Significance
The balance of mitochondrial fission and fusion, two aspects of mitochondrial dynamics, is important for mitochondrial function. In this study, we show that Caenorhabditis elegans lysosomal activity regulates mitochondrial dynamics by affecting mitochondrial fission through interfering the metabolism of a micronutrient, vitamin B12. Vitamin B12 is exclusively obtained from diets in animals, including C. elegans and humans, and its uptake is mediated by the lysosome. We show that lysosomal dysfunction causes vitamin B12 deficiency that leads to reduction of methionine and S-adenosylmethionine to in turn increase mitochondrial biogenesis and fission. Our study provides insight on the interactions between mitochondrial function and micronutrient metabolism.
Keywords: mitochondrial dynamics, vitamin B12, interorganelle communication, vacuolar V-ATPase, methionine restriction
Abstract
Mitochondrial fission and fusion are highly regulated by energy demand and physiological conditions to control the production, activity, and movement of these organelles. Mitochondria are arrayed in a periodic pattern in Caenorhabditis elegans muscle, but this pattern is disrupted by mutations in the mitochondrial fission component dynamin DRP-1. Here we show that the dramatically disorganized mitochondria caused by a mitochondrial fission-defective dynamin mutation is strongly suppressed to a more periodic pattern by a second mutation in lysosomal biogenesis or acidification. Vitamin B12 is normally imported from the bacterial diet via lysosomal degradation of B12-binding proteins and transport of vitamin B12 to the mitochondrion and cytoplasm. We show that the lysosomal dysfunction induced by gene inactivations of lysosomal biogenesis or acidification factors causes vitamin B12 deficiency. Growth of the C. elegans dynamin mutant on an Escherichia coli strain with low vitamin B12 also strongly suppressed the mitochondrial fission defect. Of the two C. elegans enzymes that require B12, gene inactivation of methionine synthase suppressed the mitochondrial fission defect of a dynamin mutation. We show that lysosomal dysfunction induced mitochondrial biogenesis, which is mediated by vitamin B12 deficiency and methionine restriction. S-adenosylmethionine, the methyl donor of many methylation reactions, including histones, is synthesized from methionine by S-adenosylmethionine synthase; inactivation of the sams-1 S-adenosylmethionine synthase also suppresses the drp-1 fission defect, suggesting that vitamin B12 regulates mitochondrial biogenesis and then affects mitochondrial fission via chromatin pathways.
The mitochondrion generates energy, supplies biosynthetic intermediates to carbohydrate, amino acid, steroid, and fat metabolic, and anabolic pathways, and produces iron sulfur clusters. The various demands of metabolic intermediates and ATP in different cell types and under different conditions has led to a complex regulation of mitochondrial production, localization, and dynamics. Mitochondria are highly dynamic and undergo regulated fusion and fission to change their abundance and localization in particular cells or subcellular locations where energy needs are high, or to scavenge other mitochondria damaged by free radicals, for example. Fusion can sustain mitochondrial function under aging and other conditions that damage mitochondria because fused mitochondria can compensate for organelle damage by sharing functional components (including mitochondrial DNA, RNAs, and proteins); fission contributes to quality control by segregating damaged mitochondria for degradation and also facilitating new mitochondria generation. The balance between fission and fusion may be disrupted during aging as age-dependent neurodegenerative diseases disrupt mitochondrial dynamics (1–3).
Mitochondrial fusion and fission are mediated by evolutionally conserved dynamin family guanosine triphosphatases (GTPases). Caenorhabditis elegans DRP-1 (DRP, dynamin-related protein) is a cytosolic dynamin that mediates mitochondrial fission, during which DRP-1 is recruited to the mitochondrial outer membrane to form spirals that constrict and sever the organelle (4–6). Two other C. elegans GTPases, FZO-1 (Mitofusion 1, or MFN1 in mammals) and EAT-3 (Optic Atrophy 1, or OPA1 in mammals), mediate the opposite activity of the fission–fusion cycle of mitochondria: fusion of mitochondrial outer and inner membranes, respectively. Other receptors and mediators have been identified by genetic analysis in yeast of the pathways for mitochondrial fusion and fission, although many are not conserved in animals (1–3).
Here we show that a dynamin mutation that disrupts mitochondrial fission causes a dramatic tangling of the normally periodic pattern of mitochondria in C. elegans muscle cells, and that many gene inactivations or drugs that cause lysosomal dysfunction strongly suppress these mitochondrial defects to a highly periodic pattern. The lysosome is also a membrane-bound subcellular organelle. It serves as the scavenger of the cell to degrade components, and in defense against bacteria and viruses. As an intracellular digestive system, its function requires the acidification of the lysosomal lumen via a membrane-anchored proton pump, the vacuolar H+-ATPase (V-ATPase) (7), to in turn activate acid proteases. These acidified vesicles contain hydrolytic enzymes that can break down various macromolecules. The lysosome also delivers small-molecule nutrients by degradation of extracellular or intracellular substrates. For example, in mammals vitamin B12 is retrieved from the diet by the lysosome (8). We find that in C. elegans, dietary vitamin B12 deficiency, similar to lysosomal dysfunction, can also strongly suppress the mitochondrial dynamics defect of a mitochondrial fission mutant. Dietary B12 supplementation effectively reduces the mitochondrial abnormalities, developmental and locomotion defects in lysosomal defective animals. Vitamin B12, or cobalamin, is an essential animal micronutrient that is exclusively synthesized by bacteria and archaea (9, 10). Within animal cells, B12 is a cofactor for methionine synthase (MTR) that catalyzes the methylation of homocysteine to methionine in the cytosol (11); and methylmalonyl-CoA mutase (MCM), that catalyzes the methylmalonyl-CoA to succinyl-CoA reaction in the catabolism of branched chain amino acids, odd-chain fatty acids, and cholesterol in the mitochondrion (12). MTR and MCM are encoded by C. elegans metr-1 and mmcm-1, respectively. We found that inactivation of metr-1 but not mmcm-1 recapitulated the suppression of dynamin fission defects by B12 deficiency. Thus, vitamin B12 deficiency caused by lysosomal dysfunction or vitamin B12 dietary deficiency decreases the activity of MTR. This decrease in methionine synthesis induces mitochondrial biogenesis and fission activity, which suppresses the loss of dynamin-mediated mitochondrial fission.
Results
Inactivation of spe-5 Vacuolar ATPase Suppresses the Lethality and Mitochondrial Fission Defects of a drp-1/Mitochondrial Dynamin Mutant.
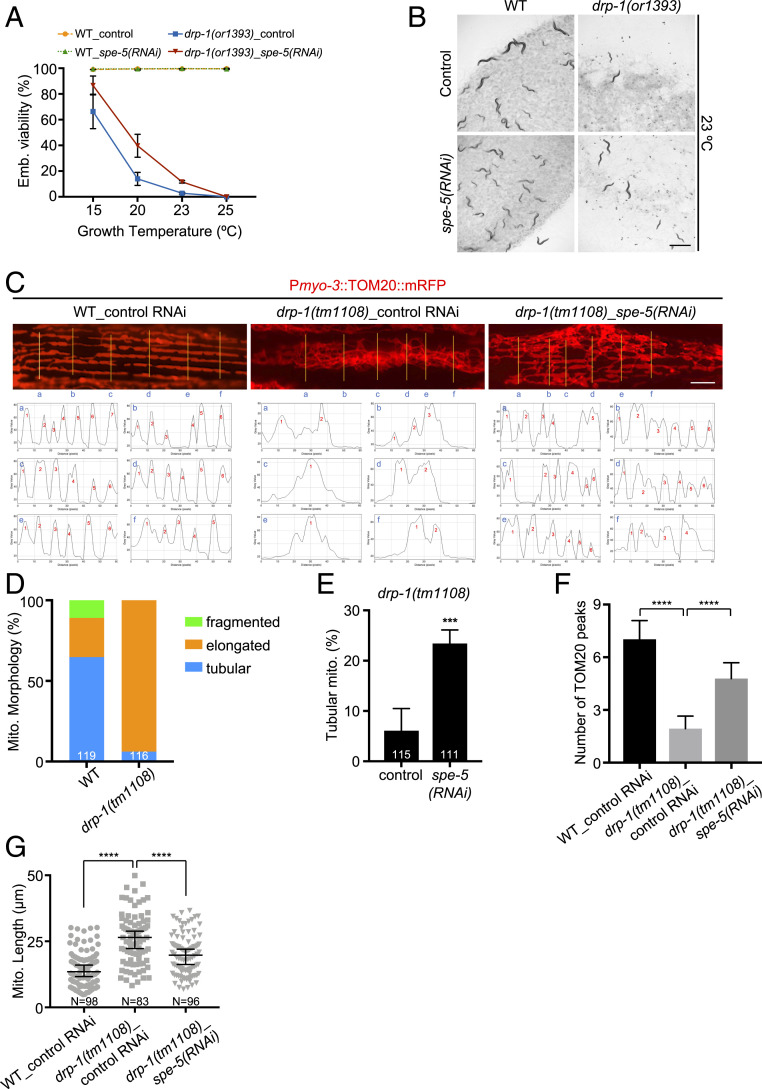
drp-1(or1393) is a temperature-sensitive allele with a conserved glycine to glutamic acid (G39E) substitution mutation in the N-terminal dynamin domain. This mutation causes high penetrance lethality at the nonpermissive temperature (13) (Fig. 1 A and B and SI Appendix, Fig. S1A). Because DRP-1 is a key mediator of mitochondrial fission, gene activities that regulate DRP-1 activity or the process of mitochondrial fission or fusion may affect the penetrance of drp-1(or1393) lethality. We performed an RNA interference (RNAi) screen to identify gene inactivations that suppress the fission defects of the drp-1(or1393) mutant (SI Appendix, Fig. S1B). We found similarity between the mitochondrial fission defects of a drp-1 mutant and a dynein heavy-chain gene dhc-1 mutant; gene inactivation of dhc-1 causes mitochondrial fission defects that disrupt the periodic pattern of muscle mitochondria as strongly as a drp-1 gene inactivation or mutation (SI Appendix, Fig. S1C), suggesting that DHC-1 also functions in mitochondrial fission. Thus, we could utilize a previous genome-scale RNAi screen for suppression of dhc-1 lethality that identified 49 gene inactivations (14); these genes were candidates to also suppress drp-1 lethality and mitochondrial fission defects. We tested the 49 dhc-1 suppressor-gene inactivations on drp-1(or1393) (SI Appendix, Fig. S1B). Of these 49 candidate genes, we found that inactivations of spe-5/vacuolar ATPase and C42C1.3 partially suppressed the lethality of drp-1(or1393) at 23 °C (SI Appendix, Fig. S1D). Inactivation of spe-5/vacuolar ATPase increased the viability of drp-1(or1393) at the nonpermissive temperature 23 °C by fivefold, but did not affect wild-type (Fig. 1 A and B). spe-5 inactivation also suppressed the lethality of another widely used loss-of-function allele, drp-1(tm1108) (SI Appendix, Fig. S1E). C42C1.3 is uncharacterized and worm-specific. We did not further study this hit.
