Abstract
The Notch pathway is critical for the development of the extracellular matrix in cartilage by regulating both anabolic and catabolic cellular activities. Similarly, Notch signaling plays a biphasic role in adult cartilage health and osteoarthritis by maintaining homeostasis and contributing to degeneration, respectively. The temporomandibular joint (TMJ) is the synovial joint of the craniofacial complex and is subject to injury and osteoarthritis. While Notch has been studied in axial skeletal joints, little is known about the role of Notch in TMJ development and disease. We identified fibrocartilage stem cells (FCSCs) localized within the TMJ condyle superficial zone niche that regenerate cartilage and repair joint injury. Here we investigate the role of Notch in regulating TMJ development and FCSC fate. Using a Notch reporter mouse, we discovered FCSCs localized within the TMJ superficial niche exhibit Notch activity during TMJ morphogenesis. We further showed that constitutively activating Notch promotes FCSC differentiation toward both cartilage and bone lineages, but inhibits adipogenesis. Using a TNF-α–induced TMJ inflammatory arthritis mouse model, we found that the expression of Notch receptors and ligands are upregulated and coupled with cells undergoing cartilage to bone transdifferentiation, which may contribute to TMJ pathogenesis. We also discovered that global Notch inhibition reduces osteogenic and chondrogenic differentiation of FCSCs. Together, these findings suggest that Notch is critical for FCSC fate specification and TMJ homeostasis, and reveal that inhibition of the Notch pathway may be a new therapeutic target for treating TMJ osteoarthritis.
Keywords: cartilage, temporomandibular disorders, regenerative medicine, bone biology, cell signaling, joint disease
Introduction
The temporomandibular joint (TMJ) is critical for chewing food and speaking. The TMJ disc divides the intra-articular joint space into the inferior joint cavity (IJC) and superior joint cavity (SJC), which facilitate rotational and translational mechanics, respectively (Okeson 2003). The condyle surface and disc are fibrocartilage (Benjamin and Evans 1990), which consists of fibrous and cartilaginous tissues (Singh and Detamore 2009). During TMJ development the condyle undergoes endochondral ossification and cartilage anlagen are remodeled into bone (Silbermann and Frommer 1972). At E14.5 the condylar primordium (CP) is established, at E16.5 the SJC is formed and disc cells separate, at E18.5 the IJC and disc are formed (Owtad et al. 2013; Liang et al. 2016). Unlike limb growth plates, the TMJ condyle functions both as a growth center for the jaw and also as an articular joint (Shibata et al. 1996). Neural crest cells and mandibular bone periosteum cells are speculated to contribute to TMJ formation (Chai et al. 2000), but the cell of origin is not well established. Given the restricted number of cells and lack of vascular supply, the TMJ has poor regenerative properties (Huey et al. 2012). Therefore, TMJ osteoarthritis (OA) causes pain, jaw dysfunction, and irreversible tissue loss (Scrivani et al. 2008). TMJ OA treatments are limited to either symptom management (Stoustrup and Twilt 2015) or surgery (Kneeland et al. 1987; Englund et al. 2003). Our lab has discovered that the TMJ condyle superficial zone (SZ) harbors fibrocartilage stem cells (FCSCs) that differentiate into chondrocytes, regenerate cartilage, and may be manipulated for the treatment of TMJ OA (Embree et al. 2016; Nathan et al. 2018). However, the signals critical for regulating FCSC fate during TMJ homeostasis and repair are not well defined.
The Notch pathway plays a key role in cartilage development and OA by performing dual functions in regulating molecules involved in both cartilage formation and degradation (Hosaka et al. 2013; Liu et al. 2015; Saito and Tanaka 2017). The mammalian Notch signaling pathway is highly conserved and has 4 different Notch receptors, Notch1–4, that exist as single transmembrane proteins (Artavanis-Tsakonas et al. 1999). A total of 5 canonical Notch ligands, including δ-like1 and Jagged1, bind to Notch receptors to initiate Notch pathway activation, and cause a series of receptor cleavage events that result in the cytoplasmic release of Notch intracellular domain (NICD) (D’Souza et al. 2010). NICD translocates to the nucleus and activates downstream target genes, such as members of the Hes/Hey families (Artavanis-Tsakonas et al. 1999; Kopan and Ilagan 2009). In axial skeletal joints, Notch receptors are expressed during development and in adult articular cartilage (Hayes et al. 2003) and increase in expression in human osteoarthritic cartilages (Mahjoub et al. 2012). Moreover, constitutively activated Notch in mice leads to progressive degenerative change in the knee joints of mice (Liu et al. 2015). However, the role of Notch regulation of craniofacial synovial joints is substantially understudied. Here we examined the role of Notch signaling in regulating TMJ FCSC fate, pathogenesis, and as a plausible therapeutic target for TMJ OA.
Materials and Methods
Animals
All preclinical animal studies conformed to Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines.
All animal procedures were performed using equal males and females with approval from the Institution of Animal Care and Use Committee at Columbia University Irving Medical Center (AC-AAAV0454; AC-AAAU6480; AC-AAAV0455). To evaluate Notch activity, CBF:H2B-Venus transgenic mice (JAX Mice, Ellsworth) (Nowotschin et al. 2013) were bred and genotyped according to recommended protocol. To collect CBF:H2B-Venus transgenic embryos, breeding pairs were placed together in the evening and females were checked for plugs after 12 h followed by separation. CBF:H2B-Venus embryos were collected at E14.5 (n = 4 mice) and E18.5 (n = 6 mice) and pups were collected at P0 (n = 3 mice) and P21 (n = 3 mice). 10-wk–old Sprague Dawley rats (n = 8 rats, Taconic Biosciences) were utilized for primary cell isolation. For TNF-α inflammatory mouse model experiments, 8-wk–old C57BL/6J mice (JAX Mice) were used (n = 6 mice).
