Significance
What explains global patterns of diversity—environmental history or ecology? Most studies have focused on latitudinal gradients—the decline of diversity from the tropics to the poles. A problem with this is that it conflates predictions of historical and ecological hypotheses: The productive tropics have also experienced high Cenozoic biome stability. Longitudinal diversity gradients can overcome this constraint. We use a longitudinal diversity gradient in the megadiverse Cape Floristic Region to model species and evolutionary diversity in terms of Pleistocene climate stability and ecological heterogeneity. We find that biome stability is the strongest predictor of diversity measures, and argue that stability, in conjunction with measures of ecological opportunity—other than productivity—may provide a general explanation for global diversity patterns.
Keywords: Cape Floristic Region, longitudinal gradient, beta diversity, phylogenetic diversity, spatial models
Abstract
Research on global patterns of diversity has been dominated by studies seeking explanations for the equator-to-poles decline in richness of most groups of organisms, namely the latitudinal diversity gradient. A problem with this gradient is that it conflates two key explanations, namely biome stability (age and area) and productivity (ecological opportunity). Investigating longitudinal gradients in diversity can overcome this problem. Here we investigate a longitudinal gradient in plant diversity in the megadiverse Cape Floristic Region (CFR). We test predictions of the age and area and ecological opportunity hypotheses using metrics for both taxonomic and phylogenetic diversity and turnover. Our plant dataset includes modeled occurrences for 4,813 species and dated molecular phylogenies for 21 clades endemic to the CFR. Climate and biome stability were quantified over the past 140,000 y for testing the age and area hypothesis, and measures of topographic diversity, rainfall seasonality, and productivity were used to test the ecological opportunity hypothesis. Results from our spatial regression models showed biome stability, rainfall seasonality, and topographic heterogeneity were the strongest predictors of taxonomic diversity. Biome stability alone was the strongest predictor of all diversity metrics, and productivity played only a marginal role. We argue that age and area in conjunction with non–productivity-based measures of ecological opportunity explain the CFR’s longitudinal diversity gradient. We suggest that this model may possibly be a general explanation for global diversity patterns, unconstrained as it is by the collinearities underpinning the latitudinal diversity gradient.
The roles of contemporary ecological factors vs. Cenozoic environmental stability in determining large-scale biodiversity patterns continue to generate lively debate (1–7). Research on this topic has been dominated by studies of the latitudinal decline in richness toward the poles of most taxa. The many hypotheses invoked to explain the latitudinal gradient have been elegantly distilled by Schluter (5) into two—one mainly ecological (ecological opportunity), and the other historical (age and area). The former argues that diversity patterns are underpinned by differences in ecological opportunity associated with gradients in habitat heterogeneity, productivity, and the intensity of biotic interactions, all of which influence the length of niche axes: This hypothesis predicts a positive relationship between diversity and speciation rate. The age and area hypothesis posits that high diversity is a consequence of areas—sufficiently large to support viable populations of the focal taxa—having high environmental stability over evolutionary timescales, which reduces extinction rates and results in the accumulation of species both in old lineages and more recent radiations (2, 5, 7). Area and stability combine to increase rates of speciation and reduce rates of extinction. Large areas, being more heterogeneous, provide longer niche axes than small areas and offer more opportunities for speciation and reduced risks of extinction, and overall will affect the total number of species (8–10). Environmental stability promotes high speciation rates owing to increased opportunities for niche differentiation in stable selective mosaics but also ensures lower rates of extinction, and will affect the total number of species and their spatial arrangement (11–13). Although these two hypotheses have primarily been tested against species richness patterns, the recent increase and availability of regional species and phylogenetic datasets have enabled the testing of predictions for other diversity metrics, such as beta and phylogenetic diversity, which are central to our understanding of global diversity patterns (7, 14–18).
The age and area hypothesis predicts that biotas would have high beta diversity (changes in species composition along ecological gradients) owing to the accumulation of habitat specialists associated with both early- and later-diverging lineages. In this case, spatial turnover (species replacement), rather than species loss (nestedness), should prevail as the driver of beta diversity (17, 19, 20) (Fig. 1 A and B). The ecological opportunity hypothesis predicts the same patterns, but for a different reason: Richness accumulates in areas of high ecological opportunity that foster rapid, ecological speciation in numerous clades (Fig. 1 A and C). Beta diversity is largely driven by recently evolved species that have subdivided the long niche axes characteristic of high-opportunity regions. Spatial turnover should be high in areas of high ecological opportunity and high stability, allowing for the evolution of numerous range-restricted, habitat-specialist species, whereas areas of high ecological opportunity and low stability should have higher nestedness due to recolonization of empty niches after events of instability (19).
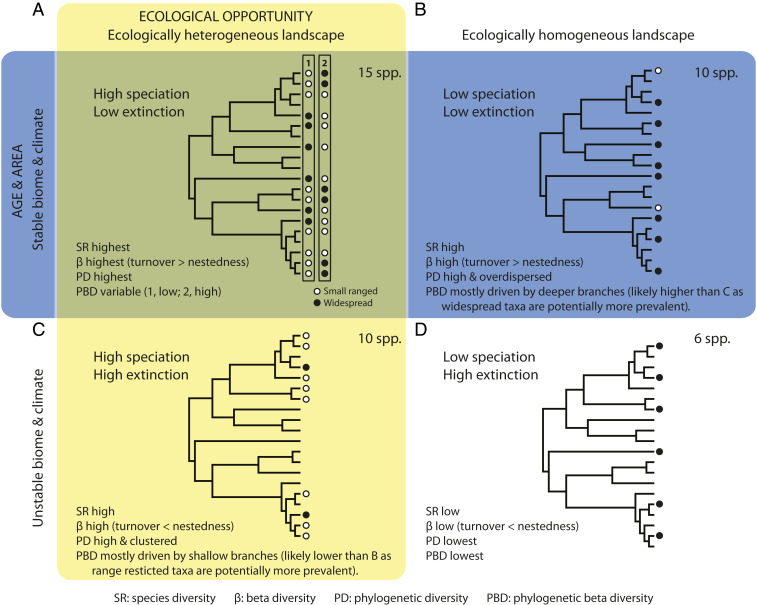
Fig. 1.
Hypothetical examples depicting the possible scenarios by which the ecological opportunity hypothesis, which focuses upon gradients in, for example, topographic diversity, seasonality, and water–energy, and/or the age and area hypothesis, here considered in terms of Late Pleistocene climate and biome stability, can explain plant diversity patterns in the CFR. Areas where both hypotheses would influence diversity achieve the highest values for all diversity metrics (A), except possibly for phylogenetic beta diversity (PBD), the value of which will vary depending on the proportion of range-restricted species and their distribution on the tree. In A, scenario 1 has a high proportion of range-restricted, recently diverged species and thus a low PBD, while in scenario 2 the range-restricted species are predominantly older, resulting in a higher PBD. The effect of the age and area hypothesis alone is shown in B, while the outcomes of the ecological opportunity hypothesis alone are depicted in C. In B and C, PBD will increase with higher proportions of range-restricted species but will be less affected by the distribution of these species (contrary to the situation in A); range-restricted taxa are expected to be more prevalent in C. An area that is ecologically homogeneous and with unstable biome and climate (D) has the lowest diversity metrics. Black dots and circles depict the distribution on the phylogenetic tree of the species present in each scenario.
The two hypotheses make different predictions for phylogenetic diversity-based metrics. For equivalent species richness, the age and area hypothesis predicts high phylogenetic diversity, owing to the preservation of older lineages, which are widely dispersed on phylogenetic trees (Fig. 1B), whereas ecological opportunity predicts lower phylogenetic diversity, owing to the preponderance of younger, recently evolved species swarms, which are mostly clustered on phylogenetic trees (Fig. 1C) (2, 7, 16, 21–23). Phylogenetic beta diversity, which measures phylogenetic turnover (i.e., turnover in branch length) (24), will vary depending on the proportion of range-restricted species present in a given area and their distribution within the phylogenetic tree (i.e., the phylogenetic distance separating them). For areas with similar species richness, phylogenetic beta diversity is predicted to be similar under the age and area hypothesis and the ecological opportunity hypothesis (15, 17, 20), although driven by different phylogenetic patterns, namely fewer deeper branches for the former (Fig. 1B) and many shallower branches for the latter (Fig. 1C). However, one would expect a larger proportion of widespread taxa to be present under the age and area hypothesis because of the longer time for range expansion to occur (Fig. 1B). Environmental stability fosters the large-scale preservation of clades (i.e., low extinction; Fig. 1 A and B), whereas in regions of high ecological opportunity, high diversification rates produce fewer, but more species-rich, phylogenetic groups (Fig. 1 C and D) (1, 5, 7, 21–23).
