Significance
NAD+ plays a critical role in redox-linked biological reactions as a cofactor or substrate. The knowledge of its de novo biosynthesis in mammals and certain bacteria is incomplete due to missing information of a nonheme iron dioxygenase mechanism and the conformation of its product that nonenzymatically produces the universal NAD+ precursor, quinolinic acid. To date, how the enzyme regulates the nonenzymatic product remains unknown. This work fills the gap by combining single-crystal spectroscopic and X-ray crystallographic studies. The results provide a comprehensive view of the dioxygenase mechanism by enabling step-by-step visualization of the catalytic cycle and the protein dynamics during catalysis. Additionally, how the enzyme regulates the pathway product distributions, including the nonenzymatic product of biologically significant compound, is proposed.
Keywords: metalloprotein, mechanistic enzymology, oxygen activation, kynurenine pathway, NAD biosynthesis
Abstract
The synthesis of quinolinic acid from tryptophan is a critical step in the de novo biosynthesis of nicotinamide adenine dinucleotide (NAD+) in mammals. Herein, the nonheme iron-based 3-hydroxyanthranilate-3,4-dioxygenase responsible for quinolinic acid production was studied by performing time-resolved in crystallo reactions monitored by UV-vis microspectroscopy, electron paramagnetic resonance (EPR) spectroscopy, and X-ray crystallography. Seven catalytic intermediates were kinetically and structurally resolved in the crystalline state, and each accompanies protein conformational changes at the active site. Among them, a monooxygenated, seven-membered lactone intermediate as a monodentate ligand of the iron center at 1.59-Å resolution was captured, which presumably corresponds to a substrate-based radical species observed by EPR using a slurry of small-sized single crystals. Other structural snapshots determined at around 2.0-Å resolution include monodentate and subsequently bidentate coordinated substrate, superoxo, alkylperoxo, and two metal-bound enol tautomers of the unstable dioxygenase product. These results reveal a detailed stepwise O-atom transfer dioxygenase mechanism along with potential isomerization activity that fine-tunes product profiling and affects the production of quinolinic acid at a junction of the metabolic pathway.
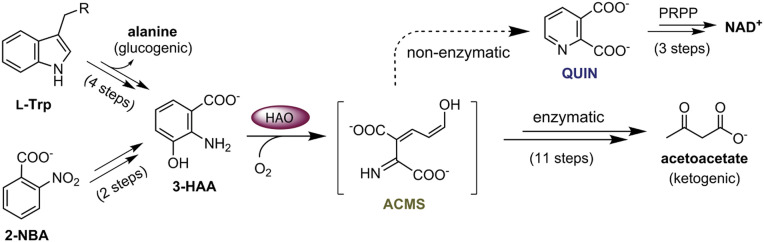
Nicotinamide adenine dinucleotide (NAD+) is an essential and ubiquitous metabolite known for its role in transferring electrons throughout metabolic networks. NAD+ is produced via de novo biosynthetic pathways, or by salvage and recycling routes (1). All kingdoms of life use quinolinic acid (QUIN) as a universal precursor for the de novo biosynthesis of NAD+. In mammals, QUIN is synthesized from the tryptophan metabolite 3-hydroxyanthranilic acid (3-HAA) (Scheme 1). The production of QUIN from 3-HAA involves O2 activation and aromatic ring cleavage of the C3–C4 bond by 3-hydroxyanthranilate-3,4-dioxygenase (HAO). Dioxygenation of 3-HAA generates an unstable acyclic product, 2-amino-3-carboxymuconate-6-semialdehyde (ACMS) (2), which either metabolizes to acetoacetate through a series of enzymatic reactions, or nonenzymatically (3) autocyclizes to produce QUIN with the release of a water molecule (Fig. 1A). The biosynthesis of NAD+ from QUIN is accomplished firstly through a condensation reaction with phosphoribosyl pyrophosphate (PRPP), creating nicotinic acid mononucleotide and subsequently converting to nicotinic acid dinucleotide. Next, amidation of nicotinic acid dinucleotide results in the synthesis of nicotinamide mononucleotide, which is further converted to NAD+ (SI Appendix, Fig. S1).
Scheme 1.
The l-tryptophan kynurenine pathway and the bacterial 2-nitrobenzoic acid (2-NBA) biodegradation pathway converge at the aromatic metabolite 3-hydroxyanthranilic acid (3-HAA). The nonheme iron-dependent enzyme, 3-hydroxyanthranilic acid 3,4-dioxygenase (HAO), sits at the critical junction that directs most of the metabolic flux to the enzyme-controlled ketogenic pathway, which branches off for de novo biosynthesis of NAD+ via quinolinic acid (QUIN). The precise conformation of the ring-cleaved product of the HAO reaction, 2-amino-3-carboxymuconic semialdehyde (ACMS), is critical for determining the partitioning of the metabolite flux at the pathway junction for enzymatic and nonenzymatic routes.
Fig. 1.
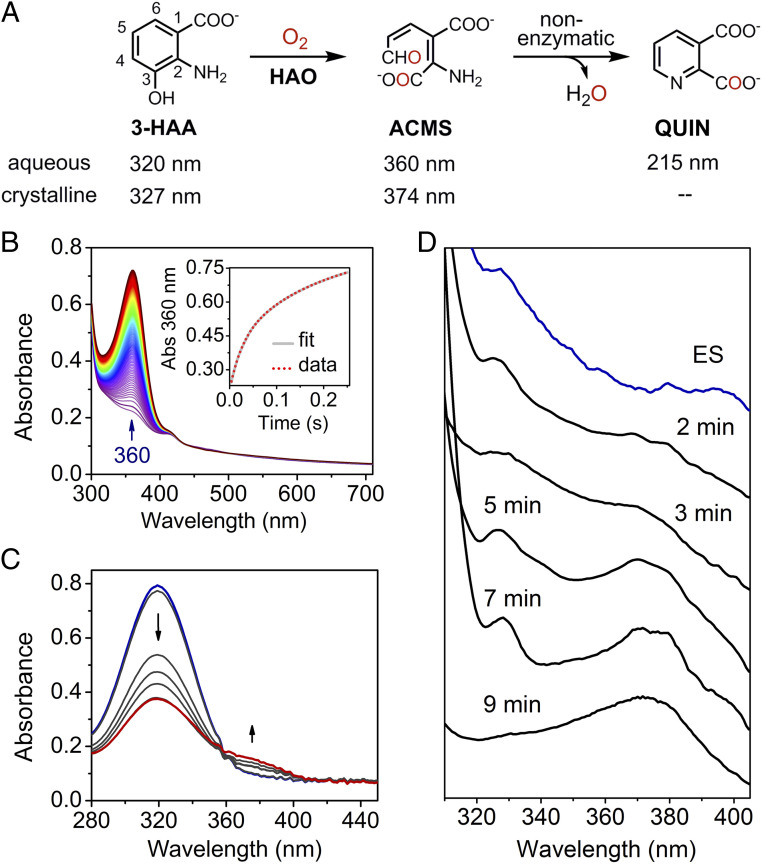
Crystalline state of HAO alters the reaction rate and populates catalytic intermediates that are kinetically unresolved in the solution state. (A) The chemical reaction catalyzed by HAO and the nonenzymatic cyclization. (B) Reaction of anaerobically premixed enzyme–substrate complex (0.10 mM) and O2 (5 mM, generated by chlorite dismutase) in solution monitored by stopped-flow photodiode array spectroscopy. The Inset shows the time course of ACMS production monitored by the single-wavelength detector at 360 nm, fitted with the double-exponential function. (C) The catalytic activity of the HAO single crystals was monitored at room temperature by a benchtop spectrophotometer. Seven crystals were incubated in crystallization mother liquor (pH 8.5) with the cryoprotectant described in Experimental Procedures, 3-HAA (0.3 mM), and O2 (1.3 mM). (D) Representative absorption spectra of time-resolved in crystallo reaction obtained from single-crystal UV-vis microspectroscopy. Each spectrum was collected from one single crystal at 100 K.
Unlike eukaryotes, prokaryotes primarily synthesize QUIN from dihydroxyacetone phosphate and l-aspartate (4, 5). The l-tryptophan–kynurenine pathway, however, is also found in a few bacteria (6, 7), and vice versa, some eukaryotes are known to use the prokaryotic pathway (8–10). Moreover, it is also known that the majority of the kynurenine pathway has been utilized by some prokaryotes to live on the toxic industrial waste, 2-nitrobenzoic acid (2-NBA) as the sole source of carbon, nitrogen, and energy (11). The kynurenine pathway and the 2-NBA biodegradation pathway unite at 3-HAA, where HAO is the first common enzyme shared in these two pathways (Scheme 1). HAO from all sources presents in a homodimeric form, and each subunit consists of a Cupin structural fold (12, 13). The catalytic machinery is orchestrated by a nonheme iron ion coordinated by a 2-His-1-Glu facial triad (14), which is located in the center of a Cupin domain. Some bacterial HAOs contain a noncatalytic, rubredoxin-like site on the protein surface for iron acquisition and storage (15). In the solution state, HAO is an efficient catalyst with a reported turnover rate of 25 s−1 (12, 16). The fast turnover rate is partially due to substrate-induced loop dynamics that help to increase O2 concentration in the hydrophilic active site by making it more hydrophobic to accommodate the sequential binding of 3-HAA and O2, two substrates with disparate polarities, at the catalytic iron center (16).
