Significance
Human cytomegalovirus (HCMV) is a β-herpesvirus that establishes life-long infection, leaving the risk of persistent reactivation and disease. Inhibition of viral entry into susceptible cells is an attractive strategy to prevent infection. MCMV is a surrogate to model host-pathogen interactions; however, the precise mechanism of MCMV entry remains incomplete. Here, we utilized a pronecroptotic form of MCMV as a strategy to perform a genome-wide forward genetic screen to identify host factors essential for infection. We characterize VEGF receptor Nrp-1 as a novel mediator of MCMV infection required for viral gene expression, replication, and necroptosis induction. Our observations define Nrp-1 as a key cellular factor that facilitates MCMV infection and may be an entry receptor for the virus.
Keywords: cytomegolovirus, necroptosis, neuropilin, CRISPR
Abstract
Herpesviruses are ubiquitous human pathogens that cause a wide range of health complications. Currently, there is an incomplete understanding of cellular factors that contribute to herpesvirus infection. Here, we report an antiviral necroptosis-based genetic screen to identify novel host cell factors required for infection with the β-herpesvirus murine cytomegalovirus (MCMV). Our genome-wide CRISPR-based screen harnessed the capacity of herpesvirus mutants that trigger antiviral necroptotic cell death upon early viral gene expression. Vascular endothelial growth factor (VEGF) and semaphorin-binding receptor Neuropilin-1 (Nrp-1) emerge as crucial determinants of MCMV infection. We find that elimination of Nrp-1 impairs early viral gene expression and reduces infection rates in endothelial cells, fibroblasts, and macrophages. Furthermore, preincubation of virus with soluble Nrp-1 dramatically inhibits infection by reducing virus attachment. Thus, Nrp-1 is a key determinant of the initial phase of MCMV infection.
Herpesviruses are a family of double-stranded DNA viruses which vest heavily in sustaining cell viability until they produce progeny, disseminate, and transmit to new hosts. Large DNA viruses have evolved to encode a range of gene products that suppress innate cell death pathways to prevent the elimination of infected cells prior to viral replication (1–3). Apoptosis is generally one of the first lines of defense activated by the host in response to viral infection. Necroptosis is unleashed as an alternative to apoptosis in settings where proapoptotic caspase-8 activity is compromised by virus-encoded caspase-8 inhibitors, contributing to the emergence of necroptosis as a host defense pathway in mammals (4, 5). Necroptosis is mediated by receptor-interacting protein kinase (RIPK) 3-dependent phosphorylation of the pseudo-kinase mixed–lineage kinase domainlike (MLKL) which oligomerizes to create pores in the cell membrane, resulting in rupture (6).
MCMV provided the first evidence for virus-induced necroptosis independent of tumor necrosis factor (TNF) (7). The M45-encoded viral inhibitor of RIP activation (vIRA) prevents induction of this pathway by blocking Z-NA-binding protein 1 (ZBP1; also known as DAI or DLM-1)-dependent oligomerization and activation of RIPK3 (8, 9). ICP6, the large subunit of the ribonucleotide reductase (R1) protein of herpes simplex virus (HSV)-1, acts similarly to vIRA. However, while ICP-6 blocks RIP homotypic interaction motif (RHIM)-dependent pronecrotic signal transduction in cells from its natural human host, it triggers necroptosis in mouse cells (10–13). Our previous studies also showed ICP6 RHIM mutant HSV-1 shares a mechanism to trigger virus-induced ZBP1–RIPK3/MLKL-dependent necroptosis similar to M45 RHIM mutant MCMV (Fig. 1A) (14). Despite significant information on these processes, a comprehensive understanding of host factors involved in herpesvirus infection remains incomplete.
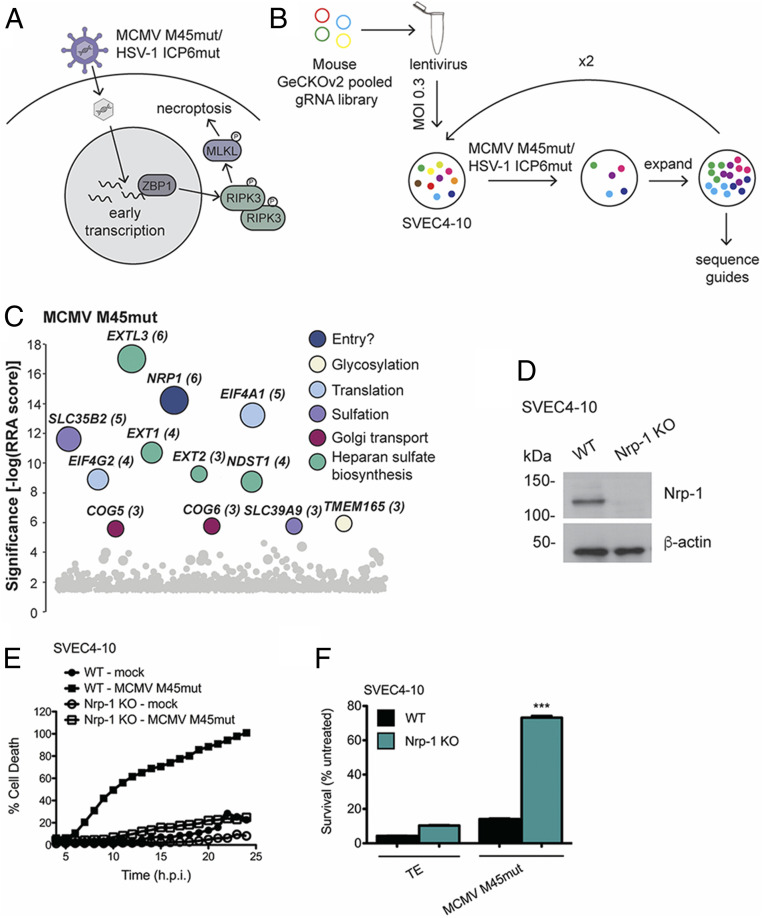
Fig. 1.
