The early 2000s brought a new age in the field of mass spectrometry (MS) with the introduction of ambient ionization MS techniques. As defined by one of its early visionaries, Prof. R. Graham Cooks, ambient ionization refers to “the ionization of unprocessed or minimally modified samples in their native environment, and it typically refers to the ionization of condensed phase samples in air.” Since its inception in 2004, many researchers and laboratories have contributed with approaches for sampling and ionization at atmospheric conditions, greatly decreasing experimental complexity and time required for MS analyses. In the last 15 years, innovations in the field of ambient ionization MS have grown expansively, pushing these technologies far past their point of conception and integrating them into the broader scientific community in creative and stimulating ways.
Ambient ionization MS was first described in the literature in October of 2004 with the introduction of the solvent-based desorption electrospray ionization (DESI) technique,1 followed closely by the publication of the plasma-based direct analysis in real time (DART) technique.2 Improvements in the design and analytical performance of both methods have been continuously pursued to enable their use in a variety of applications, from forensics to clinical analyses including tissue molecular imaging.3,4 Concomitantly, tens of new ambient ionization MS techniques and variations thereof employing other physical-chemical processes such as laser ablation, thermal desorption, and vibrational excitation were developed to directly probe and/or ionize various samples in their native environments. As the field has grown to include many methodologies, the definition of ambient ionization MS has in part evolved to include analysis of samples that are first subjected to offline preparation steps, after which they are directly analyzed by an ambient ionization MS technique.5,6 In this review, we cover methods that require none to slightly more intensive sample preparation steps prior to direct MS analysis and thus can still be widely considered within the realm of ambient ionization MS techniques.
This review primarily covers the most recent advancements and applications of ambient ionization MS, with a defined focus on research described in manuscripts published within the past 2 years (January 2016–September 2018). For a more comprehensive overview of the field since its inception, please refer to earlier editions of the Analytical Chemistry Annual Reviews on liquid extraction ambient MS,7 mechanisms of ambient MS techniques,6 and ambient sampling/ionization applications and trends,8 among other excellent reviews.9-12 We have split this review into two main parts. The first part has an emphasis on explanations of the most established ambient ionization techniques, improvements that have been implemented within these methods, as well as new platforms that have been recently developed. The second part provides an overview on how ambient ionization MS methods have been applied to address a wide range of scientific questions and solve practical problems.
DEVELOPMENT OF AMBIENT IONIZATION TECHNIQUES
Ambient ionization MS techniques can be largely categorized into three main classes primarily based on their desorption method: liquid extraction, plasma desorption, and laser ablation. Ambient ionization MS methods that do not clearly fall into one of these three categories or that couple multiple ionization techniques into one source are termed here as “alternative” and “integrated” sources, respectively. The most common desorption/ionization techniques and applications discussed in this review are illustrated in Scheme 1. In the following section, we provide brief explanations of the most established techniques and outline recent improvements and source developments. We limit the techniques included in this section to those that are most widely utilized and have appeared in more than 30 peer-reviewed publications since their first publication or to those that have been developed within the past 2 years. However, all methods mentioned in this review are included in Table 1 with relevant citations for readers’ reference. Additionally, methods developed for specific applications, i.e., single cell analysis or therapeutic drug monitoring, are covered in their corresponding application section in the second half of this review.
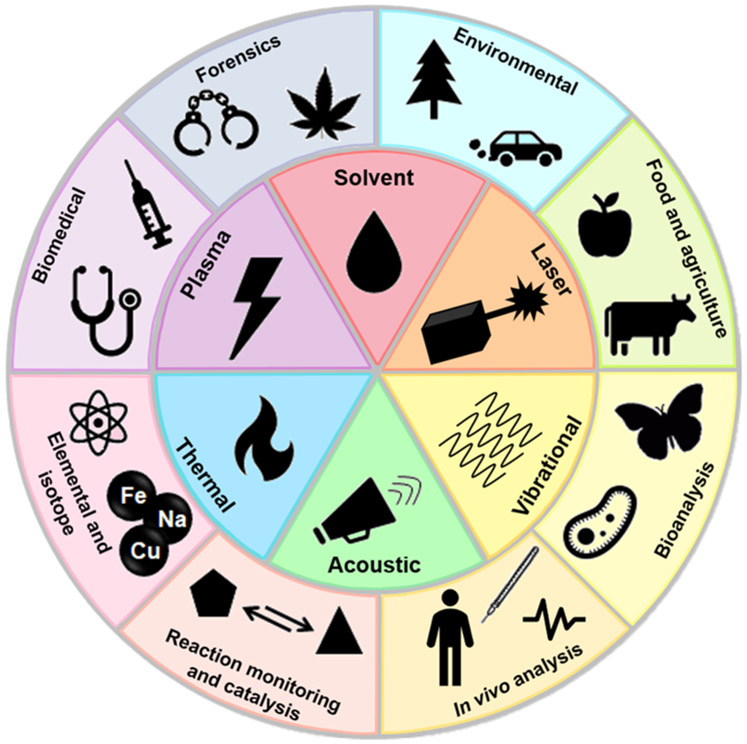
Scheme 1.