HAO has long been recognized as a potential drug target (17–22). The tryptophan–kynurenine pathway produces a series of neurologically active compounds, such as 3-HAA, 3-hydroxykynurenine, kynurenic acid, and QUIN. The latter two are the only known endogenous metabolites capable of modulating the activity of the N-methyl-d-aspartate (NMDA) receptors, which are the predominant molecular devices responsible for controlling synaptic plasticity and memory function (23–25). QUIN is also an agonist of NMDA receptors. When the QUIN concentration exceeds basal levels, neurons in the brain become overexcited, and apoptosis may follow, thereby creating undesired biological consequences (18, 25, 26). The kynurenine pathway also synthesizes a significant portion of NAD+ in critical tissues such as the brain and heart (23). Despite its biological significance and the length of time it has been known, the regulatory mechanisms of QUIN production through the kynurenine pathway have remained elusive.
ACMS is the proposed enzymatic product of HAO, which occupies a central position in both pathways (Scheme 1). However, this transient metabolite has not been characterized. Presently, the exact conformation of ACMS produced by the HAO-mediated enzymatic reaction remains unknown due to its short lifetime (2) and a large number of possible conformers. ACMS has 32 potential conformers based on the combinations of distinct tautomers, isomers, and rotamers (SI Appendix, Fig. S2). On the basis of a synthetic modeling study, ACMS is expected to be a 2Z,4E keto tautomer in solution, which undergoes facile isomerization about the C2–C3 and C4–C5 double bonds via transient formation of its enol tautomer to form QUIN by an electrocyclization (3). Although HAO has been studied for decades, the lack of solution-state intermediates renders the catalytic mechanism challenging to study. How HAO balances the catalytic activity of its downstream biomachinery and nonenzymatic QUIN production remains an unanswered question. To identify its catalytic intermediates and interrogate the mechanism of HAO, as well as understand how HAO contributes to the regulation of product partitioning at the ACMS junction, we describe an extensive effort to use the effects of protein in the crystalline state to slow the reaction of HAO-mediated catalysis, which becomes critical to understand the mechanism used for fine-tuning production of QUIN.
Results
HAO Is Catalytically Active in the Crystalline State with Significantly Altered Kinetics.
Stopped-flow UV-vis spectroscopy was used to monitor the HAO reaction in the presence of excess O2 produced by the oxygen-evolving enzyme chlorite dismutase (27). The production of ACMS was followed at 360 nm, which has an extinction coefficient (47,500 M−1⋅cm−1) over one order of magnitude greater than that of 3-HAA at 320 nm (2). The time course of ACMS formation could be described by fitting with a double-exponential function with apparent rate constants of 38.87 and 5.07 s−1 (Fig. 1B). We noticed no spectral features indicative of an intermediate in the difference spectra. One possibility is that an intermediate is present, but it does not exhibit a distinct absorbance at most wavelengths. Another possibility is that no catalytic intermediates populate in the millisecond or longer time window in the aqueous phase. The two summed exponential fitting is most likely required due to the dimeric nature of HAO with inequivalent iron centers, as found in the first enzyme of the pathway, a homotetramer of a heme-based dioxygenase (28).
It has been demonstrated that embedding an enzyme in a crystalline lattice could dramatically alter reaction kinetics, including extradiol dioxygenases and other enzymes related to HAO in the same metabolic pathway (29–33). Here, we first tested the catalytic competency of single crystals of HAO using a benchtop spectrophotometer. Single crystals suspended in an O2-saturated crystallization mother liquor with cryoprotectant in a cuvette containing 3-HAA were able to degrade the substrate. We spectroscopically followed the conversion of 3-HAA to the intermediate product ACMS, which then nonenzymatically decayed to QUIN (Fig. 1C). After reacting for 70 min, all incubated crystals were found intact by microscopy. Due to the missing knowledge of the anticipated slower diffusion rate in the crystalline lattice for both substrate and product, a detailed kinetic analysis is not warranted for the reaction in crystallo under this condition.
Next, we conducted a time-dependent, single-crystal UV-vis microspectroscopic experiment to detect whether any intermediate would accumulate during the catalytic reaction. Snapshots of crystals flash cooled to 77 K at different time points of reaction and examined at 100 K confirmed their catalytic competency (Fig. 1D). Substrate-bound HAO crystals exhibited a λmax at 327 nm. Crystals reacted for 2, 3, 5, 7, and 9 min or longer exhibited optical features distinct from those of the enzyme–substrate complex, suggesting specific intermediates populating at these time points. The considerably slower reaction speed in crystals provides a promising platform to investigate reaction intermediates and the enzyme-bound product throughout the HAO-mediated reaction. This set of experiments indicates that the catalytic rate constant is over 10,000-fold slower than that in solution (25 s−1) (12, 16).
A Free-Radical Intermediate Populated and Characterized by Single-Crystal Electron Paramagnetic Resonance Spectroscopy.
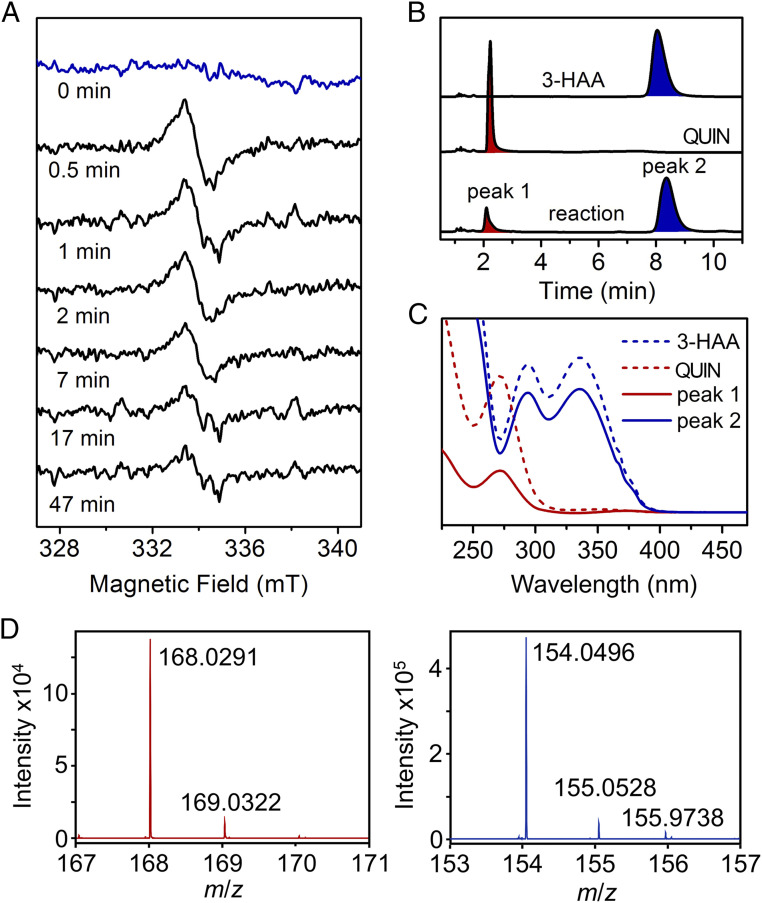
Next, intermediates were probed for the presence of a radical state in pre–steady-state conditions (Fig. 2A). A slurry of approximately 1,000 small-sized HAO single crystals was prepared in crystallization mother liquor with cryoprotectant where O2 was limited. It is worth mentioning that the term “single crystal” refers to a monocrystalline state to differentiate from the amorphous or polycrystalline state of crystals. HAO microcrystals alone had no spectroscopic features at the g = 2 region at 70 K. When substrate (5 mM) was introduced aerobically at room temperature, and freeze-quenched in liquid ethane at different times, a g = 2.005 signal with a linewidth of 1.15 mT was observed after incubation with 3-HAA for 30 s. The observed linewidth is presumably generated from a conjugated system (34–36) and is inconsistent with a superoxo anion radical (37). The sample was then thawed, aged for another 30 s, and rapidly refrozen in liquid ethane to acquire a second spectrum, resulting in the same signal with less intensity. The g = 2.005 radical signals continued decreasing over 50 min with multiple cycles of thawing and freezing. The protein crystal electron paramagnetic resonance (EPR) experiments suggest that a substrate-based radical intermediate accumulates under pre–steady-state conditions using the crystallized enzyme as a catalyst. Notably, the rate of the reaction in crystallo is affected by the dimension and shape of a crystal (38, 39). The maximal intensity of radical species at 30 s in microcrystals corresponds to a later time point of the reaction with the larger-sized crystals used in the X-ray diffraction experiments.
Fig. 2.
Single-crystal continuous-wave EPR spectroscopy of the HAO reaction with a crystal slurry reveals a substrate-based radical intermediate. (A) Time-lapse in crystallo reaction at room temperature with samples analyzed by X-band EPR spectroscopy at 70 K. (B) HPLC–UV-vis analysis of the single-crystal reaction mixture. Elution profiles are shown with the absorbance at 280 nm. (C) UV-vis spectra of the HPLC fractions eluted with solvent containing 2% acetonitrile and 0.1% formic acid. (D) High-resolution mass spectrometry of the HPLC fractions. Peak 1 (blue) was identified as 3-HAA, and peak 2 (red) was QUIN.