Fig. 1.
Inactivation of the vacuolar ATPase spe-5 suppresses drp-1 lethality and mitochondrial fission defects. (A and B) Viability of wild-type (WT) and drp-1(or1393) in control versus spe-5(RNAi) at indicated temperatures. Mean ± SD from at least three biological replicates. n = 80∼120 for each data point. (Scale bar, 1 mm.) (C) Mitochondrial morphology of wild-type and drp-1(tm1108) in control versus spe-5(RNAi) in a single body wall muscle cell. Mitochondria are visualized by a mitochondrial outer membrane localized mRFP fusion protein. (Lower) Plot profiles of the cross sections indicated by yellow lines. Criterion for a peak: Peak to trough (both sides) > 20 gray value units (y units). (Scale bar, 5 μm.) (D) Percentage of mitochondria with fragmented, elongated or tubular morphology in wild-type and drp-1(tm1108) in body wall muscles. The total numbers of mitochondria observed are indicated at the bottom of each bar. (E) Percentage of mitochondria with tubular morphology in drp-1(tm1108) body wall muscles after indicated RNAi treatments. The total numbers of mitochondria observed are indicated at the bottom of each bar. Mean ± SD ***P < 0.001. (F) Peak numbers for the plot profiles of mitochondrial morphology in indicated animals. Mean ± SD ****P < 0.0001. n = 40∼50 for each group. (G) Mitochondrial lengths in body wall muscles in indicated animals. Mitochondrial lengths were calculated by MiNA toolset. Median with 95% CI Mann–Whitney U test. ****P < 0.0001. N indicates the sample size.
Mitochondria in body wall muscles can be visualized with mitochondrial red fluorescent protein (RFP) fusion proteins that form a highly periodic 2D network in wild-type with strong fluorescent peaks ∼0.4 μm wide and ∼13.5-μm long, where mitochondria are localized along sarcomere, and nonfluorescent troughs ∼1.3-μm wide that have no mitochondria (Fig. 1C). In wild-type, muscle mitochondria are localized in multiple parallel arrays along the sarcomeres of muscles, as visualized in one muscle cell in each panel of Fig. 1C by an RFP protein fused at the N terminus of the translocase of outer mitochondrial membrane TOMM-20 (Fig. 1 C and D and SI Appendix, Fig. S1F). The drp-1 mutation causes defects in mitochondrial fission so that mitochondria fuse more than normal into an elongated and very tangled, nonparallel morphology in more than 90% of muscle mitochondria (Fig. 1 C and D). Inactivation of spe-5 strongly suppressed the mitochondrial fission defects in drp-1(tm1108); ∼25% of mitochondria were restored to a highly parallel, tubular-like morphology that are more like wild-type (Fig. 1 C and E and SI Appendix, Fig. S2).
To more precisely quantitate a periodic or aperiodic mitochondrial network in suppressed mitochondrial fission mutants or the mitochondrial fission mutant, respectively, we profiled the pattern of fluorescence across six cross-sections per muscle cell image to display the periodic placement of mitochondria. While highly parallel, tubular-like mitochondria of wild-type C. elegans displayed periodic TOMM-20 peaks from the plot profiles of the cross-sections of the morphology images, the elongated and tangled mitochondria of drp-1 fission-defective animals showed far fewer TOMM-20 peaks from the plot profiles of mitochondrial morphology images (Fig. 1C and SI Appendix, Fig. S2). drp-1(tm1108) animals had ∼1.9 TOMM-20 peaks on average in each muscle cell image, whereas wild-type animals showed ∼7.0 TOMM-20 peaks on average. Inactivation of spe-5 increased the average number of TOMM-20 peaks to almost five peaks per cell in drp-1(tm1108), or about 2.5× more than drp-1(tm1108) (Fig. 1F), suggesting the mitochondrial fission defect in drp-1(tm1108) was ameliorated by spe-5 inactivation. An independent measure of the suppression of fission defects was to determine mitochondrial lengths. The mitochondria are highly tangled in drp-1(tm1108), which makes it impossible to measure the mitochondrial lengths from simple photographic images. We used a Mitochondrial Network Analysis (MiNA) toolset to generate a morphological skeleton for the mitochondrion to calculate the mitochondrial lengths by ImageJ (15). We found that increased mitochondrial lengths by DRP-1 dysfunction was suppressed by spe-5 inactivation (Fig. 1G). These results suggest the involvement of spe-5/vacuolar ATPase in mitochondrial fission regulation.
Mitochondrial function is tightly surveilled in eukaryotes. If mitochondrial dysfunction is detected, cells activate a series of responses, including the mitochondrial unfolded protein response (mtUPR). HSP-6 is one of the mitochondrial chaperones that mediates the mtUPR (16, 17). Phsp-6::GFP was strongly induced in drp-1(tm1108) (SI Appendix, Fig. S3A). If spe-5 inactivation suppressed the mitochondrial fission defects of drp-1, it should also alleviate the mtUPR induced by drp-1 deficiency. Indeed, gene inactivation of spe-5 reduced Phsp-6::GFP induction twofold in drp-1(tm1108) (SI Appendix, Fig. S3A). In contrast, spe-5 depletion did not decrease and in fact slightly increased Phsp-6::GFP expression in wild-type (SI Appendix, Fig. S3A). Consistent with the reporter gene, the induction of endogenous mitochondrial chaperone genes hsp-6 and hsp-60 by drp-1 depletion were suppressed by spe-5 inactivation. (SI Appendix, Fig. S3B). Consistent with the lower mitochondrial stress in the drp-1(tm1108);spe-5 (RNAi) strain, we found that inactivation of spe-5 increased mitochondrial membrane potential in drp-1(tm1108) (SI Appendix, Fig. S3C).
V-ATPase Dysfunction Suppresses drp-1 Lethality and Mitochondrial Fission Defects.
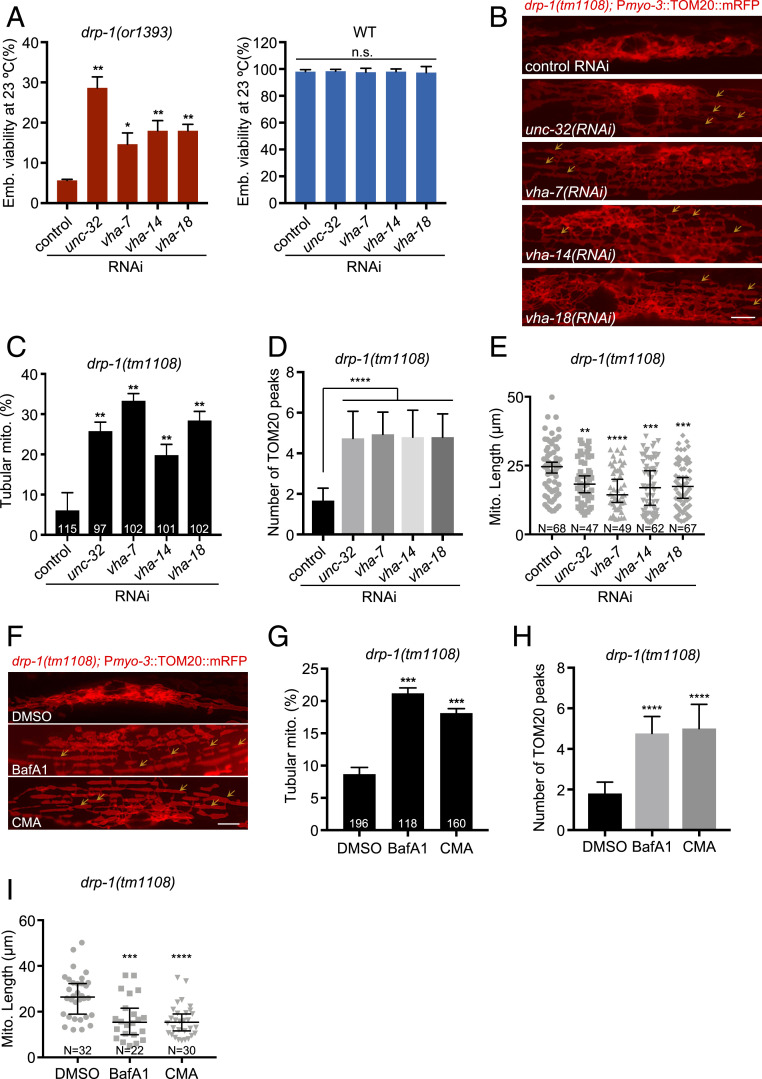
spe-5 encodes a subunit B of the vacuolar V-ATPase (18), suggesting that V-ATPase activity or the activity of the lysosome might regulate mitochondrial dynamics. Although spe-5 was identified based on its essential function in spermatogenesis, and is most strongly expressed in the male germline, spe-5 is also expressed in the fem-3 loss-of-function sex determination mutant which has nonmale soma and germline (18), and gene-expression analysis shows that spe-5 is expressed in particular neurons and intestine (19–21). Thus, SPE-5 is expressed more broadly and could act more generally in vacuolar ATPase function, in addition to its known role in spermatogenesis. Our findings that spe-5 gene inactivation suppresses the mitochondrial fission defects of drp-1(tm1108) muscle are consistent with a broader function than spermatogenesis (Fig. 1 C and E–G and SI Appendix, Fig. S2). To further test the involvement of the lysosome in mitochondrial fission and fusion regulation, we tested if inactivation of other lysosomal subunits can suppress the fission defects of drp-1. RNAi gene inactivation of multiple other V-ATPase subunit genes also suppressed drp-1 lethality (Fig. 2A). Consistent with the suppression of lethality, these V-ATPase gene inactivations restored the tangled nonparallel mitochondria of the drp-1(tm1108) mutant to a more parallel tubular morphology (Fig. 2 B and C). The suppression by V-ATPase gene inactivations was most obvious in the number of TOMM-20 peaks from the plot profiles of mitochondrial morphologies, which increased approximately threefold by V-ATPase gene inactivations in drp-1(tm1108) (Fig. 2D).
