Significance
Here we describe and utilize a model of medulloblastoma, a malignancy accounting for 20% of all childhood brain cancers. We used iPS-derived neural stem cells with a familial mutation causing aberrant SHH signaling. We show that these cells, when transplanted into mouse cerebellum, form tumors that mimics SHH-driven medulloblastoma, demonstrating the development of cancer from healthy neural stem cells in vivo. Our results show that reprogramming of somatic cells carrying familial cancer mutations can be used to model the initiation and progression of childhood cancer.
Keywords: medulloblastoma, neural stem cells, disease model
Abstract
Medulloblastoma is the most common malignant brain tumor in children. Here we describe a medulloblastoma model using Induced pluripotent stem (iPS) cell-derived human neuroepithelial stem (NES) cells generated from a Gorlin syndrome patient carrying a germline mutation in the sonic hedgehog (SHH) receptor PTCH1. We found that Gorlin NES cells formed tumors in mouse cerebellum mimicking human medulloblastoma. Retransplantation of tumor-isolated NES (tNES) cells resulted in accelerated tumor formation, cells with reduced growth factor dependency, enhanced neurosphere formation in vitro, and increased sensitivity to Vismodegib. Using our model, we identified LGALS1 to be a GLI target gene that is up-regulated in both Gorlin tNES cells and SHH-subgroup of medulloblastoma patients. Taken together, we demonstrate that NES cells derived from Gorlin patients can be used as a resource to model medulloblastoma initiation and progression and to identify putative targets.
Medulloblastoma is the most common malignant childhood brain tumor. Molecular classification has identified key developmental signaling pathways regulating tumor development and segregate medulloblastoma into at least four subgroups: wingless (WNT), sonic hedgehog (SHH), group 3, and group 4 (1). The SHH-subgroup, where SHH-pathway is constitutively active, comprises about 30% of total medulloblastoma. Common drivers for this subgroup include mutations or deletions of negative regulators PTCH1 or suppressor of fused (SUFU), as well as activating mutations of smoothened (SMO), and gene amplifications of transcription factors GLI2 and MYCN (2).
Although current treatments have significantly improved survival of affected children, they often result in devastating side effects, such as cognitive deficits, endocrine disorders, and increased incidence of secondary cancers later in life (3), highlighting the importance of developing effective therapies that will not harm the healthy brain. To identify and test therapeutic targets against medulloblastoma, we need to develop models that mimic the initiation and progression of the disease. The limitations of disease modeling in nonhuman organisms drive solutions that include humanizing animals or creating cellular models that reliably mimic key processes in healthy and/or diseased humans. However, primary tumor cell lines established from surgically removed tumors represent an end point of tumor development when cells are already transformed and genetic rearrangements have taken place. Furthermore, tumor cell lines cultured in vitro are prone to genetic drift and the molecular diversity and the tumor heterogeneity seen in the original tumor is seldom recapitulated in tumor cell lines (4). To overcome these limitations, we took advantage of cellular reprogramming to establish healthy neural stem cells carrying a germline mutation known to activate the SHH signaling pathway. Induced pluripotent stem (iPS) cells generated by expression of reprogramming factors in skin fibroblasts have demonstrated a pluripotent phenotype similar to that of embryonic stem (ES) cells (5), thus patient-derived iPS cells create a renewable cell source to model human diseases (6). In addition, iPS cells and their derivatives mimic early stages of human development, making them an attractive system for studying early onset diseases such as childhood cancers that are thought to originate in stem or progenitor cells (7).
The central nervous system develops from a small number of highly plastic progenitors called neuroepithelial cells. These cells have been shown to be competent in generating granule cells, a major cell population in the cerebellum (8). Long-term self-renewing neuroepithelial-like stem cells (NES cells) have successfully been generated from human ES cells and iPS cells (9), with similar biological properties and gene expression patterns to neuroepithelial stem cells captured from the developing human hindbrain that give rise to cerebellum, pons, and medulla oblongata (10). NES cells maintain their stem cell properties even after long-term propagation in vitro, and upon removal of growth factors, these cells differentiate into functional neurons and glial cells with a hindbrain identity (9). We hypothesized that NES cells generated from reprogrammed noncancerous somatic cells carrying a medulloblastoma driver mutation may give rise to tumors when exposed to a permissive environment. To test this, we used iPS cell technology to derive a medulloblastoma model by creating NES cells from a Gorlin syndrome patient. Gorlin syndrome is an autosomal dominant syndrome caused by germline mutation in one allele of the PTCH1 gene. Gorlin syndrome patients can suffer from early onset nevoid basal cell carcinoma, jaw keratocyts, fibromas, and multiple developmental defects (11). Importantly, 5% of Gorlin syndrome patients develop pediatric medulloblastoma (12). Although germline mutations in PTCH1 are rare, they mimic common sporadic mutations in medulloblastoma. Our results show that Gorlin syndrome NES cells, but not NES cells from healthy individuals, model the development from noncancerous to cancerous, and form tumors in vivo that closely mimic human medulloblastoma. We further identify LGALS1 as a SHH target gene in medulloblastoma.
Results
Characterization of Neuroepithelial Stem Cells with Germline PTCH1 Mutation Derived from a Gorlin Syndrome Patient.
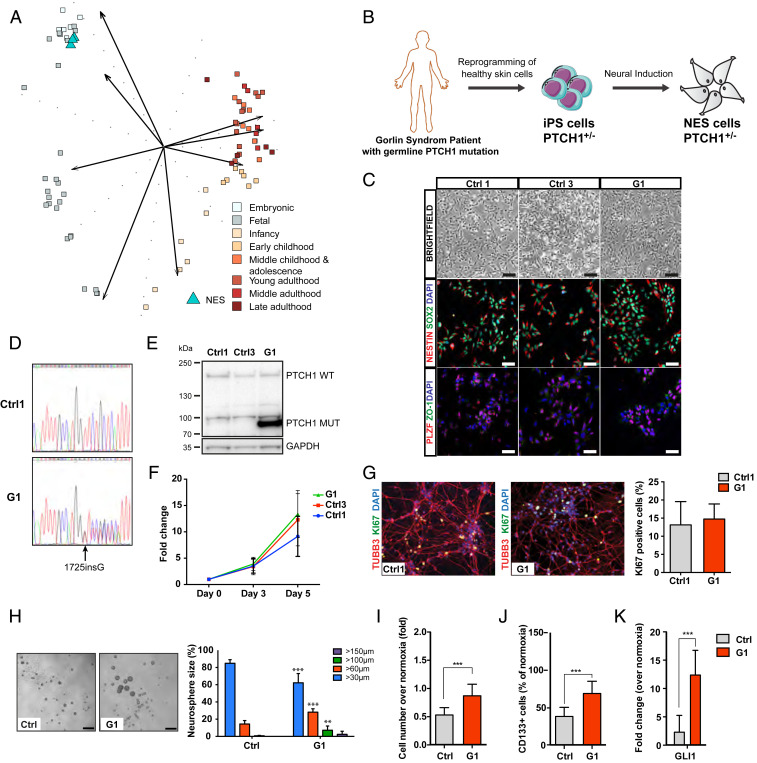
Animal models have shown that different subtypes of medulloblastoma arise from distinct neural stem cell or progenitor populations within the cerebellum, brainstem, or rhombic lip, and SHH-subgroup can develop from granular neural precursors (GNP) in the external granular layer or in multipotent neural stem cells when they commit to the granule neural lineage (13, 14). We derived NES cells from healthy iPS cells and compared NES cell gene expression profile with different stages of cerebellar development and found that NES cells closely resemble embryonic cerebellar cortex and upper rhombic lip, and fetal cerebellar cortex (Fig. 1A). In addition, NES cells express GNP markers ATOH1, MEIS1, and neural stem cell marker SOX3 (SI Appendix, Fig. S1A), indicating that NES cells represent a developmental stage when pediatric medulloblastoma arises and express GNP markers, and thus a suitable cell type for studying medulloblastoma onset and development. Next, we generated NES cells derived from noncancerous keratinocytes obtained from a Gorlin syndrome patient with a PTCH1 germline mutation 1762insG [Gorlin patient 1 (G1)], a frameshift mutation resulting in introduction of premature stop codon (V588GfsX39), leading to PTCH1 protein truncation (15–17). Patient and control iPS cells were generated by delivery of reprogramming factors, OCT4, SOX2, KLF4, and MYC into noncancerous somatic cells using nonintegrating Sendai virus and NES cells were derived by neural induction from control and G1 iPS cells (Fig. 1B), as previously described (9, 18, 19). NES cells derived from two healthy controls (Ctrl1 and Ctrl3) as well as the Gorlin patient (G1) represent a developmentally early neural stem cell population as reflected by their rosette-like organization and expression of neural stem cell markers SOX2, NESTIN, apical expression of neuroepithelial stem cell marker ZO-1, and rosette marker PLZF (Fig. 1C). The iPS reprogramming and NES cell generation did not cause any genetic abnormalities as control and G1 NES cells show a normal karyotype (SI Appendix, Fig. S1B). In addition, we detected both wild-type (WT) and mutant PTCH1 alleles in G1 NES, but only WT alleles in control NES (Fig. 1D). Western blot analysis shows the presence of both WT and truncated (Mut) PTCH1 protein in G1 NES (Fig. 1E). Intriguingly, we could not observe any significant up-regulation of SHH target genes expression in patient cells even though they express truncated PTCH1 protein (SI Appendix, Fig. S1C). This could indicate that the remaining wild-type allele of PTCH1 is still sufficiently blocking SHH signaling, or that the SHH-pathway is not active in proliferating monolayer NES cells. It has been reported that human iPS cells can acquire dominant-negative p53 mutations during reprogramming and expansion (20). To exclude that p53 mutations have been introduced during reprogramming to iPS cells or NES cell generation, we sequenced exons 4 to 10, where the majority of mutations in the TP53 gene occur (https://p53.iarc.fr/), and could not detect any mutations (SI Appendix, Fig. S1D). In accordance, we observed up-regulation of both p53 and p21 proteins upon cisplatin treatment (SI Appendix, Fig. S1E), demonstrating that the p53 protein is activated by DNA damage and is functional. Taken together, these data show that Gorlin syndrome NES cells are karyotypically and morphologically normal, retain the parental PTCH1 mutation, and express truncated PTCH1 protein.
Fig. 1.
Characterization of NES cells with germline PTCH1 mutation derived from a Gorlin syndrome patient. (A) Comparing transcriptome of normal NES cells with cerebellar developmental stages ranging from embryonic (combined cerebellar cortex and upper rhombic lip), and fetal to late adulthood (cerebellar cortex) using PCA. NES cells (n = 3) show strongest similarity to the embryonic group (4 ≤ Age < 8 post conceptual weeks). (B) Schematic showing generation of NES cells from G1 Gorlin syndrome patient. (C) NES cells organize in rosette-like structures (Top, Scale bar, 200 µm), expressed neural stem cell markers NESTIN and SOX2 (Middle), and rosette markers PLZF and ZO-1 (Bottom), (Scale bar, 100 µm.) (D) Sanger DNA sequencing showed both wild-type and mutated alleles of PTCH1 gene in G1 (1762insG) NES cells. (E) Western blot of both WT and truncated (Mut) forms of PTCH1 protein in G1 NES cells. (F) Proliferation rate of Ctrl1, Ctrl3, and G1 NES cells, evaluated by cell counting. Mean ± SD, n = 3 independent experiments. (G) After 14 d of differentiation, Ctrl1 and G1 NES cells differentiate to neurons as shown by TUBB3 staining. KI67-positive cells were quantified and normalized to 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI)-positive nuclei and presented as mean ± SD, n = 3 independent experiments, n.s, not significant. (H) G1 NES cells formed larger neurospheres compared to control NES cells, bar 250 µm. Histogram shows percentage distribution of neurospheres by size. Mean ± SD, n = 3 to 6 independent experiments, one-way ANOVA with Dunnett correction, ***P ≤ 0.001, **P ≤ 0.01. (I) G1 NES cells had higher cell numbers compared to control NES cells in hypoxia. Data are expressed as fold over cell number in normoxia. Mean ± SD, n = 3 independent experiments, ***P ≤ 0.001, Student t test. (J) CD133 Flow cytometry analysis showing that G1 NES cells had significantly higher number of CD133-positive cells compared to control NES cells in hypoxia. Data are expressed as percentage of CD133-positive cells out of live cells. Mean ± SD, n = 3 independent experiments, ***P ≤ 0.001, Student t test. (K) qRT-PCR analysis of GLI1 expression in hypoxia relative to normoxia. Data are shown as mean ± SD, n = 3 independent experiments. ***P ≤ 0.001, Student t test.
Gorlin NES Cells Form Larger Neurospheres and Are Able to Proliferate in Hypoxic Conditions.
Next, we examined the in vitro properties of the cells. We did not observe differences in the proliferation rate between G1 and control NES (Fig. 1F). NES cells are cultured in the presence of epidermal growth factor (EGF) and fibroblast growth factor 2 (FGF2); upon removal of growth factors cells exit the cell cycle in an unsynchronized manner and differentiate into mostly neurons. After 14 d in differentiating condition, all NES cell lines were able to differentiate toward a neuronal phenotype, as shown by their morphology and positive neuron-specific tubulin beta 3 class III (TUBB3) staining (Fig. 1G). In addition, we could not observe any difference in remaining KI67-positive cells between control and Gorlin NES cells (Fig. 1G), demonstrating that Gorlin NES cells are able to differentiate into postmitotic neurons similar to Controls. It has previously been shown that SHH-pathway activity is lost in cells grown as monolayer but maintained when grown as neurospheres (21, 47). To investigate the effect on neurosphere formation, cells were seeded on ultra-low adhesion plates at clonal density. Interestingly, G1 NES cells formed significantly larger neurospheres than control cells (Fig. 1H), suggesting that mutation in one PTCH1 allele gives the cells a growth advantage in a three-dimensional (3D) environment.
SHH has been shown to be induced by hypoxia in mouse neural progenitors and neurons, and hypoxia together with addition of SHH ligand induce proliferation of neural progenitors (22). To test the effect of hypoxia on NES cell proliferation, cells were cultured in 1% O2 for 4 d and counted. G1 NES cells showed increased cell numbers compared to Controls (Fig. 1I). We observed an increase in EdU (5-ethynyl-2'-deoxyuridine)-positive cells, but could not detect any significant difference in apoptosis, assessed by Annexin V/propidium iodide (PI) flow cytometry (SI Appendix, Fig. S1 F and G), demonstrating that the increase in cell numbers is due to increased cell proliferation and not reduced apoptosis in Gorlin NES compared to control NES cells. This was further confirmed by analysis of CD133 (PROM1) levels, as CD133 is expressed in actively proliferating human neural stem cells, and is down-regulated when cells exit the cell cycle (23). Flow cytometry analysis showed that a higher proportion of G1 NES cells remain CD133high in hypoxia compared to control NES cells (Fig. 1J), demonstrating that G1 NES cells are actively proliferating in hypoxia. In addition, we found increased messenger RNA (mRNA) expression of GLI1 in hypoxic Gorlin NES cells compared to control NES cells (Fig. 1K), suggesting that Gorlin NES cells activate SHH-pathway in hypoxia. Interestingly, we observed significant up-regulation of neuronal differentiation marker TUBB3 in Ctrl1 and Ctrl3 NES cells exposed to hypoxia. In contrast, Gorlin NES expressed similar TUBB3 mRNA levels as in normoxia (SI Appendix, Fig. S1H), suggesting that control NES cells exposed to hypoxic condition are exiting cell cycle and start to differentiate while Gorlin NES remain in a proliferative undifferentiated state. Taken together, NES cells derived from noncancerous somatic cells carrying one PTCH1 mutant allele do not show any overt phenotype in monolayer proliferative or differentiating conditions compared to control NES cells. However, the Gorlin NES cells have a growth advantage in 3D culture systems and hypoxia, conditions known to up-regulate SHH signaling.
