Significance
Much of our current knowledge on protein folding is based on work focused on isolated domains. In this study, using a combination of NMR and kinetic experiments, we depict the folding pathway of a multidomain construct comprising two PDZ domains in tandem, belonging to the protein Whirlin. We demonstrate the presence of a misfolded intermediate that competes with productive folding. Interestingly, we show that, unexpectedly, this misfolded state retains the native-like functional ability to bind its physiological ligand, representing a clear example of a functionally competent misfolded state. On the basis of these results and a comparative analysis of the amino acidic sequences of Whirlin from different species, we propose a possible physiological role of the misfolded intermediate.
Keywords: folding, kinetics, misfolding
Abstract
Although more than 75% of the proteome is composed of multidomain proteins, current knowledge of protein folding is based primarily on studies of isolated domains. In this work, we describe the folding mechanism of a multidomain tandem construct comprising two distinct covalently bound PDZ domains belonging to a protein called Whirlin, a scaffolding protein of the hearing apparatus. In particular, via a synergy between NMR and kinetic experiments, we demonstrate the presence of a misfolded intermediate that competes with productive folding. In agreement with the view that tandem domain swapping is a potential source of transient misfolding, we demonstrate that such a kinetic trap retains native-like functional activity, as shown by the preserved ability to bind its physiological ligand. Thus, despite the general knowledge that protein misfolding is intimately associated with dysfunction and diseases, we provide a direct example of a functionally competent misfolded state. Remarkably, a bioinformatics analysis of the amino acidic sequence of Whirlin from different species suggests that the tendency to perform tandem domain swapping between PDZ1 and PDZ2 is highly conserved, as demonstrated by their unexpectedly high sequence identity. On the basis of these observations, we discuss on a possible physiological role of such misfolded intermediate.
Although more than 75% of the eukaryotic proteome is composed of multidomain proteins (1), much of our current knowledge of protein folding relies on studies of single domain proteins. In fact, protein domains are generally assumed to be able to fold independently, as demonstrated by the ability to express them in isolation. However, the possibility that the interaction between these structural subunits may play a critical role in the formation of the native structure, as well as in dictating misfolding events, cannot be excluded (2, 3). Consequently, recent years have seen a considerable increase in efforts to understand the folding of more complex multidomain systems (2, 4–12).
One of the simplest mechanisms through which proteins increase their complexity lies in the duplication of their structural subunits (13). Thus, some proteins may be characterized by repetition of contiguous homologous domains that are often found in tandem. While tandem repeats are frequently found in the proteome, a clear relationship between the presence of these repetitions and their role in the mechanical and functional properties of proteins remains relatively elusive (14–16).
Studies of protein folding mechanisms have shown that the presence of tandem repeats may complicate folding substantially (4, 8, 11, 12). In fact, the concurrent unfolding of contiguous domains can represent an additional potential source of misfolding. In this scenario, misfolded intermediates may be stabilized by interactions involving two adjacent domains, thus competing with productive folding and resulting in an increased complexity of the observed pathway.
Whirlin is a large scaffolding protein involved in the hearing apparatus, where it participates in the transduction of sound-induced vibrations in the cochlea into electric potentials, which are then transmitted to the brain (17, 18). Whirlin is located within the actin-filled stereocilia of the hair cells, and its physiological role is exerted by mechanically transmitting, in complex with other proteins, the concerted deflection of these cilia as induced by sound waves, opening mechanotransduction channels of the hair bundle. The genes encoding these proteins are affected by mutations responsible for a hereditary sensory disease called Usher syndrome associated with deafness and progressive blindness (19). The C-terminal PDZ binding motif of Sans, a scaffolding protein mediating several protein–protein interactions and also containing ankyrin repeats and a SAM domain, interacts with the N-terminal region of Whirlin comprising two PDZ domains, PDZ1 and PDZ2 (20, 21). One mutation responsible for Usher syndrome results in a Whirlin with truncated PDZ domains (17, 19). Although the presence of two PDZ domains is almost universally conserved in Whirlin in different species, only PDZ1 interacts with Sans and other partners, and no specific physiological partners for PDZ2 has been found. The structure of the PDZ1-PDZ2 tandem of Whirlin (P1-P2) has been addressed recently (22). Using a combination of solution NMR and SAXS, it was observed that P1-P2 behaves as a highly dynamic supramodule in which the two individual domains transiently interact with each other and populate a well-defined closed formation, as well as a heterogeneous ensemble of more open conformations. This dynamic arrangement of the PDZ1-PDZ2 supramodule regulates its binding activity.
To shed light on multistate folding, we investigated the pathway of PDZ1-PDZ2 folding. As detailed below, the fortuitous differences in stability between PDZ1 and PDZ2 in the tandem P1-P2 makes this construct as an ideal system for studying multidomain folding, allowing the selective unfolding of either one domain (PDZ1, being the least stable) or both domains with increasing denaturant concentrations. We demonstrate that while the (un)folding of PDZ1 is essentially unaffected when PDZ2 is held in its native conformation, the concurrent denaturation of both domains leads to the accumulation of a misfolded kinetic trap that competes with native folding and substantially slows the productive pathway. In agreement with the view that tandem domain swapping is a potential source of transient misfolding (4, 8), we demonstrate that the kinetic trap retains a native-like functional activity, as shown by the preserved capability to bind a peptide mimicking Sans. Thus, while protein misfolding is classically associated with highly debilitating medical conditions (23–26), we provide a direct example of a functionally competent misfolded state. Moreover, a bioinformatics analysis of the amino acidic sequence of Whirlin from different species suggests that the tendency to perform tandem domain swapping between PDZ1 and PDZ2 is highly conserved, as mirrored by their unexpectedly high sequence identity. On the basis of these observations, we discuss a possible physiological role of such misfolded intermediates.
Results
Given that the binding of Whirlin to its physiological ligand Sans is mediated by its PDZ1 domain, to make our folding and binding studies possible, we engineered a fluorescent variant of PDZ1 that could be used for both folding and binding experiments. In fact, as detailed below, while the wild-type (WT) construct, previously used for structural characterization by NMR (22), contains two Trp residues in positions 237 and 375, located in the hairpin connecting PDZ1 with PDZ2 and in a C-terminal tail of the protein, respectively, these residues do not return a detectable fluorescence change on unfolding, precluding kinetic characterization. Therefore, based on our previous experiments on the (un)folding of PDZ domains (27–31), position Tyr-168 in PDZ1 was mutated into Trp. Trp168 proved to be an excellent probe for both folding and binding studies. Thus, the variant Y168W, denoted below as pP1-P2, was used as a fluorescent pseudo-WT protein in all our experiments.