Fig. 2.
V-ATPase dysfunction suppresses drp-1 lethality and mitochondrial fission defects. (A) Viability of wild-type (WT) or drp-1(or1393) after V-ATPase gene inactivations at 23 °C. Mean ± SD from at least three biological replicates. n = 80∼120 for each data point. **P < 0.01, *P < 0.05. (B) Mitochondrial morphology in drp-1(tm1108) body wall muscles after indicated RNAi treatment. Arrows indicate some examples of nice parallel mitochondrial structure. (Scale bar, 5 μm.) (C) Percentage of mitochondria with tubular morphology in drp-1(tm1108) body wall muscles after indicated RNAi treatments. The total numbers of mitochondria observed are indicated at the bottom of each bar. Mean ± SD **P < 0.01. (D) Peak numbers for the plot profiles of mitochondrial morphology in indicated animals. Mean ± SD ****P < 0.0001. n = 40∼50 for each group. (E) Mitochondrial lengths in body wall muscles in indicated animals. Mitochondrial lengths were calculated by MiNA toolset. Median with 95% CI Mann–Whitney U test. ****P < 0.0001, ***P < 0.001, **P < 0.01. N indicates the sample size. (F) Mitochondrial morphology in drp-1(tm1108) body wall muscles with DMSO, BafA1, or CMA treatment. DMSO, solvent control; BafA1, bafilomycin A1; CMA, concanamycin A. Arrows indicate some examples of nice parallel mitochondrial structure. (Scale bar, 5 μm.) (G) Percentage of mitochondria with tubular morphology in drp-1(tm1108) body wall muscles after DMSO, BafA1, or CMA treatment. The total numbers of mitochondria observed are indicated at the bottom of each bar. Mean ± SD ***P < 0.001. (H) Peak numbers for the plot profiles of mitochondrial morphology in drp-1(tm1108) body wall muscles after DMSO, BafA1, or CMA treatment. Mean ± SD ****P < 0.0001. n = 40∼50 for each group. (I) Mitochondrial lengths in drp-1(tm1108) body wall muscles after DMSO, BafA1, or CMA treatment. Mitochondrial lengths were calculated by MiNA toolset. Median with 95% CI Mann–Whitney U test. ****P < 0.0001, ***P < 0.001. N indicates the sample size.
Consistent with this periodicity analysis, V-ATPase gene inactivations decreased mitochondrial lengths in drp-1(tm1108) (Fig. 2E). Drug inhibition of vacuolar V-ATPase activity with bafilomycin A1 (BafA1) (22) or concanamycin A (CMA) (23) also suppressed drp-1 lethality but had no effect on wild-type (SI Appendix, Fig. S4 A and B). BafA1 and CMA treatment suppressed the mitochondrial fission defects in drp-1(tm1108), increasing the percentage of mitochondria with a parallel tubular morphology by approximately twofold (Fig. 2 F and G). Similarly, BafA1 and CMA treatment increased the number and periodicity of TOMM-20 peaks from the plot profiles of mitochondrial morphologies (Fig. 2H) and decreased mitochondrial lengths in drp-1(tm1108) (Fig. 2I), suggesting mitochondrial fission defects in drp-1(tm1108) were suppressed by these V-ATPase activity inhibitors. Disrupting V-ATPase activity by gene inactivation or chemical inhibitor treatment decreased the induction of Phsp-6::GFP and endogenous mitochondrial chaperone genes hsp-6 and hsp-60 in drp-1(tm1108) (SI Appendix, Fig. S4 C–F), suggesting that the mitochondrial defects normally caused by a decrease in drp-1 activity are ameliorated by also inducing vacuolar V-ATPase dysfunction. V-ATPase dysfunction by gene inactivation or chemical inhibitor treatment increased mitochondrial membrane potential in drp-1(tm1108) (SI Appendix, Fig. S5), suggesting that mitochondrial functions were improved. Overall, these data show that the activity of the key proton pump of the lysosome, the vacuolar V-ATPase, regulates the mitochondrial overfusion defect caused by DRP-1 dynamin-mediated mitochondrial fission dysfunction.
Lysosomal Activity Affects Mitochondrial Dynamics.
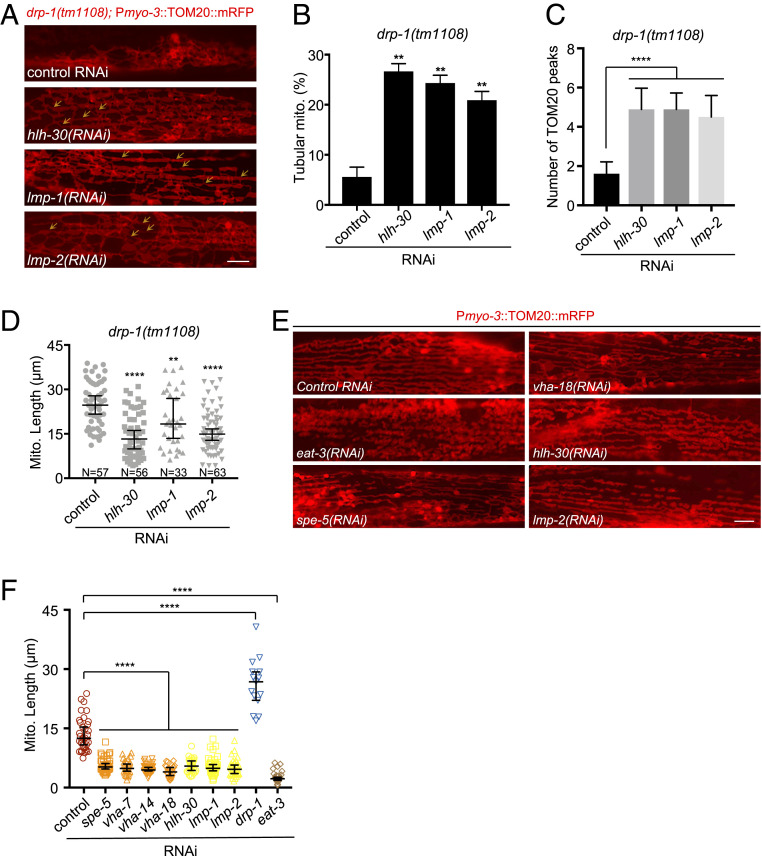
V-ATPase is the multisubunit proton pump that acidifies the lysosome. To further test the involvement of the lysosome in mitochondrial dynamics, we examined more factors that regulate lysosomal biogenesis or activity. HLH-30, the C. elegans ortholog of the transcription factor EB (TFEB), is a master regulator of lysosomal biogenesis (24). LMP-1 (LAMP-1 in mammals) and LMP-2 (LAMP-2 in mammals) (lysosomal-associated membrane protein 1 and 2, respectively) constitute ∼50% of the lysosomal membrane proteins and are required for lysosomal biogenesis and maintaining lysosome integrity (25, 26). We found that inactivations of hlh-30, lmp-1, and lmp-2 each suppressed the mitochondrial fission defects caused by drp-1 deficiency, increasing the percent of mitochondria with a parallel tubular morphology by four- to fivefold (Fig. 3 A and B). The numbers of TOMM-20 peaks from the plot profiles of mitochondrial morphologies in drp-1(tm1108) were increased by approximately threefold by inactivations of hlh-30, lmp-1, or lmp-2 (Fig. 3C). Consistent with the restoration of a periodic mitochondrial morphology by lysosomal dysfunction, these lysosomal gene inactivations decreased mitochondrial lengths in drp-1(tm1108) (Fig. 3D), suggesting that mitochondrial fission defect in drp-1(tm1108) was ameliorated by lysosomal dysfunction. Inactivations of hlh-30, lmp-1, and lmp-2 mildly suppressed the induction of Phsp-6::GFP and endogenous hsp-6 and hsp-60 expression in drp-1(tm1108) (SI Appendix, Fig. S6 A and B), suggesting that the mitochondrial homeostasis in drp-1(tm1108) were improved by disruption of lysosomal activity. Similarly, lysosomal dysfunction by inactivations of hlh-30, lmp-1, or lmp-2 increased mitochondrial membrane potential in drp-1(tm1108) (SI Appendix, Fig. S6C), suggesting improvement of mitochondrial function.
Fig. 3.
Lysosomal activity affects mitochondrial dynamics. (A) Mitochondrial morphology in drp-1(tm1108) body wall muscles after indicated RNAi treatment. Arrows indicate some examples of nice parallel mitochondrial structure. (Scale bar, 5 μm.) (B) Percentage of mitochondria with tubular morphology in drp-1(tm1108) body wall muscles after indicated RNAi treatments. Mean ± SD **P < 0.01. (C) Peak numbers for the plot profiles of mitochondrial morphology in indicated animals. Mean ± SD ****P < 0.0001. n = 40∼50 for each group. (D) Mitochondrial lengths in body wall muscles in indicated animals. Mitochondrial lengths were calculated by MiNA toolset. Median with 95% CI Mann–Whitney U test. ****P < 0.0001, **P < 0.01. N indicates the sample size. (E) Mitochondrial morphology in body wall muscles after indicated RNAi treatment. (Scale bar, 5 μm.) (F) Lengths of mitochondria after indicated gene inactivations by RNAi in N2 (wild-type). n = 15∼40, Median with 95% CI Mann–Whitney U test. ****P < 0.0001.