CRISPR knockout (KO) screen reveals novel candidate mediators of MCMV and HSV-1 infection. (A) Diagram of activation of death signaling during mutant herpesvirus infection. The Z-NA sensor ZBP-1 recognizes early transcriptional events in herpesvirus infection, triggering activation of necroptotic signaling. (B) Schematic of the genome-wide CRISPR screen for novel candidate mediators of MCMV infection. The mouse GeCKOv2 gRNA library was transfected into 293T cells to produce lentivirus. Library-containing lentivirus was then transduced at multiplicity of infection (MOI) 0.3 into SVEC4-10 cells. Transduced cells were selected based on puromycin resistance and infected at MOI 5 with MCMV M45mut for 1 h. After surviving cells had grown to 80% confluency, the infection process was repeated until cells were completely resistant. Cellular genomes were then sequenced to identify enriched gRNAs. (C) Screen results for MCMV. Hits are stratified along a vertical axis according to significance and arbitrarily spread along the horizontal axis. The size of the circle and the number next to the gene name both indicate the number of gRNAs for that gene that were found in the resistant population. Colors indicate gene ontology. (D) Western blot showing level of Nrp-1 KO in SVEC4-10 cells. (E) Cell death over time in wild-type (WT) and Nrp-1 KO SVEC4-10 cells infected with MCMV M45mut. Cells were infected in triplicate with MCMV M45mut (MOI 10) in media containing 50 nM Sytox green and 2 μM Hoechst and imaged every hour with a Cytation 5 Imaging Reader. Cell death was calculated as (GFP + cells/Hoechst + cells) × 100% and averaged from four images per well. (F) Cell survival after 20 h.p.i. in WT and Nrp-1 KO SVEC4-10 cells after infection with MCMV M45mut. Cells were infected with MCMV M45mut (MOI 10) or treated with 25 ng/mL TNFα + 5 μM emricasan or media alone. After 24 h, survival was quantified by Cell Titer Glo Luminescent Cell Viability Assay. Infections were performed in triplicate with three independent replicates. Error bars indicate SEM. Significance was determined using a Student’s t test (***P < 0.0001).
We set out to expand the understanding of host-virus interactions by utilizing antiviral necroptosis as a platform for a forward genetic screen to comprehensively define host factors essential for infection. With the availability of CRISPR guide RNA (gRNA) libraries, CRISPR forward-genetic screens are a powerful method for discovering novel players in viral infection (15). This strategy has led to the rapid identification of crucial cellular factors for many viruses. Most CRISPR screens have involved RNA viruses, such as West Nile virus, dengue virus, Zika virus, hepatitis C virus, and influenza (16–21). Extending this approach to large DNA viruses, such as herpesvirus, has been challenging due, in part, to the fact that herpesvirus infections do not result in rapid cell lysis. Selection strategies in a screen often involve either virus-induced cell death or flow-cytometry-based methods. Thus, we utilized the mutant herpesviruses described above to induce ZBP1-dependent necroptosis in cells expressing CRISPR gRNA libraries thereby selecting for cells resistant to virus-induced death.
One aim for this screen was to uncover host factors mediating early steps of herpesvirus infection. Targeting infection at early stages, particularly, virus entry, is an attractive option for preventing infections since herpesviruses establish latency in their hosts. There is a relative paucity of information on MCMV entry despite its use as a model for in vivo CMV infection, whereas HCMV has been much more extensively studied. HCMV enters cells by different mechanisms depending on the cell type being infected (22). In fibroblasts, HCMV enters predominantly by direct fusion with the plasma membrane (23). In epithelial or endothelial cells, however, entry is dependent on endocytosis and subsequent pH-dependent escape from the endocytic vesicle (24). Two different complexes of viral envelope glycoproteins define these different modes of infection. A trimer consisting of glycoproteins gH, gL, and gO mediates entry into fibroblasts while a pentameric gH/gL/UL128/UL130/UL131 complex achieves entry into epithelial, endothelial, and myeloid cells. Platelet-derived growth factor receptor α is the most widely accepted receptor for the trimer on fibroblasts (25–27). Until recently, the identity of the receptor for epithelial and endothelial cells was unknown. There have now been several receptors identified for the pentamer: Nrp-2, OR14I1, CD46, and CD147 (28–31). There is no previous report of an entry receptor for MCMV.
Here, our screen identifies Nrp-1 as a key mediator of MCMV infection. We show that loss of Nrp-1 drastically reduces viral early gene expression in multiple cell types. The external domain of Nrp-1 is found to be required for mediating MCMV infection. Additionally, MCMV infection can be blocked at the level of viral attachment either by excess soluble Nrp-1 or a Nrp-1 blocking antibody. These studies support Nrp-1 as a key host factor for early stages of MCMV infection and a potential entry receptor.
Results
CRISPR Screen for Host Functions Affecting Herpesvirus Infection.
To identify cellular genes required for herpesvirus infection, we employed the mouse genome-scale CRISPR KOv2 (GeCKOv2) genome-wide pooled CRISPR gRNA library. This library targets 20,000 genes and microRNAs with six gRNAs per target (32). The gRNAs were delivered by lentiviral transduction into SVEC4-10 endothelial cells. These cells robustly activate death signaling in response to both MCMV M45mut and HSV-1 ICP6mut (7, 14). Transduction was carried out at a MOI of 0.3 to limit delivery of multiple gRNAs per cell. Library-containing cells were then infected with the pronecroptotic herpesviruses, resulting in the elimination of cells susceptible to infection and allowing for the enrichment of cells resistant to virus-induced necroptosis (Fig. 1A). Cells were then subjected to additional rounds of infection and expansion until almost all cells were resistant to virus-induced cell death. Genomic DNA was isolated from this population and sequenced to identify gRNAs that were enriched in the virus-treated group compared to untreated library-containing cells. A schematic of our screen approach is depicted in Fig. 1B.
This screen revealed several candidate genes that have not previously been implicated in HSV-1 or MCMV infection (Fig. 1C and SI Appendix, Fig. S1A). Based on their known functions, the majority of these genes would be predicted to be necessary for transcription and translation of viral gene products and processing of the resulting proteins. Many genes required for heparan sulfate (hs) synthesis were also identified, such as EXTL-1–3. Hs is an attachment factor for MCMV as well as many other viruses (33, 34). Interestingly, several of the genes have even been identified in similar genetic screens performed with HIV and Rift Valley Fever virus. Among these are SLC35B2, B3GAT3, and several members of the COG family (35, 36). There was also overlap in hits for HSV-1 and MCMV: the two translation initiation factors EIF4A1 and EIF4G2. The identification of Pvrl-1, the known entry receptor for HSV-1 (37), as a top hit (SI Appendix, Fig. S1A), as well as several hs synthesis genes demonstrate that a virus-induced necroptosis screen has the capacity to identify host factors required for viral infection.