The two hypotheses, however, are not necessarily mutually exclusive (5). A system where both hypotheses have traction (i.e., a stable biome with high ecological heterogeneity) would show high beta diversity and both high phylogenetic diversity and phylogenetic beta diversity, a consequence of high speciation and low extinction rates (Fig. 1A). In this scenario, phylogenetic beta diversity can also be low if most narrow-ranged species are recently evolved (Fig. 1A) (1). On the other hand, a stable biome with an ecologically homogeneous environment and an unstable biome with an ecologically heterogeneous environment would both have high phylogenetic diversity, but it would be overdispersed in the former (i.e., principally formed of isolated lineages) (Fig. 1B) and clustered in the latter (i.e., generally comprising fewer, but more speciose, lineages) (Fig. 1C). Likewise, under these two scenarios, phylogenetic beta diversity would be high, although higher in the first case, driven principally by deep branches (Fig. 1B), than in the second case, which will be driven mostly by shallower branches (Fig. 1C).
The age and area and ecological opportunity hypotheses have seldom been tested simultaneously and never for a diversity gradient within an extratropical megadiversity center; most research has focused on the latitudinal gradient, which conflates the predictions of historical and ecological hypotheses: The productive tropical rainforest biomes, which offer high opportunities for ecological speciation (e.g., epiphytes in tall, multilayered forests) (4, 25, 26), have also experienced the highest stability throughout the Cenozoic (2, 5, 27, 28). This problem can be overcome by researching diversity gradients where environmental stability and ecological heterogeneity are uncoupled, as occurs along many longitudinal diversity gradients. Examples include comparisons of diversity in temperate biomes of southeastern North America and eastern Asia (3, 29), between Europe and eastern Asia/North America (30), and among the Mediterranean-climate regions across the globe (13). These studies conclude that historical events and biogeographic idiosyncrasies play a more important role in explaining diversity than ecological factors associated with contemporary environments. However, the world’s most diverse regions, the mountainous areas of tropical Asia and the Neotropics (1, 5, 7, 31), combine the environmental features predicted by both the age and area and the ecological opportunity hypotheses to be associated with megadiversity.
The Cape Floristic Region (CFR), a Mediterranean-climate region, provides an excellent opportunity to investigate simultaneously the ecological and historical drivers of diversity (32). First, the CFR flora is the richest extratropical flora in the world, comprising 9,383 species (68% endemic) in just 90,760 km2. Second, the CFR flora is well-known taxonomically, spatially, and phylogenetically. Third, biological heterogeneity is relatively homogeneous throughout the region; the diversity and structure of plant communities are relatively similar for analogous landscapes throughout the CFR. Fourth, the region shows a pronounced longitudinal gradient in regional-scale (1 to 10,000 km2) diversity: The numbers per unit area of taxa associated with clades endemic to the CFR, as well as regional-scale richness of entire floras, decline markedly in a longitudinal pattern, from southwest to southeast (32). Fifth, longitudinal gradients of Pleistocene climate and biome stability are evident across the CFR, with more stable climates in the west, where Mediterranean climates persisted over much of the region, and less stable climates in the east, where the CFR flora was replaced at times by a subtropical flora (33–35).
Here, we use the longitudinal plant diversity gradient in the CFR to test the predictions of the age and area and ecological opportunity hypotheses by modeling several key diversity metrics, incorporating both species richness and evolutionary history, in relation to variables reflecting ecological and historical phenomena. Our analysis was conducted at the regional scale; our mapping unit is a 2-minute grid cell (ca 12 km2), sufficiently large to include, in all parts of the CFR, substantial environmental gradients and several floristically distinct plant communities. Since our focus is on the evolution of CFR plant diversity, we included in our analysis only species associated with “Cape clades,” groups largely endemic to the CFR and which have their diversity centered within the region (36). Our comprehensive dataset includes modeled occurrences across 8,347 2-minute grid cells for 4,813 species (∼51% of total CFR species) and dated molecular phylogenies for 21 Cape clades. Patterns of Cape clade species richness are strongly correlated with overall CFR plant richness (SI Appendix, Fig. S1) and we therefore consider them reflective of taxonomic patterns for the entire flora. We used measures of topographical heterogeneity, productivity (evapotranspiration), and rainfall seasonality as surrogates for ecological opportunity (4, 6, 25, 37). For historical measures, climate and biome stability were assessed using an ensemble of general circulation model experiments to calculate climate variability and biome persistence over the last 140 ka (kiloannum) (35). This time span is appropriate for our study since many Cape clades have speciated massively during the Pleistocene (38); almost half (48.6%) of all divergence events in the current study took place in the last 2 Ma.
If the ecological opportunity hypothesis explains the CFR’s species and evolutionary diversity gradients, we would expect significant positive relationships between richness and both topographical heterogeneity and productivity and a negative relationship between richness and rainfall seasonality (more seasonal environmental precipitation becomes limiting in different seasons [i.e., precipitation only during the cool season vs. precipitation only during the warm season], whereas less seasonal environments provide greater opportunities for niche specialization to warm- and cool-season precipitation) (32). We also expect similar relationships for beta diversity because rapid, ecological speciation should result in high spatial turnover of ecological specialists along habitat gradients. For evolutionary diversity, we expect richness hotspots to be correlated with low phylogenetic diversity per species (made up of fewer but more speciose lineages) and relatively low phylogenetic beta diversity, owing to the predominance of recently radiating clades likely comprising range-restricted species. On the other hand, for the age and area hypothesis, we expect richness, the spatial turnover component of beta diversity, and phylogenetic diversity all to be associated with areas of high climate and biome stability, owing to the preservation of clades, a consequence of low extinction rates. For the same reason, phylogenetic beta diversity is more likely to be positively associated with climate and biome stability because of the prevalence of deeper branches, despite species being also more likely to exhibit wider distributions. We also predict that in regions with stable biomes and climates and with ecologically heterogeneous landscapes, both hypothesized mechanisms will have influenced diversity patterns.
Results
Ecological and Stability Predictors.
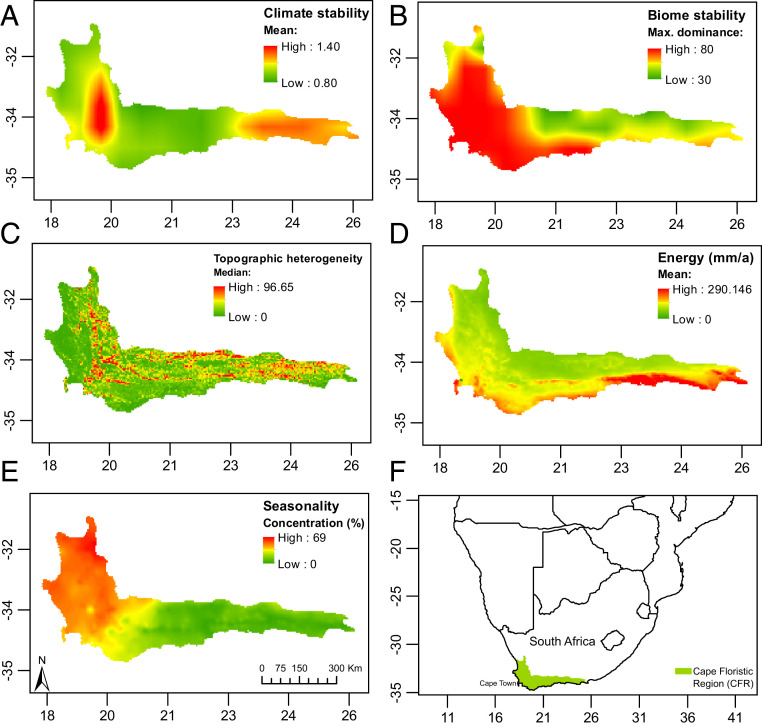
The spatial patterns for the five covariates used to test our predictions are shown in Fig. 2. Two nodes of high Late Pleistocene climate stability were identified, one in the west and a less pronounced one in the east CFR (Fig. 2A). However, a clear west–east gradient of biome stability was retrieved (Fig. 2B). The node of high climate stability in the east does not translate into high biome stability since eastern climates are currently marginal for Cape vegetation (32, 34) so that even small climatic shifts can cause biome replacement; thus, biome persistence was lower in the eastern CFR. There is little evidence of a topographic heterogeneity gradient across the CFR; areas of high and low values are evenly spread across the region (Fig. 2C). Productivity was highest in the southeastern and southwestern CFR, and medium to low in the central and interior regions (Fig. 2D). A strong west–east seasonality gradient exists (Fig. 2E), with the west showing predominance of a winter seasonal moisture regime (SI Appendix, Fig. S1), whereas precipitation seasonality was less pronounced in the southwest and low in the east, where rainfall occurs throughout the year.
