Significance
KRAS, NRAS, and BRAF mutations that activate MAPK signaling occur in half of myeloma patients and confer a poor prognosis. Our studies link activating RAS and RAF mutations with enhanced proteasome assembly and capacity through the MAPK intermediate ELK1. These downstream changes reduce cellular stress, thereby promoting myeloma cell survival, and confer proteasome inhibitor resistance, which can be overcome by combinations with BRAF or MEK inhibitors. They provide a possible reason for the frequency of MAPK pathway mutations in myeloma, and support use of these combinations specifically in patients with BRAF- or RAS-mutated disease.
Keywords: BRAF, KRAS, NRAF, proteasome capacity, proteasome inhibitor sensitivity
Abstract
KRAS, NRAS, and BRAF mutations which activate p44/42 mitogen-activated protein kinase (MAPK) signaling are found in half of myeloma patients and contribute to proteasome inhibitor (PI) resistance, but the underlying mechanisms are not fully understood. We established myeloma cell lines expressing wild-type (WT), constitutively active (CA) (G12V/G13D/Q61H), or dominant-negative (DN) (S17N)-KRAS and -NRAS, or BRAF-V600E. Cells expressing CA mutants showed increased proteasome maturation protein (POMP) and nuclear factor (erythroid-derived 2)-like 2 (NRF2) expression. This correlated with an increase in catalytically active proteasome subunit β (PSMB)-8, PSMB9, and PSMB10, which occurred in an ETS transcription factor-dependent manner. Proteasome chymotrypsin-like, trypsin-like, and caspase-like activities were increased, and this enhanced capacity reduced PI sensitivity, while DN-KRAS and DN-NRAS did the opposite. Pharmacologic RAF or MAPK kinase (MEK) inhibitors decreased proteasome activity, and sensitized myeloma cells to PIs. CA-KRAS, CA-NRAS, and CA-BRAF down-regulated expression of endoplasmic reticulum (ER) stress proteins, and reduced unfolded protein response activation, while DN mutations increased both. Finally, a bortezomib (BTZ)/MEK inhibitor combination showed enhanced activity in vivo specifically in CA-NRAS models. Taken together, the data support the hypothesis that activating MAPK pathway mutations enhance PI resistance by increasing proteasome capacity, and provide a rationale for targeting such patients with PI/RAF or PI/MEK inhibitor combinations. Moreover, they argue these mutations promote myeloma survival by reducing cellular stress, thereby distancing plasma cells from the apoptotic threshold, potentially explaining their high frequency in myeloma.
Multiple myeloma is the second most commonly diagnosed hematologic malignancy (1), and the number of cases may grow by ∼60% between 2010 and 2030 (2). Recent therapeutic advances, including PIs (3), have doubled median overall survival (4, 5). This has been paralleled by an increased understanding of the myeloma mutational spectrum, which was noted three decades ago to include KRAS and NRAS mutations (6). More recent studies confirmed the RAS/MAPK pathway is the most frequently mutated (7–9). For example, one 463 patient study found mutated KRAS, NRAS, or BRAF in 21.2%, 19.4%, and 6.7%, respectively, and KRAS and NRAS tended to be mutually exclusive (9). Most mutations clustered in codons 12, 13, and 61 for KRAS, NRAS, and codon 600 for BRAF (9), which are associated with MAPK activation. Moreover, in relapsed/refractory disease, marrow (10) or circulating (11) tumor DNA sequencing revealed activating RAS mutations in up to 70%.
Beyond the possibility that myeloma is a RAS pathway driven disease, some studies, although not all (9), suggest these mutations impact prognosis. Activating mutations have been implicated in transitions from monoclonal gammopathy of undetermined significance (MGUS) to myeloma and from intramedullary to extramedullary disease (12). RAS mutations are found in more advanced and aggressive clinical scenarios (10, 11, 13–15) and in the relapsed/refractory setting were associated with a decreased survival of 2.1 versus 4.0 y for WT-RAS (16). In particular, NRAS mutations correlated with decreased PI sensitivity (17, 18), and these raised interest in using MEK inhibitors alone (19, 20) or in combinations, such as with AKT inhibitors (21). Notably, while two responses were seen out of 36 unselected patients with selumetinib (SEL) (19), trametinib showed a 40% response rate among 40 patients with MAPK pathway-activated myeloma (20).
RAS signaling has been intensively studied since RAS mutations are common in cancer (22). Through downstream effectors, including RAF, phosphoinositide 3-kinase, Ral guanine nucleotide dissociation stimulator, and phospholipase C ε, RAS influences cell proliferation and tumor progression (22). In myeloma, RAS mutations may induce cytokine independence (23), cooperate with AKT to promote survival (24), and contribute to adhesion and chemoresistance (25). Given that RAS mutations may inhibit PI efficacy (17), and PI resistance is linked to the proteasome load/capacity balance (26–29), we sought to study the possibility that RAS/RAF mutations influenced proteasome activity. We, herein, demonstrate that activating KRAS, NRAS, and BRAF mutations enhanced proteasome activity through a RAS/RAF/MEK/MAPK/ETS domain-containing protein (ELK)-1/POMP pathway. Also, ER stress was reduced in myeloma cells with activating RAS and RAF mutations. Finally, RAS signaling inhibitors enhanced ER stress and sensitized myeloma to PIs, and their combinations were synergistic. Our data provide insights into the role of RAS and RAF mutations in myeloma biology, a rationale for why they are common, and support clinical translation of MAPK pathway/PI combinations.
Results
Activating MAPK Mutants Modulate PI Sensitivity.
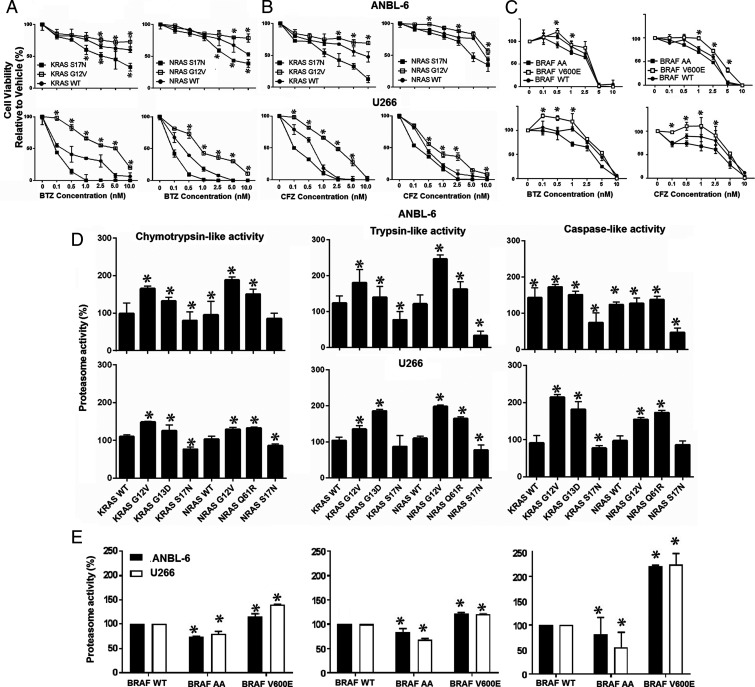
ANBL-6 and U266 myeloma cells harbor WT-KRAS, WT-NRAS, and WT-BRAF, and stable lines were prepared overexpressing WT-, CA-G12V-, or DN-S17N-KRAS or -NRAS. CA mutations induced increased proliferation, while DNs tended to slow cell growth (SI Appendix, Fig. S1). When exposed to BTZ, CA-KRAS and -NRAS conferred decreased sensitivity versus WT controls (Fig. 1A), while DNs enhanced sensitivity. For example, in U266 cells, BTZ’s median inhibitory concentrations were 4.5, 12, and 1.3 nM for the WT-KRAS, G12V-KRAS, and DN-KRAS cells, respectively (Table 1). When challenged with carfilzomib (CFZ) (Fig. 1B), similar trends were seen (Table 1). MAPK pathway activation by CA-V600E-BRAF also suppressed PI sensitivity (Fig. 1C), while DN-AA-BRAF enhanced sensitivity. As proteasome capacity is a major determinant of PI sensitivity, we evaluated the chymotrypsin-like (ChT-L), trypsin-like (T-L), and postglutamyl peptide hydrolyzing (PGPH; caspase-like) activities. Activating KRAS mutants, including G12V and G13D (SI Appendix, Table S1), increased these activities in ANBL-6 and U266 cells (Fig. 1D). In U266 cells, for example, these increased by 1.25- and 1.48-fold for the ChT-L, 1.35- and 1.89-fold for the T-L, and 1.81- and 2.14-fold for the caspase-like activities in G13D and G12V cells, respectively. Similarly, G12V- and Q61R-NRAS mutations enhanced these activities, while DN-S17N-KRAS and DN-S17N-NRAS reduced them (Fig. 1D). Also, BRAF-V600E increased these activities, while DN-BRAF reduced them (Fig. 1E). Interestingly, even in the presence of BTZ or CFZ, KRAS- and NRAS-G12V expressing ANBL-6 (SI Appendix, Fig. S2) and U266 (SI Appendix, Fig. S3) cells retained more proteasome activity, while S17N (SI Appendix, Figs. S2 and S3) and DN-BRAF reduced them (Fig. 1E).