After completion of the in-tube EPR catalytic reaction with HAO single crystals, the crystals and aqueous solution were separated by gentle centrifugation. Those crystals were dissolved, resulting in a 200-μL aqueous solution with a protein concentration of 185 μM. The aqueous portion of EPR sample was subjected to high-performance liquid chromatography (HPLC) for product analysis. As shown in Fig. 2B, two significant peaks were separated by chromatography. The peak at 2.1 min had the same retention time and absorption spectrum (λmax at 272 nm) as a QUIN standard, the nonenzymatically cyclized product of ACMS, while the peak at 8.4 min had the same retention time and absorbance spectrum (λmax at 294, 335 nm) as the 3-HAA standard (Fig. 2C). These absorbance maxima were observed under acidic HPLC conditions, which are distinct from those observed at pH 8.5. We further characterized the eluted fractions using high-resolution mass spectrometry (Fig. 2D). The results confirmed that the 2.1-min fraction was the newly formed QUIN (m/z = 168.0292, mass accuracy of 0.60 ppm), and the 8.4-min fraction was the unreacted substrate 3-HAA (m/z = 154.0499, mass accuracy of 1.95 ppm). Based on the integrated peak area, 0.15 mM QUIN was formed during the in-tube reaction with the single-crystal slurry, which corresponds to approximately one catalytic turnover.
Observing Substrate Binding to the Catalytic Iron Via X-Ray Crystallography.
The slower turnover rate in crystals provides an opportunity to probe the reaction cycle by X-ray crystallography to trap and characterize catalytic intermediates that are otherwise not populated in the solution reaction. To this end, we performed time-resolved reactions in crystallo, first in the absence (for substrate binding) and then in the presence of O2. The substrate-free single crystals of FeII-HAO were grown in an anaerobic chamber. A resting-state structure at 1.60-Å resolution reveals two solvent-derived ligands completing a distorted octahedral complex (Fig. 3A). To study substrate binding, HAO crystals grown anaerobically were incubated in mother liquor containing 3-HAA (1 mM). Turnover conditions were studied by soaking crystals in air-saturated solutions with 1 mM 3-HAA. In all cases, similarly sized crystals were used, and after various incubation periods between 20 and 1,800 s, they were flash cooled by submersion in liquid nitrogen. Representative high-quality crystal structures of distinct reaction cycle intermediates (≥80% ligand occupancy) are presented (for details, see SI Appendix, Table S1). The optimal time points were determined by screening several thousand crystals, from which hundreds of datasets were collected with good reproducibility. Although the time points obtained from UV-vis absorbance microspectroscopy and X-ray diffraction are independent of each other, the catalytic turnover rate matches well because crystals of similar size were used. For this reason, single-crystal UV-vis microscopy presumably features the structurally defined intermediates populated at the same reaction times.
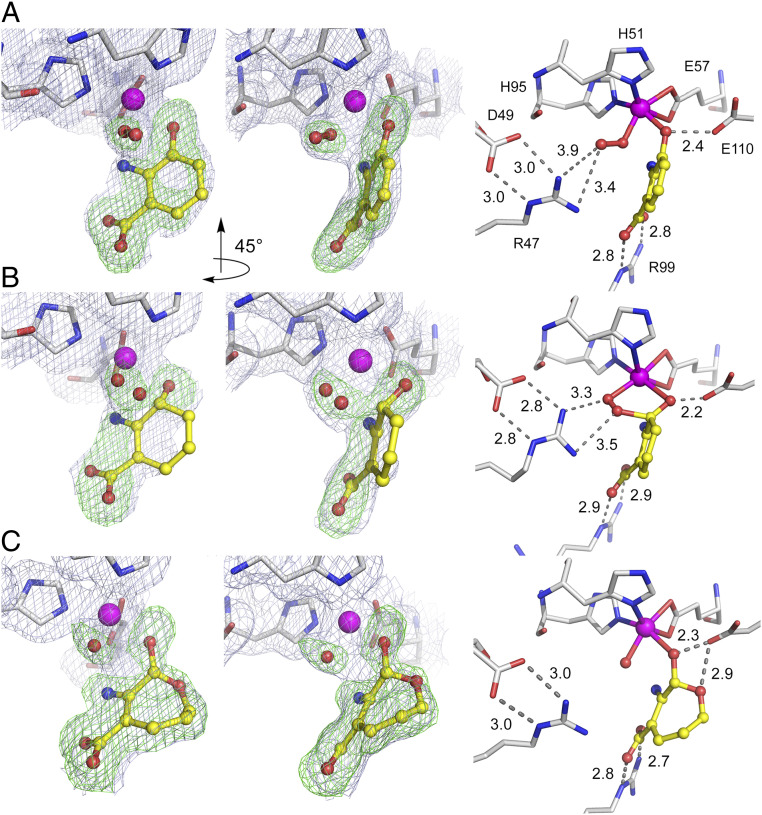
Fig. 3.
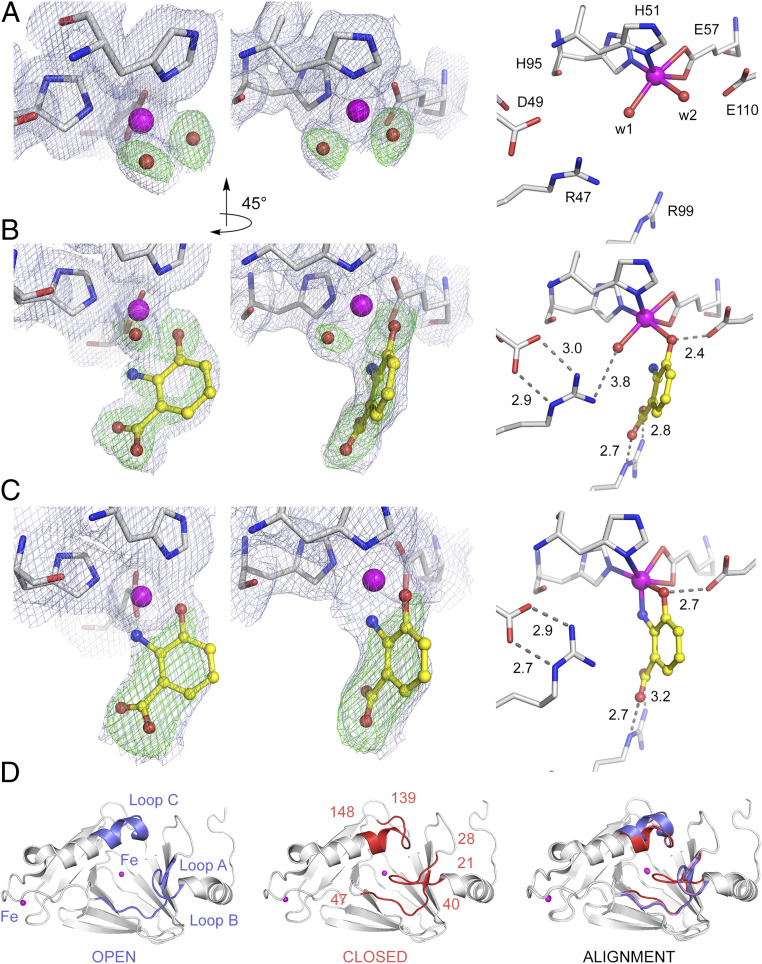
A two-step 3-HAA binding process. (A) The ligand-free structure in resting enzyme, (B) the transient (E•S)I intermediate after ∼20-s exposure to 3-HAA, and (C) the equilibrium (E•S)II complex observed after incubation periods of 30 s with 3-HAA. The light blue 2Fobs – Fcalc maps and green omit Fobs – Fcalc maps are contoured at 1.0 σ and 3.0 σ, respectively. Atom color code: gray, carbon (protein residues); yellow, carbon (ligand); blue, nitrogen; red, oxygen; magenta, iron. (D) The crystal structure of the substrate-free enzyme is in a fully open form (blue), and the transient monodentate (E•S)I intermediate exhibits a partially closed conformation (pink), whereas the structure of the bidentate (E•S)II complex is in a fully closed conformation (red). Significant loop flexibility is observed at three specific loop regions consisting of highly conserved residues 21 to 28 of loop A, 40 to 47 of loop B, and 139 to 148 of loop C.
Under anaerobic conditions, the earliest enzyme–substrate complex (E•S) structure was obtained after approximately 20 s by hand-mixing 3-HAA with HAO crystals and flash cooling in liquid nitrogen. In this E•S structure refined to 1.90-Å resolution (Protein Data Bank [PDB] entry: 6VI6), 3-HAA is a monodentate ligand with its C3-hydroxyl moiety bound to the catalytic iron (Fig. 3B). The bond distance between FeII and C3-hydroxyl group is 2.3 Å, while the C2-amino group is 3.2 Å from the FeII ion (SI Appendix, Table S2). The previously observed loop movements that enhance the hydrophobicity of the Fe center and facilitate O2 binding upon bidentate chelation of the substrate are not observed in this intermediate (16). Only one of three loops composed of residues 139 to 148 partially shift to the previously defined “closed” form (Fig. 3D and Movie S1). Thus, this is a previously unobserved E•S structure. We interpret this state as a transient enzyme–substrate intermediate and abbreviate it as (E•S)I. The capture of this intermediate provides information as to how 3-HAA interacts with the catalytic Fe center step-by-step at the initial stages of catalysis. Anaerobic incubation of 3-HAA for 30 s or longer yielded the expected bidentate E•S complex structure (Fig. 3C), which was originally reported at 3.20-Å resolution (12), and now updated to 2.61-Å resolution (PDB entry: 6VI7). The bidentate structure is referred to as the (E•S)II intermediate, and the reported loop movements relative to the ligand-free enzyme structure that increase the hydrophobicity of the iron center are observed, which, in turn, increases the binding affinity of dioxygen (16) (Fig. 3D).
Oxygen Activation at the Nonheme Iron Center.