One of the unanticipated hits for MCMV M45mut virus infection included Nrp-1. This receptor that has not previously been implicated in MCMV infection was the only cell surface protein among the hits and the second highest overall hit for MCMV M45mut. Nrp-1 is a receptor for VEGF and semaphorin and, consequently, plays crucial roles in angiogenesis and axonal growth (38). Intriguingly, Nrp-1 has previously been reported to function as an entry factor for the γ-herpesvirus Epstein–Barr virus (EBV) and human T cell lymphotropic virus type 1 (39, 40). Additionally, Nrp-2 was recently shown to contribute to HCMV infection in some cell types (28). To test whether Nrp-1 may be an essential cellular factor required for MCMV infection, we generated Nrp-1 CRISPR KO SVEC4-10 cells. In alignment with the results of our screen, cells deficient in Nrp-1 expression were highly resistant to MCMV M45mut-induced death (Fig. 1 D and E).
Nrp-1 Is Required for Early Viral Gene Expression.
Our necroptosis-based screening design would, in theory, allow for identification of cellular factors that are necessary for all aspects of MCMV infection from virus binding and entry, to IE3-dependent activation of necroptosis that normally occurs beginning 8 h postinfection (h.p.i.) (Fig. 1A) (9). To determine whether Nrp-1 influenced the cell death machinery, we treated Nrp-1 KO cells with a variety of necroptotic stimuli. We found that Nrp-1 KO cells were as susceptible to TNF-induced necroptosis as WT cells (Fig. 1F). Furthermore, we found no difference in ZBP1-mediated necroptosis triggered by HSV-1 ICP6mut infection between WT and Nrp-1 KO cells (SI Appendix, Fig. S1B). These data indicate that the cellular necroptotic program induced by either the death receptors or the viral sensor ZBP1 remains unaffected by the loss of Nrp-1. These observations suggest that Nrp-1 facilitates MCMV infection at a step apical to the death signals generated during infection.
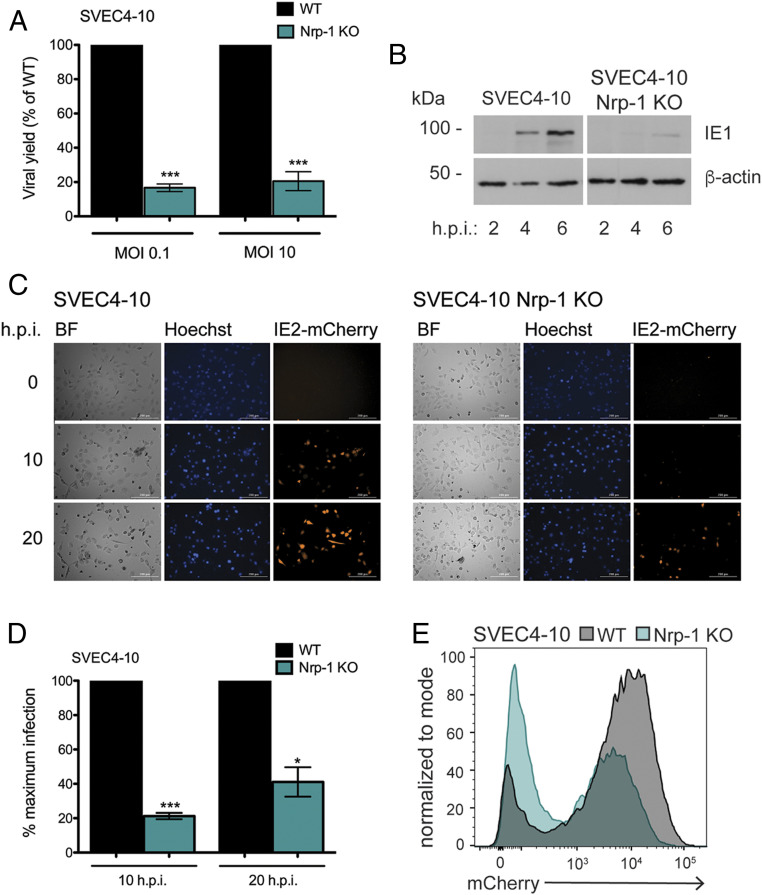
We next evaluated the infectious capacity of WT MCMV. When infected with WT virus at either a low (0.1) or a high (10) MOI, Nrp-1 KO cells produced less progeny, 20–60% of the viral yield of WT cells at 72 (MOI 0.1) and 48 (MOI 10) h.p.i. (Fig. 2A). ZBP1 is activated concomitant with early transcription (9), and, thus, perturbation of viral gene expression could, therefore, potentially contribute to the reduction in cell death. Immediate early gene expression during WT virus infection was assessed by immunoblot blot for the viral protein IE1. Nrp-1 KO greatly reduced IE1 expression at 4 h.p.i. compared to WT cells (Fig. 2B). Viral gene expression was further assessed using a reporter virus that expressed mCherry under the control of the IE2 promoter (MCMV IE2-mCherry) which is expressed with similar kinetics as IE1. We found that loss of Nrp-1 resulted in diminished reporter activation (Fig. 2 C–E). Furthermore, time-lapse imaging revealed that detection of the reporter virus was dramatically delayed in Nrp-1 KO cells and that, overall, fewer Nrp-1 KO cells express the reporter (Fig. 2C). Nrp-1 KO cells expressing the reporter also reach lower levels of mCherry fluorescence intensity when assessed by flow cytometry (Fig. 2E), consistent with a restriction of viral gene expression. These results indicate Nrp-1 functions at an early stage of MCMV infection.
Fig. 2.
Loss of Nrp-1 reduces viral gene expression. (A) MCMV yield in WT and Nrp-1 KO SVEC4-10 cells. WT and Nrp-1 KO SVEC4-10 cells were infected at the indicated MOI with WT MCMV for 1 h. Cells were harvested after 48 (MOI 10) or 72 (MOI 0.1) h.p.i., and viral yield determined by plaque assay. Values shown are viral yield relative to that of WT cells. Infections were performed in triplicate. (B) Western blot for viral immediate early gene expression. (C) Fluorescence microscopy of infected WT or Nrp-1 KO SVEC4-10 cells. Cells were infected in triplicate at MOI 1 with MCMV IE2-mCherry in media containing 2 μM Hoechst and imaged with a Cytation 5 Imaging Reader every hour for 24 h. Shown are representative images. (D) Quantification of mCherry+ cells in C. Shown is the average of four images. Rates of infection in Nrp-1 KO cells were normalized to rates of infection in WT cells. (E) Flow cytometry for IE2-mCherry expression. Cells were infected as in C, and red fluorescence quantified after 20 h by flow cytometry. At least, two replicates were performed per experiment. Error bars indicate SEM. Significance was determined using a Student’s t test (***P < 0.0001, *P < 0.01).
