Significance
Many brain regions, notably the hippocampus, contain a record of the recent past with time cells, neurons that fire in sequence, each at a specific time after a triggering event. The origin of this neural timeline has been unclear. This paper reports a timing signal in the entorhinal cortex (EC), which provides input to the hippocampus. Rather than firing sequentially, all EC neurons activated shortly after a stimulus and then decayed at a variety of rates. Because different neurons decay at different rates, one can reconstruct how far in the past the stimulus was presented by noting which neurons are still active. These results align well with the theoretical proposal that the brain represents the real Laplace transform of the past.
Keywords: time, entorhinal cortex, memory, Laplace transform
Abstract
Episodic memory is believed to be intimately related to our experience of the passage of time. Indeed, neurons in the hippocampus and other brain regions critical to episodic memory code for the passage of time at a range of timescales. The origin of this temporal signal, however, remains unclear. Here, we examined temporal responses in the entorhinal cortex of macaque monkeys as they viewed complex images. Many neurons in the entorhinal cortex were responsive to image onset, showing large deviations from baseline firing shortly after image onset but relaxing back to baseline at different rates. This range of relaxation rates allowed for the time since image onset to be decoded on the scale of seconds. Further, these neurons carried information about image content, suggesting that neurons in the entorhinal cortex carry information about not only when an event took place but also, the identity of that event. Taken together, these findings suggest that the primate entorhinal cortex uses a spectrum of time constants to construct a temporal record of the past in support of episodic memory.
Episodic memory, the vivid recollection of an event situated in a specific time and place (1), depends critically on medial temporal lobe (MTL) structures, including the hippocampus and entorhinal cortex (EC) (2–5). Building on pioneering work demonstrating a spatial code in the hippocampus and EC (6, 7), recent research has shown that hippocampal representations also carry information about the time at which past events took place, suggesting that the MTL maintains a representation of spatiotemporal context in support of episodic memory (8–10). Although a great deal is known about the temporal coding properties of neurons in the hippocampus, the temporal code in the EC, which provides the majority of the cortical input to the hippocampus, is less understood, but see refs. 11–14.
Hippocampal time cells provide a record of recent events including explicit information about when an event occurred. Analogous to hippocampal place cells that fire when an animal is in a circumscribed region of physical space (6, 15), hippocampal time cells fire during a circumscribed period within unfilled delays (8, 9, 16). Across studies, there is a remarkable consistency in the properties of hippocampal time cells. Hippocampal time cells peak at a range of times during the delay interval and typically code time with decreasing accuracy as the delay unfolds, as manifest by fewer neurons with peak firing late in the delay and wider time fields later in the delay (12, 17, 18). Hippocampal time cells have been observed in a wide range of behavioral paradigms, including tasks with and without explicit memory demands during the delay (17) and experiments in which the animal is fixed in space (19, 20). In addition, it has been shown that different stimuli trigger different time cell sequences (19, 20). Taken together, time cells provide an explicit record of how far in the past an event took place (i.e., the amount of time that has passed since the beginning of a delay period or since the presentation of a to-be-remembered stimulus). By examining which time cells are active at a particular time, we can easily determine not only what event took place but how far in the past that event occurred.
Many of the properties of hippocampal time cells have been observed in other brain regions including prefrontal cortex (21–24) and striatum (24–26), suggesting that the hippocampus is part of a widespread network that carries episodic information. A recent report from the rat lateral EC adds important data to this growing body of literature regarding the representation of time in the brain. Tsao et al. (14) observed a population of neurons that changed slowly and reliably enough to decode time within the experiment over a range of timescales. Unlike time cells, which respond a characteristic time since the event that triggers their firing, lateral EC neurons respond immediately upon entry into a new environment and then relax slowly. The relaxation times across individual neurons were very different, ranging from tens of seconds to thousands of seconds. To distinguish this population from time cells, we will refer to neurons that are activated by an event and then relax their firing gradually as temporal context cells. The designation “temporal context cells” is not meant to indicate some intrinsic biological property but is simply intended, much like the terms “place cell” or “time cell,” as a convenient shorthand to describe the functional properties of these neurons. Because these temporal context cells code for time, but with very different properties than time cells, these two populations provide a potentially important clue about the nature of temporal coding in the brain and the neural mechanisms that may support episodic memory.
Here, we identified temporal context cells in monkey EC during a free-viewing task (27). We examined EC neuron responses in a 5-s period after presentation of an image. In the time after presentation of the image, a representation of what happened when should carry both time-varying information about when the image was presented as well as information that discriminates the identity of the image. To anticipate the results, the data demonstrate that neurons in monkey EC are activated shortly after a visual stimulus and then decay with a variety of rates, enabling reconstruction of when the image was presented. This form of temporal coding is similar to temporal context cells observed in rat lateral EC (14) but is qualitatively different from time cells that have been observed in the hippocampus and other regions. Because each image was shown twice over the course of the experiment, we were able to assess whether the pattern of activation over neurons depends on the identity of the image presented. Taken together, these data suggest that these temporal context cells carry information about what happened in addition to when it happened.
Results
A total of 349 neurons were recorded from the EC in two macaque monkeys during performance of a visual free-viewing task. Each trial began with a required fixation on a small cross, followed by the presentation of a large, complex image that remained on the screen for 5 s of free viewing (Fig. 1A). Unlike canonical hippocampal time cells, which are activated at a variety of points within a time interval (e.g., Fig. 1C), most entorhinal neurons changed their firing a short time after the presentation of the image. Fig. 2A shows three representative neurons that responded to image presentation (more examples are shown in SI Appendix, Fig. S3). While most of these neurons increased their firing rate after the image was presented, some neurons decreased their firing rate in response to image presentation. Although behavior was not controlled during the 5-s free-viewing period, the response of these neurons was consistent across trials, which can be seen by examination of the trial rasters, indicating that the temporal responsiveness was unlikely to be a correlate of behavior.
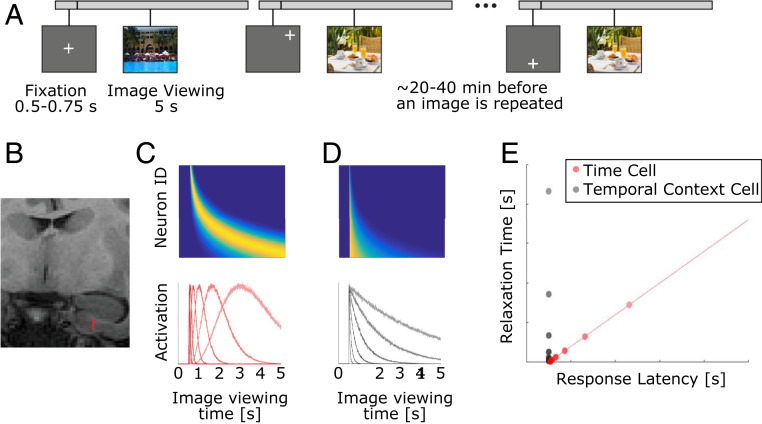
Fig. 1.
