Mechanisms ensuring faithful reproduction are enforced for viruses, as for all other organisms, by natural selection. As a virus particle is a package containing the viral genome (RNA or DNA, as the case may be) for replication and transmission to the next generation, it is essential that the genome be packaged into the assembling virus particle with high fidelity. In turn, viruses use a wide variety of mechanisms for this selective packaging.
HIV type 1 (HIV-1), the causative agent of AIDS, is a retrovirus. The genome within a retrovirus particle is composed of RNA. When it infects a cell, this RNA is copied into double-stranded DNA, which is then integrated into the chromosomal DNA of the cell. How the genomic RNA (viral RNA [vRNA]) is selected for packaging during virus particle assembly is not well understood, and a paper by Ding et al. (1) in PNAS now adds a piece to this intriguing puzzle.
Retrovirus particles are roughly spherical, ∼100 to 120 nm in diameter, and are initially assembled primarily from several thousand molecules of the building block, the Gag polyprotein. Thus, it is the Gag polyprotein that selects the vRNA for incorporation into the nascent particle. However, after the virus particle is released from the virus-producing cell, Gag is cleaved by the virus-coded protease into a series of fragments. This “virus maturation” event is a wholesale reorganization of the structure of the virus and is essential for the particle’s ability to undertake an infection of a new host cell. Therefore, while Gag is the principal protein in the immature particle, the predominant proteins in the mature, infectious virus particle are fragments of Gag.
There are a number of remarkable aspects to the selective packaging of vRNA during HIV assembly. vRNA is selectively packaged because it contains a “packaging signal” or “ψ” [also designated the “core encapsidation signal” (2)]. If Gag is expressed in a cell without any ψ-containing RNA, it still assembles efficiently into virus-like particles, packaging an unselected population of cellular mRNA molecules (3). Thus, vRNA is normally in competition with a vast excess of cellular RNAs for inclusion into the particles, and its ψ moiety provides the crucial advantage in this competition.
What is ψ? ψ is a stretch of ∼150 to 250 bases near the 5′ end of vRNA. It is highly structured, with several conserved stem-loops. This is obviously large enough to bind multiple Gag molecules. In fact, the RNA in nascent virus particles is actually a dimer of vRNA, with two ∼10,000-base genomic RNA molecules joined by intermolecular base-pairing between their ψ regions. A further twist is that the ψ region of vRNA can assume either of two alternative conformations, and that only one of these is capable of dimerization and packaging; molecules with the other conformation presumably function as mRNA in the virus-producing cell (4). Moreover, it has recently become clear that not all of the HIV RNA molecules in the cell are initiated at exactly the same base, and that their start site strongly influences their choice between these conformers and hence their ability to be packaged (5, 6).
Michael Summers and his colleagues have worked for many years on the extremely challenging problem of determining the three-dimensional structure of ψ (specifically, of the ψ conformer capable of dimerization and packaging). They recently reported a novel model structure in which two three-way junctions are positioned one after the other in the RNA (7). These provocative findings are an important step in bridging the gap between biology and biochemistry, and raise the possibility of ultimately developing new therapeutic strategies, as discussed below.
As noted, Gag is a polyprotein, composed of distinct domains that are separated into discrete proteins when the virus matures into an infectious particle. Of these domains, the one most important with respect to interactions with nucleic acids is the nucleocapsid (NC) domain. NC is very small (only 55 amino acids) and is strongly positively charged. The entire vRNA within the mature particle is coated with NC molecules. It is largely flexible, except for two 14-amino acid “zinc fingers,” which are structured because their cysteine and histidine residues coordinate a zinc ion. Replacements of these zinc-coordinating residues with other amino acids yields a Gag protein that can assemble into virus-like particles, but has lost the ability to specifically package vRNA (8, 9); this observation shows that the zinc fingers play an essential role in packaging vRNA.