The Extracellular Domains of Nrp-1 Are Critical for MCMV Infection.
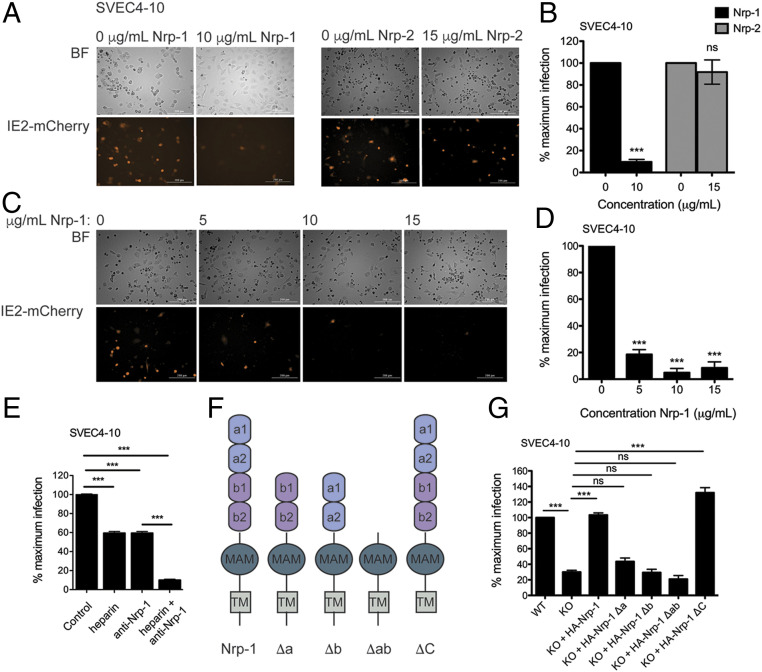
We next assessed whether excess soluble Nrp-1 was able to block MCMV infection of SVEC4-10 cells. Purity and integrity of soluble recombinant Nrp-1 were verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) Coomassie staining (SI Appendix, Fig. S2A). MCMV was preincubated with soluble recombinant Nrp-1 for 1 h prior to infection. In the presence of soluble Nrp-1, few cells were infected by MCMV as measured by the IE2-mCherry reporter, and rates of infection were reduced in a dose-dependent fashion (Fig. 3 A and B). In alignment with these results, preincubation of MCMV M45mut with soluble Nrp-1 also prevented virus-induced necroptosis (SI Appendix, Fig. S2C). A recent study demonstrated that soluble Nrp-2 prevents HCMV infection (28). We, therefore, tested the capacity of soluble Nrp-2 to affect MCMV infection. In contrast to robust inhibition by soluble Nrp-1, soluble Nrp-2 had no effect on MCMV infection (Fig. 3 C and D and SI Appendix, Fig. S2B), consistent with Nrp-1 but not Nrp-2 being identified by our genetic screen. These results support Nrp-1 as a primary mediator of infection and establish a possible binding interaction between Nrp-1 and MCMV. To test this further, we infected cells in the presence of a Nrp-1 blocking antibody. As a comparison, we also infected cells in the presence of excess heparin, a known MCMV-binding molecule (34). Infection rates were equally reduced in the presence of either heparin or anti-Nrp-1 antibody (Fig. 3E). When both heparin and anti-Nrp-1 antibody were present, infection was reduced to an even greater extent (Fig. 3E). This indicates that both heparin and Nrp-1 promote MCMV infection of endothelial cells.
Fig. 3.
The extracellular domains of Nrp-1 mediate MCMV infection. MCMV IE2-mCherry was incubated with soluble recombinant Nrp-1, Nrp-2, or media alone for 1 h at 37 °C before being added to WT SVEC4-10 cells at MOI 1 in triplicate. Cells were imaged 20 h postinfection with a Cytation 5 Imaging Reader (A), and mCherry+ cells quantified (B). Significance was determined using a Student’s t test. (C) Fluorescence microscopy for IE2-mCherry expression. Cells were infected in triplicate with virus pretreated as in A with varying concentrations of soluble Nrp-1 and imaged as in A. (D) Quantification of mCherry+ cells from (C). Significance was determined by using a one-way ANOVA with Dunnett’s multiple comparisons test. (E) Infection rates in SVEC4-10 cells pretreated with anti-Nrp-1 antibody or heparin. Cells were incubated for 30 min with 20 μg/mL heparin, 4 μg/mL anti-Nrp-1 antibody, or both. After 6 h of infection with WT MCMV, cells were fixed and stained for IE1 expression. Significance was determined by one-way ANOVA with Bonferroni’s multiple comparisons test. (F) Structure of Nrp-1 and domain deletion mutants. (G) Quantification of mCherry+ cells in SVEC4-10 WT or Nrp-1 KO cells reconstituted with full-length and deletion Nrp-1 mutants. Cells were infected with MCMV IE2-mCherry (MOI 1) in triplicate and imaged after 20 h.p.i. Significance was determined using a one-way ANOVA with Bonferroni’s multiple comparisons test. All microscopy data were quantified by taking the average from four images. At least, two replicates were performed per experiment. Error bars indicate SEM. (***P < 0.0001).
To identify the region(s) of the Nrp-1 protein that contribute to MCMV infection, we generated a series of Nrp-1 deletion mutants. The extracellular region of Nrp-1 consists of two N-terminal CUB domains (a1 and a2), followed by the b1 and b2 domains homologous to coagulation factors V and VIII, and a MAM domain which allows oligomerization (Fig. 3F). The a1a2 domains bind class III secreted semaphorins, i.e., Sema3A, and the b1b2 region binds heparin, VEGF, and Sema3 (41–43). Nrp-1 also has a transmembrane domain and a short intracellular domain that mediates signaling upon ligand binding. We generated N-terminal 6×-His and HA epitope-tagged deletion mutants with either the a1a2 (Δa), b1b2 (Δb), or both a1a2 and b1b2 (Δab) domains deleted (Fig. 3F). Nrp-1 KO SVEC4-10 cells were reconstituted with either WT or mutant Nrp-1 (SI Appendix, Fig. S3 A and B), and susceptibility to infection was monitored by IE2-mCherry positivity. Full-length Nrp-1 fully restored levels of infection, whereas removal of both the a1a2 and the b1b2 domains (Δab) completely ablated sensitivity (Fig. 3G and SI Appendix, Fig. S3C). Deletion of the a1a2 domains or the b1b2 domains individually reduced infection similar to that of Nrp-1 KO cells (Fig. 3G and SI Appendix, Fig. S3C). In alignment with these results, MCMV M45mut-induced necroptosis was also dependent on the extracellular a1a2 and b1b2 domains (SI Appendix, Fig. S3D). Thus, both external domains appear to be critical for mediating MCMV infection.