The first oxygenated intermediate structure (E•S•O2)I was obtained with crystals flash-cooled at 30 to 50 s after air exposure of the anaerobically substrate-soaked crystals. At this time interval, we analyzed multiple similar structures; the best X-ray diffraction dataset for this intermediate was determined to 1.95-Å resolution (PDB entry: 6VI8). Refinement against either the (E•S)I or (E•S)II yielded positive Fobs − Fcalc omit electron density in the anticipated O2 binding site adjacent to the metal center and the 3-HAA molecule (SI Appendix, Fig. S3), indicating that only 3-HAA and one oxygen (H2O or OH) coordination cannot satisfy all of the electron density observed at the active site. The positive density contoured at 3.0 σ is the same site occupied by nitric oxide in the ternary complex structure of HAO with 3-HAA, as well as the O2 in the ternary complex structure of HAO with inhibitor 4-Cl-3-HAA (12) (SI Appendix, Fig. S4). The electron density around the Fe center obtained with (E•S•O2)I crystals cannot be modeled with the structures derived from crystals incubated for longer time intervals (SI Appendix, Fig. S3). Therefore, this first reactive intermediate after O2 exposure is most consistent with the active-site iron of HAO coordinated by 3-HAA and an O2 ligand (Fig. 4A). Starting from the (E•S•O2)I, the aforementioned three loops adopt a partially open conformation, and the active site is no longer in the fully closed state (Movie S1).
Fig. 4.
Dioxygen binding to the enzyme–substrate complex and O–O bond cleavage. (A) A ferric superoxo (E•S•O2)I or ferrous substrate radical (E•S•O2)II (30 to 50 s), (B) alkylperoxo (E•S•O2)III (1 min), and (C) seven-membered lactone (E•Int)II (3 to 5 min). The light blue 2Fobs – Fcalc maps and green Fobs – Fcalc omit maps are contoured at 1.0 σ and 3.0 σ, respectively. Atom color code: gray, carbon (protein residues); yellow, carbon (ligand); blue, nitrogen; red, oxygen; magenta, iron.
In the (E•S•O2)I intermediate, 3-HAA is a monodentate ligand, which binds the Fe ion with its hydroxyl group in the position opposite to His95. The O2 binds to the Fe in the position opposite Glu57 in an end-on manner. Compared to the bidentate (E•S)II complex, the C3-hydroxyl group of 3-HAA in the (E•S•O2)I structure is slightly rotated out of the plane of the ring, moving toward Glu110. The O–O bond distance was refined to 1.4 Å, which is consistent with a superoxo species (37). Depending on the oxidation state of the Fe, this species can adopt two resonance structures, either the (E•S•O2)I or (E•S•O2)II complex for a ferric superoxide anion or a ferrous-bound substrate radical, as previously proposed in the study of homoprotocatechuate 2,3-dioxygenase (HPCD) (37). Since the two resonance forms cannot be structurally distinguished at this resolution, one or both species could populate in the crystal. The out-of-plane rotation of the hydroxyl group of 3-HAA indicates that the organic substrate is partially activated for a superoxide attack.
The next oxygenated intermediate (E•S•O2)III was trapped in crystals flash cooled 1 min after mixing, and the structure was refined to 2.30-Å resolution (PDB entry: 6VI9). A notable difference between the (E•S•O2)I, II and (E•S•O2)III species is that the latter structure exhibits a much closer distance between the distal oxygen of the iron-bound O2 and the C3 of 3-HAA, shortened from 3.1 to 1.8 Å (SI Appendix, Table S4), suggesting the formation of a nascent C–O bond (Fig. 4B). (E•S•O2)III is consistent with an FeII-alkylperoxo intermediate in which the distal oxygen atom of O2 forms a covalent bond with the organic substrate. In this structure, the C1-carboxyl and the C3-hydroxyl groups of the substrate become more extensively puckered relative to the phenyl ring, suggesting a loss of aromaticity. Additionally, compared with the end-on binding mode in (E•S•O2)I and (E•S•O2)II, the iron–oxygen molecule geometry is more bent, and the Fe–O–O angle decreases from 128.1° to 101.6°. The refinement of models with 3-HAA and other reaction pathway intermediates resulted in obvious negative and positive densities around the catalytic iron center (SI Appendix, Fig. S5). In the structure of the (E•S•O2)III intermediate, the substrate amino group is in H-bond distance from the proximal iron-bound oxygen atom, potentially indicating a role as an acid catalyst to facilitate the O–O bond cleavage.
Probing the Catalytic Mechanism by Viewing O–O Bond Cleavage and O-Atom Transfer: Observation of a Monooxygenated, Seven-Membered Lactone Intermediate.
In the 3- to 5-min reaction time range, a high-resolution intermediate structure was obtained and refined to 1.59-Å resolution (PDB entry: 6VIA). Structures obtained in this time range show extended, nonplanar, excess electron density in the active site, which is not amenable to fitting by 3-HAA or the intermediates described above. The two anticipated candidates from computational studies of aromatic dioxygenation of extradiol dioxygenases are an epoxide (40), labeled as (E•Int)I, and a seven-membered lactone, labeled as (E•Int)II (41–43). In modeling this intermediate, the seven-membered lactone (E•Int)II provided substantially better fit into the observed electron density than (E•Int)I or any other intermediates (SI Appendix, Fig. S6). (E•Int)II and (E•Int)III were proposed as resonance structures with a low energy barrier for transition (42); however, the observed electron density was better fitted by (E•Int)II (Fig. 4C). Accordingly, the intermediate in (E•Int)II coordinates the iron ion in a monodentate fashion with its hydroxyl group at a distance of 2.3 Å to the Fe ion and 2.3 Å to Glu110. The proximal oxygen from O2 remains bound to the Fe with an FeIII–OH bond length of 2.2 Å. The lactone intermediate is stabilized by H-bonding interactions between the newly incorporated oxygen and Glu110. Notably, Arg47 is H-bonded with the distal oxygen atom of the O2 during catalysis until this step. Previous mutant studies have shown that Arg47, along with Arg99 and Glu110, plays a critical role in catalysis (12). The intermediate structures in Fig. 4 show that Arg47 is crucial to stabilizing the bound oxygen and directing it toward C3 until O–O bond cleavage. Asp49 forms a salt bridge with Arg47, thereby limiting its free rotation. Additional views of the seven-membered lactone intermediate with Fobs – Fcalc omit maps are shown in SI Appendix, Fig. S7. It is worth noting that this seven-membered ring-containing intermediate was obtained from multiple crystals with a resolution between 1.59 and 1.75 Å, suggesting that this is structurally the most accessible intermediate during the catalytic reaction in crystallo.
The radical form of the lactone intermediate (E•Int)II is a possible candidate for the species observed in single-crystal EPR experiments. Fig. 2A shows that the maximal intensity of the free radical signal is observed at 30 s after O2 exposure in the EPR experiments using microcrystals, while (E•Int)II occurs at 3 to 5 min. However, the time difference correlates well with the proposed lactone intermediate due to distinct crystal size utilized in EPR and X-ray diffraction experiments. We noticed that the smaller the protein crystal was, the faster the reaction took place in crystallo when the reaction was monitored by an individual single crystal of HAO under microspectroscope. A radical intermediate with an FeIII-bound hydroxyl anion also explains the lower electron density observed at C3. Also, the possibility of electron transfer between two resonance states may cause some electron deficiency at the catalytic Fe center. Hence, the occupancy for Fe and ε-lactone was reduced to 0.8, resulting in a better fit for the overall density. This high-resolution HAO intermediate structure provides unambiguous experimental evidence for the involvement of an ε-lactone intermediate in the extradiol dioxygenase catalytic cycle.
Visualizing the Formation of the Dioxygenase Product Enol Tautomers and Isomerization.
The subsequent step in the dioxygenation is ring opening by the Fe-bound hydroxyl, attacking the C3 position of the lactone intermediate (42). This step is thought to be an energetically favorable step (44). We obtained the first product-bound structure at 2.10-Å resolution (PDB entry: 6X11), after aerobic soaking for 9 min (SI Appendix, Table S1). As described below, this intermediate, abbreviated as (E•P)I, is best described as a 3E,5Z,2t,4c-enol tautomer of ACMS (Fig. 5A). The dioxygenated product is a monodentate ligand attached to the iron ion with its nascent carboxyl group. The electron density of (E•P)I is distinct from all other intermediates described above (SI Appendix, Fig. S8). The torsion angles of the ACMS ligand requires assignment of the C2–C3 and C4–C5 bonds as single, and the trending of electron density reveals the best fit with a 3E,5Z,2t,4c-enol tautomer (SI Appendix, Fig. S2). Fitting with keto tautomers of ACMS or bidentate enol tautomers could not satisfy the density maps (SI Appendix, Fig. S8). The first oxygen atom derived from dioxygen is 3.1 Å from the previous covalently linked carbon in (E•Int)II (O2 and C3; SI Appendix, Table S3), indicating the C–O bond is cleaved. However, the conformation of (E•P)I retains some cyclic structural feature, suggesting an intermediate formed immediately following the ring opening from the lactone intermediate (E•Int)II. This oxygenated intermediate also inherits the monodentate coordination status found in the previous lactone intermediate. These observations are consistent with findings in other extradiol dioxygenases that the newly formed carboxyl group binds to the iron in a monodentate fashion with the products retaining a more cyclized conformation (29, 45, 46).