FCSC Isolation and Culture
The TMJ condylar cartilage superficial zone is a niche for FCSCs (Embree et al. 2016; Nathan et al. 2018). FCSCs were isolated from 10-wk–old Sprague Dawley rats as previously described (Embree et al. 2016). Briefly, superficial zone tissues were physically separated from the condyle using fine tipped forceps and digested in dispase II/collagenase I (4mg/mL, 3mg/mL). Single cell suspensions of FCSCs were cultured (5% CO2, 37°C) in basal medium consisting of DMEM (Invitrogen) supplemented with 20% lot-selected fetal bovine serum (FBS, Hyclone), glutamax (Invitrogen), penicillin-streptomycin (Invitrogen), and 100 mM 2-mercaptoethanol (Gibco) for 4 to 6 d. Heterogeneous single cell FCSCs colonies were detached with trypsin-EDTA (Gibco) and plated at P1 for the in vitro experiments.
Bacterial Strains and Growth Conditions
Escherichia coli strains were grown aerobically at 37°C in SOC medium (Super Optimal Broth with Catabolic repressor, ThermoFisher). All cloning experiments were performed using the electrocompetent recA mutant strain E. coli Stellar (Clontech), plated on campenicillin 1.5% agar plates (ThermoFisher), and incubated overnight at 37°C. 5 ng of NICD pCCL DNA was used for transformation. Colonies were digested using the restriction enzyme EcoR1 and underwent PCR using NICD forward and reverse primers. QIAfilter Plasmid Maxi Kit (Qiagen) was used to isolate plasmid DNA and was sequenced at Columbia University Medical Center to ensure the correct product was made (Appendix Fig. 1A).
Lentiviral Mediated Transduction
293T cells were transfected with either the pCCL.pkg vector encoding GFP (vector control) or constitutively active NICD, which encodes the entire cytoplasmic domain and is ligand independent, as previously described (Murtomaki et al. 2013). Rat FCSCs were infected with lentivirus and after 48 h of transduction the cells were imaged (Appendix Fig. 1B).
TMJ Condyle Explant Model
TMJ condyles were dissected from male and female C57Bl/6 mice, age 6 wk (n = 7) under sterile conditions. The TMJ condyle and ~3 mm of the ramus was dissected from the mandible and cultured in a 24-well plate using BGJb media (Thermo Fisher) supplemented with 1% penicillin/streptomycin (Invitrogen). Condyle explants were incubated overnight (5% CO2, 37°C) to normalize to culture conditions. After overnight incubation, 2 ng/mL or 10 ng/mL rhTNF-alpha (R&D 210-TA-020/CF) was added to the media, while the contralateral explant was treated with the vehicle control (Henry Schein). Explants were subjected to 3 freeze/thaw cycles for control. After 48 h, condyle explants were snap-frozen in liquid nitrogen for RNA isolation.
RNA Isolation and qRT-PCR
Total RNA was purified from FCSCs (Invitrogen) and treated with DNAse I (Ambion) to remove genomic DNA. RNA quantity and purity was determined using spectrophotometer (DeNovix, Inc.). RNA samples (260/280≥1.8) were used to obtain cDNA (Biorad). qRT-PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) and rat primers (Appendix Table 1) (Integrated DNA Technologies). Gene expression levels were normalized to the housekeeping gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh).
Multi-lineage Differentiation
Multi-lineage differentiation was tested in vitro using chemically defined media for chrondrogenesis, osteogenesis, and adipogenesis (Embree et al. 2016). For chrondrogenesis, cells (1 × 106 per pellet) were pelleted in 15 mL polypropylene tubes by centrifugation and cultured (5% CO2, 37°C) for 3 wk in high glucose Dulbecco’s Modified Eagle medium (Gibco) supplemented with 10-8 M dexamethasone, 100 μM L-ascorbic acid, 1% insulin, transferrin, selenium (ITS), 1 mM pyruvate, and 10 ng/ml TGF-β1 (R&D Systems). After 3 wk, pellets were processed for histology and immunohistochemistry. To induce osteogenesis, FCSCs (5 × 104) were cultured in a 12-well plate for 1 wk in media containing αMEM supplemented with 20% FBS, dexamethasone (10−8 M), 100 μM L-ascorbic acid, and 2 mM β-glycerophosphate (ThermoFisher). To induce adipogenesis, cells (5 × 104) were cultured in a 12-well plate using commercial adipogenic media for 1 wk (Gibco). Calcium nodules and fat were visualized by staining with alizarin red and Oil Red O, respectively.
Histology and Immunohistochemistry
Tissue samples were fixed in 4% paraformaldehyde, decalcified in ethylenediaminetetraacetic acid (Sigma Aldrich), and prepared for either paraffin or frozen embedded sections. Serial tissue sections were stained with hematoxylin and eosin (H&E). For immunohistochemistry tissue sections were enzymatically treated with Chondroitinase ABC (ThermoFisher) and immunolabeled with primary antibodies (Appendix Table 2) at 4°C overnight followed by secondary antibody (Invitrogen, 1:1000) to detect immunoreactivity. Isotype-matched negative control antibodies were used under the same conditions.
Inflammatory Arthritis Mouse Models
rhTNF-α (R&D 210-TA-020/CF) (0.020 mL, 0.5 μg/mL, 8 wk, n = 6 mice) was injected unilaterally into the TMJ intra-articular space of C57Bl/6 mice twice at 3-d intervals as previously described (Morel et al. 2019). Saline was injected on the contralateral side as control. Animals were euthanized after 14 d and prepared for histology. Treated TMJs were processed for histology after 2 and 14 d and examined by immunohistochemistry.
Statistical Analysis
All statistics were calculated using Prism8 (GraphPad Software). The statistical significance between 2 groups was determined using paired Student’s t test assuming Gaussian distribution. The normality of distribution was confirmed using Kolmogorov-Smirnov test and the resulting two-tailed P value ≤0.05 was regarded as statistically significant. Among 3 groups, 1-way ANOVA followed by Tukey’s post hoc test was used for statistical comparisons. For multiple comparisons, a 2-way ANOVA followed by Tukey’s post hoc or multiple t tests followed by Bonferroni correction was used for statistical comparisons.