The interaction of a virus with a surface receptor has the potential to elicit an intracellular signaling response. Nrp-1 couples with a number of cellular receptors, such as VEGFR and receptor tyrosine kinases (RTKs) for intracellular signaling (38). Signaling is mediated by the cytoplasmic domain of Nrp-1 through an interaction with the PDZ domain of G α interacting protein (GAIP)-interacting protein, C terminus-1 (44). A report of Nrp-1 as an entry receptor for EBV implicated RTK signaling pathways in infection (40). To determine whether Nrp-1 signaling functions in a similar role during MCMV infection, we generated a Nrp-1 mutant lacking the cytoplasmic domain (Nrp-1 ΔC, Fig. 3F and SI Appendix, Fig. S3 A and B). Despite relatively low expression (SI Appendix, Fig. S3B), Nrp-1 KO SVEC4-10 cells expressing Nrp-1 ΔC were as susceptible to MCMV infection as WT Nrp-1-expressing cells (Fig. 3G and SI Appendix, Fig. S3C). Thus, the ectodomain of Nrp-1 functions independently of the cytoplasmic signaling domain to mediate MCMV infection.
Nrp-1 Facilitates MCMV Infection in Multiple Cell Types.
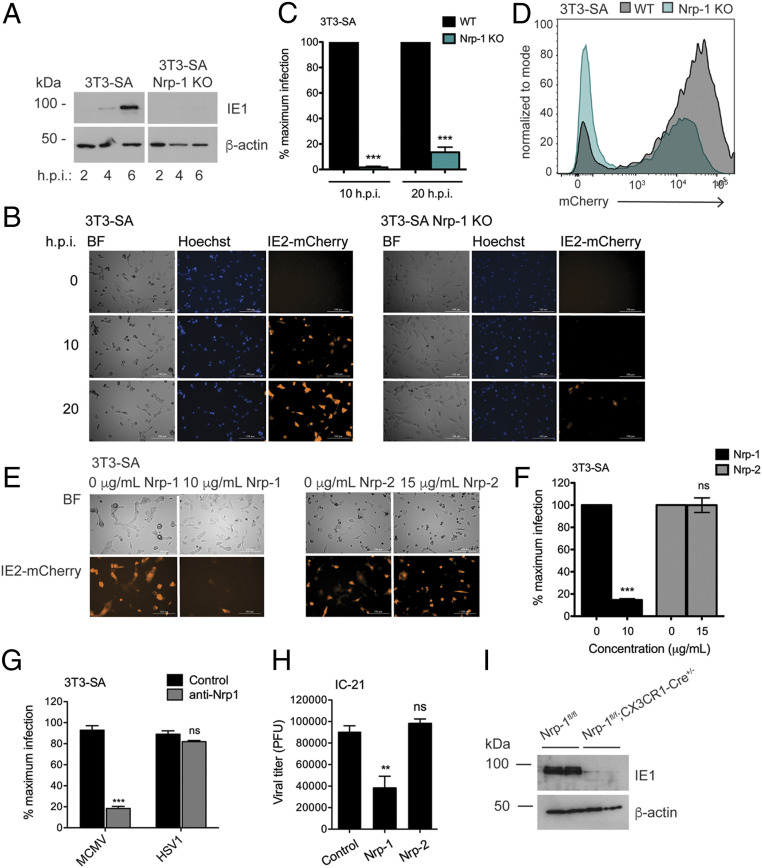
To determine whether Nrp-1 mediated infection in additional cell types, we generated Nrp-1 KO 3T3-SA fibroblasts. Similar to SVEC4-10 cells, 3T3-SA Nrp-1 KO cells were highly resistant to MCMV M45mut-induced death and demonstrated a reduced capacity to support viral replication (SI Appendix, Fig. S4 A–C). The 3T3-SA Nrp-1 KO cells also showed reduced viral gene expression as measured by IE1 and mCherry reporter levels (Fig. 4 A–D). Furthermore, soluble Nrp-1 but not Nrp-2 blocked virus infection of 3T3-SA fibroblasts (Fig. 4 E and F and SI Appendix, Fig. S5 A–D). Preincubation with Nrp-1 blocking antibody was also able to greatly reduce MCMV infection but had no effect on HSV-1 infection (Fig. 4G). Unlike the SVEC4-10 cells, Nrp-1 blocking antibody alone was sufficient to block infection in these fibroblast cells without an additional requirement for exogenous heparin.
Fig. 4.
Nrp-1 mediates MCMV infection of multiple cell types. (A) Western blot for viral immediate early gene expression in 3T3-SA cells. (B) Fluorescence microscopy for IE2-mCherry expression. Cells were infected in triplicate at MOI 1 with MCMV IE2-mCherry in media containing 2 μM Hoechst and imaged with a Cytation 5 Imaging Reader every hour for 24 h. Shown are representative images. (C) Quantification of mCherry+ cells from B. (D) Cells were infected as in B and red fluorescence quantified after 20 h by flow cytometry. (E) Fluorescence microscopy for IE2-mCherry expression. MCMV IE2-mCherry was incubated with soluble recombinant Nrp-1, Nrp-2, or media alone for 1 h at 37 °C before being added to WT SVEC4-10 and 3T3-SA cells in triplicate at MOI 1. Cells were imaged 20 h postinfection with a Cytation 5 Imaging Reader. (F) Quantification of mCherry+ cells from E. (G) Infection rates in the presence of anti-Nrp-1 antibody. The 3T3-SA cells cultured in coverslips were preincubated with anti-Nrp1 antibody (4 μg/mL) for 30 min and then infected for 6 h with MCMV or HSV-1 (MOI 10). The percentage of infected cells was assessed by staining for IE1 (MCMV) or ICP0 (HSV-1). (H) Quantification of viral attachment at the cell surface in the presence of soluble neuropilins. MCMV was incubated with soluble recombinant Nrp-1, Nrp-2, or media alone for 1 h before being added to IC-21 cells in triplicate at MOI 1 and incubated for 30 min at 4 °C. Cells were harvested, and unbound virions washed away. The amount of adhered virus was then quantified by plaque assay. Significance was determined by using a one-way ANOVA with Dunnett’s multiple comparisons test. (I) Western blot for IE1 expression in BMDMs lacking Nrp-1 and infected with MCMV (MOI 1) for 6 h. BMDMs were generated from two mice per genotype and treated with 2 μM 4-hydroxy-tamoxifen for 7 d to induce Cre activity. All microscopy data were quantified by taking the average from four images. At least, two replicates were performed per experiment. Error bars indicate SEM. Significance was determined using a Student’s t test unless otherwise stated (***P < 0.0001, **P < 0.001).