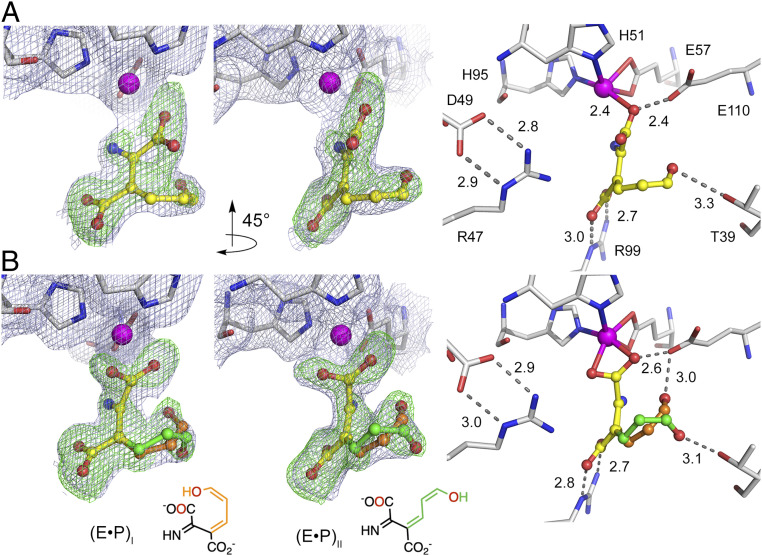
Fig. 5.
Visualizing the oxygenated ACMS product in two distinct enol tautomers bound to the catalytic iron ion in the monodentate and bidentate modes. (A) Monodentate product complex (E•P)I (9 min) and (B) bidentate (E•P)I and (E•P)II (9 to 12 min) product complexes in one structure. The light blue 2Fobs – Fcalc maps and green Fobs – Fcalc omit maps are contoured at 1.0 σ and 3.0 σ, respectively. Atom color code: gray, carbon (protein residues); yellow, carbon (ligand); orange, carbon (3E,5Z,2t,4c-enol tautomer of ACMS); green, carbon (3E,5Z,2t,4t-enol tautomer); blue, nitrogen; red, oxygen; magenta, iron.
The last intermediate characterized is also a product-bound complex. A 1.84-Å resolution structure of enol tautomers of ACMS with two distinct conformations in one crystal was obtained (PDB entry: 6VIB) when the crystal reacted with substrates for a longer time (9 to 12 min). All previous intermediates, including monodentate (E•P)I, cannot satisfy the strong electron density maps near the catalytic Fe center with a reasonable fit (SI Appendix, Fig. S9). When the electron density was modeled as the oxygenated product in this structure, the newly formed carboxyl group no longer binds to the Fe ion in a monodentate fashion, as found in the previous (E•P)I structure. Rather, the product is in a bidentate metal-ion coordination state. Fitting with a bidentate ligand of 3E,5Z,2t,4c-enol tautomer resulted in positive density maps around the enol group (SI Appendix, Fig. S8G), suggesting a facile rotation of the enol tail. Upon fitting with both the (E•P)I and a conformation with a rotated enol tail, the extra positive electron density was resolved (Fig. 5B). The keto tautomers of ACMS were also examined for this intermediate but, again, did not yield an acceptable fit for the electron density. Therefore, not only a monodentate-to-bidentate coordination transition was found, but also a distinct conformer from (E•P)I was also observed in this structure. This conformer is best described as 3E,5Z,2t,4t-enol tautomer of ACMS, abbreviated as (E•P)II. The final refinement gave the best fitting of (E•P)I and (E•P)II with an occupancy ratio of 9:7. Chemically, ACMS has four geometric isomers, each with its own enol tautomers. The conversion of these isomers was demonstrated in an NMR study of an analog compound (3). Among these isomers and tautomers, there are viable possibilities for multiple rotamers. Thus, the different conformational combinations lead to the 32 potential ACMS structures, which include the above two conformations observed in crystal structures (SI Appendix, Fig. S2). From (E•P)I to (E•P)II, there is a single bond (C4–C5) rotation of 60° from 4-cis to 4-trans conformations. Notably, (E•P)II represents a more linear product conformation compared to (E•P)I and corresponds to an intermediate of the later catalytic stage. In the bidentate product, the enol group of (E•P)I is stabilized by Glu110, while in (E•P)II, the oxygen interacts with Thr39 (Fig. 5).
Discussion
The results presented here show that the HAO reaction occurs at a much slower rate in crystallo than in the aqueous phase, leading to a near-complete view of the HAO catalytic cycle (Fig. 6). The catalytic pathway is complex, including the events of monodentate and then bidentate 3-HAA coordination, binding and activation of dioxygen, O–O bond cleavage, oxygen insertion, Criegee rearrangement causing ring expansion, a second oxygen transfer with ring opening, and finally, an enzyme-assisted change of the product conformation. These events are observed through a series of high-quality crystal structures, six of which are not previously observed intermediate structures of the HAO reaction.
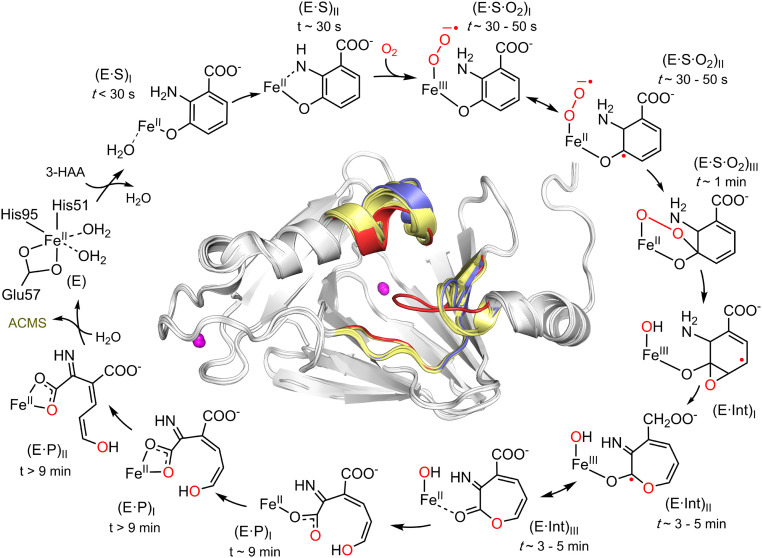
Fig. 6.
HAO catalytic reaction cycle defined by the characterized intermediates. The time windows for observing the in crystallo reaction intermediates are highlighted. The structurally characterized intermediates include monodentate E•S complex, bidentate E•S complex, Fe-bound superoxo, alkylperoxo, ε-lactone, monodentate and then bidentate 3E,5Z,2t,4c-enol tautomer of ACMS, as well as bidentate 3E,5Z,2t,4t-enol tautomer of ACMS. The superposition of the crystal structures of the substrate-free HAO and the above seven intermediates is shown as an Inset to highlight the conformational change of loops A, B, and C during the catalytic reactions. Loops of substrate-free, bidentate E•S complex, and subsequent intermediates after dioxygen binding are colored in blue, red, and yellow, respectively.
The observation of a monodentate (E•S)I followed by a bidentate (E•S)II substrate binding has previously been reported in type I, but not type III, extradiol dioxygenases (47, 48). This finding in a type III enzyme suggests that majority of extradiol dioxygenases may employ a similar multistep substrate-binding mechanism as first found in type I dioxygenases (49). The substrate 3-HAA binds to the Fe center with a deprotonated hydroxyl group. An active site histidine in type I enzymes deprotonates the catechol substrate and induces an asymmetric monoanionic ligand (50–52). This histidine residue is absent in HAO. Glu110, however, is found in a similar position.
Dioxygen binds to the Fe ion in an end-on mode in HAO. Based on the structure, the distal oxygen of the superoxide is directed correctly to proceed via radical recombination with the radical on the C3 carbon of the substrate, resulting in an alkylperoxo intermediate (E•S•O2)III with its oxy bridge trans to the glutamate metal ligand. The alkylperoxo closely resembles the bond lengths and orientations of the equivalent intermediate trapped in HPCD (29). One apparent variation between HAO and HPCD is the metal–ligand interaction during oxygen activation. The organic substrate is a bidnetate ligand in the superoxo and alkylperoxo complexes in HPCD but monodentate in HAO. The possible reasons for the discrepancy include the difference between a ketone and an amine of the respective substrate molecules, and the transient coordination of the amine/imine in the HAO-catalyzed reaction, which is likely due to the absence of a supporting second coordination sphere residue.
The monooxygenated lactone intermediate, (E•Int)II, results from heterolytic O–O bond cleavage from the alkylperoxo intermediate, followed by Criegee rearrangement. Notably, a gem diol and an epoxide are anticipated prior to the formation of the lactone intermediate, but they are not captured in the HAO reaction. The gem diol intermediate has been captured in HPCD and characterized by X-ray crystallography (53). The epoxide has not yet been trapped and characterized. We have obtained a few structural datasets that appear to be best modeled for the anticipated epoxide intermediate. However, the resolution and ligand occupancy do not meet the standards applied to the other intermediates. Thus, it remains inconclusive whether the proposed epoxide is an intermediate in the extradiol dioxygenase pathway.
Nevertheless, the capture of the lactone intermediate indicates that dioxygen insertion by HAO takes place stepwise, rather than concerted. Of note, HAO is an established drug target (17, 18), which has attracted significant attention due to its participation in the tryptophan–kynurenine pathway, which plays a role in exercise, inflammation, and mental health (54). Enzymes in this pathway are also checkpoint proteins regulating the immune response. Cancer cells overexpress this pathway to bypass immune surveillance. The successful trapping and characterization of the lactone intermediate may facilitate structure-aided drug design as the ε-lactone may be the most specific target for this dioxygenase.
