Abstract
A pathogenic mutation in BRCA2 significantly increases the risk of breast and ovarian cancers making it imperative to examine the functional consequences of variants of uncertain clinical significance. Variants that are predicted to result in a truncated protein are unambiguously classified as pathogenic. We have previously shown how a pathogenic splice site variant known to generate a premature termination codon (PTC) in exon9 and a nonsense mutation at exon7, can generate functional BRCA2 by skipping exons4–7 and restoring the reading frame. Using a well-established mouse embryonic stem-based assay, we functionally characterize here one splice site mutation and 11 pathogenic BRCA2 variants that are either nonsense mutation or generate PTC in different exons ranging from exons 4 to 7. Our study shows that 5 variants can restore the open reading frame by exon skipping and generate functional protein. This suggests further need to exercise prudence when classifying clearly pathogenic variants.
Keywords: BRCA2, variants, splicing, PTC bypass, functional assay
Mutations in breast cancer susceptibility gene, BRCA2, contribute to increased risk of early onset familial breast and ovarian cancers (1). Sequencing based genetic tests are now being used to identify individuals at risk of developing the disease (2). However, determining the disease risk associated with a BRCA2 variant is challenging, especially for variants that do not clearly disrupt the gene product. Such variants are called variants of uncertain clinical significance (VUS) (3). Given the increased risk of breast, ovarian and other cancers in BRCA2 mutation carriers, functional evaluation of VUS is clinically very relevant. One of the consortiums that is dedicated to functional classification of BRCA2 VUS is Evidence based Network for the Interpretation of Germline Mutant Alleles (ENIGMA, https://enigmaconsortium.org/) (4). BRCA exchange is another global repository focused on improving our understanding of BRCA variants (5).
While most functional studies are focused on determining the pathogenicity of VUS, functional interpretation of some variants that are predicted to be pathogenic may not also be clear-cut. A splice site variant of BRCA2 in intron 7, NM_000059.3(BRCA2):c.631+2T>G (IVS7+2T>G), is predicted to cause skipping of exon 7 and generate a PTC in exon 9 (6). We uncovered that in addition to skipping of exon 7, the variant also generates transcripts that lack exons 4–7. Skipping of exons 4–7 restores the open reading frame (ORF) and results in a full-length protein with an internal deletion of 105 amino acids (7). Another pathogenic variant NM_000059.3(BRCA2):c.581G>A (p.Trp194Ter) which results in a premature stop codon in exon 7, also restores the ORF by skipping exons 4–7 (7).
Using a mouse embryonic stem cell (mESC)-based assay, we demonstrated that these 105-amino acids are dispensable for the DNA repair function of BRCA2. Furthermore, we have generated mice lacking exons 4–7 and shown that they are viable, fertile and do not exhibit any increase in tumor predisposition (8). We describe here one splice site mutation and four pathogenic variants that bypass the PTC and generate a functional protein by skipping different exons in this region (exons 4–7). Our findings highlight the need to thoroughly characterize all potentially pathogenic (frame-shift, nonsense and splice site) variants.