Results
Notch Activity in FCSCs During TMJ Morphogenesis
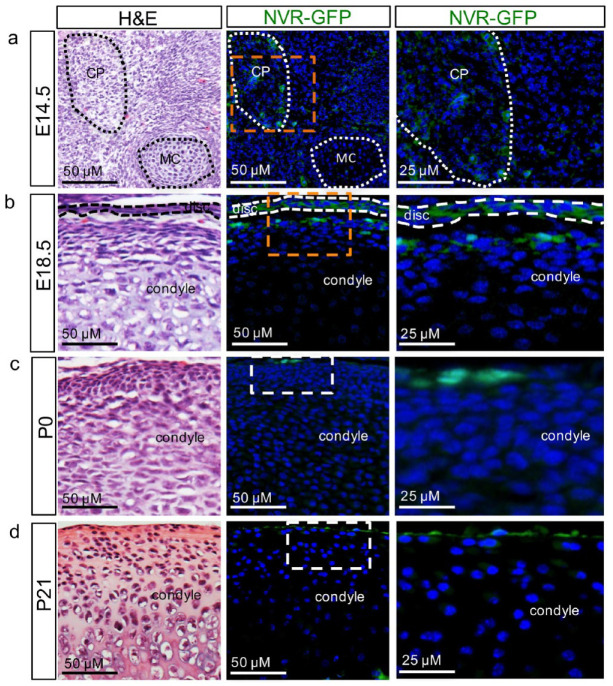
While Notch signaling has been shown to play a critical role in axial skeletal development (Canalis 2018), the role of Notch signaling in the formation of the craniofacial synovial joint or the TMJ is not well defined. To determine whether cells exhibit Notch activity during TMJ morphogenesis, we used the CBF:H2B-Venus reporter mouse strain, whereby individual cells transducing a Notch signal can be visualized by green fluorescent protein (GFP) (Nowotschin et al. 2013) (Fig. 1). Embryonic mammalian TMJ is developed from 2 distinct blastemas, including the glenoid blastema, which forms the glenoid fossa, and the CP blastema, which forms the condyle and disc (Yamaki et al. 2005; Liang et al. 2016). In the CBF:H2B-Venus reporter mouse or Notch venus reporter-GFP (NVR-GFP) mouse strain at E14.5, the CP is formed adjacent to Meckel’s cartilage (MC) (Owtad et al. 2013; Liang et al. 2016) and GFP+ cells exhibiting Notch activity can be visualized within the CP (Fig. 1A), which will eventually form the TMJ disc and condyle. At E18.5, following the separation of the disc from the condyle and formation of the SJC and IJC, GFP+ cells with Notch activity are localized within the TMJ disc and the superficial zone niche harboring FCSCs with potent chondrogenic potential (Embree et al. 2016) (Fig. 1B). GFP+ cells exhibiting Notch activity remained localized to the condylar cartilage superficial zone niche in postnatal mice prior to weaning at P0 (Fig. 1C) and at weaning at P21 (Fig. 1D). We next evaluated Notch and Notch ligands correlated with Notch reporter GFP+ cells at E18.5 in the TMJ by immunohistochemistry (Appendix Fig. 1). Both Notch1 and Notch2 were co-localized with GFP+ cells, where Notch4 was not expressed in the TMJ (Appendix Fig. 1), suggesting that Notch1 and Notch2 may activate Notch signaling in NVR GFP+ cells during TMJ formation. Downstream Notch target Hes1 and Notch ligands DLL4 and Jagged were not co-localized with NVR GFP+ cells (Appendix Fig. 1), suggesting that other targets and ligands may be implicated during TMJ development. Taken together, these data suggest that Notch activity may play a role in mediating FCSC fate during TMJ morphogenesis.
Figure 1.
Fibrocartilage stem cells (FCSCs) exhibit Notch activity during temporomandibular joint (TMJ) morphogenesis. CBF:H2B-Venus reporter mouse strain or the Notch venus reporter-green fluorescent protein (NVR-GFP), whereby individual cells transducing a Notch signal are visualized by GFP in mice at (A) E14.5 within the condylar primordium (CP) adjacent to Meckel’s cartilage (MC), (B) E18.5 within the TMJ disc and superficial zone lining the mandibular condyle, (C) P0 within the superficial zone (SZ) harboring FCSCs, (D) P21 within the SZ harboring FCSCs.