A denticity change, from one to two, is observed in the product bound complex. Most of the products observed through in crystallo reaction in extradiol dioxygenases maintain a cyclized conformation (29, 45, 46), as seen in (E•P)I of HAO. A linearized product like (E•P)II is less commonly seen bound at the active site in dioxygenases in the degradation of aromatic molecules (40). Among the forms of ACMS, QUIN is produced via a pericyclic reaction from the 3E,5Z,2c,4c form of the enol tautomer. In (E•P)I, the iron-bound ACMS intermediate is observed to be in its 3E,5Z,2t,4c conformation, which has easy access to the QUIN forming reaction trajectory via one single bond (2,3-bond) rotation (SI Appendix, Fig. S2). The 3E,5Z,2t,4t state in (E•P)II creates a higher energy barrier to QUIN formation. Glu110 and Thr39 may facilitate such a rotation. Without the aid of the Fe ion and second sphere residues, the 4,5-bond rotation may not occur as readily. Thr39 becomes involved in the product-bound complex only at the end of the reaction pathway. Unlike other active-site residues presented in the structural figures, Thr39 is not strictly conserved in type III extradiol dioxygenases. Methionine is found in the same position in some HAO during sequences alignment (16). This variation may be derived from a need to tune the efficiency of the metal- and active site residue-assisted conversion of the product conformation.
After release from the active site, the enol tautomer of ACMS either spontaneously cyclizes to the essential NAD+ precursor, QUIN, or is further metabolized by ACMS decarboxylase (55–59) (SI Appendix, Fig. S1). Interestingly, a previous study has shown that the autocyclization of QUIN from ACMS released from HAO is impeded by the presence of HAO compared to the cyclization rate in the absence of HAO (3). Our product-bound complex structures provide a rationale for the underlying mechanism of how the presence of the enzyme slows the spontaneous formation of QUIN. We propose that HAO possesses in situ isomerase activity. By maintaining ACMS in a more stable conformation, HAO raises the bar for autocyclization and hence reserves a portion of the metabolic flux for other downstream enzymes of the kynurenine pathway. Similar isomerase activity has been found in the dehydrogenase enzyme two steps down in the pathway (31, 33). It has been found that the ratio of QUIN relative to the enzyme-controlled metabolism is 1:72 in normal healthy individuals (60). Overproduction of QUIN within cells should be avoided since it is excitotoxic at high concentrations and causes excessive stimulation of neuronal cells (61, 62). The potential isomerization of HAO may represent a natural strategy to maintain some noncyclized ACMS, highlighting a mechanism for this bifurcated pathway to fine-tune the conformation of an unstable metabolic intermediate.
Single-crystal EPR studies have been made to characterize the stable tyrosyl radical in ribonucleotide reductase, photosystem II, and the oxidized state of [FeFe]-hydrogenase by using special sample containers (63–65). Previous methods used a limited number of crystals, which creates spectra with orientation dependence yielding additional information such as g-tensor parameters. However, orientation dependence complicates data interpretation, especially when identifying an unknown intermediate during a time-resolved reaction. In this study, we tested the minimal number of equivalent single crystals needed in a regular, quartz X-band EPR tube to surmount the orientation dependency and created an updated method for detecting intermediates in crystallo by EPR spectroscopy during enzymatic reactions.
A common feature during enzymatic action is that many enzymes adopt an open conformation at the catalytic center, and upon substrate binding, the catalytic center becomes closed. However, little is known whether it is in the open or closed conformation during the intermediary steps of catalysis. Our success in obtaining seven intermediate crystal structures provided an outstanding opportunity to address such a question in an iron-based dioxygenase. Previously, we have shown that the closed conformation due to 3-HAA binding increases the hydrophobicity of the iron center and causes a substantial increase of the O2 affinity of the active site (16). The structural studies in this work show how the dioxygenase modulates the active-site conformation throughout the catalytic reaction. Fig. 6, Inset, and Movie S1 show that the active site changes quickly from the closed to a primarily, but not wholly, open form after the arrival of dioxygen at the iron center. While the active site gradually moves to the fully open form along the reaction pathway, the loop B of the alkylperoxo intermediate is an outlier which moves to the closed conformation. Thus, the superoxo and alkylperoxo intermediates not only differ from each other in the iron-bound substrates, 3-HAA and O2; they also have a significantly different conformational state in residues between 40 and 47 (loop B). In the alkylperoxo intermediate, Arg99 of loop B moves toward the iron center and is responsible for stabilizing the oxy bridge. This requirement from the second coordination sphere may explain the closed conformation of loop B. From the alkylperoxo to the monooxygenated lactone intermediate, a significant movement of loop B to the near open form takes place, presumably because the intermediate stabilization by Arg99 is no longer required. Subsequent changes from the lactone to the enol tautomers of the dioxygenated product are minor, and most loop movement for opening is focused on loop C (residues 139 to 148). It is worth noting that these loop dynamics during catalysis are rarely observed and difficult to predict. It is also challenging to determine the active-site dynamics during catalysis in the solution state of the HAO reaction due to its high turnover rate. Thus, the results presented in this work provide a case study and comprehensive view regarding both the active-site chemistry and the protein conformational dynamics during an enzymatic action.
Experimental Procedures
The materials and routine methods are described in SI Appendix, including the source of reagents, protein expression, isolation, enzyme activity assays, kinetic analyses, product analysis by HPLC and high-resolution mass spectrometry, crystallization, and X-ray data collection and refinement.
Single-Crystal UV-Vis Spectroscopy.
The catalytic activity was initially demonstrated by using an Agilent 8453 UV-vis spectrophotometer in a reaction mixture containing seven single crystals in mother liquor with 25% PEG 8000, 200 mM MgCl2, and 30% glycerol, 0.3 mM 3-HAA, and 1.3 mM O2. The reaction was monitored at room temperature in a quartz cuvette. The λmax numbers for 3-HAA and ACMS in solution at pH 8.5 are 320 and 360 nm, respectively. At this pH, ACMS is less stable and forms QUIN more rapidly with much less ACMS observed at 375 nm than expected by the loss of 3-HAA (55). After 70 min, all seven crystals were found intact by microscopy (Leica Microsystems).
In situ single-crystal UV-vis absorption spectroscopy was used to characterize the reaction intermediates and turnovers. A fully automated, remote accessible microscope spectrophotometer at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 9-2 was employed to collect the UV-vis absorption spectra of HAO single crystals during reaction or alongside diffraction measurements. A default 50-μm light spot at the sample was used to monitor the catalytic reaction in crystal. All spectra were measured at 100 K. Reference of the spectroscopy was taken in the absence of crystal samples. In order to minimize artifacts from the frozen mother liquor, ice, or the nylon loop, different orientations of a single crystal were scanned by phi-angle rotation from 0 to 360° with a step of 10°. The optimal absorption spectra exhibiting reproducible spectral features were selected for presentation. The λmax for 3-HAA and ACMS in crystals are 325 and 374 nm, respectively.
X-band Continuous-Wave EPR Spectroscopy of HAO Single Crystals.
Anaerobically crystallized HAO microcrystals (∼1,000 single crystals) were prepared in mother liquor containing 20% glycerol. EPR samples were prepared in 4-mm quartz EPR tubes and rapidly frozen by liquid ethane. X-band EPR spectra were recorded by a Bruker E560 spectrometer at 9.4-GHz microwave frequency with an SHQE high-Q resonator at 100-kHz modulation frequency. The temperature was maintained with a cryogen-free 4 K temperature system. All spectra were collected at 70 K and 16-µW microwave power, with an average of nine scans. The g values and linewidths reported were obtained by inspection of the EPR line shape.
The in-tube reaction was prepared by the addition of 5 mM 3-HAA into HAO crystals to a final volume of 200 µL. The minimum time for sample handling is 30 s before freezing. After acquiring each spectrum, the EPR sample was thawed and incubated at room temperature before being rapidly frozen. The selected time points for spectral acquisition are 0.5, 1, 2, 7, 17, and 47 min. After completion of the reaction, crystals and aqueous solution were separated by gentle centrifugation (2,200 rpm). Crystals were dissolved in buffer (200 μL of 50 mM Tris at pH 8.5) with continuous vortexing. Protein concentrate was estimated as 185 μM based on the absorbance at 280 nm. The aqueous portion of the in-tube reaction was retained for analysis by HPLC and mass spectrometry.
Preparation of the Reaction Intermediates.
In order to study the formation of the binary E•S complex, single HAO crystals were incubated with mother liquor (0.1 M Tris⋅HCl, pH 8.5, 0.2 M MgCl2, 20% PEG 8000) supplemented with 1 mM 3-HAA (about 45-fold larger than the Km value) in an anaerobic chamber. The monodentate E•S complex was trapped by flash-cooling crystals 20 to 30 s after they were briefly dipped in mother liquor augmented with 20% glycerol as a cryoprotectant. The chelated, equilibrium E•S complex was flash-cooled after 30 or 300 s. Turnover was initiated at room temperature by incubating crystals in 1 mM 3-HAA and then air-saturated mother liquor. The crystals were flash-cooled after 50-s, 1-, 3-, 5-, 7-, 9-, or 13-min incubation periods. Crystals were prepared separately, and flash-cooled directly in liquid nitrogen after being dipped into mother liquor with 20% glycerol. The diffraction datasets and refined structures with the highest resolution are presented here. The corresponding data collection and refinement statistics are summarized in SI Appendix, Table S1. The coordination distances and exogenous ligand occupancy are shown in SI Appendix, Tables S2–S4. Each dataset was obtained on a distinct single crystal from one continuous data collection at the beamline. No datasets were merged during analysis because each crystal and time point are unique.
Ligand Refinement and Molecular Modeling.