Fig. 2.
Spatial patterns of the five predictor variables (A–E) plotted for the Cape Floristic Region (F).
Species and Evolutionary Diversity Patterns.
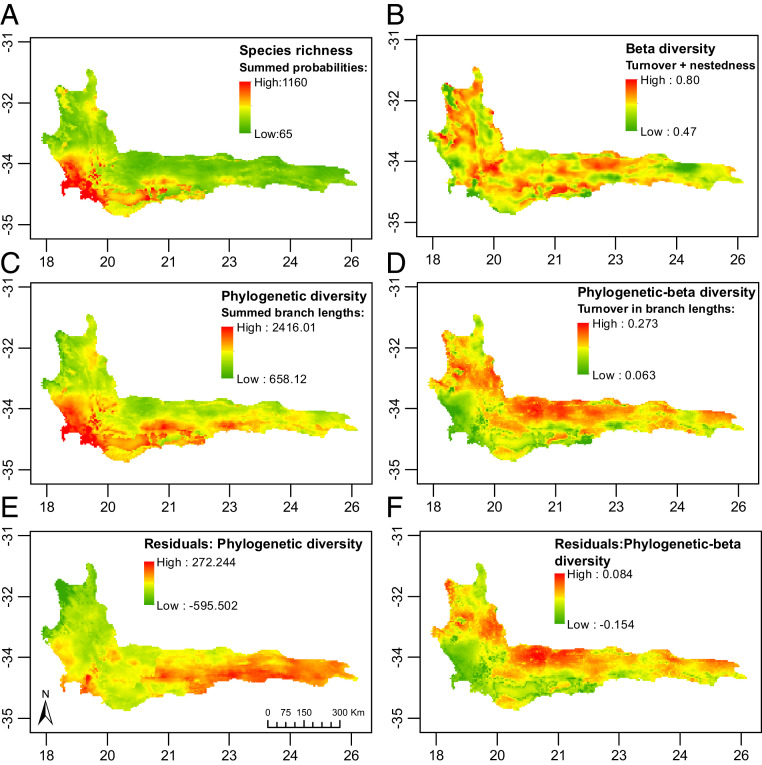
The spatial patterns across the region for species and evolutionary diversity of CFR-centered plant clades are shown in Fig. 3. We recovered a marked west–east gradient in species richness across the southern CFR, with the highest concentrations of species in the southwest (>380 species per grid cell) (Fig. 3A). Species richness declined eastward into the year-round rainfall region (SI Appendix, Fig. S2), where we recorded 65 to 100 species per grid cell. Total taxonomic beta diversity showed consistently high values (∼0.65) across almost the entire CFR (Fig. 3B) and was predominantly the result of species turnover (SI Appendix, Fig. S3). Nodes of high beta diversity were associated with lower mountain slopes and adjacent lowlands, areas of rapid transition of the CFR’s major vegetation types, namely fynbos, renosterveld, and succulent karoo (39).
Fig. 3.
Spatial patterns of the four diversity variables (A–D) and of residuals from linear regressions of phylogenetic diversity on species richness (E) and of phylogenetic beta diversity on taxonomic beta diversity (F), plotted for the Cape Floristic Region.
Highest values of phylogenetic diversity were concentrated in the southwestern CFR (Fig. 3C) and were broadly concordant with the patterns of species richness. Residuals of phylogenetic diversity over species richness showed a clear concentration of positive residuals in the eastern CFR (Fig. 3E), indicating that phylogenetic diversity is generally overdispersed in the east and more clustered in the west. High values of phylogenetic beta diversity were somewhat patchily distributed across the CFR (Fig. 3D) but showed an obverse pattern to phylogenetic diversity; the southwestern CFR had comparatively low phylogenetic beta diversity, most likely caused by a concentration of closely related and narrow-ranged endemics (40) (as in Fig. 1A, scenario 1). Positive residuals of phylogenetic beta diversity over taxonomic beta diversity were mostly concentrated in northern parts of the CFR (Fig. 3F), where high phylogenetic beta diversity occurs without high taxonomic beta diversity (Fig. 3B). Areas of high positive residuals indicate high phylogenetic beta diversity associated with turnover of deeper branches on the phylogenetic tree (as in Fig. 1A, scenario 2). This suggests that these areas hold a high proportion (but a low absolute number) of small-ranged species belonging to older clades.
Spatial Regression Models.
A separate full model including all covariates was run for each of the four metrics of diversity, removing one covariate at a time, and covariate support was assessed using CIs and widely applicable information criterion (wAIC) statistics (Materials and Methods, Table 1, and SI Appendix, Table S1). The direction of the relationship and the strength of the effect the covariate has on a diversity variable are summarized in Table 1 and Fig. 4 (full details are in SI Appendix, Fig. S4 and Tables S1 and S2).
Table 1.
Raw mean effects of the INLA analysis for raw diversity variables and controlling for the effects of species richness
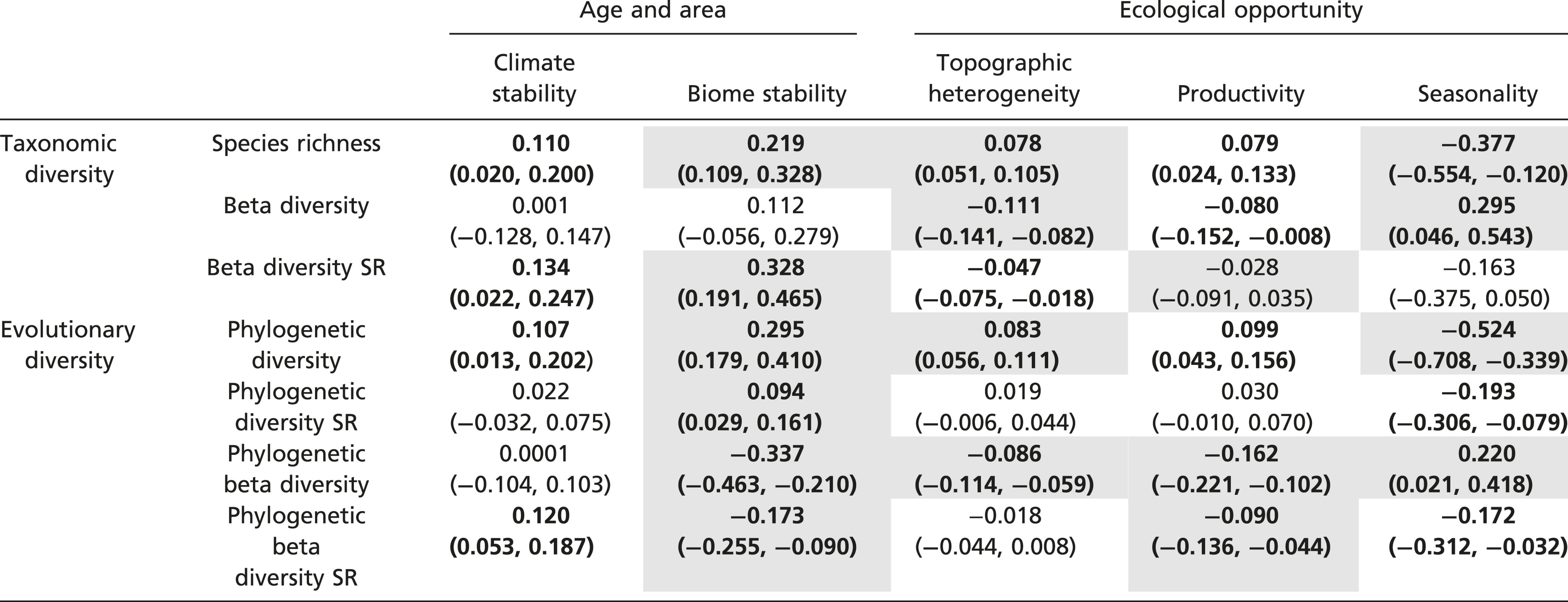 |
The sets of historical and ecological covariates best explaining the spatial diversity patterns are shown by well-supported effects (in bold font) and wAIC values: Shaded cells indicate a wAIC value increase of ≥3 when a covariate is removed from a model with a full set of covariates (see SI Appendix, Tables S1 and S2 for full model results). Medians with lower (0.025) and upper (0.975) quantiles are shown in parentheses. SR, species richness.