Gorlin NES Cells Form Tumors In Vivo that Mimic Human SHH-Driven Medulloblastoma.
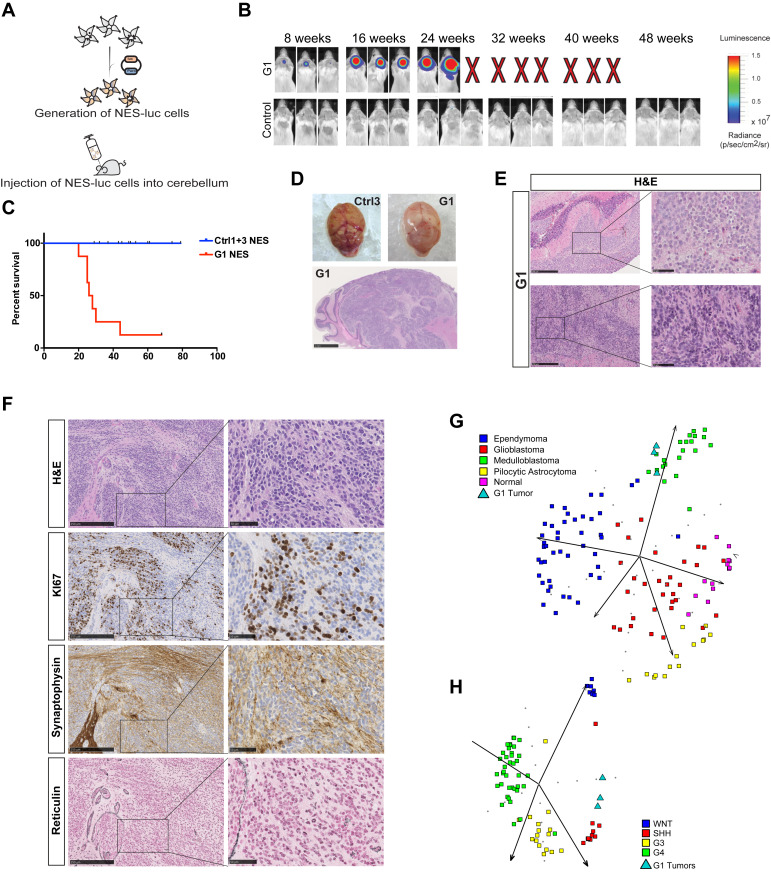
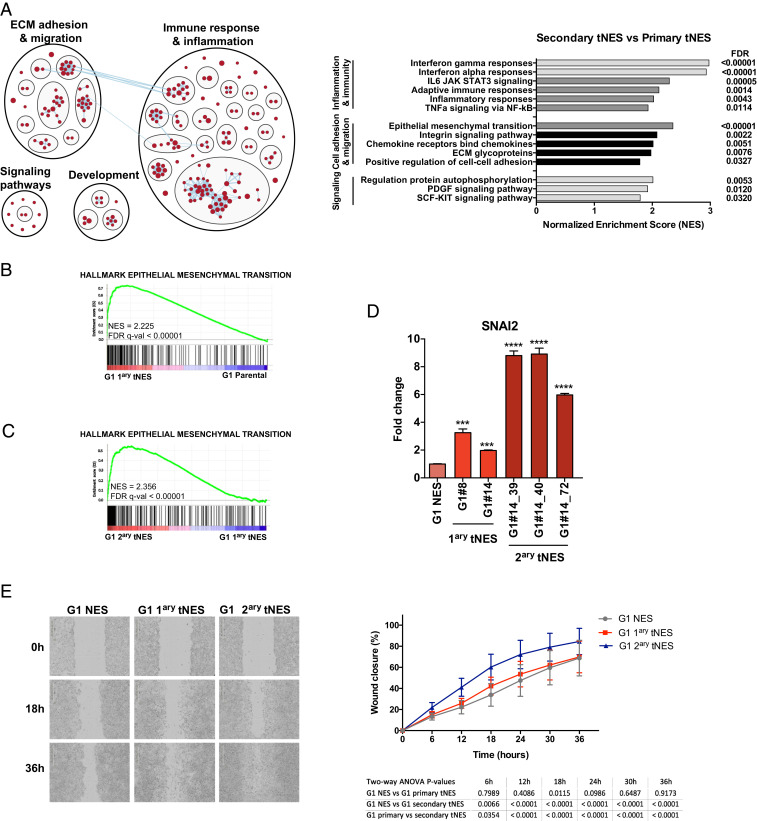
Next, we tested whether Gorlin NES cells could give rise to tumors in vivo. Cells were transduced with a firefly luciferase gene reporter (Luc) to allow in vivo tumor growth monitoring (Fig. 2A and SI Appendix, Fig. S2A). Both Gorlin and control NES cells were orthotopically transplanted into the cerebellum of immunodeficient mice and luciferase activity could be detected in mice injected with Gorlin NES cells 8 wk after injection with increasing signal intensity over time, while no luciferase activity was observed in mice injected with either Ctrl1luc or Ctrl3luc cells (Fig. 2B). Mice were killed when they exhibited physical deterioration, domed heads, and/or significant weight loss. The median survival of mice injected with G1luc NES cells were 27 wk (Fig. 2C), and we observed that 87% of mice injected with G1 cells developed tumors. Similar findings were observed with an additionally derived G1 NES cell line (SI Appendix, Fig. S2 A and B). Brains removed from mice injected with Gorlin NES cells revealed large mass on the cerebellum surface of the brain (Fig. 2D). Histological analysis showed tumors of mainly classical medulloblastoma histology with small dense nuclei (Fig. 2 E and F). Some tumors presented with a nodular appearance; however, we only observed limited reticulin-positive areas (Fig. 2F). The tumors were SYNAPTOPHYSIN (SYP) and KI67 positive, showing that they are of neuronal origin and actively proliferating in vivo (Fig. 2F). Furthermore, tumors stained positive for neural stem cell marker NESTIN, a common marker for all medulloblastoma subtypes (8), and the SHH-subgroup marker GAB1 (24), but only few cells positively stained for NEUN, a marker for more differentiated and mature neurons (SI Appendix, Fig. S2C). In addition, tumors cells were positive for anti-human nuclear antigen marker, showing that they are of human origin (SI Appendix, Fig. S2C). Human cells were successfully isolated from the tumors formed in the cerebellum of tumor-bearing mice injected with G1 NES cells (hereafter called primary tNES cells), and they could be cultured in neural stem cell media in vitro over many passages (SI Appendix, Fig. S2D). In contrast, no cell lines could be established from mice injected with either Ctrl1luc or Ctrl3luc NES cells. The primary G1 tNES cells expressed SOX2 and NESTIN, displayed NES-like rosette morphology, and had normal karyotype (SI Appendix, Fig. S2 D and E). We analyzed isolated tNES cells by RNA sequencing, and principal component analysis (PCA) clustered G1 primary tNES cells with human medulloblastoma tumors and not with other types of central nervous system (CNS) tumors (Fig. 2G). Further analysis revealed that primary tNES cells had a gene expression profile resembling the SHH-subgroup (Fig. 2H), demonstrating that Gorlin patient NES cells form tumors in vivo that mimic human SHH-driven medulloblastoma. To understand the biological changes occurring during tumor development, we analyzed the expression changes and found 1,138 genes to be significantly changed (false discovery rate [FDR] < 0.05), 740 genes were found to be up-regulated and 398 genes down-regulated in tumor cells compared to parental G1 NES cells (Dataset S1). Gene set enrichment analysis (GSEA) showed that extracellular matrix (ECM) adhesion and migration, metabolism, development, signaling pathways, and immune regulatory pathways were significantly up-regulated in tNES cells compared to parental NES cells (SI Appendix, Fig. S2F and Dataset S2). Among most significantly up-regulated genes in tNES compared to parental NES are genes regulating migration and invasion of cells (POSTN, TNC, EGFLAM, SNAI2) (Dataset S1).