Fig. 1.
CA MAPK mutants induce proteasome inhibitor resistance and proteasome capacity. ANBL-6 (Upper) or U266 cells (Lower) expressing CA (CA-G12V), DN (DN-S17N), or WT-KRAS or WT-NRAS were treated with vehicle, BTZ (A), or CFZ (B) for 72 h. Cell viability was assayed, and data from one of three independent experiments, each performed in triplicate, are presented as mean ± SD (asterisks indicate P ≤ 0.05 versus WT). ANBL-6 or U266 cells expressing DN-AA-, CA-V600E-, or WT-BRAF (C) were similarly evaluated. The chymotrypsin-, trypsin-, and caspase-like activities were measured in ANBL-6 (Upper) and U266 cells (Lower) expressing KRAS, NRAS (D), or BRAF variants (E).
Table 1.
Median inhibitory concentrations for BTZ and CFZ in KRAS and NRAS WT and mutant ANBL-6 and U266 myeloma cells
| KRAS variants | NRAS variants | |||||
| WT (nM) | G12V (nM) | S17N (nM) | WT (nM) | G12V (nM) | S17N (nM) | |
| ANBL-6 | ||||||
| BTZ | 7 ± 0.25 | 16 ± 0.85 | 2.5 ± 0.19 | 11.5 ± 0.05 | 18 ± 0.25 | 3.5 ± 0.057 |
| P = 0.031* | P = 0.052* | |||||
| CFZ | 4.5 ± 0.28 | 12 ± 0.57 | 1.3 ± 0.21 | 6 ± 0.82 | 11.8 ± 1.06 | 3.6 ± 0.20 |
| P = 0.031* | P = 0.031* | |||||
| U266 | ||||||
| BTZ | 0.9 ± 0.03 | 6.5 ± 0.33 | 0.1 ± 0.01 | 0.56 ± 0.02 | 1.8 ± 0.21 | 0.11 |
| P = 0.041* | P = 0.031* | |||||
| CFZ | 0.82 ± 0.03 | 3.5 ± 0.12 | 0.32 ± 0.02 | 0.42 ± 0.027 | 1.2 ± 0.034 | 0.2 ± 0.018 |
| P = 0.031* | P = 0.031* | |||||
P values are for the comparison between the G12V variants and the WT NRAS or KRAS.
KRAS, NRAS, and BRAF Enhance Proteasome Subunit Expression and Assembly.
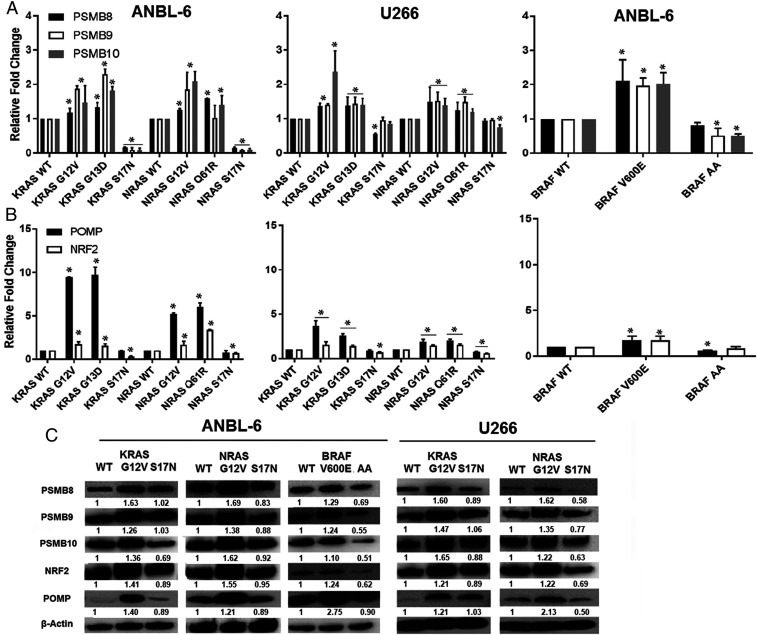
Increases in proteasome activity imply greater production and assembly of catalytically active proteasome complexes, and we first evaluated proteasome subunits β 8 (PSMB8), PSMB9, and PSMB10. These encode the ChT-L, PGPH, and T-L activities, respectively, when incorporated into mature immunoproteasomes, which are the major variants in myeloma (30, 31). PSMB8, PSMB9, and PSMB10 transcription increased (Fig. 2A) in ANBL-6 (left panel) and U266 (middle) cells with G12V- and G13D-KRAS, and G12V- and Q61R-NRAS. For example, in ANBL-6 cells, G12V- and Q61R-NRAS enhanced transcription by 1.3- and 1.6-fold for PSMB8, 1.2- and 2.2-fold for PSMB9, and 1.4- and 2.1-fold for PSMB10, respectively. Similarly, V600E-BRAF enhanced PSMB8, PSMB9, and PSMB10 transcription (Fig. 2 A, Right). In contrast, DN-S17N-KRAS and DN-S17N-NRAS, and DN-BRAF suppressed their expression especially compared to the CA mutants. Since these subunits are expressed as precursors which are cleaved into active proteases after incorporation into the proteasome, we also looked at the key assembly chaperone POMP and its upstream regulator NRF2 (29). POMP and NRF2 were induced by CA-KRAS and CA-NRAS in ANBL-6 and U266 (Fig. 2 B, Left and Middle) and in ANBL-6 cells with BRAF-V600E (Fig. 2 B, Right). Consistent with the DN-RAS data, DN-BRAF suppressed POMP and NRF2 especially compared to the V600E cells. At the protein level (SI Appendix, Table S2), CA-KRAS-G12V, NRAS-G12V, or BRAF-V600E increased PSMB8, PSMB9, PSMB10, NRF2, and POMP (Fig. 2C). DN-KRAS, DN-NRAS, or DN-BRAF, on the other hand, showed expression comparable to or lower than the WTs, and especially versus the CA constructs. Notably, PSMB8, PSMB9, PSMB10, NRF2, and POMP, were increased by CA-KRAS-G13D and NRAS-Q61R (SI Appendix, Fig. S4).