We speculated that lysosomal activity might act in mitochondrial fission, for example, by lysosomal-mediated mitophagy. However, we found that inactivations of the mitophagy pathway genes including the mitochondrial phosphatase and tensin-induced kinase 1 (PINK-1), the cytosolic E3 ubiquitin ligase Parkin (PDR-1), or the Atg6/Vps30/Beclin1 homolog BEC-1 did not suppress the lethality or mitochondrial fission defects in drp-1-defective animals (SI Appendix, Fig. S6 D–F). It makes sense that a mitophagy defect does not affect mitochondrial morphology in drp-1-depleted animals because DRP-1–dependent mitochondrial fission is a prerequisite for mitophagy (27). Thus, dysfunction in mitophagy could not rescue a drp-1 mutant defect in mitochondrial fission.
Because either inhibition of vacuolar V-ATPase activity or disruption of lysosomal biogenesis/integrity suppresses the mitochondrial fission defects of drp-1/dynamin mutant, a reasonable model is that lysosomal dysfunction promotes mitochondrial fission so as to rebalance the fusion–fission events in the drp-1 mutant. Indeed, we found that inactivations of V-ATPase subunit genes or lysosomal biogenesis/integrity genes caused a modest increase in mitochondrial fragmentation in an otherwise wild-type background (Fig. 3 E and F). However, the mitochondrial fragmentation caused by lysosomal dysfunction was less severe than that caused by disruption of eat-3, which encodes the homolog of the mitochondrial inner membrane fusion dynamin OPA1 (Fig. 3 E and F), suggesting that lysosome does not directly function in mitochondrial fission–fusion, or other regulatory pathways are also involved.
Vitamin B12 Deficiency Increases Mitochondrial Fission and Suppresses drp-1 Defects.
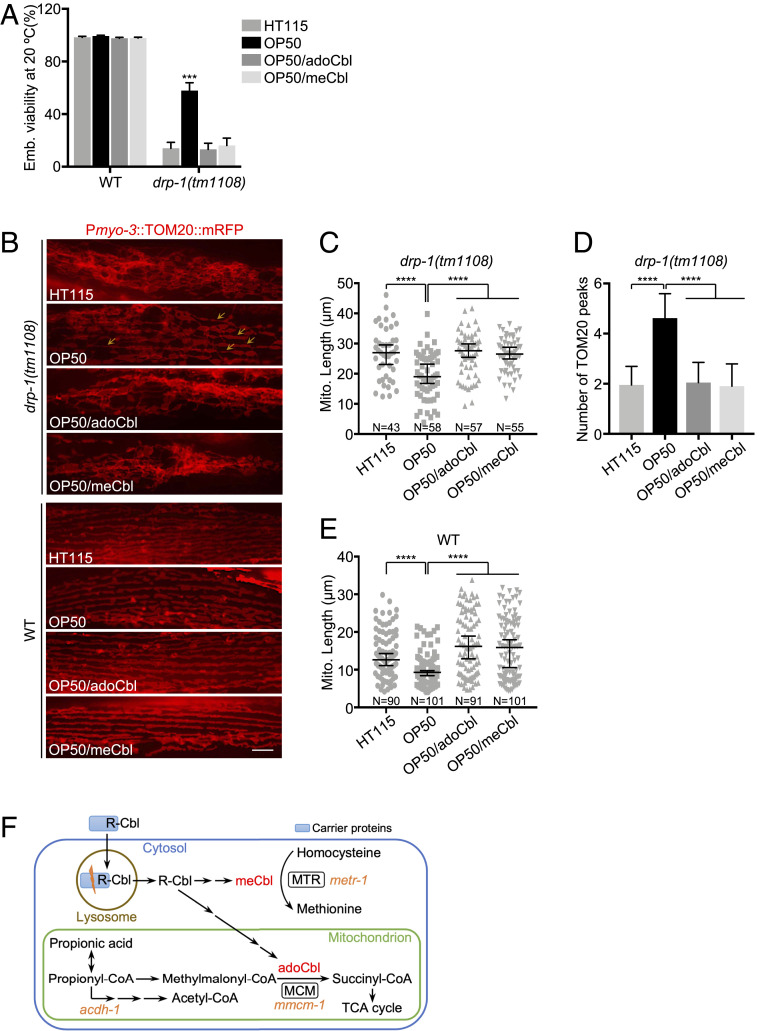
Escherichia coli bacteria are the standard laboratory food source for C. elegans. The most common C. elegans diet in the laboratory is OP50, an E. coli B-type strain (28), whereas an E. coli K12-derived RNase III-deficient strain HT115 is used for RNAi screens. Most C. elegans genetic analysis and day-to-day maintenance of strains is done on OP50 E. coli B, which is then switched to HT115 for feeding RNAi experiments. We noticed that the drp-1 loss-of-function (lf) mutant, drp-1(tm1108) [drp-1(lf) hereafter] grew faster and with more viability on E. coli OP50 than on E. coli HT115 (Fig. 4A and SI Appendix, Fig. S7A). drp-1(lf) grown on E. coli OP50 induced Phsp-6::GFP less than those grown on E. coli HT115 (SI Appendix, Fig. S7 B, Upper), suggesting that E. coli B supplies or lacks a key molecule that mediates the mitochondrial abnormalities in drp-1(lf). Consistent with the improved growth and viability on E. coli OP50, the highly tangled mitochondria in drp-1(lf) was suppressed by feeding on E. coli OP50 compared to E. coli HT115 (Fig. 4 B, Upper, and Fig. 4C). The TOMM-20 peaks from the plot profiles of mitochondrial morphologies in drp-1(lf) were increased by 2.4-fold by feeding on E. coli OP50 compared to E. coli HT115 (Fig. 4D), suggesting that mitochondrial fission defect in drp-1(lf) was ameliorated by feeding on E. coli OP50 diet compared to E. coli HT115.
Fig. 4.
Vitamin B12 deficiency increases mitochondrial fission and suppresses drp-1 defects. (A) Viability of wild-type (WT) or drp-1(tm1108) fed on E. coli HT115 (B12-proficient) or E. coli OP50 (B12-deficient) with or without B12 supplementation. adoCbl, adenosylcobalamin; meCbl, methylcobalamin. The final concentration of adoCbl or meCbl supplemented to the culture plates is 12.8 nM. Mean ± SD from at least three biological replicates. n = 90∼120 for each data point. ***P < 0.001. (B) Mitochondrial morphology in body wall muscles in drp-1(tm1108) (Upper) or wild-type (Lower) fed on E. coli HT115 or E. coli OP50 with or without B12 supplementation. Arrows indicate some examples of nice parallel mitochondrial structure in drp-1(tm1108) on E. coli OP50. (Scale bar, 5 μm.) (C and E) Mitochondrial lengths in body wall muscles in drp-1(tm1108) (C) or wild-type (E) animals fed on E. coli HT115 or E. coli OP50 with or without B12 supplementation. Mitochondrial lengths were calculated by MiNA toolset. Median with 95% CI Mann–Whitney U test. ****P < 0.0001. N indicates the sample size. (D) Peak numbers for the plot profiles of mitochondrial morphology in drp-1(tm1108) animals fed on E. coli HT115 or E. coli OP50 with or without B12 supplementation. Mean ± SD ****P < 0.0001. n = 40∼50 for each group. (F) Vitamin B12 metabolic pathways. C. elegans genes are in orange italics.
There are many genetic differences between E. coli B and E. coli K12, but one difference that has been noted is that E. coli B supplies low amounts of vitamin B12 to C. elegans (29–31). E. coli strains do not carry the biosynthetic operons for B12 synthesis but hundreds of disparate bacterial species synthesize vitamin B12 (32). In many animals, including C. elegans and humans, vitamin B12 is exclusively obtained from bacteria or diets and is necessary for a variety of methyl transfer reactions. E. coli strains in natural ecosystems obtain vitamin B12 from other organisms. In the laboratory, E. coli transports vitamin B12 from the growth media. However, the E. coli B-type strain has greatly reduced vitamin B12 transport and uptake from the growth medium, which is usually supplemented with peptone to provide vitamins and other nutrients essential for growth (33). E. coli K12 appears to take up higher levels of vitamin B12 from the bactopeptone growth media on worm plates to supply to the animals (29–31).
As a micronutrient, the endogenous vitamin B12 level is very low and thus hard to quantify directly and accurately. However, B12 deficiency compromises the MCM-mediated mitochondrial propionic acid breakdown pathway and activates a propionate shunt; the first step of this pathway is carried out by acyl-CoA dehydrogenase ACDH-1 (Fig. 4F) (34). Pacdh-1::GFP has been used as a reporter of cellular B12 supply (30, 31). C. elegans grown on E. coli OP50 strongly induced Pacdh-1::GFP, but this gene fusion is much more weakly expressed in animals feeding on E. coli HT115, a K12 strain (SI Appendix, Fig. S8 A, Upper and SI Appendix, Fig. S8C). This demonstrates the dietary B12 deficiency for animals on E. coli OP50 but not on E. coli HT115.
To test whether the dramatic difference in the mitochondrial defects of drp-1(lf) on the E. coli K12 HT115 (B12-proficient) and E. coli B OP50 (B12-deficient) diets is caused by the B12 supply rather than by other differences between these two E. coli strains, we supplemented the E. coli OP50 diet with vitamin B12. Vitamin B12 adenosylcobalamin (adoCbl) or methylcobalamin (meCbl) are interconvertible within animal cells (31, 35, 36), and we found that either form of B12 supplementation effectively complemented the dietary B12 deficiency on E. coli OP50, as indicated by Pacdh-1::GFP expression (SI Appendix, Fig. S8 A, Upper and SI Appendix, Fig. S8 C and D). Feeding on E. coli HT115 (B12-proficient) diet or on E. coli OP50 supplemented by B12 also suppressed the Pacdh-1::GFP induction, suggesting that possible differences in metabolizing B12 in these E. coli strains are not the cause of the different phenotypes we observed above. We found that either adoCbl or meCbl supplementation abolished the suppression of drp-1(lf) lethality by an E. coli OP50 diet (Fig. 4A). In addition, B12 supplementation suppressed the decrease of Phsp-6::GFP induction in drp-1(lf) grown on E. coli OP50 (SI Appendix, Fig. S7 B, Upper). Consistent with suppression of mtUPRs, we observed that the suppression of the drp-1(lf) mitochondrial disorganization and fission defect by feeding on E. coli OP50 was abrogated by B12 supplementation (Fig. 4 B–D), indicating that the E. coli OP50 diet-caused B12 deficiency leads to the better growth and mitochondrial improvement in drp-1(lf).