Monocytes and macrophages are key cell types for facilitating viral dissemination in vivo (45, 46). We, therefore, tested the ability of soluble Nrp-1 to block infection of IC-21 macrophages. Infection rates in these cells were notably decreased as measured by IE1 levels or quantification of viral yield (SI Appendix, Fig. S5 E and F). Treatment of IC-21 cells with the Nrp-1 blocking antibody resulted in almost complete elimination of infection (SI Appendix, Fig. S5G). Similar to fibroblast cells, exogenous heparin had no influence on MCMV infection of IC-21 cells and did not exhibit any synergistic effects when combined with Nrp-1 blocking antibody (SI Appendix, Fig. S5G). The low level of Nrp-1-independent infection in this cell type led us to investigate how soluble Nrp-1 blocks MCMV infection. To measure virus attachment at the cell surface, we infected IC-21 cells in the presence of soluble Nrp-1 or Nrp-2 at 4 °C in order to prevent virus internalization. After allowing the virus to adsorb for 30 min, cells were immediately harvested, and unbound virions washed away. The amount of bound virus was then determined by plaque assay. We found that soluble Nrp-1 but not Nrp-2 diminished virus attachment, further supporting a role for Nrp-1 in MCMV entry (Fig. 4H). A similar effect on virus attachment was seen with the Nrp-1 blocking antibody (SI Appendix, Fig. S5H).
After footpad injection in mice, MCMV travels to the popliteal lymph nodes where it gains access to the blood stream and infects CX3CR1+ patrolling monocytes to disseminate to distal tissues (45). To determine whether Nrp-1 functions in these cells, we crossed Nrp-1fl/fl mice to mice expressing a tamoxifen-inducible Cre under the control of the CX3CR1 promoter. Tissue-specific elimination of Nrp-1 is necessary as Nrp-1 KO mice are perinatal lethal. We generated bone marrow-derived macrophages (BMDMs) from Nrp-1fl/fl and Nrp-1fl/flCx3cr1-Cre mice and stimulated Cre activity in vitro by treatment with 4-hydroxy-tamoxifen. BMDMs from Nrp-1fl/flCx3cr1-Cre mice showed reduced Nrp1 expression following treatment with tamoxifen (SI Appendix, Fig. S5I). In line with the requirement of Nrp-1 for MCMV infection in immortalized cells, primary BMDMs with Cre-mediated deletion of Nrp-1 were protected from infection as indicated by resistance to MCMV M45mut-induced necroptosis (SI Appendix, Fig. S5J) and reduced WT MCMV IE1 expression (Fig. 4I). Overall, these results show Nrp-1 is a broad mediator of MCMV infection in fibroblasts, endothelial cells, and macrophages.
The finding that a cell surface protein is partially required for entry into both endothelial cells and fibroblasts is contrary to the established model for HCMV where different cellular receptors mediate entry into these cell types. The gH/gL/gO trimer is conserved in MCMV, but there is no clear homolog for the HCMV pentamer in MCMV. The MCMV chemokine MCK-2 is important for infection of macrophages and dissemination in vivo (45–48). Using a bacmid derivative of the Smith strain of MCMV, it has been proposed that MCK-2 forms a complex with gH and this is required for MCMV entry into macrophages but not fibroblasts and may promote an alternative entry pathway (49). However, another study using an MCK-2-deficient K181 strain of MCMV, the strain used here, did not find a requirement for MCK-2 in infection of macrophages (45). We tested the involvement of MCK-2 by infecting cells with WT or MCK-2-deficient virus and measuring infectivity by staining for IE1+ cells. We find that, in vitro, a MCK-2-deficient virus had very little difference in infectivity in SVEC4-10 endothelial cells, 3T3-SA fibroblasts, or IC-21 macrophages (SI Appendix, Fig. S5K). This suggests that, for K181 MCMV infection, there is no strict requirement for MCK-2 for infection for a range of different cell types.
Discussion
MCMV and HSV-1 naturally block antiviral necroptosis by antagonizing ZBP1 activation of RIPK3 via an evolutionally conserved process dependent on their respective ribonucleotide reductases, M45 and ICP6 (50). During infection, ZBP1 senses viral and/or cellular Z-form NA that accumulates during early gene expression (9, 51). Viruses harboring RHIM domain mutations induce ZBP1–RIPK3 signaling that eliminates virally infected cells and in the case of MCMV, M45mut virus is completely attenuated in vivo (7, 8). In this study, we have applied this potent host defense mechanism as a means to map the cellular machinery essential for these herpesviruses to establish a foothold in the host. This strategy has the potential to identify genes essential for multiple steps during infection, including virus entry, trafficking, gene expression, and necroptosis induction.
Here, we identified several genes required for MCMV and HSV-1 infection, including genes that have been previously identified as important for infection, such as Pvrl-1 for HSV-1 or hs synthesis genes necessary for MCMV attachment. Therefore, using engineered pronecroptotic viruses similar to those described here has the potential to enable identification of genes unique to a particular virus infection (e.g., Nrp-1 for MCMV) as well as identify cellular genes necessary for infection by a wide variety of viruses. The core set of viral genes shared among herpesviruses that are required for replication has long been described; however; to date, the compendium of cellular genes broadly required for herpesvirus infection remains undefined. We anticipate necroptosis-based strategies similar to those used here will facilitate this process with the potential to guide pan-anti-viral therapeutic development.