Equilibrium Denaturation of P1-P2: Evidence for Sequential Independent Unfolding of the Two Individual Domains.
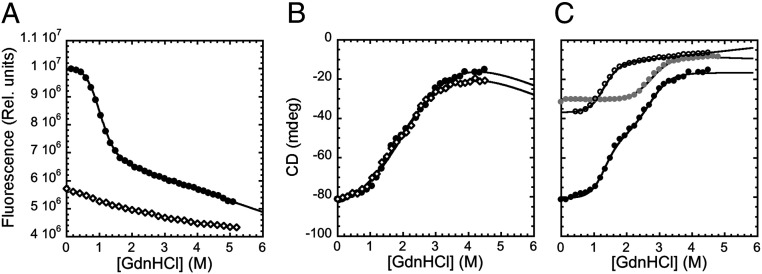
The GdnHCl-induced equilibrium denaturation of pP1-P2 monitored by fluorescence and circular dichroism (CD) is depicted in Fig. 1. A qualitative comparison between the equilibrium transitions obtained using the two different spectroscopic techniques highlights a clear difference, with the data obtained by CD returning a broader transition. A two-state analysis of the fluorescence data displays a denaturation midpoint of 0.96 ± 0.02 M. The calculated mD-N value, which reflects the change in surface area becoming accessible to the solvent on unfolding (32), is 2.53 ± 0.07 kcal mol−1 M−1, consistent with the value expected for an individual PDZ domain. Conversely, the apparent mD-N value obtained from CD experiments was 0.74 ± 0.03 kcal mol−1 M−1, indicating that the transition observed by CD is characterized by an inherent complexity, possibly detecting the unfolding of the two individual PDZ domains of the construct. Importantly, while no detectable change could be observed on unfolding on WT P1-P2, the CD transitions of pP1-P2 and P1-P2 were superposable (Fig. 1), indicating that mutation of Tyr-168 to Trp had a negligible effect on the stability of the construct.
Fig. 1.
Equilibrium denaturation experiments. (A) GdnHCl-induced equilibrium denaturation of WT P1-P2 (open diamonds) and pP1-P2 (black circles) monitored by fluorescence. The change in the intrinsic fluorescence of the tryptophan residue vs. GdnHCl concentrations is consistent with a two-state transition. No detectable change could be observed in the case of WT P1-P2. (B) GdnHCl-induced equilibrium denaturation of WT P1-P2 (open diamonds) and pP1-P2 (black circles) monitored by CD. As discussed in the text, the denaturation obtained by CD experiments does not fit well to a two-state transition, revealing an additional complexity. (C) GdnHCl-induced equilibrium denaturation of pP1-P2 (black circles), pPDZ1 (empty circles), and PDZ2 (gray circles) monitored by CD. The curves obtained for pPDZ1 and PDZ2 are consistent with a two-state transition. The pP1-P2 denaturation curve was satisfactorily fitted by the sum of the individual curves obtained for pPDZ1 and PDZ2 in isolation.
To interpret the data obtained by CD, we compared the denaturation curves obtained for pP1-P2 with those obtained for PDZ1 and PDZ2, expressed and purified in isolation (Fig. 1C). Inspection of Fig. 1 indicates that in both cases, equilibrium unfolding was consistent with an apparent two-state transition, returning similar values of mD-N, 2.33 ± 0.09 for PDZ1 and 2.61 ± 0.12 kcal mol−1 M−1 for PDZ2. Furthermore, a global fit of the equilibrium unfolding obtained for the tandem pP1-P2, with shared thermodynamic parameters, indicates that the denaturation transition of this construct followed by CD is perfectly consistent with the values expected from the sum of the individual curves obtained for PDZ1 and PDZ2 in isolation. This finding suggests that the two domains of the construct pP1-P2 unfold as independent units, and there are no evident interactions among them, with the two behaving as the same domain in isolation.
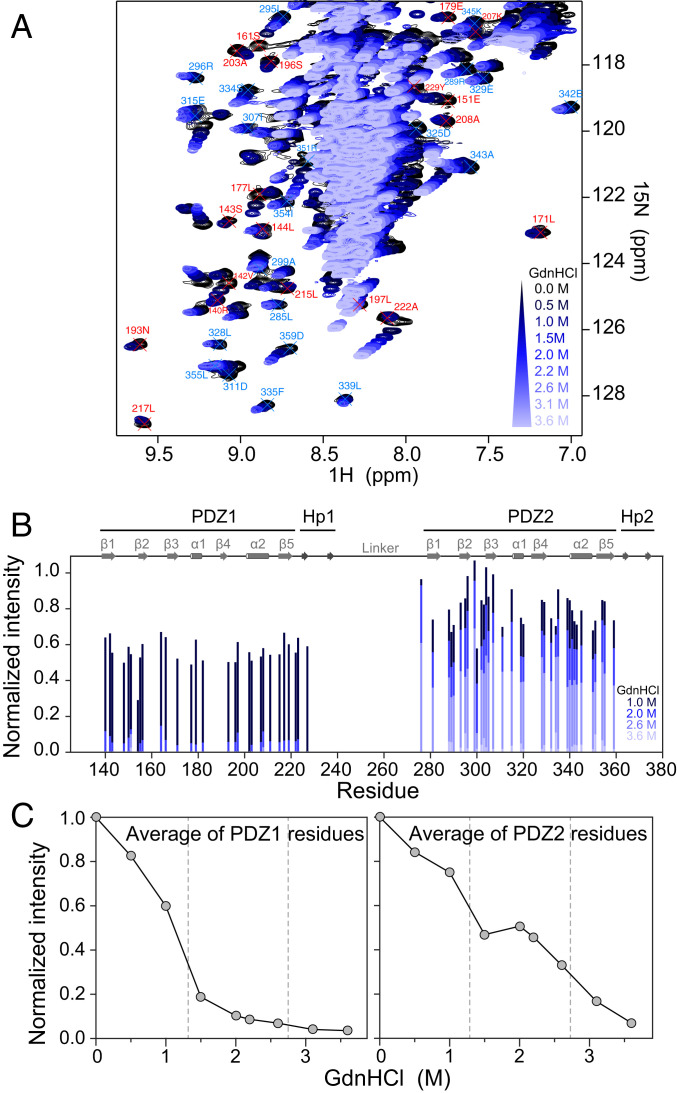
From the data reported in Fig. 1, it is possible to identify a GdnHCl concentration at which only PDZ1 is denatured, whereas PDZ2 should maintain its native conformation, i.e., 2.2 M. To further support this observation, we monitored the chemical denaturation of pP1-P2 by NMR. 1H-15N heteronuclear single quantum correlation (HSQC) spectra were recorded on 15N-labeled pP1-P2 at increasing GdnHCl concentrations, ranging from 0 to 3.6 M (Fig. 2A). The addition of GdnHCl induces overall changes in the pP1-P2 spectrum, with the progressive disappearance of the well-dispersed folded resonances of pP1-P2 (black spectrum) and the appearance of a fully unfolded state with 1H chemical shifts around 8 to 8.5 ppm (light blue). Due to the nontrivial dependence of chemical shifts on denaturant concentration (33), we chose to monitor pP1-P2 unfolding by studying the changes in intensities of the nonambiguous and isolated peaks. As illustrated in Fig. 2B, peak intensities in PDZ1 decrease at a lower GdnHCl concentration than in PDZ2, indicating earlier unfolding of PDZ1. From the decay of average peak intensities in each PDZ, PDZ1 is totally unfolded at 2.2 M GdnHCl, and PDZ2 is totally unfolded at 3.5 M GdnHCl (Fig. 2C), consistent with fluorescence and CD measurements. Of additional interest, while the denaturation of PDZ1 conforms to a two-state transition when monitored by NMR, we observed additional complexity for PDZ2. In particular, the calculated average of PDZ2 displays a broader transition. This effect may arise from the unfolding of the previously described supertertiary structure of the tandem (22).