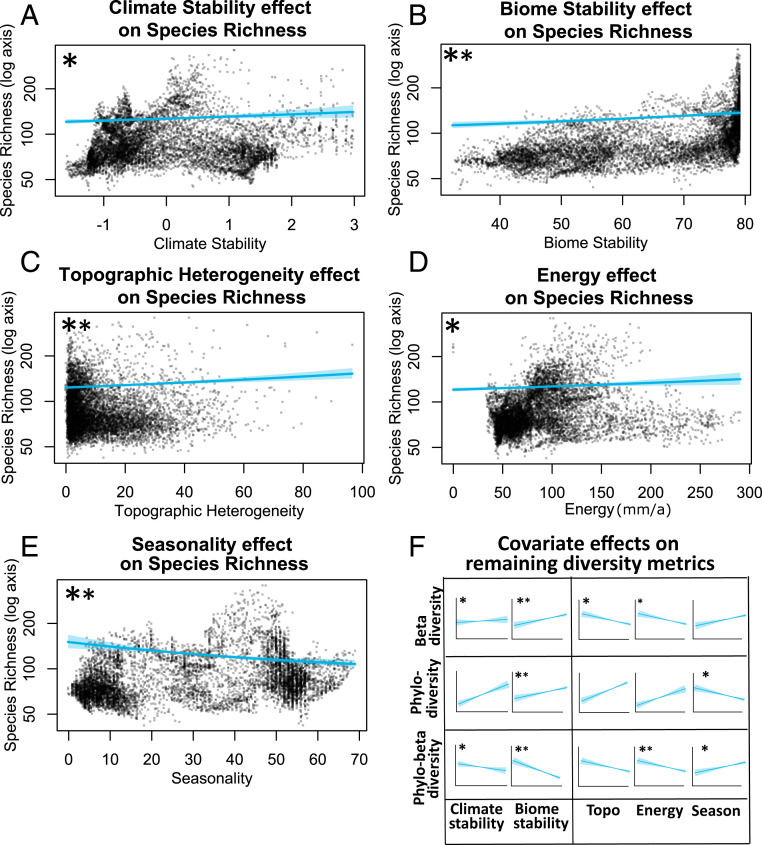
Fig. 4.
(A–E) Relationships between species richness predicted from models with (A) climate stability, (B) biome stability, (C) topographic heterogeneity, (D) energy, and (E) seasonality. (F) Simplified plots of the relationship of these covariates with the remaining diversity variables controlling for species richness (species turnover, phylogenetic diversity, and phylogenetic beta diversity; see SI Appendix, Fig. S4 for detailed plots). Within each plot, the results are shown with the median estimate and 95% confidence intervals (shaded). Confidence intervals are computed from models that include all fixed and spatially explicit random effects: The presence of strong spatial effects generates wider scatter in the points than may be expected from plotted confidence intervals. A large asterisk indicates well-supported effects with confidence intervals that exclude 0; a small asterisk indicates that models excluding a specific covariate received more support from wAIC statistics than a full model including all covariates. For example, excluding climate stability or energy received more support from wAIC statistics than the full model, suggesting the positive effects of biome stability and topographic heterogeneity and the negative effects of seasonality on species richness are the most robust.
For species richness, we found strong evidence (support both from CIs and wAIC statistics) for a positive relationship with both biome stability and topographic heterogeneity and a negative relationship with seasonality (areas with moderate seasonality in the southwestern and southern CFR generally had higher richness whereas high-seasonality areas in the northwestern CFR were relatively species-poor, as were the areas of lowest seasonality in the east) (Fig. 2). Species richness showed marginal positive relationships with productivity and climate stability.
Before controlling for species richness, we found that ecological covariates were the best predictors for taxonomic beta diversity; however, the direction of these relationships did not all match the direction of our predictions (Fig. 1). We recorded a negative effect with topographic heterogeneity and productivity and a positive effect with seasonality; topographic heterogeneity and seasonality also received support from wAIC statistics. Controlling for species richness altered these relationships and only topographic heterogeneity (negative relationship) was retained as a marginally significant ecological predictor, whereas both historical stability predictors showed well-supported positive effects. Biome stability received additional support from wAIC statistics and therefore emerged as the most robust predictor of taxonomic beta diversity.
For metrics of evolutionary diversity, we found a similar pattern for phylogenetic diversity to that observed for species richness, with all covariates having a strong effect (Table 1). Other than seasonality, which was negatively related to phylogenetic diversity, all covariates showed positive relationships with this metric. As was the case for species richness, models excluding climate stability or productivity received more support from wAIC statistics than the full model, indicating that the positive effects of biome stability and topographic heterogeneity and the negative effects of seasonality are best at predicting phylogenetic diversity. However, when controlling for species richness, almost all of the strong effects of covariates disappeared, except for the positive relationship with biome stability.
For phylogenetic beta diversity, we found well-supported negative relationships with all covariates, except for seasonality. Seasonality showed a well-supported positive relationship, with areas of high seasonality (the strongly winter-rainfall, northwestern CFR) having high phylogenetic beta diversity. After accounting for species richness, the model retained a well-supported negative relationship between phylogenetic beta diversity and biome stability and productivity. Climate stability offered marginal support for a negative relationship with phylogenetic beta diversity, while seasonality retained marginal support for a positive relationship. Phylogenetic beta diversity, therefore, appears highest in less stable and low-productivity environments such as the northern fringes of the eastern CFR.
Overall, results from our spatial regression models supported our predictions of greater species and phylogenetic diversity (Fig. 1A) and lower phylogenetic beta diversity (Fig. 1A, scenario 1) associated with the areas of high biome stability, namely the southwestern CFR. These areas support the highest numbers of taxa, many of which are range-restricted and recently diversified (SI Appendix, Fig. S3 and Table S3). We also found well-supported evidence consistent with the prediction that the turnover component of taxonomic beta diversity would be positively related to biome stability (Fig. 1 A and B). We found mostly marginal support for the role of ecological predictors in patterns of diversity, and the directions of the individual diversity–covariate relationships did not always follow expected predictions. Although topographical heterogeneity showed a strong positive relationship with species richness (Fig. 1A), it had a strong negative relationship with beta diversity, contrary to our predictions (Fig. 1 A and C). Our prediction that topographical heterogeneity would have a strong, positive relationship with evolutionary diversity metrics (Fig. 1 A and C) was also rejected. Our prediction that productivity would be positively related to species richness was only marginally supported, and we retrieved little support for our prediction of a positive relationship between phylogenetic diversity and productivity (Fig. 1 A and C). We also did not find support for the prediction that taxonomic beta diversity would be positively related to productivity; instead, we found some support for a negative relationship. Contrary to our predictions (Fig. 1A, scenario 2), phylogenetic beta diversity was negatively associated with climate and biome stability and productivity.
Discussion
As an extratropical center of plant megadiversity, the CFR has puzzled evolutionary biologists for decades. A relatively recent model for predicting global plant diversity patterns, which used measures of productivity and topographic heterogeneity as explanatory variables while explaining diversity patterns for other bioregions, predicted half the observed species richness of the CFR (37). Here we show that biome stability (age and area) in combination with low seasonality and high topographic heterogeneity (ecological opportunity) were the best predictors of taxonomic plant richness in the CFR (Fig. 1A). Importantly, productivity, widely invoked as a key driver of global patterns of richness (4, 25, 37), played only a marginal role in explaining these patterns (see also ref. 7). We recognize, however, that we have presented a set of verbal predictions that may not fully capture how different processes map to patterns. Further testing of our predictions by simulation with a wider range of parameters would help to confirm the importance of biome stability in shaping regional diversity patterns.
Our results go to the heart of one of the most enduring patterns in ecology and evolution: Areas of high productivity (such as the humid tropics) are repositories of large amounts of diversity. While the CFR has long been seen as an exception to this rule (13, 32), we demonstrate this analytically. The relationship between energy and diversity is largely the historical legacy of a warm and wet world during the Cenozoic (2, 5, 7), which was disrupted since the Middle Miocene by progressive aridification and cooling. Tropical areas may well be diverse not primarily because of high water–energy regimes but because of age and area; their biotas have persisted in vast equatorial regions for the past 60 My, resulting in a far greater accumulation of species than in the younger temperate and arctic zones (1, 5, 6, 41). In this sense, the CFR appears not to be an exception but an example of a general model for explaining regional-scale taxonomic diversity gradients: Richness patterns can be best predicted by measures of Cenozoic environmental stability.
