Significance
We have used cryogenic electron microscopy (cryo-EM) to achieve direct, probe-free imaging of lateral domains in biomimetic lipid membranes under native conditions and to characterize differences in their structures. First, measurements of membrane thickness in laterally uniform single-component membranes show that cryo-EM is capable of subangstrom resolution of interleaflet membrane thickness. All-atom simulations are used to predict the cryo-EM appearance of submicron domains in vesicles with coexisting liquid domains and these are quantitatively validated by direct imaging of phase-separated membranes. We then extend this approach to observe nanoscopic domains in isolated cellular membranes, demonstrating direct imaging of nanodomains in biomimetic and biological membranes.
Keywords: nanodomains, cryo-EM, liquid-ordered, liquid-disordered, GPMVs
Abstract
The nanoscale organization of biological membranes into structurally and compositionally distinct lateral domains is believed to be central to membrane function. The nature of this organization has remained elusive due to a lack of methods to directly probe nanoscopic membrane features. We show here that cryogenic electron microscopy (cryo-EM) can be used to directly image coexisting nanoscopic domains in synthetic and bioderived membranes without extrinsic probes. Analyzing a series of single-component liposomes composed of synthetic lipids of varying chain lengths, we demonstrate that cryo-EM can distinguish bilayer thickness differences as small as 0.5 Å, comparable to the resolution of small-angle scattering methods. Simulated images from computational models reveal that features in cryo-EM images result from a complex interplay between the atomic distribution normal to the plane of the bilayer and imaging parameters. Simulations of phase-separated bilayers were used to predict two sources of contrast between coexisting ordered and disordered phases within a single liposome, namely differences in membrane thickness and molecular density. We observe both sources of contrast in biomimetic membranes composed of saturated lipids, unsaturated lipids, and cholesterol. When extended to isolated mammalian plasma membranes, cryo-EM reveals similar nanoscale lateral heterogeneities. The methods reported here for direct, probe-free imaging of nanodomains in unperturbed membranes open new avenues for investigation of nanoscopic membrane organization.
Membrane physiology is intrinsically dependent on the structural and dynamical properties of lipid bilayers. Lipid collective behavior influences membrane fluidity, lipid packing, bilayer thickness, elastic moduli, and surface charge density, which in turn affect protein interactions, dynamics, and functions (1, 2). A striking example is the variation in membrane physical properties in the eukaryotic secretory system: endoplasmic reticulum (ER) membranes are thinner and more fluid than those of the Golgi, which are themselves thinner and more fluid than plasma membranes (PMs) (3). These thickness differences have been implicated in protein sorting through the secretory pathway, with proteins containing shorter transmembrane regions being preferentially retained in the ER while longer transmembrane domains (TMDs) are sorted to the PM (4–6).
In addition to variations between different membranes, the particular lipid compositions of many eukaryotic cells may be poised for variations within a given membrane. Preferential interactions between specific lipid classes (e.g., saturated acyl chains, sphingolipids, glycolipids, and sterols) can combine to produce lateral heterogeneities with unique structures, compositions, and putative cellular functions (7). This capacity for functional lipid-driven lateral segregation of biomembranes is known as the “lipid raft hypothesis” and has been one of the most widely studied (8) and controversial (9) topics of modern membrane biology.
The physicochemical underpinning of the raft hypothesis is the fact that biomimetic mixtures of synthetic or isolated lipids often form coexisting liquid phases (reviewed in refs. 10 and 11), with the cholesterol- and saturated-lipid-rich liquid-ordered (Lo) phase serving as the model for biological lipid rafts. Phase-separated model membranes have yielded a wealth of insights into the principles underlying the organization of biological membranes; however, their lack of proteins and compositional complexity prevent straightforward translation to living membranes. Within that context, the direct observation of liquid–liquid phase separation in PMs isolated from living cells as giant PM vesicles (GPMVs) was a key confirmation of the capacity of biological membranes to separate into coexisting lipid domains (12), which was later confirmed in yeast vacuoles (13, 14). Since then, GPMVs have been widely used to assay the lipid and protein compositions of raft domains, their resulting physical properties, modulation of these properties by external inputs, and the nature of the miscibility phase transition in biomembranes. For example, it was shown that proteins with longer TMDs preferentially sort into the more tightly packed raft phase (15), implying that this raft phase is also relatively thick. Most importantly, protein partitioning in GPMVs is strongly related to the subcellular localization of proteins in live cells, suggesting a functional role for rafts in membrane traffic (15). Similar connections between GPMVs and cell physiology have been reported for the diffusion of lipids (16), nanoscale protein organization in live cells (17), and the assembly of viral proteins at the PM (18, 19).
Despite these and other significant advances supporting the functional relevance of ordered membrane domains, the nature and function of lipid rafts remain controversial. Studies using imaging mass spectrometry failed to observe cholesterol-rich domains in cells (20). Similarly, a single-molecule tracking study reported no effects of enriching putatively raft-associated glycosylphosphatidylinositol-anchored proteins on other proteins’ localization or diffusion (21). Ultimately, the controversy persists because direct, microscopic detection of domains in mammalian cells has proven elusive, though it should be emphasized that such domains have been directly observed in living yeast (13, 14).
Seeing is believing, and therefore some of the most compelling studies of liquid–liquid phase separation have involved direct imaging of micrometer-sized domains in giant unilamellar vesicles and GPMVs (12, 22–25). However, such large, thermodynamically stable domains are generally not observed in mammalian PMs, where active processes and cytoskeletal interactions (26, 27) may be expected to produce more dynamic and/or nanoscopic structures. Thus, tools to investigate the nanoscale organization of biological and biomimetic membranes are essential. Several methods, including electron spin resonance (28), NMR (23, 29), small-angle neutron scattering (SANS) (30), fluorescence resonance energy transfer (31), and interferometric scattering microscopy (32) have proven useful for indirectly probing nanoscale phase separation across temporal and spatial scales. In principle, both atomic force microscopy (AFM) and superresolution microscopy are capable of directly detecting domains below the optical resolution limit, but both require immobilizing membranes on solid supports, which inherently affects bilayer structure and dynamics (33, 34). To date, there has been no direct imaging of coexisting nanoscopic phases in unsupported membranes, including synthetic biomimetic models, isolated biomembranes, or cell membranes in situ. Consequently, major questions remain about the sizes, morphologies, physical properties, composition, and very existence of membrane nanodomains.
To address some of these questions, we have applied cryogenic electron microscopy (cryo-EM), a revolutionary development in structural biology that allows direct, probe-free, angstrom-level resolution of biological samples in situ (35, 36). Previous observations have suggested that EM is sensitive enough to resolve the two bilayer leaflets, with cell membranes appearing as a double line separated by about 3 to 5 nm in EM images (36, 37). What has not been demonstrated is whether cryo-EM is capable of resolving the known thickness difference between coexisting liquid-disordered (Ld) and Lo phases in model membranes or analogously hypothesized thickness differences in biomembranes. Here, we show that cryo-EM can provide subangstrom resolution of membrane thickness differences and that these can be used to directly image nanodomains in biomimetic and biological membranes.
Results
Cryo-EM Detects Bilayer Thickness Differences with Subangstrom Resolution.