Representation of the Different Modes of Desorption/Ionization Typically Used in Ambient Ionization MS Techniques (Center Wheel) and Examples of Their Applications (Outer Wheel)
Table 1.
| technique | acronym | desorption mechanism | ionization mechanism | spatial resolution |
refs |
|---|---|---|---|---|---|
| Established Ambient Ionization Platforms (Developed Prior to 2016) | |||||
| air-flow assisted desorption electrospray ionization | AFADESI | liquid extraction | electrospray | 100–200 μm | 124,228 |
| coated blade spray | CBS | liquid extraction | electrospray | N/A | 121,122 |
| desorption atmospheric pressure chemical ionization | DAPCI | plasma desorption | corona discharge | —200 μm | 45,229 |
| direct analysis in real time | DART | plasma desorption | corona discharge | N/A | 2,169,179,184,230 |
| dielectric barrier discharge ionization | DBDI | plasma desorption | dielectric barrier discharge | <200 μm | 50,54,231 |
| desorption electrospray ionization | DESI | liquid extraction | electrospray | 30–200 μm | 1,17,133,141,193 |
| easy ambient sonic-spray ionization | EASI | liquid extraction | sonic spray | 30–200 μm | 14,180,208 |
| extractive electrospray ionization | EESI | liquid extraction | electrospray | N/A | 15,232 |
| electrospray-assisted laser desorption/ionization | ELDI | laser ablation | electrospray | 100–200 μm | 61,227 |
| flowing atmospheric pressure afterglow | FAPA | plasma desorption | corona discharge | N/A | 44,52 |
| field-induced droplet ionization | FIDI | vibrational | field induced | N/A | 209,210,233 |
| internal extractive electrospray ionization | iEESI | liquid extraction | electrospray | N/A | 129,234 |
| laser ablation electrospray ionization | LAESI | laser ablation | electrospray | —70 μm | 64,68,235 |
| liquid extraction surface analysis | LESA | liquid extraction | electrospray | 1 mm | 22,23,111,159 |
| liquid microjunction surface sampling probe | LMJSSP | liquid extraction | electrospray | 600 μm | 20,103,113 |
| liquid surface-atmospheric pressure glow discharge | LS-APGD | laser ablation | glow discharge | N/A | 223,236 |
| low temperature plasma | LTP | plasma desorption | dielectric barrier discharge | 150 μm | 48,56 |
| matrix assisted ionization | MAI | sublimation | inlet ionization | N/A | 89,90 |
| matrix assisted laser desorption electrospray ionization | MALDESI | laser ablation | electrospray | 50–200 μm | 65,73 |
| microwave plasma torch | MPT | plasma desorption | microwave plasma | N/A | 203,222,237 |
| nano desorption electrospray ionization | nano-DESI | liquid extraction | electrospray | 12–150 μm | 21,96,114,156 |
| probe electrospray ionization | PESI | liquid extraction | electrospray | ~700 nm | 37,127,158,174 |
| picosecond infrared laser desorption | PIRL | laser ablation | vibrational excitation of matrix | 1–5 mm | 76,145 |
| paper cone spray ionization | PCSI | liquid extraction | electrospray | N/A | 238 |
| pressure probe electrospray ionization | PPESI | direct analysis | electrospray | 2–5 μm | 154,239 |
| paper spray ionization | PSI | liquid extraction | electrospray | N/A | 24,152,181,183,219 |
| rapid evaporative ionization mass spectrometry | REIMS | thermal desorption | chemical/thermal evaporation | 82,84-86,162 | |
| solvent assisted inlet ionization | SAII | liquid extraction | inlet ionization | N/A | 91 |
| surface acoustic wave nebulization | SAWN | acoustic desorption | N/A | 164,165 | |
| secondary electrospray ionization | SESI | liquid extraction | electrospray | N/A | 16,142,143,170 |
| single probe | liquid extraction | electrospray | 10 μm | 99,100 | |
| touch spray | liquid extraction | electrospray | ~6 mm | 139,176 | |
| New Ambient Ionization Platforms (Published January 2016–September 2018) | |||||
| droplet assisted inlet ionization | DAII | atomization | inlet ionization | N/A | 92 |
| extractive atmospheric pressure photoionization | EAPPI | ultrasonic nebulization extraction | photoionization | N/A | 87 |
| laser ablation-aerosol mass spectrometry-chemical ionization mass spectrometry | LA-AMS-CMS | laser ablation | electron ionization and/or chemical ionization | 25–50 μm | 79 |
| laser ablation dielectric barrier discharge | LA DBD | laser ablation | dielectric barrier discharge | 40 μm | 57 |
| laser desorption corona beam ionization | LD-CBI | laser ablation | corona discharge | ~1 mm | 58 |
| laser desorption/ionization droplet delivery | LDIDD | laser ablation | photoionization and electrospray | 2–3 μm | 157 |
| laser diode thermal desorption atmospheric pressure ionization | LDTD-APPI | laser thermal | photoionization | N/A | 78 |
| laser-induced acoustic desorption atmospheric pressure photoionization | LIAD-APPI | laser-induced stresses on molecular crystals | photoionization | N/A | 63 |
| MasSpec Pen | MasSpec Pen | liquid extraction | solvent vaporization/ionization | 0.5–5 mm | 40 |
| nanotip ambient ionization mass spectrometry | NAIMS | plasma desorption | corona discharge | 5 μm | 55 |
| robotic surface analysis | RoSA | liquid extraction | electrospray | 1 mm | 42 |
| SpiderMass | SpiderMass | laser ablation | vibrational excitation of matrix | <1 mm | 75 |
| sponge spray ionization | SSI | liquid extraction | electrospray | N/A | 119 |
| triboelectric nanogenerator | TENG | liquid extraction | electrospray | N/A | 39 |
| T-probe | T-probe | liquid extraction | electrospray | 5–8 μm | 155 |
| vibrating sharp edge spray ionization | VSSI | vibrational | solvent vaporization/ionization | N/A | 88 |
Liquid Extraction Techniques.
