Significance
The genetic characterization of a common phenotype for an entire population reveals both the genetic epidemiology of that phenotype and the power of family-based, population-wide genomic analysis. We characterized the genetics of hearing loss throughout the Palestinian population of the West Bank and Gaza. In families with no prior history of hearing loss, we estimate that 56% of hearing loss is genetic and 44% is not genetic. For most families with inherited hearing loss, causal genes and mutations were identified. Most inherited hearing loss in the population was attributable to consanguinity. Given the ongoing decline in consanguineous marriage, inherited hearing loss will likely be much rarer in the next generation.
Keywords: hearing loss, human genetics, genomics, deafness, consanguinity
Abstract
The genetic characterization of a common phenotype for an entire population reveals both the causes of that phenotype for that place and the power of family-based, population-wide genomic analysis for gene and mutation discovery. We characterized the genetics of hearing loss throughout the Palestinian population, enrolling 2,198 participants from 491 families from all parts of the West Bank and Gaza. In Palestinian families with no prior history of hearing loss, we estimate that 56% of hearing loss is genetic and 44% is not genetic. For the great majority (87%) of families with inherited hearing loss, panel-based genomic DNA sequencing, followed by segregation analysis of large kindreds and transcriptional analysis of participant RNA, enabled identification of the causal genes and mutations, including at distant noncoding sites. Genetic heterogeneity of hearing loss was striking with respect to both genes and alleles: The 337 solved families harbored 143 different mutations in 48 different genes. For one in four solved families, a transcription-altering mutation was the responsible allele. Many of these mutations were cryptic, either exonic alterations of splice enhancers or silencers or deeply intronic events. Experimentally calibrated in silico analysis of transcriptional effects yielded inferences of high confidence for effects on splicing even of mutations in genes not expressed in accessible tissue. Most (58%) of all hearing loss in the population was attributable to consanguinity. Given the ongoing decline in consanguineous marriage, inherited hearing loss will likely be much rarer in the next generation.
The discovery of genes responsible for inherited hearing loss has revealed molecular mechanisms essential to the development and maintenance of hearing that will ultimately enable prevention and treatment of its loss (1). In the short term, identification of the genetic cause of hearing loss in a child enables parents to anticipate possible progression or syndromic effects (2), judge the potential effectiveness of cochlear implants (3), and, with pregestational diagnosis, to opt to preclude hearing loss in future children.
Families from regions with traditions of consanguinity have been particularly informative for discovery of genes responsible for recessive traits. In particular, over the past 20 y, families from Palestinian communities with traditions of consanguineous marriages and large families have been instrumental to the discovery and characterization of genes responsible for hearing loss (4–11). For this project, we characterized the genetics of inherited hearing loss throughout the Palestinian population, ascertaining children with hearing loss throughout the Palestinian West Bank and Gaza and evaluating responsible genes and mutations. Characterization of hearing loss for an entire population both reveals the epidemiology of hearing loss for that region and provides valuable information to address the genetics of hearing loss worldwide.
Results
Genetic Epidemiology of Hearing Loss.
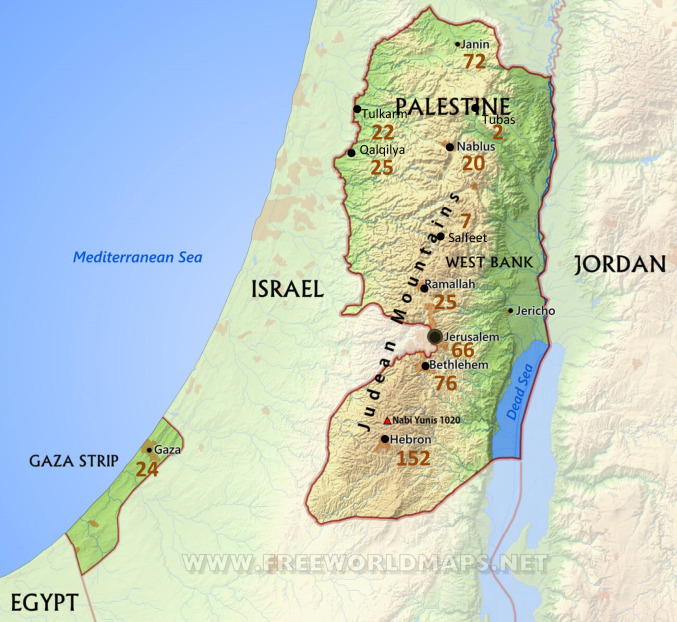
The project enrolled and sequenced 2,198 participants from 491 families (Table 1). The families live in all parts of the West Bank and Gaza (Fig. 1). Of all families, 52% (254/491) included more than one child with hearing loss (multiplex families) and 48% (237/491) included only one child with hearing loss (singleton families). In 9.1% (23/254) of multiplex families and 4.2% (10/237) of singleton families, children with hearing loss also presented with syndromic features.
Table 1.
Demographic and clinical features of Palestinian families with hearing loss
| N | Proportion | |
| Residence | ||
| Bethlehem | 76 | 0.15 |
| Gaza | 24 | 0.05 |
| Hebron | 152 | 0.31 |
| Jenin | 72 | 0.15 |
| Jerusalem | 66 | 0.13 |
| Nablus | 20 | 0.04 |
| Qalqilya | 25 | 0.05 |
| Ramallah | 25 | 0.05 |
| Salfeet | 7 | 0.01 |
| Tubas | 2 | 0.004 |
| Tulkarem | 22 | 0.04 |
| Clinical features | ||
| Nonsyndromic hearing loss | 458 | 0.93 |
| Syndromic hearing loss | 33 | 0.07 |
| 2+ affected children (multiplex) | 254 | 0.52 |
| 1 affected child (singleton) | 237 | 0.48 |
| Prelingual onset | 482 | 0.98 |
| Postlingual onset | 9 | 0.02 |
| Total | 491 | 1.00 |
Fig. 1.
