300 kV with magnification of 22,500×. Superresolution mode was used for a Gatan K2 Summit camera to record the cryo-EM micrographs with a pixel size of 0.6535 Å. Defocus values were set from −2.6 to −1.8 μm. Then, 32 dose-fractionated frames were taken for each micrograph, and the electron dose rate was set at ∼6.12 e−/s/Å2 with a total exposure time of 8 s (0.25 s per frame). Hence, the total dose was ∼49 e−/Å2 per micrograph. Data were collected by the Serial EM software.
As for image preprocessing, MotionCorr2 was used to correct beam-induced sample motion and implement dose weighting for all 32 frames (50). The pixel size was further binned to 1.307 Å. CTFFIND4.1.8 was used to estimate the defocus values of dose-weighted micrographs (51).
RELION3.0 was used for manual picking, particle extraction, 2D classification, 3D classifications, 3D refinement, and postprocessing (52). Then, 10,460 filaments were picked manually from 3,177 micrographs. Particles were extracted into 686- and 320-pixel boxes using the 90% overlap scheme. The fibrils of pY39 straight, twist-dimer, and twist-trimer were separated by 2D classification.
Helical Reconstruction.
AFM measurement, cryo-EM micrographs, and 2D classification were used together to calculate an initial twist angle. Then, 4.8 Å was used as an initial value of helical rise. Local searches of symmetry in 3D classification were used to find the final twist angle and rise value. By 2D class averages of the twist-dimer filament segments, a staggered arrangement was observed between the subunits on the two sides of the filament. Therefore, the two protofilaments of twist-dimer were related by a pseudo-21 axis. Accordingly, the rise value of pY39 twist-dimer was reset to 4.8/2 = 2.4 Å, and the twist angle was reset to (360° − 0.7°)/2 = 179.65°.
Several iterations of 2D classification and 3D classification were performed to select particles which belong to the same polymorph. Then, a 3D initial model was built by selected 2D classes. The 3D classification was performed several times to generate a proper reference map for 3D refinement. The 3D refinement of the selected 3D classes with appropriate reference was performed to obtain final reconstruction.
The final map of the pY39 twist-dimer was convergence with the rise of 2.41 Å and the twist angle of 179.65°. Postprocessing was performed to sharpen the map with a B-factor of −83.6159 Å2. Based on the gold-standard Fourier shell correlation (FSC) = 0.143 criteria, the overall resolution was reported as 3.22 Å. The final map of pY39 twist-trimer was convergence with the rise of 4.80 Å and the twist angle of −0.69°. Postprocessing was performed to sharpen the map with a B-factor of −85.8426 Å2. The overall resolution was reported as 3.37 Å.
Atomic Model Building and Refinement.
Phenix.auto_sharpen (53) was used to sharpen the map of both pY39 twist-dimer and twist-trimer. Atomic models were subsequently built into the refined maps with COOT (54). The model of the pY39 twist-dimer was built based on the model of the WT α-syn fibril (PDB ID: 6A6B). To build the pY39 twist-trimer model, a single chain from the twist-dimer structure was extracted and fitted into the density map of the twist-trimer. For both twist-dimer and twist-trimer fibrils, a five-layer model was generated for structure refinement. Structure models were refined by using the real-space refinement program in PHENIX (55).
Protease Digestion of α-Syn PFFs.
Ten micrograms of α-syn PFFs in PBS (PH 7.4) were incubated for 30 min at 37 °C in the presence of trypsin (Promega) or proteinase K (Invitrogen). Digestion reaction was terminated by adding 1 mM phenylmethylsulfonyl fluoride (PMSF). The samples were boiled with sodium dodecyl sulfate (SDS)-loading buffer for 15 min and then resolved on 4 to 20% Bis-Tris gels (GenScript). The gels were stained with Coomassie brilliant blue and imaged with Image Lab 3.0 (Bio-Rad).
In-Gel Digestion and LC-MS/MS Analysis.
Target bands were cut from the Coomassie blue-stained SDS/polyacrylamide gel electrophoresis (PAGE) gel. Proteins in the band were digested by trypsin, and then extracted from the gel following the protocol developed by the laboratory of Mann and coworkers (56). Peptides extracted from the gel were analyzed using an on-line EASY-nL-LC 1000 coupled with an Orbitrap Q-Exactive HF mass spectrometer (Thermo Fisher).