(A and B) Summary of experimental procedures. (A) Trial schematic for three trials. On each trial, the monkey freely viewed an image. After the monkey viewed an image for 5 s, the image disappeared. Following every trial, the monkey performed multiple gaze calibration trials and received a fruit slurry reward (Methods has details). Images were presented twice during an experimental session. Between 20 and 40 min passed before an image was repeated. (B) Estimated position of recording channels in the EC in one recording session is shown in red on a coronal MRI. (C–E) Two hypotheses for neural representations of time. (C and D) Heat plot (Upper) and tuning curves (Lower) for two hypotheses for how a time interval of image viewing could be coded in neural populations. In the heat plots, cooler colors correspond to low activity, while warmer colors correspond to higher activity. (C) Hypothetical activity for sequentially activated time cells, like those observed in the hippocampus. In this population, different neurons exhibit peak responses at different times indicating different firing fields. Because the time of peak response across neurons covaries with the spread of the firing field, neurons with later firing fields display wider firing fields. (D) Hypothetical activity for monotonically decaying temporal context cells, like those observed in rodent EC. Neurons in this population reach their peak at about the same time. However, different neurons decay at different rates. (E) Properties of neurons representing time passage by the hypothesis shown in C (red) or the hypothesis shown in D (gray). A population of time cells (red) should exhibit responses that occur at different times across a time interval, and these neurons should show a robust correlation between when response occurs and the time it takes the response to return to baseline. Conversely, a population of exponentially decaying temporal context cells (gray) should exhibit responses that occur in a more restricted time range shortly after the start of a time interval, and these neurons should show no correlation between when peak response occurs and the time it takes to relax back to baseline.
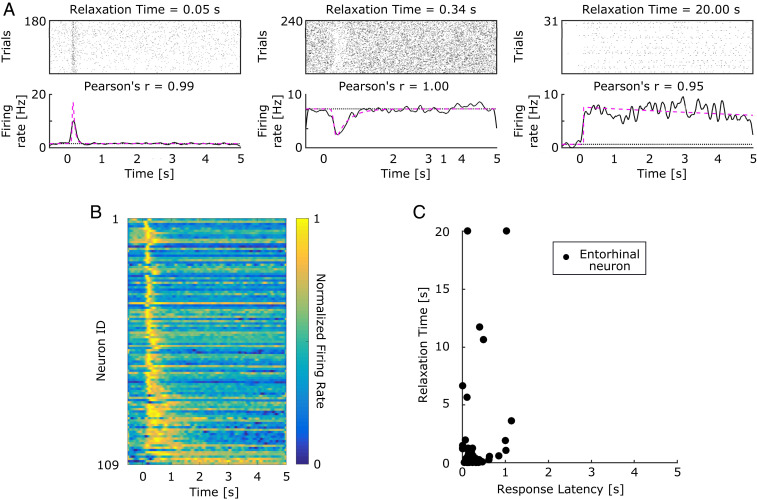
Fig. 2.
Temporal context cells in monkey EC respond shortly after image presentation and then return to baseline at a spectrum of rates. (A) Three representative context cells that responded to image onset and decayed at different rates (SI Appendix, Fig. S3 shows more examples). Each pair of plots indicates the activity of one neuron relative to image onset in a raster plot (Upper) and peristimulus time histogram (PSTH) (Lower). In each raster, a tick mark indicates when the neuron fired an action potential. In the PSTH, the solid black line indicates the smoothed firing rate, and the pink line indicates the model estimate of firing rate. The estimated baseline firing rate is indicated by the black dotted line. Relaxation Time refers to the duration between response peak and when the neurons returned 63% of the way to baseline. Pearson’s is the correlation between the fits for even and odd trials. (B) A heat plot of the normalized firing rate of 109 temporal context cells, relative to image onset, sorted by their Relaxation Time. The color scheme is the same as in Fig. 1 C and D. The majority of neurons responded within 1 s of image onset, relaxing back to baseline with a spectrum of decay rates; some neurons relaxed back to baseline much sooner than other neurons that relaxed more slowly. (C) A scatterplot of the joint distribution of each neuron’s Response Latency (the time at which a cell begins to respond) and Relaxation Time. Response Latencies did not span the entire 5 s, unlike Relaxation Time, and a neuron’s Response Latency and Relaxation Time were not correlated. SI Appendix, Fig. S2 shows the marginal distributions of these parameters.
Although the image-responsive neurons in EC responded at about the same time poststimulus, they relaxed back to their baseline firing at different rates. Whereas some neurons relaxed back to baseline quickly (Fig. 2A, Left), some relaxed much more slowly. For instance, the neuron shown in Fig. 2A, Right did not return to baseline even after 5 s.
Temporal Receptive Field Analysis Revealed a Population of Temporal Context Cells in EC.
To quantify the qualitative results from examination of the individual units, we classified neurons that changed their firing in synchrony with image presentation and measured their temporal receptive fields with a model with three parameters. Each neuron’s firing rate in the time from 0.5 s before image onset to 5 s after was quantified using a model-based approach. The firing field was estimated as a convolution of a Gaussian (latency in neuronal response) and an exponential decay (SI Appendix, Fig. S1A). This approach builds on previous work to estimate time cell activity as a Gaussian firing field (17, 22, 23). The method for estimating parameters is described in detail in Methods. In this model, we were able to quantify apparent response properties using two key parameters: 1) the parameter , which describes the mean of the Gaussian, estimates the time at which each neuron begins to respond (Response Latency), and 2) the parameter , which describes the time constant of the exponential term, estimates how long each neuron takes to relax back to 63% of its maximum deviation from baseline firing (Relaxation Time) (SI Appendix, Fig. S1B). A third parameter controls the SD of the Gaussian term. Using this model, we tested whether a neuron had a time-locked response to image onset by quantifying the extent a model with a temporal response field fit the neuron’s data better than a model with only constant firing including the prestimulus period. Neurons that were better fit by including a temporal firing field were referred to as “visually responsive.” Importantly, this method would identify populations of either hippocampal time cells or temporal context cells as visually responsive. However, as shown in Fig. 1E, these two different forms of temporal coding would produce distinguishable distributions of parameters.
A substantial fraction of entorhinal neurons changed their firing in response to image presentation.
In order to minimize the noise and obtain the most accurate distribution of response parameters across neurons, we used a conservative criterion to identify neurons that responded to image presentation. This method identified 109 of 349 neurons as visually responsive. Of those 109 responsive neurons, 84 neurons showed an increase in their firing rate in response to image onset, whereas 25 showed a decrease in their firing rate. Fig. 2B summarizes the temporal response properties of these 109 neurons. Each row of the figure shows the averaged response of one neuron over the course of a trial. The data demonstrate that almost all of the neurons reached their maximum deviation from baseline within a few hundred milliseconds of the image presentation. This can be appreciated from the vertical yellow strip along the left edge of the heat map. These results are in striking contrast to the typical responses of hippocampal time cells (e.g., Fig. 1C). Analogous plots for hippocampal time cells, which vary smoothly in their peak times, result in a curved ridge extending from the upper left to the lower right. In contrast, the variability across neurons in this entorhinal population was not in the time point at which the neurons reached their maximum deviation from baseline but rather, in the time course over which each neuron relaxed. This can be appreciated in the progressive widening of the ridge in Fig. 2B from top to bottom.
Visually responsive entorhinal units showed short Response Times but a broad distribution of Relaxation Times.
Fig. 2C shows the Response Latency and Relaxation Time for the 109 entorhinal neurons that were categorized as visually responsive (SI Appendix, Fig. S2 shows the marginal distributions for each parameter). Response Latency values were clustered tightly at small values (median = 0.16 s, interquartile range = 0.13 to 0.24 s). For 90% of neurons, the Response Latency was less than 0.40 s. In contrast, Relaxation Times showed a wider distribution (median = 0.23 s, interquartile range = 0.10 to 0.61 s, percentile = 1.29 s) and even included values longer than the 5-s duration of the viewing period. Lastly, the third parameter , which controls the SD of the Gaussian, was small and tightly clustered across neurons (median = 0.02 s, interquartile range = 0.001 to 0.06 s, percentile = 0.31 s). The consistently small value of this parameter indicates that the shape of the temporal receptive fields was well described by a delayed exponential function.