Cities and towns of Palestinian families with hearing loss. The number of families enrolled from each place is indicated in red.
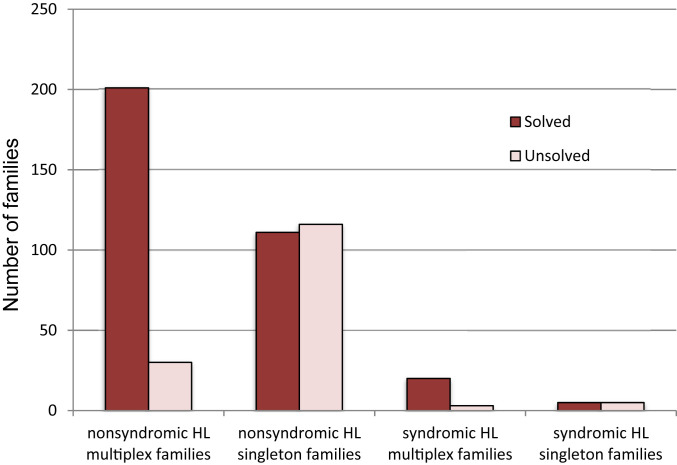
Genetic diagnoses were obtained for 87% (201/231) of multiplex families and 49% (111/227) of singleton families with nonsyndromic hearing loss, and for 87% (20/23) of multiplex families and 50% (5/10) of singleton families with syndromic hearing loss (Fig. 2). Of the 337 families with genetic diagnoses, hearing loss was autosomal recessive in 323 families, X-linked recessive in 3 families, and autosomal dominant in 11 families (SI Appendix, Tables S1, S2A, and S2B). Severity and onset of hearing loss were associated with recessive versus dominant inheritance. Hearing loss was severe to profound and prelingual in onset in most children in families with recessive hearing loss, but mild to moderate and progressive in families with dominant hearing loss.
Fig. 2.
Families with hearing loss (HL), with and without genetic diagnoses.
For families with nonsyndromic hearing loss, the difference in proportions of multiplex versus singleton families with genetic diagnoses was highly significant: relative risk (RR) = 1.78, 95% CI [1.54, 2.05], P = 5.02E-19. This difference likely reflects a higher prevalence of nongenetic hearing loss among singleton families and can be used to estimate the proportion of hearing loss in singleton families caused by nongenetic factors. That is, if hearing loss in all multiplex families has a genetic basis, of which 87% was identifiable by the gene panel, then an estimate of the proportion of nongenetic hearing loss among singleton families is [227 − (111/0.87)]/227, or 44%.
For 13 families with children with apparently nonsyndromic hearing loss, damaging mutations were identified in genes associated with syndromic conditions: ADGRV1, CLPP, GPSM2, HSD17B4, LARS2, SLC26A4, USH1C, and USH2A. Syndromic features due to mutations in some of these genes may appear years after the onset of hearing loss. In contrast, hearing loss of the five unresolved singleton cases of syndromic hearing loss all appeared in the context of developmental delay. These five cases may be caused by de novo mutations in genes not in the hearing-loss panel.
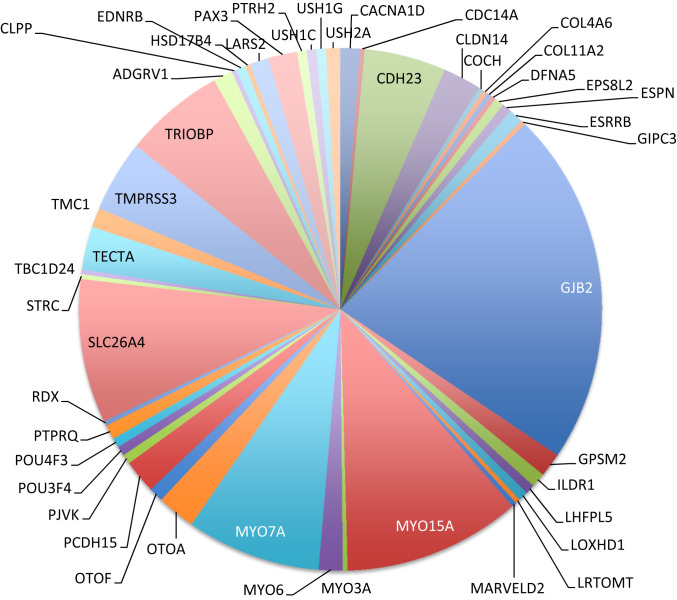
Heterogeneity of the genetic causes of inherited hearing loss has been recognized since its first genetic diagnoses (12). Genetic heterogeneity with respect to both genes and alleles was striking in the Palestinian population. The 337 solved families harbored 143 different mutations in 48 different genes (Table 2 and Fig. 3). SI Appendix, Table S2B indicates the phenotypes and genotypes of each of these 337 families.
Table 2.
Genetic heterogeneity of inherited hearing loss in Palestinian families
| Hearing loss of family | No. of families | No. of different genes* | No. of different mutations* |
| Syndromic | 66 | 19 | 43 |
| Nonsyndromic | 271 | 34 | 104 |
| All families | 337 | 48 | 143 |
Sums are greater than numbers for all families, because some genes and mutations appeared both in families with syndromic hearing loss and in families with nonsyndromic hearing loss.
Fig. 3.
Genes responsible for hearing loss in Palestinian families.
Effect of Consanguinity on the Prevalence of Hearing Loss.