Notch Promotes FCSC Fate Specification Toward Cartilage and Bone Lineages
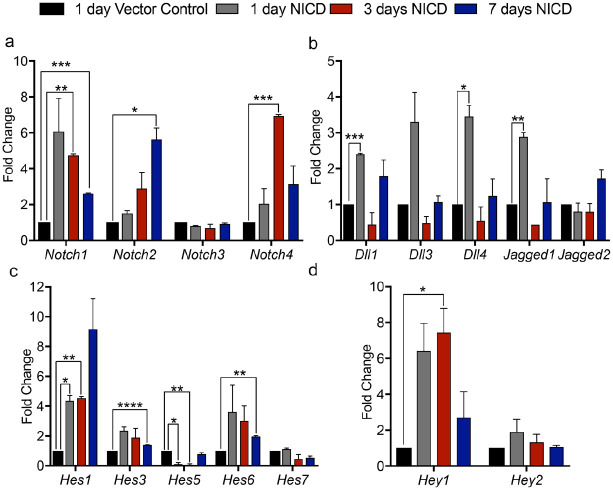
The Notch pathway plays a crucial role in stem cell fate specification during embryogenesis, including modulating stem cell differentiation and self-renewal (Calvi et al. 2003; Shawber et al. 2007). Our data show that Notch activity is present in cells within the TMJ superficial zone niche harboring FCSCs (Fig. 1). To investigate the putative role of Notch signaling in modulating FCSC lineage, we created a FCSC line with ligand-independent constitutively activated Notch signaling (Appendix Fig. 2, Figs. 2 and 3). To isolate FCSCs, the mandibular condyle superficial zone was surgically removed from adult Sprague Dawley rats, enzymatically digested, and heterogeneous single-cell FCSC colonies were expanded and plated at P1 for transduction (Embree et al. 2016; Nathan et al. 2018). We transduced FCSCs with a lentivirus encoding NICD to ectopically express a constitutively active form of the Notch1 cytoplasmic domain (Shawber et al. 2007; Murtomaki et al. 2013). We also transduced FCSCs with a control lentivirus encoding only GFP. Transduction of NICD and control plasmids with lentiviruses in FCSCs was confirmed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and visualization of GFP signal (Appendix Fig. 2B). Transcripts for Notch receptors (Notch1, Notch2, Notch3, Notch4), ligands (DLL1, DLL3, DLL3, Jagged1, Jagged2) and downstream targets were evaluated in transduced NICD FCSCs at 1, 5, and 7 d following infection and compared to GFP FCSCs at 1 d by qRT-PCR (Fig. 2). Transduced NICD FCSCs exhibited marked increase in Notch1 and Notch2 and ligands Dll1, Dll4, and Jagged1, but not Notch3 receptor or Jagged2 ligand similar to expression in adult articular cartilage (Fig. 2A and B) (Ustunel et al. 2008; Grogan et al. 2009; Hosaka et al. 2013). Downstream Notch targets Hes1, Hes3, Hes5, Hes6, and Hey1 were significantly upregulated in transduced NICD FCSCs relative to transduced GFP FCSC control, confirming Notch pathway activation in NICD FCSC line (Fig. 2C and D).
Figure 2.
Transduced fibrocartilage stem cells (FCSCs) with a Notch1 intracellular domain (NICD) constitutively activates Notch pathway. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) of FCSCs transduced with a lentivirus encoding for a NICD after 1, 3, and 7 d relative to FCSCs transduced with an empty vector control of transcripts related to (A) Notch receptors (Notch1–4), (B) ligands (Dll1, Dll3, Dll4, Jagged1, Jagged2), and (C, D) downstream targets (Hes1, Hes3, Hes5, Hes6, Hes7, Hey1, Hey2). Data represented are mean fold change relative to 1 d vector control and normalized to Glyceraldehyde 3-phosphate dehydrogenase (Gapdh); n = 3; **** P ≤ 0.0001; ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
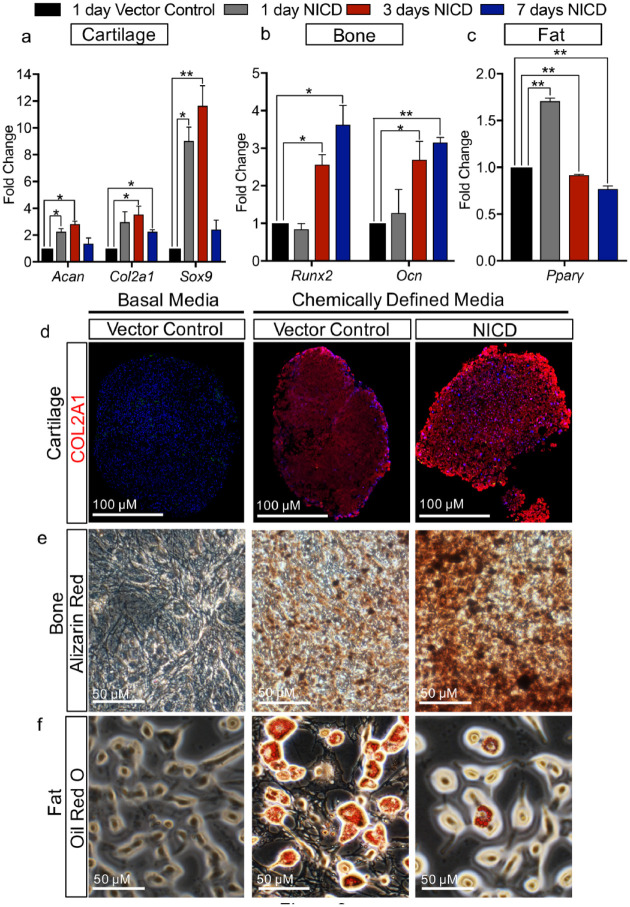
Figure 3.
Notch1 promotes fibrocartilage stem cell (FCSC) differentiation toward cartilage and bone lineages, but not fat lineage. (A–C) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) of FCSCs transduced with a lentivirus encoding for a Notch1 intracellular domain (NICD) after 1, 3, and 7 d relative to FCSCs transduced with an empty vector control of transcripts related to (A) cartilage (Acan, Col2a1, Sox9), (B) bone (Runx2, Ocn), and (C) fat (Pparγ). Data represented are mean fold change relative to 1 d vector control and normalized to Gapdh; n = 3; **P ≤ 0.01; * P ≤ 0.05. (D–F) FCSCs transduced with a lentivirus encoding for a NICD were subject to chemically defined media in comparison to basal media control and FCSCs transduced with vector control. Differentiation toward (D) cartilage lineage was confirmed by immunohistochemistry for COL2A1, (E) bone lineage was confirmed by Alizarin red staining for calcium deposition, and (F) fat lineage was confirmed by Oil Red O staining for lipid droplets.