Since ψ is the ticket for selective packaging of an RNA by Gag, one might imagine that Gag binds with a uniquely high affinity to ψ. However, tests with recombinant Gag protein show that at physiological ionic strength, Gag binds very tightly to any RNA (10, 11). This tight binding is not surprising, as Gag has a significant net positive charge and its “matrix” (MA) domain, as well as the NC domain, is positively charged. The role of electrostatics in this nonspecific RNA–Gag interaction is revealed by its sensitivity to ionic strength: Raising the salt concentration in the assay buffer readily detaches bound RNA from Gag. Notably, however, this analysis also shows that the binding of Gag to ψ is far more salt-resistant than its binding to control RNAs. In other words, even though the tight binding at physiological salt concentrations is predominantly electrostatic and thus essentially independent of nucleotide sequence, there is a unique, nonelectrostatic component in the interaction of Gag with ψ (10, 11). A crucial unanswered question, then, is how this relates to selective packaging of ψ-containing RNAs, which obviously occurs at physiological ionic strength. One possible explanation could be that interaction with ψ initiates particle assembly more efficiently than interaction with other RNAs (4, 12, 13).
The paper by Ding et al. (1) now adds exciting information to this story. They have used NMR and calorimetry in an extremely detailed investigation of the interaction of free NC protein with ψ. (The small size of NC makes it tractable for structural studies; these would be impossible with Gag, which is somewhat over 500 amino acids in length and contains several highly flexible stretches.) They report that on the order of 20 NC molecules can bind to ψ at physiological ionic strength. Unexpectedly, the tightest binding events are endothermic, meaning that they possess a major nonelectrostatic component. It thus seems almost certain that these NC–ψ interactions Ding et al. (1) describe are responsible for the nonelectrostatic interactions between Gag and ψ discussed above.
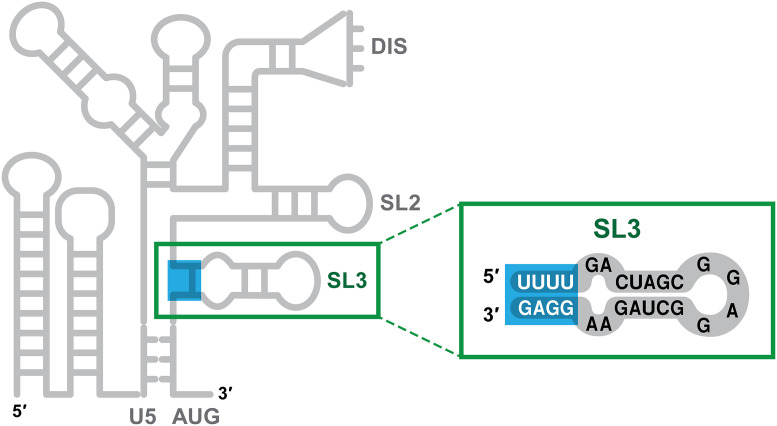
By a painstaking analysis of the interactions of NC with fragments of ψ, Ding et al. successfully identified the target within ψ of these nonelectrostatic binding events. It turns out to be a stretch of four base pairs near the connection point of the two three-way junctions mentioned above. These base pairs are not canonical Watson–Crick base pairs and have largely been overlooked in the analysis of ψ structure: They are pairs between nucleotides 306 to 309 (sequence UUUU) and 328 to 331 (sequence GGAG) of HIV RNA. Thus, three of the four pairs are G:U pairs (while G normally pairs with C, it can also pair with U in RNA; G:U base pairs are less stable than G:C base pairs and are frequently called “wobble” base pairs). The specific binding to these bases is consistent with the known preference of NC for unpaired or weakly paired G bases in nucleic acids (14–16). These base pairs, near the base of a well-characterized stem-loop (“stem-loop 3”) within ψ, are highlighted in Fig. 1.
Fig. 1.
Labile base pairs within the HIV-1 packaging signal. The left-hand portion of the figure shows, in cartoon form, the secondary structure of the 5′ 344 bases of HIV-1 vRNA. “ψ” extends roughly from nucleotide 105 (indicated as “U5”) to the end of the cartoon. U5, DIS, SL2, SL3, and AUG are landmarks within ψ. The figure highlights the four base pairs near the base of stem-loop 3 (SL3), which are the targets of high-affinity, endothermic binding by the viral NC protein.
To investigate the mechanism of NSC 260594 action, Ding et al. incubated a ψ fragment containing the UUUU:GGAG base-paired motif with the compound and found that it specifically altered the NMR signals from bases adjacent to the motif. Thus, it evidently binds ψ at this motif.