Fig. 2.
Gorlin NES cells are able to form tumors mimicking medulloblastoma in vivo. (A) Schematic outline of NES cells transduced with luciferase reporter and transplanted in the cerebellum of mice. (B) Mice were imaged for bioluminescence. Representative images at 8 wk interval showed G1luc NES cells were able to proliferate while neither Ctrl1 nor Ctrl3 (shown as control) NES cells could. (C) Overall survival analysis of mice injected with Ctrl1luc or Ctrl3luc (combined as Controls, Ctrl1 n = 5, Ctrl3 n = 9), G1luc (n = 8). Kaplan-Meier curves depict differences in survival and statistical differences determined using the log rank test. G1luc P ≤ 0.0001. (D) Gross appearance of a tumor (Top Right) from G1luc NES injection compared to a brain injected with Ctrl3luc NES cells (Top Left). (H and E) staining of a tumor grown in the cerebellum (Bottom), (Scale bar, 1 mm.) (E) Representative hematoxylin and eosin (H&E) staining of tumors from Gorlin NES cells showing classical (Top) and a more nodular (Bottom) MB histology, (scale bar, 250 µm [Left] or 50 µm [Right]). (F) Cerebellar tumor sections stained for H&E, reticulin, neural marker synaptophysin, and proliferation marker KI67, (scale bar, 250 µm.) (G) PCA showing the spread of four different brain tumor types and normal brain based on metagene signatures. G1 primary tNES cells displayed the strongest similarity with medulloblastoma. (H) PCA showing that G1 primary tNES cells clustered with human SHH medulloblastoma in the spread of the four subgroups based on metagene signatures.
Isolated Tumor NES Cells Show Accelerated Tumor Formation In Vivo.
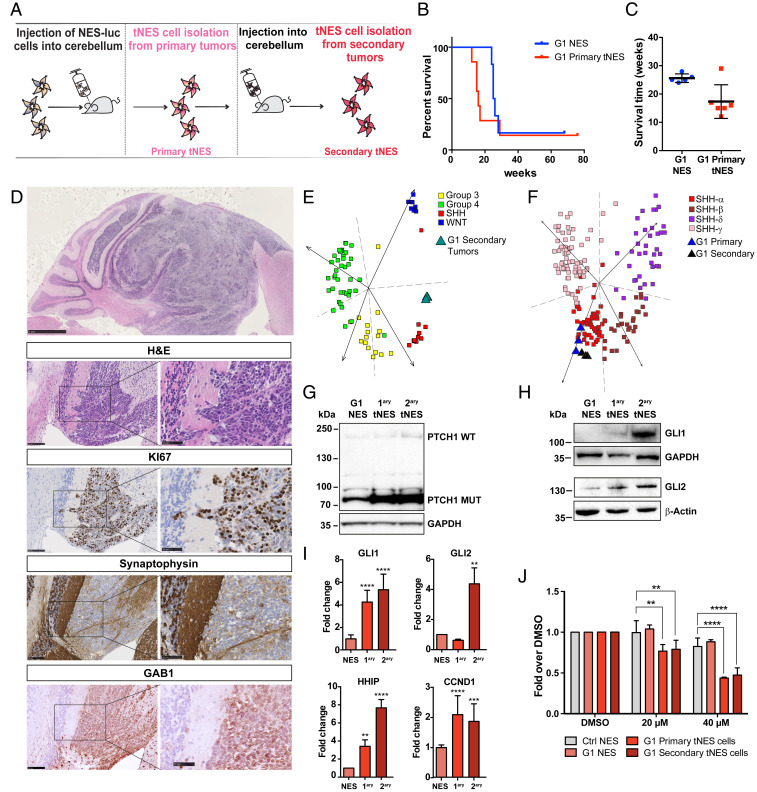
To investigate whether primary tNES cells have acquired features that can re-establish tumor growth, G1 primary tNES cells were retransplanted into cerebellum of immunodeficient mice (Fig. 3A). We observed faster tumor formation compared to the parental NES line, with a median survival of 16 wk (Fig. 3 B and C). Histologically the tumors resembled the primary tumors, with highly proliferative areas that were positive for KI67, GAB1, and SYP (Fig. 3D). Human cells were isolated from these secondary tumors, to establish NESTIN and SOX2 positive secondary tNES cell lines (SI Appendix, Fig. S3A). Again, gene expression profile analysis of isolated G1 secondary tNES cells clustered them together with human SHH-subgroup medulloblastoma (Fig. 3E). In addition, both primary and secondary tNES clustered with the SHH-α subgroup (Fig. 3F), the most prevalent SHH-subgroup in children aged 3 to 16 (25). In patients, PTCH1 mutations are often, but not always, found with somatic loss of heterozygosity (LOH) of the remaining wild-type allele (26–28). We did not observe PTCH1 LOH in either primary or secondary tNES (Fig. 3G). However, mutant PTCH1 protein was expressed at higher levels than the wild-type protein in parental G1 NES (Figs. 1E and 3G), with further increase in primary and secondary tNES (Fig. 3G and SI Appendix, Fig. S3B). In addition, both GLI1 and GLI2 protein and mRNA levels gradually increased from parental NES to secondary tNES (Fig. 3 H and I), as well as GLI target genes HHIP and CCND1 (Fig. 3I).
Fig. 3.
Isolated tumor cells show accelerated tumor formation and tumor progression in vivo. (A) Schematic overview of isolation and reinjection of tNES cells forming tumors in secondary hosts. (B) Shortened tumor onset was observed for mice injected with primary tNES (G1luc#14, n = 7) cells compared to those that were injected with the parental line, G1luc (n = 6). (C) Histogram showing significant reduced time of tumor onset in tumor-bearing mice injected with primary tNES compared to parental G1 NES. *P ≤ 0.05, Student t test. (D, Top) Representative H&E staining of tumors arising from orthotopic transplantation of primary tNES cells, (scale bar, 1 mm). (Lower) Cerebellar tumor sections were stained for H&E, proliferation marker KI67, neural marker synaptophysin, and SHH-subgroup marker GAB1, (Left) (scale bar, 100 µm), (Right) (scale bar, 50 µm). (E) PCA showing G1 secondary tNES cells grouped with human SHH-subgroup in the spread of the four medulloblastoma subtypes based on metagene signatures. (F) PCA showing G1 primary and secondary tNES grouped with human SHH-alpha subgroup in the spread of the four SHH subtypes based on metagene signatures. (G) Western blot of both WT and truncated (Mut) forms of PTCH1 protein in G1 NES and tNES cells. (H) Western blot of GLI1 and GLI2 protein expression in G1 NES and tNES cells (G1, G1luc#14, G1luc#14_40). (I) Gene expression analysis using qRT-PCR showing relative fold mRNA expression of GLI1, GLI2, HHIP1, CCND1 in G1 NES and tNES cells. Data are shown as mean ± SD, n = 3 biological replicates, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, Student t test. (J) Resazurin assay showing decreased viability in primary and secondary G1 tNES cells on Vismodegib treatment. Mean ± SD, n = 3 independent experiments, **P ≤ 0.01, ****P ≤ 0.0001, two-way ANOVA with Dunnett correction.