We tested the effects of B12 deficiency on mitochondrial dynamics. We found that wild-type animals grown on E. coli OP50 had mildly increased mitochondrial fragmentation, which could be complemented by exogenous B12 supplementation (Fig. 4 B, Lower), as has been observed (37). Moreover, Phsp-6::GFP was slightly induced on an E. coli OP50 diet, indicating mild mitochondrial dysfunction caused by B12 deficiency (SI Appendix, Fig. S7 B, Lower). In addition, synchronized animals grown on E. coli OP50 were shorter than those on B12 plus diets (SI Appendix, Fig. S7 B, Lower), suggesting development is mildly compromised under B12 deficiency, as noted previously (29, 31).
Lysosomal Retrieval of Vitamin B12.
In mammals, dietary vitamin B12 is first internalized by carrier proteins, including intrinsic factor and transcobalamin. The B12 bound to carrier protein is transported into the lysosome, where the carrier proteins are degraded by acid proteases to liberate B12 into the cytoplasm, where it is modified to adoCbl or meCbl (Fig. 4F) (8). The enzymes that use vitamin B12 as a cofactor are broadly conserved, MTR and MCM (Fig. 4F).
Lysosomal function is required for B12 supply in C. elegans. Treating animals with the V-ATPase inhibitors CMA or BafA1 induced Pacdh-1::GFP on an E. coli HT115 B12-proficient diet (SI Appendix, Fig. S9A). Lysosomal dysfunction by genetic mutations or inactivations of V-ATPase subunit genes or lysosomal biogenesis/integrity genes by RNAi (on E. coli HT115) also induced Pacdh-1::GFP (SI Appendix, Fig. S9 B and C). The Pacdh-1::GFP induction by V-ATPase inhibitors or lysosomal dysfunction on E. coli HT115 suggested cellular B12 deficiency, probably due to disruption of vitamin B12 release from the lysosome. The lmp-1(lf) mutation caused developmental delay and induction of Phsp-6::GFP on an E. coli OP50 B12-deficient diet (SI Appendix, Fig. S10A), but vitamin B12 supplementation rescued the developmental defect and hsp-6 induction on an E. coli OP50 diet (SI Appendix, Fig. S10A). The lmp-1(lf) lysosomal-defective animals grown on E. coli OP50 had more fragmented mitochondria and decreased average mitochondrial lengths in body wall muscles than those grown on E. coli HT115; dietary supplementation of B12 increased the mitochondrial connectivity and reduced the fragmentation defect in lmp-1(lf) grown on E. coli OP50 (SI Appendix, Fig. S10B).
In humans, lysosomal storage diseases (LSDs) caused by mutations in lysosomal proteins are associated with fragmented mitochondria and manifest as neurodegeneration. We found that lmp-1(lf) lysosomal-defective animals showed locomotion defects, including decreased movement velocity and increased movement tortuosity, especially on the B12-deficient E. coli OP50 diet (SI Appendix, Fig. S10 C and D). Dietary B12 supplementation greatly reduced the locomotion defects in lmp-1(lf) (SI Appendix, Fig. S10 C and D), consistent with the observation of improved mitochondrial health in lmp-1(lf) with dietary B12 supplementation (SI Appendix, Fig. S10 A and B).
Methionine Restriction Mediates B12 Deficiency-Caused Mitochondrial Fragmentation.
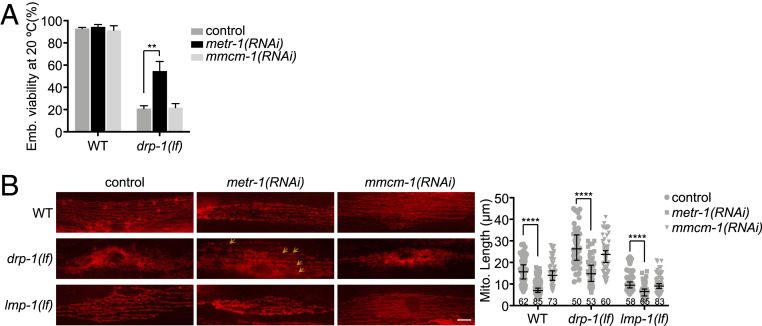
Vitamin B12 is a cofactor for cytosolic MTR and mitochondrial MCM that are encoded by C. elegans metr-1 and mmcm-1, respectively (Fig. 4F). We examined which enzyme mediates the B12 deficiency-caused mitochondrial fragmentation. Inactivation of metr-1 but not mmcm-1 completely recapitulated the mitochondrial phenotypes caused by B12 deficiency. In particular, inactivation of metr-1 suppressed the lethality and slow growth in drp-1(lf) but delayed development in lmp-1(lf), whereas RNAi of mmcm-1 did not affect growth or mitochondrial morphology of drp-1(lf) or lmp-1(lf) (Fig. 5A and SI Appendix, Fig. S11 A and B, Upper). Inactivation of metr-1 in a wild-type background caused modest mitochondrial fragmentation, similar to lysosomal dysfunction or dietary B12 deficiency, while RNAi of metr-1 reduced the mitochondrial fission defect in drp-1(lf) but enhanced the mitochondrial fragmentation defect in lmp-1(lf) fed on E. coli HT115 RNAi bacteria (Fig. 5B). Consistent with the mitochondrial status, Phsp-6::GFP induction is decreased in drp-1(lf) but increased in lmp-1(lf) by metr-1 gene inactivation, while RNAi of metr-1 mildly induced Phsp-6::GFP in an otherwise wild-type background (SI Appendix, Fig. S11 B, Lower). Gene inactivation of mmcm-1 did not affect mitochondrial morphology or hsp-6 expression more than control RNAi (Fig. 5B and SI Appendix, Fig. S11 B, Lower). Overall, these data indicate that B12 deficiency-caused mitochondrial fragmentation is mediated by the vitamin B12-requiring enzyme MTR.
Fig. 5.
Inactivation of MTR increases mitochondrial fission. (A) Viability of wild-type (WT) and drp-1(lf) grown on indicated RNAi clones. Mean ± SD from at least three biological replicates. n = 90∼120 for each data point. **P < 0.01. (B) Mitochondrial morphologies and lengths in body wall muscles in wild-type, lmp-1(lf) and drp-1(lf) grown on indicated RNAi clones. Arrows indicate some examples of nice parallel mitochondrial structure. Mitochondrial lengths were calculated by MiNA toolset. Median with 95% CI Mann–Whitney U test. ****P < 0.0001. N indicates the sample size. (Scale bar, 5 μm.)
MTR converts homocysteine to methionine that is required for the synthesis of methyl donor S-adenosylmethionine (SAM). Disruption of MTR enzymatic activity reduces methionine biosynthesis and leads to methionine restriction. We reasoned that the reduction of methionine mediated the increased mitochondrial fragmentation observed in lysosomal-defective or B12-deficient animals. Indeed, dietary supplementation of methionine greatly reduced the Phsp-6::GFP induction by metr-1 gene inactivation or dietary B12 deficiency in lmp-1(lf) (SI Appendix, Fig. S11 C, Upper). Methionine supplementation also reduced the mild hsp-6 induction by metr-1 RNAi or dietary B12-deficiency in wild-type animals (SI Appendix, Fig. S11 C, Lower). Methionine restriction leads to decrease of SAM levels. To test if low SAM causes the mitochondrial phenotypes observed in B12-deficient or lysosomal-deficient animals, we inactivated sams-1 that encodes the SAM synthetase and found that SAM reduction increased mitochondrial fission in wild-type as well as suppressed the mitochondrial fission defects in drp-1(lf) animals (SI Appendix, Fig. S12A), similar to that caused by B12 deficiency or lysosomal dysfunction. Inactivation of sams-1 mildly induced Phsp-6::GFP similar to metr-1 RNAi (SI Appendix, Fig. S12B). Mitochondrial fragmentation caused by sams-1 inactivation could not be rescued by exogenous B12 supplementation or methionine supplementation (SI Appendix, Fig. S12C). Combined, these results indicate that methionine restriction by B12 deficiency leads to reduction of SAM, which causes an increase of mitochondrial fission.
Lysosomal Dysfunction Augments Mitochondrial Biogenesis.