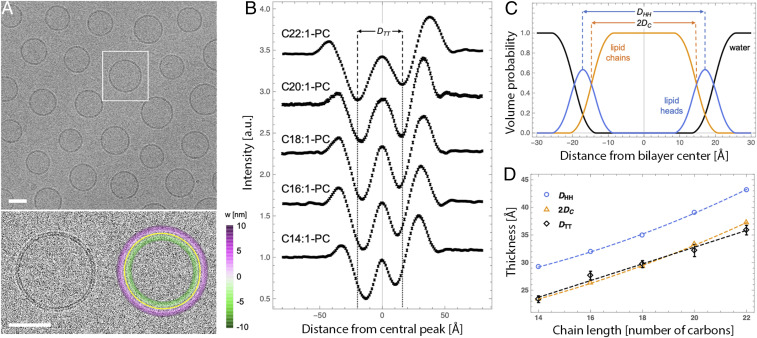
To assess the capability of cryo-EM to measure bilayer thickness, we directly imaged unstained synthetic lipid membranes. Fig. 1A shows a cryo-EM image of vesicles composed of dioleoyl phosphatidylcholine (DOPC; di18:1-PC), a phospholipid that spontaneously forms bilayers upon hydration. Extrusion through a 50-nm polycarbonate filter produces largely monodisperse vesicles with an average diameter of 74 ± 10 nm as determined from the images [unless noted, all synthetic vesicles experimentally examined in this paper contain 5 mol % of the negatively charged lipid 1-palmitoyl-2-oleoyl phosphatidylglycerol (POPG) to minimize multilamellar vesicles (38)]. A large majority of the extruded vesicles consisted of a single bilayer, the structure of which is evident as two dark concentric rings (referred to here and throughout as troughs) flanked by brighter bands (Fig. 1 A, Lower Left). This intensity variation arises from phase contrast between the transmitted and scattered electron waves, as previously described (39).
Fig. 1.
Cryo-EM is sensitive to subangstrom variation in bilayer thickness. (A) Cryo-EM image of a field of di18:1-PC vesicles prepared at room temperature (Upper) and an expanded view of a vesicle without (Lower Left) and with (Lower Right) an overlay of the fitted vesicle contour. (Scale bars, 50 nm.) The color scale in the lower right image corresponds to the length of the shortest line connecting each pixel to the contour (i.e., its distance from the bilayer center). (B) Radially averaged intensity profiles from cryo-EM images for a series of fluid-phase PC vesicles. The trough-to-trough distance DTT is shown for the C22:1 bilayer and extended to the x axis to show relative differences between the profiles (negative numbers on the x axis correspond to features closer to the vesicle center). Profiles are vertically offset for clarity and error bars on individual data points are 95% confidence intervals. (C) The volume probability profile for a di18:1-PC bilayer obtained from analysis of SAXS data at 25 °C. Shown are two standard reference thicknesses defined by SAXS analysis: the headgroup-to-headgroup distance DHH and the hydrocarbon thickness 2DC. (D) Bilayer thicknesses determined from SAXS (DHH and 2DC) and cryo-EM (DTT) for the PC series (dashed lines are a visual guide). DTT is shown as average ± SEM of ∼50 vesicles. The uncertainty of SAXS thicknesses are estimated to be ± 2%. All vesicles contained 5 mol % POPG to minimize multilamellar structures.
To quantify these features, we designed and implemented an algorithm to determine the intensity profile along the bilayer normal (see SI Appendix for details). Briefly, the vesicle contour is arbitrarily identified as the bright central peak between the dark troughs (Fig. 1 A, Lower Right, solid yellow line). Then, individual pixel intensities within 10 nm of the contour are binned by distance from the nearest contour point to generate radially integrated intensity profiles I(w) (Fig. 1B), shown for individual di18:1-PC vesicles in SI Appendix, Fig. S1. To test how such profiles reflect differences in membrane thickness, we imaged vesicles comprising a series of PC lipids with different acyl chains ranging from 14 to 22 carbons in length, all containing one unsaturation per acyl chain to ensure fluid phase at room temperature (at which all samples were cryopreserved). Fig. 1B shows I(w) profiles (averaged over 5 nm arc-length segments from multiple vesicles) for this lipid series. The trough-to-trough distance DTT systematically increases with increasing chain length (Table 1 and black symbols in Fig. 1D), fully consistent with the expected increase in bilayer thickness.
Table 1.
Structural parameters derived from analysis of experimental cryo-EM images
| Lipid | DTT, Å | ||
| di14:1 | 56 | 2,244 | 23.4 ± 0.6 |
| di16:1 | 50 | 2,211 | 27.7 ± 0.8 |
| di18:1 | 98 | 4,256 | 29.7 ± 0.6 |
| di20:1 | 48 | 1,489 | 32.2 ± 1.1 |
| di22:1 | 51 | 2,820 | 35.9 ± 0.9 |
Samples are fluid-phase bilayers composed of PC lipids with two monounsaturated chains and cryopreserved at room temperature. DTT corresponds to the measured distance between the troughs of the I(w) profile obtained by averaging the profiles of 5-nm arc-length bilayer segments from multiple vesicles (Nves is the number of analyzed vesicles and Nseg is the number of 5-nm segments). Uncertainty in DTT was estimated using standard Monte Carlo methods as described in SI Appendix.
We hypothesized that, as an electron scattering technique, cryo-EM should be especially sensitive to the electron-rich phosphates, and that DTT should therefore roughly correspond to the distance between lipid headgroups. To test this hypothesis, we compared cryo-EM profiles to different bilayer thicknesses (i.e., corresponding to different reference planes within the lipid interfacial region) derived from small-angle X-ray scattering (SAXS) for the same PC series (Fig. 1D and SI Appendix, Table S2). X-rays scatter from the electron cloud and are thus particularly sensitive to the location of the headgroup phosphates, the most electron-dense region of a phospholipid bilayer. Fitting scattering curves (shown in SI Appendix, Fig. S2) to a model of the lipid volume probability profile along the bilayer normal (shown in Fig. 1C for DOPC) yields several useful bilayer structural parameters including the headgroup–headgroup distance DHH and the hydrocarbon thickness 2DC (SI Appendix, Table S2). Somewhat surprisingly, we observed excellent correspondence between DTT and the lipid hydrophobic thickness 2DC, rather than the expected distance between the electron-rich phosphate groups DHH. Possible explanations for these discrepancies are discussed below, but these data clearly demonstrate that DTT is strongly correlated with bilayer thickness and that cryo-EM can detect variation in bilayer thickness with subangstrom sensitivity.
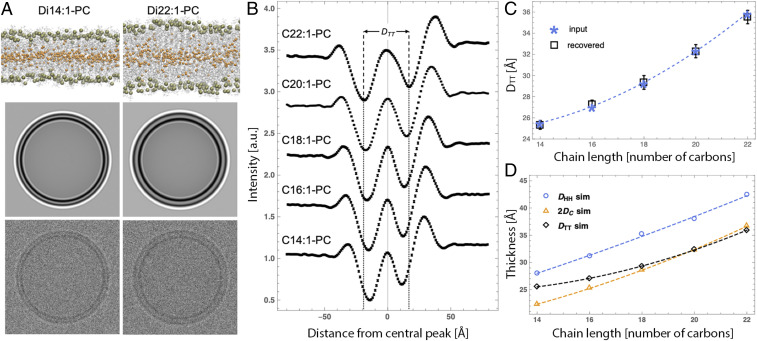
Trends in Cryo-EM-Derived Thickness Are Recapitulated in Simulated Images.