Supplementary Material
Acknowledgments
We thank the Center of Cryo-Electron Microscopy (Zhejiang University School of Medicine) for help with data collection. This work was supported by the Major State Basic Research Development Program (2016YFA0501902), the National Natural Science Foundation of China (91853113, 81661148047, and 31872716), the National Key R & D Program of China (2018YFA0507600 and 2019YFA0904200), the Science and Technology Commission of Shanghai Municipality (18JC1420500), the “Eastern Scholar” project supported by the Shanghai Municipal Education Commission, the Shanghai Pujiang Program (18PJ1404300), and the Shanghai Municipal Science and Technology Major Project (Grant 2019SHZDZX02).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Density maps of the pY39 α-syn fibrils are available at the Electron Microscopy Data Bank with entry codes EMD-0801 (twist-dimer) and EMD-0803 (twist-trimer). The structure models are deposited in the Protein Data Bank with entry codes 6L1T (twist-dimer) and 6L1U (twist-trimer).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922741117/-/DCSupplemental.
Data Availability.
Density maps of the pY39 α-syn fibrils are available at the Electron Microscopy Data Bank (EMDB) with entry codes EMD-0801 (twist-dimer) and EMD-0803 (twist-trimer). The structure models are deposited into the Worldwide Protein Data Bank with entry codes PDB 6L1T (twist-dimer) and PDB 6L1U (twist-trimer).
References
- 1.Goedert M., Spillantini M. G., Del Tredici K., Braak H., 100 years of Lewy pathology. Nat. Rev. Neurol. 9, 13–24 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Woerman A. L. et al., α-Synuclein: Multiple system atrophy prions. Cold Spring Harb. Perspect. Med. 8, a024588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spillantini M. G. et al., Alpha-synuclein in Lewy bodies. Nature 388, 839–840 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Braak H., Sandmann-Keil D., Gai W., Braak E., Extensive axonal Lewy neurites in Parkinson’s disease: A novel pathological feature revealed by alpha-synuclein immunocytochemistry. Neurosci. Lett. 265, 67–69 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Kim S. et al., Transneuronal propagation of pathologic alpha-synuclein from the gut to the brain models Parkinson’s disease. Neuron 103, 627–641.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng C. et al., Cellular milieu imparts distinct pathological α-synuclein strains in α-synucleinopathies. Nature 557, 558–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prusiner S. B. et al., Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl. Acad. Sci. U.S.A. 112, E5308–E5317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo J. L., Lee V. M., Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nat. Med. 20, 130–138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine P. M. et al., α-Synuclein O-GlcNAcylation alters aggregation and toxicity, revealing certain residues as potential inhibitors of Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 116, 1511–1519 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenreiro S., Eckermann K., Outeiro T. F., Protein phosphorylation in neurodegeneration: Friend or foe? Front. Mol. Neurosci. 7, 42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson J. P. et al., Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 281, 29739–29752 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Theillet F. X. et al., Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature 530, 45–50 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara H. et al., Alpha-synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4, 160–164 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Paleologou K. E. et al., Phosphorylation at S87 is enhanced in synucleinopathies, inhibits alpha-synuclein oligomerization, and influences synuclein-membrane interactions. J. Neurosci. 30, 3184–3198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negro A., Brunati A. M., Donella-Deana A., Massimino M. L., Pinna L. A., Multiple phosphorylation of alpha-synuclein by protein tyrosine kinase Syk prevents eosin-induced aggregation. FASEB J. 16, 210–212 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Mahul-Mellier A. L. et al., c-Abl phosphorylates α-synuclein and regulates its degradation: Implication for α-synuclein clearance and contribution to the pathogenesis of Parkinson’s disease. Hum. Mol. Genet. 23, 2858–2879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brahmachari S. et al., Activation of tyrosine kinase c-Abl contributes to α-synuclein-induced neurodegeneration. J. Clin. Invest. 126, 2970–2988 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S. et al., The c-Abl inhibitor, Radotinib HCl, is neuroprotective in a preclinical Parkinson’s disease mouse model. Hum. Mol. Genet. 27, 2344–2356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dikiy I. et al., Semisynthetic and in vitro phosphorylation of alpha-synuclein at Y39 promotes functional partly helical membrane-bound states resembling those induced by PD mutations. ACS Chem. Biol. 11, 2428–2437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao X. et al., Pathological α-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science 353, aah3374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volpicelli-Daley L. A., Luk K. C., Lee V. M., Addition of exogenous α-synuclein preformed fibrils to primary neuronal cultures to seed recruitment of endogenous α-synuclein to Lewy body and Lewy neurite-like aggregates. Nat. Protoc. 