Fig. 2.
CA KRAS, NRAS, and BRAF increase NRF2, POMP, and PSMB8-10. PSMB8, PSMB9, and PSMB10 mRNAs were assessed by qRT-PCR in cells expressing RAS and BRAF mutants (A). Data are representative of three independent experiments, presented as mean ± SD of triplicates, and asterisks indicate P ≤ 0.05 versus the WT controls. POMP and NRF2 expression were then evaluated in these same cell lines by qPCR (B). Lysates from ANBL-6 (Left) and U266 cells (Right) expressing MAPK pathway mutants were analyzed for POMP, NRF2, PSMB8, PSMB9, and PSMB10 levels (C).
POMP Induction Occurs through RAS/RAF and ELK1.
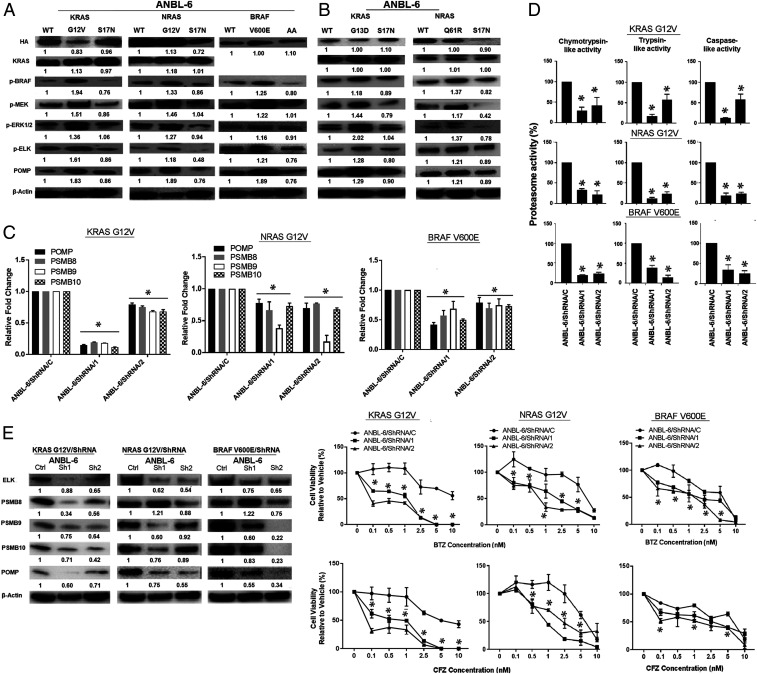
Oxidative stress induces NRF2 and POMP as an adaptive response to increase proteasome capacity (32, 33). Since RAS oncoproteins modulate reactive oxygen species (ROS) (34), we looked at these but found no consistent association between ROS and CA- or DN-mutants, so we evaluated downstream signaling. As expected, KRAS- and NRAS-G12V increased phospho(p)-BRAF, p-MEK, and p-p44/42 MAPK (extracellular signal-regulated kinase [ERK]), while BRAF-V600E increased p-MEK and p-ERK (Fig. 3A and SI Appendix, Figs. S5 and S6A). KRAS- and NRAS-S17N decreased p-BRAF, p-MEK, and p-ERK (Fig. 3A), and BRAF-AA reduced p-MEK and p-MAPK levels, while KRAS-G13D and NRAS-Q61R had opposite effects (Fig. 3B). Downstream of ERK-1/2, activating KRAS, NRAS, and BRAF enhanced p-ELK1 (Fig. 3 A and B and SI Appendix, Figs. S5 and S6A). Since ELK1 binding sites are found in the POMP, PSMB8, PSMB9, and PSMB10 promoters (SI Appendix, Fig. S7), we used short hairpin RNAs (shRNAs) to suppress ELK1 in KRAS-G12V, NRAS-G12V, and BRAF-V600E cells. This reduced POMP, PSMB8, PSMB9, and PSMB10 messenger RNA (mRNA) (Fig. 3 C, Upper and SI Appendix, Fig. S6 B, Left and Middle) and protein levels (Fig. 3 C, Lower and SI Appendix, Fig. S6 B, Right). ELK1 suppression reduced ChT-L, PGPH, and T-L activities in ANBL-6 and U266 cells (Fig. 3D and SI Appendix, Fig. S6C). Notably, this enhanced BTZ and CFZ sensitivity (Fig. 3E and SI Appendix, Fig. S6D), indicating a link between proteasome function and RAS/RAF/MEK/ERK/ELK1 signaling.
Fig. 3.
CA mutants activate downstream signaling and proteasome capacity in an ELK1-dependent fashion. ANBL-6 cells expressing WT-, G12V-, or S17N-RAS mutants or WT-, V600E-, or AA-BRAF were analyzed for the activation status of MAPK pathway intermediates (A). A similar approach compared ANBL-6 cells expressing CA-KRAS-G13D or NRAS-Q61R (B) mutants to their DN or WT controls. ELK1 knockdown was obtained by two different shRNAs, and the impact on POMP, PSMB8, PSMB9, and PSMB10 was evaluated by qPCR (C, Upper) and Western blotting (C, Lower). The impact of ELK1 suppression was examined on the chymotrypsin-, trypsin-, and caspase-like activities (D) in ANBL-6 cells. Finally, sensitivity of cells with ELK1 knockdown to BTZ and CFZ (E) was studied.
RAF or MEK Inhibitors Enhance PI Sensitivity in CA-NRAS or CA-KRAS Cells.