The utility of implementing viruses engineered to induce necroptosis is illustrated here by the unveiling of Nrp-1 as a key host factor necessary for the initial steps in MCMV infection. Nrp-1 was the second most significant hit with all six gRNAs in the library significantly enriched. We find Nrp-1 is required for efficient immediately early gene expression and subsequent steps of the virus lifecycle, including activation of ZBP1-dependent necroptosis. The extracellular domains of Nrp-1 were important for its function. Interestingly, the intracellular domain was dispensable, suggesting that virus-induced signaling via Nrp-1 is not required, in contrast to the role of Nrp-1 signaling for EBV entry (40). Furthermore, infectivity in the presence of soluble Nrp-1 or Nrp-1 blocking antibody but not soluble Nrp-2 was abrogated, suggesting that Nrp-1 may bind directly to components of the MCMV envelope. Indeed, these treatments reduced the amount of virus adhering to the cell surface. These lines of evidence support that Nrp-1 may function as an entry receptor for MCMV. Of note, loss of Nrp-1 did not ultimately abolish viral replication or the emergence of IE2-mCherry expression in cells. Cell infection occurs in the absence of Nrp-1, albeit with much slower kinetics, revealing less efficient processes for initiation of MCMV infection. Our data demonstrate that, at least, in endothelial cells, one alternate/collaborative pathway is heparin dependent. Thus, Nrp-1 primarily functions to facilitate efficient MCMV infection.
The identification of Nrp-1 as a mediator of early MCMV infection is supported by the independent findings of Martinez-Martin et al. who recently identified Nrp-2 as the entry receptor for the HCMV pentamer (28). Nrp-2 was found to bind to the pentamer but not the trimer, making it essential for entry into epithelial cells but dispensable for entry into fibroblasts. Here, we have identified Nrp-1 as a putative receptor for MCMV but find no role for Nrp-2. This may represent an evolutionary divergence of MCMV and HCMV with MCMV specific for Nrp-1 and HCMV for Nrp-2. While Nrp-1 and Nrp-2 only share 44% amino acid identity, the overall structure is similar, and both bind ligands by a common modality (42, 43). Of note, human and murine Nrp-1 share 93% amino acid identity.
A major challenge for studies of HCMV entry is that propagation of the virus in tissue culture leads to viral genome changes that often result in loss or mutation of genes critical for the production of envelope glycoproteins. Consequently, many laboratory strains of HCMV exhibit different tropism and entry mechanisms than clinical strains (52). MCMV closely mimics the pathogenesis of HCMV, exhibits similar tropism, and elicits a comparable immunological response but is genetically more stable (53). MCMV infection appears to deviate from HCMV in that there is a common Nrp-1-dependent pathway necessary for efficient infection of multiple cell types. Furthermore, the viral MCK-2 chemokine appears to be dispensable for entry of K181 MCMV, suggesting Nrp-1 interacts with the gH/gL/gO trimer or a yet unidentified complex to facilitate infection of epithelial cells, macrophages, and fibroblasts. Altogether, harnessing pronecroptotic MCMV has revealed Nrp-1 as a critical host determinant of infection.
Materials and Methods
WT, M45mut, and IE2-mCherry MCMV K181 viruses were propagated and titered as previously described (7). HSV-1 F ICP6mut virus was grown as previously described (10). Recombineering for MCMV IE2-mCherry was generated using techniques previously described (54). mCherry-N1 (Clontech) was inserted into the HpaI sites flanking the IE2exon1 as described in the SI Appendix, Materials and Methods. For the CRISPR screens, the mouse GeCKOv2 pooled CRISPR gRNA library (Addgene, No. 1000,000,052) was packaged using a lentivirus expression system as previously described (55). Lentivirus containing the library was then used to transduce SVEC4-10 endothelial cells at MOI 0.3. 2 × 108, library-containing cells were then infected with the MCMV M45mut or HSV-1 ICP-6mut viruses (MOI 5) for 1 h, then cultured in the presence of 200 μg/mL phosphonoformic acid, an inhibitor of the herpes DNA polymerase. Genomic DNA was isolated from cells resistant to virus-induced necroptosis. gRNA sequences were amplified and then sequenced. The cutoff for candidate gene hits was performed using a published computational tool (MAGeCK version 0.5.4) (56). Cell viability assays were determined indirectly by either measuring intracellular ATP levels using the Cell Titer-Glo Luminescent Cell Viability Assay kit (Promega) or by directly monitoring cell membrane integrity on a Cytation 5 Imaging Reader (Biotek). To generate Nrp-1fl/fl; Cx3cr1-CreER+/− mice, Nrp-1fl/fl C57BL/6 (JAX No. 005247) and Cx3cr1-CreER C57BL/6 (JAX No. 020940) mice were intercrossed, and genotypes determined by PCR from tail snips. Mice were housed in facilities at the University of Texas Health Science Center at San Antonio (UTHSCSA). All animal experiments were performed in accordance with protocols by the Institutional Animal Care and Use Committee at UTHSCSA.
Supplementary Material
Acknowledgments
Data were generated in the Flow Cytometry Shared Resource Facility, which is supported by University of Texas Health San Antonio, Grant NIH-National Cancer Institute (NCI) P30 CA054174-20 (Cancer Therapy & Research Center at UTHSCSA), UL1 TR001120 (Clinical and Translational Science Award), and the Genome Sequencing Facility, which is supported by UTHSCSA, NIH-NCI P30 CA054174 (Cancer Center Support Grant to UTHSCSA), and Cancer Prevention and Research Institute of Texas Core Facility Award (RP160732). This work was also supported by NIH grants R01 AI141970 to J.C., DP5 OD012198 to W.J.K., and the Craniofacial Oral-biology Student Training in Academic Research fellowship award (T32DE014318-16) and National Institute of Dental and Craniofacial Research F31 predoctoral fellowship F31DE029395 to R.K.L.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921315117/-/DCSupplemental.
Data Availability.
All materials, reagents, and methods are included in the paper or further detailed in the SI Appendix.