As one approach to identifying the site of this high-affinity, endothermic binding by NC, Ding et al. made a series of mutations in these paired bases and in their immediate surroundings. These studies showed that the lability of the base-paired structure is essential for the binding, since the binding is blocked either if the G:U pairs are replaced by G:C pairs or if the stem containing these G:U pairs is extended by bases that can form stable Watson–Crick pairs with each other. The NMR analysis also showed that binding by NC induces the unfolding or rearrangement of the critical GGAG:UUUU base-paired structure.
It is particularly interesting that NC disrupts this rather labile structure. One of the remarkable properties of NC is its activity as a nucleic acid chaperone (17). These chaperones catalyze the rearrangement of nucleic acids into the most thermodynamically stable structures, i.e., the structures with the greatest number of base pairs. (This activity is essential during the reverse transcription step of HIV infection, in which the copying of the RNA into double-stranded DNA involves several hybridization events.) One property contributing to the chaperone activity of NC is its modest preference for single-stranded over double-stranded nucleic acids; this enables it to destabilize base-paired structures, as required for their rearrangement into more stable structures.
To test the significance of the UUUU:GGAG motif for selective packaging of vRNA, Ding et al. also replaced it with UUCC:GGAG in an HIV-derived vector. This alteration replaces a pair of G:U pairs with more stable G:C pairs. The mutant and control wild-type vectors were expressed in mammalian cells together with HIV structural proteins, and the effect of the mutation upon the packaging of the vector RNA was measured. It was found that the mutant RNA was packaged at a significantly lower level than the control. This result demonstrates that these bases are important elements in the functional HIV packaging signal.
The fact that selective packaging of vRNA is essential for efficient viral replication makes it a potential target for antiviral intervention. In fact, Ingemarsdotter et al. (18) recently described a compound, NSC 260594, that binds within ψ and interferes with vRNA packaging and the production of infectious virus. To investigate the mechanism of NSC 260594 action, Ding et al. incubated a ψ fragment containing the UUUU:GGAG base-paired motif with the compound and found that it specifically altered the NMR signals from bases adjacent to the motif. Thus, it evidently binds ψ at this motif. These results are a proof of the principle that therapies disrupting RNA structure and blocking the protein–RNA interactions involved in vRNA packaging are possible. This is an exciting approach to antiviral therapy. Obviously, developing such therapies will require a thorough understanding of these interactions; the results of Ding et al. are an important step in this direction.
Acknowledgments
I thank Siddhartha Datta and Saraswati Sukumar for helpful comments on the manuscript.
Footnotes
The author declares no competing interest.
See companion article, “Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal,” 10.1073/pnas.2008519117.
References
- 1.Ding P., et al. , Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc. Natl. Acad. Sci. U.S.A. 117, 17737–17746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heng X., et al. , Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 417, 224–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rulli S. J., Jr, et al. , Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 81, 6623–6631 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rein A., RNA packaging in HIV. Trends Microbiol. 27, 715–723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown J. D., et al. , Structural basis for transcriptional start site control of HIV-1 RNA fate. Science 368, 413–417 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masuda T., et al. , Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci. Rep. 5, 17680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keane S. C., et al. , RNA structure. Structure of the HIV-1 RNA packaging signal. Science 348, 917–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldovini A., Young R. A., Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 64, 1920–1926 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorelick R. J., et al. , Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64, 3207–3211 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comas-Garcia M., et al. , Dissection of specific binding of HIV-1 Gag to the “packaging signal” in viral RNA. eLife 6, e27055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webb J. A., Jones C. P., Parent L. J., Rouzina I., Musier-Forsyth K., Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: Implications for viral genomic RNA packaging. RNA 19, 1078–1088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comas-Garcia M., et al. , Efficient support of virus-like particle assembly by the HIV-1 packaging signal. eLife 7, e38438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikolaitchik O. A., et al. , Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 9, e1003249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amarasinghe G. K., et al. , NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301, 491–511 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson K. A., et al. , High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6, e96 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Guzman R. N., et al. , Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279, 384–388 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Levin J. G., Guo J., Rouzina I., Musier-Forsyth K., Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80, 217–286 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Ingemarsdotter C. K., Zeng J., Long Z., Lever A. M. L., Kenyon J. C., An RNA-binding compound that stabilizes the HIV-1 gRNA packaging signal structure and specifically blocks HIV-1 RNA encapsidation. Retrovirology 15, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]