For the proband of each family, the proportion of homozygous loci among all loci in the panel was calculated based on the proband’s genotypes at all single-nucleotide polymorphisms (SNPs) (common or rare) at each locus. Then, for each proband, a Z score was calculated to normalize this proportion vis-à-vis all families in the cohort (Methods). Means and SDs of the Z distributions were 0.48 ± 1.03 for families with documented consanguinity and −0.88 ± 0.63 for families with documented nonconsanguinity (P = 3.67E-5, two-tailed t test). Using these distributions to assess the likelihood of consanguinity for families with incomplete pedigree information yielded estimates for the cohort as a whole of 90% (440/491) consanguineous families and 10% (51/491) nonconsanguineous families.
The effect of consanguinity on the frequency of hearing loss in the Palestinian population can be estimated by considering how the profile of hearing loss would change if all families were nonconsanguineous (SI Appendix, Table S3). In a fully nonconsanguineous population, the number of families with dominant hearing loss, X-linked hearing loss, recessive hearing loss due to compound heterozygous mutations, or hearing loss due to nongenetic causes would not necessarily change. In contrast, the number of families with recessive hearing loss due to homozygous mutations would decrease by more than 99%. Overall, then, in a fully nonconsanguineous population, the decrease in the number of families with hearing loss would be ∼58%.
Functional Analysis of Candidate Mutations.
A population-wide survey offers unique tools to evaluate the consequences of candidate mutations. Pedigree analysis from families representing the population as a whole (not only severely affected families) and population-wide estimates of candidate allele frequencies can be integrated with in silico evaluation and experimental studies to evaluate coding sequence mutations and identify and characterize mutations in noncoding genomic regions.
Mutations Specific to the Critical Isoform of a Deafness Gene.
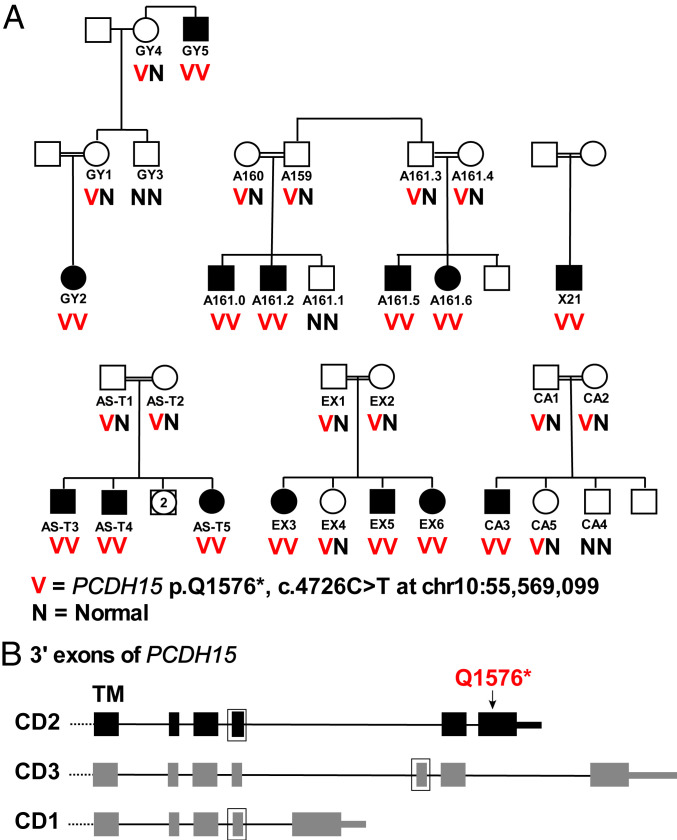
Mutations in protocadherin 15 (PCDH15) are the cause of hearing loss and vestibular dysfunction in the Ames waltzer mouse (13) and can lead to either Usher syndrome or nonsyndromic hearing loss in humans (14). Genomic structure of PCDH15 is complex, with multiple alternate transcripts converging in three isoforms (CD1, CD2, CD3) defined by alternate C-terminal cytoplasmic domains. Based on molecular and functional characterization of isoform-specific mouse models of the three isoforms, only CD2 contributes to development and maintenance of hearing (15). A founder mutation in Palestinian families supports the critical role of PCDH15 isoform CD2. Children from six Palestinian families, all with congenital severe-to-profound nonsyndromic hearing loss, were homozygous for PCDH15 p.(Gln1576*) (c.4726C>T; NM_001142769.3; chr10:55,569,099) (Fig. 4A). This site maps to an exon transcribed only in isoform CD2 of PCDH15, where it creates a stop in the last coding exon and is predicted to lead to loss of 215 amino acid residues of the PCDH15 protein (Fig. 4B). Pedigree analysis of the six families carrying PCDH15 p.(Gln1576*) indicates that the mutation cosegregates with hearing loss, and that the probability of this degree of cosegregation appearing by chance is 5.09E-7. The families are from four different towns in the central and northern West Bank, and the mutation does not appear in any of 1,309 Palestinian controls from the West Bank and Gaza. (Nor does it appear in the Genome Aggregation Database [gnomAD], although gnomAD may include few controls from this population.) Given all these factors, we interpreted PCDH15 p.(Gln1576*) as likely pathogenic. Interpretation of this mutation illustrates the power of population-wide information to complement experimental results from mouse models.
Fig. 4.
Families with hearing loss due to nonsense mutation in PCDH15 isoform CD2. (A) Families with hearing loss and homozygosity for PCDH15 p.(Q1756*). The chance of this degree of cosegregation occurring by chance is <10E-6. (B) PCDH15 isoforms, indicating exons encoding the transmembrane (TM) domain and cytoplasmic domains C-terminal to the TM domain. Exons outlined in boxes are present in some but not all transcripts. Of the three isoforms of PCDH15, only CD2 is critical to the development and maintenance of hearing. PCDH15 p.(Q1576*) is transcribed only on isoform CD2.