Fig. 2.
Chemical denaturation of pP1-P2 followed by NMR. (A) Region of 1H-15N HSQC spectra recorded on 15N-labeled pP1-P2 at increasing GdnHCl concentrations from 0 (black) to 3.6 M (light blue). Unambiguous assignment of isolated peaks is indicated in red for PDZ1 and in blue for PDZ2. (B) Normalized peak intensities for each unambiguously assigned residue plotted at 1, 2, 2.6, and 3.6 M GdnHCl from dark to light blue. Domains of pP1-P2 and secondary structure elements are indicated at the top. (C) Average of the normalized peak intensities plotted as a function of GdnHCl concentration for PDZ1 (Left) and PDZ2 (Right) residues, highlighting the denaturation midpoints measured for PDZ1 and PDZ2, 1.3 M and 2.7 M GdnHCl, respectively.
The Folding Mechanism of PDZ1 in Tandem with Native PDZ2 Is Marginally Perturbed.
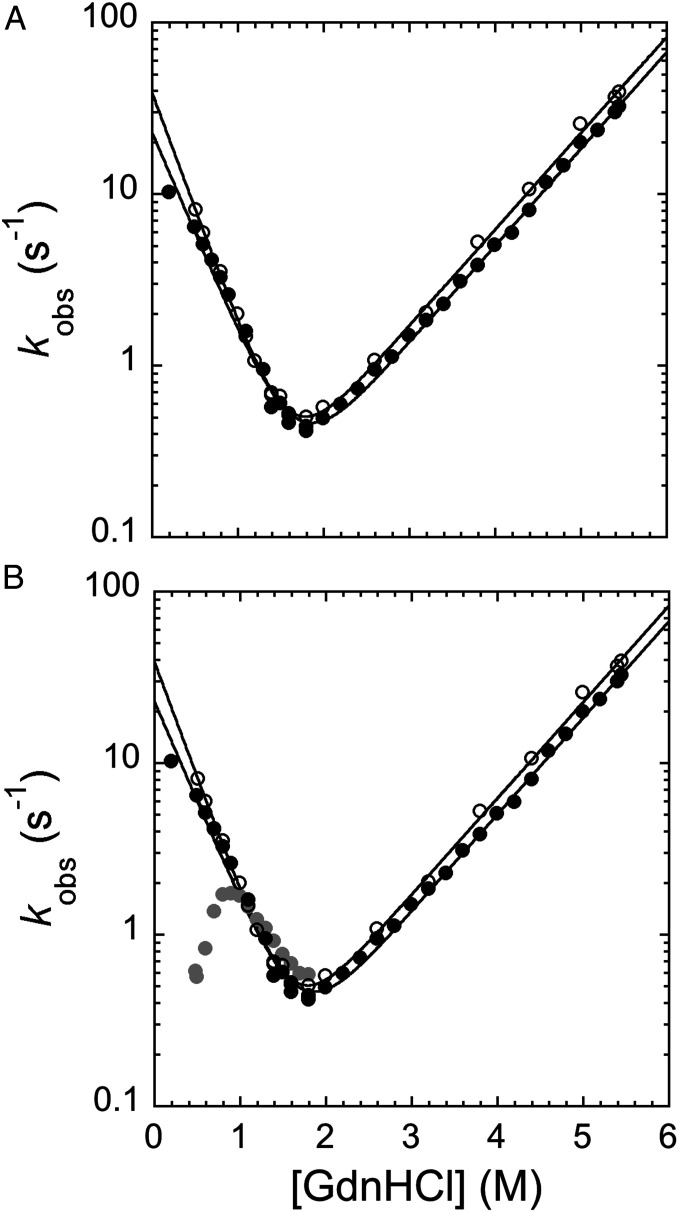
As highlighted above, the difference in stability between the two domains in the pP1-P2 construct allows the selective unfolding of PDZ1 or both domains at different denaturant concentrations. Therefore, pP1-P2 offers the unique opportunity to compare the folding of PDZ1 in both the presence and the absence of a flanking folded or unfolded domain. For this purpose, we first characterized the folding and unfolding of PDZ1 in isolation and then in comparison with that observed in the pP1-P2 construct. The unfolding and refolding kinetics of PDZ1 were monitored by stopped-flow kinetics, by rapidly diluting the protein into solutions containing different GdnHCl concentrations. In all experiments, the time course of the emission fluorescence was satisfactorily fitted to a single-exponential equation. The measured chevron plot of PDZ1 is shown in Fig. 3A. The data are consistent with a simple two-state V-shaped chevron, with no clear deviation from linearity in the folding and unfolding branches. Furthermore, both the mD-N value and the calculated midpoint are consistent with the same parameters obtained from equilibrium experiments, confirming the two-state nature of the reaction (34).
Fig. 3.