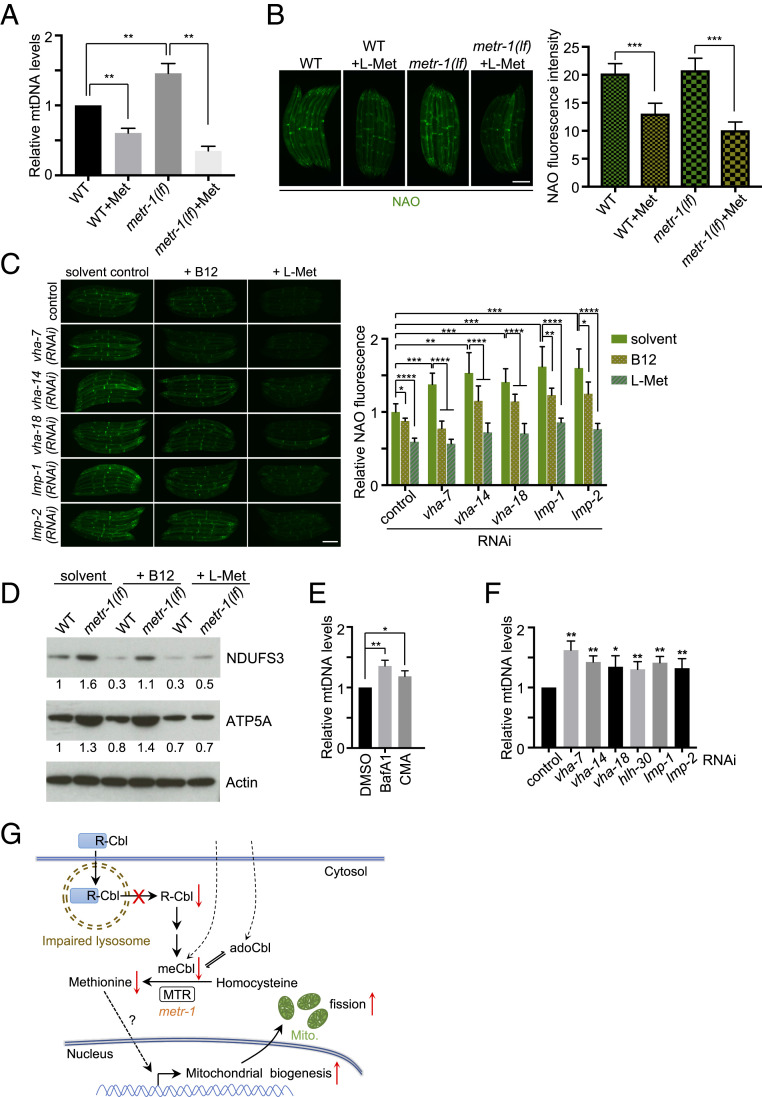
Methionine restriction increased mitochondrial DNA (mtDNA) copy number and mitochondrial density in some rat tissues (38, 39). We found that deletion of C. elegans metr-1 increased mtDNA level 1.5-fold compared to wild-type. Dietary methionine supplementation reduced mtDNA levels either in wild-type or metr-1(lf) (Fig. 6A). Nonyl acridine orange (NAO) is a fluorescent marker that is retained in mitochondria independent of the mitochondrial membrane potential, making it an ideal probe to measure mitochondrial mass (40). Mitochondrial mass measured by NAO is comparable between wild-type and metr-1(lf). The discrepancy might be due to differential growth rates, as metr-1(lf) showed mild developmental delay compared to wild-type. However, methionine supplementation significantly reduced the NAO-stained fluorescence in either wild-type or metr-1(lf), indicating that methionine supplementation suppresses the increase of mitochondrial mass (Fig. 6B).
Fig. 6.
Lysosomal dysfunction leads to increase mitochondrial mass. (A) Relative mitochondrial DNA levels in wild-type and metr-1(lf) animals with or without methionine supplementation. Mean ± SD from at least three biological replicates. **P < 0.01. (B and C) Animals with indicated treatment were stained with NAO. Mean ± SD ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05. (Scale bars, 0.2 mm.) (D) Immunoblots of lysates from animals with indicated treatment. Relative protein levels are indicated below the gel lanes. (E and F) Relative mitochondrial DNA levels in animals with indicated treatment. Mean ± SD from at least three biological replicates. **P < 0.01, *P < 0.05. (G) A model for lysosomal activity affects mitochondrial fission through vitamin B12 metabolism.
We reasoned that lysosomal dysfunction caused B12 deficiency to then cause methionine restriction, which could lead to an increase of mitochondrial mass. Indeed, depletions of vacuolar V-ATPase subunits or lysosomal membrane proteins LMP-1 and LMP-2 caused an ∼1.5-fold increase in mitochondrial mass as measured by NAO staining (Fig. 6C). Moreover, supplementation of B12 or methionine suppressed the increase of NAO staining by lysosomal gene inactivations (Fig. 6C), suggesting that lysosomal dysfunction induced mitochondrial mass is mediated by B12 deficiency and methionine restriction. Consistent with these data, detection of protein levels of conserved mitochondrial electron transport proteins using antibodies raised to highly conserved mammalian complex I component NDUFS3 (C. elegans ortholog NUO-2) and complex V component ATP5A (C. elegans ortholog NUO-2) (41) showed increased mitochondrial mass in metr-1(lf) (Fig. 6D), as well as in lysosomal mutants hlh-30(lf) and lmp-1(lf) (SI Appendix, Fig. S13A), or in animals with V-ATPase inhibitor treatment (SI Appendix, Fig. S13B). B12 supplementation reduced these oxidative phosphorylation protein levels in all lysosomal-defective animals as well as in wild-type, but not in metr-1(lf) (Fig. 6D and SI Appendix, Fig. S13 A and B). Supplementation of methionine readily reduced the protein levels of NDUFS3 and ATP5A in both wild-type and metr-1(lf) (Fig. 6D). These results indicate that lysosomal dysfunction increases mitochondrial mass mediated by B12 deficiency and methionine restriction. Moreover, inhibition of lysosomal function by V-ATPase inhibitor drugs or gene inactivations induced an increase in mtDNA copy number by ∼50% (Fig. 6 E and F). In addition, RNAi of hlh-30, lmp-1, lmp-2, and V-ATPase subunit genes increased the expression of several mitochondrial biogenesis genes (SI Appendix, Fig. S13C). These results suggest that lysosomal dysfunction increased mitochondrial biogenesis.
Mitophagy is selective macroautophagy that degrades damaged mitochondria. Lysosomal dysfunction could impair mitophagy and lead to accumulating damaged mitochondria to increase the mitochondrial abundance. However, we found that inactivations of the mitophagy pathway genes pink-1, pdr-1, or bec-1 did not increase, but in the cases of pdr-1 and bec-1 RNAi slightly decreased NAO staining in animals (SI Appendix, Fig. S13D). In addition, inactivations of mitophagy genes did not change mtDNA levels (SI Appendix, Fig. S13E), consistent with a previous observation (42). Moreover, mitophagy deficiency did not increase the expression of the tested mitochondrial biogenesis genes (SI Appendix, Fig. S13F). Taken together, these results indicate that lysosomal dysfunction triggers an increase in mitochondrial mass by increasing mitochondrial biogenesis rather than by decreasing mitophagy.
Discussion
Mitochondrial dynamics are regulated by the balance of fission and fusion; imbalances in fission and fusion cause mitochondrial abnormalities that lead to many diseases, including neurodegeneration. From a screen of gene inactivations that suppress the mitochondrial fission defects of a drp-1/dynamin mutant, we identified a vacuolar V-ATPase subunit gene of the lysosome, spe-5. Testing other gene inactivations that cause lysosomal dysfunction showed that increased mitochondrial fission is a consequence of compromised lysosomal function. In yeast, age-dependent acidity decline in the lysosome-like vacuole is accompanied by mitochondrial fragmentation (43). In humans, LSDs are associated with mitochondrial fragmentation (44). These observations suggest a coupling between lysosomal function and mitochondrial fission across phylogeny. Our work further reveals that lysosomal dysfunction causes B12 deficiency by interrupting the release of vitamin B12 from the lysosome. This B12 deficiency compromises the enzymatic activity of MTR, a cytoplasmic enzyme that uses meCbl as cofactor to convert homocysteine to methionine. Decreased methionine level, or methionine restriction, transduces the signal to the nucleus by yet to be identified factors to augment mitochondrial biogenesis, which may be coupled to an increase of fission activity for new mitochondrial generation (Fig. 6G).
The V-ATPase proton pump is a multisubunit molecular motor that includes three A and three B subunits, which form a hexamer that hydrolyzes ATP to in turn induce conformational changes in membrane-spanning protein subunits that pump protons across the membrane (45). C. elegans encodes two very closely related V-ATPase subunit B isoforms, SPE-5 and VHA-12, which have 88% identical protein sequences over the entire 500 amino acids of the protein but divergent DNA sequences, so that RNAi with a 1-kb vha-12 double-stranded RNA (dsRNA) is lethal, whereas RNAi with a 1-kb spe-5 dsRNA is viable (46, 47). But while spe-5 was the first indication of a mechanistic connection between lysosomal function and mitochondrial fission, other V-ATPase subunit gene inactivations also suppressed the drp-1 lethality and mitochondrial fission defects (Fig. 2 A–E), validating the mechanistic coupling between lysosomal function and mitochondrial fission. This suggests that SPE-5 and other V-ATPase gene subunits function in muscle to suppress the mitochondrial fission defects of drp-1(tm1108) (Fig. 1 C and E–G and SI Appendix, Fig. S2). Although VHA-12 is thought to be a more general isoform of subunit B, it is possible that SPE-5 may assemble as one or more of the three subunits B per vacuolar ATPase in some tissues, including in muscle where the suppression of drp-1 suggests it functions.
DRP-1/Drp1 dynamin mediates key cell biological steps in mitochondrial fission. However, our study shows that lysosomal dysfunction causes B12 deficiency and methionine restriction to increase mitochondrial fission, so that the fission–fusion cycles are rebalanced and the mitochondrial fission defect in a drp-1/dynamin mutant is strongly suppressed (SI Appendix, Fig. S13G). These data show that there are pathways to mitochondrial fission that are independent of DRP-1 in C. elegans. Interestingly, previous studies also indicated that Drp1-independent fission exists in mammalian cells as mitochondria still undergo fragmentation in Drp1−/− murine embryonic fibroblasts (48); and expression of a dominant-negative mutant of Drp1, Drp1(K38A), could not completely inhibit hFis1-induced mitochondrial fragmentation (49). DRP-1/Drp1-independent fission, perhaps also via methionine biosynthetic pathways, may also operate in mammals.
The lysosome is required for carrier protein-mediated B12 transport and uptake in humans (8, 50) (Fig. 4F). Our data show that lysosomal dysfunction also causes vitamin B12 deficiency in C. elegans (SI Appendix, Fig. S9). However, exogenous supplementation of metabolic active B12 reduced hsp-6 induction and mitochondrial fragmentation defects even in a lysosomal mutation background (SI Appendix, Fig. S10 A and B), suggesting that B12 might be also absorbed by a carrier protein-independent/lysosome-independent method. As a small molecule, B12 may be passively transported, which is inefficient, but in our experiments we used a high dosage (13 nM is relatively high for a micronutrient, such as vitamin B12). There is also passive transport for B12 in humans (51, 52).
B12 deficiency may disrupt the balance of mitochondrial dynamics under normal conditions (SI Appendix, Fig. S13G). Mild B12 deficiency affected mitochondrial homeostasis and increased susceptibility to pathogenic infections in animals (37). However, in contrast, in a drp-1/dynamin mutation background, B12 deficiency improves C. elegans mitochondrial health by rebalancing mitochondrial fission–fusion events (SI Appendix, Fig. S13G), demonstrating the elaborate regulation of mitochondrial dynamics.