Response Time and Relaxation Time were not correlated across neurons.
Across the 109 neurons, Response Latency and Relaxation Time were not significantly correlated with one another: Kendall’s and . To assess whether this null effect was reliable, we computed the Bayes factor, which enables an estimate of the likelihood of the null hypothesis. This analysis yielded a Bayes factor of , providing support that neuron Response Latency and Relaxation Time values are uncorrelated. Across neurons, Response Latency and were also not correlated with one another: Kendall’s , and . Unlike hippocampal time cells, there was no evidence that temporal context cells that peaked later in the time interval showed broader firing fields. In contrast to hippocampal time cells, which show a systematic relationship between the peak time of firing and the width of the temporal firing field (16, 17, 28), the overarching conclusion from these analyses is that the firing of entorhinal neurons deviated from background firing shortly after the presentation of the stimulus and then relaxed exponentially at a variety of rates.
Populations of Entorhinal Temporal Context Cells Carry Graded Information about Time.
It is well understood that hippocampal time cells can be used to decode the time since the beginning of a time interval (18) (SI Appendix, Fig. S4). To assess the temporal information present in the population of entorhinal neurons, with special attention to the population of temporal context cells identified by the model-based analysis above, we trained a linear discriminant analysis (LDA) decoder to estimate time following presentation of an image. To the extent the predicted time bin for out-of-sample data is close to the actual time bin, one can conclude that the population response carried information about time. We first describe results from the entire population and then focus on the subpopulation of temporal context cells.
Time was decoded better than chance from the population of entorhinal neurons.
Fig. 3 shows the results of the LDA on all 349 neurons from monkey EC. Our first question was whether or not the population contains information about time. For each time bin in Fig. 3A, the confidence of the decoder (the posterior distribution) is shown across the range of possible time estimates. Perfect prediction would correspond to a bright diagonal; random decoding would correspond to a uniform gray square. Qualitatively, the nonuniformity of Fig. 3A suggests that elapsed time can be decoded from the population of EC neurons. To quantitatively assess this, we found that the posterior distribution from the test data was reliably different from a uniform distribution using a χ2 goodness of fit test: and .
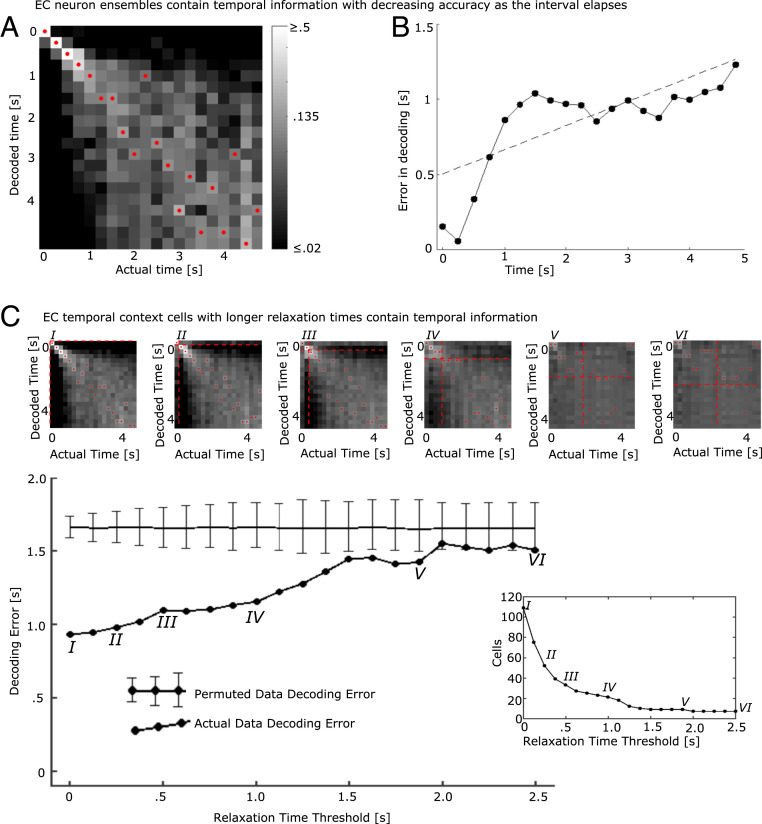
Fig. 3.
The population of entorhinal neurons encodes time with decreasing accuracy as the interval elapses. An LDA decoder was trained to decode time since presentation of the image and then tested on excluded trials. (A) Decoder performance trained on the entire population of EC neurons. The x axis indicates the actual time bin that the LDA attempted to decode; the y axis indicates the decoded time; and the shading indicates the log of the posterior probability, with lighter shading for higher probabilities (see color bar). The decoded time with the highest posterior for each actual time is marked with a red dot. The increasing spread of the diagonal and increasing dispersion of the red dots toward the lower right of the figure suggest that decoding accuracy decreases with the passage of time. (B) Decoding error increases with the passage of time. The x axis gives the actual time since image onset; the y axis gives the mean of the absolute error produced by the decoder in A. The dotted black line is a fitted regression line. The absolute error increases with the passage of time. (C) The temporal code was distributed along many temporal context cells, including those with slow relaxation times. The LDA decoder was first trained with the entire population of temporal context cells. To determine how the temporal code was distributed across the population of temporal context cells, we reran the classifier but only allowing neurons with progressively longer Relaxation Times to contribute to the analysis. For a Relaxation Time threshold of zero, all temporal context cells were included in the analysis, leading to results very comparable with the entire population (point labeled I). Then, the Relaxation Time threshold was increased from zero. For each value of the threshold, only temporal context cells with Relaxation Times greater than or equal to the Relaxation Time threshold were included in the analysis. The large line graph in Lower shows absolute error averaged over all time bins as a function of Relaxation Time threshold. Heat maps showing the entire posterior distribution for selected points labeled by Roman numerals are shown in Upper (same color bar and convention as in A). The threshold for Relaxation Time for each of the heat maps is shown as dashed red lines. Note the earlier times that were most accurate (0 to 0.75 s) dropped substantially in accuracy as the faster Relaxation Times were removed from the analysis. The horizontal line in the main line graph shows the absolute error in decoding from a permuted dataset; error bars show the 0.025 and 0.975 quantiles. (Lower Right) The number of temporal context cells remaining in the analysis as a function of Relaxation Time threshold. The gradual decline in accuracy suggests that temporal information was distributed smoothly throughout the population of temporal context cells.
Supporting this result, the mean absolute value of decoding error from the cross-validated LDA was reliably lower than the decoding error from training with a permuted dataset. In each of 1,000 permutations, we randomly reassigned the time bin labels of the training events used to train the classifier. The absolute value of the decoding error for the original data was 0.923 s, which was more accurate than the mean absolute value of the decoding error for all 1,000 permutations. As shown in SI Appendix, Fig. S5A, the values for the permuted data were approximately normal with mean 1.65 s and SD 0.04 s, resulting in a z score of more than 18 (). These analyses demonstrate that time since image presentation could be decoded from populations of neurons in monkey EC.
The precision of the time estimate decreased as the interval unfolded.