The lack of agreement between DTT and the well-understood headgroup–headgroup distance DHH derived from scattering measurements prompted further investigation into the relationships between bilayer structure and features observed in cryo-EM images. To that end, we used all-atom molecular dynamics (MD) simulations to predict I(w) profiles for the same PC bilayer series as experimentally evaluated above. Simulated membranes were slightly less densely packed than corresponding experimental bilayers, resulting in slightly larger lipid areas and thinner bilayers for all compositions (SI Appendix, Table S3). (We note that this discrepancy, which is likely due to limitations in simulation force fields, does not impact the ensuing analysis since experimental and simulated images were analyzed independently of each other.) Importantly, trends of bilayer thickness as a function of chain length were in excellent agreement with those derived from experimental scattering data (compare Figs. 1D and 2D). Following the procedure described by Wang et al. (39), cryo-EM images were computed from all-atom simulations as described in SI Appendix. Briefly, the image intensity is proportional to the phase shift experienced by an electron passing through the sample. Flat bilayer electron phase shift profiles [, with corresponding to the bilayer center] were computed from time-averaged atomic number density and charge density distributions along the bilayer normal. The flat bilayer profiles were converted to radial electron phase shift profiles by adding an arbitrary vesicle radius (taken to be 60 nm) to the w-coordinate. The radial profile was then analytically projected onto a plane and convolved with a contrast transfer function (CTF) to produce a synthetic cryo-EM image.
Fig. 2.
Simulated cryo-EM images generated from all-atom bilayer models corresponding to experiments. (A) Snapshots of all-atom molecular dynamics simulations of di14:1-PC and di22:1-PC bilayers (Upper) with corresponding simulated cryo-EM images before (Middle) and after (Lower) addition of noise. (B) Average intensity profiles obtained from noisy simulated cryo-EM images (n = 60) for a series of PC lipids. The vertical dashed lines show the trough-to-trough distance DTT for the di22:1-PC bilayer to highlight differences between the profiles. Profiles are offset vertically for clarity and error bars on individual data points are 95% confidence intervals. (C) DTT calculated from analytical I(w) profiles used as input to generate the images (asterisks) are compared to DTT (open squares with error bars) recovered from analysis of noisy simulated images. (D) Comparison of the chain length dependence of bilayer thicknesses (defined in Fig. 1 caption) and DTT, all derived from simulations. Dashed lines in C and D are visual guides.
Simulated cryo-EM images (without and with added Gaussian noise), shown in Fig. 2A for the thinnest (di14:1-PC) and thickest (di22:1-PC) bilayers in the series, faithfully reproduce the pattern of alternating light and dark rings observed in experimental images and reveal a distinctly larger DTT for the thicker bilayer. Average I(w) profiles obtained from analysis of noisy images (n = 60 vesicles) are shown in Fig. 2B (values reported in SI Appendix, Table S4), demonstrating very good overall agreement with the experimentally determined profiles (compare Figs. 1B and 2B), most relevantly a systematic increase in DTT with increasing lipid chain length. Importantly, the custom image processing algorithm, used to independently analyze both the experimental and simulation image data, yields an unbiased estimate of the true DTT that is accurate to ± 0.5 Å (Fig. 2C and SI Appendix, Table S4).
The comparison between DTT calculated from simulated images and the various bilayer thicknesses derived directly from the all-atom simulations is shown in Fig. 2D. Simulated DTT increases monotonically but not linearly with lipid chain length, in contrast to the trend observed in experimental data (Fig. 1D). In particular, simulated DTT for the shorter chain lengths 14:1 and 16:1 is slightly thicker than the hydrophobic thickness 2DC, while in experiments DTT and 2DC are nearly identical (Fig. 1D). Possible reasons for the discrepancy are discussed below, but generally these results confirm the origin of the structural features recovered from the analysis of the cryo-EM data, making the approach applicable to the visualization and analysis of nanoscale bilayer organization.
All-Atom Simulations Predict Two Sources of Contrast Between Coexisting Bilayer Phases.
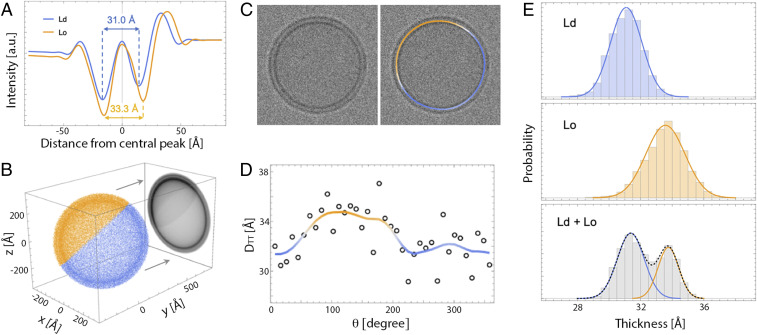
Having demonstrated the sensitivity of cryo-EM to bilayer thickness through a combination of experiment and simulation, we next sought to determine if cryo-EM can resolve coexisting phases within an individual vesicle by differences in their thickness. Previous work has established that Lo phases enriched in cholesterol and high-melting saturated lipids are 6 to 10 Å thicker than Ld phases enriched in low-melting lipids such as DOPC or 1-palmitoyl-2-oleoyl phosphatidylcholine (POPC) (30, 40, 41). In good agreement with experimental data, a bilayer thickness difference of ∼8 Å was observed in MD simulations of Ld and Lo phases composed of dipalmitoyl phosphatidylcholine (DPPC), DOPC, and cholesterol (42). We used simulation trajectories of pure Ld and Lo phases obtained from ref. 42 to investigate the ability of cryo-EM to detect two environments of different thickness within one vesicle (see SI Appendix for details of the simulations). Analytical projection and convolution of electron phase shift profiles for the two simulated pure Ld and Lo bilayers yielded a DTT difference of 2.3 Å (Fig. 3A), substantially smaller than the 6.7-Å hydrophobic thickness difference between the two bilayers calculated directly from the simulation trajectories (SI Appendix, Table S5).
Fig. 3.
Analysis of simulated cryo-EM images of phase-separated bilayers. (A) Analytical I(w) profiles calculated from all-atom MD simulation trajectories of Lo and Ld phases composed of DPPC/DOPC/cholesterol (42). (B) A 2D image of a vesicle with coexisting Ld (blue) and Lo (orange) domains is obtained by projecting a pointillist representation of the 3D spatial variation in electron phase shift density (given by the radial profiles in A) onto the xz plane. (C) A simulated cryo-EM image obtained from the projection in B after convolution with a CTF and addition of noise (Left) and the same image color-coded by the smoothed thickness values of individual 5-nm arc-length segments around the vesicle contour (Right). (D) Variation in the thickness of 5-nm segments (open circles) around the circumference of the vesicle in C. Local Gaussian smoothing (solid line) reveals the location of Ld and Lo domains. (E) Thickness distribution of 5-nm segments obtained from 100 simulated images of an Ld vesicle (Top), an Lo vesicle (Center), and a vesicle with a 60/40 Ld/Lo phase area fraction (Bottom). Solid lines are fits to Gaussians.
After analyzing the two phases individually, we computationally constructed vesicles that contained coexisting Lo and Ld domains to determine the feasibility of detecting differences between phases within a single vesicle. The analytical procedure described above is not applicable to bilayers that exhibit spatial variation in lipid composition, as in a phase-separated vesicle. We therefore constructed images of vesicles with coexisting Ld and Lo phases by modifying a previously described Monte Carlo method for computing scattering curves from such vesicles (43) (full details in SI Appendix). Briefly, a three-dimensional (3D) pointillist representation of a spherical vesicle with a 60/40 Ld/Lo phase fraction was created by generating random points in proportion to the 3D spatial variation in electron phase shift contrast (Fig. 3B). The set of points representing the phase-separated vesicle was then rotated around a randomly oriented vector originating at the center of the vesicle and projected onto the xz plane. The projected (two-dimensional, 2D) points were binned into pixels of 2.5-Å edge length (Fig. 3B), and the corresponding image was created by assigning to each pixel an intensity proportional to the total number of points in the bin. The image was then convolved with a CTF and subjected to Gaussian noise to simulate a cryo-EM projection image of a phase-separated vesicle (Fig. 3 C, Left). Fig. 3 A–C reveal that differences in the electron phase shift profiles of Ld and Lo bilayers manifest in images as differences in the relative intensities of Ld and Lo troughs (i.e., Lo is darker than Ld) as well as differences in the trough–trough distance (i.e., ) that are visible even in noisy images (Fig. 3 C, Left).