Liquid extraction techniques encompass methods that utilize solvent to extract or desorb molecules from a sample surface. The majority of liquid extraction techniques utilize electrospray ionization (ESI) or related ionization mechanisms as a means for ion generation.13 As such, liquid extraction ambient ionization MS techniques are typically used to analyze polar molecules that are more susceptible to ionization by ESI-based mechanisms. Liquid extraction ambient ionization MS techniques are often divided into three distinguished categories: spray-based extraction, direct liquid extraction, and substrate spray.7 Spray-based techniques use a plume of droplets generated by a solvent spray to desorb molecules from the sample surface, which are then analyzed by the mass spectrometer. These techniques include desorption electrospray ionization (DESI), easy ambient sonic-spray ionization (EASI), extractive electrospray ionization (EESI), and secondary electrospray ionization (SESI). DESI and EASI are similar in that both employ a solvent spray directed at a condensed phase sample to desorb molecules but differ in that DESI utilizes a voltage to generate an electrospray1 to extract and ionize analytes from the sample surface, whereas EASI does not employ a voltage bias and thus generates ions through sonic-spray mechanisms.14 Computational fluid dynamics performed to simulate the DESI process revealed that the primary DESI spray forms a thin film of solvent on the sample surface, from which secondary microdroplets containing the desorbed analytes are sequentially formed in a splashing process and propelled to the mass spectrometer for analysis. Similar to ESI, the resulting gas-phase ions are commonly protonated/deprotonated molecules or often adducts with alkali, alkaline, or halogen ions. Differently from DESI and EASI, EESI is most often used to extract molecules from liquid samples using two colliding aerosol plumes, one containing the sample solution and the other containing the extractive electrospray solvent.15 The interaction between the spray plumes allows extraction and ionization of analytes from the sample, which are then directed to the mass spectrometer for analysis. SESI is similar to EESI as it utilizes an ESI solvent plume to extract and ionize analytes, but the analyte is in the gas phase prior to this interaction.16
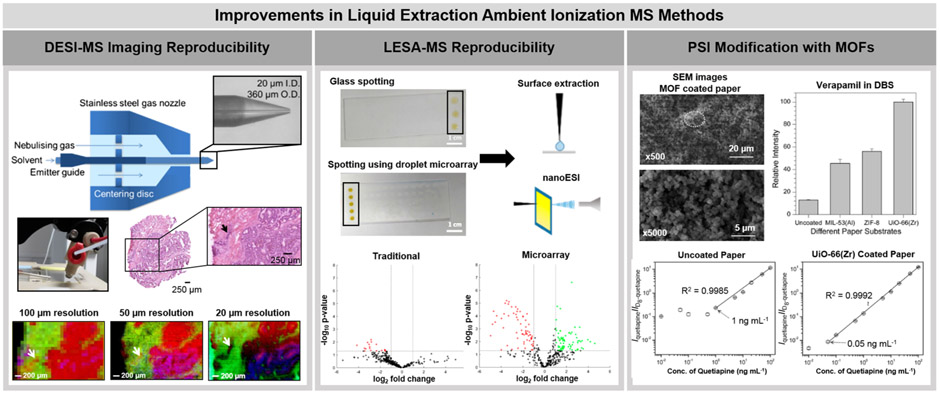
Technical refinements of spray-based ambient ionization MS techniques have been recently pursued to improve their analytical reproducibility. DESI performance, in particular, is dependent on source and geometrical optimization, with slight alterations in source parameters greatly affecting ion signal intensity.17,18 To address source irreproducibility, Tillner et al. showed that positioning of the solvent capillary within the outer gas capillary in the DESI sprayer was a major contributor to spray-to-spray variability.19 By increasing the outer diameter of the spray capillary and using a positioning disk within the DESI source (left panel, Figure 1), the relative standard deviation (RSD) was reduced by a factor of 9.1 within individual sprayers and by a factor of 9.7 across sprayers when compared to the commercial source.17 Additionally, this source design allowed for imaging at an improved spatial resolution of 20 μm, which is approximately an order of magnitude higher than what is typically achieved with DESI (~150–250 μm). Using the optimized spray design, visualization of a 50 μm × 250 μm tumor region was achieved by DESI within a colorectal cancer tissue section.
Figure 1.