Although the population response in EC could be used to reconstruct time, inspection of Fig. 3A suggests that the precision of this reconstruction was not constant throughout the interval. Fig. 3B shows the average absolute value of the decoding error at each time bin. These data suggest that this error increased as a function of time. A linear regression of decoding error as a function of time showed a reliable slope, , as well as intercept , both , , and . The information in the entorhinal population about the time of image presentation decreases in accuracy as the image presentation recedes into the past.
Time can be decoded well past the peak firing of temporal context cells.
Theories that proposed the existence of temporal context cells argue that they convey information about time via their gradual decay. Another possibility is that the temporal context cells only carry decodable information about time because of their rapid deflection near time 0. If that is the case, then the population of entorhinal neurons should only carry information about time in the period close to the Response Latency. To assess how far into the interval time could be reconstructed, we repeated the LDA analysis excluding progressively more time bins starting from zero. If the LDA can reconstruct time above chance using only bins corresponding to times , then we can conservatively conclude that time can be reconstructed at least time into the interval. To assess this quantitatively, the actual data were compared with permuted data for each repetition of the LDA using absolute error to assess performance (Methods has details; SI Appendix, Fig. S5). This analysis showed that time more than 2.25 s after the image onset can be reliably decoded (). This conservative estimate is an order of magnitude longer than the median value of the peak time (0.160 s), suggesting that the gradual decay of temporal context cells could be used to reconstruct information about time. Decoder performance varies later into the delay, with the performance of the decoder actually improving as noisier bins are eliminated from the analysis. For instance, time at 3 s can be reliably decoded (). Three seconds is the largest amount of time that can be excluded from the LDA, while still allowing for time to be decoded significantly better than chance (below the line marked with ** in SI Appendix, Fig. S5).
Information about time was distributed throughout the population of temporal context cells.
Theories that proposed the existence of temporal context cells argue that they convey information about time via their gradual decay. To determine how temporal information was distributed across the population of temporal context cells, we performed a decoding analysis restricting our attention to temporal context cells. The analysis was performed initially using all temporal context cells and then progressively removed cells with Relaxation Times shorter than a Relaxation Time threshold. The Relaxation Time threshold ranged from 0 s—including all 109 temporal context cells—to 2.5 s—at which point only 7 temporal context cells remained in the analysis. For each Relaxation Time threshold, performance was summarized by averaging decoding error across all bins. If only a subpopulation of temporal context cells with fast Relaxation Times contributed to the temporal information in the ensemble, we would expect an abrupt decrease in performance as the Relaxation Time threshold passed through that critical value.
These results are shown in Fig. 3C, with selected decoder posteriors shown for various Relaxation Time thresholds (Fig. 3C, I–VI). The population at each Relaxation Time threshold was used to generate its own permuted dataset. The first observation is that the performance of the decoder changed gradually, suggesting that temporal context cells with a range of Relaxation Times conveyed useful information about the time of image presentation. Examination of the heat maps in Fig. 3C suggests that excluding temporal cells with Relaxation Times below a particular value (indicated by dashed red lines) disrupts the ability to distinguish times below that value. However, the ability to decode time above the threshold is relatively intact. Statistically, the decoder performed better than chance even with a Relaxation Time threshold of 1.875 s (with nine cells remaining). This analysis suggests that temporal information is distributed throughout the population of temporal context cells. Further, the population conveys information about a range of timescales because the population has a variety of Relaxation Times.
EC Neurons Conveyed Information about Image Identity.
In this experiment, each image was presented twice. Although it was not practical to assess image coding using a classifier, it was possible to exploit the repetition of images to determine whether EC neurons contained information about image identity. This question was addressed using both single-cell analyses and population analyses, which showed convergent results. In both cases, we compare the first and second presentations of the same image with the first and second presentations of different images. Note that because these analyses always compare a first presentation with a second presentation, they are not confounded by repetition effects that have been observed in entorhinal neurons (29–31).
Firing rate of individual neurons was correlated for same presentation of images.
For each neuron, we assembled an array giving the firing rate during the first presentation of each image (averaged over 5 s) and asked whether this array was correlated with the firing rate of second presentations of the same images. Many individual EC neurons responded to several images, as reported in more detail in SI Appendix, Supplementary Text (especially SI Appendix, Fig. S7). If the firing rate of a neuron depends on the identity of the image, we would expect to see a positive correlation using this measure. For this analysis, we restricted our attention to the neurons () in the entorhinal population that were recorded long enough to be observed for both first and second presentations of a block of stimuli (repetitions were separated by 20 to 40 min).
The mean correlation coefficient (Kendall’s ) across neurons was significantly greater than 0 [, , ], indicating that the spiking activity of many neurons depended on image identity (SI Appendix, Fig. S6B). This comparison was confirmed by a Wilcoxon signed rank test on the values of Kendall’s , , and . This finding was also observed for the subset of visually responsive neurons () that we describe as temporal context cells. Taken in isolation, the temporal context neurons showed a mean correlation coefficient significantly greater than 0 as measured by t test [] and Wilcoxon signed rank test (). Neurons that were not temporal context cells () also had a mean correlation coefficient different from 0 []. These results are consistent with a population that contains information about stimulus identity.
The population of EC neurons was more similar for repeated presentations of the same image.
The preceding analyses show that the firing of many EC neurons distinguished image identity above chance. If the response of the entire population contained information about stimulus identity, we would expect, all things equal, that pairs of population vectors corresponding to presentations of the same image would be more similar to one another than pairs of population vectors corresponding to presentations of different images. To control for any potential repetition effect, these analyses compared the similarity between the repetition of an image with its original presentation with the similarity between the second presentation of an image with the first presentation of a different image. To control for any potential recency effects, we swapped images adjacent to the original presentation of the target image. To be more concrete, if we label a sequence of images initially presented in sequence as A, B, and C, we would separately compare the population response to the repetition of B with the response to the initial presentation of A, B, and C. We refer to the similarity of the second presentation of B to the first presentation of B as lag 0. The similarity of the second presentation of B to the first presentation of A is referred to as lag ; the similarity to the first presentation of C is lag . To the extent that the similarity at lag 0 is greater than lag and , we can conclude that the population vector distinguishes image identity.
SI Appendix, Fig. S6D shows the results of this population analysis. The similarity for lag 0 pairs (comparing population response to an image with its repetition) was greater than the similarity for lags (comparing the response to neighbors of its original presentation). Statistical comparisons with lags each showed a reliable difference. A paired t test comparing population similarity at the level of blocks ( trial blocks of repeated images) showed that similarity at lag 0 was reliably larger than both lag [, , Cohen’s ] and lag [, , Cohen’s ]. To evaluate the same hypothesis using a nonparametric method, we performed a permutation analysis by randomly swapping within-session pairs of lag 0 and lag and calculating the mean difference between the pairs 100,000 times. The observed value exceeded the value of 100,000/100,000 permuted values for both lags and . We conclude that the population response was more similar for presentations of the same image than for presentations of different images. This analysis, which controlled for repetition and recency, demonstrates that the response of the population of EC neurons reflected image identity. Coupled with the other results reported here, this means that the population carried information about what happened when.
Discussion
Episodic memory requires information about both the content of an event as well as its temporal context (1, 3, 10). In this study, many EC neurons responded to the onset of the image. These temporal context cells responded to image onset at about the same time, within about 300 ms of image onset. However, different temporal context cells showed variable rates of relaxation back to baseline (Fig. 2). Information about time since the image was presented could be decoded due to gradually decaying firing rates over a few seconds (Fig. 3). Notably, the relaxation rate was not constant across neurons but rather, showed a spectrum of time constants. The population vectors following repeated presentations of the same image were more similar to one another than to presentations of different images. This, coupled with several control analyses, shows that the firing of entorhinal neurons also distinguished stimulus identity. Taken together, the results demonstrate that in the time after image presentation, the population of EC neurons contained information about what happened when.