Next, we treated cells with Vismodegib, a SMO inhibitor that has shown positive results in a Phase II clinical trial for adult recurrent SHH-subgroup medulloblastoma (29). Vismodegib treatment decreased viability of both G1 primary and secondary tNES cells compared to G1 parental and control NES cells (Fig. 3J), which may suggest that the tNES cells have become dependent on an active SHH-pathway.
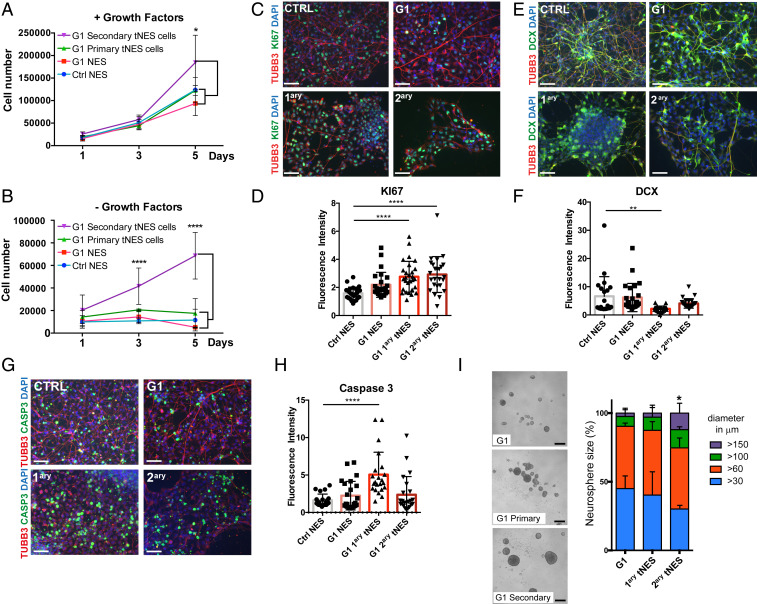
Further characterization of isolated tNES showed increased proliferation rate in secondary tNES cells compared to primary tNES, parental G1, and control NES cells (Fig. 4A). Removal of growth factors from the media should induce growth arrest and differentiation. Interestingly, G1 secondary tNES cells continued to proliferate in media without EGF and FGF2 (Fig. 4B), suggesting that they have a reduced dependence on growth factors. To further investigate the underlying cellular mechanism for the continued growth in absence of growth factors we analyzed the cells for proliferation, differentiation, and apoptosis markers. We observed an increase of secondary tNES cells remaining KI67 positive after removal of growth factors (Fig. 4 C and D), together with a decrease of the neuronal differentiation marker doublecortin (DCX) (Fig. 4 E and F), suggesting that the tNES cells have a reduced ability to exit cell cycle and differentiate to neurons. Surprisingly, we similarly observed up-regulation of KI67-positive cells and down-regulation in DCX-positive cells in G1 primary tNES during differentiation (Fig. 4 C–F), which did not correspond to an increase in cell numbers (Fig. 4B); however, we found a significant increase in cleaved Caspase 3-positive cells in G1 primary tNES cells, compared to parental and G1 secondary tNES cells (Fig. 4 G and H), suggesting enhanced apoptosis in primary G1 tNES cells when cultured without growth factors EGF and FGF2. Furthermore, G1 secondary tNES cells formed significantly larger neurospheres compared to G1 parental and primary tNES cells (Fig. 4I). Taken together, our data suggest that secondary G1 tNES cells have acquired features of classical hallmarks of cellular transformation, such as continued proliferation and evading differentiation and apoptosis cues.
Fig. 4.
Gorlin tNES cells show an increase in proliferation and a decrease in neural differentiation. (A and B) Proliferation of Ctrl3 NES, G1 NES, G1 primary tNES cells (G1luc#14), and G1 secondary tNES cells (G1luc#14_40) in the presence or absence of EGF and FGF2 for 5 d was analyzed by cell counting. Mean ± SD, n = 3 independent experiments, *P ≤ 0.05, ****P ≤ 0.0001, two-way ANOVA with Tukey correction. (C) Immunofluorescence analysis of KI67 (green) expressing cells after 7 d in differentiating condition, (Scale bar, 50 μm.) (D) Quantification of KI67-positive signal measured by fluorescence intensity normalized to DAPI-positive cells. Mean ± SD, ****P ≤ 0.0001, Student t test. (E) Immunofluorescence analysis of DCX (green) expressing cells after 7 d in differentiating condition, (Scale bar, 50 μm.) (F) Quantification of DCX-positive signal measured by fluorescence intensity normalized to DAPI-positive cells. Mean ± SD, **P ≤ 0.01, Student t test. (G) Immunofluorescence analysis of cleaved Caspase 3 (green) expressing cells after 7 d in differentiating condition, (scale bar, 50 μm.) (H) Quantification of cleaved Caspase 3-positive signal measured by fluorescence intensity normalized to DAPI-positive cells. Mean ± SD, ****P ≤ 0.0001 Student t test. (I) G1 primary (G1luc#14) and secondary tNES cells (G1luc#14_40) formed larger neurospheres compared to the parental G1 NES cells after 6 d in culture, (scale bar, 250 µm). Histogram show percentage distribution of neurospheres by sizes of parental, primary, and secondary tumor cells, n = 4 to 6 independent experiments, *P ≤ 0.05 for secondary tumor cells neurospheres >150 µm diameter compared to parental cells, one-way ANOVA with Dunnett correction.
Progressive Activation of Inflammatory and Metastatic Pathways during Medulloblastoma Development.
Comparison of gene expression changes between G1 primary and secondary tNES cells showed 1955 genes to be significantly changed (FDR < 0.05), with 1,214 genes up-regulated and 741 genes down-regulated in G1 secondary tNES cells (Dataset S3). GSEA showed a significant enrichment of mainly inflammatory and ECM adhesion and migration pathways in G1 secondary tNES compared to primary G1 tNES (Fig. 5A and Dataset S4). Comparing parental NES with primary tNES (Fig. 5B) and primary tNES with secondary tNES (Fig. 5C), we observed a progressive up-regulation of genes regulating Epithelial-to-Mesenchymal transition (EMT), migration and invasion (SNAI2, BST2, TNC), as well as genes involved in both inflammatory responses and EMT (PLAUR, OSMR, LGALS1, and LGALS3) (Datasets S3 and S4). In addition, we found a significant enrichment of genes involved in focal and cell adhesion, as well as extracellular matrix components, including Collagens (COL1A1, COL6A2, COL11A1), Integrins (ITGA8, ITGA11), and Laminins (LAMA2, LAMC3) (SI Appendix, Fig. S3C), all of which are found highly expressed in human SHH-subgroup medulloblastoma patients (SI Appendix, Fig. S3D). Therefore, we tested whether the secondary tNES cells acquired higher cell migration and invasion abilities. First, we confirmed increasing expression of SNAI2 in G1 secondary tNES compared to primary tNES and parental NES (Fig. 5D). Next, using wound closure assay we found that G1 secondary tNES cells have higher migration rates compared to both primary tNES and parental cells (Fig. 5E). Furthermore, secondary tNES was found to be more invasive as determined by the cells’ ability to invade through a layer of basement membrane proteins in transwell chamber invasion assay (SI Appendix, Fig. S3F). Taken together, these results demonstrate that secondary tNES have acquired a more aggressive phenotype.