Recent improvements pursued in liquid extraction ambient ionization MS technologies. The left panel shows the development and use of a newly designed DESI-MS spray source for improved analytical reproducibility and spatial resolution. A center disk within the spray nozzle is used to position the emitter more centrally within the source. A colorectal cancer tissue section was imaged with this source at three distinct spatial resolutions. A spatial resolution of 20 μm allowed for clear visualization of a small tumor region within the tissue (designated by an arrow). Adapted from Faster, More Reproducible DESI-MS for Biological Tissue Imaging (ref 17), J. Am. Soc. Mass Spectrom., Vol. 28, Issue 10, Tillner, J.; Wu, V.; Jones, E. A.; Pringle, S. D.; Karansci, T.; Dannhorn, A.; Veselkov, K.; McKenzie, J. S.; Takats, K. pp. 2090–2098, Copyright 2017, with permission from Elsevier. The center panel shows how a superhydrophobic-superhydrophilic patterning approach on a glass slide improved both liquid sample deposition and extraction with LESA-MS by preventing droplet dispersion. Improved analytical reproducibility was achieved with this system. Using this approach, an increased number of detected species that exhibited significantly altered signal intensity between the urine of participants before and after tea consumption were detected. Reproduced from Meurs, J.; Alexander, M. R.; Levkin, P. A.; Widmaier, S.; Bunch, J.; Barret, D. A.; Kim, D. Anal. Chem. 2018, 90, 6001–6005 (ref 23). Copyright 2018 American Chemical Society. The right panel shows how the modification of a paper substrate with MOFs, specifically UiO-66(Zr), can improve the detection of the blood pressure medication Verapamil and the antipsychotic Quetiapine in dried blood spots (DBS) using PSI. Reproduced from Wang, X.; Zheng, Y.; Wang, T.; Xiong, X.; Fang, X.; Zhang, Z. Anal. Methods 2016, 8, 8004–8014 (ref 24), with permission of The Royal Society of Chemistry.
Direct liquid extraction techniques use the basic chemical principles of solid–liquid extraction for sample analysis. Generally, solvent is allowed to interact directly with the sample to extract analytes that are then transferred and ionized for MS analysis. The liquid microjunction-surface sampling probe (LMJ-SSP) was the first liquid extraction technique developed. In LMJ-SSP, a robotic probe comprised of two coaxial tubes is precisely positioned at a vertical distance from the sample surface, allowing a continuous flow of solvent to protrude from the probe and come in contact with the surface.20 The solvent flow arising from the outer probe tube interacts with and extracts molecules from the sample surface, while continuously being reaspirated through the inner probe tube by a venturi ESI source for analysis. Nano-DESI is another continuous flow liquid extraction method, in which the liquid junction is generated by two silica capillaries positioned at an angle to each other.21 The capillaries create a solvent bridge at the sample surface to extract molecules from the sample and spray the solvent containing molecules into the mass spectrometer. Liquid extraction surface analysis (LESA) differs from LMJ-SSP and nano-DESI in that a solvent droplet supplied by a nano-ESI tip held at a distance from the surface by a robotic arm is used to extract molecules from a sample surface, rather than a continuous flow.22 The droplet interacts with the sample for a period of time (typically seconds) and is then reaspirated into the nano-ESI tip. The robotic arm then moves and repositions the tip containing the solvent and extracted molecules onto an ESI chip placed within the mass spectrometer interface for ionization and MS analysis. Direct liquid extraction techniques often offer higher sensitivity compared to spray-based techniques due to the direct contact of the extraction solvent with the sample surface for a longer period of time as well as full introduction of the analyte into the mass spectrometer. However, liquid extraction techniques often have lower spatial resolution (600–2000 μm, with the exception of nano-DESI) than spray-based techniques (20–200 μm, please see Table 1) and are susceptible to diffusion of the extraction solvent outside the desired liquid microjunction, potentially reducing spatial resolution and reproducibility and thus decreasing overall data quality. To prevent solvent spreading and improve LESA reproducibility, Meurs et al. recently implemented a droplet microarray of super hydrophilic spots surrounded by a superhydrophobic border into the LESA workflow.23 The center panel of Figure 1 shows the use of the droplet microarray applied to the analysis of dried liquid samples, with both the human urine sample and the extraction solvent remaining confined to the desired sample location. This improvement in experimental workflow for liquid sample analysis resulted in a 3-fold decrease in the RSD of the ion signal intensity compared to traditional LESA and improved principal component analysis (PCA) separation based on the metabolic profiles of urine samples before and after tea consumption.
Substrate spray techniques differ from spray and liquid extraction methods in that the former generates ions directly from the sample or the substrate in which the sample is contained. The most widely used form of substrate spray is paper spray ionization (PSI), in which a liquid sample is applied to a triangular-shaped piece of filter paper and allowed to dry, followed by application of a spray solvent and voltage to the paper piece for introduction into the mass spectrometer.24 This method often allows interfering species from complex matrixes, such as salts, to adhere to the paper while soluble analytes are released and analyzed, improving sensitivity and reproducibility. In PSI, internal standards can be spiked into the liquid sample prior to deposition on the paper substrate, allowing for more precise quantitative assays. Thus, PSI in particular has shown great promise for quantitative analysis of analytes in clinical and forensic samples compared to other liquid extraction ambient ionization methods. Due to the hydrophilic nature of the paper substrate, however, analysis of hydrophilic compounds has presented a challenge when using traditional PSI.25 Additionally, analysis of analytes in the negative ion mode has proven difficult with PSI due to the increased potential for corona discharge.26 Several groups have aimed to increase the sensitivity of PSI,27-34 more specifically for the analysis of hydrophilic molecules.35,36 The most common strategy employed is physical modification of the substrate to increase sensitivity for targeted analytes. For example, Wang et al. explored coating the paper substrate with metal–organic frameworks (MOFs) to reduce interactions between the hydrophilic paper and five common therapeutic drugs within dried blood spot samples.36 Coating the paper in UiO-66(Zr), a commercially available MOF material, reduced the lower limit of quantitation by 8.5–46.6-fold to values that are well below the therapeutic ranges of the drugs analyzed (right panel, Figure 1). Other research groups have developed methods to improve PSI performance through modifications of the technique itself. These variations include focusing a laser at the paper spray tip to enhance ionization of polycyclic aromatic hydrocarbons32 and integrating a sheath gas and focusing lens within a paper spray cartridge to enhance overall sensitivity,30 among others.31,33 Alternatively to PSI, probe electrospray ionization (PESI) uses a sharp needle to collect material from a sample, which is then sprayed directly from the needle upon application of a spray solvent and voltage.37 In a similar fashion, triboelectric nanogenerator (TENG) wooden-tip MS is a substrate spray technique recently reported in which a liquid sample is loaded onto a wooden tip, such as a toothpick, and then sprayed into the mass spectrometer by applying a high voltage to the tip.38 The high voltage source in TENG-MS is supplied by the electrostatic energy generated through the rubbing of objects together, also known as the triboelectric effect. In the case of TENG-MS, human-powered mechanical energy is generated from a sliding freestanding TENG for ionization.39 The use of mechanical energy makes TENG-MS more useful for portable mass spectrometers as it removes the necessity for a high-voltage supply that is required in most substrate spray systems.