Sequentially activated time cells, such as have been observed in the hippocampus (8, 9, 17), medial EC (12), and many other brain regions (23–25, 32), also contain information about what happened when in the past. However, entorhinal temporal context cells have very different firing properties than sequentially activated time cells. As a population, time cells convey the amount of time that has passed since the occurrence of an event by firing at different temporal delays after the triggering event. In contrast, EC temporal context cells all responded at about the same time but relaxed at different rates. The range of relaxation times enables the population to convey information about different timescales. For instance, a temporal context cell that returns to baseline firing within 1 s would not be effective in distinguishing a 5-s interval from a 10-s interval. In contrast, a cell that decays back to baseline around 7 s would be effective in distinguishing this longer time period. In this way, a range of decay rates enables the population of temporal context cells to decode time over a wide range of timescales.
Relationship to Findings from Rodent and Monkey EC.
The pattern of results observed here aligns well with a recent report from rodent lateral EC (14). In that study, lateral EC neurons changed their firing in response to a salient event (i.e., the animal entering a new environment) and then relaxed back to baseline monotonically. Notably, different neurons relaxed at different rates with time constants ranging from tens of seconds to many minutes. However, despite the many methodological differences between that study and this one—rats moving through a series of open enclosures vs. seated monkeys observing a series of images—the response properties shared striking similarities, suggesting a common computational function for EC across species.
The results in this paper are consistent with studies that have shown long-lasting responses in EC neurons in different preparations in rodents. For example, sustained responses have been observed in both in vitro (33–37) and anesthetized (38, 39) approaches. Similarly, slow changes in firing rate were observed in the EC of rats during trace eyeblink conditioning across distinct environmental contexts (40). Computational modeling studies have suggested that properties of a calcium nonspecific cation current observed in slice are sufficient to generate a spectrum of response decay periods, ranging from brief to prolonged (41, 42). Juxtaposed with work showing spatial responses in navigating rodents (43–45), these findings suggest EC neurons code for “position” in both temporal and spatial domains.
The current results are also consistent with previous findings from the monkey MTL. A recent investigation of single-neuron activity across shorter timescales (within a 1-s delay) identified time-varying responses in the hippocampus but not the EC (13). However, in earlier work with longer trial durations, entorhinal neurons exhibited response dynamics across several seconds (46). The current results also mirror previous studies showing coding for temporal information in monkey prefrontal cortex attributable to slow ramping activity (47–49). It remains to be seen if those findings reflect similar or distinct computational mechanisms to those observed in EC in this study.
Exponentially Decaying Neurons with a Spectrum of Time Constants Are the Laplace Transform of Time.
Why would the brain use two distinct coding schemes—time cells vs. temporal context cells—to represent time? One proposed answer is that there might be a local circuit processing advantage from having time cells that can signal a specific moment at the single-neuron level, instead of having that information exist in the collective responses of different temporal context cells. Mechanistically, the brain may achieve creating a time cell response by combining the responses of temporal context cells (50). The mathematical description of this process would begin with the brain estimating a temporal record of the past as neural activity that is the real Laplace transform of a function of past time (28, 50, 51).
Cells coding for the real Laplace transform have exponential receptive fields with a variety of rate constants, very much like the results observed here in entorhinal neurons (Fig. 2). In this proposal, the activity representing the contents of the past changes in the time after an image presentation. At a time after image presentation, the neural representation of the past is a function with the presentation of the image at time . As increases, this function changes smoothly. A population of neurons coding the real Laplace transform of time should thus change shortly after image presentation and then relax exponentially, with different neurons relaxing at different rates.
Time cells like those in the hippocampus, in contrast, can be described by performing an additional computation upon the activity described above. The inverse Laplace transform—which can be approximated using a set of feed-forward connections with center-surround weights on the activity described above—directly estimates what happened when. Instead of exponential receptive fields, cells coding for the inverse transform have circumscribed receptive fields that tile the time axis. As the image presentation recedes into the past, neural response to its presentation resembling the inverse transform would generate a series of sequentially activated time cells.
Laplace transforms of other variables in the MTL.
This computational framework for representing functions over continuous variables using the Laplace transform and its inverse can be generalized from time to other variables as well (28). For instance, border cells in EC (52) code for distance from an environmental landmark with monotonically decaying receptive fields. If the firing profile of border cells is exponential and if the parameter controlling the spatial sensitivity of this profile differs across neurons, analogous to the differing relaxation times in Fig. 2, then a population of border cells would code for the real Laplace transform of distance to the border. Applying the inverse transform to such a population would generate boundary vector cells (53), which have been observed in the subiculum and have been argued to drive classic hippocampal place fields (54). Analogously, trajectory coding cells and splitter cells observed in the EC and hippocampus can be understood as coding for the Laplace transform and inverse of functions over ordinal position—the sequence of movements leading up to the present (55, 56). More broadly, this computational framework can be used to generate functions over a variety of spatiotemporal trajectories, which has been proposed to be a basic function of the MTL (57, 58). In this view, time cells and place cells are just two cases of a more general computational function (12, 16, 59).
Laplace transforms of time throughout the brain.
If the brain contains a compressed record of the past (60, 61) as a neural representation across many different “kinds” of memory (62, 63), then one might expect the existence of neurons with conjunctive receptive fields for what happened when across many different brain regions. Indeed, stimulus-specific time cells coding for what happened when have been found in not only regions believed to be important for episodic memory (19, 64) but also, regions that support working memory (23, 65) and classical conditioning (66).
Because computational work using the Laplace transform has shown that a population of time cells can be constructed from temporal context cells (28, 50, 51), we may speculate about the pervasiveness of this phenomenon across the brain. Perhaps temporal context cells may be found in other brain regions outside the EC to support this computation across separate regions of the brain. Alternatively, especially given the high sensory convergence within the EC, perhaps temporal context cells in the EC are utilized for generating time cells across the brain.
Methods
Subjects, Training, and Surgery.
Two male rhesus macaques (Macaca mulatta), 10 and 11 y old weighing 13.8 and 16.7 kg, respectively, were trained to sit in a primate chair (Crist Instrument Company, Inc.) and to release a touch bar for fruit slurry reward delivered through a tube. The monkeys were trained to perform various tasks by releasing the touch bar at appropriate times relative to visual stimuli presented on a screen. Magnetic resonance images of each monkey’s head were made both before and after surgery to plan and confirm implant placement. Separate surgeries were performed to implant a head post; then months later, a recording chamber; and finally, a craniotomy within the chamber. All experiments were performed in accordance with protocols approved by the Emory University and the University of Washington Institutional Animal Care and Use Committees.
Electrophysiology.