In order to minimize bias, ligand refinement was initiated at the final stage of structure refinement by examining ligand-omit difference maps. In every case, 3-HAA was the initial ligand model. As warranted, subsequent ligand models in the reaction cycle were fit into the 2Fobs – Fcalc electron density maps and by examining the Fobs – Fcalc maps. Phenix eLBOW was applied to generate the geometry restraint information of 3-HAA–derived superoxo, alkylperoxo, epoxide, ε-lactone, and enol tautomers of ACMS. With the exception of two conformations of the product in one crystal structure, only the intermediates with ≥80% ligand occupancy are included. The light blue 2Fobs – Fcalc maps around the metal-ligands (His51, Glu57, and His95) and the exogenous ligands are contoured at 1.0 σ. Finally, the exogenous ligands were removed, and the resultant ligand-free structures were refined to obtain the Fobs – Fcalc omit maps (green) contoured at 3.0 σ, which are in agreement with the active-site ligands in the full models. The detailed ligand refinement and model fitting are described in SI Appendix, Materials and Method, under X-ray data collection and refinement.
Supplementary Material
Acknowledgments
We thank Drs. Jennifer L. DuBois and J. Marty Bollinger Jr. for providing the chlorite dismutase plasmid used in the solution-state kinetics studies, and Dr. Jiafeng Geng for participating in the kinetic studies. We also thank Dr. Allen M. Orville and Dr. Ana Gonzalez for assistance with our initial attempts to perform the single-crystal microscopy spectroscopic studies. This work was supported by National Science Foundation Grants CHE-1623856 and CHE-1808637, and partially by the National Institutes of Health Grants GM108988, GM133721, and MH107985, and the Lutcher Brown endowment. We thank the staff scientists for assistance with remote data collections at beamline 9-2 of the SSRL and beamlines 19-BM, 19-ID, 22-ID, and 22-BM in the Argonne National Laboratory of Advanced Photon Source (APS). Use of the SSRL and APS is supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under Contracts DE-AC02-76SF00515 and DE-AC02-06CH11357, respectively. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including Grant P41GM103393).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005327117/-/DCSupplemental.
Data Availability.
The structure models fitted to the electron density maps have been deposited in the RCSB Protein Data Bank (https://www.rcsb.org). These data are available through the following PDB entries: 6VI5, substrate-free enzyme (1.60-Å resolution) (66); 6VI6, monodentate substrate–enzyme complex (1.90-Å resolution) (67); 6VI7, bidentate substrate–enzyme complex (2.61-Å resolution) (68); 6VI8, superoxo intermediate (1.95-Å resolution) (69); 6VI9, alkylperoxo intermediate (2.31-Å resolution) (70); 6VIA, lactone intermediate (1.59-Å resolution) (71); 6X11, monodentate 3E,5Z,2t,4c-enol tautomer of ACMS–enzyme complex (2.10-Å resolution) (72); and 6VIB, enzyme–product complex in 3E,5Z,2t,4c-enol tautomer and 3E,5Z,2t,4t-enol tautomer conformations (1.84-Å resolution) (73). A video clip, which was constructed by using the University of California, San Francisco, ChimeraX software (74), is available in Movie S1.
References
- 1.Magni G., Di Stefano M., Orsomando G., Raffaelli N., Ruggieri S., NAD(P) biosynthesis enzymes as potential targets for selective drug design. Curr. Med. Chem. 16, 1372–1390 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Li T., Ma J. K., Hosler J. P., Davidson V. L., Liu A., Detection of transient intermediates in the metal-dependent nonoxidative decarboxylation catalyzed by α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase. J. Am. Chem. Soc. 129, 9278–9279 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Colabroy K. L., Begley T. P., The pyridine ring of NAD is formed by a nonenzymatic pericyclic reaction. J. Am. Chem. Soc. 127, 840–841 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Saunders A. H. et al., Characterization of quinolinate synthases from Escherichia coli, Mycobacterium tuberculosis, and Pyrococcus horikoshii indicates that [4Fe-4S] clusters are common cofactors throughout this class of enzymes. Biochemistry 47, 10999–11012 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esakova O. A. et al., Structure of quinolinate synthase from Pyrococcus horikoshii in the presence of its product, quinolinic acid. J. Am. Chem. Soc. 138, 7224–7227 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Kurnasov O. et al., NAD biosynthesis: Identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 10, 1195–1204 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Colabroy K. L., Begley T. P., Tryptophan catabolism: Identification and characterization of a new degradative pathway. J. Bacteriol. 187, 7866–7869 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster J. W., Moat A. G., Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol. Rev. 44, 83–105 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazzaniga F., Stebbins R., Chang S. Z., McPeek M. A., Brenner C., Microbial NAD metabolism: Lessons from comparative genomics. Microbiol. Mol. Biol. Rev. 73, 529–541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esakova O. A. et al., An unexpected species determined by X-ray crystallography that may represent an intermediate in the reaction catalyzed by quinolinate synthase. J. Am. Chem. Soc. 141, 14142–14151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muraki T., Taki M., Hasegawa Y., Iwaki H., Lau P. C. K., Prokaryotic homologs of the eukaryotic 3-hydroxyanthranilate 3,4-dioxygenase and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase in the 2-nitrobenzoate degradation pathway of Pseudomonas fluorescens strain KU-7. Appl. Environ. Microbiol. 69, 1564–1572 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Colabroy K. L., Begley T. P., Ealick S. E., Structural studies on 3-hydroxyanthranilate-3,4-dioxygenase: The catalytic mechanism of a complex oxidation involved in NAD biosynthesis. Biochemistry 44, 7632–7643 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Pidugu L. S. et al., Crystal structures of human 3-hydroxyanthranilate 3,4-dioxygenase with native and non-native metals bound in the active site. Acta Crystallogr. D Struct. Biol. 73, 340–348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegg E. L., Que L. Jr., The 2-His-1-carboxylate facial triad–an emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur. J. Biochem. 250, 625–629 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Liu F. et al., An iron reservoir to the catalytic metal: The ruberoxin iron in an extradiol dioxygenase. J. Biol. Chem. 290, 15621–15634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y., Liu F., Liu A., Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities. J. Biol. Chem. 293, 10415–10424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarcz R., Okuno E., White R. J., Bird E. D., Whetsell W. O. Jr., 3-Hydroxyanthranilate oxygenase activity is increased in the brains of Huntington disease victims. Proc. Natl. Acad. Sci. U.S.A. 85, 4079–4081 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarcz R., Whetsell W. O. Jr., Mangano R. M., Quinolinic acid: An endogenous metabolite that produces axon-sparing lesions in rat brain. Science 219, 316–318 (1983). [DOI] [PubMed] [Google Scholar]
- 19.Eastman C. L., Urbańska E. M., Chapman A. G., Schwarcz R., Differential expression of the astrocytic enzymes 3-hydroxyanthranilic acid oxygenase, kynurenine aminotransferase and glutamine synthetase in seizure-prone and non-epileptic mice. Epilepsy Res. 18, 185–194 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Fornstedt-Wallin B., Lundström J., Fredriksson G., Schwarcz R., Luthman J., 3-Hydroxyanthranilic acid accumulation following administration of the 3-hydroxyanthranilic acid 3,4-dioxygenase inhibitor NCR-631. Eur. J. Pharmacol. 386, 15–24 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Nandi D. et al., Purification and inactivation of 3-hydroxyanthranilic acid 3,4-dioxygenase from beef liver. Int. J. Biochem. Cell Biol. 35, 1085–1097 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Colabroy K. L. et al., The mechanism of inactivation of 3-hydroxyanthranilate-3,4-dioxygenase by 4-chloro-3-hydroxyanthranilate. Biochemistry 44, 7623–7631 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Stone T. W., Darlington L. G., Endogenous kynurenines as targets for drug discovery and development. Nat. Rev. Drug Discov. 1, 609–620 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Stone T. W., Mackay G. M., Forrest C. M., Clark C. J., Darlington L. G., Tryptophan metabolites and brain disorders. Clin. Chem. Lab. Med. 41, 852–859 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Schwarcz R., The kynurenine pathway of tryptophan degradation as a drug target. Curr. Opin. Pharmacol. 4, 12–17 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Beal M. F. et al., Replication of the neurochemical characteristics of Huntington’s disease by quinolinic acid. Nature 321, 168–171 (1986). [DOI] [PubMed] [Google Scholar]
- 27.Dassama L. M. et al., O2-evolving chlorite dismutase as a tool for studying O2-utilizing enzymes. Biochemistry 51, 1607–1616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta R., Fu R., Liu A., Hendrich M. P., EPR and Mössbauer spectroscopy show inequivalent hemes in tryptophan dioxygenase. J. Am. Chem. Soc. 132, 1098–1109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovaleva E. G., Lipscomb J. D., Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science 316, 453–457 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knoot C. J., Purpero V. M., Lipscomb J. D., Crystal structures of alkylperoxo and anhydride intermediates in an intradiol ring-cleaving dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 112, 388–393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huo L. et al., Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action. Nat. Commun. 6, 5935 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis-Ballester A. et al., Molecular basis for catalysis and substrate-mediated cellular stabilization of human tryptophan 2,3-dioxygenase. Sci. Rep. 6, 35169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y. et al., A pitcher-and-catcher mechanism drives endogenous substrate isomerization by a dehydrogenase in kynurenine metabolism. J. Biol. Chem. 291, 26252–26261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kivelson D., Theory of ESR linewidths of free radicals. J. Chem. Phys. 33, 1094–1106 (1960). [Google Scholar]