From the simulated images, we next calculated local DTT for annular segments of ∼5-nm arc length around the circumference of each vesicle (Fig. 3D). Although DTT of individual 5-nm segments is somewhat noisy, a clear trend emerges after four-point Gaussian smoothing (solid line) that is consistent with the location of Ld and Lo domains in the projection (Fig. 3 B and C). Fig. 3E shows DTT histograms obtained from 100 simulated images of either an Ld vesicle (Fig. 3 E, Top), an Lo vesicle (Fig. 3 E, Center), or different random projections of the phase-separated Ld/Lo vesicle shown in Fig. 3B (Fig. 3 E, Bottom). Histograms of the simulated Ld or Lo bilayers show unimodal distributions centered at the expected DTT value for each phase, while a clear bimodal distribution is observed in the analysis of phase-separated vesicles. By fitting the histogram data to two Gaussians, we recovered a DTT difference (i.e., ) of 2.5 Å (Fig. 3 E, Bottom), in excellent agreement with the 2.3 Å calculated from an analytical projection of the Ld and Lo phase shift profiles and subsequent convolution with the CTF (Fig. 3A). To summarize, analysis of simulated images suggests that coexisting fluid domains of different thickness within a single nanoscopic vesicle can be readily detected and quantified by cryo-EM.
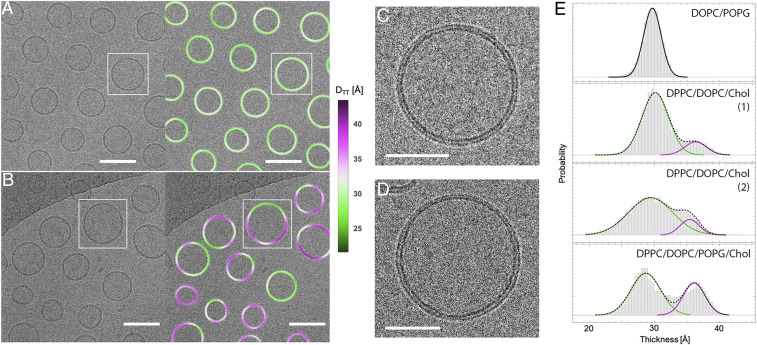
Cryo-EM Images Reveal Coexisting Nanoscale Liquid Domains in Biomimetic Lipid Mixtures.
A cryo-EM image of vesicles composed of the quaternary mixture DPPC/DOPC/POPG/cholesterol 40/35/5/20 is shown in Fig. 4B, with an expanded view of a vesicle shown in Fig. 4D. For comparison, an image of DOPC vesicles (also with 5% POPG) is shown in Fig. 4A, with an expanded view of one vesicle shown in Fig. 4C. Much greater variation in DTT within individual vesicles is observed in the quaternary mixture, consistent with the expected phase behavior of these lipid compositions. Specifically, DOPC forms a uniform fluid Ld phase at room temperature, while ternary mixtures similar to our POPG-doped quaternary mixture separate into coexisting Ld and Lo phases (22).
Fig. 4.
Direct visualization of coexisting phases in cryo-EM images of multicomponent lipid mixtures. (A) DOPC vesicles (with 5 mol % POPG) extruded through 50-nm pores (Left) and the same image color-coded by variation in trough–trough distance DTT (Right). (B) DPPC/DOPC/POPG/cholesterol 40/35/5/20 vesicles extruded through 50-nm pores (Left) and the same image color-coded by variation in DTT (Right), revealing significantly greater thickness variation within individual vesicles as compared to DOPC. (Scale bars in A and B, 100 nm.) (C and D) Zoom-in of boxed vesicles in A and B, respectively. (Scale bars in C and D, 50 nm.) (E) Thickness distribution of 5-nm segments obtained from vesicles composed of DOPC with 5% POPG (Top), two replicate samples of DPPC/DOPC/cholesterol with no POPG (Middle), and DPPC/DOPC/cholesterol with 5% POPG (Bottom). Solid lines are individual Gaussians and dashed lines are the total profiles for two-Gaussian fits.
DTT histograms obtained from 5-nm bilayer segments of >100 vesicles reveal the expected unimodal distribution for DOPC/DOPG 95/5 (Fig. 4 E, Upper) and a bimodal distribution for the DPPC/DOPC/POPG/cholesterol 40/35/5/20 (Fig. 4 E, Lower) with and an Ld phase area fraction of 0.64 (Table 2). Ternary mixtures composed of DPPC/DOPC/cholesterol 40/40/20 (i.e., without POPG) also showed a bimodal thickness distribution with and Ld phase area fraction of 0.80 (each calculated as the average of two replicate samples; see Fig. 4 E, Middle and Table 2), confirming that the contrast between phases was observable without charged lipids. The obtained area fractions are in reasonable agreement with values of 0.64 to 0.71 calculated from 2H-NMR phase diagrams (44, 45), assuming Ld and Lo molecular areas measured for a similar composition (40). The values are in good agreement with previously reported thickness differences measured by ensemble-averaged scattering approaches (30, 40) or in supported bilayers by AFM (41). Interestingly, was notably larger in experiments than in simulated bilayers (Fig. 3E), which may be related to approximations used to calculate the electron phase shift profiles from simulation trajectories or to deficiencies in the simulation force fields themselves (Discussion).
Table 2.
Thickness distribution analysis of multicomponent model membrane vesicles and GPMVs
| Ternary 1* | 131 | 4,153 | 30.13 ± 0.05 | 2.05 ± 0.04 | 36.3 ± 0.1 | 1.7 ± 0.1 | 0.85 ± 0.01 | 0.15 ± 0.01 | 1.06 |
| Ternary 2* | 134 | 4,047 | 29.7 ± 0.1 | 3.0 ± 0.1 | 36.1 ± 0.1 | 1.6 ± 0.1 | 0.75 ± 0.02 | 0.25 ± 0.02 | 0.87 |
| Quarternary† | 219 | 9,282 | 28.69 ± 0.04 | 2.27 ± 0.03 | 36.14 ± 0.04 | 1.68 ± 0.03 | 0.64 ± 0.01 | 0.36 ± 0.01 | 8.67 |
| GPMV | 91 | 9387 | 33.0 ± 0.5 | 3.5 ± 0.2 | 37.3 ± 0.1 | 2.2 ± 0.1 | 0.5 ± 0.1 | 0.5 ± 0.1 | 1.04 |
Parameter uncertainties are calculated from the covariance matrix at the solution of a two-Gaussian fit of the histogram data and do not reflect potential sample-to-sample variation.
DPPC/DOPC/cholesterol 40/40/20 replicates.
DPPC/DOPC/POPG/cholesterol 40/35/5/20.
Cryo-EM Imaging of Nanoscale Heterogeneity in Isolated Plasma Membrane Vesicles.