Fig. 5.
Progressive activation of inflammatory and migratory pathways during tumor development. (A, Left) Enrichment map generated by Broad Institute GSEA tool (FDR < 0.05) and visualized by Cytoscape EnrichmentMap and AutoAnnotate application, showing biological pathways enriched in G1 secondary tNES cells compared to primary NES cells. Red nodes represent up-regulated biological pathways. (Right) Histogram of most significantly deregulated pathways in G1 secondary tNES compared to primary G1 tNES. (B) Enrichment plot showing over-representation of genes involved in EMT in G1 primary tNES compared to G1 parental NES. (C) Enrichment plot showing over-representation of genes involved in EMT in G1 secondary tNES tumors compared to G1 primary tNES. (D) Gene expression analysis by qRT-PCR showing increasing levels of SLUG (SNAI2) in G1 primary and secondary tNES compared to parental NES cells. Data are shown as mean ± SD, n = 3 biological replicates, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, Student t test. (E) Increased migration rate of G1 secondary tNES cells (G1luc#14_40) compared to primary tNES (G1luc#14) and parental G1 NES cells was observed in wound closure assay. Mean ± SD, combining three independent experiments. Statistical analysis by two-way ANOVA with Tukey correction is presented in a table below the graph.
GALECTIN-1 Is Highly Expressed in SHH-Subgroup Medulloblastoma and Is a GLI-Target Gene.
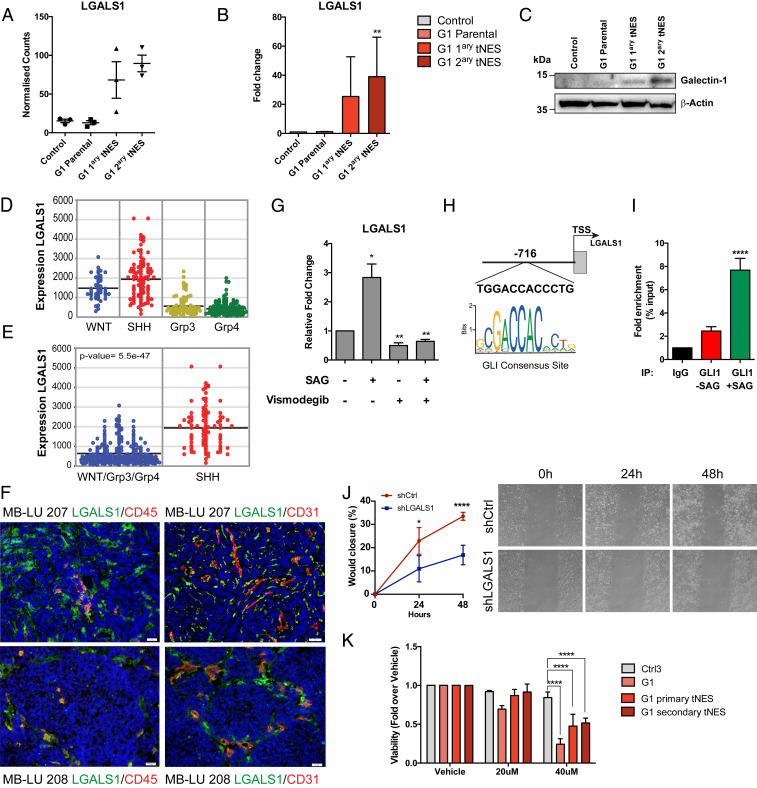
The tumor microenvironment plays a fundamental role in tumor progression and therapy responses. Interestingly, higher expression of inflammatory-related genes have been found in SHH-subroup medulloblastoma patients compared to other subgroups (30), similarly to what we observe in our model. We identified LGALS1 as a gene up-regulated in both G1 primary and secondary tNES cells compared to parental G1 NES (Fig. 6A), In addition, LGALS1 is a gene commonly found in both inflammatory and ECM/EMT enriched pathways in secondary G1 tNES (Fig. 5A and Dataset S4). LGALS1 encodes GALECTIN-1 (GAL-1), a beta-galactoside binding lectin that has been correlated with poor prognosis in many cancer types (reviewed in ref. 31). GAL-1 has been shown to increase migration, invasion, immune evasion, and chemoresistance of glioblastoma and neuroblastoma cells (32–34); however, its role in medulloblastoma development has not been studied. We confirmed the RNA sequencing data observing increasing levels of LGALS1 mRNA expression in G1 primary and secondary tNES cells (Fig. 6B); in addition, we observed increasing GAL-1 protein levels in secondary tNES (Fig. 6C). Next, we examined array data from human medulloblastoma patients (27, 35–37) to investigate LGALS1 expression levels in different subgroups of human medulloblastoma (Fig. 6D). LGALS1 has significantly higher expression in human SHH-subgroup medulloblastoma samples compared to the other three subgroups (Fig. 6E), confirming a positive correlation in the expression of LGALS1 between our model and human patients. Using immunohistochemistry, we found that GAL-1 was expressed in human SHH-subgroup tumor samples (Fig. 6F). In addition to tumor cells, we also found cells that stained positive for both GAL-1 and the endothelial cell marker CD31, or the immune marker CD45 (Fig. 6F), indicating that many GAL-1 expressing cells are stromal or immune cells and not only tumor cells. Our data suggest a connection between the SHH-pathway and LGALS1 expression. This was further supported by finding a significant positive correlation between LGALS1 expression and SMO or GLI1 (SI Appendix, Fig. S4 A and B) in cohorts of human medulloblastoma. To further test if LGALS1 is directly activated by the SHH-pathway, we treated the medulloblastoma cell line, DAOY, with smoothened agonist (SAG) and observed a significant up-regulation of LGALS1 that was reversed on Vismodegib treatment, again suggesting a direct link between the SHH-pathway and LGALS1 expression (Fig. 6G). Therefore, we scanned the LGALS1 gene promoter region and found a potential GLI consensus site located 716 nt from the transcription start site (Fig. 6H and SI Appendix, Fig. S4C). Using chromatin immunoprecipitation, we found GLI1 binding to this region and that the binding increased on SAG treatment (Fig. 6I). Furthermore, we also detected GLI2 binding to this region (SI Appendix, Fig. S4D), albeit at lower levels, and that activation of SHH-pathway resulted in a significant increase of the transcriptional activation marker H3K9Ac at the GLI consensus site (SI Appendix, Fig. S4E), altogether demonstrating that LGALS1 is a GLI target gene.
Fig. 6.