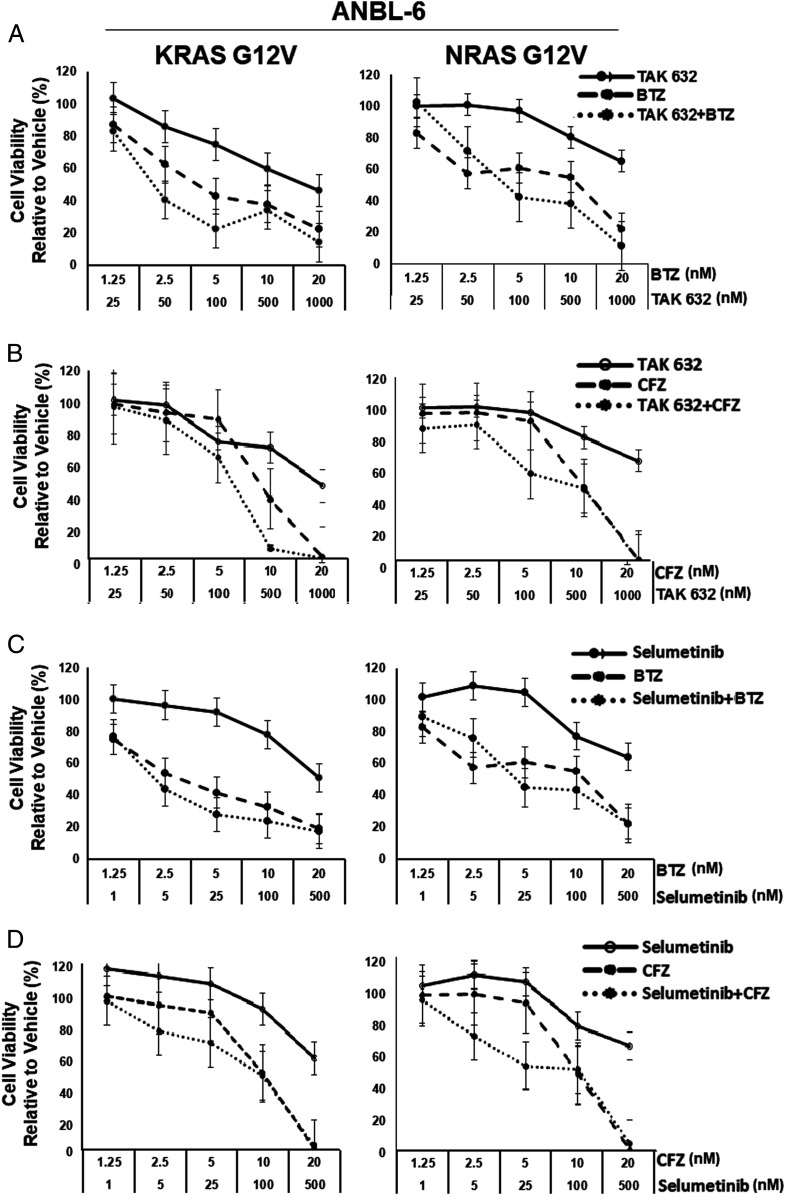
Since CA-NRAS and CA-KRAS contributed to PI resistance, we considered that RAF or MEK inhibition could enhance sensitivity. We exposed ANBL-6- and U266-based cells to BTZ or CFZ with or without the pan-RAF inhibitor TAK-632 (35) or the MEK inhibitor SEL (36). In WT cells, TAK-632/BTZ (SI Appendix, Fig. S8A and Table S3) or SEL/BTZ (SI Appendix, Fig. S8C and Table S4), did occasionally show an additive-to-synergistic impact. However, G12V-KRAS or G12V-NRAS cells more consistently showed additive-to-synergistic effects with TAK-632/BTZ (Fig. 4A and SI Appendix, Fig. S9A and Table S3) or SEL/BTZ (Fig. 4C and SI Appendix, Fig. S9C and Table S4). These produced predominantly antagonistic effects in DN-S17N, ANBL-6, or U266 cells (SI Appendix, Fig. S10 and Tables S3 and S4). When CFZ was used, qualitatively similar findings were noted (Fig. 4 B and D and SI Appendix, Figs. S8–S10 and Tables S3 and S4) with more consistent additive-to-synergistic effects in CA-RAS cells. To confirm the data with a second MEK inhibitor, trametinib was used and showed occasional synergy in WT-KRAS or WT-NRAS cells, consistent synergy in G12V cells, and antagonism in S17N cells (SI Appendix, Fig. S11 and Table S5). The differences between the combinations and the single agents in CA-KRAS and CA-NRAS cells were statistically significant (P < 0.05) compared to WT and DN models.
Fig. 4.
BTZ or CFZ with TAK-632 or SEL produce synergistic anti-myeloma activity in CA RAS cells. ANBL-6 cells expressing CA-RAS mutants were incubated with TAK-632, BTZ, or the combination (A), and cellular viability was determined. Data were from triplicate experiments and were plotted as the mean ± SD, while combination indices were provided in SI Appendix, Tables S3 and S4. CFZ was also combined with TAK-632 in the same cell line (B). Next, BTZ (C) or CFZ (D) were added to SEL, and the data were collected, analyzed, and presented as above.
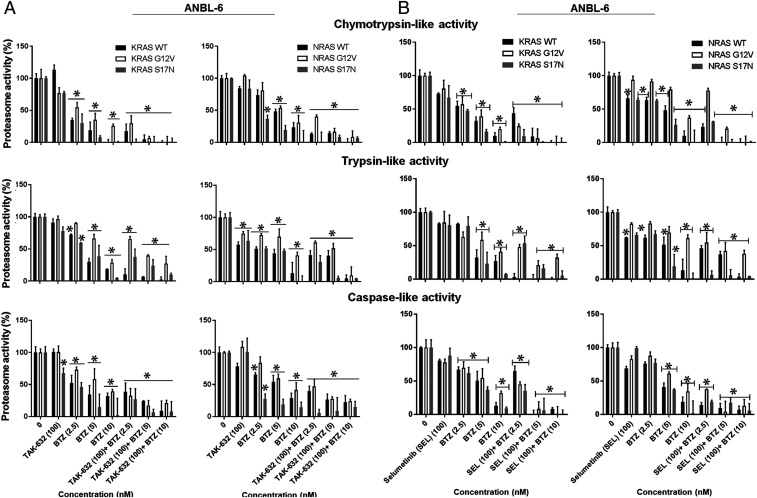
It was also of interest to evaluate the impact of BTZ with RAF or MEK inhibitors on proteasome activity. In ANBL-6 (Fig. 5A) or U266 cells (SI Appendix, Fig. S12A), TAK-632 had no consistent impact on ChT-L activity, which is the rate-limiting step in proteasome-mediated proteolysis (37). Similarly, no consistent T-L or PGPH inhibition was seen with TAK-632 (Fig. 5A and SI Appendix, Fig. S12A) or in purified proteasome preparations. BTZ produced a progressive increase in proteasome inhibition which was most pronounced for the ChT-L activity (Fig. 5A and SI Appendix, Fig. S12A). As noted earlier (Fig. 1D), proteasome activity was relatively preserved in cells harboring CA-RAS mutants. When TAK-632 was combined with BTZ, this produced greater proteasome inhibition (Fig. 5A and SI Appendix, Fig. S12A), consistent with the greater ability of this doublet to reduce cell viability (Fig. 4 and SI Appendix, Fig. S9). Similarly, SEL alone had modest if any effects (Fig. 5B and SI Appendix, Fig. S12B), but, with BTZ, there was enhanced and, in some cases, complete proteasome inhibition.
Fig. 5.
BTZ with TAK-632 or SEL enhances proteasome inhibitory activity in myeloma cells. The chymotrypsin-, trypsin-, and caspase-like activities were measured in ANBL-6 cells expressing the noted RAS mutants after treatment with TAK-632, BTZ, or both (A). These activities were then measured in ANBL-6 cells with the indicated concentrations of SEL, BTZ, or both (B). Incubations were for 24 h in all of the panels, and data were presented as the mean ± SD of triplicates with asterisks indicating P < 0.05.
Activating MAPK Mutants Attenuates the Unfolded Protein Response.
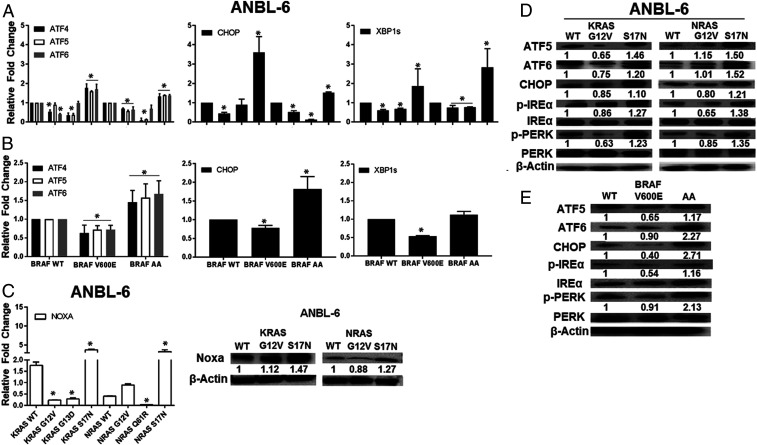
Enhanced proteasome capacity due to CA-KRAS, CA-NRAS, or CA-BRAF could allow plasma cells to reduce their reliance for survival on the ER unfolded protein response (UPR) induced by proteotoxic stress (38). To evaluate this, we first examined expression of activating transcription factor (ATF)-4, a downstream effector of the protein kinase RNA-like ER kinase (PERK) UPR arm, and ATF6, a sensor of a second UPR arm. KRAS-G12V and KRAS-G13D, and NRAS-G12V and NRAS-Q61R, reduced ATF4 and ATF6 mRNA levels in ANBL-6 (Fig. 6A) and U266 cells (SI Appendix, Fig. S13A). In contrast, S17N-KRAS or S17N-NRAS enhanced ATF4 and ATF6 expression, suggesting their reduction of proteasome capacity enhanced ER stress, and a similar pattern was seen for the mitochondrial UPR mediator ATF5 (39). BRAF-V600E also reduced ATF4, ATF5, and ATF6 in ANBL-6 cells (Fig. 6B), while the DN-AA mutant enhanced these. Beyond ATF4, two other downstream ER UPR effectors are CHOP and the spliced variant of X-box binding protein 1 (XBP1s). mRNAs of both were decreased by CA-KRAS, CA-NRAS, or CA-BRAF and increased by their DN counterparts (Fig. 6 A and B and SI Appendix, Fig. S13A).