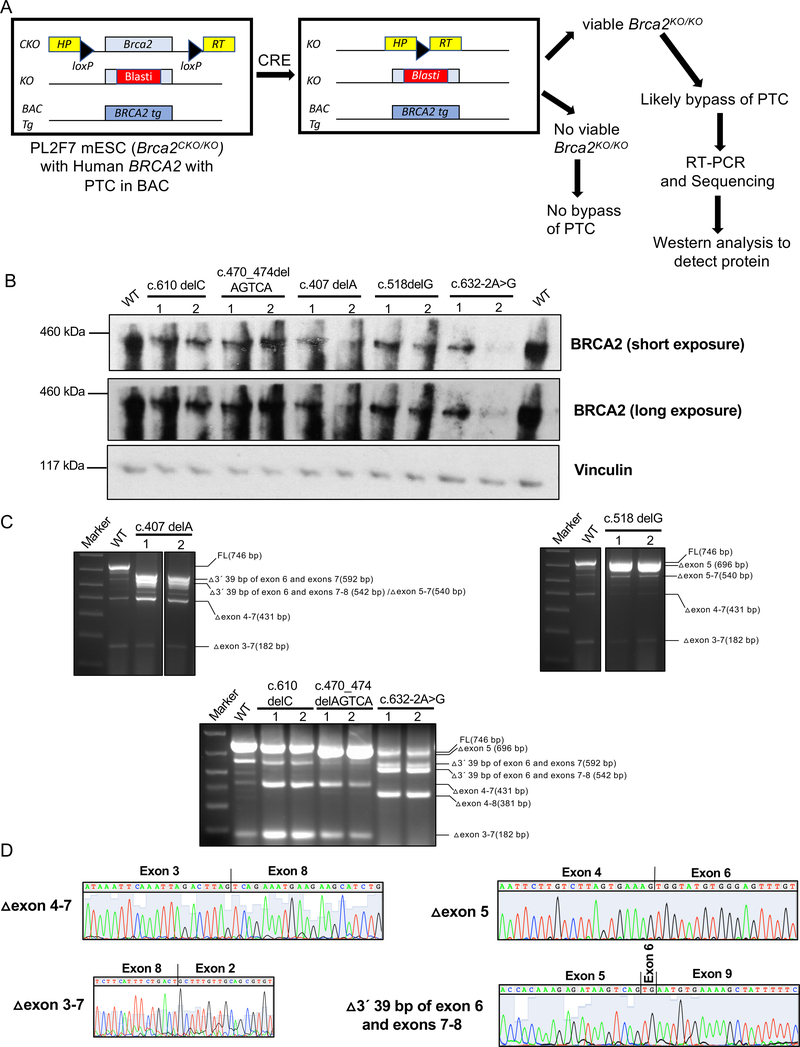
To examine other pathogenic variants that may generate partially functional protein, we selected 12 potential pathogenic mutations (NM_000059.3:c.396T>A (p.Cys132Ter); NM_000059.3:c.407delA (p.Asn136fs); NM_000059.3:c.462_463delAA (p.Arg155_Asp156insTer); NM_000059.3:c.470_474delAGTCA (p.Lys157fs); NM_000059.3:c.489_490insG (p.Leu164fs), NM_000059.3:c.518delG (p.Gly173Valfs), NM_000059.3:c.538_539delAT (p.Ile180fs); NM_000059.3:c.539_540insAT (p.Ser181fs); NM_000059.3:c.541delT (p.Ser181fs); NM_000059.3:c.574_575delAT (p.Met192fs), NM_000059.3:c.610delC (p.Ser205fs), NM_000059.3:c.632–2A>G) that map to exon/introns 4–7 (Table 1). We hypothesized that these mutations may affect the splicing of BRCA2 mRNA. We examined the impact of these variants on splicing regulatory sequences using the Human Splicing Finder (http://www.umd.be/HSF/), an in silico tool. We found several variants to result in a number of potential changes such as activation of cryptic donor site, creation of exonic splicing silencer (ESS) sites, alteration of exonic splicing enhancer (ESE) sites as summarized in Table 1. To directly assess the impact of these alterations on splicing that may generate a functional protein, we tested their ability to support the viability of Brca2KO/KO ES cells(9). We generated these mutations in human BRCA2 cloned in a bacterial artificial chromosome (BAC) and electroporated individual BACs into PL2F7 mouse ES cells (Figure 1A). PL2F7 ES cells contain a knockout allele (KO) of Brca2 and a conditional allele (CKO) where the two loxP sites are flanked by two halves of the human HPRT minigene(9). Cre-mediated deletion of the conditional allele generates a functional HPRT minigene that can be selected for in hypoxanthine-aminopterin thymidine (HAT) media (Figure 1A). In the absence of functional BRCA2, HAT resistant Brca2KO/KO mESCs fail to survive. However, cell lethality is rescued when a BAC containing WT BRCA2 or a neutral variant is expressed in the cells. We found 5 variants (c.407delA, c.470_474delAGTCA, c.518delG, c.610delC, c.632–2A>G) to support cell viability, albeit at a frequency that was much reduced than the cell viability conferred by WT BRCA2 (Table 1). We confirmed the genotype of the viable clones by Southern blot analysis which showed the loss of the conditional allele in some of the HAT resistant colonies (Supplementary Figure 1). Survival of Brca2KO/KO mESC suggests that these variants are likely to encode a partially functional protein (Table 1).
Table 1.