A few technologies based on liquid extraction principles have also been developed as easy-to-use and automated platforms for sampling of three-dimensional or uneven objects and surfaces, followed by MS analysis. The MasSpec Pen, for example, was recently described as a hand-held tool coupled to a mass spectrometer for gentle analysis and diagnosis of tissue samples.40 The hand piece includes a flexible and biocompatible polydimethylsiloxane tip containing three channels, one for solvent delivery, one for gas delivery, and one for droplet transport to the extended transfer tube of the mass spectrometer, which merge at a small reservoir where the water droplet is contained and exposed to the sample for extraction of analytes. Once the device is in contact with a sample surface, the entire process from solvent delivery to MS analysis is automatically processed and triggered by a single press of a foot pedal. The droplet is transported directly into the mass spectrometer, where the solvent is vaporized and gas phase ions are formed through a process likely similar to that proposed as inlet ionization.41 The MasSpec Pen has been demonstrated as a fast (~10 s/analysis) and nondestructive method to acquire diagnostic MS profiles from in vivo and ex vivo cancer tissues, making it suitable for clinical use in tissue evaluation and disease prediction. Another technology, the robotic surface analysis (RoSA), aims to address the inability of liquid extraction ambient MS techniques to analyze 3-dimensional or uneven objects like a football.42 The sampling process of RoSA is similar to PESI, in a which a blunt needle attached to a robotic arm is directed toward the surface of a sample, guided by laser scanning of the object prior to analysis. The robotic arm utilizes force-sensing to prevent extensive damage to the sample, allowing the needle to come into brief contact with the sample to collect analytes. The probe is then inserted into a port of a T-shaped junction in which solvent is flowing to extract the analytes from the probe and introduce them into the mass spectrometer by ESI. RoSA was used to analyze drug tablets and a football, with minimal human intervention.
Plasma Desorption Techniques.
Plasma desorption ambient ionization techniques are derived from the same chemical principles as atmospheric pressure chemical ionization (APCI). In APCI, a plasma generated by an electrical discharge electrode interacts with and ionizes gas molecules near the sample region, which in turn ionizes gas phase analytes of interest through a series of ion–molecule reactions. The gas-phase analytes are often produced by rapid vaporization of a liquid chromatography effluent.43 Plasma desorption ambient ionization MS techniques utilize a similar process of plasma discharge to desorb and ionize molecules but are tailored for the direct analysis of unmodified samples rather than coupled to separation techniques.
DART was the first plasma-based ambient ionization technique reported and has remained the most commonly used.2 DART sources expose a carrier gas, typically helium, argon, or nitrogen, to a corona discharge needle to excite the gas molecules into reagent ions, typically protonated water clusters. The excited gas molecules flow out of the source to desorb and ionize molecules from a sample that is placed between the source and the mass spectrometer. The reagent gas can also be heated to improve analyte desorption from the sample surface and thus increase the sensitivity of the method. Flowing atmospheric pressure afterglow (FAPA) utilizes the plasma afterglow from helium or argon to generate charge-transfer reagent ions which can then interact with and ionize analytes from a sample.44 FAPA utilizes a higher current when compared to DART to improve desorption of the sample without the addition of a heated reagent gas. This higher current can also result in a wider variety of reagent gas ions, including NO+• and O2+•. Another plasma-based technique, desorption atmospheric pressure chemical ionization (DAPCI), also utilizes a corona discharge as the ionization source, but heated gaseous solvents are used as reagent ions that interact with the sample surface.45
Plasma ionization methods can be used to ionize a variety of molecules in both the positive and the negative ion modes. However, as the analytes need to be in the gas phase prior to ionization, DART is limited to somewhat volatile compounds at a narrow molecular weight range (<1000 Da). Additionally, plasma ionization methods typically require a heated gas to thermally desorb analytes, often leading to ion fragmentation and/or thermal damage of the sample. Increasing the overall sensitivity of plasma methods has been a focus of recent research. For example, Li et al. have implemented an external ion funnel prior to the MS inlet to focus the ions generated by DART from the sample and direct them more efficiently into the mass spectrometer.46 Using the ion funnel, an increase in the ion signal intensities from three organic acid compounds were observed, suggesting that this method could be used to improve DART detection limits.