Fig. 6.
CA KRAS, NRAS, and BRAF mutants attenuate the unfolded protein response. ATF4, ATF5, ATF6, and C/EBP homologous protein TF (CHOP), and spliced XBP-1 expression were assessed by qRT-PCR in ANBL-6 cells (A) with the indicated RAS mutants. These were also evaluated in ANBL-6 cells expressing BRAF mutants (B) relative to their WT controls. NOXA mRNA and protein expression were then assessed by qRT-PCR (C, Left) and Western blotting (C, Right) in ANBL-6 cells. Lysates from ANBL-6 cells expressing KRAS and NRAS mutants were analyzed for the UPR proteins ATF5, ATF6, and CHOP, and phosphorylation/activation of UPR receptors IREα and PERK (D). Abundance of these same proteins was next evaluated in ANBL-6 cells with BRAF mutants (E).
NOXA is a terminal effector that induces cell death if UPR activation does not attenuate proteotoxic stress (40). Its expression generally followed the UPR genes in that NOXA decreased in CA-KRAS, CA-NRAS, or CA-BRAF cells, while the DNs enhanced NOXA (Fig. 6C and SI Appendix, Fig. S13B). ATF5, ATF6, and CHOP proteins decreased in the presence of KRAS- or NRAS-G12V (Fig. 6D and SI Appendix, Fig. S13C) and BRAF-V600E (Fig. 6E). The same was true for activated inositol-requiring protein-1, p-PERK protein (Fig. 6D and SI Appendix, Fig. S13C), and mRNA levels (SI Appendix, Fig. S14) which, in addition to ATF6, are the three upstream ER UPR sensors. Finally, in ANBL-6 and U266 cells with KRAS- or NRAS-G12V, NOXA was similar to, or lower than the WT controls (Fig. 6C and SI Appendix, Fig. S13B), and higher in the S17N-DNs.
If ELK1 is a key signaling intermediate, its expression should parallel cell viability and proteasome activity changes. Consistent with this possibility, cells harboring CA mutations treated with BTZ/SEL (SI Appendix, Fig. S15A) or BTZ/TAK-632 (SI Appendix, Fig. S16) had the greatest reduction in p-ELK1, whereas smaller or no changes were seen in WT or DN cells. Notably, similar trends were seen when POMP (SI Appendix, Figs. S15A and S16) and PSMB8, PSMB9, and PSMB10 (SI Appendix, Fig. S15A) were evaluated. Also, these were accompanied by the reverse changes in ATF6, CHOP, XBP1s, p-PERK, and BiP (SI Appendix, Fig. S15B), which increased when p-ELK1 and PSMB levels decreased, consistent with greater proteotoxic stress levels. To more directly evaluate ELK1's role, we suppressed it with two different shRNAs, and this enhanced ATF6, CHOP, XBP1s, p-PERK, and BiP levels (SI Appendix, Fig. S17). Finally, compared to WT controls, CA-ELK1 enhanced BTZ and CFZ resistance, while DN-ELK1 increased sensitivity (SI Appendix, Fig. S18A). This was accompanied by increased ChT-L, T-L, and C-L activities in CA-ELK1 cells and a decrease in DN cells (SI Appendix, Fig. S18B). CA-ELK1 expression was sufficient to increase POMP and proteasome subunit expression (SI Appendix, Fig. S18C) and reduce UPR intermediates (SI Appendix, Fig. S18D).
Enhanced Antitumor Activity of a MEK/Proteasome Inhibitor Regimen.
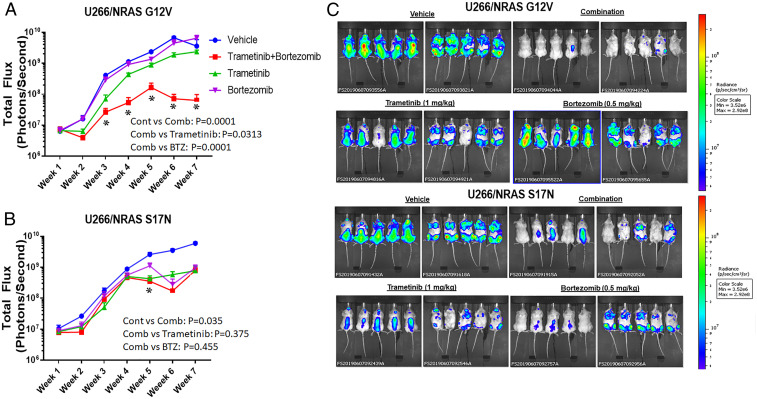
To test whether trametinib and BTZ enhanced activity in vivo, we developed systemic models. In mice with NRAS-G12V cells, BTZ and trametinib alone showed modest activity, while the combination showed enhanced efficacy (Fig. 7 A and C). BTZ showed greater activity in the NRAS-S17N xenograft (Fig. 7 B and C), consistent with earlier data showing resistance with the CA mutation. However, the BTZ/trametinib combination did not consistently show increased antitumor efficacy compared with BTZ or trametinib (Fig. 7 B and C) as it had in the G12V model. Finally, human immunoglobulin E (IgE) levels (SI Appendix, Fig. S19) also showed the combination was most effective in reducing disease burden in the G12V model.
Fig. 7.
Antitumor activity of a BTZ/trametinib regimen in vivo. Mice with U266 cells expressing CA-G12V-NRAS (A) and treated with vehicle, trametinib, BTZ, or the combination were subjected to whole animal in vivo imaging. Tumor burden as represented by mean total flux (emitted photons per second) was plotted according to time ± SD of triplicates with asterisks indicating P < 0.05 versus the vehicle controls. A similar experiment but with mice bearing DN-S17N-NRAS (B) was also shown as are whole animal images for both groups (C) from week 7.
Discussion
Sequencing of plasma cells from newly diagnosed myeloma patients confirms this disease is dominated by RAS (43% of patients) and Nuclear factor kappa B (NF-κB) pathway (17%) mutations (9). Interestingly, their distribution is not uniform among different gene expression profiling-defined subgroups. For example, NRAS-Q61 mutations are common in hyperdiploid disease and with translocation t(11;14), but less common in the MMSET and MAF subtypes (41). Most studies suggest NRAS, KRAS, or BRAF mutations activate MAPK signaling and may associate with reduced dual specificity phosphatase (DUSP)-6 (41). Since DUSP6 is a negative MAPK signaling regulator, its loss would further enhance this pathway’s activation. Also, RAS–RAF mutations may be inversely associated with NF-κB activation in all subgroups excluding MAF, suggesting that MAPK or NF-κB activation play important roles in myelomagenesis, but these have not all been well defined.