Summary of BRCA2 variants with PTC examined and their impact on ES cell survival,sensitivity to DNA damaging agents
| VARIANT Nucleotide change (HGVS) | Protein | Exon/Intron | Distance from splice site | PTC in exon | In-silico prediction on splicing | ES cell rescue (%) | Drug sensitivity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | Clone 1 | Clone 2 | Mitomycin C | Methyl Methane sulfonate | Camptothecin | Olaparib | Cisplatin | ||||||
| c.396T>A | p.Cys132Ter | Ex4 | −30 | 4 | 0 | 0 | NA | NA | NA | NA | NA | ||
| c.407delA | p.Asn136fs | Ex4 | −19 | 5 | 7 | 1.02 | 1.62 | Yes | Yes | Yes | Yes | Yes | |
| c.462_463delAA | p.Asp156Ter | Ex5 | −13 | 5 | Activation of cryptic donor site, Creation of ESS site, Alteration of ESE site | 0 | 0 | NA | NA | NA | NA | NA | |
| c.470_474delAG TCA | p.Lys157fs | Ex5 | −2 | 6 | Alteration of WT splice donor site, Creation of ESS site, Alteration of ESE site | 9 | 0.87 | 1.16 | Mild | Yes | Yes | Yes | Yes |
| c.489_490insG | p.Leu164fs | Ex6 | 14 | 7 | Creation of ESS site | 0 | 0 | NA | NA | NA | NA | NA | |
| c.518delG | p.Gly173fs | Ex7 | 2 | 7 | Alteration of ESE site | 21 | 0.85 | 1.8 | Yes | Yes | Mild | Yes | Yes |
| c.538_539delAT | p.Ile180fs | Ex7 | 22 | 7 | Alteration of ESE site | 0 | 0 | NA | NA | NA | NA | NA | |
| c.539_540insAT | p.Ser181fs | Ex 7 | 23 | 7 | Creation of ESS site, Alteration of ESE site | 0 | 0 | NA | NA | NA | NA | NA | |
| c.541delT | p.Ser181fs | Ex 7 | 25 | 7 | Alteration of ESE site | 0 | 0 | NA | NA | NA | NA | NA | |
| c.574_575delAT | p.Met192fs | Ex 7 | −57 | 7 | Activation of cryptic acceptor site, Creation of ESS site, Alteration of ESE site | 0 | 0 | NA | NA | NA | NA | NA | |
| c.610delC | p.Ser205fs | Ex 7 | −22 | 7 | Creation of ESS site, Alteration of ESE site | 9 | 2.3 | 4.8 | Mild | No | Mild | No | Mild |
| c.632–2A>G | Int7 | −2 | Alteration of WT splice acceptor site | 9 | 0.4 | 0.4 | Yes | Yes | Yes | Yes | Yes | ||
In-silico splicing prediction was performed using the tool Human Splicing Finder-Version 3.1 (http://www.umd.be/HSF/). ESE: Exonic splicing enhancer; ESS: Exonic splicing silencer. ES cell rescue percentage was calculated by comparing the cells that lost conditional BRCA2 (confirmed by Southern analysis) to the total number of cells plated on M15 medium.
Figure 1: Functional evaluation of BRCA2 variants in mouse ES cells.
A. Overview of the ES cell-based functional assay where mutations are expressed in mESC containing a null allele (KO) and a conditional allele (CKO) of Brca2, which is flanked by two loxP sites containing the 5´ and 3´ halves of human HPRT minigene. Upon Cre expression the conditional allele is deleted and generates a functional HPRT minigene. Recombinant clones are selected in the presence of hypoxanthine-aminopterin-thymidine (HAT). In the absence of a functional BRCA2, such cells are not viable. Viable clones were further evaluated by RT-PCR and Western blot analysis.
B. Western blot analysis of Brca2KO/KO mESC showing expression of full-length protein by variants with a premature stop codon.
C. RT-PCR analysis to identify transcripts responsible for viability of Brca2KO/KO mESC expressing BRCA2 mutant alleles predicted to result in premature protein truncation. Primers used for RT-PCR are 5’GACACGCTGCAACAAAGCA3’ (exon 2)and 5’CATGACTTGCAGCTTCTCTTTGA3’ (exon 9).