Methods that generate low-temperature plasmas have also been developed as ambient ionization MS techniques. Dielectric barrier discharge (DBD), for example, generates an electrical discharge by applying a high-voltage alternating current between two electrodes separated by an insulating barrier.47 The low-temperature nature of the plasma generated is advantageous as it typically results in less sample damage and molecular fragmentation.48,49 Dielectric barrier discharge ionization (DBDI) and low-temperature plasma (LTP) ionization methods are examples of ambient ionization MS methods that employ low-temperature plasmas for sample analysis. In DBDI, the plasma is generated at the tip of a needle electrode and contacted with the sample that has been deposited on a glass slide. A copper counter electrode is placed beneath the glass slide to allow generation of ions.50 Unlike DBDI, LTP utilizes a hand-held probe configuration with the high voltage electrode mounted outside the dielectric barrier and the grounded electrode located within the dielectric barrier with the discharge gas.48 As this configuration does not require the sample to be located between the two electrodes, LTP is more suitable for in vivo analysis and fieldable applications.
Plasma-based ambient ionization MS techniques can be tuned for analysis of nonpolar compounds via electron-transfer ionization mechanisms, which is an advantage over solvent-based techniques.51 Optimization of experimental conditions including gas flow and composition, discharge current, and relative humidity, have been investigated to evaluate ionization efficiency of nonpolar analytes.52-54 For instance, Huba et al. showed that dopants such as gas additives greatly increase the ionization efficiency of polycyclic aromatic hydrocarbons using DBDI.54 In particular, addition of fluorobenzene and chlorobenzene to the nitrogen reagent gas yielded the greatest positive effects on the ionization efficiency of hydrocarbons. This improvement was attributed to a shift toward radical ionization pathways, indicated by the increased formation of radical cations likely facilitated by the addition of the gas dopants.
Recent developments in plasma-based ambient ionization MS methods have largely focused on expanding their use in MS imaging. The desorption area in plasma-based sources are typically larger (600–3000 μm) than what is commonly achieved using solvent-based ambient ionization MS sources (~20–300 um) and are often difficult to finely tune, resulting in relatively low lateral spatial resolution. To improve spatial control and resolution, Zhou et al. developed a MS imaging platform termed nanotip ambient ionization mass spectrometry (NAIMS), in which high voltage is applied between a tungsten tip and a conductive metal plate to generate a corona discharge plasma.55 The relatively high spatial resolution of ~5 μm was achieved by confining the plasma between the tungsten nanotip and the metal plate. The NAIMS source orientation is most similar to DBDI and LTP sources,50,56 although it generates an APCI-like plasma rather than DBD plasma and thus does not require a dielectric barrier. NAIMS was shown to be ideal for imaging nonpolar substances, including phenanthrene deposited on a copper substrate, although imaging of more polar small metabolites such as acetaldehyde within mouse brain tissue was also achieved. Another approach that has been explored for imaging applications is to couple plasma ionization with laser desorption techniques. The laser is utilized to ablate the sample surface to generate gas-phase neutrals which are then ionized by a plasma-based ionization technique prior to mass analysis.57,58 For example, Fowble et al. integrated an ultraviolet (UV) laser with a DART ionization source, such that the laser desorbed molecules from discrete portions of the sample, which were then ionized by DART and introduced into the mass spectrometer.59 Imaging of small molecules with varying polarities within a Datura leichhardtii seed was achieved with this system at a spatial resolution of 50 μm. Similar approaches utilizing laser ablation will be further discussed in the following section.
Laser Ablation Techniques.
Laser ablation approaches typically employing UV or infrared (IR) laser sources have also been explored as ambient ionization MS techniques. The use of lasers to promote sample desorption is very appealing as lasers can be optically focused to provide highly efficient desorption at superior spatial resolution and pulse frequencies than what is achieved by solvent- and plasma-based desorption approaches. However, the ionization efficiency achieved by laser sources is low, as the majority of desorbed molecules generated by the laser ablation process are neutrals.60 Therefore, most laser-based ambient ionization MS techniques are coupled to a secondary ionization source, such as ESI, DART, APCI, and atmospheric pressure photoionization (APPI), to enhance ionization efficiency and thus sensitivity.57-59,61-63 Among these laser ablation techniques, two have remained popular: laser ablation electrospray ionization (LAESI) and matrix assisted laser desorption electrospray ionization (MALDESI).64,65
LAESI utilizes a mid-IR laser to ablate a sample surface, generating a plume of mostly neutral molecules. The spatial resolution of LAESI is dictated by the diameter of the focused laser beam and is routinely operated at ~200 μm.66 The sample is placed on an x–y–z translational stage within a few centimeters of the mass spectrometer ionization inlet. After laser ablation, the plume of the molecules generated is intercepted by an electrospray beam, which ionizes the molecules and directs them toward the mass spectrometer inlet. As with other ESI based methods, ionization is favored toward polar molecules. While LAESI does not require sample pretreatment or any matrix addition, the samples must be rich in their water content in order to absorb the IR laser and properly excite the target.64 Improvements in method sensitivity have been a recent focus of research in LAESI.67,68 For example, LAESI analysis of polar and less-polar analytes including verapimil and arginine during the same experiment has been achieved by optimizing a solvent gradient in the ESI source.68 Optimization of the LAESI ablation chamber orientation has also been pursued to improve sensitivity during remote LAESI, a variation of the technique in which the sample analyzed is further from the mass spectrometer inlet, which often results in loss of sensitivity compared to traditional LAESI.67,69 The new design implemented a gas flow coaxially to the ablation plume rather than orthogonally, allowing for similar performance to traditional LAESI without the operational limitation of placing the sample directly in front of the mass spectrometer orifice.