Each recording session, a laminar electrode array (AXIAL array with 13 channels; FHC, Inc.) mounted on a microdrive (FHC, Inc.) was slowly lowered into the brain through the craniotomy. Magnetic resonance images along with the neural signal were used to guide the penetration. Spikes and local field potentials were recorded using hardware and software from Blackrock, Inc., and neural data were sampled at 30 kHz. A 500-Hz high-pass filter was applied, as well as an electric line cancellation at 60 Hz. Spikes were sorted offline into distinct clusters using principal components analysis (Offline Sorter; Plexon, Inc.). Sorted clusters were then processed further by custom code in MATLAB to eliminate any data where minimum interspike interval was less than 0.001 s and to identify any missed changes in signal (e.g., decreased amplitude in the waveform of interest, a new waveform appearing) using raster plots and plots of waveforms across the session for each neuron. When change in signal was identified, appropriate cuts were made to exclude compromised spike data from before or after a change point; 455 potential single neurons originally isolated in Offline Sorter were reduced to 357 single neurons. To further ensure recording location within the EC and identify from which cortical layers neurons were recorded, we examined each session’s data for the stereotypical electrophysiological signature produced across EC layers at the onset of saccadic eye movement (27, 67, 68). Recording sessions took place in both anterior and posterior regions of the EC. One recording session, which other electrode placement metrics suggest was conducted above the EC within the hippocampus, lacked this electrophysiological signature and was excluded from further analysis (eight single neurons were excluded from this session). No recording sessions showed the current source density electrophysiological signature of adjacent perirhinal cortex (69) at stimulus onset.
Experimental Design and Behavioral Task.
For all recordings, the monkey was seated in a dark room, head fixed and positioned so that the center of the screen (54.1 29.9-cm liquid crystal display screen, 120-Hz refresh rate, 1,280 720 pixels; BenQ America Corp.) was aligned with his neutral gaze position and 60 cm away from the plane of the his eyes (equating to 25 screen pixels per degree of visual angle or /cm). Stimulus presentation was controlled by a personal computer running Cortex software (National Institute of Mental Health). Gaze location was monitored at 240 Hz with an infrared eye-tracking system (I-SCAN, Inc.). Gaze location was calibrated before and during each recording session with calibration trials in which the monkey held a touch-sensitive bar while fixating a small () gray square presented at various locations on the screen. The square turned yellow after a brief delay chosen uniformly from the interval from 0.40 to 0.75 s. The monkey was required to release the bar in response to the color change for delivery of the fruit slurry reward. The subtlety of the color change forced the monkey to fixate the location of the small square to correctly perform those trials, therefore allowing calibration of gaze position to the displayed stimuli. Specifically, the gain and offset of the recorded gaze position were adjusted so that gaze position matched the position of the fixated stimulus. Throughout the session, intermittent calibration trials enabled continual monitoring of the quality of gaze position data and correction of any drift.
Before each image presentation, a crosshair () appeared in 1 of 18 possible screen locations. After gaze position registered within a window around the crosshair and was maintained within that spatial window for between 0.50 and 0.75 s (chosen uniformly), the image was presented. Images were large, complex images downloaded from the public photo-sharing website, Flickr (https://www.flickr.com/). If necessary, images were resized by the experimenter for stimulus presentation (sized for Monkey WR and for Monkey MP). Monkeys freely viewed the image, and then, the image vanished after gaze position had registered within the image frame for a cumulative 5 s. No food reward was given during image-viewing trials. Each image presentation was followed by three calibration trials.
Image stimuli were unique to each session, and each image was presented twice within a session about 20 to 40 min apart. Images were presented in a block design so that novel and previously seen images were presented throughout the session. Within a trial block, novel images (30 or 60) would first be shown and then presented again in pseudorandom order. After completing a block of trials, a new block of trials would begin. In the first 16 sessions, a three-block design of 60 image presentations (30 novel) per block was used, with a total maximum of 180 image presentations per session. In the rest of the sessions (), there was a total maximum of 240 image presentations across two trial blocks (120 image presentations of which 60 were novel within each trial block).
Analysis of Neural Firing Fields.
In order to determine temporal firing fields, spikes were analyzed using a custom maximum likelihood estimation script run in MATLAB 2016a. We calculated model fits across all trials available for each particular neuron considering the time from 500 ms before image presentation to 5 s after image presentation. Fits of nested models were compared using a likelihood ratio test. In the present paper, we considered three models: a constant firing model, a model adding a Gaussian time term, and an ex-Gaussian model for which the time term was given by the convolution of a Gaussian and an exponential time term. The constant model,
| [1] |
consisted of a single parameter that predicted the constant probability of a spike at each time .
The ex-Gaussian model describes the temporal modulation of the firing field as the convolution of a Gaussian function with an exponentially decaying function:
| [2] |
The ex-Gaussian distribution has been used extensively in studies of human response time data for many years (70). In the limit as , the exponential function becomes a delta function, and the result of the convolution in Eq. 2 is a Gaussian function. Similarly, in the limit as , the Gaussian function becomes a delta function, and the result of the convolution is an exponential function starting at . As such, this model is able to describe a range of peak firing times as well as varying degrees of skew (Fig. 1).
Two terms, and , describe the contributions of the constant and time-modulated terms. Three parameters describe the shape of the temporally modulated term (SI Appendix, Fig. S1). and describe the mean and SD of the Gaussian distribution, which estimate the time that a neuron’s response maximally deviates from baseline and the variability in that response time, respectively. In the text, is referred to as the Response Time. measures the time constant of the exponential decay and captures the time that a neuron has returned 63% of the way back to baseline. In the text, is referred to as the Relaxation Time.
To estimate parameters of Eq. 2 numerically, we used an explicit form for the solution of the convolution in Eq. 2:
| [3] |
where erfc is the complementary error function. was allowed to take values between 0 and 5 s. was allowed to take values between 0 and 20 s. was allowed to take values between 0 and 1 s. Likelihood was estimated in a 5.5-s-long window that included the 0.5 s prior to presentation of the image and the 5 s after presentation of the image.
We evaluated the models for each neuron via a likelihood ratio test and counted the number of neurons that 1) were better fit by the ex-Gaussian model at the 0.05 level, Bonferonni corrected by the total number of 349 neurons; 2) changed their firing by at least 1 Hz; and 3) reached a firing rate of at least 3 Hz. In addition, we required that both even and odd trials for a neuron were significantly fit by the model and that those fits had a Pearson’s correlation coefficient greater than 0.4.
LDA.
An LDA classifier was used to decode time since onset of the image from the population using information from all neurons.
LDA implementation.
Even and odd trials were used for training and testing, respectively. The number of available trials varied for each neuron. To mitigate any problems from this, several steps were taken. First, four neurons with less than 30 trials each were entirely excluded from this analysis. Neurons with less than 200 trials were bootstrapped to 200 trials, while neurons with more than 200 trials were randomly down sampled. Time was discretized into 0.25-s bins. For each bin of each trial, the firing rate was calculated across neurons. To avoid errors due to a singular covariance matrix, a small amount of uniform noise (between 0 and Hz) was added to the firing rate in each time bin. The averaged firing rate of each time bin for each training trial across all neurons made up an element of the training data. The averaged firing rate of each time bin for each testing trial across all neurons made up an element of the testing data. LDA was implemented using the MATLAB function “classify.” This function takes in the training data, testing data, labels for the training data, and a selection of the method of estimation for the covariance matrix (the option “linear” was used) and returns a posterior distribution across bins for each test trial.
Estimating the duration of temporal coding.
To assess the quality of temporal information at different points within the interval, the LDA was repeated for successively fewer bins, at each step removing the earliest time bin. If time since presentation of the image can be decoded above chance using only information after time , one can conclude that the population contained temporal information about time at least a time after presentation of the image. For each repetition, the decoder was tested by training it on data with permuted time labels. We compared the absolute error of the actual data with the distribution generated from 1,000 permutations. The classifier’s performance was considered significantly better than chance if fewer than 10 of 1,000 permutations gave a better result than the unpermuted data (corresponding roughly to ).