GALECTIN-1 is up-regulated in SHH medulloblastoma and a direct GLI target gene. (A) RNA sequencing results showed increased expression of LGALS1 in G1 primary and secondary tNES cells compared to parental G1 NES cells. Mean ± SD, n = 3 biological replicates. (B) Confirmation of the increased expression of LGALS1 was performed using qRT-PCR. Mean ± SD, n = 3 biological replicates. **P ≤ 0.01, Student t test compared to control NES. (C) Western blot of GALECTIN-1 protein expression in G1 NES and tNES cells. (D) Expression of LGALS1 in human samples of WNT, SHH, group 3 (Grp3), and group 4 (Grp4) medulloblastoma. (E) Expression of LGALS1 in human SHH-subgroup was significantly higher than in the other three medulloblastoma subgroups. (F) Immunofluorescence analysis of GALECTIN-1 (green), CD31 (red), and CD45 (red) expression, in human SHH medulloblastoma samples, (Scale bar, 20 µm). (G) Analysis of LGALS1 mRNA expression levels by qRT-PCR of DAOY cells treated with vehicle, SAG, or Vismodegib alone, or in combination. Data are shown as mean ± SD, n = 3 independent experiments, *P ≤ 0.05, **P ≤ 0.01, Student t test. (H) Schematic of LGALS1 promoter showing putative GLI-binding site. (I) GLI1 chromatin immunoprecipitation (ChIP) of DAOY cells treated with vehicle or SAG and qPCR analysis using primers flanking GLI-bindings site in LGALS1 promoter. Mean ± SD, n = 3 independent experiments, ****P ≤ 0.0001, Student t test. (J) shRNA-mediated knockdown of LGALS1 results in decreased migration rate of secondary tNES cells (G1luc#14_40) compared to shCtrl. Mean ± SD, showing one representative experiment out of three independent experiments. (K) Treatment of G1 parental, G1luc#14 primary and G1luc#14_40 secondary tNES cells with OTX008 for 72 h and measuring viability using Resazurin assay. Data are shown as mean ± SD, n = 3 independent experiments, ****P ≤ 0.0001, two-way ANOVA with Dunnett correction.
Considering GAL-1 has been shown to increase cell migration, we tested the effect of small hairpin RNA (shRNA)-mediated LGALS1 inhibition (SI Appendix, Fig. S4F) on secondary tNES migration and found that down-regulation of LGALS1 reduced the migration capacity of secondary tNES cells (Fig. 6J). To further test the effect of GAL-1 inhibition on Gorlin tNES cells, we treated cells with an allosteric GAL-1 inhibitor, OTX008 (38) and found that inhibition of GAL-1 significantly suppressed cell viability of not only G1 tNES cells but also parental Gorlin NES cells compared to control NES cells (Fig. 6K). We also observed a down-regulation in viability in the Vismodegib-resistant medulloblastoma cell lines DAOY and UW228-3 (39) on treatment with OTX008, although at high concentrations (SI Appendix, Fig. S4 G and H). Taken together, we show here that healthy neural stem cells derived from Gorlin syndrome patients carrying germline PTCH1 mutations can be used to model onset and progression of SHH-subgroup medulloblastoma. Furthermore, we demonstrate that this model can be used to identify SHH-pathway target genes and have the potential to be further explored to identify therapeutic targets for SHH-subgroup medulloblastoma patients.
Discussion
Disease models are useful in understanding the initiation and progression of specific diseases and to help identify and test relevant therapeutic targets. Murine models have greatly contributed to elucidating the role of SHH signaling in medulloblastoma pathogenesis [reviewed in (40)]. However, species variability between mouse and human may lead to unsuccessful translation of successful treatment in mouse models into cure for human disease (41, 42). Cellular reprogramming techniques have made it possible to create models for diseases with scarce sources of human cells. The advantages of our model are that the “renewable” NES cells express mutant PTCH1 at normal pathophysiological level and they are transcriptionally similar to cerebellum during embryonic and fetal development stages, which are also the age groups when medulloblastoma is thought to originate (2). By using normal cells carrying a germline mutation as a cellular source instead of using cancer cell lines derived from an established tumor that carries many mutations, it is possible to follow the stepwise tumor development resulting from the original mutation.
SHH signaling is important during normal development of the cerebellum and aberrant SHH signaling promotes transformation of granule neuron precursor (GNP) cells and tumor formation within the cerebellar hemisphere (43). Previous studies have shown that in order for medulloblastoma to develop, SHH needs to be activated in GNP cells or a neural stem cells committed to a granular cellular lineage (13, 14). However, it has been shown that quiescent SOX2+ mouse neural stem cells can recapitulate different stages of stem cell hierarchy to drive relapse of SHH medulloblastoma in mice, suggesting that medulloblastoma can be derived from a more immature neural stem cell than a cell already committed to the granular cellular lineage (44). In this study, we show that patient-derived NES cells, an immature neural stem cell type, are capable of generating tumors that histologically and transcriptionally mimic human SHH-subgroup medulloblastoma. One possible explanation could be that once injected into the cerebellar microenvironment, a permissive niche is generated where the Gorlin NES cells aberrantly activate the SHH-pathway, triggering proliferation and tumor development. Another explanation could be that the lower oxygen level inside the cerebellum allows the Gorlin NES cells to maintain their progenitor identity and to continue to proliferate and not differentiate as we found when exposing cells to hypoxic condition.
We observed up-regulation of several inflammatory pathways during both primary and secondary tumor development, indicating again the importance of the crosstalk between the patient NES cells and the surrounding tumor microenvironment. It has been reported that the SHH medulloblastoma (MB) subgroup has an inflammatory signature that coincides with increased infiltration of tumor-associated macrophages that further fuel tumor development (30). In addition, inducible production of interferon gamma (IFNγ) in the developing brain activates SHH signaling in GNP cells, leading to an autocrine loop supporting proliferation and resulting in hyperplasia of the external germinal layer in the cerebellum (45). The importance of the microenvironment was also demonstrated in a glioma mouse model where SHH signaling was dependent on the cross talk between tumor cells and neighboring astrocytes (46). Taken together, these data show the close relationship between SHH signaling and inflammatory responses, which was also observed in our model.
The increased ability of secondary tumor-derived cells to migrate and invade in vitro as well as up-regulation of EMT-related genes in these cells suggest that these cells have attained a more aggressive phenotype and may have an increased ability to metastasize. Several of the genes found up-regulated in the secondary tumors, including LGALS1, have key roles in both inflammation and metastasis, but have not previously been reported to play a role in medulloblastoma development. Considering GAL-1 is expressed by infiltrating immune cells, stromal cells, and tumor cells, inhibiting GAL-1 may be a double-edged sword that will target both the tumor cells as well as the tumor stroma. Here we show that LGALS1 mRNA is up-regulated by activation of the SHH-pathway, that the LGALS1 gene is a direct target of GLI proteins, and that inhibition of LGALS1 inhibit secondary tNES migration, suggesting that GAL-1 may play a role in medulloblastoma development and should be further studied. However, for this more specific inhibitors will need to be developed that can cross the blood-brain-barrier and are active at lower concentrations to fully evaluate its potential as a therapeutic target in medulloblastoma.
In conclusion, we demonstrate that human neural stem cells generated from reprogrammed noncancerous somatic cells carrying a PTCH1 germline mutation can faithfully model SHH-driven medulloblastoma creating a valuable resource for studying both medulloblastoma initiation and progression. In addition, Gorlin NES and tNES cells can be used as tools for screening for therapeutic targets. Another advantage of using NES cells as screening tools is that both proliferating NES and differentiated neurons from the same patients could be included in early-stage screens to assess neurotoxicity or to define the therapeutic window for putative drugs.
Material and Methods
All animal experiments were conducted in accordance with guidelines of Karolinska Institutet and approved by Stockholm’s North Ethical Committee of Animal Research. Detailed information about experimental design including iPS and NES cell culture conditions, orthotopic injections, proliferation and differentiation assays, oligonucleotide sequences, antibodies, bioinformatic analyses, and statistical methods can be found in SI Appendix.
Material and Data Availability.
Data has been deposited at Mendeley, DOI: 10.17632/syhdb33jpt.1. RNA sequencing data have been deposited at Gene Expression Omnibus (GEO) with accession number GSE106718.