Liquid–liquid phase separation has been directly observed in isolated mammalian PMs (i.e., GPMVs) (12), providing an essential connection between the well-studied phase behavior of simple synthetic membranes and vastly more complex biological membranes (46). While the capacity of isolated PMs to separate into macroscopic Lo/Ld domains has been directly demonstrated, such phase separation is typically observed below room temperature, while at room temperature isolated PM vesicles appear uniform (12, 47–49). How the capacity for phase separation manifests itself under physiological conditions remains unresolved. One experimentally supported hypothesis suggests that PMs are poised for higher-order phase transitions, which produce fluctuations that become smaller (nanoscopic) as temperature increases beyond the macroscopic phase transition (50). To directly evaluate the possibility that isolated PMs retain lateral heterogeneity beyond their macroscopic phase transition temperature, we imaged membrane thickness in GPMVs by cryo-EM.
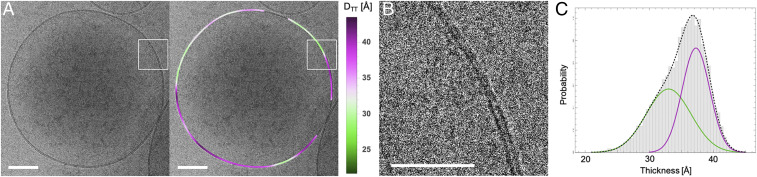
GPMVs were isolated from rat basophilic leukemia (RBL) cells as previously described, concentrated by centrifugation, and cryopreserved at room temperature, similar to the synthetic liposomes above. Although such preparations contain micrometer-scale vesicles that are typically imaged by light microscopy (12, 51), cryo-EM also revealed many 100- to 500-nm-diameter vesicles (Fig. 5). Despite the presence of transmembrane proteins, the bilayers of these vesicles appeared as dark concentric circles with bright halos (Fig. 5 A and B), similar in appearance to synthetic liposomes. We applied the above-described methodology to obtain a DTT distribution for 5-nm membrane segments in these isolated PMs that could be compared directly to distributions from synthetic membranes. GPMVs imaged by light microscopy at this temperature appear uniform, as they are above the Lo/Ld miscibility transition temperature (12, 48, 50, 52, 53). However, we observed clearly non-Gaussian DTT distributions in GPMV preparations, distinct from all single-component liposome preparations and similar to the phase-separated liposomes (Fig. 5C). The distribution was well described by a two-Gaussian fit (Table 2 and Fig. 5C) with relatively thick and thin regions. The thicker areas (37.3 Å) had DTT comparable to the Lo phase, while the relatively thin regions (33.0 Å) were approximately intermediate between the Lo and Ld phase in synthetic membranes. Thus, these images reveal nanoscopic lateral heterogeneities in biomembranes above the macroscopic phase separation temperature, qualitatively consistent with physical predictions (50).
Fig. 5.
Heterogeneity in GPMVs revealed by cryo-EM. (A) GPMV image (Left) and the same image color-coded by variation in trough–trough distance DTT (Right). (B) Zoom-in of boxed regions in A showing a domain boundary. (Scale bars, 100 nm in A and 50 nm in B.) (C) Thickness distribution of 5-nm segments obtained from 91 GPMVs. Solid lines are fits to Gaussians.
Discussion
Direct Probe-Free Imaging of Nanoscopic Domains from Thickness Differences.
We and the jointly submitted article by Cornell et al. report direct images of coexisting Ld and Lo nanodomains in unsupported lipid vesicles. No probes were added to the vesicles in this study. Instead, contrast in cryo-EM images derives from thickness differences between the coexisting phases. Notably, thickness differences between phases are not essential, as differences in electron density of the two phases also provide intensity contrast that is apparent in images. Very few techniques are capable of resolving liquid–liquid coexistence in free-standing membranes without extrinsic probes. SANS (30, 54) and 2H-NMR (23, 29, 55) rely on deuterated lipids, which—while not found in living organisms—are nevertheless true mixture components rather than trace probes. X-ray scattering of oriented bilayer stacks can resolve coexisting Ld and Lo by differences in either the average orientation of chain tilt at wide scattering angles (56) or lamellar repeat distance at small scattering angles (57). While the evidence of coexisting phases provided by these spectroscopic/scattering techniques is convincing, it is also indirect and ensemble-averaged. More importantly, information about domain size and properties is either entirely lacking or relies on modeling (30, 43). The direct imaging of nanodomains demonstrated here can provide insights into domain morphologies and size distributions that may be important for the biological functions of membrane domains. We have previously shown that vesicles containing a mixture of low-melting temperature (Tm) lipids such as DOPC and POPC (together with cholesterol and a saturated lipid) can have Lo/Ld thickness mismatches as small as ∼6 Å (30), but even these small thickness differences are much greater than the thickness resolution of the above-described method (e.g., Fig. 1D).
We note that the nanoscopic size of domains in our images of synthetic vesicles (Fig. 4B) is not an intrinsic property of the lipid mixture: micrometer-sized vesicles of the same composition have micrometer-sized Ld and Lo domains (22). Instead, domain size in these samples is constrained by the nanoscopic dimensions of the extruded vesicles themselves. It remains to be determined whether the sources of contrast we report will also be observed in mixtures with intrinsically nanoscopic domains, such as those near a critical point (44) or having high fractions of a mixed-chain low-Tm lipid (58, 59). We note that while our method can readily resolve small overall differences in membrane thickness between preparations (e.g., Fig. 1D), we have not yet explored in detail the resolution limits for how large coexisting membrane domains, and the differences between them, must be within a single vesicle. These issues will be the subject of continued investigation; however, observations of thickness heterogeneity in GPMVs that appear uniform by light microscopy (Fig. 5) suggest that intrinsically nanoscopic domains can indeed be observed.
Heterogeneous Thickness Distribution in Cell-Derived PMs.
GPMVs derived from the PM of living cells show a broad thickness distribution that is well approximated by a sum of two Gaussians (Fig. 5). The thicker component has a mean DTT that is notably similar to that of the Lo phase in DPPC/DOPC/cholesterol liposomes (37.3 Å in the GPMV compared to 36.2 Å in the ternary mixture). In contrast, the DTT of the thinner regions (33.0 Å) is notably larger than the Ld phase of the model membranes evaluated here (29.9 Å). These observations are fully consistent with previous reports that the ordered phase in GPMVs has properties similar to synthetic Lo phases, whereas differences between the coexisting phases are significantly smaller in natural membranes compared to synthetic systems (48, 60). These differences could be attributable to the transmembrane protein content in GPMVs (52) or the high abundance of “hybrid” lipids (one saturated and one unsaturated acyl chain), all of which may attenuate the differences between coexisting ordered and disordered lipid phases compared to synthetic, lipid-only mixtures (46). More importantly, the observation that GPMVs contain nanoscopic lateral domains under conditions at which they appear macroscopically uniform may inform on the mechanisms by which the principles of Lo/Ld phase separation are functionalized by living cells. Detailed comparisons to various existing models (e.g., critical fluctuations) are beyond the scope of the current study but may be accessible with existing methodologies.
Discrepancies between Experimental and Simulated Images.