Unfolding and refolding kinetics of pP1-P2 and pPDZ1. (A) Chevron plot of pPDZ1 (empty circles) and pP1-P2 (black circles) obtained when starting the refolding experiment by diluting the protein with mild denaturant concentrations (i.e., 2.2 M GdnHCl). The two chevron plots are perfectly superimposable, indicating that in this condition, the folding of PDZ1 is not affected by the presence of a flanking domain when PDZ2 is held in its native conformation (see the text for details). (B) Chevron plot of pPDZ1 (empty circles) and pP1-P2 obtained when starting the refolding by diluting the protein with a mild denaturant concentration (i.e., 2.2 M GdnHCl; black circles) and a high denaturant concentration (i.e., 5.37 M GdnHCl; gray circles). As explained in detail in the text, when the refolding starts at higher GdnHCl concentrations, both PDZ1 and PDZ2 domains are denatured, leading to accumulation of a kinetic trap (represented by the presence of a pronounced rollover in the refolding arm) that competes with the productive folding of the protein. The buffer used in both experiments was 50 mM Tris⋅HCl pH 7.5 and 0.3 M NaCl.
To measure the folding kinetics of PDZ1 in the pP1-P2 construct in the presence of native PDZ2, we first denatured pP1-P2 in 2.2 M GdnHCl and then triggered refolding by rapid mixing with buffer. Analogously, unfolding was initiated by mixing the native protein with solutions containing GdnHCl at different concentrations. In analogy to what was observed with PDZ1 in isolation, both folding and unfolding time courses were satisfactorily fitted to a single exponential decay at any final denaturant concentration. A comparison between the chevron plots of PDZ1 in isolation and in the tandem is reported in Fig. 3A. It is evident that when PDZ2 is held in its native conformation, the folding of PDZ1 is essentially unaffected by the presence of a neighboring domain, with both folding and unfolding rate constants remaining unaltered.
A Hidden Kinetic Trap in the Folding of pP1-P2.
The experiments shown in Figs. 1–3 demonstrate that when pP1-P2 is denatured at mild denaturant concentrations, PDZ2 retains its native conformation, and its presence has a marginal effect on the folding and stability of PDZ1. To further investigate the effect of the presence of a neighboring domain on PDZ1, we conducted additional refolding experiments in which pP1-P2 was denatured at high denaturant concentrations, i.e., with both PDZ domains denatured. The refolding kinetics of pP1-P2 measured in these experiments are reported in Fig. 3B. It is evident that when both PDZ domains are denatured, productive folding at low denaturant concentration is slowed remarkably, as mirrored by the presence of a pronounced rollover (35). Moreover, the refolding rate constants increase with increasing denaturant concentrations, indicating that the intermediate is more structured than the transition state and thus must unfold for productive folding to occur (36, 37). On the basis of these observations, we conclude that the concurrent denaturation of both PDZ domains causes the rapid accumulation of a kinetic trap, which competes with productive folding.
Also of interest, the apparent slope of the refolding rate constant at low denaturant concentrations appears to be steeper than that of the unfolding branch by nearly twofold. This finding indicates that the misfolded intermediate may contain more folded structure than a single PDZ domain. This hypothesis is explored further below.
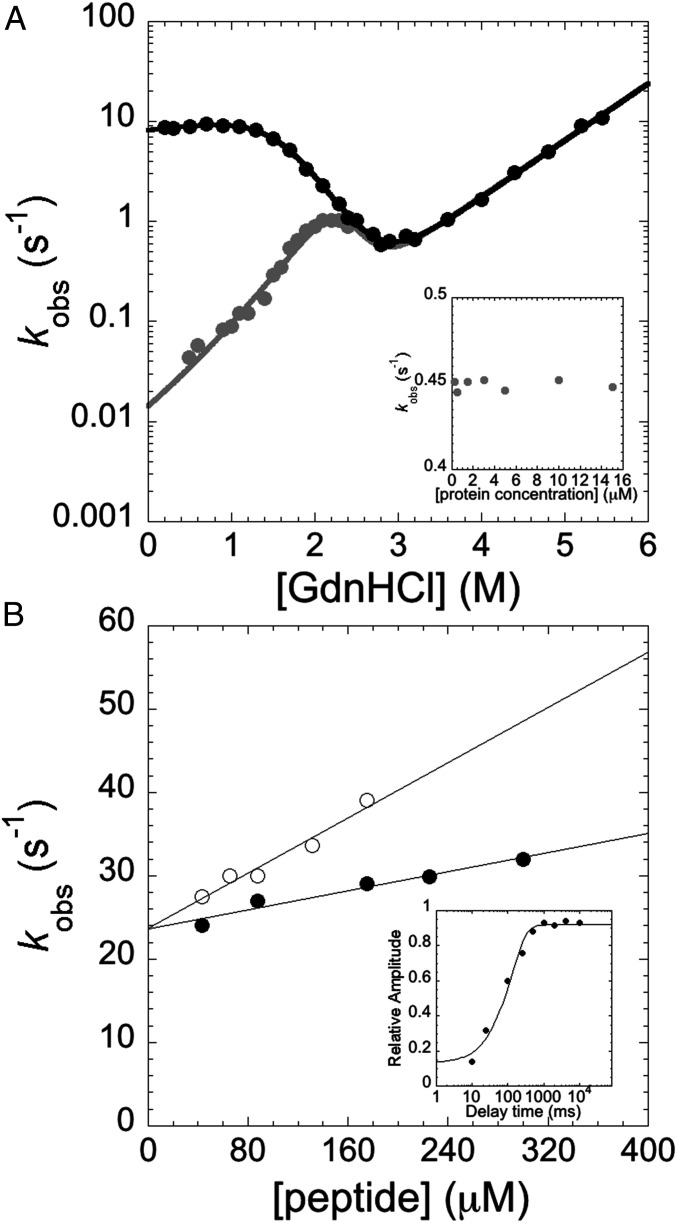
To further highlight the presence of an intermediate in the folding of pP1-P2, we repeated the folding and unfolding experiments in the presence of a stabilizing agent, 0.5 M sodium sulfate. Fig. 4A compares the folding rate constants obtained in stabilizing conditions when pP1-P2 is denatured in mild or high denaturant concentrations. In agreement with our previous observations for other PDZ domains (31, 36), the presence of sodium sulfate emphasizes the presence of a misfolded kinetic trap and, at low denaturant concentration, folding is slowed by more than two orders of magnitude. To exclude the presence of intermolecular interactions, refolding experiments were also conducted at different protein concentrations, ranging from 250 nM to 15 μM. As expected with a monomolecular folding reaction, rate constants were found to be insensitive to protein concentration (Fig. 4A). Inspection of data in Fig. 4 also reveals that the folding kinetics starting from 2.2 M GndHCl in the presence of sodium sulfate display a pronounced rollover, inconsistent with a two-state mechanism even when misfolding is avoided. A likely interpretation of this effect is that the salt stabilizes a marginally stable folding intermediate, which is barely detectable at low salt concentrations but well populated in the presence of sulfate.