Supplementary Material
Acknowledgments
We thank Professor Austin Smith, Cambridge, for providing G1 iPS cells; Professor David Lane’s and Dr. Sonia Lain’s laboratories for technical assistance; especially Dr. Nicolas Fritz and Dr. Gergana Popova; Professor Marie Arsenian Henriksson’s laboratory for helpful discussions; and the iPS core facility at Karolinska Institutet. This work was supported by grants from the Swedish Childhood Cancer Foundation (PR2018-0133, NCP2016-0022, PR2014-0046), the Swedish Cancer Society (CAN2016/823, CAN2014/864), and the Swedish Research Council (2016/00753). E.S. was supported by a postdoctoral fellowship from the Swedish Childhood Cancer Society (TJ2015-0067). M.W. is supported by a Young Investigator Award from the Swedish Cancer Society (CAN2012/1330). A.F. is supported by the Swedish Foundation for Strategic Research (SSF). T.M. was supported by the French Government (National Research Agency, ANR; CNRS; INSERM) through the “Investments for the Future” LABEX SIGNALIFE: program reference No. ANR-11-LABX-0028-01, UCA (Université Côte d’Azur), and by the Fondation ARC (SFI201212055859), the Fondation del’Avenir, Société Française de Dermatologie, and The Institut National du Cancer.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Data has been deposited at Mendeley, DOI: 10.17632/syhdb33jpt.1. RNA sequencing data have been deposited at Gene Expression Omnibus (GEO) with accession number GSE106718.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920521117/-/DCSupplemental.
References
- 1.Northcott P. A. et al., Medulloblastomics: The end of the beginning. Nat. Rev. Cancer 12, 818–834 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kool M. et al., Molecular subgroups of medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, group 3, and group 4 medulloblastomas. Acta Neuropathol. 123, 473–484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald T. J., Aguilera D., Castellino R. C., The rationale for targeted therapies in medulloblastoma. Neuro-oncol. 16, 9–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivanov D. P., Coyle B., Walker D. A., Grabowska A. M., In vitro models of medulloblastoma: Choosing the right tool for the job. J. Biotechnol. 236, 10–25 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K. et al., Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Lee D. F. et al., Modeling familial cancer with induced pluripotent stem cells. Cell 161, 240–254 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marin Navarro A., Susanto E., Falk A., Wilhelm M., Modeling cancer using patient-derived induced pluripotent stem cells to understand development of childhood malignancies. Cell Death Discov. 4, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbertson R. J., Ellison D. W., The origins of medulloblastoma subtypes. Annu. Rev. Pathol. 3, 341–365 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Falk A. et al., Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS One 7, e29597 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tailor J. et al., Stem cells expanded from the human embryonic hindbrain stably retain regional specification and high neurogenic potency. J. Neurosci. 33, 12407–12422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo Muzio L., Nevoid basal cell carcinoma syndrome (Gorlin syndrome). Orphanet J. Rare Dis. 3, 32 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amlashi S. F., Riffaud L., Brassier G., Morandi X., Nevoid basal cell carcinoma syndrome: Relation with desmoplastic medulloblastoma in infancy. A population-based study and review of the literature. Cancer 98, 618–624 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Yang Z. J. et al., Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 14, 135–145 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schüller U. et al., Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14, 123–134 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brellier F. et al., Ultraviolet responses of Gorlin syndrome primary skin cells. Br. J. Dermatol. 159, 445–452 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Valin A. et al., PTCH1 +/- dermal fibroblasts isolated from healthy skin of Gorlin syndrome patients exhibit features of carcinoma associated fibroblasts. PLoS One 4, e4818 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright A. T. et al., Deficient expression of aldehyde dehydrogenase 1A1 is consistent with increased sensitivity of Gorlin syndrome patients to radiation carcinogenesis. Mol. Carcinog. 54, 473–484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shahsavani M. et al., An in vitro model of lissencephaly: Expanding the role of DCX during neurogenesis. Mol. Psychiatry 23, 1674–1684 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Huang M. et al., Engineering genetic predisposition in human neuroepithelial stem cells recapitulates medulloblastoma tumorigenesis. Cell Stem Cell 25, 433–446.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merkle F. T. et al., Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 545, 229–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasai K. et al., Shh pathway activity is down-regulated in cultured medulloblastoma cells: Implications for preclinical studies. Cancer Res. 66, 4215–4222 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Sims J. R. et al., Sonic hedgehog regulates ischemia/hypoxia-induced neural progenitor proliferation. Stroke 40, 3618–3626 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y. et al., CD133 (Prominin) negative human neural stem cells are clonogenic and tripotent. PLoS One 4, e5498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellison D. W. et al., Medulloblastoma: Clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 121, 381–396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavalli F. M. G. et al., Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31, 737–754.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raffel C. et al., Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 57, 842–845 (1997). [PubMed] [Google Scholar]
- 27.Northcott P. A. et al., The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waszak S. M. et al., Spectrum and prevalence of genetic predisposition in medulloblastoma: A retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol. 19, 785–798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson G. W. et al., Vismodegib exerts targeted efficacy against recurrent sonic hedgehog-subgroup medulloblastoma: Results from Phase II pediatric brain tumor consortium studies PBTC-025B and PBTC-032. J. Clin. Oncol. 33, 2646–2654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margol A. S. et al., Tumor-associated macrophages in SHH subgroup of medulloblastomas. Clin. Cancer Res. 21, 1457–1465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou F. C., Chen H. Y., Kuo C. C., Sytwu H. K., Role of galectins in tumors and in clinical immunotherapy. Int. J. Mol. Sci. 19, 430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puchades M. et al., Proteomic investigation of glioblastoma cell lines treated with wild-type p53 and cytotoxic chemotherapy demonstrates an association between galectin-1 and p53 expression. J. Proteome Res. 6, 869–875 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Cimmino F. et al., Galectin-1 is a major effector of TrkB-mediated neuroblastoma aggressiveness. Oncogene 28, 2015–2023 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Gao J., Wang W., Knockdown of galectin-1 facilitated cisplatin sensitivity by inhibiting autophagy in neuroblastoma cells. Chem. Biol. Interact. 297, 50–56 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Kool M. et al., Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One 3, e3088 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fattet S. et al., Beta-catenin status in paediatric medulloblastomas: Correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J. Pathol. 218, 86–94 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Robinson G. et al., Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Astorgues-Xerri L. et al., OTX008, a selective small-molecule inhibitor of galectin-1, downregulates cancer cell proliferation, invasion and tumour angiogenesis. Eur. J. Cancer 50, 2463–2477 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Pambid M. R. et al., Overcoming resistance to Sonic Hedgehog inhibition by targeting p90 ribosomal S6 kinase in pediatric medulloblastoma. Pediatr. Blood Cancer 61, 107–115 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Wu X., Northcott P. A., Croul S., Taylor M. D., Mouse models of medulloblastoma. Chin. J. Cancer 30, 442–449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller J. A., Horvath S., Geschwind D. H., Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc. Natl. Acad. Sci. U.S.A. 107, 12698–12703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dragunow M., The adult human brain in preclinical drug development. Nat. Rev. Drug Discov. 7, 659–666 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Gibson P. et al., Subtypes of medulloblastoma have distinct developmental origins. Nature 468, 1095–1099 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanner R. J. et al., Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell 26, 33–47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J., Lin W., Popko B., Campbell I. L., Inducible production of interferon-gamma in the developing brain causes cerebellar dysplasia with activation of the Sonic hedgehog pathway. Mol. Cell. Neurosci. 27, 489–496 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Becher O. J. et al., Gli activity correlates with tumor grade in platelet-derived growth factor-induced gliomas. Cancer Res. 68, 2241–2249 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Po Agnese et al., Hedgehog controls neural stem cells through p53‐independent regulation of Nanog. The EMBO Journal 29, 2646–2658, 10.1038/emboj.2010.131 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.