Other important metrics of diversity also appear best explained by measures of stability, with positive correlations retrieved for all but one diversity metric, namely phylogenetic beta diversity. High values of species turnover (∼60% changes in species composition) were recorded throughout the CFR and showed a strong positive correlation with biome stability. Contrary to our predictions (e.g., Fig. 1A), greater ecological opportunity did not necessarily equate to higher values of species turnover. This pattern is likely a consequence of biome stability allowing the persistence in and generation of habitat specialists (greater niche filling) in the southwestern CFR, from both young and old lineages. The pattern cannot be attributed to topographical heterogeneity per se since this is essentially invariant across the CFR (32, 42). The low ratio of species loss (the nestedness component of beta diversity) in the less stable areas of the eastern CFR is surprising, considering the findings by other studies where high nestedness was associated with areas experiencing climate instability (e.g., refs. 17, 21, 43, 44). However, by focusing only on Cape clades, which tend to be habitat specialists, we do not fully capture the many habitat generalists associated with widespread clades that are best represented in the eastern CFR (33, 39), and which may contribute more to nestedness.
Phylogenetic diversity in the CFR shows patterns similar to species richness, with a concentration of high values in the western part of the region. Our results confirm that overall, phylogenetic diversity is more evenly distributed in the phylogenetic tree, and generally on longer branches (i.e., overdispersed), in the eastern CFR (45, 46). Our finding of a strong positive relationship of phylogenetic diversity with biome stability (Table 1) supports this pattern, which can be explained by the presence in the western part of the CFR of a high number of closely related taxa that accumulated over time in a relatively stable environment (45). The strong relationship of phylogenetic diversity with biome stability may suggest high speciation rates coupled to lower extinction rates for the southwestern CFR (e.g., ref. 42) (Fig. 1A). However, owing to the high incidence of range-restricted taxa in the western CFR (40, 42), extinction rates may likely be high (47). On the other hand, the eastern CFR has experienced greater biome instability, leading to limited speciation and increased extinction compared with the western part of the region, as exemplified by the presence of fewer species from more disparate lineages positioned on long branches in the phylogenetic tree (e.g., ref. 45) (Fig. 1D). Importantly, paleoecological data-modeling studies suggest more stable biomes and environments in the western than eastern CFR during the Late Pleistocene; during glacial periods, CFR biomes persisted or even expanded in the west; in the east, large areas were replaced by subtropical grassland (e.g., refs. 44, 48–50).
The phylogenetic beta diversity patterns revealed here are somewhat more difficult to explain and need to be considered in parallel with taxonomic beta diversity (24). High levels of phylogenetic beta diversity and positive residuals (i.e., excess phylogenetic beta diversity above and beyond that expected from taxonomic beta diversity) were found mostly in the north of the CFR, with low levels of phylogenetic beta diversity (and negative residuals) concentrated in the southwest corner of the region. This suggests that these areas hold a high proportion (but a low absolute number) of small-ranged species (40, 42) belonging to older clades (Fig. 1A, scenario 2). However, some species near the northern boundaries of the CFR may be present in only a few localities within the CFR but have a much wider range extending outside of the region. This would bias the results toward higher phylogenetic beta diversity values in the northern part of the CFR because these potentially wider ranges would not be accounted for in the present calculations. On the other hand, the coastal regions of the CFR are mostly characterized by negative residuals and high taxonomic beta diversity (Fig. 3 B and E), which indicates the presence of a high proportion of range-restricted species, mostly from recently diversified clades (Fig. 1A, scenario 1).
Using a region of extraordinarily high plant richness and endemism, we suggest that age and area best explain large-scale patterns of plant diversity. We further argue that far from being the exception, the CFR model suggests that environmental stability may be the primary predictor of plant megadiversity. This explanation, retrieved for a longitudinal gradient, appears equally applicable to the intensively researched latitudinal diversity gradient (1, 2, 5, 21). Our use of a longitudinal gradient of diversity is important in that it allowed us to explore predictors of regional-scale diversity not necessarily concordant with gradients of productivity. Given sufficient biome stability in combination with high ecological opportunity, we see no reason why megadiversity should not evolve in low-production bioregions. An illustrative example is the extraordinarily high biodiversity of South Africa’s winter-rainfall desert—the Succulent Karoo—which, like the adjacent CFR, enjoyed a relatively stable Pleistocene climate (51).
Materials and Methods
Cape Plant Database.
We built a plant species distributional database for South African angiosperms incorporating data from national plant atlas and citizen science projects and databased herbarium specimens (52–55). The final database comprised 19,622 taxa (ca 96% of South African taxa) (56) and just over 1.8 million point locality records. In order to account for the inherent biases in such presence-only or “atlas-type” data, we employed a geospatial modeling technique (57) to interpolate the distribution records for each plant species and to calculate a continuous probability of occurrence surface for each species at a 2-minute grid cell scale (∼12 km2), with an associated measure of uncertainty. We followed the same modeling procedures (“spatial model 1”) described in detail by ref. 57, using code provided in spatial model 1 that built on earlier models by ref. 58. For each species, we built a model at 2-minute resolution combining point pattern analysis methods with environmental niche information to account for ecological similarity, inferred observer effort, and geographical distance. Briefly, this process involved two stages, each consisting of a number of separate steps. The first stage involved selecting a sample of nonfocal species records to act as pseudoabsences (reflecting the pattern of observation in the dataset), and the second stage involved interpolating distributions based on presence and pseudoabsence records. In slightly more detail, the first stage required 1) mapping all records of the focal species and generating a kernel density estimate for records of this species; 2) identifying all records of all other plant species (not just representatives of Cape clades) >100 m from records of the focal species and generating similar kernel density estimates; 3) computation of the difference in density estimates between focal and nonfocal species (an approximate index of the probability of encountering the focal species); 4) computation of an environmental envelope within a principal-component analysis of rainfall (mean annual rainfall and rainfall season) (59) and temperature variables (mean winter and mean summer temperature) (59) and soil covariates (60) (means taken from aggregating original soil data resolution of ca 1 km2 to our ca 12-km2 grid cell size; soil properties: % calcium carbonate, % clay, % silt, % sand; and pH); 5) computing the environmental distance between all 2-minute raster cells and the centroid of the environmental envelope occupied by the focal species; and 6) sampling records of the nonfocal species using the environmental distance and geographic probability of encountering the focal species to bias selection toward locations where absence was most likely. With pseudoabsence records selected, the second stage of analysis involved regression kriging of the presence/absence points onto the 2-minute raster surface, using the rainfall, temperature, and soil covariates. For species recorded from <5 locations in the database, we were unable accurately to interpolate distribution and simply generated a raster map with presence (1) and assumed absence (0) directly from the recorded data. We sought to verify distributions for well-known species, sending maps to colleagues with detailed knowledge of the species groups concerned and asking for expert opinion on the map quality. Our estimated species richness patterns were consistent with expert opinion. Once the surfaces for probability of occurrence of all species were calculated, we then selected only those species associated with predefined Cape clades (following the criteria of ref. 36: CFR origin and >50% of species native to the CFR) and for which phylogenetic data were available (SI Appendix, Table S3). Finally, the calculated probability of occurrence surfaces for all Cape clade species was clipped to the extent of the CFR as defined by ref. 61. Our final Cape clade database consisted of modeled occurrences across 8,347 2-minute grid cells for 4,813 taxa [51% of total CFR species (62)]. These probabilities of occurrence surfaces were used in all our metrics of contemporary and evolutionary diversity. All data analyses and geospatial modeling were undertaken in R (63) using the packages spatstat (64), sp (65, 66), rgdal (67), and gstat (68).
Taxonomic Plant Diversity.
We calculated two measures of taxonomic species diversity: species richness and beta diversity. Species richness was calculated for each grid cell as the summed probability surfaces for all our Cape clade species. Three different measures of beta diversity were calculated using the indices presented by ref. 19: Sorenson’s beta diversity [βsor = b + c/(2a + b + c)] and its two component parts of Simpson’s spatial turnover, βsim = min(b,c)/[a + min(b,c)], and nestedness, βnes = βsor − βsim. Variable a is the number of species common to a focal and neighbor grid cell, b is the number of species that occur only in the focal grid cell, and c is the number of species that occur only in the adjacent cell. In each case, we computed a, b, and c based on probabilities of presence: a is simply the sum of the probability of presence of all species; b is the sum of the product of the probabilities that a species was present in the focal cell but absent in a neighbor; and c is the sum of the product of the probabilities that a species was absent in the focal cell but present in a neighbor. Using interpolated species distributions offered advantages over and above raw presence-only data, as our beta diversity indices were not overly biased by gaps in the data (i.e., false absences). Calculated beta diversity for each grid cell represented the mean value of probabilities between the focal cell and all its neighbors (maximum of eight). We specifically partitioned beta diversity into its two component parts across the CFR, as the processes associated with species loss and gain (nestedness) and replacement (turnover) can be fundamentally different and can offer contrasting insights into the generation of diversity (17, 19, 43).