Fig. 4.
(A) Unfolding and refolding kinetics of pP1-P2 in the presence of a stabilizing agent (0.5 M Na2SO4). pP1-P2 was denatured in a moderate (2.2 M; black circles) or high (5.37 M; gray circles) GdnHCl concentration. The refolding of pP1-P2 in the latter condition was slowed by more than two orders of magnitude by the presence of Na2SO4. Data were fitted as described in Materials and Methods. (Inset) Refolding kinetics of pP1-P2 measured at 0.5 M GdnHCl, starting from high denaturant concentrations, at protein concentrations ranging from 250 nM to 15 μM. As expected from a monomolecular folding reaction, rate constants were found to be insensitive to protein concentration. (B) Binding experiments measured by FRET between the Trp (donor) and dansyl (acceptor) attached to the Sans peptide. Empty circles represent the pseudo-first-order plot of the binding experiment between pP1-P2 and the peptide, performed at 25 °C in buffer composed of 50 mM Tris⋅HCl, 0.5 M Na2SO4, and 0.54 M GdnHCl. The binding between the peptide and the misfolded intermediate of pP1-P2 was obtained by a double-jump stopped-flow experiment (black circles). (Inset) Dependence of the relative amplitude of native-like binding competent species as a function of delay time between the first (refolding) and second (binding) mixes. In this case, pP1-P2 denatured in 2.2 M GdnHCl was mixed with refolding buffer and then challenged with 200 μM peptide.
The Misfolded Intermediate of pP1-P2 Retains the Ability to Bind the Physiological Partner of PDZ1.
To address the functional properties of the intermediate observed in pP1-P2 folding, we designed a double-jump stopped-flow experiment. In detail, the fully denatured pP1-P2 (i.e., in the presence of 5.37 M GdnHCl) was first mixed with the refolding buffer in the presence of 0.5 M sodium sulfate to populate the misfolded intermediate. Then, in a second mix, the solution was challenged with different concentrations of a peptide mimicking the C-terminal sequence of Sans, the physiological partner of Whirlin (21). The interval between the first and second mixings was held below 2 s. In fact, since under these experimental conditions, formation of the native state of pP1-P2 displays a rate constant of 0.04 ± 0.002 s−1, any faster binding events to the Sans peptide might be assigned to the misfolded intermediate. Importantly, it was previously shown that only PDZ1 binds Sans, and no physiological partners for PDZ2 have been identified (22).
In analogy to previous work on other PDZ domains (38–40), binding was followed by monitoring fluorescence resonance energy transfer (FRET) between the tryptophan, acting as a donor, and a dansyl group covalently attached to the N terminus of the peptide, acting as an acceptor. Under all the investigated conditions, binding time courses were consistent with a single exponential decay. The pseudo-first-order plot of the binding experiment obtained by double jump is reported in Fig. 4B. Surprisingly, we observe that the intermediate binds Sans with a similar, but not identical, affinity to that of native pP1-P2, with an apparent kon of 0.030 ± 0.004 s−1 μM−1 and a koff of 23.6 ± 0.7 s−1 vs. the values of 0.07 ± 0.01 s−1 μM−1 and 24 ± 1 s−1 measured for native pP1-P2 at the same experimental conditions. On the basis of these observations, we conclude that while misfolded, the intermediate in pP1-P2 retains its ability to bind the physiological ligand of PDZ1.
The foregoing double-jump experiment relies on the assumption that only PDZ1, and not PDZ2, is capable of binding to the peptide. Therefore, we tested the binding of isolated PDZ2 against the peptide and did not observe any transition. Likewise, no binding could be detected when pP1P2 was challenged with the peptide at mildly denaturing conditions, i.e., 2.2 M GdnHCl, where PDZ1 is essentially denatured while PDZ2 is still folded. To provide an additional control in this set of experiments, we also performed an additional double-jump experiment in which pP1P2, kept at mildly denaturing conditions, was mixed with refolding buffer to the same the final denaturant concentration of the double-jump experiment reported above and then, after a controlled delay time, the mixture was mixed with Sans peptide. Importantly, at a peptide concentration of 200 μM and long delay times, we observed a rate constant of 45 s−1, consistent with the value measured for the native state. Conversely, at very short delay times, we observed a binding rate constant of 10 s−1, which is in agreement with the folding rate constant observed in the refolding experiments starting from the mildly denaturing conditions. Importantly, the dependence of the observed amplitude of the fast binding phase as a function of the delay time between the first and the second mixes also showed a rate constant of ∼10 s−1 (Fig. 4B). This finding indicates that when the misfolded intermediate does not accumulate, only the fully folded pP1P2 can bind the peptide.
An Unexpectedly High Sequence Identity between PDZ1 and PDZ2.
A plausible scenario in which tandem repeats may form metastable kinetic traps involves the formation of domain-swapped intermediates (4, 8, 11). Domain swapping occurs when two neighboring domains form intertwined interactions, thereby reciprocally exchanging structural subunits. In these conditions, interdomain rather than intradomain interactions may satisfy the structural architecture of the native domain topology. It has been previously shown that tandem domain swapping is promoted when the sequence identity of continuous domains is high (12). Furthermore, a large bioinformatic analysis has demonstrated that consecutive homologous domains almost exclusively have sequence identities of <40%, such that misfolding is minimized (12).
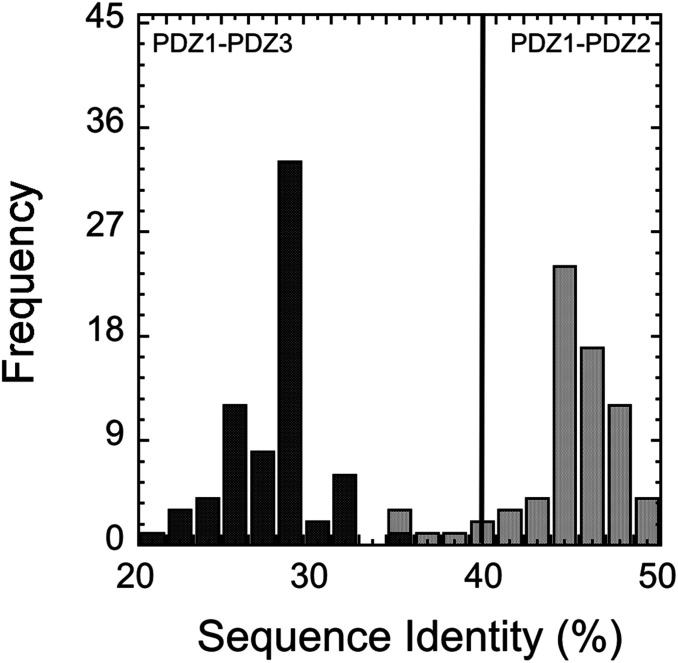
We analyzed the sequence identity of the PDZ domains in Whirlin from 70 species obtained from the UniProt database. In particular, we compared the pairwise sequence identities between the contiguous PDZ1 and PDZ2, found in the pP1-P2 construct, and a third PDZ domain, PDZ3, located downstream in the sequence of Whirlin. Contrary to expectations, we found that the contiguous PDZ1 and PDZ2 had an unusually high level of sequence identity, ranging from 34% to 53%, while PDZ1 displays a lower sequence identity to PDZ3 (Fig. 5).