Our data show that MAPK-activating NRAS, KRAS, and BRAF mutations increase proteasome activity (Fig. 1) by enhancing expression of proteasome subunits and of chaperones involved in their assembly (Fig. 2). This is dependent on the TF ELK1 (Fig. 3 and SI Appendix, Figs. S3, S5, and S6), which has multiple binding sites in these genes’ promoters. Importantly, downstream effects reduce plasma cell stress as measured by lower expression or activation of UPR sensors and UPR effectors (Fig. 6 and SI Appendix, Fig. S13). These findings provide a mechanistic basis for prior studies showing that RAS may inhibit ER stress in cancer cell lines, including H929 myeloma cells (42). Moreover, they provide a possible reason for why NRAS, KRAS, and BRAF mutations are common in myeloma. Plasma cells are protein producing factories that rely on the UPR for survival due to their chronic exposure to proteotoxic stress. Mutations that would help them cope with this stress could promote preferential outgrowth of those subclones. Indeed, some studies have suggested that MAPK mutations are associated with transformation of MGUS to active myeloma (43–45). One could hypothesize that RAS mutations, beyond their known effects on cell proliferation, differentiation, and tumor progression (22), would contribute by enabling greater cell survival. Interestingly, our findings are consistent with previously published data showing BTZ sensitivity is reduced in patients with NRAS mutations (17), but the latter study did not find this with KRAS mutations. As this effort involved only eight patients having codon 12 KRAS mutations split among seven different variants, this sample size may have been too small to detect a difference. Alternatively, some variants may have a greater impact on BTZ sensitivity in vivo, and further studies are needed to differentiate between these possibilities.
During our studies, we identified ELK1 as a novel intermediate linking RAS/RAF/MEK/MAPK signaling to the ubiquitin-proteasome pathway. Notably, we did not study other downstream effectors, such as RAS-associated domain family members (RASSFs), which can link RAS to ubiquitination (46). Indeed, De Smedt et al. reported that RASSF4 is epigenetically down-regulated during myeloma progression and that its expression sensitized myeloma cells to BTZ and trametinib (47). Thus, their findings provide an additional rationale for a BTZ/MEK inhibitor combination and indicate that further studies are needed to delineate all of the links between RAS signaling and the ubiquitin-proteasome pathway.
Beyond the newly diagnosed setting, NRAS, KRAS, and BRAF mutations occur with increased frequency in relapsed/refractory disease (10, 11). The same survival mechanisms that support early events during myelomagenesis could enable persistence of myeloma clones later, especially if they can better survive in the marrow microenvironment where hypoxia contributes to ER stress (48). Of note, since hypoxia is a general feature of the cancer microenvironment, this same mechanism could contribute to pathobiology of NRAS-, KRAS-, and BRAF-driven solid tumors. Also, our data show that the enhanced proteasome capacity supported by activating MAPK mutations confers BTZ and CFZ resistance (Figs. 1 and 3). Given the general adoption of PI-containing regimens into our chemotherapeutic armamentarium for newly diagnosed myeloma (49, 50), one might expect that such combinations would spare cells with relative PI resistance. This could, in part, explain the increased prevalence of these mutations in later disease stages, with likely contributions from other processes, including genomic instability.
Finally, our data indicate that proteasome and MAPK pathway inhibitor combinations show synergy. The enhanced effectiveness of SEL with BTZ has been reported (51), but our findings extend these to include CFZ and a pan-RAF inhibitor. More importantly, the finding of greater synergy in models driven by NRAS, KRAS, and BRAF activating mutations provide a pathway to the clinic for this molecularly defined patient subset. Targeting of BRAF and MEK as single-agent approaches has already been evaluated clinically. For example, the RAF inhibitor vemurafenib showed activity in three cases with the V600E mutation (52, 53) and with the MEK inhibitor cobimetinib in another (54). Single-agent SEL was studied in unselected relapsed/refractory patients, but only 5.6% responded and experienced short response durations (19). When selection was performed in another study to identify patients with KRAS, NRAS, or BRAF mutations, or MAPK pathway activation, trametinib showed a 58% response rate (20). While progression-free survival data were not reported, the time-to-next treatment was a respectable 186 d. However, of 58 patients treated, 36 started with a trametinib-based combination or had other drugs added at progression to single-agent trametinib, and only a small minority of these were PIs. Our data strongly argue that patients with activating KRAS, NRAS, or BRAF mutations, or MAPK pathway activation by gene expression profiling, should be treated with a regimen containing a MEK and a proteasome inhibitor for optimal efficacy.
Materials and Methods
Reagents and Cell Lines.
Cell culture reagents, DNA restriction, and modifying enzymes, TRIzol total RNA isolation reagent, and lipofectamine were from Invitrogen Corp. (Carlsbad, CA). BTZ, CFZ, SEL, and trametinib were from Selleck Chemical (Houston, TX). Stock solutions were prepared in dimethyl-sulfoxide (Fisher Scientific; Pittsburgh, PA). TAK-632 was from Takeda Pharmaceutical Company (Cambridge, MA). U266 and ANBL-6 cells were from the American Type Culture Collection (Manassas, VA) and Dr. Diane F. Jelinek (Mayo Clinic; Rochester, MN), respectively. Cells were cultured in Roswell Park Memorial Institute medium 1640 (Corning Cellgro; Manassas, VA) with 10% fetal bovine serum, l-glutamine, and 1% penicillin/streptomycin, and ANBL-6 cells received 1 ng/mL of Interleukin-6. Cell lines were validated through our Characterized Cell Line Core Facility.
Cell Viability Assays.
Viability was evaluated using the WST-1 tetrazolium reagent (Clontech Laboratories; Mountain View, CA) (55, 56). Cells were plated in triplicate and exposed to the indicated drug concentrations for 72 h. Cell proliferation was evaluated by cell counting after staining with Trypan blue.
Generation of Stable Cell Lines.
Vectors containing WT-, G12V-, S17N-KRAS, and S17N-NRAS were from the UMR cDNA Resource Center (Rolla, MO). Plasmids encoding WT- and AA-BRAF mutants were gifts from Dr. Kun-Liang Guan (University of California, San Diego) and Dr. Yasuharu Nishimura (Kumamoto University, Japan), respectively. KRAS-G13D, NRAS-Q61R, and BRAF-V600E were generated using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA, USA) with primers (SI Appendix, Table S1) synthesized by Sigma-Aldrich (St. Louis, MO). Coding sequences were subcloned into the lentiviral transfer vector pCDH-CMV-MCS-EF1-co green fluorescent protein (GFP) and verified by sequencing. Plasmids with confirmed insertions and the control vector pCDH-CMV-coGFP were transfected into 293T cells with the packaging vectors psPAX2 and pMD2.G. Recombinant lentivirus particles were harvested at 24 and 48 h, and supernatants were concentrated by polyethylene glycol precipitation. ANBL-6 and U266 cells were transduced with comparable amounts of recombinant lentiviruses in growth medium containing polybrene and sorted by flow.
ELK1-targeted shRNAs lentiviral constructs were from Sigma-Aldrich. The template vectors carrying ELK1(1-428), ELK1(1-428)-EN(2-298), and ELK1(1-405)-VP16(410-490) were provided by Dr. Andrew D. Sharroks (57). ELK1, ELK1-EN, or ELK1-VP16 were cloned into the pCDH-CMV-MCS-EF1α-copGFP lentivirus vector (System Biosciences) using Xba1 and NheI (New England BioLabs) and primers listed in SI Appendix, Table S1. VP16 (P06492) amino acids 410–490 from the human herpes simplex virus 1 were selected for ELK1 (P19419) activation and EN (P02836) amino acids 2–298 from a fruit fly for ELK1 repression. The final ELK1 up-/down-regulation vectors, ELK1-VP16, and ELK1-EN were obtained after transformation in One Shot Stbl3 Chemically Competent Escherichia coli (Thermo Fisher Scientific), clone selection by C-AMP antibiotic, and validation by sequencing.
Immunoblotting.