Missense Mutations.
Of the 143 different damaging mutations in families in the cohort, 45 (31%) were missense mutations (SI Appendix, Table S4). Among these are four alleles that are founder mutations for the Palestinian population or for the Arab population more generally. Specifically in Palestinian families, CACNA1D p.(Ala376Val) is responsible for moderate hearing loss associated with cardiac anomalies, including prolonged atrioventricular conduction on an electrocardiogram; LARS2 p.(Asn153His) is responsible for Perrault syndrome, characterized by recessive moderate-to-severe hearing loss and neurological anomalies in females and males and ovarian dysgenesis in females; and CLDN14 p.(Pro28Leu) is responsible for recessive congenital severe-to-profound nonsyndromic hearing loss in multiple families. In the wider Mediterranean and west Asian region, MYO7A p.(Gly2163Ser) is responsible for recessive nonsyndromic hearing loss; this allele has been previously reported from Iran, Pakistan, Turkey, and Algeria. Overall, based on evolutionary conservation, pedigree analysis, and allele frequencies among Palestinian controls, we interpreted 16 of 18 previously unreported missense mutations as likely pathogenic (SI Appendix, Table S4A) and suggested reclassification of 6 others from variants of unknown significance to be likely pathogenic (SI Appendix, Table S4B). Evidence for causality is summarized for each of these 24 alleles. The other 21 missense mutations had been previously reported from multiple sources as pathogenic or likely pathogenic for hearing loss (SI Appendix, Table S4C). Missense mutations likely damaging to protein function were responsible for hearing loss in 24% (80/337) of solved families.
Splice-Site and Enhancer Mutations.
Transcriptional effects were characterized by integrating experimental analysis of patient RNA, cosegregation analysis in pedigrees, allele frequencies in Palestinian controls, and bioinformatics. Of the 143 different mutations responsible for hearing loss in the cohort, 33 (23%) were predicted to cause hearing loss by altering transcription (SI Appendix, Table S5). Two mutations in MYO15A and two in SLC26A4 are founder mutations. MYO15A c.3609+985G>A, the deeply intronic mutation described below, and MYO15A c.7207G>T, which introduces a cryptic splice donor and stop in MYO15A exon 35, are the most common non-GJB2 hearing-loss mutations in the Palestinian population. SLC26A4 c.1001G>T and SLC26A4 c.1341+1delG are each responsible for recessive severe-to-profound hearing loss in multiple families throughout west Asia and the Mediterranean, in some children accompanied by an enlarged vestibular aqueduct characteristic of Pendred syndrome. Mutations with transcriptional effects were responsible for hearing loss in 24% (80/337) of solved families.
Noncoding Mutations.
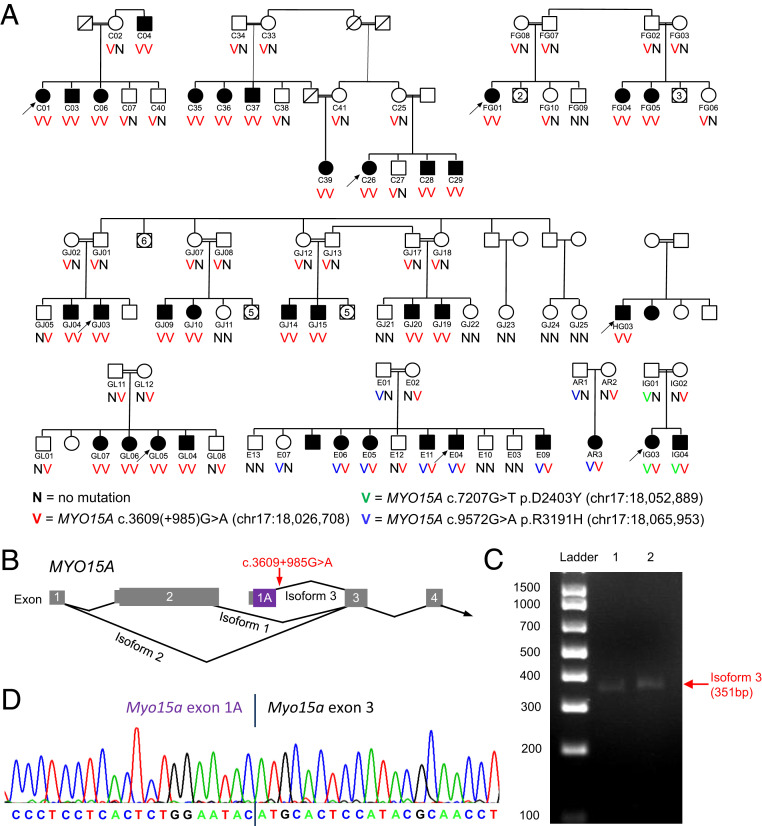
Disease-causing mutations in noncoding genomic regions are difficult to find and characterize, particularly for genes not expressed in accessible tissue. Few pathogenic noncoding mutations have been identified for any phenotype. In the Palestinian population, highly informative families enabled identification of a noncoding mutation in MYO15A as the cause of hearing loss in multiple families. In family C, with congenital profound hearing loss in 11 affected individuals, homozygosity mapping had revealed a region of 14.2 Mb (chr17:15,271,973 to 29,466,722; hg19) shared by all affected individuals (16). The shared homozygous region includes MYO15A, but exome sequencing did not reveal any damaging coding sequence mutations in MYO15A or any of the other 167 genes in the region. Subsequent whole-genome sequencing of affected relatives of family C yielded several previously undocumented noncoding variants in the homozygous region, including chr17:18,026,708G>A, in MYO15A intron 2, at MYO15A c.3609(+985) (Fig. 5A). Several lines of evidence supported MYO15A c.3609(+985)A>G as a candidate causal mutation for the hearing loss of family C. Loss-of-function mutations of MYO15A are responsible for recessive congenital profound hearing loss (17), consistent with the family C phenotype. The variant cosegregated with hearing loss in all of family C, as expected given the homozygosity mapping. The variant was not present in any public database or in Palestinian controls. Although deeply intronic, the reference base pair was conserved as guanine throughout mammals (gerp score 5.1). The 150-bp genomic sequence immediately proximal to the variant site (chr17:18,026,558 to 18,026,707; hg19) was predicted to have exonic potential by the Exonify program, based on conservation in human, dog, rat, and mouse (18) (Fig. 5B). The mutation of family C was predicted to destroy the donor splice site of this hypothetical exon; NNSPLICE scores were 0.96 and 0.00, and MaxEnt scores were 7.77 and −0.41 for wild-type and mutant sequences, respectively. And, finally, a transcript corresponding to this predicted exon had been identified by others by 5′ rapid amplification of cDNA ends from mouse pituitary (19).