D. Chromatogram of alternatively spliced transcripts that are predicted to encode functional BRCA2.
We examined the BRCA2 protein expression in the rescued clones by Western blotting. In spite of the potential to generate a PTC in every case, we detected BRCA2 protein that was apparently similar in size to the full-length protein (Fig. 1B). The level of BRCA2 protein in those variants were low compared to the cells that express WT BRCA2 (Fig. 1B). To uncover how these variants are able to bypass the PTC, we performed RT-PCR analysis of total RNA from the viable clones followed by Sanger sequence analysis (Figure 1C, D Supplementary Table 1). Among various transcripts that can restore ORF, we identified the transcripts lacking exons 4–7 in c.407delA, c.518delG, c.610delC and c.470_474delAGTCA, (Figure 1C, D, Supplementary Table 1). A novel transcript lacking 3’ 39 bp of exon 6 and entire exons 7 and 8 was detected in c.407delA and in c.632–2A>G (Fig. 1C, D and Supplementary Table 1). This transcript is predicted to encode protein with an internal deletion of 68 amino acids in c.632–2A>G whereas in c.407delA the transcript has PTC in exon 5. A transcript lacking exon 5 was observed in c.518delG and in c.610delC (Fig. 1C, D and Supplementary Table 1). Although exon 5 deletion is predicted to generate PTC in exon 7, deletion of an additional base in exon 7 due to the c.518delG mutation will restore the ORF and generate a protein lacking 17 amino acids. In c.610delC, Δexon 5 transcripts have the PTC before the one bp deletion, resulting in a truncated protein. A transcript lacking exons 5, 6 and 7 was observed in c.407delA (Fig. 1C, D and Supplementary Table 1). Deletion of exons 5, 6 and 7 generates a PTC in exon 8 but this transcript in c.407delA also has single base deletion in exon 4, which restores the ORF and generates a protein with an internal deletion of 69 amino acids.
We next examined if the protein expressed by these alternatively spliced transcripts is proficient in DNA repair functions by challenging the Brca2KO/KO ES clones with five different DNA damaging agents. We selected genotoxins that affect essential DNA metabolic processes such as strand separation, replication or repair using various mechanisms (cisplatin and mitomycin C (MMC ) are DNA inter-strand cross-linking agent, methyl methane-sulfonate (MMS) is an alkylating agent, camptothecin is a topoisomerase inhibitor, and olaparib is a poly ADP-ribose polymerase (PARP) inhibitor). A defect in BRCA2 function results in hypersensitivity to these agents (10–12). Cells expressing c.610delC exhibited no or mild sensitivity to these agents including a PARP inhibitor, suggesting the presence of a functional BRCA2 (Table 1). In these cells, we detected the expression of transcripts lacking exons 4–7 (Δ exon 4–7), which we have shown before to encode a functional protein (Figure 1C and Supplementary table 1). In contrast, we found Brca2KO/KO ESC rescued by c.470_474delAGTCA, c.407delA and c.518delG and c.632–2A>G variants to be hypersensitive to these DNA damaging suggesting the BRCA2 is not fully functional. This is surprising based on our previous finding that exons 4–7 are dispensable for the DNA repair function of BRCA2. In addition, although the expression level of BRCA2 is reduced compared to the WT BRCA2 levels, it is similar in all the clones except one of the clones expressing c.632–2A>G (Figure 1B). While we do not precisely understand the cause of hypersensitivity of these cells, we speculate that this may be attributed to expression of other alternatively spliced transcripts that may restore the reading frame but may not generate a functional protein such as the transcripts lacking exons 3–7 (Δ exon 3–7) (Figure 1B and C, Supplementary Table 1). Exon3, which encodes the PALB2 binding domain of BRCA2, has been shown to be essential for BRCA2 function (12, 13). This is further supported by the finding that a BRCA2 variant that results in complete skipping of exon 3, c.316+5G>C, confers high risk of breast and ovarian cancer (14). Therefore, it is possible that although Western analysis showed the levels of BRCA2 to be comparable between different variants, the levels of functional BRCA2 may be low in Brca2KO/KO ESC rescued by c.470_474delAGTCA, c.407delA and c.518delG and c.632–2A>G variants.