- 35.Gräslund A., Sahlin M., Electron paramagnetic resonance and nuclear magnetic resonance studies of class I ribonucleotide reductase. Annu. Rev. Biophys. Biomol. Struct. 25, 259–286 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Stubbe J., van Der Donk W. A., Protein radicals in enzyme catalysis. Chem. Rev. 98, 705–762 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Mbughuni M. M. et al., Trapping and spectroscopic characterization of an FeIII-superoxo intermediate from a nonheme mononuclear iron-containing enzyme. Proc. Natl. Acad. Sci. U.S.A. 107, 16788–16793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margolin A. L., Navia M. A., Protein crystals as novel catalytic materials. Angew. Chem. Int. Ed. Engl. 40, 2204–2222 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Stoddard B. L., Farber G. K., Direct measurement of reactivity in the protein crystal by steady-state kinetic studies. Structure 3, 991–996 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., Li J., Liu A., Oxygen activation by mononuclear nonheme iron dioxygenases involved in the degradation of aromatics. J. Biol. Inorg. Chem. 22, 395–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegbahn P. E. M., Haeffner F., Mechanism for catechol ring-cleavage by non-heme iron extradiol dioxygenases. J. Am. Chem. Soc. 126, 8919–8932 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Georgiev V., Borowski T., Blomberg M. R. A., Siegbahn P. E. M., A comparison of the reaction mechanisms of iron- and manganese-containing 2,3-HPCD: An important spin transition for manganese. J. Biol. Inorg. Chem. 13, 929–940 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Deeth R. J., Bugg T. D. H., A density functional investigation of the extradiol cleavage mechanism in non-heme iron catechol dioxygenases. J. Biol. Inorg. Chem. 8, 409–418 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Dong G., Lu J. R., Lai W. Z., Insights into the mechanism of aromatic ring cleavage of noncatecholic compound 2-aminophenol by aminophenol dioxygenase: A quantum mechanics/molecular mechanics study. ACS Catal. 6, 3796–3803 (2016). [Google Scholar]
- 45.Li D. F. et al., Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor. Acta Crystallogr. D Biol. Crystallogr. 69, 32–43 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Jeoung J.-H., Bommer M., Lin T.-Y., Dobbek H., Visualizing the substrate-, superoxo-, alkylperoxo-, and product-bound states at the nonheme Fe(II) site of homogentisate dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 110, 12625–12630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groce S. L., Miller-Rodeberg M. A., Lipscomb J. D., Single-turnover kinetics of homoprotocatechuate 2,3-dioxygenase. Biochemistry 43, 15141–15153 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Cho H. J. et al., Substrate binding mechanism of a type I extradiol dioxygenase. J. Biol. Chem. 285, 34643–34652 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lipscomb J. D., Mechanism of extradiol aromatic ring-cleaving dioxygenases. Curr. Opin. Struct. Biol. 18, 644–649 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaillancourt F. H. et al., Definitive evidence for monoanionic binding of 2,3-dihydroxybiphenyl to 2,3-dihydroxybiphenyl 1,2-dioxygenase from UV resonance Raman spectroscopy, UV/Vis absorption spectroscopy, and crystallography. J. Am. Chem. Soc. 124, 2485–2496 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Viggiani A. et al., The role of the conserved residues His-246, His-199, and Tyr-255 in the catalysis of catechol 2,3-dioxygenase from Pseudomonas stutzeri OX1. J. Biol. Chem. 279, 48630–48639 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Cho J.-H., Jung D.-K., Lee K., Rhee S., Crystal structure and functional analysis of the extradiol dioxygenase LapB from a long-chain alkylphenol degradation pathway in Pseudomonas. J. Biol. Chem. 284, 34321–34330 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kovaleva E. G., Lipscomb J. D., Intermediate in the O–O bond cleavage reaction of an extradiol dioxygenase. Biochemistry 47, 11168–11170 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cervenka I., Agudelo L. Z., Ruas J. L., Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science 357, eaaf9794 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Li T., Walker A. L., Iwaki H., Hasegawa Y., Liu A., Kinetic and spectroscopic characterization of ACMSD from Pseudomonas fluorescens reveals a pentacoordinate mononuclear metallocofactor. J. Am. Chem. Soc. 127, 12282–12290 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Huo L. et al., Human α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling. Proteins 83, 178–187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y., Davis I., Matsui T., Rubalcava I., Liu A., Quaternary structure of α-amino-β-carboxymuconate-ϵ-semialdehyde decarboxylase (ACMSD) controls its activity. J. Biol. Chem. 294, 11609–11621 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huo L., Davis I., Chen L., Liu A., The power of two: Arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in α-amino-β-carboxymuconate-epsilon-semialdehyde decarboxylase. J. Biol. Chem. 288, 30862–30871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li T. et al., α-Amino-β-carboxymuconic-epsilon-semialdehyde decarboxylase (ACMSD) is a new member of the amidohydrolase superfamily. Biochemistry 45, 6628–6634 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Patterson J. I., Brown R. R., Linkswiler H., Harper A. E., Excretion of tryptophan-niacin metabolites by young men: Effects of tryptophan, leucine, and vitamin B6 intakes. Am. J. Clin. Nutr. 33, 2157–2167 (1980). [DOI] [PubMed] [Google Scholar]
- 61.Blight A. R., Cohen T. I., Saito K., Heyes M. P., Quinolinic acid accumulation and functional deficits following experimental spinal cord injury. Brain 118, 735–752 (1995). [DOI] [PubMed] [Google Scholar]
- 62.Pérez-De La Cruz V., Carrillo-Mora P., Santamaría A., Quinolinic acid, an endogenous molecule combining excitotoxicity, oxidative stress and other toxic mechanisms. Int. J. Tryptophan Res. 5, 1–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hofbauer W. et al., Photosystem II single crystals studied by EPR spectroscopy at 94 GHz: The tyrosine radical Y(D)(*). Proc. Natl. Acad. Sci. U.S.A. 98, 6623–6628 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sidabras J. W. et al., Extending electron paramagnetic resonance to nanoliter volume protein single crystals using a self-resonant microhelix. Sci. Adv. 5, y1394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Högbom M. et al., Displacement of the tyrosyl radical cofactor in ribonucleotide reductase obtained by single-crystal high-field EPR and 1.4-Å x-ray data. Proc. Natl. Acad. Sci. U.S.A. 100, 3209–3214 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y., Liu K. F., Yang Y., Liu A., Probing extradiol dioxygenase mechanism in NAD+ biosynthesis by viewing reaction cycle intermediates - A resting state structure. RCSB Protein Data Bank. 10.2210/pdb6VI5/pdb. Deposited 12 January 2020. [DOI]
- 67.Wang Y., Liu K. F., Yang Y., Liu A., Observing a ring-cleaving dioxygenase in action through a crystalline lens - A substrate monodentately bound structure. RCSB Protein Data Bank. 10.2210/pdb6VI6/pdb. Deposited 12 January 2020. [DOI]
- 68.Wang Y., Liu K. F., Yang Y., Liu A., Probing extradiol dioxygenase mechanism in NAD+ biosynthesis by viewing reaction cycle intermediates - A substrate bidentately bound structure. RCSB Protein Data Bank. 10.2210/pdb6VI7/pdb. Deposited 12 January 2020. [DOI]
- 69.Wang Y., Liu K. F., Yang Y., Liu A., Observing a ring-cleaving dioxygenase in action through a crystalline lens - A superoxo bound structure. RCSB Protein Data Bank. 10.2210/pdb6VI8/pdb. Deposited 12 January 2020. [DOI]
- 70.Wang Y., Liu K. F., Yang Y., Liu A., Observing a ring-cleaving dioxygenase in action through a crystalline lens - An alkylperoxo bound structure. RCSB Protein Data Bank. 10.2210/pdb6VI9/pdb. Deposited 12 January 2020. [DOI]
- 71.Wang Y., Liu K. F., Yang Y., Liu A., Observing a ring-cleaving dioxygenase in action through a crystalline lens - A seven-membered lactone bound structure. RCSB Protein Data Bank. 10.2210/pdb6VIA/pdb. Deposited 12 January 2020. [DOI]
- 72.Wang Y., Liu K. F., Yang Y., Liu A., Observing a ring-cleaving dioxygenase in action through a crystalline lens - An enol tautomer of ACMS monodentately bound structure. RCSB Protein Data Bank. 10.2210/pdb6X11/pdb. Accessed 17 May 2020. [DOI]
- 73.Wang Y., Liu K. F., Yang Y., Liu A., Observing a ring-cleaving dioxygenase in action through a crystalline lens - Enol tautomers of ACMS bidentately bound structure. RCSB Protein Data Bank. 10.2210/pdb6VIB/pdb. Deposited 12 January 2020. [DOI]
- 74.Goddard T. D. et al., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure models fitted to the electron density maps have been deposited in the RCSB Protein Data Bank (https://www.rcsb.org). These data are available through the following PDB entries: 6VI5, substrate-free enzyme (1.60-Å resolution) (66); 6VI6, monodentate substrate–enzyme complex (1.90-Å resolution) (67); 6VI7, bidentate substrate–enzyme complex (2.61-Å resolution) (68); 6VI8, superoxo intermediate (1.95-Å resolution) (69); 6VI9, alkylperoxo intermediate (2.31-Å resolution) (70); 6VIA, lactone intermediate (1.59-Å resolution) (71); 6X11, monodentate 3E,5Z,2t,4c-enol tautomer of ACMS–enzyme complex (2.10-Å resolution) (72); and 6VIB, enzyme–product complex in 3E,5Z,2t,4c-enol tautomer and 3E,5Z,2t,4t-enol tautomer conformations (1.84-Å resolution) (73). A video clip, which was constructed by using the University of California, San Francisco, ChimeraX software (74), is available in Movie S1.