To evaluate the role of the Notch pathway in defining FCSC lineage specification, we evaluated cartilage, bone, and fat transcripts in transduced NICD FCSCs relative to GFP FCSCs by qRT-PCR. The expression of both cartilage (Fig. 3A, Sox9, Acan, Col2a1) and bone transcripts (Fig. 3B, Runx2, Ocn) were significantly higher in NICD FCSCs, while the expression of the fat transcript Pparγ (Fig. 3C) was significantly decreased in NICD FCSCs relative to GFP FCSCs. To further confirm the role of Notch in mediating FCSC fate, we performed multilineage differentiation assays using NICD FCSCs in chemically defined media. Chondrogenic differentiation was assessed in pellet cultures and revealed that NICD FCSCs showed abundant expression of Aggrecan relative to control GFP FCSCs cultured in basal media or chondrogenic media (Fig. 3D). Upon culture in osteogenic media, NICD FCSCs showed intense alizarin red staining relative to GFP FCSCs cultured in basal and osteogenic media (Fig. 3E). On the other hand, NICD FCSCs cultured in adipogenic media revealed reduction in Oil O Red relative to GFP FCSC controls (Fig. 3F). Taken together, these data suggest that the Notch pathway promotes FCSCs to differentiate toward cartilage and bone lineages, but not toward the fat lineage.
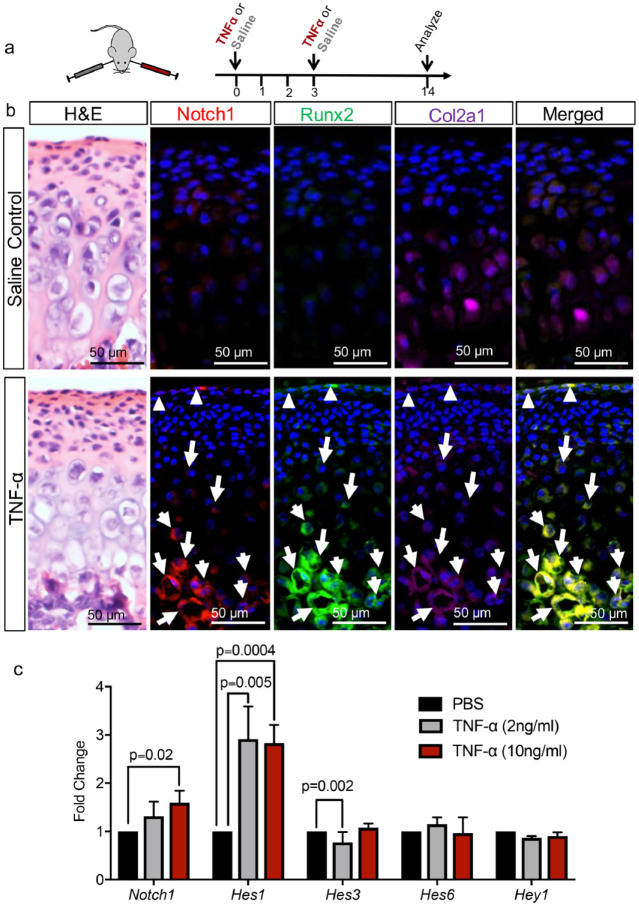
Notch 1 Is Coupled with Cartilage to Bone Transformation in a TMJ Arthritis Mouse Model
We showed Notch activity during TMJ morphogenesis in FCSCs and also found that constitutively activated Notch in FCSCs mediates both osteogenesis and chondrogenesis. The Notch pathway has also been shown to induce transdifferentiation in multiple mature cell types during disease pathogenesis, including epithelial to mesenchymal transformation in cancer (Shao et al. 2015; Choi et al. 2019). Recent mouse genetic studies have showed that chondrocytes directly transdifferentiate into osteoblasts and osteocytes during endochondral osteogenesis and disease in the TMJ mandibular condyle (Yang et al. 2014; Zhou et al. 2014; Jing et al. 2015; Park et al. 2015). However, whether Notch mediates cartilage to bone transformation is unknown. Therefore, we evaluated whether Notch played a role in cartilage to bone transformation in TMJ pathology using a TNF-α–induced TMJ arthritis mouse model that exhibits acute inflammation, including increased Adamts5 and IL1-β expression, and increased extracelluar matrix turnover (Morel et al. 2019) (Fig. 4). To induce inflammatory arthritis, TNF-α (0.020 mL of 0.5 μg/mL) was injected unilaterally into the TMJ intra-articular space in 6-wk–old mice twice every 3 d and saline was injected into the contralateral TMJ as a control (Fig. 4A). After 14 d, we evaluated Notch1 ligand expression by immunohistochemistry in chondrocytes transdifferentiating into osteoblasts dually marked by Col2a1+/Runx2+ cells (Zhou et al. 2014; Jing et al. 2015). Immunohistochemistry showed in saline control Runx2 expression (purple) in maturation and hypertrophic zones, with some Col2a1 expressing chondrocytes (green) (Fig. 4B). Cells dually marked with Col2a1+/Runx2+ (arrows) were localized at the cartilage/bone interphase, suggesting cartilage to bone transdifferentiation, similar to other studies (Jing et al. 2015) (Fig. 4B, arrows). However, Col2a1+/Runx2+ cells did not express Notch1 (red) in saline control (Fig. 4B). Upon local delivery of TNF-α, a cytokine upregulated in OA (Morel et al. 2019), Col2a1+/Runx2+ cells were co-localized with Notch1 in the superficial zone harboring FCSCs (Fig. 4B, triangles) and within the cartilage to bone interphase (Fig. 4B, arrows). These data suggest that Notch1 may mediate cartilage to bone transformation in inflammatory TMJ OA. We further evaluated other Notch ligands that may also promote cartilage to bone transformation upon exposure to TNF-α by immunohistochemistry. Both Notch2 and Notch4 were upregulated in TMJ condyles exposed to TNF-α after 2 and 14 d (Appendix Fig. 3), while DDL4, JAGGED1, and HES1 were upregulated after 14 d following 2 intra-articular injections of TNF-α (Appendix Fig. 4). To investigate whether ectopic Notch1 expression during TNF-α–induced TMJ arthritis activates Notch signaling, we evaluated Notch downstream targets in TMJ condyle explants treated with 2 and 10 ng/mL TNF-α (Fig. 4C). qRT-PCR using RNA derived from TMJ explants showed that Notch1 expression was significantly increased in TNF-α–treated explants, while the expression of downstream targets Hes1 and Hes3 were also significantly increased in TNF-α–treated condyle explants (Fig. 4C). Taken together, these data suggest that Notch activation also plays a role in TMJ arthritis, where Notch ligands and downstream targets are upregulated in TMJ arthritis and may promote cartilage to bone transformation.