References
- 1.Brune W., Andoniou C. E., Die another day: Inhibition of cell death pathways by cytomegalovirus. Viruses 9, 249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Upton J. W., Chan F. K., Staying alive: Cell death in antiviral immunity. Mol. Cell 54, 273–280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber G. N., Host defense, viruses and apoptosis. Cell Death Differ. 8, 113–126 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Kaiser W. J., Upton J. W., Mocarski E. S., Viral modulation of programmed necrosis. Curr. Opin. Virol. 3, 296–306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mocarski E. S., Upton J. W., Kaiser W. J., Viral infection and the evolution of caspase 8-regulated apoptotic and necrotic death pathways. Nat. Rev. Immunol. 12, 79–88 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H. et al., Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell 54, 133–146 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Upton J. W., Kaiser W. J., Mocarski E. S., Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7, 302–313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Upton J. W., Kaiser W. J., Mocarski E. S., DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11, 290–297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sridharan H. et al., Murine cytomegalovirus IE3-dependent transcription is required for DAI/ZBP1-mediated necroptosis. EMBO Rep. 18, 1429–1441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo H. et al., Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe 17, 243–251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X. et al., Direct activation of RIP3/MLKL-dependent necrosis by herpes simplex virus 1 (HSV-1) protein ICP6 triggers host antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 111, 15438–15443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Z. et al., RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host Microbe 17, 229–242 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Yu X., He S., The interplay between human herpes simplex virus infection and the apoptosis and necroptosis cell death pathways. Virol. J. 13, 77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo H. et al., Species-independent contribution of ZBP1/DAI/DLM-1-triggered necroptosis in host defense against HSV1. Cell Death Dis. 9, 816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDougall W. M., Perreira J. M., Reynolds E. C., Brass A. L., CRISPR genetic screens to discover host-virus interactions. Curr. Opin. Virol. 29, 87–100 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Ma H. et al., A CRISPR-based screen identifies genes essential for West-Nile-virus-induced cell death. Cell Rep. 12, 673–683 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson R. B. et al., A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat. Microbiol. 3, 1214–1223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savidis G. et al., Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Rep. 16, 232–246 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Ren Q. et al., A Dual-reporter system for real-time monitoring and high-throughput CRISPR/Cas9 library screening of the hepatitis C virus. Sci. Rep. 5, 8865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heaton B. E. et al., A CRISPR activation screen identifies a pan-avian influenza virus inhibitory host factor. Cell Rep. 20, 1503–1512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puschnik A. S., Majzoub K., Ooi Y. S., Carette J. E., A CRISPR toolbox to study virus-host interactions. Nat. Rev. Microbiol. 15, 351–364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanarsdall A. L., Johnson D. C., Human cytomegalovirus entry into cells. Curr. Opin. Virol. 2, 37–42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Compton T., Nepomuceno R. R., Nowlin D. M., Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology 191, 387–395 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Ryckman B. J., Jarvis M. A., Drummond D. D., Nelson J. A., Johnson D. C., Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 80, 710–722 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabanova A. et al., Platelet-derived growth factor-α receptor is the cellular receptor for human cytomegalovirus gHgLgO trimer. Nat. Microbiol. 1, 16082 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu K., Oberstein A., Wang W., Shenk T., Role of PDGF receptor-α during human cytomegalovirus entry into fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 115, E9889–E9898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y. et al., Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry. PLoS Pathog. 13, e1006281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Martin N. et al., An unbiased screen for human cytomegalovirus identifies neuropilin-2 as a central viral receptor. Cell 174, 1158–1171.e19 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Xiaofei E. et al., OR14I1 is a receptor for the human cytomegalovirus pentameric complex and defines viral epithelial cell tropism. Proc. Natl. Acad. Sci. U S A 116, 7043–7052 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein K. R. et al., CD46 facilitates entry and dissemination of human cytomegalovirus. Nat. Commun. 10, 2699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanarsdall A. L. et al., CD147 promotes entry of pentamer-expressing human cytomegalovirus into epithelial and endothelial cells. MBio 9, e00781-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spillmann D., Heparan sulfate: Anchor for viral intruders? Biochimie 83, 811–817 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Price P., Allcock R. J., Coombe D. R., Shellam G. R., McCluskey J., MHC proteins and heparan sulphate proteoglycans regulate murine cytomegalovirus infection. Immunol. Cell Biol. 73, 308–315 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Riblett A. M. et al., A haploid genetic screen identifies heparan sulfate proteoglycans supporting Rift Valley fever virus infection. J. Virol. 90, 1414–1423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park R. J. et al., A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat. Genet. 49, 193–203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montgomery R. I., Warner M. S., Lum B. J., Spear P. G., Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87, 427–436 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Guo H. F., Vander Kooi C. W., Neuropilin functions as an essential cell surface receptor. J. Biol. Chem. 290, 29120–29126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghez D. et al., Neuropilin-1 is involved in human T-cell lymphotropic virus type 1 entry. J. Virol. 80, 6844–6854 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H. B. et al., Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 6, 6240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker M. W., Xu P., Guo H. F., Vander Kooi C. W., Mechanism of selective VEGF-A binding by neuropilin-1 reveals a basis for specific ligand inhibition. PLoS One 7, e49177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker M. W., Xu P., Li X., Vander Kooi C. W., Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 287, 11082–11089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vander Kooi C. W. et al., Structural basis for ligand and heparin binding to neuropilin B domains. Proc. Natl. Acad. Sci. U.S.A. 104, 6152–6157 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Mukhopadhyay D., Xu X., C terminus of RGS-GAIP-interacting protein conveys neuropilin-1-mediated signaling during angiogenesis. FASEB J. 20, 1513–1515 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Daley-Bauer L. P., Roback L. J., Wynn G. M., Mocarski E. S., Cytomegalovirus hijacks CX3CR1(hi) patrolling monocytes as immune-privileged vehicles for dissemination in mice. Cell Host Microbe 15, 351–362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahl F. R. et al., Mck2-dependent infection of alveolar macrophages promotes replication of MCMV in nodular inflammatory foci of the neonatal lung. Mucosal Immunol. 8, 57–67 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Jordan S. et al., Virus progeny of murine cytomegalovirus bacterial artificial chromosome pSM3fr show reduced growth in salivary Glands due to a fixed mutation of MCK-2. J. Virol. 85, 10346–10353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleming P. et al., The murine cytomegalovirus chemokine homolog, m131/129, is a determinant of viral pathogenicity. J. Virol. 73, 6800–6809 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner F. M. et al., The viral chemokine MCK-2 of murine cytomegalovirus promotes infection as part of a gH/gL/MCK-2 complex. PLoS Pathog. 9, e1003493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nailwal H., Chan F. K., Necroptosis in anti-viral inflammation. Cell Death Differ. 26, 4–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takaoka A. et al., DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Wilkinson G. W. et al., Human cytomegalovirus: Taking the strain. Med. Microbiol. Immunol. 204, 273–284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sweet C., The pathogenicity of cytomegalovirus. FEMS Microbiol. Rev. 23, 457–482 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Hilterbrand A. T., Boutz D. R., Marcotte E. M., Upton J. W., Murine cytomegalovirus deubiquitinase regulates viral chemokine levels to control inflammation and pathogenesis. MBio 8, e01864-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joung J. et al., Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W. et al., MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All materials, reagents, and methods are included in the paper or further detailed in the SI Appendix.