Fig. 5.
Mutation at MYO15A c.3609(+985), predicted to alter transcription of a cochlear and pituitary-specific isoform of myosin 15A. (A) Families with mutations at MYO15A c.3609(+985) (red V), MYO15A c.7207G>T (green V), and MYO15A c.9572G>A (blue V). MYO15A c.3609(+985) disrupts the splice donor of isoform-specific exon 1A, predicted to lead to complete loss of transcript for this isoform. Splice effects of MYO15A c.7207G>T and MYO15A c.9572G>A are described in SI Appendix, Table S5. The chance of this degree of cosegregation of MYO15A c.3609(+985) with hearing loss occurring by chance is less than 10E-20. (B) MYO15A isoform 1 (NM_010862.2), isoform 2 (NM_182698.2), and proposed isoform 3. Locations in isoform 3 of the alternate first exon and of MYO15A c.3609(+985) are indicated. (C) Expression of the alternate first exon in mouse cochlear RNA. (D) Transcript sequence from mouse cochlear RNA indicating expression of exon 1A and splicing to exon 3.
Because MYO15A is expressed only in the cochlea and pituitary gland, blood-derived patient RNA was not informative for evaluating effects of MYO15A c.3609(+985)G>A on splicing. Therefore, we first demonstrated expression of the predicted exon in mouse cochlea (Fig. 5C), in order to confirm its relevance to hearing loss. We hypothesized that loss of the donor site of this exon would preclude transcription. At the same time, we genotyped the variant site in all study participants still without a genetic diagnosis for their hearing loss. In four other families with the same phenotype, deaf individuals were homozygous for MYO15A c.3609(+985)G>A and, in three families with the same phenotype, deaf individuals were compound heterozygous for MYO15A c.3609(+985)G>A and another MYO15A splice or enhancer mutation. In each family, MYO15A c.3609(+985)G>A cosegregated perfectly with the phenotype; the likelihood of this degree of cosegregation by chance was less than 10E-20.
More noncoding mutations leading to hearing loss likely remain to be found. From the 154 families with no genetic diagnosis, 11 probands were heterozygous for a pathogenic or likely pathogenic allele of a known hearing-loss gene, but with no second allele present in the coding or proximal intronic sequence (SI Appendix, Table S6A). (One proband was heterozygous for pathogenic alleles in two different genes.) Noncoding second alleles may explain hearing loss in some of these individuals. However, all but one of these 11 probands are singleton cases, so several may have hearing loss for nongenetic reasons and heterozygosity for damaging alleles may be by chance. The frequency of chance occurrence of unpaired damaging heterozygous alleles is illustrated by such alleles of GJB2 in several families with hearing loss due to another gene (SI Appendix, Table S6B).
Finally, hearing loss in multiplex families with no candidate causal gene or mutation was more likely than hearing loss in solved families to be dominantly inherited, progressive, and/or moderate in severity, rather than recessive, congenital, and severe to profound. Pedigree analysis of the largest unsolved families using all variants (common and rare) in the gene panel suggests that none have hearing loss linked to a known deafness gene. We suspect, therefore, that hearing loss in most unresolved multiplex families will prove to be due to as-yet-undiscovered genes for hearing loss.
Discussion
Genomic analysis of hearing loss for the Palestinian population reveals insights of interest for this phenotype in this population, the genetics of hearing loss worldwide, and the interpretation of genomic data for any common, highly heterogeneous condition.
The 2020 Palestinian population of the West Bank and Gaza is ∼5.1 million persons, including 3.2 million residents of the West Bank and 1.9 million residents of Gaza (20). Consanguineous marriage (that is, marriage between second cousins or closer relatives) has been historically common in this population, but is declining, from more than 40% of marriages between 1948 and 1959 compared with 24% of marriages between 2005 and 2009 (21, 22). The decline is most significantly associated with the increasing education of girls (19, 20), a trend that continues, with literacy rates of Palestinian girls and women estimated at 79% in 1995 and 94% in 2015 (23).