In both experimental and simulated cryo-EM images, DTT increases monotonically with lipid chain length. However, the trends are markedly different at shorter chain lengths, with simulated values changing less steeply than reference thicknesses derived from SAXS measurements (compare Figs. 1D and 2D). Thus, while images simulated from all-atom MD configurations appear to correctly recapitulate the trend of increasing DTT with increasing bilayer thickness, the sources of contrast in the images may not be completely captured by the simulations or their analysis. This limitation is also evidenced in the analysis of the simulated pure Ld and pure Lo bilayers, where 2DC and DTT are almost identical for the Ld bilayer (31.2 Å compared to 31.0 Å) but very different for the Lo bilayer (37.9 Å versus 33.3 Å). One possible source for these discrepancies is the presence of a small amount of POPG in experimental vesicles that was not included in simulated bilayers. We previously showed that the addition of up to 5 mol % POPG or 1-palmitoyl-2-oleoyl phosphatidylserine does not change the structure of the bilayer (38), but we cannot exclude the possibility that this small fraction of charged lipid could influence the interaction of electrons with the bilayer and hence the observed I(w) profiles. Another possible source for these discrepancies may be the lack in simulated images of detailed electrostatic interactions that determine electron scattering, namely the use of the shielded Coulomb potential of isolated neutral atoms for calculating the electron phase shift profile. This approximation neglects changes in the electron distribution due to covalent bonds in the lipids. A third potential discrepancy is in the analytical approach for calculating DTT from simulated bilayers. The simulated bilayer patches are flat and therefore their electron phase shift profiles are symmetric about the bilayer midplane. In contrast, vesicles are slightly curved, possibly resulting in subtle differences in lipid packing between the two leaflets that are not present in the simulated images. Indeed, we observed a weak dependence of measured thickness on vesicle size for di18:1-PC vesicles, which may indicate a slight packing asymmetry in smaller extruded vesicles (SI Appendix, Fig. S6.1D and SI Appendix). The packing differences between the two leaflets may be larger for shorter chain lipids, which may explain the subtle discrepancies between DTT and 2DC observed for the di14:1-PC and di16:1-PC bilayers. Yet another likely source of error is that the simulation force field used here does not account for electron cloud polarizability. Future developments in simulation methodology, including the use of polarizable force fields, may further improve the quantitative agreement between DTT in simulations and experiments.
Relationship Between Bilayer Thickness and Cryo-EM DTT.
Bilayer thickness is a fundamentally important membrane parameter. Thickness is a determinant of membrane permeability (61), provides a connection between the membrane’s area compressibility and bending modulus (62), and defines the relationship between lipid volume and lateral area (63). Knowledge of bilayer thickness—defined in symmetric membranes as twice the distance between the bilayer midplane and some reference plane within the lipid molecule, typically in the interfacial region—is crucial for many biophysical models that underlie our understanding of membrane function. Despite this central importance, there are few methods to directly measure bilayer thicknesses, with most relying on ensemble averaging and models to interpret scattering behavior. We show here that cryo-EM provides the necessary spatial resolution at the single-vesicle level to reliably measure relative bilayer thickness and, further, can resolve thickness differences between Lo and Ld phases.
It is also important to point out that while DTT is clearly correlated with bilayer thickness and is sensitive to angstrom-level thickness variation (Fig. 1), it does not appear to directly correspond to a reference plane, such as those determined by X-ray and neutron scattering (64). Specifically, DTT reported here is derived from an image formed by a 2D projection of the electrostatic potential of a 3D vesicle and subsequent modification of the projected intensity by a CTF. Each of these processes has the effect of smearing the features related to the matter density distribution. Moreover, the spacing of interference fringes in a cryo-EM image, from which DTT is measured, arises from a complex interplay between the underlying bilayer structure and the CTF, which in turn depends on specific instrument parameters. In other words, changing the experimental settings (most notably, the defocus length) can change DTT and thus the apparent bilayer thickness. Therefore, while trends in DTT are reliable when samples are imaged and analyzed with the same protocols, DTT values as obtained represent a bilayer thickness convolved with the exact parameters of imaging conditions. We are presently pursuing a more detailed theoretical and experimental analysis that will ultimately be necessary to derive absolute bilayer structural parameters.
Limitations of Cryo-EM for Detecting Domains.
While the methods we describe allow ultrastructural insights into biomimetic and biological membranes, a number of limitations have become evident in the course of these investigations. Unsurprisingly, DTT measurements in small bilayer arcs (as in Figs. 3–5) are quite noisy and accurate estimates of population averages require numerous samples, in analogy with single-particle averaging in cryo-EM. This approach relies on the assumption of homogeneity, both within and between individual vesicles. The uniformity of any given vesicle population is likely preparation-dependent and rarely well characterized. Especially in multicomponent mixtures, significant vesicle-to-vesicle heterogeneities are possible.
While images of single-component vesicles suggest that cryo-EM can resolve population thickness differences as small as 0.5 Å, this analysis requires significant averaging. Thus, smaller domains will inherently be more difficult to define and measure. The thickness contrast and domain size limitations of this technique remain to be defined. However, we note that all measurements described here have necessitated relatively small numbers of vesicles, with larger sample sizes likely facilitating higher resolution. On the other hand, there are other sources of noise that are more difficult to control, such as variations in ice thickness or sample degradation due to beam radiation.
Methods
Materials.
A list of materials is available in SI Appendix.
Preparation of Large Unilamellar Vesicles.
Aqueous lipid dispersions at 20 mg/mL were prepared by first mixing appropriate volumes of lipid stocks in organic solvent with a glass Hamilton syringe. The solvent was evaporated with an inert gas stream followed by high vacuum overnight. The dry lipid film was hydrated with ultrapure water above the lipid’s main transition temperature (or for multicomponent mixtures, above the main transition of the highest melting component DPPC) for at least 1 h with intermittent vortex mixing. The resulting multilamellar vesicle suspension was subjected to at least five freeze/thaw cycles using a −80 °C freezer then extruded through a polycarbonate filter using a handheld miniextruder (Avanti Polar Lipids) by passing the suspension through the filter 31 times. Unsaturated lipids were extruded at room temperature, and multicomponent mixtures were extruded at 45 °C. Because different measurement techniques have different optimal concentration ranges, samples were diluted with water or buffer prior to measurement as described in SI Appendix. Samples for cryo-EM measurements were typically cryopreserved and measured within a day of preparation. Samples for SAXS were measured within a few days of preparation.
SAXS.
SAXS measurements were performed using a Rigaku BioSAXS-2000 home source system (Rigaku Americas) equipped with a HF007 copper rotating anode, a Pilatus 100K 2D detector, and an automatic sample changer. SAXS data were collected at a fixed sample-to-detector distance using a silver behenate calibration standard, with a data collection time of 3 h. The one-dimensional scattering intensity [, where is the X-ray wavelength and is the scattering angle relative to the incident beam] was obtained by radial averaging of the corrected 2D image data, an operation that was performed automatically using Rigaku SAXSLab software. Data were collected in 10-min frames with each frame processed separately to assess radiation damage; there were no significant changes in the scattering curves over time. Background scattering from water collected at the same temperature was subtracted from each frame, and the background-corrected from the individual frames was then averaged, with the SD taken to be the measurement uncertainty and used in weighted least-squares analyses described in SI Appendix.
Cryo-EM.
To cryopreserve vesicles, 4 μL of a 1 mg/mL sample were applied to a Quantifoil 2/2 carbon-coated 200-mesh copper grid that was glow-discharged for 1 min at 15 mA in a Pelco Easi-Glow discharge device. After manual blotting, the grids were plunged from room temperature into liquid ethane cooled with liquid N2. Cryopreserved grids were stored in liquid N2 until use. Samples were coded prior to preservation and all sample preparation and image collection was accomplished by an investigator blind to the sample composition. Image collection was accomplished at ∼2 μm under focus on a FEI Polara G2 operated at 300 kV equipped with a Gatan K2 Summit direct electron detector operated in counting mode. Data collection was performed in a semiautomated fashion using Serial EM software operated in low-dose mode (65). Briefly, areas of interest were identified visually and 8 × 8 montages were collected at low magnification (2,400×) at various positions across the grid and then individual areas were marked for automated data collection. Data were collected at 2.7 Å per pixel. Movies of 30 dose-fractionated frames were collected at each target site with the total electron dose being kept to <20 e−/Å2. Dose-fractionated movies were drift-corrected with MotionCor2 (66).