Evaluating the distribution of temporal information over temporal context cells with different Relaxation Times.
To assess the distribution of temporal information over temporal context cells as a function of their Relaxation Time, neurons with small Relaxation Times were progressively omitted from the LDA. First, all temporal context cells were used (corresponding to a Relaxation Time threshold of zero); then, only cells with Relaxation Time longer than 0.125 s, only cells with Relaxation Time longer than 0.25 s, and so forth. The longest Relaxation Time threshold evaluated was 2.5 s. Performance was parametrized by averaging the absolute value of decoding error across all time bins. As a control, for each pseudo-subpopulation of cell, bins with permuted labels were also trained on and decoded from 1,000 times.
Stimulus Sparsity Analysis.
To assess stimulus specificity in a way that facilitates comparison with previous human work, we followed the analysis method of ref. 71. In order to determine how many images a given neuron responded to, for each neuron we formed a distribution of baseline firing rates from the 500 ms prior to image onset on each trial. For each image, we took the trials in which they appeared and binned the firing rates of the 1 s following image onset into 19 overlapping bins, each of which was 100-ms long. We then compared these binned firing rates with the baseline distribution via a two-tailed Mann–Whitney U test using the Simes procedure and a conservative significance threshold of . For each neuron, we counted the number of images that exceeded this threshold.
Population Vector Analysis of Stimulus Specificity.
We constructed population vectors to evaluate the degree to which the entire population of entorhinal neurons was sensitive to the identities of visual images. For each repeated image, we created two population vectors, one corresponding to the first presentation and one corresponding to the second presentation. Each vector was created from the mean firing activity of all neurons recorded in a session during the 5 s of free viewing. Mean firing rates were normalized by each neuron’s maximum average firing rate so that firing rates ranged from zero to one. Only blocks where all images were presented twice were considered. In order to control for different block lengths between sessions, only the first 30 images presented in each block were used. All neurons that were recorded during first and second presentations of an image were included in this analysis (). The average number of simultaneously recorded neurons in a block was 8.51, with an SD of 4.04 and a range from 2 to 19. Similarity was measured by the cosine similarity of the two population vectors. We compared the cosine similarity of two presentations of the same image with the first presentation of one image and the second presentation of a different image. As a control, we instead compared the population vector from the repetition of an image with the adjacent near neighbors of the original image presentation. Near neighbors were required to be the first presentation of an image. Within session error bars represent the 95% CI (72).
Supplementary Material
Acknowledgments
This work was supported by NIH Grants 2R01MH080007 (to E.A.B.) and R01MH093807 (to E.A.B.), National Institute of Mental Health (NIMH) Grant P51 OD010425 (to E.A.B.), National Institute of Biomedical Imaging and Bioengineering Grant R01EB022864 (to M.W.H.), NIMH Grant R01MH112169 (to M.W.H.), and Office of Naval Research, Multidisciplinary University Research Initiative Grant N00014-16-1-2832.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917197117/-/DCSupplemental.
Data Availability.
Code and data are available at GitHub, https://github.com/tcnlab/TemporalContextCells-ECmonkey.
References
- 1.Tulving E., Elements of Episodic Memory (Oxford, New York, NY, 1983). [Google Scholar]
- 2.Milner B., The memory defect in bilateral hippocampal lesions. Psychiatr. Res. Rep. Am. Psychiatr. Assoc. 11, 43–58 (1959). [PubMed] [Google Scholar]
- 3.Eichenbaum H., Yonelinas A., Ranganath C., The medial temporal lobe and recognition memory. Annu. Rev. Neurosci. 30, 123–152 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dede A. J., Frascino J. C., Wixted J. T., Squire L. R., Learning and remembering real-world events after medial temporal lobe damage. Proc. Natl. Acad. Sci. U.S.A. 113, 13480–13485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Squire L. R., Stark C. E., Clark R. E., The medial temporal lobe. Annu. Rev. Neurosci. 27, 279–306 (2004). [DOI] [PubMed] [Google Scholar]
- 6.O’Keefe J., Dostrovsky J., The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34, 171–175 (1971). [DOI] [PubMed] [Google Scholar]
- 7.Fyhn M., Molden S., Witter M. P., Moser E. I., Moser M. B., Spatial representation in the entorhinal cortex. Science 305, 1258–1264 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Pastalkova E., Itskov V., Amarasingham A., Buzsaki G., Internally generated cell assembly sequences in the rat hippocampus. Science 321, 1322–1327 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacDonald C. J., Lepage K. Q., Eden U. T., Eichenbaum H., Hippocampal “time cells” bridge the gap in memory for discontiguous events. Neuron 71, 737–749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichenbaum H., On the integration of space, time, and memory. Neuron 95, 1007–1018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naya Y., Suzuki W., Integrating what and when across the primate medial temporal lobe. Science 333, 773–776 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Kraus B. J., et al. , During running in place, grid cells integrate elapsed time and distance run. Neuron 88, 578–589 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naya Y., Chen H., Yang C., Suzuki W. A., Contributions of primate prefrontal cortex and medial temporal lobe to temporal-order memory. Proc. Natl. Acad. Sci. U.S.A. 114, 13555–13560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsao A., et al. , Integrating time from experience in the lateral entorhinal cortex. Nature 561, 57–62 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Wilson M. A., McNaughton B. L., Dynamics of the hippocampal ensemble code for space. Science 261, 1055–1058 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Kraus B. J., Robinson R. J. 2nd, White J. A., Eichenbaum H., Hasselmo M. E., Hippocampal “time cells”: Time versus path integration. Neuron 78, 1090–1101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salz D. M., et al. , Time cells in hippocampal area CA3. J. Neurosci. 36, 7476–7484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mau W., et al. , The same hippocampal CA1 population simultaneously codes temporal information over multiple timescales. Curr. Biol. 28, 1499–1508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacDonald C. J., Carrow S., Place R., Eichenbaum H., Distinct hippocampal time cell sequences represent odor memories immobilized rats. J. Neurosci. 33, 14607–14616 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terada S., Sakurai Y., Nakahara H., Fujisawa S., Temporal and rate coding for discrete event sequences in the hippocampus. Neuron 94, 1–15 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Bolkan S. S., et al. , Thalamic projections sustain prefrontal activity during working memory maintenance. Nat. Neurosci. 20, 987–996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiganj Z., Kim J., Jung M. W., Howard M. W., Sequential firing codes for time in rodent mPFC. Cerebr. Cortex 27, 5663–5671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiganj Z., Cromer J. A., Roy J. E., Miller E. K., Howard M. W., Compressed timeline of recent experience in monkey lPFC. J. Cognit. Neurosci. 30, 935–950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin D. Z., Fujii N., Graybiel A. M., Neural representation of time in cortico-basal ganglia circuits. Proc. Natl. Acad. Sci. U.S.A. 106, 19156–19161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mello G. B., Soares S., Paton J. J., A scalable population code for time in the striatum. Curr. Biol. 25, 1113–1122 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Akhlaghpour H., et al. , Dissociated sequential activity and stimulus encoding in the dorsomedial striatum during spatial working memory. eLife 5, e19507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meister M. L., Buffalo E. A., Neurons in primate entorhinal cortex represent gaze position in multiple spatial reference frames. J. Neurosci. 