Fig. 5.
Analysis of the sequence identity (%) between PDZ1 and PDZ2 (gray bars) and PDZ1 and PDZ3 (black bars) of Whirlin from 70 species obtained from the Uniprot database.
Remarkably, nearly all pairwise sequence identities between PDZ1 and PDZ2 in Whirlin were exceeded 40%, and approximately 80% of the sequences displayed a pairwise identity >45%. This finding suggests that these two domains might be under an evolutionary pressure to maintain their unusual similarities.
Discussion
The general knowledge of protein misfolding is intimately linked to dysfunction and disease. In fact, for decades it has been well established that the aberrant incorrect structures are a recurrent trigger of protein aggregation, leading to disorders ranging from neurologic disorders, such Alzheimer’s disease and Parkinson’s disease, to systemic amyloidosis (23–25). Consequently, nature has evolved several systems to solve incorrect protein folding or to degrade irreversibly misfolded polypeptides.
A peculiar protein misfolding event is represented by tandem domain swapping (4, 8). In this case, if the increasing complexity of protein architecture demands the presence of contiguous homologous domains, then the concurrent unfolding of two neighboring subunits may induce intertwined interactions. This phenomenon causes two adjacent domains to form a “central domain,” equivalent to a circular permutation, formed from the central region of the two-domain sequence, and a “terminal domain” comprising the remaining parts of the sequence. Notably, by following this scenario, a misfolded intermediate may appear to contain more folded structure than would be expected when studying a single domain, as in the case of pP1-P2.
Studies of titin and Ig domains have shown that evolution tends to minimize domain-swapped misfolding by lowering the sequence identity of adjacent domains, which is almost universally <40% in tandem repeat proteins (8, 11, 12). In this context, it is of interest that all the sequences of Whirlin in the UniProt database display unusually high sequence identities between adjacent PDZ1 and PDZ2. Therefore, also considering that PDZ domains are naturally prone to circular permutation (36) and domain swapping (41), and that previous protein engineering experiments suggested that PDZ domains may be successfully expressed as circularly permutated variants (42, 43), it is somewhat surprising to find that the tendency to perform domain swapping might be evolutionary conserved in Whirlin. Can the misfolded intermediate play a physiological role in the function of Whirlin?
Mechanical proteins are exposed to tensile forces that might unfold them and lead to the accumulation of long-lived states displaying multiple unfolded domains (12). In these conditions, there could be a tendency to form aberrant structures, leading to a loss of function. In the case of Whirlin, the anchoring to Usher protein via its PDZ1 domain guarantees mechanical communication from the surface of the stereocilia to the actin filaments, ensuring the hearing of sound waves. It is intriguing that the presence of two PDZ domains is almost universally conserved in Whirlin (17, 21), yet no physiological partners could be found for PDZ2 (22, 44). Thus, on the basis of our findings, we speculate that the second PDZ domain, enhancing the possibility of accumulating a functionally competent domain-swapped state, may represent a structural buffer to increase the repertoire of structures to conserve the anchoring to Sans, even in the presence of mechanical unfolding.
In summary, our work highlights an example of a functionally competent misfolded intermediate. Despite expectations, the sequence patterns in natural proteins highlight that the tendency to form this metastable state appears to be conserved. Future work on other PDZ-containing tandem repeats will shed light on the generality of these findings.
Materials and Methods
Site-Directed Mutagenesis.
The construct P1-P2 is identical to that described previously (22). The variant pP1-P2 was obtained by substituting the tyrosine residue in position 168 with a tryptophan using the QuikChange Lightning Mutagenesis Kit (Agilent). The DNA encoding for the pseudo-WT fluorescent PDZ1 Y168W (pPDZ1) domain was obtained using the QuikChange Lightning Mutagenesis Kit to insert a stop codon in the pP1-P2 sequence downstream the region encoding for the PDZ1 domain (residues 140 to 224). The sequence was confirmed by DNA sequencing. PDZ2 was produced from a DNA library at NZYTech. The primers oligo were purchased from Eurofins Genomics.
Protein Expression and Purification.
DNA encoding for pP1-P2, pPDZ1, and PDZ2 were transformed into Escherichia coli BL21 (DE3) cells for protein expression. The protein expression was induced with isopropyl β-d-1-thiogalactopyranoside to a final concentration of 100 μg/mL at an OD600 of 0.8, and the cells were incubated for 3 h at 30 °C. The cells were then cultured after an overnight incubation at 20 °C. Each construct and mutant was purified from the soluble fraction in 50 mM NaPho at pH 7.2 with 0.3 mM NaCl with a HiTrap Chelating High-Performance column (GE Healthcare) and then eluted with a gradient to 1 M imidazole. The imidazole was removed using a HiPrep 26/10 Desalting column (GE Healthcare). The purity of the protein sample was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. The peptide mimicking the C-terminal sequence of Sans was purchased from JPT Peptide Technologies.
Equilibrium Experiments.
Fluorescence equilibrium (un)folding experiments were performed on a standard spectrofluorometer (FluoroMax single photon counting spectrofluorometer; Horiba). The protein pP1-P2 was excited at 280 nm, and emission spectra were recorded between 300 and 400 nm at increasing denaturant concentrations. The experiment was performed at 25 °C using a quartz cuvette with a path length of 1 cm. The far-UV CD denaturation spectra were measured using a Jasco 810 dichrograph, which was flushed with N2 and equipped with a Peltier thermoregulation system. One-mm-thick quartz cuvettes were used. Spectra were measured between 200 and 250 nm at 25 °C. The scanning speed was 50 nm/min, with a data pitch of 0.5 nm. CD ellipticities were selected at a single wavelength (225 nm) and plotted against denaturant concentration. The spectra of single PDZ1 and PDZ2 were analyzed quantitatively following a two-state model. The spectra of pP1P2 were fitted with the sum of two two-state models.