Cryo-EM Data Analysis.
A detailed description of analysis procedures is found in SI Appendix. Briefly, the vesicle contour, arbitrarily chosen to be the bright central peak between the dark troughs, was identified from filtered images; from the analysis of simulated images, we conservatively estimate the uncertainty in locating the contour to be ±2 Å. Individual pixel intensities within 10 nm of the contour were then binned by distance from the nearest contour point to generate a radially integrated intensity profile I(w) for the vesicle, from which the trough–trough distance DTT was determined. For analysis of phase-separated vesicles, the contour was divided into segments of ∼5-nm width and DTT determined separately for each segment.
MD Simulations.
All-atom MD simulations of five single-component bilayers (di14:1-PC, di16:1-PC, di18:1-PC, di20:1-PC or di22:1-PC) were performed with NAMD (67) using the CHARMM36 force field parameters (68, 69). All bilayers contained 64 lipids per leaflet, 45 waters per lipid and no ions, and except for di14:1-PC were constructed and equilibrated with the CHARMM-GUI protocols (70–72). Since di14:1-PC was not available in CHARMM-GUI at the time of construction, special steps were taken to construct and equilibrate it as described in SI Appendix. The production runs for all bilayers were performed at constant temperature of 298 K and constant pressure of 1 atm using the same simulation parameters as in ref. 73. Number and electron density profiles of each lipid and water atom in the systems were calculated from the last 680 to 700 ns of the centered bilayer trajectories with the Density Profile tool in VMD (74).
GPMV Isolation and Preparation.
RBL cells were cultured in medium containing 60% Eagle’s minimum essential medium, 30% RPMI, 10% fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in humidified 5% CO2. GPMVs were isolated from RBLs as described previously (12, 51). Briefly, GPMV formation was induced by 25 mM paraformaldehyde + 2 mM DTT in isotonic GPMV buffer containing NaCl, 10 mM Hepes, and 2 mM CaCl2, pH 7.4. All GPMVs from one 60-mm cultured dish were combined into a 1.5-mL microcentrifuge tube and concentrated into a loose pellet by centrifugation at 20,000 × g for 20 min. The pellet was resuspended by gentle pipetting in 20 μL of GPMV buffer immediately prior to cryopreservation. Small vesicles present in this preparation were abundant by cryo-EM. These may represent submicroscopic vesicles formed in the same manner as micrometer-scale GPMVs or fragmentation of larger GPMVs into smaller ones during preparation. Both possibilities could conceivably lead to differences in composition or properties of GPMVs. We also note that these GPMVs do not retain all features of native PMs. While their composition is likely to be representative of the PM (47, 75), the phospholipid asymmetry, cytoskeletal interactions, and energy-dependent processes of live cell PMs are lost (46, 51).
Supplementary Material
Acknowledgments
This work was supported by NSF Grant MCB-1817929 (to F.A.H.). Funding for I.L. was provided by the NIH/National Institute of General Medical Sciences (GM124072, GM134949, and GM120351), the Volkswagen Foundation (Grant 93091), and the Human Frontiers Science Program (RGP0059/2019). Funding for M.N.W. was provided by NIH/National Institute of Neurological Disease and Stroke (R01NS101686) and from the William Wheless III professorship. M.D. was supported by NIH F32GM134704. SAXS measurements were supported by Department of Energy scientific user facilities at Oak Ridge National Laboratory. The computational work used the Extreme Science and Engineering Discovery Environment, account TG-MCB180168. The Polara electron microscope was supported, in part, through the Structural Biology Imaging Center at The University of Texas Health Science Center at Houston. The Gatan K2 Summit detector was funded by the NIH (S10OD016279). We thank Alex Sodt and Ed Lyman for providing simulation data. We gratefully acknowledge the members of the I.L. laboratory for extensive discussions and support on this manuscript, as well as the Sarah Keller laboratory for their critical feedback on our manuscript and collegial/collaborative approach to concurrent publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002200117/-/DCSupplemental.
Data Availability.
The data supporting the findings of this study are available within the paper and SI Appendix.
References
- 1.Janmey P. A., Kinnunen P. K., Biophysical properties of lipids and dynamic membranes. Trends Cell Biol. 16, 538–546 (2006). [DOI] [PubMed] [Google Scholar]
- 2.van Meer G., Voelker D. R., Feigenson G. W., Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitra K., Ubarretxena-Belandia I., Taguchi T., Warren G., Engelman D. M., Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad. Sci. U.S.A. 101, 4083–4088 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharpe H. J., Stevens T. J., Munro S., A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142, 158–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretscher M. S., Munro S., Cholesterol and the Golgi apparatus. Science 261, 1280–1281 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Lorent J. H. et al., Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 8, 1219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levental I., Levental K. R., Heberle F. A., Lipid rafts: Controversies resolved, mysteries remain. Trends Cell Biol. 30, 341–353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levental I., Veatch S., The continuing mystery of lipid rafts. J. Mol. Biol. 428, 4749–4764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munro S., Lipid rafts: Elusive or illusive? Cell 115, 377–388 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Marsh D., Cholesterol-induced fluid membrane domains: A compendium of lipid-raft ternary phase diagrams. Biochim. Biophys. Acta 1788, 2114–2123 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Feigenson G. W., Phase boundaries and biological membranes. Annu. Rev. Biophys. Biomol. Struct. 36, 63–77 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumgart T. et al., Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 104, 3165–3170 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rayermann S. P., Rayermann G. E., Cornell C. E., Merz A. J., Keller S. L., Hallmarks of reversible separation of living, unperturbed cell membranes into two liquid phases. Biophys. J. 113, 2425–2432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toulmay A., Prinz W. A., Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 202, 35–44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz-Rohrer B. B., Levental K. R., Simons K., Levental I., Membrane raft association is a determinant of plasma membrane localization. Proc. Natl. Acad. Sci. U.S.A. 111, 8500–8505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinoshita M. et al., Raft-based sphingomyelin interactions revealed by new fluorescent sphingomyelin analogs. J. Cell Biol. 216, 1183–1204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stone M. B., Veatch S. L., Steady-state cross-correlations for live two-colour super-resolution localization data sets. Nat. Commun. 6, 7347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ono A., Relationships between plasma membrane microdomains and HIV-1 assembly. Biol. Cell 102, 335–350 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sengupta P. et al., A lipid-based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat. Cell Biol. 21, 452–461 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Frisz J. F. et al., Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 110, E613–E622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sevcsik E. et al., GPI-anchored proteins do not reside in ordered domains in the live cell plasma membrane. Nat. Commun. 