B12 deficiency is common in the human population, particularly the elderly. Neurological disorders are significant clinical manifestations of B12 deficiency in humans (53–55). In yeast, lysosomal-like vacuolar acidity declines and mitochondria become gradually fragmented during aging (43). Increased mitochondrial fragmentation is widespread in LSDs as well as age-related neurodegeneration, such as Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease (56–59). Mutations affecting lysosomal function cause a class of metabolic diseases, or LSDs, which are associated with neurologic symptoms and neurodegeneration (44). Mitochondrial abnormalities are a common feature for most LSDs (44), suggesting cross-talk between the two organelles. Based on our findings, we speculate that B12 deficiency caused by declines of lysosomal function may cause mitochondrial fragmentation during the aging process to in turn cause age-related neurological disorders and neurodegeneration. Dietary supplementation of B12 suppressed this mitochondrial fragmentation defect and the locomotion movement abnormalities in lysosomal-defective animals (SI Appendix, Fig. S10), suggesting the potential for vitamin B12 as a supplementary treatment in age-related neurological disorders and LSDs associated with mitochondrial fragmentation. Interference with the mitochondrial dynamics to rebalance the fission–fusion events could be a new therapeutic strategy for diseases with mitochondrial dysregulation.
Materials and Methods
C. elegans Strains and Maintenance.
C. elegans were grown on standard nematode growth medium (NGM) at 20 °C, or other temperatures when indicated. To synchronize animals, embryos were harvested from gravid hermaphrodites treated with bleach solution containing 0.5 M NaOH, allowed to hatch in M9 buffer overnight. The following strains were used in this study: N2 Bristol: wild-type, CU6372: drp-1(tm1108)IV, RB2375: lmp-1(ok3228)X, JIN1375: hlh-30(tm1978)IV, RB755: metr-1(ok521)II, RB1434: mmcm-1(ok1637)III, PS6192: syIs243[Pmyo-3::TOM20::mRFP], SJ4100: zcIs13[Phsp-6::GFP]V, VL749: wwIs24[Pacdh-1::GFP]. drp-1(or1393)IV was generated by out-crossing EU2706: ruIs32[Ppie-1::GFP::H2B]III; drp-1(or1393)IV provided by B. Bowerman (University of Oregon, Eugene, Oregon) into N2. drp-1(lf); Pmyo-3::TOM20::mRFP was generated by crossing drp-1(tm1108) into Pmyo-3::TOM20::mRFP. drp-1(lf); Phsp-6::GFP was generated by crossing drp-1(tm1108) into Phsp-6::GFP. lmp-1(lf); Pmyo-3::TOM20::mRFP was generated by crossing lmp-1(ok3228) into Pmyo-3::TOM20::mRFP. lmp-1(lf); Phsp-6::GFP was generated by crossing lmp-1(ok3228) into Phsp-6::GFP.
RNAi.
E. coli HT115 bacteria expressing targeted dsRNAs were cultured in LB containing 50 μg/mL ampicillin at 37 °C overnight and seeded 150 μL per 60 mm or 100 μL per well of a 24-well RNAi plate containing 5 mM isfopropyl-β-d-thiogalactopyranoside (IPTG) and 100 μg/mL carbenicillin. Plates were allowed to dry and induce at room temperature overnight. Synchronized animals were transferred to the RNAi plates and cultured at 20 °C or at indicated temperatures depending on the experiments. For all RNAi experiments, an empty L4440 vector was used as the control.
For the RNAi screen for drp-1 suppressors, ∼15 synchronized L1 drp-1(or1393) were dropped on each well on 24-well RNAi plates and kept at 23 °C for 3 d when the animals laid plenty of eggs. This was continued in the culture at 23 °C for another 48 h and we then scored the hatched progeny on each well.
Viability Assays.
Synchronized L1 animals were grown on indicated RNAi plates at 20 °C, 23 °C, or 25 °C for 3 d, or at 15 °C for 5 d when they reached adults. Approximately 20 gravid adult animals were transferred to new RNAi plates and allowed to lay eggs for 2 h and then removed. Egg numbers were counted. The eggs continued to keep at corresponding temperatures for another 48 h. Hatched progeny was then scored for viability. Brightfield images representing the viability were obtained by a Zeiss AxioZoom V16 microscope.
Mitochondrial Morphology and Length.
After the indicated treatment, synchronized adult animals containing the Pmyo-3::TOM20::mRFP transgene were mounted on 2% agarose pads for microscopic imaging. Each treatment scored mitochondrial morphology in 8∼10 body wall muscles in the middle of the worm body and 10∼15 animals in total. Mitochondria exhibiting tubular structure in most parts of a single muscle cell were defined as ones with “tubular morphology,” while those exhibiting elongated structure in most parts of a muscle was defined as ones with “elongated morphology”; “fragmented morphology” indicated that mitochondria in a muscle cell displayed many fragmented pieces. Fluorescence images representing mitochondrial morphology were obtained by a Zeiss Axio Image Z1 microscope.
Mitochondrial length was quantified by ImageJ software as described previously (15) with modification. Briefly, the images were preprocessed (Unsharp mask → Enhance local contrast → Median), then converted to binary and generated a morphological skeleton for calculating the “branch lengths” as the mitochondrial lengths. Statistical analyses with Mann–Whitney U test were performed with Prism 7.
Induction of GFP Reporters.
Synchronized L1 animals were grown on indicated culture plates at 20 °C for 3 to 4 d. We dropped ∼2 μL of 5 mM levamisole (Sigma) onto a new NGM plate and transferred several animals into it. Images were captured by a Zeiss AxioZoom V16 microscope with the same exposure time and magnification. Fluorescence intensities or pixel intensities were measured by ImageJ software. Statistical analyses were done from at least three biological replicates.
Chemical Treatment.
To inhibit V-ATPase activity, synchronized L1 animals were grown at 20 °C for 2 d until they reached late L4s. Next, 100 μL M9 buffer containing 10 μg/mL BafA1 (LC Laboratories) dissolved in DMSO or 1 μg/mL CMA (AG Scientific) dissolved in DMSO was directly added onto the bacterial lawn. Plates were air-dried and kept at 20 °C for another 48 h.
For B12 supplementation, animals were directly grown on culture plates containing 12.8 nM adoCbl or meCbl (Sigma). For methionine supplementation, animals were grown on culture plates containing 13.4 mM l-methionine (Sigma).
For propionic acid and homocysteine hypersensitivity assays, ∼100 embryos for each genetic mutation were transferred to culture plates containing solvent control, 100 mM propionic acid (Sigma), or 15 mM homocysteine (Sigma). Two days later we scored the hatched animals that had developed into L3 or later stages. Statistical analyses were done from at least three biological replicates.
NAO and TMRE Staining.
Synchronized L1 N2 were grown on indicated plates at 20 °C for 2 d until they reached late L4. Next, 5 μM NAO (acridine Orange 10-nonyl bromide; Life Technologies) or 1 μM TMRE (tetramethylrhodamine, ethyl ester; Life Technologies) in 100 μL M9 buffer was then directly added onto the bacterial lawn. Plates were air dried and kept at 20 °C overnight. Animals were then picked and transferred to new RNAi plates without NAO or TMRE to grow for 8 h to clean any nonuptake fluorescent dye. Images were then obtained with a Zeiss AxioZoom V16 microscope using the same exposure time and magnification and then processed by ImageJ software.
mtDNA Levels.
Synchronized L1 N2 were grown on standard NGM plates seeded with E. coli OP50 at 20 °C for 2 d until they reached late L4s, then incubated with desired drugs or transferred to the desired RNAi plates and kept at 20 °C for another 48 h. Animals were then harvested and total DNA was extracted and quantified using quantitative real-time PCR, as described previously (60). Primers were designed to quantify nd1 (for mtDNA) and act-3 (for genomic DNA) as previously described (61), and mtDNA levels were then normalized to genomic DNA.
RNA Isolation and Quantitative RT-PCR.
Synchronized L1 animals were grown on standard NGM plates seeded with E. coli OP50 at 20 °C for 2 d until they reached late L4s, then incubated with desired drugs or transferred to the desired RNAi plates and kept at 20 °C for another 48 h. Animals were then harvested and total RNA was extracted by RNeasy Plus Universal Kit (Qiagen) following the manufacturer’s instructions. One microgram of total RNA was then used for first-strand cDNA synthesis using QuantiTect Reverse Transcription Kit (Qiagen). Quantitative real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad). Quantification of transcripts was normalized to act-4 (62) and relative mRNA levels were calculated by ΔΔCt. Primers for desired targets were designed to span exon/exon boundaries.
atp-5: 5′-ACGTCTTCCGTGAACTGGTCGAAG-3′ and 5′-GCAAAGTCGATCTTGGGAAGATCG-3′; cox-4: 5′-TCGTCGGATACGGAGCAAATG-3′ and 5′-TCGAATTCGGACAGCGTTTGAC-3′; hmg-5: 5′-GAAGTTGTCTGGAGCTGGAATG-3′ and 5′-GGAGGACGACATGGTATTCATC-3′; gas-1: 5′-CAATTGAGGCTCCAAAGGGAGAG-3′ and 5′-GACATGTAGCACACATCGTGGATG-3′; gst-4: 5′-GTCTATCACAAGATACTTGGCAAG-3′ and 5′-ATCACGGGCTGGTTCAACAACTTC-3′; gst-10: 5′-TGCAGACTGGAGCAATTATGCGTC-3′ and 5′-TCCCTCGTCGTAGTAAATGTAACG-3′.
Western Blotting and Antibodies.
Synchronized L1 animals were grown on plates with indicated treatment at 20 °C for 3 d. Animals were then harvested and washed with M9 buffer for 2∼3 times, then resuspended with 1× NuPAGE LDS Sample Buffer (Invitrogen) containing 5% β-mercaptoethanol and heated at 70 °C for 10 min. Lysates were loaded onto NuPAGE 4 to 12% Bis-Tris Protein Gels (Invitrogen) and transferred onto Nitrocellulose membrane (Invitrogen). The membrane was then blocked with 5% nonfat milk and probed with designated primary and secondary antibodies.