Phylogenetic Plant Diversity.
Phylogenetic diversity metrics were computed for 21 Cape clades for which molecular data were available (SI Appendix, Table S3). Phylogenetic trees were compiled from one of three data sources: 1) trees acquired directly from the publication or provided by the authors; 2) matrices obtained from the publication or from the authors; and 3) sequence data downloaded from GenBank. Trees acquired directly from their published source were made ultrametric using the function chronos (69) as implemented in the R package APE (70), which implements the penalized likelihood method (71). The “correlated” model of substitution rate variation among branches was applied and the root of the tree was assigned a value of 1.0. If an ultrametric tree was obtained directly from the original publication, it was standardized so that its root was given a value of 1.0. For cases for which either matrices or sequence data were obtained, the software RAxML (version 8.2.8), as implemented on the CIPRES portal (https://www.phylo.org), was used to reconstruct a phylogenetic tree under the maximum-likelihood (ML) criterion, with 1,000 rapid bootstrap replicates followed by the search of the best ML tree; the GTRCAT model was used and all other parameters were set up with their default settings. DNA sequence data were retrieved from GenBank using Geneious (version 7.1.2) (72) and aligned using the MUSCLE (73) algorithm. The approach used for each Cape clade is described in SI Appendix, Table S3.
The 21 individual species-level Cape clade trees were grafted onto a previously published genus-level phylogeny of the Cape flora (45). This approach was favored for several reasons. First, accurately calibrating phylogenetic trees from Cape groups is particularly difficult due to the limited information available in the fossil record for the vast majority of these clades (e.g., ref. 36). Second, the comparison of phylogenetic diversity metrics between clades would be invalid if all clades were in effect assigned the same age, as performed here (i.e., all root ages assigned a value of 1.0), which they are evidently not (e.g., refs. 38, 49, 74). Third, embedding all 21 Cape clades in a flora-wide tree allows us to compile overall phylogenetic diversity metrics for all clades and account for their deep history, which is particularly important in the case of phylogenetic beta diversity because the age of a group will significantly affect turnover in branch lengths (i.e., shallow vs. deep branches).
The function paste.tree from the R package phytools (75) was used to graft the individual trees onto the Cape flora genus-level tree. For clades comprising more than one genus (e.g., Bruniaceae, Podalyrieae, Restionaceae), all genera except one (randomly selected) were first pruned so that all 21 clades were represented by only one branch in the Cape flora tree. For each clade, the crown node was grafted in the middle of the corresponding branch in the Cape flora tree. Phylogenetic diversity and phylogenetic beta diversity metrics were calculated with the resulting Cape flora genus-level tree comprising the grafted Cape clades, considering only the species found in the Cape clades in the calculations (i.e., the other genera included in the Cape flora tree were not considered here).
Phylogenetic diversity was calculated for each grid as the sum of all branches connecting all members of a set of taxa, including the root of the tree. Branch lengths were weighted using the same probabilistic computations used for species diversity (see above), with a terminal branch weighted by the probability of occurrence in a given cell of the species it represents, while all internal branches were weighted by the joint probability of occurrence in a given cell of all of the species it subtends. Phylogenetic beta diversity was compiled using Sorenson’s index, similar to taxonomic beta diversity as described above, where variable a is the sum of the branch lengths common to a given grid cell and an adjoining grid cell, b is the sum of the branch lengths that only occur in a given grid cell, and c is the sum of the branch lengths that occur only in the adjacent cell. As for the phylogenetic diversity calculation, branch lengths were weighted using their probability of occurrence in each grid cell.
Surrogate Variables for Ecological Opportunity.
We calculated topographic heterogeneity from the Shuttle Radar Topography Mission digital elevation model (DEM; available from earthexplorer.usgs.gov/) computing the mean absolute difference in altitude between the focal pixel and its eight neighbors at the native 30-m resolution (76), and then calculating the median value per 2-minute grid cell (SI Appendix, Fig. S5). As beta diversity was measured at 2-minute resolution, we further compared this measure of topographic heterogeneity with the somewhat cruder analysis generated by first aggregating the DEM data to 2-minute resolution and computing the mean altitude, and then computing roughness on this using the same algorithm. These two alternative surfaces were correlated at r = 0.632, so we used the first in all analyses (SI Appendix, Fig. S5). Seasonality was calculated using a measure of rainfall concentration (ranging between 0% for zero seasonality and 100% for all rainfall in a single month) (59). We used as a measure of productivity, annual actual evapotranspiration obtained from satellite data [the MOD16A2 Version 6 Evapotranspiration/Latent Heat Flux product is an 8-d composite product produced at 500-m-pixel resolution (77)]. Actual evapotranspiration is a measure of water–energy balance closely associated with plant productivity (4). We used 8-d values to generate an annual value (mm/a) and aggregated this to our 2-minute grid taking the median value for each 2-minute cell.
Surrogate Variables for Environmental Stability.
We investigated climate and biome changes over the past 140 ka, a period spanning two major glacial–interglacial cycles (Marine Oxygen Isotope Stages 6 to 1) (35). Results from 78 paleoclimate experiments and a preindustrial experiment made with a consistent configuration of the Hadley Centre unified model (78), a fully coupled atmosphere–ocean general circulation model (79), were used to compute anomalies for monthly mean temperature, precipitation, and cloudiness. Thin-plate splines fitted to these anomalies (80) were used to interpolate them to a 0.5° grid. Paleoclimate scenarios at 0.5° grid resolution were then generated for the 78 time slices by applying the interpolated anomalies to observed recent (1961 to 1990) values in the CRU CL 1.0 dataset (81). Nine bioclimatic variables were computed for each grid cell and time slice, including 1961 to 1990: annual thermal sums above 0 and 5 °C; mean temperatures of the coldest and warmest months; an estimate of the annual ratio of actual to potential evapotranspiration; annual total intensity of the wet and dry season(s); and maximum wet and dry season intensity (for details, see ref. 35). Values for each bioclimatic variable were then standardized to 0 mean and unit SD across all grid cells and time slices, the standardized values being used to compute Euclidean distances between all 3,081 possible time-slice pairs for each grid cell. Finally, the mean of the Euclidean distances for a grid cell was used as the metric of climate stability, with smaller values indicating greater stability. No two covariates were particularly strongly correlated (all r < 0.6; SI Appendix, Fig. S6).
The relationships between the relative extents in each 0.5° grid cell of each of the nine regional biomes (39) and present climate were modeled using quantitative climatic response surfaces (78). Details of the modeling approach are given by ref. 35. These models were used to simulate the relative extent of each biome in each grid cell for each of the 79 time slices. The frequency with which each biome dominated each grid cell (i.e., had the greatest relative extent) across time slices was counted and the biome with the highest frequency of dominance in a grid cell was identified and its frequency was used as the metric of biome stability for that grid cell. After computation, we downscaled predictions to our 2-minute raster using bilinear interpolation.
Spatial Regression Models.
To test predictions about drivers of diversity, we fitted spatial regression models to each of the taxonomic and phylogenetic diversity surfaces, using covariates (topographic heterogeneity, actual evapotranspiration, rainfall seasonality, and biome and climate stability) representing the primary hypotheses to predict diversity patterns. Specifically, we fitted intrinsic continuous autoregressive [iCAR (82)] models using integrated nested Laplace approximation [INLA (83)] via the R-INLA package (84). iCAR models have been shown to perform well in a variety of spatial regression situations (85) and INLA provides a fast, Bayesian approach to fitting these computationally demanding models. As components of beta diversity (taxonomic and phylogenetic) and phylogenetic diversity measures are strongly influenced by local gradients in species richness (19, 45), we fitted further models to predict these variables that also included species richness as a covariate, expecting that including this covariate would remove relationships that are due primarily to drivers of species richness rather than beta and phylogenetic diversity per se. We expect the models with species richness to be both more conservative and more reliable, but included models without them to facilitate understanding of the simpler relationships. As INLA provides a Bayesian approach to model fitting, we assessed support for parameter estimates by identifying whether or not 95% credible intervals overlapped 0 and compared models using Watanabe-Akaike information criterion (86). Although there appears to be potential for a degree of circularity in our use of environmental variables to model species distributions and then relating modeled species data to environmental data in our spatial regression models, it will not necessarily do so, and previous work demonstrates that covariates predicting richness can be markedly different from covariates predicting individual distributions (57). If this potential circularity was problematic, we would expect the environmental data to outperform the other covariates but, as our results did not support this, we can be confident our results are not an artifact.