In conclusion, we have demonstrated that alternatively spliced transcripts can restore the function of variants that are predicted to result in PTC. Previously, we have confirmed the expression of BRCA2 transcript skipping exons 4–7 that encodes fully functional protein in human EBV-lymphoblastoid cells with c.581G>A (p.Trp194Ter) variant (7). Such alternatively spliced transcripts may explain the lack of genetic data to support the pathogenicity of some truncating variants identified in BRCA2 (c.9699_9702del (p.Cys3233Trpfs)) and also in BRCA1 and MSH2 (15). Although we cannot rule out the differences in the DNA damage response between mouse and human cells as reported previously by Biechonski and Milyavsky (2013), our findings further bolster the need to carefully examine all BRCA2 variants, including those that are apparently functionally null and considered pathogenic (16).
Supplementary Material
ACKNOWLEDGEMENTS
The research was sponsored by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, U.S. National Institutes of Health.
Footnotes
Conflict of interest statement
On behalf of all authors, the corresponding author states that there is no conflict of interest.
REFERENCES
- 1.Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017;317(23):2402–16. [DOI] [PubMed] [Google Scholar]
- 2.Toland AE, Brody LC, Committee BICS. Lessons learned from two decades of BRCA1 and BRCA2 genetic testing: the evolution of data sharing and variant classification. Genet Med. 2019;21(7):1476–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toland AE, Andreassen PR. DNA repair-related functional assays for the classification of BRCA1 and BRCA2 variants: a critical review and needs assessment. J Med Genet. 2017;54(11):721–31. [DOI] [PubMed] [Google Scholar]
- 4.Spurdle AB, Healey S, Devereau A, Hogervorst FB, Monteiro AN, Nathanson KL, et al. ENIGMA--evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat. 2012;33(l):2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cline MS, Liao RG, Parsons MT, Paten B, Alquaddoomi F, Antoniou A, et al. BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 2018;14(12):el007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pyne MT, Brothman AR, Ward B, Pruss D, Hendrickson BC, Scholl T. The BRCA2 genetic variant IVS7 + 2T->G is a mutation. J Hum Genet. 2000;45(6):351–7. [DOI] [PubMed] [Google Scholar]
- 7.Biswas K, Das R,Alter BP,Kuznetsov SG,Stauffer S,North SL,et al. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood. 2011;118(9):2430–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thirthagiri E, Klarmann KD, Shukla AK,Southon E,Biswas K,Martin BK,et al. BRCA2 minor transcript lacking exons 4–7 supports viability in mice and may account for survival of humans with a pathogenic biallelic mutation. Hum Mol Genet. 2016;25(10):1934–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuznetsov SG, Liu P,Sharan SK.Mouse embryonic stem cell-based functional assa yto evaluate mutations in BRCA2. Nat Med. 2008;14(8):875–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant HE,Schultz N,Thomas HD,Parker KM,Flower D,Lopez E,et al. Specific killin gof BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–7. [DOI] [PubMed] [Google Scholar]
- 11.Farmer H,McCabe N,Lord CJ,Tutt A, Johnsonn DA,Richardson TB,et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–21. [DOI] [PubMed] [Google Scholar]
- 12.Biswas K,Das R,Eggington JM,Qiao H, North SL,Stauffer S,et al. Functiona levaluation of BRCA2 variants mapping to the PALB2-binding and C-terminal DNA-binding domains using a mouse ES cell-based assay. Hum Mol Genet. 2012;21(18):3993–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartford SA,Chittela R,Ding X,Vyas A,Martin B,Burkett S, et al. Interaction with PALB2 Is Essential for Maintenance of Genomic Integrity by BRCA2. PLoS Genet 2016;12(8):e1006236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caputo SM,Leone M,Damiola F,Ehlen A, Carreira A,Gaidrat P, et al. Ful lin-frame exon 3 skipping of BRCA2 confers high risk of breast and/or ovarian cancer. Oncotarget. 2018;9(25):17334–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenthal ET, Bowles KR, Pruss D, van Kan A, Vail PJ, McElroy H, et al. Exceptions to the rule: case studies in the prediction of pathogenicity for genetic variants in hereditary cancer genes. Clin Genet. 2015;88(6):533–41. [DOI] [PubMed] [Google Scholar]
- 16.Biechonski S, Milyavsky M. Differences between human and rodent DNA-damage response in hematopoietic stem cells: at the crossroads of self-renewal, aging and leukemogenesis. Transl Cancer Res 2013;2(5):372–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.