NMR Experiments.
15N-labeled pP1-P2 was expressed and purified as described previously (42). The 15N-labeled NMR sample of pP1-P2 was prepared at an initial concentration of 266 μM in 240 μL of a buffer containing 50 mM sodium phosphate, 150 mM NaCl, and 2% D2O, pH 3.7. Chemical denaturation of pP1-P2 was followed by GdnHCl titration. Aliquots of a 6 M GdnHCl stock solution were successively added to the pP1-P2 sample. For each GdnHCl point (0.5, 1, 1.5, 2, 2.2, 2.6, 3.1, and 3.6 M), a 2D 1H-15N HSQC spectrum was recorded at 25 °C on an 800-MHz Bruker AVANCE NEO spectrometer equipped with a TCI cryoprobe. From the series of HSQC, the resonance peak intensities were measured for each residue as a function of GdnHCl concentration, using CcpNmr Analysis 2.4 software (45). The intensities were corrected to account for the dilution effect at each titration point and normalized to the intensities without GdnHCl. Assignment of most of the isolated resonances could be deduced from those at pH 7.5, 6.5, and 5.9.
Kinetic Experiments.
Kinetic folding experiments.
Rapid mixing for kinetic folding and unfolding experiments were carried out on a stopped-flow device (PiStar; Applied Photophysics). Fluorescence emissions of pP1P2 and pPDZ1 were measured with a 320-nm bandpass filter using an excitation wavelength of 280 nm. The kinetic folding experiments were conducted with a 1.5 μM final protein concentration and denaturant concentrations ranging from 0.2 to 5.27 M. Refolding experiments of pP1P2 were performed under two different starting conditions, with the protein diluted in either a mild denaturant concentration (i.e., 2.2 M GndHCl) or a strong denaturant concentration (i.e., 5.37 M GndHCl). The temperature was set at 25 °C. Buffers were 50 mM Tris⋅HCl pH 7.5 and 0.3 M NaCl, and 50 mM Tris⋅HCl pH 7.5 and 0.5 M Na2SO4. Kinetic traces were fitted with a single exponential decay using Applied Photophysics software to obtain the observed rate constant, kobs. The logarithmic values of kobs were plotted vs. GndHCl concentration (chevron plot). The traces that did not show a rollover effect were fitted to a standard two-state chevron.
Owing to the complexity of the observed kinetics, observed data were fitted as follows. For the chevron arising from the unfolding and the refolding data starting from mildly denaturing conditions, data were fitted to a three-state mechanism involving the accumulation of a transient intermediate (35). Subsequently, the chevron arising from the data at strong denaturing conditions was fitted by applying the analytical solution to an off-pathway scenario, obtained from the eigenvalues of a three-state matrix, as described previously (46). Since only the slow λ2 phase could be observed, to allow the fitting procedure to converge, we forced the parameters for productive folding to those obtained from the fit at mildly denaturing conditions.
Kinetic binding experiments.
Kinetic binding between pP1-P2 and the peptide mimicking the C-terminal region of Sans was measured by monitoring FRET between the tryptophan, acting as a donor, and a dansyl group covalently attached to the N terminus of the peptide, acting as an acceptor. The experiment was performed on a single-mixing SX18 stopped-flow instrument (Applied Photophysics), recording the changes in fluorescence emission. An excitation wavelength of 280 nm was used, and fluorescence emissions were collected using a 320-nm cutoff glass filter. The binding experiments were carried out at pseudo-first-order conditions by mixing a constant concentration of pP1-P2 (4 μM) vs. increasing concentrations of Sans (ranging from 40 to 200 μM). The buffer was 50 mM Tris⋅HCl and 0.5 M Na2SO4, pH 7.5. The kobs values were calculated from the average of three to six single traces and by fitting the time course for binding using a single exponential equation.
Double-jump kinetic experiments.
Refolding and rapid-binding experiments were carried out on an Applied Photophysics stopped-flow instrument with double-jump capability at an excitation wavelength of 280 nm. Fluorescence emissions were measured using a 320-nm cutoff glass filter. Refolding and binding were initiated by symmetric mixing of the denatured and Sans peptides with the appropriate buffer. In the first mix, unfolded pP1P2 (4 μM) was first mixed against a refolding buffer (0.54 M GdnHCl at 50 mM sodium phosphate, pH 7.5 and 0.5 M sodium sulfate). Unfolded pP1P2 was obtained by incubation in GdnHCl 5.37 M at pH 7.5. In the second mix., after a 2-s delay, binding was initiated by transferring the solution into a buffer composed of 0.54 M GdnHCl, 50 mM sodium phosphate pH 7.5, and 0.5 M sodium sulfate in the presence of Sans concentrations ranging from 40 to 300 μM. A control experiment (Fig. 4B, Inset) was also run under the same conditions described above but starting under mildly denaturing conditions, with 2.2 M GdnHCl at 50 mM sodium phosphate pH 7.5, and measuring binding at different delay times between the first and second mixing events.
Data Availability Statement.
All data used to support the results and conclusions of this work are included in the paper.