Figure 4.
Notch1 is coupled with cartilage to bone transformation in TNF-α–induced temporomandibular joint (TMJ) arthritis mouse model. (A) TNF-α (0.020 mL of 0.5 μg/mL) was injected unilaterally into the TMJ intra-articular space of 6-wk–old mice twice 3 d apart to induce inflammatory TMJ arthritis, while saline was injected into the contralateral TMJ as a control. TMJ condyles were examined by immunohistochemistry after 14 d. (B) Immunohistochemistry was performed for Notch1 (red), Runx2 (green), Col2a1 (purple) using TMJ condyles following exposure to saline control and TNF-α. In TNF-α treated condyles, triangles indicate Notch1+/Col2a1+/Runx2+ cells in the superficial zone harboring fibrocartilage stem cells, and arrows indicate Notch1+/Col2a1+/Runx2+ cells in maturation and hypertrophic zones. (C) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) of TMJ condyle explants treated with either TNF-α (2 and 10 ng/mL) or vehicle controls for 48 h for Notch1, Hes1, Hes3, Hes6, and Hey1. Data represented are mean fold change relative to vehicle control explants and normalized to Gapdh; n = 3 explants.
Notch Inhibition Suppresses Osteogenic Differentiation of FCSCs
Our data suggest that Notch activation in FCSCs induces their differentiation into bone and promotes cartilage to bone transformation during TMJ arthritis. Therefore, Notch inhibition may also suppress osteogenic differentiation of FCSCs. To induce osteogenesis, we cultured FCSCs in osteogenic media in the presence of global Notch inhibitor gamma-secretase inhibitor IX or DAPT (Hosaka et al. 2013). We evaluated Notch, bone, and cartilage transcripts in FCSCs relative to FCSCs treated with DAPT by qRT-PCR (Fig. 5) after 1 and 3 wk in osteogenic media. Notch1 and bone transcripts (Runx2, Ocn) were significantly upregulated in control FCSCs at 1 wk relative to 3 wk, confirming osteogenic differentiation of FCSCs is coupled with Notch1 expression (Fig. 5). However, ectopic addition of the global Notch inhibitor DAPT to FCSCs significantly reduced the expression of Notch1, Runx2, and Ocn at 3 wk relative to control FCSCs (Fig. 5). These data suggest that Notch inhibition suppresses FCSC fate toward bone and may be used to block cartilage to bone transformation for the treatment of TMJ arthritis (Luo et al. 2018).
Figure 5.
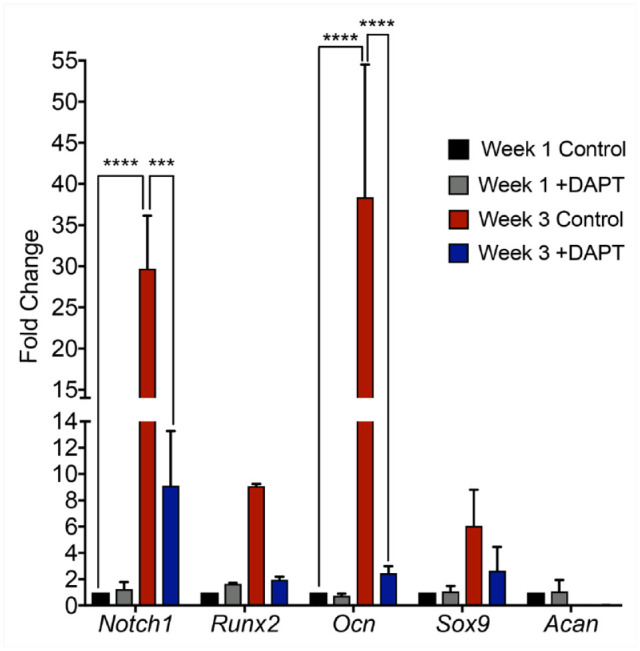
Global Notch inhibitor DAPT suppresses osteogenic differentiation of fibrocartilage stem cells (FCSCs). qRT-PCR of FCSCs cultured in osteogenic media in the absence and presence of DAPT after 1 and 3 wk for Notch1, Runx2, Ocn, Sox9 and Acan. Data represented are mean fold change relative to 1 wk control FCSCs and normalized to Gapdh; n = 2; **** P ≤ 0.0001; ***P ≤ 0.001.
Discussion
This study underscores the importance of the Notch pathway in mediating cell fate decisions during TMJ development and disease. First, we show that Notch activity is present in the superficial zone niche harboring FCSCs during TMJ morphogenesis, suggesting that Notch regulates FCSC fate decisions during TMJ development. To further investigate the role of Notch in mediating FCSC fate, we created a FCSC line with ligand independent constitutively activated Notch activity and showed that Notch activation promotes osteogenic and chondrogenic differentiation of FCSCs, but not adipogenesis. These data suggest that Notch induces FCSCs to differentiate toward cartilage and bone lineages. Our study is in line with others demonstrating a key role of Notch in regulating chondrogenesis and endochondral ossification during axial skeletal development (Hosaka et al. 2013). Differential Notch signaling has been studied as a key modulator of chondrogenesis, by both inhibiting and promoting mesenchymal stem cells and chondrocytes (Karlsson and Lindahl 2009). Thus, the effect of Notch on cells in cartilage is both ligand-dependent and cell-type dependent.