MALDESI was originally developed as a method that combined the MALDI sample preparation and laser ablation steps with an ESI source for ionization. As such, samples had to be cocrystallized with an organic matrix for laser absorption, followed by ablation using an UV laser, and sequential ionization of the plume with an orthogonal ESI beam.65 Recent implementations of MALDESI, however, use a thin layer of ice as the matrix and a mid-IR laser for ablation, thus precluding the need for matrix deposition and facilitating its use as an ambient ionization MS technique.70,71 Developments of MALDESI have majorly been focused on technical improvements for imaging applications,72 most notably its spatial resolution.73 Previously, the spatial resolution in IR-MALDESI was restricted to ~150 μm, mostly due to limitations in the IR laser spot size. Implementation of a multielement optical system allowed narrowing the laser focal point to a 50 μm spot size, thus enabling higher lateral resolution for tissue imaging applications.
Recent developments in laser ablation ambient ionization MS include the picosecond infrared laser (PIRL) and Spidermass techniques, both of which utilize an IR laser source to promote desorption and ionization.74,75 These methods employ resonant infrared laser ablation (RIR-LA) to excite the O–H stretching band of water within hydrated samples, commonly biological tissues, causing desorption and ionization of molecules. The ions generated are then transported to the mass spectrometer through a transfer tube for analysis. Note that in PIRL, a picosecond laser is used while the SpiderMass employs a nanosecond IR laser, thus potentially resulting in different heat depositions on the sample surface as well as different molecular ions.76 In both techniques, the desorption/ionization mechanisms are similar to those in IR-MALDI, in which endogenous water serves as a matrix to excite and ionize the surrounding molecules.77
New developments in laser-based ambient ionization MS techniques have also focused on integrations of laser ablation with various postionization methods other than ESI. These methods include laser diode thermal desorption coupled to atmospheric pressure photoionization (LDTD-APPI),78 laser-induced acoustic desorption-atmospheric pressure photoionization (LIAD-APPI),63 and laser ablation-aerosol mass spectrometry-chemical ionization mass spectrometry (LA-AMS-CIMS).79 Of these, both LDTD-APPI and LIAD-APPI utilize photoionization as the ionization method, a process in which high energy UV photons interact with and ionize neutral gas-phase molecules that present low ionization potentials, such as low-polarity molecules.80 For example, Benham et al. showed the use of LIAD-APPI to analyze six low-polarity molecules, including cholesterol, at limits of detection at or below molar amounts reported by desorption atmospheric pressure photoionization, the first photoionization-based ambient ionization MS method.63
Alternative Sources and Ionization Mechanisms.
In addition to liquid extraction, plasma desorption, and laser ablation, thermal, vibrational, acoustic, and evaporative desorption have been shown as alterative and effective methods to desorb and ionize molecules from complex samples in ambient conditions. For example, rapid evaporative ionization mass spectrometry (REIMS), developed in 2009, is an ambient ionization MS technique that utilizes thermal energy produced by a surgical electrocautery device to induce vaporization and concurrent ionization of molecules from various samples. REIMS has been broadly used for the analysis of complex samples, including bacteria cultures81,82 and food products,83 with a key focus on analysis and diagnosis of human cancer tissues in both in vivo and ex vivo applications.84-86
Ultrasonic or vibrational processes have also been implemented as means to nebulize a sample prior to MS analysis in extractive atmospheric pressure photoionization (EAPPI) and vibrating sharp-edge spray ionization (VSSI) ambient ionization MS techniques. EAPPI uses ultrasonic waves to generate an aerosol from a sample solution, producing gas-phase neutral molecules, which are then driven by a carrier gas toward a photoionization source for subsequent ionization and mass spectrometry analysis.87 Alternatively, in VSSI the liquid sample is placed on a glass microscope slide, which is then subjected to a high-frequency mechanical vibration generated by a piezoelectric transducer. This vibration/nebulization process produces a spray from the sharp edge of the slide,88 which is directly introduced into the inlet of a mass spectrometer for analysis. Within the inlet, ionization of the molecules occurs through solvent evaporation and ESI-like mechanisms, a process similar to what has been described as inlet ionization. An advantage of VSSI over similar substrate spray techniques such as PSI and PESI is its ability to produce a spray without a high voltage, which is desirable for fieldable applications.