38, 2430–2441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard M. W., et al. , A unified mathematical framework for coding time, space, and sequences in the hippocampal region. J. Neurosci. 34, 4692–707 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiang J. Z., Brown M., Differential neuronal encoding of novelty, familiarity and recency in regions of the anterior temporal lobe. Neuropharmacology 37, 657–676 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Meyer T., Rust N. C., Single-exposure visual memory judgments are reflected in inferotemporal cortex. eLife 7, e32259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jutras M. J., Buffalo E. A., Recognition memory signals in the macaque hippocampus. Proc. Natl. Acad. Sci. U.S.A. 107, 401–406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tiganj Z., Gershman S. J., Sederberg P. B., Howard M. W., Estimating scale-invariant future in continuous time. Neural Comput. 31, 681–709 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klink R., Alonso A., Muscarinic modulation of the oscillatory and repetitive firing properties of entorhinal cortex layer ii neurons. J. Neurophysiol. 77, 1813–1828 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Egorov A. V., Hamam B. N., Fransén E., Hasselmo M. E., Alonso A. A., Graded persistent activity in entorhinal cortex neurons. Nature 420, 173–178 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Tahvildari B., Fransén E., Alonso A. A., Hasselmo M. E., Switching between “On” and “Off” states of persistent activity in lateral entorhinal layer III neurons. Hippocampus 17, 257–63 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Yoshida M., Fransén E., Hasselmo M. E., mGluR-dependent persistent firing in entorhinal cortex layer III neurons. Eur. J. Neurosci. 28, 1116–26 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyde R. A., Strowbridge B. W., Mnemonic representations of transient stimuli and temporal sequences in the rodent hippocampus in vitro. Nat. Neurosci. 15, 1430–1438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahn T. T., McFarland J. M., Berberich S., Sakmann B., Mehta M. R., Spontaneous persistent activity in entorhinal cortex modulates cortico-hippocampal interaction in vivo. Nat. Neurosci. 15, 1531–1538 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leitner F. C., et al. , Spatially segregated feedforward and feedback neurons support differential odor processing in the lateral entorhinal cortex. Nat. Neurosci. 19, 935–944 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Pilkiw M., et al. , Phasic and tonic neuron ensemble codes for stimulus-environment conjunctions in the latereral entorhinal cortex. eLife 6, e28611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiganj Z., Hasselmo M. E., Howard M. W., A simple biophysically plausible model for long time constants in single neurons. Hippocampus 25, 27–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Tiganj Z., Hasselmo M. E., Howard M. W., A neural microcircuit model for a scalable scale-invariant representation of time. Hippocampus 29, 260–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deshmukh S. S., Knierim J. J., Representation of non-spatial and spatial information in the lateral entorhinal cortex. Front. Behav. Neurosci. 5, 69 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C., et al. , Egocentric coding of external items in the lateral entorhinal cortex. Science 362, 945–949 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Høydal Ø. A., Skytøen E. R., Andersson S. O., Moser M. B., Moser E. I., Object-vector coding in the medial entorhinal cortex. Nature 568, 400–404 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Suzuki W. A., Miller E. K., Desimone R., Object and place memory in the macaque entorhinal cortex. J. Neurophysiol. 78, 1062–1081 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Machens C. K., Romo R., Brody C. D., Functional, but not anatomical, separation of what and when in prefrontal cortex. J. Neurosci. 30, 350–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossi-Pool R., et al. , Emergence of an abstract categorical code enabling the discrimination of temporally structured tactile stimuli. Proc. Natl. Acad. Sci. U.S.A. 113, E7966–E7975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi-Pool R., et al. , Temporal signals underlying a cognitive process in the dorsal premotor cortex. Proc. Natl. Acad. Sci. U.S.A. 116, 7523–7532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shankar K. H., Howard M. W., A scale-invariant internal representation of time. Neural Comput. 24, 134–193 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Shankar K. H., Howard M. W., Optimally fuzzy temporal memory. J. Mach. Learn. Res. 14, 3753–3780 (2013). [Google Scholar]
- 52.Boccara C. N., et al. , Grid cells in pre- and parasubiculum. Nat. Neurosci. 13, 987–94 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Lever C., Burton S., Jeewajee A., O’Keefe J., Burgess N., Boundary vector cells in the subiculum of the hippocampal formation. J. Neurosci. 29, 9771–9777 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burgess N., O’Keefe J., Neuronal computations underlying the firing of place cells and their role in navigation. Hippocampus 6, 749–762 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Frank L. M., Brown E. N., Wilson M., Trajectory encoding in the hippocampus and entorhinal cortex. Neuron 27, 169–178 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Wood E. R., Dudchenko P. A., Robitsek R. J., Eichenbaum H., Hippocampal neurons encode information about different types of memory episodes occurring in the same location. Neuron 27, 623–33 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Hasselmo M. E., How We Remember: Brain Mechanisms of Episodic Memory (MIT Press, Cambridge, MA, 2012). [Google Scholar]
- 58.Dannenberg H., Kelley C., Hoyland A., Monaghan C. K., Hasselmo M. E., The firing rate speed code of entorhinal speed cells differs across behaviorally relevant time scales and does not depend on medial septum inputs. J. Neurosci., 1450–1418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howard M. W., Eichenbaum H., Time and space in the hippocampus. Brain Res. 1621, 345–354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.James W., The Principles of Psychology (Holt, New York, NY, 1890). [Google Scholar]
- 61.Husserl E., The Phenomenology of Internal Time-Consciousness (Indiana University Press, Bloomington, IN, 1966). [Google Scholar]
- 62.Chater N., Brown G. D. A., From universal laws of cognition to specific cognitive models. Cognit. Sci. 32, 36–67 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Howard M. W., Shankar K. H., Aue W., Criss A. H., A distributed representation of internal time. Psychol. Rev. 122, 24–53 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Taxidis J., et al. , Emergence of stable sensory and dynamic temporal representations in the hippocampus during working memory. bioRxiv:474510 (20 November 2018).
- 65.Cruzado N. A., Tiganj Z., Brincat S. L., Miller E. K., Howard M. W., Conjunctive representation of what and when in monkey hippocampus and lateral prefrontal cortex during an associative memory task. bioRxiv:709659 (26 June 2020). [DOI] [PubMed] [Google Scholar]
- 66.Adler A., et al. , Temporal convergence of dynamic cell assemblies in the striato-pallidal network. J. Neurosci. 32, 2473–2484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Killian N. J., Jutras M. J., Buffalo E. A., A map of visual space in the primate entorhinal cortex. Nature 491, 761–764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Killian N. J., Potter S. M., Buffalo E. A., Saccade direction encoding in the primate entorhinal cortex during visual exploration. Proc. Natl. Acad. Sci. U.S.A. 112, 15743–15748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takeuchi D., Hirabayashi T., Tamura K., Miyashita Y., Reversal of interlaminar signal between sensory and memory processing in monkey temporal cortex. Science 331, 1443–1447 (2011). [DOI] [PubMed] [Google Scholar]
- 70.Ratcliff R., Murdock B. B., Retrieval processes in recognition memory. Psychol. Rev. 83, 190–214 (1976). [Google Scholar]
- 71.Mormann F., et al. , Latency and selectivity of single neurons indicate hierarchical processing in the human medial temporal lobe. J. Neurosci. 28, 8865–8872 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morey R. D., Confidence intervals from normalized data: A correction to Cousineau (2005). Tutor. Quant. Methods Psychol. 4, 61–64 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code and data are available at GitHub, https://github.com/tcnlab/TemporalContextCells-ECmonkey.