Supplementary Material
Acknowledgments
J.F.C. was supported by a South African National Research Foundation Research Career Advancement Fellowship (Grant 91442). J.F.C. and C.M.B. were supported by travel funding through a British Council Researcher Links Travel Grant. R.A. was supported by funding from the South African National Research Foundation (Grants 85802 and 119125). B.H. was supported by a Leverhulme Trust Research Grant (F/00 128/BI) and a Durham University Matariki Partnership Travel Grant. We acknowledge the Centre for High Performance Computing, South Africa, for providing computational resources to this research project, and for the assistance of Kevin Colville. We thank John Manning, Toney Rebello, and Les Powrie for their expert opinion on species distribution maps.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915646117/-/DCSupplemental.
Data Availability.
Plant species and phylogenetic data are available from the published sources and online repositories listed in Materials and Methods and SI Appendix.
References
- 1.Mittelbach G. G. et al., Evolution and the latitudinal diversity gradient: Speciation, extinction and biogeography. Ecol. Lett. 10, 315–331 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Ricklefs R. E., Evolutionary diversification and the origin of the diversity-environment relationship. Ecology 87 (suppl. 7), S3–S13 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Qian H., Ricklefs R. E., Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 407, 180–182 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Hawkins B. A. et al., Energy, water, and broad-scale geographic patterns of species richness. Ecology 84, 3105–3117 (2003). [Google Scholar]
- 5.Schluter D., Speciation, ecological opportunity, and latitude. Am. Nat. 187, 1–18 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Latham R. E., Ricklefs R. E., Global patterns of tree species richness in moist forests: Energy-diversity theory does not account for variation in species richness. Oikos 67, 325 (2006). [Google Scholar]
- 7.Fine P. V. A., Ecological and evolutionary drivers of geographic variation in species diversity. Annu. Rev. Ecol. Evol. Syst. 46, 369–392 (2015). [Google Scholar]
- 8.Rosenzweig M. L., Species Diversity in Space and Time, (Cambridge University Press, 1995). [Google Scholar]
- 9.Pyron R. A., Wiens J. J., Large-scale phylogenetic analyses reveal the causes of high tropical amphibian diversity. Proc. Biol. Sci. 280, 20131622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Losos J. B., Schluter D., Analysis of an evolutionary species-area relationship. Nature 408, 847–850 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Hopper S. D., OCBIL theory: Towards an integrated understanding of the evolution, ecology and conservation of biodiversity on old, climatically buffered, infertile landscapes. Plant Soil 322, 49–86 (2009). [Google Scholar]
- 12.Morueta-Holme N. et al., Habitat area and climate stability determine geographical variation in plant species range sizes. Ecol. Lett. 16, 1446–1454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowling R. M. et al., Variation in plant diversity in Mediterranean-climate ecosystems: The role of climatic and topographical stability. J. Biogeogr. 42, 552–564 (2015). [Google Scholar]
- 14.Qian H., Ricklefs R. E., A latitudinal gradient in large-scale beta diversity for vascular plants in North America. Ecol. Lett. 10, 737–744 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Qian H., Swenson N. G., Zhang J., Phylogenetic beta diversity of angiosperms in North America. Glob. Ecol. Biogeogr. 22, 1152–1161 (2013). [Google Scholar]
- 16.Massante J. C. et al., Contrasting latitudinal patterns in phylogenetic diversity between woody and herbaceous communities. Sci. Rep. 9, 6443 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto-Ledezma J. N., Larkin D. J., Cavender-Bares J., Patterns of beta diversity of vascular plants and their correspondence with biome boundaries across North America. Front. Ecol. Evol. 6, 1–13 (2018). [Google Scholar]
- 18.Lamanna C. et al., Functional trait space and the latitudinal diversity gradient. Proc. Natl. Acad. Sci. U.S.A. 111, 13745–13750 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baselga A., Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 19, 134–143 (2010). [Google Scholar]
- 20.McFadden I. R. et al., Temperature shapes opposing latitudinal gradients of plant taxonomic and phylogenetic β diversity. Ecol. Lett. 22, 1126–1135 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Mannion P. D., Upchurch P., Benson R. B. J., Goswami A., The latitudinal biodiversity gradient through deep time. Trends Ecol. Evol. (Amst.) 29, 42–50 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Qian H., Jin Y., Ricklefs R. E., Phylogenetic diversity anomaly in angiosperms between eastern Asia and eastern North America. Proc. Natl. Acad. Sci. U.S.A. 114, 11452–11457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian H., Zhang Y., Zhang J., Wang X., Latitudinal gradients in phylogenetic relatedness of angiosperm trees in North America. Glob. Ecol. Biogeogr. 22, 1183–1191 (2013). [Google Scholar]
- 24.Graham C. H., Fine P. V. A., Phylogenetic beta diversity: Linking ecological and evolutionary processes across space in time. Ecol. Lett. 11, 1265–1277 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Currie D. J., Energy and large-scale patterns of animal- and plant-species richness. Am. Nat. 137, 27–49 (1991). [Google Scholar]
- 26.Cirtwill A. R., Stouffer D. B., Romanuk T. N., Latitudinal gradients in biotic niche breadth vary across ecosystem types. Proc. Biol. Sci. 282, 20151589 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Hammen T., Absy M. L., Amazonia during the last glacial. Palaeogeogr. Palaeoclimatol. Palaeoecol. 109, 247–261 (1994). [Google Scholar]
- 28.van der Hammen T., Hooghiemstra H., Neogene and Quaternary history of vegetation, climate, and plant diversity in Amazonia. Quat. Sci. Rev. 19, 725–742 (2000). [Google Scholar]
- 29.Qian H., Ricklefs R. E., White P. S., Beta diversity of angiosperms in temperate floras of eastern Asia and eastern North America. Ecol. Lett. 8, 15–22 (2005). [Google Scholar]
- 30.Huntley B., Species-richness in north-temperate zone forests. J. Biogeogr. 20, 163 (1993). [Google Scholar]
- 31.Hughes C., Eastwood R., Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. U.S.A. 103, 10334–10339 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowling R. M., Bradshaw P. L., Colville J. F., Forest F., Levyns’ law: Explaining the evolution of a remarkable longitudinal gradient in Cape plant diversity. Trans. R. Soc. S. Afr. 72, 184–201 (2017). [Google Scholar]
- 33.Engelbrecht F. A. et al., Downscaling Last Glacial Maximum climate over southern Africa. Quat. Sci. Rev. 226, 105879 (2019). [Google Scholar]
- 34.Bar-Matthews M. et al., A high resolution and continuous isotopic speleothem record of paleoclimate and paleoenvironment from 90 to 53 ka from Pinnacle Point on the South Coast of South Africa. Quat. Sci. Rev. 29, 2131–2145 (2010). [Google Scholar]
- 35.Huntley B. et al., Explaining patterns of avian diversity and endemicity: Climate and biomes of southern Africa over the last 140,000 years. J. Biogeogr. 43, 874–886 (2016). [Google Scholar]
- 36.Linder H. P., The radiation of the Cape flora, southern Africa. Biol. Rev. Camb. Philos. Soc. 78, 597–638 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Kreft H., Jetz W., Global patterns and determinants of vascular plant diversity. Proc. Natl. Acad. Sci. U.S.A. 104, 5925–5930 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linder H. P., Evolution of diversity: The Cape flora. Trends Plant Sci. 10, 536–541 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Mucina L., Rutherford M. C., The Vegetation of South Africa, Lesotho and Swaziland, (Strelitzia 19, South African National Biodiversity Institute, 2006). [Google Scholar]