While endochondral ossification is critical for bone development, it also plays a key role during OA (Hosaka et al. 2013; Liu et al. 2015). The central dogma of endochondral ossification relies on hypertrophic differentiation and apoptosis of chondrocytes, degradation of the cartilage ECM by proteinases, vascularization, and replacement with bone. However, recent evidence suggests a new model, whereby chondrocytes directly transdifferentiate into osteocytes in the jaw bone (Yang et al. 2014; Zhou et al. 2014; Jing et al. 2015; Park et al. 2015). The role of Notch signaling in promoting epithelial to mesenchymal transdifferentiation of cancer cells induces metastasis in breast cancer (Shao et al. 2015; Choi et al. 2019). Here, we also show a role for Notch in promoting transdifferentiation in TMJ chondrocytes using a TNF-α–induced TMJ arthritis mouse model (Morel et al. 2019). We showed that Notch1 expression is upregulated and in both Col2a1+/Runx2+ FCSCs and in mature chondrocytes localized in the cartilage/bone interphase. Moreover, both Notch receptors and ligands are upregulated in a TNF-α–induced TMJ arthritis mouse model, further confirming that Notch activity is upregulated in TMJ arthritis. These data suggest that Notch activity promotes cartilage to bone transformation during TMJ arthritis.
Our studies are in line with other studies implicating high Notch activity in OA (Hosaka et al. 2013; Liu et al. 2015; Luo et al. 2018). Specifically, Notch1 activity is significant in osteoarthritic cartilage relative to healthy cartilage, with abundant expression in the most damaged areas (Mahjoub et al. 2012; Saito and Tanaka 2017; Canalis 2018). Notch activation in OA has been shown to induce the expression and activation of degradative proteinases and inflammatory cytokines that ultimately contribute to cartilage demise, including MMP13, IL-1β, and IL-6 (Hosaka et al. 2013; Liu et al. 2015). We used a TNF-α–induced TMJ arthritis mouse model with high levels of IL-1β and ECM turnover and showed that Notch receptors and ligands were also upregulated, suggesting that Notch may be inducing ECM turnover in our models (Morel et al. 2019). These studies suggest that inhibitors of Notch could be a beneficial therapeutic in the treatment of TMJ arthritis. As such, similar to studies in knee OA (Hosaka et al. 2013; Luo et al. 2018), we showed that application of small molecule inhibitor of Notch DAPT reduced transcripts for Notch1 and inflammation. In addition to blocking inflammation, DAPT also reduced cartilage and bone transcripts in FCSCs. These data suggest a second therapeutic mechanism, whereby Notch inhibition blocks cartilage to bone transformation in TMJ arthritis. Taken together, we show that inhibition of Notch pathway may be a new therapeutic target for treating TMJ OA.
Author Contributions
A. Ruscitto, V. Scarpa, M. Morel, contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript; S. Pylawka, contributed to data acquisition, critically revised the manuscript; C.J. Shawber, contributed to the data interpretation, critically revised the manuscript; M.C. Embree, contributed to conception, design, and data interpretation, drafted the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034520924656 for Notch Regulates Fibrocartilage Stem Cell Fate and Is Upregulated in Inflammatory TMJ Arthritis by A. Ruscitto, V. Scarpa, M. Morel, S. Pylawka, C.J. Shawber and M.C. Embree in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
This investigation was supported by National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) 5R00DE0220660 (M.C.E.) and NIH/NIDCR 1R01DE 029068-01A1 (M.C.E.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iD: M.C. Embree  https://orcid.org/0000-0003-2256-0001
https://orcid.org/0000-0003-2256-0001
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284(5415):770–776. [DOI] [PubMed] [Google Scholar]
- Benjamin M, Evans EJ. 1990. Fibrocartilage. J Anat. 171:1–15. [PMC free article] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. 2003. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 425(6960):841–846. [DOI] [PubMed] [Google Scholar]
- Canalis E. 2018. Notch in skeletal physiology and disease. Osteoporos Int. 29(12):2611–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr., Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. 2000. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 127(8):1671–1679. [DOI] [PubMed] [Google Scholar]
- Choi S, Yu J, Park A, Dubon MJ, Do J, Kim Y, Nam D, Noh J, Park KS. 2019. BMP-4 enhances epithelial mesenchymal transition and cancer stem cell properties of breast cancer cells via Notch signaling. Sci Rep. 9(1):11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza B, Meloty-Kapella L, Weinmaster G. 2010. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 92:73–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embree MC, Chen M, Pylawka S, Kong D, Iwaoka GM, Kalajzic I, Yao H, Shi C, Sun D, Sheu TJ, et al. 2016. Exploiting endogenous fibrocartilage stem cells to regenerate cartilage and repair joint injury. Nat Commun. 7:13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund M, Roos EM, Lohmander LS. 2003. Impact of type of meniscal tear on radiographic and symptomatic knee osteoarthritis: a sixteen-year followup of meniscectomy with matched controls. Arthritis Rheum. 48(8):2178–2187. [DOI] [PubMed] [Google Scholar]
- Grogan SP, Miyaki S, Asahara H, D’Lima DD, Lotz MK. 2009. Mesenchymal progenitor cell markers in human articular cartilage: normal distribution and changes in osteoarthritis. Arthritis Res Ther. 11(3):R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes AJ, Dowthwaite GP, Webster SV, Archer CW. 2003. The distribution of Notch receptors and their ligands during articular cartilage development.J Anat. 202(6):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka Y, Saito T, Sugita S, Hikata T, Kobayashi H, Fukai A, Taniguchi Y, Hirata M, Akiyama H, Chung UI, et al. 2013. Notch signaling in chondrocytes modulates endochondral ossification and osteoarthritis development. Proc Natl Acad Sci USA. 110(5):1875–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey DJ, Hu JC, Athanasiou KA. 2012. Unlike bone, cartilage regeneration remains elusive. Science. 338(6109):917–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Zhou X, Han X, Jing J, von der Mark K, Wang J, de Crombrugghe B, Hinton RJ, Feng JQ. 2015. Chondrocytes directly transform into bone cells in mandibular condyle growth. J Dent Res. 94(12):1668–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson C, Lindahl A. 2009. Notch signaling in chondrogenesis. Int Rev Cell Mol Biol. 275:65–88. [DOI] [PubMed] [Google Scholar]