9, 2135–2146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y. et al., Amyloid fibril structure of α-synuclein determined by cryo-electron microscopy. Cell Res. 28, 897–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B. et al., Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 9, 3609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerrero-Ferreira R. et al., Cryo-EM structure of alpha-synuclein fibrils. eLife 7, e36402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuttle M. D. et al., Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 23, 409–415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerrero-Ferreira R. et al., Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife 8, e48907 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni X., McGlinchey R. P., Jiang J., Lee J. C., Structural insights into α-Synuclein fibril polymorphism: Effects of Parkinson’s disease-related C-terminal truncations. J. Mol. Biol. 431, 3913–3919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyer D. R. et al., Structures of fibrils formed by α-synuclein hereditary disease mutant H50Q reveal new polymorphs. Nat. Struct. Mol. Biol. 26, 1044–1052 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y. et al., Cryo-EM structure of full-length α-synuclein amyloid fibril with Parkinson’s disease familial A53T mutation. Cell Res. 30, 360–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyer D. R. et al., The α-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure. Proc. Natl. Acad. Sci. U.S.A. 117, 3592–3602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao K., et al. , Parkinson’s disease associated mutation E46K of α-synuclein triggers the formation of a novel fibril structure. https://www.biorxiv.org/content/10.1101/870758v2 (11 December 2019). [DOI] [PMC free article] [PubMed]
- 32.Knowles T. P. J. et al., An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Ono K., Takahashi R., Ikeda T., Yamada M., Cross-seeding effects of amyloid β-protein and α-synuclein. J. Neurochem. 122, 883–890 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Zhang M. et al., Molecular understanding of Aβ-hIAPP cross-seeding assemblies on lipid membranes. ACS Chem. Neurosci. 8, 524–537 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Moreno-Gonzalez I. et al., Molecular interaction between type 2 diabetes and Alzheimer’s disease through cross-seeding of protein misfolding. Mol. Psychiatry 22, 1327–1334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griner S. L. et al., Structure-based inhibitors of amyloid beta core suggest a common interface with tau. eLife 8, e46924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falcon B. et al., Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falcon B. et al., Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 561, 137–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiang W., Yau W. M., Lu J. X., Collinge J., Tycko R., Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature 541, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gremer L. et al., Fibril structure of amyloid-β(1-42) by cryo-electron microscopy. Science 358, 116–119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fitzpatrick A. W. P. et al., Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peelaerts W. et al., α-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 522, 340–344 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Fitzpatrick A. W., Saibil H. R., Cryo-EM of amyloid fibrils and cellular aggregates. Curr. Opin. Struct. Biol. 58, 34–42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweighauser M., et al. , Structures of α-synuclein filaments from multiple system atrophy. https://www.biorxiv.org/content/10.1101/2020.02.05.935619v1 (6 February 2020). [DOI] [PMC free article] [PubMed]
- 45.Luo F. et al., Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat. Struct. Mol. Biol. 25, 341–346 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Hu Z. W. et al., Molecular structure of an N-terminal phosphorylated β-amyloid fibril. Proc. Natl. Acad. Sci. U.S.A. 116, 11253–11258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia C. et al., Different heat shock proteins bind α-Synuclein with distinct mechanisms and synergistically prevent its amyloid aggregation. Front. Neurosci. 13, 1124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang G. M. et al., Protein chemical synthesis by ligation of peptide hydrazides. Angew. Chem. Int. Ed. Engl. 50, 7645–7649 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Mossessova E., Lima C. D., Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Zheng S. Q. et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zivanov J. et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terwilliger T. C., Sobolev O. V., Afonine P. V., Adams P. D., Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr. D Struct. Biol. 74, 545–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams P. D. et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M., In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Density maps of the pY39 α-syn fibrils are available at the Electron Microscopy Data Bank (EMDB) with entry codes EMD-0801 (twist-dimer) and EMD-0803 (twist-trimer). The structure models are deposited into the Worldwide Protein Data Bank with entry codes PDB 6L1T (twist-dimer) and PDB 6L1U (twist-trimer).