- 40.Bradshaw P. L., Colville J. F., Linder H. P., Optimising regionalisation techniques: Identifying centres of endemism in the extraordinarily endemic-rich Cape Floristic Region. PLoS One 10, e0132538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jansson R., Dynesius M., The fate of clades in a world of recurrent climatic change: Milankovitch oscillations and evolution. Annu. Rev. Ecol. Syst. 33, 741–777 (2002). [Google Scholar]
- 42.Cowling R. M., Lombard A. T., Heterogeneity, speciation/extinction history and climate: Explaining regional plant diversity patterns in the Cape Floristic Region. Divers. Distrib. 8, 163–179 (2002). [Google Scholar]
- 43.Dobrovolski R., Melo A. S., Cassemiro F. A. S., Diniz-Filho J. A. F., Climatic history and dispersal ability explain the relative importance of turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 21, 191–197 (2012). [Google Scholar]
- 44.Cowling R. M. et al., Describing a drowned ecosystem: Last Glacial Maximum vegetation reconstruction of the Palaeo-Agulhas Plain. Quat. Sci. Rev. 235, 105866 (2020). [Google Scholar]
- 45.Forest F. et al., Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445, 757–760 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Forest F., Colville J. F., Cowling R. M., “Evolutionary diversity patterns in the Cape flora of South Africa” in Phylogenetic Diversity: Applications and Challenges in Biodiversity Science, Scherson R. A., Faith D. P., Eds. (Springer, 2018), pp. 167–187. [Google Scholar]
- 47.Buerki S. et al., Contrasting biogeographic and diversification patterns in two Mediterranean-type ecosystems. PLoS One 7, e39377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marean C. W. et al., “Stone Age people in a changing South African Greater Cape Floristic Region” in Fynbos: Ecology, Evolution, and Conservation of a Megadiverse Region, Allsopp N., Colville J. F., Verboom G. A., Eds. (Oxford University Press, 2014), pp. 164–199. [Google Scholar]
- 49.Verboom G. A., Linder H. P., Hoffmann V., Bergh N. G., Cowling R. M., “Cenozoic assembly of the Greater Cape flora” in Fynbos: Ecology, Evolution, and Conservation of a Megadiverse Region, Allsopp N., Colville J. F., Verboom G. A., Eds. (Oxford University Press, 2014), pp. 93–118. [Google Scholar]
- 50.Braun K. et al., A climate and environment record dating between ∼444 and 41 ka from Pinnacle Point (South Coast, South Africa) as derived from speleothem stable isotopic compositions. Quat. Res. 91, 265–288 (2019). [Google Scholar]
- 51.Cowling R. M., Desmet P. G., Rundel P. W., Esler K. J., Extraordinary high regional-scale plant diversity in southern African arid lands: Subcontinental and global comparisons. Divers. Distrib. 4, 27–36 (1998). [Google Scholar]
- 52.South African National Biodiversity Institute, The Protea Atlas Project (1991). https://www.proteaatlas.org.za/. Deposited 16 April 2018.
- 53.Germishuizen G., Meyer N. L., Plants of Southern Africa: An Annotated Checklist, (Strelitzia 14, National Botanical Institute, Pretoria, South Africa, 2003). [Google Scholar]
- 54.South African National Biodiversity Institute , Botanical Database of Southern Africa (BODATSA) (2016). http://newposa.sanbi.org/. Accessed 3 May 2018.
- 55.South African National Biodiversity Institute , CREW/TSP/MSB/ISEP—Threatened Plant Localities Database (2020). http://redlist.sanbi.org/. Accessed 22 January 2018.
- 56.Raimondo D., The Red List of South African plants—A global first. S. Afr. J. Sci. 107, 1–2 (2011). [Google Scholar]
- 57.Mecenero S., Altwegg R., Colville J. F., Beale C. M., Roles of spatial scale and rarity on the relationship between butterfly species richness and human density in South Africa. PLoS One 10, e0124327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hengl T., Sierdsema H., Radović A., Dilo A., Spatial prediction of species’ distributions from occurrence-only records: Combining point pattern analysis, ENFA and regression-kriging. Ecol. Modell. 220, 3499–3511 (2009). [Google Scholar]
- 59.Schulze R. E., “South African atlas of climatology and agrohydrology” (Tech. Rep. WRC Report 1489/1/06, Water Research Commission, Pretoria, South Africa, 2007).
- 60.FAO/IIASA/ISRIC/ISSCAS/JRC, Harmonized World Soil Database (Version 1.2, 2012). http://www.fao.org/land-water/databases-and-software/hwsd/en/. Deposited 21 December 2016.
- 61.Goldblatt P., Manning J., Cape Plants: A Conspectus of the Cape Flora of South Africa, (Strelitzia 9, Missouri Botanical Garden, 2000). [Google Scholar]
- 62.Manning J. C., Goldblatt P., Plants of the Greater Cape Floristic Region 1: The Core Cape Flora, (South African National Biodiversity Institute, 2012). [Google Scholar]
- 63.R Core Team , R: A Language and Environment for Statistical Computing, (R Foundation for Statistical Computing, 2018). [Google Scholar]
- 64.Baddeley A., Rubak E., Turner R., Spatial Point Patterns: Methodology and Applications with R, (Chapman and Hall/CRC Press, 2015). [Google Scholar]
- 65.Pebesma E. J., Bivand R. S., Classes and methods for spatial data in R. R News 5, 9–13 (2005). [Google Scholar]
- 66.Bivand R. S., Pebesma E., Gomez-Rubio V., Applied Spatial Data Analysis with R, (Springer, ed. 2, 2013). [Google Scholar]
- 67.Bivand R., Keitt T., Rowlingson B., rgdal: Bindings for the “Geospatial” Data Abstraction Library (2018).
- 68.Graler B., Pebesma E., Heuvelink G., Spatio-temporal interpolation using gstat. R J. 8, 204–218, 10.32614/RJ-2016-014 (2016). [DOI] [Google Scholar]
- 69.Paradis E., Molecular dating of phylogenies by likelihood methods: A comparison of models and a new information criterion. Mol. Phylogenet. Evol. 67, 436–444 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Sanderson M. J., Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol. Biol. Evol. 19, 101–109 (2002). [DOI] [PubMed] [Google Scholar]
- 72.Kearse M. et al., Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verboom G. A. et al., Origin and diversification of the Greater Cape flora: Ancient species repository, hot-bed of recent radiation, or both? Mol. Phylogenet. Evol. 51, 44–53 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 76.Wilson A. J., O’Connell M. F. J., Brown B., Guinan C., Grehan J. C., Multiscale terrain analysis of multibeam bathymetry data for habitat mapping on the continental slope. Mar. Geod. 30, 3–35 (2007). [Google Scholar]
- 77.Running S. W., Mu Q., Zao M., Moreno A., User’s Guide MODIS Global Terrestrial Evapotranspiration (ET) Product (NASA MOD16A2/A3) NASA Earth Observing System MODIS Land Algorithm, (NASA, 2017). [Google Scholar]
- 78.Singarayer J. S., Valdes P. J., High-latitude climate sensitivity to ice-sheet forcing over the last 120 kyr. Quat. Sci. Rev. 29, 43–55 (2010). [Google Scholar]
- 79.Gordon C. et al., The simulation of SST, sea ice extents and ocean heat transports in a version of the Hadley Centre coupled model without flux adjustments. Clim. Dyn. 16, 147–168 (2000). [Google Scholar]
- 80.Hutchinson M., A New Objective Method for Spatial Interpolation of Meteorological Variables from Irregular Networks Applied to the Estimation of Monthly Mean Solar Radiation, Temperature, Precipitation and Windrun, (CSIRO Division of Water Resources, 1989). [Google Scholar]
- 81.New M. G., Hulme M., Jones P., Representing twentieth century space-time climate variability. Part 1: Development of a 1961–90 mean monthly terrestrial climatology. J. Clim. 12, 829–856 (1999). [Google Scholar]
- 82.Besag J., York J., Mollié A., A Bayesian image restoration with two applications in spatial statistics. Ann. Inst. Stat. Math. 43, 1–20 (1991). [Google Scholar]
- 83.Rue H., Martino S., Chopin N., Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. J. R. Stat. Soc. B 71, 319–392 (2009). [Google Scholar]
- 84.Lindgren F., Rue H., Bayesian spatial modelling with R-INLA. J. Stat. Softw. 63, 1–25 (2015). [Google Scholar]
- 85.Beale C. M., Lennon J. J., Yearsley J. M., Brewer M. J., Elston D. A., Regression analysis of spatial data. Ecol. Lett. 13, 246–264 (2010). [DOI] [PubMed] [Google Scholar]
- 86.Gelman A., et al. , Understanding predictive information criteria for Bayesian models. Stat. Comput. 24, 997–1016 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plant species and phylogenetic data are available from the published sources and online repositories listed in Materials and Methods and SI Appendix.