The high frequency of early-onset hearing loss among children of the West Bank and Gaza is clear from the many schools and clinics devoted to them. A quantitative estimate of congenital and early-onset hearing loss in the population of Palestinian ancestry was provided by a Canadian-Jordanian-Israeli survey in 2000 to 2003 of more than 16,000 Jordanian-Palestinian and Israeli-Jewish newborns (24). All infants were tested by otoacoustic emissions and auditory brainstem response. Frequency of hearing loss was significantly higher among Jordanian-Palestinian infants compared with Israeli-Jewish infants. The difference was due entirely to differences in rates of bilateral sensorineural hearing loss, the form of hearing loss most likely to be inherited: 10.2 per 1,000 Jordanian-Palestinian infants versus 1.5 per 1,000 Israeli-Jewish infants. Our results indicate that most (58%) of the hearing loss in the present-day Palestinian population is attributable to consanguinity. We anticipate that the incidence of inherited hearing loss in the Palestinian population will decrease very substantially in the next generation as fewer marriages are consanguineous. Frequencies of other, even more devastating, recessive conditions are likely to decrease as well.
Consanguinity is associated both with high overall prevalence of hearing loss and with the distribution of genes responsible for that loss. The genes most frequently responsible for hearing loss in the Palestinian population are those most frequently encountered worldwide: GJB2, SLC26A4, MYO15A, MYO7A, and CDH23. However, as indicated by Table 2 and Fig. 2, almost half the families (45%) with hearing loss in the Palestinian population harbored mutations in genes rarely encountered among families with hearing loss. The major contribution of otherwise rarely encountered genes is due to very rare, often family-specific alleles that appear as homozygotes in consanguineous families.
Our results indicate that genetic diagnoses can be provided to >85% of families with inherited hearing loss. That is, if a child’s hearing loss is genetic, chances are very high that the critical gene and mutation(s) can be identified. Such diagnoses can anticipate syndromic effects (or lack thereof) and hence guide treatment and educational plans, and can offer parents the opportunity for pregestational diagnosis for future pregnancies. This high yield of genomic analysis applies to families from other populations as well, including small families for whom it is not possible to have a clear idea whether a child’s hearing loss is genetically based only on family history. Conversely, for the Palestinian population, the estimate that ∼44% of childhood-onset hearing loss in the absence of family history is not genetic suggests public health measures, particularly against congenital viral infection, that could help reduce this burden.
Other conclusions from the project bear on gene and mutation discovery generally. First, DNA and RNA from large and informative families and from population-specific controls, evaluated with high-coverage sequencing of targeted genomic regions and appropriate bioinformatics and statistics, are highly informative for mutation discovery and characterization. A survey of the breadth and depth undertaken here highlights these strengths. For example, analysis of large and numerous pedigrees, coupled with experimentally calibrated in silico analysis of transcriptional effects, yielded inferences of high confidence for effects on splicing even of mutations in genes not expressed in accessible tissue.
Second, regulatory mutations altering transcription contribute in a major way to hearing loss. One in four (80/337) families with a genetic diagnosis for their hearing loss had a splice-altering mutation as the responsible allele. Many of these mutations were cryptic, either exonic alterations of splice enhancers or silencers or deeply intronic events. Based on our experience with other genes (25), we anticipate that some mutations that alter exonic splice enhancers and silencers may prove hypomorphic, with variable and stochastic effects on phenotype even among different individuals with the same allele.
Third, integration of human genetics with transcript analysis in a model organism was a powerful combination of approaches. This combination was particularly useful for identification and characterization of a deeply intronic allele that proved to be among the most common deafness-causing mutations in the population.
The project had several limitations. While audiologic evaluation was good throughout the region, capacity to evaluate non–hearing-related syndromic features was more limited. This limitation constrained efforts to evaluate differences in syndromic features across families with different alleles of the same genes. Documentation of viral exposure during pregnancy was also limited. In addition, because ascertainment was through schools and clinics for the deaf, the cohort likely underrepresented children with hearing loss in the context of severe developmental disorders who were too disabled to attend school. Also, while almost all West Bank schools for the deaf were included, only some of the schools caring for deaf children in Gaza were accessible.
Finally, inherited hearing loss in this population was explained by many different, individually rare mutations, each of severe effect. These rare, severe-effect mutations were revealed by a study design based on families. We anticipate that newly discovered genes for hearing loss in this population will each explain one or a few families. Noncoding mutations in known hearing-loss genes will likely explain a few more. Our results suggest that hearing loss in many as-yet-unexplained families in this population may not be genetic. Control of infectious causes of deafness, coupled with the continuing decline in consanguineous marriage, is very likely to lead to a decrease in the prevalence of hearing loss in the Palestinian population in the next generation.
Methods
Participants.
The project was approved by the human subjects committees of Bethlehem University, the University of Washington, Tel Aviv University, and the Palestinian and Israeli Ministries of Health. With the cooperation of teachers, social workers, and health care providers, families with hearing loss were ascertained from schools, rehabilitation centers, and clinics dedicated to deaf children on the West Bank and in Gaza. Families who expressed an interest in the project were visited and informed consent was obtained from parents and assent from older children. From each family, parents and all affected children were enrolled. Hearing children were enrolled only if older than the oldest onset age of hearing loss in their sibship. That is, potentially presymptomatic children were not enrolled. Family medical history was collected by interview with parents and by review of medical records. Audiologic examinations were carried out at Dar Al-Kalima School in Bethlehem, the Emirates Hearing Center in Hebron, the Palestine Red Crescent Society in Ramallah, and the Atfaluna Society for Deaf Children in Gaza City. Examinations included pure tone thresholds (air and bone); speech audiometry, including when possible speech reception and detection thresholds and discrimination; tympanometry with an admittance meter to evaluate the compliance of the tympanic membrane and the status of the middle ear; and acoustic reflex measurements, including when possible acoustic decay. Hearing controls were drawn from clinics on the West Bank and in Gaza serving pregnant women, from students at Bethlehem University, and from Palestinian participants in other projects in our laboratory with normal hearing. Analysis of identity by descent was carried out so as to include only exomes from controls more distantly related than second cousins.