- Kneeland JB, Ryan DE, Carrera GF, Jesmanowicz A, Froncisz W, Hyde JS. 1987. Failed temporomandibular joint prostheses: MR imaging. Radiology. 165(1):179–181. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 137(2):216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Li X, Gao B, Gan H, Lin X, Liao L, Li C. 2016. Observing the development of the temporomandibular joint in embryonic and post-natal mice using various staining methods. Exp Ther Med. 11(2):481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chen J, Mirando AJ, Wang C, Zuscik MJ, O’Keefe RJ, Hilton MJ. 2015. A dual role for Notch signaling in joint cartilage maintenance and osteoarthritis. Sci Signal. 8(386):ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Jiang Y, Bi R, Jiang N, Zhu S. 2018. Inhibition of Notch signaling pathway temporally postpones the cartilage degradation progress of temporomandibular joint arthritis in mice. J Craniomaxillofac Surg. 46(7):1132–1138. [DOI] [PubMed] [Google Scholar]
- Mahjoub M, Sassi N, Driss M, Laadhar L, Allouche M, Hamdoun M, Romdhane KB, Sellami S, Makni S. 2012. Expression patterns of Notch receptors and their ligands in human osteoarthritic and healthy articular cartilage. Tissue Cell. 44(3):182–194. [DOI] [PubMed] [Google Scholar]
- Morel M, Ruscitto A, Pylawka S, Reeve G, Embree MC. 2019. Extracellular matrix turnover and inflammation in chemically-induced TMJ arthritis mouse models. PLoS One. 14(10):e0223244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtomaki A, Uh MK, Choi YK, Kitajewski C, Borisenko V, Kitajewski J, Shawber CJ. 2013. Notch1 functions as a negative regulator of lymphatic endothelial cell differentiation in the venous endothelium. Development. 140(11):2365–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan J, Ruscitto A, Pylawka S, Sohraby A, Shawber CJ, Embree MC. 2018. Fibrocartilage stem cells engraft and self-organize into vascularized bone. J Dent Res. 97(3):329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotschin S, Xenopoulos P, Schrode N, Hadjantonakis AK. 2013. A bright single-cell resolution live imaging reporter of Notch signaling in the mouse. BMC Dev Biol. 13:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeson JP. 2003. Management of temporomandibular disorders and occlusion. St. Louis: Mosby. [Google Scholar]
- Owtad P, Park JH, Shen G, Potres Z, Darendeliler MA. 2013. The biology of TMJ growth modification: a review. J Dent Res. 92(4):315–321. [DOI] [PubMed] [Google Scholar]
- Park J, Gebhardt M, Golovchenko S, Perez-Branguli F, Hattori T, Hartmann C, Zhou X, deCrombrugghe B, Stock M, Schneider H, et al. 2015. Dual pathways to endochondral osteoblasts: a novel chondrocyte-derived osteoprogenitor cell identified in hypertrophic cartilage. Biol Open. 4(5):608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Tanaka S. 2017. Molecular mechanisms underlying osteoarthritis development: Notch and NF-kappab. Arthritis Res Ther. 19(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrivani SJ, Keith DA, Kaban LB. 2008. Temporomandibular disorders. N Engl J Med. 359(25):2693–2705. [DOI] [PubMed] [Google Scholar]
- Shao S, Zhao X, Zhang X, Luo M, Zuo X, Huang S, Wang Y, Gu S, Zhao X. 2015. Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a slug-dependent manner. Mol Cancer. 14:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawber CJ, Funahashi Y, Francisco E, Vorontchikhina M, Kitamura Y, Stowell SA, Borisenko V, Feirt N, Podgrabinska S, Shiraishi K, et al. 2007. Notch alters VEGF responsiveness in human and murine endothelial cells by direct regulation of VEGFR-3 expression. J Clin Invest. 117(11):3369–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Suzuki S, Tengan T, Ishii M, Kuroda T. 1996. A histological study of the developing condylar cartilage of the fetal mouse mandible using coronal sections. Arch Oral Biol. 41(1):47–54. [DOI] [PubMed] [Google Scholar]
- Silbermann M, Frommer J. 1972. The nature of endochondral ossification in the mandibular condyle of the mouse. Anat Rec. 172(4):659–667. [DOI] [PubMed] [Google Scholar]
- Singh M, Detamore MS. 2009. Biomechanical properties of the mandibular condylar cartilage and their relevance to the TMJ disc. J Biomech. 42(4):405–417. [DOI] [PubMed] [Google Scholar]
- Stoustrup P, Twilt M. 2015. Therapy: intra-articular steroids for tmj arthritis-caution needed. Nat Rev Rheumatol. 11(10):566–567. [DOI] [PubMed] [Google Scholar]
- Ustunel I, Ozenci AM, Sahin Z, Ozbey O, Acar N, Tanriover G, Celik-Ozenci C, Demir R. 2008. The immunohistochemical localization of Notch receptors and ligands in human articular cartilage, chondroprogenitor culture and ultrastructural characteristics of these progenitor cells. Acta Histochem. 110(5):397–407. [DOI] [PubMed] [Google Scholar]
- Yamaki Y, Tsuchikawa K, Nagasawa T, Hiroyasu K. 2005. Embryological study of the development of the rat temporomandibular joint: highlighting the development of the glenoid fossa. Odontology. 93(1):30–34. [DOI] [PubMed] [Google Scholar]
- Yang L, Tsang KY, Tang HC, Chan D, Cheah KS. 2014. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci USA. 111(33):12097–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, von der Mark K, Henry S, Norton W, Adams H, de Crombrugghe B. 2014. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 10(12):e1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034520924656 for Notch Regulates Fibrocartilage Stem Cell Fate and Is Upregulated in Inflammatory TMJ Arthritis by A. Ruscitto, V. Scarpa, M. Morel, S. Pylawka, C.J. Shawber and M.C. Embree in Journal of Dental Research