Cells were lysed in buffer containing complete protease inhibitor mixture (Roche Diagnostics, Indianapolis, IN) and phenyl-methyl-sulfonyl-fluoride (Sigma-Aldrich). Lysates were clarified, and equal protein amounts were loaded on 4–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels (58). Separated proteins were transferred onto membranes and probed using primary (SI Appendix, Table S2) and secondary antibodies, developed by enhanced chemiluminescence, and exposed to Hyperfilm-ECL (GE Healthcare Biosciences, Pittsburgh, PA).
TF Binding Analysis.
JASPAR (http://jaspar.genereg.net), an open access database storing curated, nonredundant TF binding profiles representing TF binding preference as position frequency matrices for multiple species in six taxonomic groups was used to predict binding to POMP, PSMB8, PSMB9, and PSMB10.
Proteasome Activity.
The chymotrypsin-, trypsin-, and caspase-like activities were determined by release of 7-amino-4-methylcoumarin from the Suc-LLVY-AMC, Ac-RLR-AMC, and Z-LLE-AMC substrates, respectively, and quantified (31).
Real Time-PCR.
Real time (RT)-PCR was carried out as described (56) with minor modifications. Complementary DNA (cDNA) was synthesized from cellular RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Quantitative (q) RT-PCR was performed using the TaqMan Gene Expression Master Mix and the ATF4 (FAMTM), ATF5 (FAMTM), ATF6 (FAMTM), CHOP (FAMTM), PSMB8, PSMB9, PSMB10 (FAMTM), POMP (FAMTM), NRF2 (FAMTM), NOXA (FAMTM), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH, VIC) probes on a StepOnePlus PCR system (Applied Biosystems). Spliced and unspliced XBP1 mRNAs were detected by Syber Green PCR (SI Appendix, Table S1), and quantification was by the comparative CT method with GAPDH as the control.
Xenograft Model.
Six-week-old nonobese diabetic mice with severe combined immunodeficiency (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) were from The Jackson Laboratory (Bar Harbor, ME), and mice were irradiated with a sublethal dose of 250 rad of total body irradiation 4–6 h prior to implantation of myeloma cells. U266 NRAS-G12V and NRAS-S17N cells (2 × 106/mouse) were intravenously injected under an Institutional Animal Care and Use Committee approved protocol. Mice were divided into four groups: Control (cohort-1) received polyethylene glycol 30 by intraperitoneal (IP) injection every other day; MEK inhibitor (cohort-2) received trametinib (3 mg/kg) IP thrice weekly (59); PI (cohort-3) received BTZ (0.5-mg/kg) IP twice weekly; and combination (cohort-4) received trametinib and BTZ. Tumor size/burden was evaluated by bioluminescence imaging after IP d-luciferin (Caliper Life Sciences) injection using the IVIS Imaging System (Caliper Life Sciences, Hanover, MD) and Living Image 4.4.SP2 software (Caliper Life Sciences). An enzyme-linked immunosorbent assay measured human IgE levels (Bethyl Laboratories, Montgomery, TX).
Statistical Analyses.
Data are expressed as the mean plus SD (for triplicate data from the same experiment) or SEMs (for multiple independent experiments). The significance of drug-effect relationships was determined by one-tailed unpaired t tests or ANOVA using Graph-Pad Prism. Bonferroni multiplicity adjustment was applied for multiple comparisons.
Supplementary Material
Acknowledgments
The authors thank the MD Anderson Flow Cytometry and Cellular Imaging and the Characterized Cell Line Core Facilities supported by the Cancer Center Support Grant (P30 CA16672). R.Z.O., the Florence Maude Thomas Cancer Research Professor, acknowledges support from the National Cancer Institute (Grants R01 CA194264, R01 CA184464, P50 CA142509, U10 CA032102), and the Leukemia and Lymphoma Society (Grant SCOR-12206-17). Additional support came from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the MD Anderson Cancer Center High Risk Multiple Myeloma Moon Shot, the Brock Family Myeloma Research Fund, the Yates Ortiz Myeloma Fund, and the Diane and John Grace Family Foundation.
Footnotes
Competing interest statement: The authors declare a competing interest. H.C.L. has provided consultancy services to Amgen, Inc., Celgene, a wholly owned subsidiary of Bristol-Myers Squibb, GlaxoSmithKline, Janssen Pharmaceutical, Sanofi-Aventis, and Takeda Pharmaceutical and has received research funding from Amgen, Inc., Celgene, a wholly owned subsidiary of Bristol-Myers Squibb, Daiichi Sankyo, GlaxoSmithKline, Janssen Pharmaceutical, and Takeda Pharmaceuticals. L.D. and N.C. are employees of Takeda Pharmaceuticals U.S.A., Inc. R.Z.O. declares laboratory research funding from BioTheryX and clinical research funding from CARsgen Therapeutics, Celgene, Exelixis, Janssen Biotech, Sanofi-Aventis, Takeda Pharmaceuticals North America, Inc. Also, R.Z.O. has served on advisory boards for Amgen, Inc., Bristol-Myers Squibb, Celgene, EcoR1 Capital LLC, Forma Therapeutics, Genzyme, GSK Biologicals, Ionis Pharmaceuticals, Inc., Janssen Biotech, Juno Therapeutics, Kite Pharma, Legend Biotech USA, Molecular Partners, Regeneron Pharmaceuticals, Inc., Sanofi-Aventis, Servier, and Takeda Pharmaceuticals North America, Inc. and as a consultant for STATinMED Research. Finally, R.Z.O. is a Founder of Asylia Therapeutics, Inc., with associated patents and an equity interest, although this technology does not bear on the current paper. The remaining authors have no conflicts of interest to declare.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005052117/-/DCSupplemental.
Data Availability.
All of the data needed to interpret these studies are included in the main paper’s figures and table and in the Supplementary figures and tables.