Among participants in the project, hearing loss could be prelingual or postlingual in onset and nonsyndromic or syndromic. All possible modes of inheritance were included. All children in the study had long-term chronic hearing loss; the study did not enroll children with transient hearing loss. For affected children, older unaffected siblings, and parents, 8.5 mL blood (less for young children) was drawn into acid citrate dextrose and genomic DNA was extracted by a simple salting-out procedure (26). Participants later discovered to carry mutations with possible transcript effects were visited again and an additional blood sample was requested for direct extraction of RNA.
Genomics.
Genomic DNA was evaluated by hybridization to a gene panel including 181 known and candidate genes for hearing loss (SI Appendix, Table S1), updated from HearSeq gene panel v4 (27). Hybridized barcoded libraries were sequenced on an Illumina HiSeq 2500 to an average coverage of 543× with 99.34% of targeted bases covered at >8×. For each family, DNA from all participating relatives was sequenced on the same run.
A bioinformatics pipeline was developed to evaluate sequence data from the panels. Sequence data were converted into fastq files, processed from real-time base calls (RTA1.8 software; Bustard), and converted to qseq.txt files. Following demultiplexing, reads were aligned to the reference human genome (hg19) using the Burrows–Wheeler Aligner (v0.7.12) (28). PCR duplicates were removed using SAMtools v0.1.18 (29). Indels were realigned and base quality score was recalibrated with the Genome Analysis Tool Kit (GATK v3.0-0-g6bad1c6) using the recommended parameters (30). Genotypes were called and filtered using GATK Unified Genotyper and Variant Filtration tools. Large insertions, deletions, and inversions were called by using Pindel (31) and BreakDancer (32). Misalignments due to duplicated regions were removed by filtering against data from >5,000 exomes sequenced in Seattle or Bethlehem, including 2,618 exomes from Palestinian subjects with normal hearing. Copy-number variants (CNVs) were called using CoNIFER (33), XHMM (34), and our in-house CNV detection pipeline (35).
Variants were annotated with respect to genomic position, genic location, and predicted function using in silico tools. Variant allele frequencies were noted from gnomAD (36) and from the Palestinian controls. Variants were interpreted using the guidelines of the American College of Medical Genetics and Genomics (37), with additional information from transcriptional analysis and cosegregation analysis in large families. Mutations were interpreted as potentially damaging if they were truncating or led to loss of transcript in patient RNA, completely deleted a critical gene, or were missense mutations with a predicted functional effect. Variants that were potentially damaging by any of these criteria were evaluated for cosegregation with hearing loss in their host family. Additional informative relatives were recruited from large kindreds for cosegregation analyses. The strength of cosegregation between candidate mutation genotype and hearing-loss phenotype was evaluated by likelihood analysis, with the strength of association measured by the probability of occurrence, by chance, of the observed degree of cosegregation of genotype with phenotype among all fully informative relatives in each family. All candidate mutations have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
Transcriptional Analysis.
Interpretation of mutations potentially altering transcription was carried out using the targeted RNA-seq approach that we developed for analysis of inherited mutations in tumor suppressor genes (25). For this approach, patients’ RNA is sequenced with the same gene panels used to evaluate the patients’ genomic DNA, yielding quantitative measures of transcriptional effects of candidate splice-site and enhancer mutations. Calibration of experimental and in silico results reveals classes of mutations, defined by changes in NNSPLICE and MaxEnt scores or changes in exonic splice enhancers or exonic splice silencers, for which in silico tools provide reliable predictions. We applied this approach to analysis of possible transcriptional effects of mutations in hearing-loss genes. All variants with minor-allele frequency <0.01, whether at canonical splice sites or other exonic or intronic sites, including deep intronic regions, were scored using NNSPLICE and MaxEnt to predict disruption or activation of splice sites (38, 39) and by Spliceman and Human Splicing Finder to predict effects of the variant on splice enhancer and silencer motifs (40–44). Experimental validation with patient RNA was carried out for genes expressed in blood or lymphoblast cell lines.
Consanguinity.
Consanguineous marriage was defined as a marriage of second cousins or closer relatives. Consanguinity was evaluated both from pedigree information provided by families and from genomic data from panel sequencing. For the latter measurement, for each locus in the panel, informative SNPs were collected, regardless of relevance to hearing loss. For each participant, the proportion of loci homozygous at all SNPs at that locus was calculated, and the proportion was expressed as a Z score normalized to proportions of homozygous loci for all participants. Distributions of Z scores were compared for families with documented consanguinity versus families with documented nonconsanguinity. Based on means and SDs of these distributions, the likelihood of consanguinity versus nonconsanguinity was estimated for each family with genomic data but incomplete pedigree information.
Supplementary Material
Acknowledgments
We thank the families of Palestine for their dedication to this project. We thank the teachers of the Princess Basma Centre in Jerusalem, Ephpheta Paul VI School in Bethlehem, Al Amal Society for Deaf Children Schools in Qalqilya and Hebron, Al Hanan Primary Deaf School in Jenin, Qaqoon Charitable Society Deaf School in Tulkarem, Palestinian Charitable Society for the Deaf in Nablus, and Atfaluna Society for Deaf Children in Gaza, and colleagues Michel Rahil in Bethlehem, Suhail Ayesh in Gaza, and Sarah Pierce, Sunday Stray, and Mary Eng in Seattle for technical help and advice. This work was performed in partial fulfillment of the requirements for a PhD degree by A.A.R., Faculty of Medicine, Tel Aviv University. This project was supported by NIH Grant R01DC011835, travel support from the Virginia Merrill Bloedel Hearing Research Center, and fellowship support to A.A.R. from the Mauerberger Foundation Fund.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009628117/-/DCSupplemental.