6, 6969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veatch S. L., Keller S. L., Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 85, 3074–3083 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veatch S. L., Polozov I. V., Gawrisch K., Keller S. L., Liquid domains in vesicles investigated by NMR and fluorescence microscopy. Biophys. J. 86, 2910–2922 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond A. T. et al., Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 102, 6320–6325 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baumgart T., Hess S. T., Webb W. W., Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature 425, 821–824 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Sharma P. et al., Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116, 577–589 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Plowman S. J., Muncke C., Parton R. G., Hancock J. F., H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 102, 15500–15505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ionova I. V., Livshits V. A., Marsh D., Phase diagram of ternary cholesterol/palmitoylsphingomyelin/palmitoyloleoyl-phosphatidylcholine mixtures: Spin-label EPR study of lipid-raft formation. Biophys. J. 102, 1856–1865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juhasz J., Sharom F. J., Davis J. H., Quantitative characterization of coexisting phases in DOPC/DPPC/cholesterol mixtures: Comparing confocal fluorescence microscopy and deuterium nuclear magnetic resonance. Biochim. Biophys. Acta 1788, 2541–2552 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Heberle F. A. et al., Bilayer thickness mismatch controls domain size in model membranes. J. Am. Chem. Soc. 135, 6853–6859 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Heberle F. A., Wu J., Goh S. L., Petruzielo R. S., Feigenson G. W., Comparison of three ternary lipid bilayer mixtures: FRET and ESR reveal nanodomains. Biophys. J. 99, 3309–3318 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Wit G., Danial J. S., Kukura P., Wallace M. I., Dynamic label-free imaging of lipid nanodomains. Proc. Natl. Acad. Sci. U.S.A. 112, 12299–12303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scomparin C., Lecuyer S., Ferreira M., Charitat T., Tinland B., Diffusion in supported lipid bilayers: Influence of substrate and preparation technique on the internal dynamics. Eur Phys J E Soft Matter 28, 211–220 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Goodchild J. A., Walsh D. L., Connell S. D., Nanoscale substrate roughness hinders domain formation in supported lipid bilayers. Langmuir 35, 15352–15363 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Egelman E. H., The current revolution in cryo-EM. Biophys. J. 110, 1008–1012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer T. D., Dash P. K., Liu J., Waxham M. N., Morphology of mitochondria in spatially restricted axons revealed by cryo-electron tomography. PLoS Biol. 16, e2006169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis J. D., et al. , The desmosome is a mesoscale lipid raft-like membrane domain. bioRxiv:10.1101/401455 (12 September 2018). [DOI] [PMC free article] [PubMed]
- 38.Scott H. L. et al., On the mechanism of bilayer separation by extrusion, or why your LUVs are not really unilamellar. Biophys. J. 117, 1381–1386 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Bose P. S., Sigworth F. J., Using cryo-EM to measure the dipole potential of a lipid membrane. Proc. Natl. Acad. Sci. U.S.A. 103, 18528–18533 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heftberger P., Kollmitzer B., Rieder A. A., Amenitsch H., Pabst G., In situ determination of structure and fluctuations of coexisting fluid membrane domains. Biophys. J. 108, 854–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bleecker J. V., Cox P. A., Keller S. L., Mixing temperatures of bilayers not simply related to thickness differences between Lo and Ld phases. Biophys. J. 110, 2305–2308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sodt A. J., Sandar M. L., Gawrisch K., Pastor R. W., Lyman E., The molecular structure of the liquid-ordered phase of lipid bilayers. J. Am. Chem. Soc. 136, 725–732 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heberle F. A., Anghel V. N. P., Scattering from phase‐separated vesicles. I. An analytical form factor for multiple static domains. J. Appl. Cryst. 48, 1391–1404 (2015). [Google Scholar]
- 44.Veatch S. L., Soubias O., Keller S. L., Gawrisch K., Critical fluctuations in domain-forming lipid mixtures. Proc. Natl. Acad. Sci. U.S.A. 104, 17650–17655 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis J. H., Clair J. J., Juhasz J., Phase equilibria in DOPC/DPPC-d62/cholesterol mixtures. Biophys. J. 96, 521–539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levental K. R., Levental I., Giant plasma membrane vesicles: Models for understanding membrane organization. Curr. Top. Membr. 75, 25–57 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Levental K. R. et al., omega-3 polyunsaturated fatty acids direct differentiation of the membrane phenotype in mesenchymal stem cells to potentiate osteogenesis. Sci. Adv. 3, eaao1193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levental K. R. et al., Polyunsaturated lipids regulate membrane domain stability by tuning membrane order. Biophys. J. 110, 1800–1810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burns M., Wisser K., Wu J., Levental I., Veatch S. L., Miscibility transition temperature scales with growth temperature in a zebrafish cell line. Biophys. J. 113, 1212–1222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veatch S. L. et al., Critical fluctuations in plasma membrane vesicles. ACS Chem. Biol. 3, 287–293 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Sezgin E. et al., Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 7, 1042–1051 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Levental I., Grzybek M., Simons K., Raft domains of variable properties and compositions in plasma membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 108, 11411–11416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gray E., Karslake J., Machta B. B., Veatch S. L., Liquid general anesthetics lower critical temperatures in plasma membrane vesicles. Biophys. J. 105, 2751–2759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marquardt D., Heberle F. A., Nickels J. D., Pabst G., Katsaras J., On scattered waves and lipid domains: Detecting membrane rafts with X-rays and neutrons. Soft Matter 11, 9055–9072 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vist M. R., Davis J. H., Phase equilibria of cholesterol/dipalmitoylphosphatidylcholine mixtures: 2H nuclear magnetic resonance and differential scanning calorimetry. Biochemistry 29, 451–464 (1990). [DOI] [PubMed] [Google Scholar]
- 56.Mills T. T. et al., Liquid-liquid domains in bilayers detected by wide angle X-ray scattering. Biophys. J. 95, 682–690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uppamoochikkal P., Tristram-Nagle S., Nagle J. F., Orientation of tie-lines in the phase diagram of DOPC/DPPC/cholesterol model biomembranes. Langmuir 26, 17363–17368 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goh S. L., Amazon J. J., Feigenson G. W., Toward a better raft model: Modulated phases in the four-component bilayer, DSPC/DOPC/POPC/CHOL. Biophys. J. 104, 853–862 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konyakhina T. M. et al., Control of a nanoscopic-to-macroscopic transition: Modulated phases in four-component DSPC/DOPC/POPC/Chol giant unilamellar vesicles. Biophys. J. 101, L8–L10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaiser H. J. et al., Order of lipid phases in model and plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 106, 16645–16650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagle J. F., Mathai J. C., Zeidel M. L., Tristram-Nagle S., Theory of passive permeability through lipid bilayers. J. Gen. Physiol. 131, 77–85 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rawicz W., Olbrich K. C., McIntosh T., Needham D., Evans E., Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 79, 328–339 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kucerka N. et al., The effect of cholesterol on short- and long-chain monounsaturated lipid bilayers as determined by molecular dynamics simulations and X-ray scattering. Biophys. J. 95, 2792–2805 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heberle F. A. et al., Model-based approaches for the determination of lipid bilayer structure from small-angle neutron and X-ray scattering data. Eur. Biophys. J. 41, 875–890 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Zheng S. Q. et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Phillips J. C. et al., Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klauda J. B. et al., Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klauda J. B., Monje V., Kim T., Im W., Improving the CHARMM force field for polyunsaturated fatty acid chains. J. Phys. Chem. B 116, 9424–9431 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Lee J. et al., CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Jo S., Lim J. B., Klauda J. B., Im W., CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doktorova M., LeVine M. V., Khelashvili G., Weinstein H., A new computational method for membrane compressibility: Bilayer mechanical thickness revisited. Biophys. J. 116, 487–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 (1996). [DOI] [PubMed] [Google Scholar]
- 75.Symons J. L. et al., Lipidomic atlas of mammalian cell membranes reveals hierarchical variation induced by culture conditions, subcellular membranes, and cell lineages. Soft Matter, 10.1039/d0sm00404a (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the paper and SI Appendix.