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2015. CA Cancer J. Clin. 65, 5–29 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Smith B. D., Smith G. L., Hurria A., Hortobagyi G. N., Buchholz T. A., Future of cancer incidence in the United States: Burdens upon an aging, changing nation. J. Clin. Oncol. 27, 2758–2765 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Manasanch E. E., Orlowski R. Z., Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 14, 417–433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi G., Anderson K. C., Understanding biology to tackle the disease: Multiple myeloma from bench to bedside, and back. CA Cancer J. Clin. 64, 422–444 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Moreau P., Attal M., Facon T., Frontline therapy of multiple myeloma. Blood 125, 3076–3084 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Neri A. et al., Ras oncogene mutation in multiple myeloma. J. Exp. Med. 170, 1715–1725 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman M. A. et al., Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolli N. et al., Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 5, 2997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker B. A. et al., Mutational spectrum, copy number changes, and outcome: Results of a sequencing study of patients with newly diagnosed Myeloma. J. Clin. Oncol. 33, 3911–3920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kortüm K. M. et al., Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood 128, 1226–1233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mithraprabhu S. et al., Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia 31, 1695–1705 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Rasmussen T., Kuehl M., Lodahl M., Johnsen H. E., Dahl I. M., Possible roles for activating RAS mutations in the MGUS to MM transition and in the intramedullary to extramedullary transition in some plasma cell tumors. Blood 105, 317–323 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Portier M. et al., p53 and RAS gene mutations in multiple myeloma. Oncogene 7, 2539–2543 (1992). [PubMed] [Google Scholar]
- 14.Corradini P. et al., Mutational activation of N- and K-ras oncogenes in plasma cell dyscrasias. Blood 81, 2708–2713 (1993). [PubMed] [Google Scholar]
- 15.Corradini P. et al., Inactivation of tumor suppressor genes, p53 and Rb1, in plasma cell dyscrasias. Leukemia 8, 758–767 (1994). [PubMed] [Google Scholar]
- 16.Liu P. et al., Activating mutations of N- and K-ras in multiple myeloma show different clinical associations: Analysis of the eastern cooperative oncology group phase III trial. Blood 88, 2699–2706 (1996). [PubMed] [Google Scholar]
- 17.Mulligan G. et al., Mutation of NRAS but not KRAS significantly reduces myeloma sensitivity to single-agent bortezomib therapy. Blood 123, 632–639 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Y. T. et al., Integrated phosphoproteomics and transcriptional classifiers reveal hidden RAS signaling dynamics in multiple myeloma. Blood Adv. 3, 3214–3227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holkova B. et al., A phase II trial of AZD6244 (Selumetinib, ARRY-142886), an oral MEK1/2 inhibitor, in relapsed/refractory multiple Myeloma. Clin. Cancer Res. 22, 1067–1075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuck C. J. et al., Inhibiting MEK in MAPK pathway-activated myeloma. Leukemia 30, 976–980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tolcher A. W. et al., Phase I study of the MEK inhibitor trametinib in combination with the AKT inhibitor afuresertib in patients with solid tumors and multiple myeloma. Cancer Chemother. Pharmacol. 75, 183–189 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Fey D., Matallanas D., Rauch J., Rukhlenko O. S., Kholodenko B. N., The complexities and versatility of the RAS-to-ERK signalling system in normal and cancer cells. Semin. Cell Dev. Biol. 58, 96–107 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Crowder C. et al., An unusual H-Ras mutant isolated from a human multiple myeloma line leads to transformation and factor-independent cell growth. Oncogene 22, 649–659 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Steinbrunn T. et al., Mutated RAS and constitutively activated Akt delineate distinct oncogenic pathways, which independently contribute to multiple myeloma cell survival. Blood 117, 1998–2004 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Hoang B. et al., Oncogenic RAS mutations in myeloma cells selectively induce cox-2 expression, which participates in enhanced adhesion to fibronectin and chemoresistance. Blood 107, 4484–4490 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bianchi G. et al., The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood 113, 3040–3049 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Leung-Hagesteijn C. et al., Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell 24, 289–304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X. D. et al., Tight junction protein 1 modulates proteasome capacity and proteasome inhibitor sensitivity in multiple Myeloma via EGFR/JAK1/STAT3 signaling. Cancer Cell 29, 639–652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B. et al., The nuclear factor (Erythroid-derived 2)-like 2 and proteasome maturation protein Axis mediate bortezomib resistance in multiple Myeloma. J. Biol. Chem. 290, 29854–29868 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn D. J. et al., Targeted inhibition of the immunoproteasome is a potent strategy against models of multiple myeloma that overcomes resistance to conventional drugs and nonspecific proteasome inhibitors. Blood 113, 4667–4676 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh A. V. et al., PR-924, a selective inhibitor of the immunoproteasome subunit LMP-7, blocks multiple myeloma cell growth both in vitro and in vivo. Br. J. Haematol. 152, 155–163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickering A. M. et al., The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 432, 585–594 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickering A. M., Linder R. A., Zhang H., Forman H. J., Davies K. J., Nrf2-dependent induction of proteasome and Pa28αβ regulator are required for adaptation to oxidative stress. J. Biol. Chem. 287, 10021–10031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chong S. J. F., Lai J. X. H., Eu J. Q., Bellot G. L., Pervaiz S., Reactive oxygen species and oncoprotein signaling-A dangerous Liaison. Antioxid. Redox Signal. 29, 1553–1588 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Okaniwa M. et al., Discovery of a selective kinase inhibitor (TAK-632) targeting pan-RAF inhibition: Design, synthesis, and biological evaluation of C-7-substituted 1,3-benzothiazole derivatives. J. Med. Chem. 56, 6478–6494 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Yeh T. C. et al., Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin. Cancer Res. 13, 1576–1583 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Dick L. R., Moomaw C. R., DeMartino G. N., Slaughter C. A., Degradation of oxidized insulin B chain by the multiproteinase complex macropain (proteasome). Biochemistry 30, 2725–2734 (1991). [DOI] [PubMed] [Google Scholar]
- 38.Hetz C., Papa F. R., The unfolded protein response and cell fate control. Mol. Cell 69, 169–181 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Fiorese C. J. et al., The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol. 26, 2037–2043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pihán P., Carreras-Sureda A., Hetz C., BCL-2 family: Integrating stress responses at the ER to control cell demise. Cell Death Differ. 24, 1478–1487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein C. K. et al., The varied distribution and impact of RAS codon and other key DNA alterations across the translocation cyclin D subgroups in multiple myeloma. Oncotarget 8, 27854–27867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yaari-Stark S. et al., Ras inhibits endoplasmic reticulum stress in human cancer cells with amplified Myc. Int. J. Cancer 126, 2268–2281 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Matozaki S., Nakagawa T., Nakao Y., Fujita T., RAS gene mutations in multiple myeloma and related monoclonal gammopathies. Kobe J. Med. Sci. 37, 35–45 (1991). [PubMed] [Google Scholar]
- 44.Bezieau S. et al., High incidence of N and K-Ras activating mutations in multiple myeloma and primary plasma cell leukemia at diagnosis. Hum. Mutat. 18, 212–224 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Davies F. E. et al., Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood 102, 4504–4511 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Donninger H., Schmidt M. L., Mezzanotte J., Barnoud T., Clark G. J., Ras signaling through RASSF proteins. Semin. Cell Dev. Biol. 58, 86–95 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Smedt E. et al., Loss of RASSF4 expression in multiple myeloma promotes RAS-driven malignant progression. Cancer Res. 78, 1155–1168 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Wang M., Law M. E., Castellano R. K., Law B. K., The unfolded protein response as a target for anticancer therapeutics. Crit. Rev. Oncol. Hematol. 127, 66–79 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Orlowski R. Z., Lonial S., Integration of novel agents into the care of patients with multiple Myeloma. Clin. Cancer Res. 22, 5443–5452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunacheewa C., Orlowski R. Z., New drugs in multiple myeloma. Annu. Rev. Med. 70, 521–547 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Tai Y. T. et al., Targeting MEK induces myeloma-cell cytotoxicity and inhibits osteoclastogenesis. Blood 110, 1656–1663 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Andrulis M. et al., Targeting the BRAF V600E mutation in multiple myeloma. Cancer Discov. 3, 862–869 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Sharman J. P. et al., Vemurafenib response in 2 patients with posttransplant refractory BRAF V600E-mutated multiple myeloma. Clin. Lymphoma Myeloma Leuk. 14, e161–e163 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Mey U. J. M., Renner C., von Moos R., Vemurafenib in combination with cobimetinib in relapsed and refractory extramedullary multiple myeloma harboring the BRAF V600E mutation. Hematol. Oncol. 35, 890–893 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Jones R. J. et al., HDM-2 inhibition suppresses expression of ribonucleotide reductase subunit M2, and synergistically enhances gemcitabine-induced cytotoxicity in mantle cell lymphoma. Blood 118, 4140–4149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bjorklund C. C. et al., Evidence of a role for activation of Wnt/beta-catenin signaling in the resistance of plasma cells to lenalidomide. J. Biol. Chem. 286, 11009–11020 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boros J. et al., Overlapping promoter targeting by Elk-1 and other divergent ETS-domain transcription factor family members. Nucleic Acids Res. 37, 7368–7380 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhuang J. et al., Ubiquitin-activating enzyme inhibition induces an unfolded protein response and overcomes drug resistance in myeloma. Blood 133, 1572–1584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kerstjens M. et al., Trametinib inhibits RAS-mutant MLL-rearranged acute lymphoblastic leukemia at specific niche sites and reduces ERK phosphorylation in vivo. Haematologica 103, e147–e150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data needed to interpret these studies are included in the main paper’s figures and table and in the Supplementary figures and tables.