Data Availability.
All candidate mutations reported in this paper are included in the SI Appendix and have been submitted to ClinVar. All study data are included in the article and SI Appendix.
References
- 1.Richardson G. P., de Monvel J. B., Petit C., How the genetics of deafness illuminates auditory physiology. Annu. Rev. Physiol. 73, 311–334 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Morton C. C., Nance W. E., Newborn hearing screening—A silent revolution. N. Engl. J. Med. 354, 2151–2164 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Miyagawa M., Nishio S. Y., Usami S., A comprehensive study on the etiology of patients receiving cochlear implantation with special emphasis on genetic epidemiology. Otol. Neurotol. 37, e126–e134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shahin H., et al. , Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive nonsyndromic hearing loss. Am. J. Hum. Genet. 78, 144–152 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shahin H., et al. , Nonsense mutation of the stereociliar membrane protein gene PTPRQ in human hearing loss DFNB84. J. Med. Genet. 47, 643–645 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Walsh T., et al. , Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am. J. Hum. Genet. 87, 90–94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shahin H., et al. , Genetics of congenital deafness in the Palestinian population: Multiple connexin 26 alleles with shared origins in the Middle East. Hum. Genet. 110, 284–289 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Walsh T., et al. , Genomic analysis of a heterogeneous Mendelian phenotype: Multiple novel alleles for inherited hearing loss in the Palestinian population. Hum. Genomics 2, 203–211 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dossena S., et al. , Functional characterization of pendrin mutations found in the Israeli and Palestinian populations. Cell. Physiol. Biochem. 28, 477–484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brownstein Z., et al. , Novel myosin mutations for hereditary hearing loss revealed by targeted genomic capture and massively parallel sequencing. Eur. J. Hum. Genet. 22, 768–775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhonker Y., et al. , The GPSM2/LGN GoLoco motifs are essential for hearing. Mamm. Genome 27, 29–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azaiez H., et al. , Genomic landscape and mutational signatures of deafness-associated genes. Am. J. Hum. Genet. 103, 484–497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alagramam K. N., et al. , The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat. Genet. 27, 99–102 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Ahmed Z. M., et al. , PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum. Mol. Genet. 12, 3215–3223 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Pepermans E., et al. , The CD2 isoform of protocadherin-15 is an essential component of the tip-link complex in mature auditory hair cells. EMBO Mol. Med. 6, 984–992 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahin H., et al. , Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur. J. Hum. Genet. 18, 407–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y., et al. , Characterization of the human and mouse unconventional myosin XV genes responsible for hereditary deafness DFNB3 and shaker 2. Genomics 61, 243–258 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Siepel A., Haussler D., Combining phylogenetic and hidden Markov models in biosequence analysis. J. Comput. Biol. 11, 413–428 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Rehman A. U., et al. , Mutational spectrum of MYO15A and the molecular mechanisms of DFNB3 human deafness. Hum. Mutat. 37, 991–1003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palestinian Central Bureau of Statistics , www.pcbs.gov.ps/site/lang__en/803/default.aspx. Accessed 20 June 2020.
- 21.Na’amnih W., et al. , Continuous decrease of consanguineous marriages among Arabs in Israel. Am. J. Hum. Biol. 27, 94–98 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Sharkia R., et al. , Changes in marriage patterns among the Arab community in Israel over a 60-year period. J. Biosoc. Sci. 48, 283–287 (2016). [DOI] [PubMed] [Google Scholar]
- 23.United Nations Development Programme , “The 2014 Palestine Human Development Report. Chapter 4.3 Education.” https://www.un.org/unispal/document/auto-insert-204688/. Accessed 20 June 2020.
- 24.Attias J., et al. , The prevalence of congenital and early-onset hearing loss in Jordanian and Israeli infants. Int. J. Audiol. 45, 528–536 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Casadei S., et al. , Characterization of splice-altering mutations in inherited predisposition to cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 26798–26807 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller S. A., Dykes D. D., Polesky H. F., A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16, 1215 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brownstein Z., et al. , Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 12, R89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanger Institute , SAMtools. www.htslib.org/. Accessed 20 June 2020.
- 30.DePristo M. A., et al. , A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye K., Schulz M. H., Long Q., Apweiler R., Ning Z., Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen K., et al. , BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6, 677–681 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krumm N. et al.; NHLBI Exome Sequencing Project , Copy number variation detection and genotyping from exome sequence data. Genome Res. 22, 1525–1532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fromer M., et al. , Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am. J. Hum. Genet. 91, 597–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nord A. S., Lee M., King M.-C., Walsh T., Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics 12, 184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.gnomAD Gene Aggregation Database , https://gnomad.broadinstitute.org/. Accessed 20 June 2020.
- 37.Richards S. et al.; ACMG Laboratory Quality Assurance Committee , Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reese M. G., Eeckman F. H., Kulp D., Haussler D., Improved splice site detection in Genie. J. Comput. Biol. 4, 311–323 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Yeo G., Burge C. B., Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11, 377–394 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R., ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31, 3568–3571 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith P. J., et al. , An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 15, 2490–2508 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Lim K. H., Ferraris L., Filloux M. E., Raphael B. J., Fairbrother W. G., Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc. Natl. Acad. Sci. U.S.A. 108, 11093–11098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim K. H., Fairbrother W. G., Spliceman—A computational web server that predicts sequence variations in pre-mRNA splicing. Bioinformatics 28, 1031–1032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desmet F. O., et al. , Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All candidate mutations reported in this paper are included in the SI Appendix and have been submitted to ClinVar. All study data are included in the article and SI Appendix.