Acknowledgments
This study received funding from the European Union’s Horizon 2020 Research and Innovation Program under Marie Sklodowska-Curie Grant Agreement PDZnet 675341, the Italian Ministero dell’Istruzione dell’Università e della Ricerca (Progetto di Interesse “Invecchiamento”, to S.G.), Sapienza University of Rome (B52F16003410005, RP11715C34AEAC9B, and RM1181641C2C24B9, to S.G.), the Associazione Italiana per la Ricerca sul Cancro (Individual Grant MFAG 2016, 18701 to S.G.), and the Istituto Pasteur Italia (Teresa Ariaudo Research Project 2018, to A.T.). F.M. was supported by a fellowship from the Associazione Italiana per la Ricerca sul Cancro. Financial support was also provided by the Conseil Régional d’Ile de France through the Soutien Aux Equipes Scientifiques Pour L’acquisition De Moyens Experimentaux 2014 NMR Center for Health Research Program (14014526; 800-MHz spectrometer).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
References
- 1.Batey S., Clarke J., Apparent cooperativity in the folding of multidomain proteins depends on the relative rates of folding of the constituent domains. Proc. Natl. Acad. Sci. U.S.A. 103, 18113–18118 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian P., Best R. B., Structural determinants of misfolding in multidomain proteins. PLoS Comput. Biol. 12, e1004933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han J. H., Batey S., Nickson A. A., Teichmann S. A., Clarke J., The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 8, 319–330 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Lafita A., Tian P., Best R. B., Bateman A., Tandem domain swapping: Determinants of multidomain protein misfolding. Curr. Opin. Struct. Biol. 58, 97–104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantaev R. et al., Manipulating the folding landscape of a multidomain protein. J. Phys. Chem. B 122, 11030–11038 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Kumar V., Chaudhuri T. K., Spontaneous refolding of the large multidomain protein malate synthase G proceeds through misfolding traps. J. Biol. Chem. 293, 13270–13283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruszka D. T. et al., Disorder drives cooperative folding in a multidomain protein. Proc. Natl. Acad. Sci. U.S.A. 113, 11841–11846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgia A. et al., Transient misfolding dominates multidomain protein folding. Nat. Commun. 6, 8861 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valle-Orero J. et al., The elastic free energy of a tandem modular protein under force. Biochem. Biophys. Res. Commun. 460, 434–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inanami T., Terada T. P., Sasai M., Folding pathway of a multidomain protein depends on its topology of domain connectivity. Proc. Natl. Acad. Sci. U.S.A. 111, 15969–15974 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borgia M. B. et al., Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins. Nature 474, 662–665 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright C. F., Teichmann S. A., Clarke J., Dobson C. M., The importance of sequence diversity in the aggregation and evolution of proteins. Nature 438, 878–881 (2005). [DOI] [PubMed] [Google Scholar]
- 13.McLachlan A. D., Repeating sequences and gene duplication in proteins. J. Mol. Biol. 64, 417–437 (1972). [DOI] [PubMed] [Google Scholar]
- 14.Pena-Francesch A., Demirel M. C., Squid-inspired tandem repeat proteins: Functional fibers and films. Front Chem. 7, 69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Riba A., Synakewicz M., Itzhaki L. S., Folding cooperativity and allosteric function in the tandem-repeat protein class. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 1749 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobe B., Kajava A. V., When protein folding is simplified to protein coiling: The continuum of solenoid protein structures. Trends Biochem. Sci. 25, 509–515 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Ebermann I. et al., A novel gene for Usher syndrome type 2: Mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum. Genet. 121, 203–211 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Maerker T. et al., A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 17, 71–86 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Mathur P. D., Yang J., Usher syndrome and non-syndromic deafness: Functions of different whirlin isoforms in the cochlea, vestibular organs, and retina. Hear. Res. 375, 14–24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorusch N., Wunderlich K., Bauss K., Nagel-Wolfrum K., Wolfrum U., Usher syndrome protein network functions in the retina and their relation to other retinal ciliopathies. Adv. Exp. Med. Biol. 801, 527–533 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Sorusch N. et al., Characterization of the ternary Usher syndrome SANS/ush2a/whirlin protein complex. Hum. Mol. Genet. 26, 1157–1172 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Delhommel F. et al., Structural characterization of whirlin reveals an unexpected and dynamic supramodule conformation of its PDZ tandem. Structure 25, 1645–1656.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Chiti F., Dobson C. M., Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Dobson C. M., Knowles T. P., Vendruscolo M., The amyloid phenomenon and its significance in biology and medicine. Cold Spring Harb. Perspect. Biol. 12, a033878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knowles T. P., Vendruscolo M., Dobson C. M., The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014). [DOI] [PubMed] [Google Scholar]
- 26.O’Brien E. P., Christodoulou J., Vendruscolo M., Dobson C. M., New scenarios of protein folding can occur on the ribosome. J. Am. Chem. Soc. 133, 513–526 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Calosci N. et al., Comparison of successive transition states for folding reveals alternative early folding pathways of two homologous proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 19241–19246 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chi C. N. et al., A conserved folding mechanism for PDZ domains. FEBS Lett. 581, 1109–1113 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Hultqvist G. et al., An expanded view of the protein folding landscape of PDZ domains. Biochem. Biophys. Res. Commun. 421, 550–553 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Ivarsson Y. et al., An on-pathway intermediate in the folding of a PDZ domain. J. Biol. Chem. 282, 8568–8572 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Ivarsson Y., Travaglini-Allocatelli C., Brunori M., Gianni S., Folding and misfolding in a naturally occurring circularly permuted PDZ domain. J. Biol. Chem. 283, 8954–8960 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Myers J. K., Pace C. N., Scholtz J. M., Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 4, 2138–2148 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plaxco K. W. et al., The effects of guanidine hydrochloride on the “random coil” conformations and NMR chemical shifts of the peptide series GGXGG. J. Biomol. NMR 10, 221–230 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Jackson S. E., Fersht A. R., Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry 30, 10428–10435 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Parker M. J., Spencer J., Clarke A. R., An integrated kinetic analysis of intermediates and transition states in protein folding reactions. J. Mol. Biol. 253, 771–786 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Gianni S. et al., Structural characterization of a misfolded intermediate populated during the folding process of a PDZ domain. Nat. Struct. Mol. Biol. 17, 1431–1437 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Otzen D. E., Oliveberg M., Salt-induced detour through compact regions of the protein folding landscape. Proc. Natl. Acad. Sci. U.S.A. 96, 11746–11751 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi C. N., Bach A., Strømgaard K., Gianni S., Jemth P., Ligand binding by PDZ domains. Biofactors 38, 338–348 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Chi C. N., Engström A., Gianni S., Larsson M., Jemth P., Two conserved residues govern the salt and pH dependencies of the binding reaction of a PDZ domain. J. Biol. Chem. 281, 36811–36818 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Gianni S. et al., The kinetics of PDZ domain-ligand interactions and implications for the binding mechanism. J. Biol. Chem. 280, 34805–34812 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Fanning A. S., Lye M. F., Anderson J. M., Lavie A., Domain swapping within PDZ2 is responsible for dimerization of ZO proteins. J. Biol. Chem. 282, 37710–37716 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivarsson Y., Travaglini-Allocatelli C., Brunori M., Gianni S., Engineered symmetric connectivity of secondary structure elements highlights malleability of protein folding pathways. J. Am. Chem. Soc. 131, 11727–11733 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Hultqvist G. et al., Energetic pathway sampling in a protein interaction domain. Structure 21, 1193–1202 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Delhommel F., Wolff N., Cordier F., (1)H, (13)C and (15)N backbone resonance assignments and dynamic properties of the PDZ tandem of Whirlin. Biomol. NMR Assign. 10, 361–365 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Vranken W. F. et al., The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Jemth P. et al., Demonstration of a low-energy on-pathway intermediate in a fast-folding protein by kinetics, protein engineering, and simulation. Proc. Natl. Acad. Sci. U.S.A. 101, 6450–6455 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used to support the results and conclusions of this work are included in the paper.