The primary antibodies used in this study included anti-NDUFS3 (Abcam, ab14711), anti-ATP5A (Abcam, ab14748), and anti-Actin (Abcam, ab179467).
Statistical Analysis.
No statistical methods were used to predetermine sample sizes. Statistical tests with appropriate underlying assumptions on data distribution and variance characteristics were used. All samples were randomly selected and from at least three biological replicates when applied. Data were analyzed with one-factor ANOVA (for mitochondrial morphology experiments) and two-tailed unpaired Student’s t tests (for the other experiments). All statistical analyses and graphing were performed with Prism 7 (GraphPad software). Statistically significant differences are indicated as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and n.s. as not significant.
Supplementary Material
Acknowledgments
We thank Bruce Bowerman for providing the ruIs32[Ppie-1::GFP::H2B]III; drp-1(or1393)IV strain. Other strains were provided by the Caenorhabditis Genetics Center funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank G.R. laboratory members for discussions and comments. This work was supported by an NIH Grant AG16636 (to G.R.). W.W. is supported by a Human Frontier Science Program Postdoctoral Fellowship.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008021117/-/DCSupplemental.
Data Availability.
All data supporting this study are available in the main text figures and supplementary information. The materials involved in this study are available upon request to the corresponding author (G.R.).
References
- 1.Westermann B., Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11, 872–884 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Chen H., Chan D. C., Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum. Mol. Genet. 18, R169–R176 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Youle R. J., van der Bliek A. M., Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smirnova E., Shurland D. L., Ryazantsev S. N., van der Bliek A. M., A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143, 351–358 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Labrousse A. M., Zappaterra M. D., Rube D. A., van der Bliek A. M., C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815–826 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Otsuga D. et al., The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143, 333–349 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohkuma S., Moriyama Y., Takano T., Identification and characterization of a proton pump on lysosomes by fluorescein-isothiocyanate-dextran fluorescence. Proc. Natl. Acad. Sci. U.S.A. 79, 2758–2762 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen M. J., Rasmussen M. R., Andersen C. B. F., Nexø E., Moestrup S. K., Vitamin B12 transport from food to the body’s cells—A sophisticated, multistep pathway. Nat. Rev. Gastroenterol. Hepatol. 9, 345–354 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Roth J. R., Lawrence J. G., Bobik T. A., Cobalamin (coenzyme B12): Synthesis and biological significance. Annu. Rev. Microbiol. 50, 137–181 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S., Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278, 41148–41159 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Koutmos M., Datta S., Pattridge K. A., Smith J. L., Matthews R. G., Insights into the reactivation of cobalamin-dependent methionine synthase. Proc. Natl. Acad. Sci. U.S.A. 106, 18527–18532 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury S., Banerjee R., Role of the dimethylbenzimidazole tail in the reaction catalyzed by coenzyme B12-dependent methylmalonyl-CoA mutase. Biochemistry 38, 15287–15294 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Lowry J. et al., High-throughput cloning of temperature-sensitive Caenorhabditis elegans mutants with adult syncytial germline membrane architecture defects. G3 (Bethesda) 5, 2241–2255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Rourke S. M., Dorfman M. D., Carter J. C., Bowerman B., Dynein modifiers in C. elegans: Light chains suppress conditional heavy chain mutants. PLoS Genet. 3, e128 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valente A. J., Maddalena L. A., Robb E. L., Moradi F., Stuart J. A., A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 119, 315–326 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Yoneda T. et al., Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 117, 4055–4066 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Samuel B. S., Breen P. C., Ruvkun G., Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508, 406–410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gleason E. J. et al., Developmental genetics of secretory vesicle acidification during Caenorhabditis elegans spermatogenesis. Genetics 191, 477–491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oranth A. et al., Food sensation modulates locomotion by dopamine and neuropeptide signaling in a distributed neuronal network. Neuron 100, 1414–1428.e10 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Topalidou I., Chalfie M., Shared gene expression in distinct neurons expressing common selector genes. Proc. Natl. Acad. Sci. U.S.A. 108, 19258–19263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jean Beltran P. M., Federspiel J. D., Sheng X., Cristea I. M., Proteomics and integrative omic approaches for understanding host-pathogen interactions and infectious diseases. Mol. Syst. Biol. 13, 922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y., Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266, 17707–17712 (1991). [PubMed] [Google Scholar]
- 23.Kataoka T. et al., Acidification is essential for maintaining the structure and function of lytic granules of CTL. Effect of concanamycin A, an inhibitor of vacuolar type H(+)-ATPase, on CTL-mediated cytotoxicity. J. Immunol. 153, 3938–3947 (1994). [PubMed] [Google Scholar]
- 24.Lapierre L. R. et al., The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4, 2267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuda M., Lysosomal membrane glycoproteins. Structure, biosynthesis, and intracellular trafficking. J. Biol. Chem. 266, 21327–21330 (1991). [PubMed] [Google Scholar]
- 26.Eskelinen E.-L., Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Aspects Med. 27, 495–502 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Twig G. et al., Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bito T., Matsunaga Y., Yabuta Y., Kawano T., Watanabe F., Vitamin B12 deficiency in Caenorhabditis elegans results in loss of fertility, extended life cycle, and reduced lifespan. FEBS Open Bio 3, 112–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacNeil L. T., Watson E., Arda H. E., Zhu L. J., Walhout A. J. M., Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153, 240–252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watson E. et al., Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell 156, 759–770 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence J. G., Roth J. R., Evolution of coenzyme B12 synthesis among enteric bacteria: Evidence for loss and reacquisition of a multigene complex. Genetics 142, 11–24 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Girolamo P. M., Kadner R. J., Bradbeer C., Isolation of vitamin B 12 transport mutants of Escherichia coli. J. Bacteriol. 106, 751–757 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson E. et al., Metabolic network rewiring of propionate flux compensates vitamin B12 deficiency in C. elegans. eLife 5, 2807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bito T., Watanabe F., Biochemistry, function, and deficiency of vitamin B12 in Caenorhabditis elegans. Exp. Biol. Med. (Maywood) 241, 1663–1668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yilmaz L. S., Walhout A. J. M., Worms, bacteria, and micronutrients: An elegant model of our diet. Trends Genet. 30, 496–503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Revtovich A. V., Lee R., Kirienko N. V., Interplay between mitochondria and diet mediates pathogen and stress resistance in Caenorhabditis elegans. PLoS Genet. 15, e1008011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrone C. E., Mattocks D. A. L., Jarvis-Morar M., Plummer J. D., Orentreich N., Methionine restriction effects on mitochondrial biogenesis and aerobic capacity in white adipose tissue, liver, and skeletal muscle of F344 rats. Metabolism 59, 1000–1011 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Hasek B. E. et al., Remodeling the integration of lipid metabolism between liver and adipose tissue by dietary methionine restriction in rats. Diabetes 62, 3362–3372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maftah A., Petit J. M., Ratinaud M. H., Julien R., 10-N nonyl-acridine orange: A fluorescent probe which stains mitochondria independently of their energetic state. Biochem. Biophys. Res. Commun. 164, 185–190 (1989). [DOI] [PubMed] [Google Scholar]
- 41.Nargund A. M., Fiorese C. J., Pellegrino M. W., Deng P., Haynes C. M., Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol. Cell 58, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palikaras K., Lionaki E., Tavernarakis N., Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Hughes A. L., Gottschling D. E., An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plotegher N., Duchen M. R., Mitochondrial dysfunction and neurodegeneration in lysosomal storage disorders. Trends Mol. Med. 23, 116–134 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Lee S.-K., Li W., Ryu S.-E., Rhim T., Ahnn J., Vacuolar (H+)-ATPases in Caenorhabditis elegans: What can we learn about giant H+ pumps from tiny worms? Biochim. Biophys. Acta 1797, 1687–1695 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Kamath R. S. et al., Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Knight A. J., Behm C. A., Minireview: The role of the vacuolar ATPase in nematodes. Exp. Parasitol. 132, 47–55 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Ishihara N. et al., Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958–966 (2009). [DOI] [PubMed] [Google Scholar]
- 49.James D. I., Parone P. A., Mattenberger Y., Martinou J.-C., hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 278, 36373–36379 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Kozyraki R., Cases O., Vitamin B12 absorption: Mammalian physiology and acquired and inherited disorders. Biochimie 95, 1002–1007 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Doscherholmen A., Hagen P. S., A dual mechanism of vitamin B12 plasma absorption. J. Clin. Invest. 36, 1551–1557 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herbert V., Streiff R. R., Sullivan L. W., Notes ON vitamin B12 absorption; autoimmunity and childhood pernicious anemia; relation OF intrinsic factor to blood group substance. Medicine (Baltimore) 43, 679–687 (1964). [DOI] [PubMed] [Google Scholar]
- 53.Allen L. H., How common is vitamin B-12 deficiency? Am. J. Clin. Nutr. 89, 693S–696S (2009). [DOI] [PubMed] [Google Scholar]
- 54.Gröber U., Kisters K., Schmidt J., Neuroenhancement with vitamin B12-underestimated neurological significance. Nutrients 5, 5031–5045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dali-Youcef N., Andrès E., An update on cobalamin deficiency in adults. QJM 102, 17–28 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Wang X. et al., Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 19318–19323 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho D.-H. et al., S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science 324, 102–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knott A. B., Perkins G., Schwarzenbacher R., Bossy-Wetzel E., Mitochondrial fragmentation in neurodegeneration. Nat. Rev. Neurosci. 9, 505–518 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Exner N. et al., Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 27, 12413–12418 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirienko N. V., Ausubel F. M., Ruvkun G., Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112, 1821–1826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bratic I. et al., Mitochondrial DNA level, but not active replicase, is essential for Caenorhabditis elegans development. Nucleic Acids Res. 37, 1817–1828 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedland A. E. et al., Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10, 741–743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting this study are available in the main text figures and supplementary information. The materials involved in this study are available upon request to the corresponding author (G.R.).