While the majority of ambient ionization MS techniques use an external ionization source to generate ions prior to introduction into the mass spectrometer, a subset operates by directly introducing the sample into the mass spectrometer inlet for analysis, without utilizing an ionization source. Despite ionization technically occurring within the mass spectrometer manifold and thus nonambient conditions, methods employing this analysis process are typically considered ambient ionization techniques if sampling occurs at ambient conditions and require minimal sample preparation. In matrix assisted ionization (MAI), for example,89 solid samples are cocrystallized within a semivolatile matrix and placed near the mass spectrometer inlet. Sublimation of the sample releases matrix–analyte clusters that enter the mass spectrometer for ionization and analysis. In an effort to improve this process, Lu et al. have developed a simplistic MAI system that can be integrated to multiple instrument platforms. This MAI system features an adapted inlet tube that can be placed 1 mm above the sample surface to assist analyte transfer to the mass spectrometer.90 A variation of MAI, termed solvent assisted inlet ionization (SAII), uses a similar mechanism to ionize analytes dissolved in a liquid matrix through direct introduction of the liquid sample into the mass spectrometer.91 The thermal energy provided by the mass spectrometer inlet in addition to the drop in pressure within this region facilitates sample vaporization and ionization. A sequential variation of SAII, termed droplet assisted inlet ionization (DAII), was more recently developed for the analysis of aerosols.92 The chemical composition of ambient aerosols can impact the climate, environment, and human health. Thus, technologies that can analyze aerosol particles with high sensitivity and accuracy are needed to better understand their chemical composition. In the study performed, DAII uses a condenser to generate aqueous droplets from aerosol particles (<100 nm) containing analytes, which were then directly introduced into the mass spectrometer. Using this approach, detection of both small and large molecules from the aerosols, such as polypropylene glycol, bovine serum albumin, and angiotensin II, was achieved using DAII.
Integrated Sources.
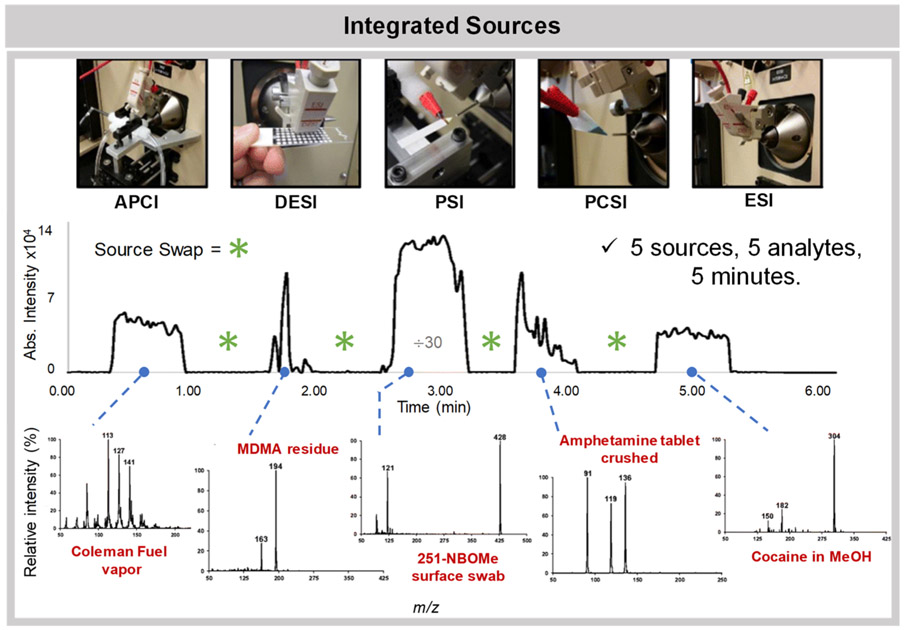
Integrated platforms have been developed combining multiple ionization techniques into an easily interchangeable, all-in-one systems that can be used to address many analytical problems at once. One such source, named integrated ambient ionization source (iAmIS), combines DART/FAPA, LTP/DBDI, and DESI into one system to reduce analysis costs and improve laboratory efficiency.93 An advantage of this platform is that the different sources can be operated individually but also simultaneously to analyze a wider variety of polar and nonpolar analytes from the same sample in a single experiment. For example, the authors operate the system using both FAPA and DESI concurrently for the analysis of a traditional Chinese medicine, Ligusticum wallichii, allowing detection of distinct chemical species by DESI and FAPA within the same experiment. Another integrated system, introduced by Lawton et al. (Figure 2), was developed with a miniature mass spectrometer and designed specifically for on-site drug evidence screening.94 The system allows for rapid interchange and use of multiple ionization sources including DESI, PSI, paper cone spray ionization (PCSI), as well as traditional ESI and APCI. To illustrate the speed of the source swapping, the authors showed that five analytes, Coleman fuel, 3,4-methylenedioxymethamphetamine, 25I-NBOMe, amphetamine, and cocaine, could be analyzed by the five different ionization techniques in under 6 minutes. The portability, ease of use, and the versatility of this platform greatly illustrates how ambient ionization MS techniques can be integrated into a single platform to enable translation into the field for forensic and potentially other applications.
Figure 2.
Platform integrating five ionization techniques including three ambient ionization MS methods, DESI, PSI, and PCSI, was developed to allow high versatility and speed of analysis. As the source interchange is quick, the system was used to analyze five unique analytes within 5 min. Reprinted from the J. Am. Soc. Mass Spectrom., Vol. 28, Lawton, Z. E.; Traub, A.; Fatigante, W. L.; Mancias, J.; O’Leary, A. E.; Hall, S. E.; Wieland, J. R.; Oberacher, H.; Gizzi, M. C.; Mulligan, C. C. Analytical Validation of a Portable Mass Spectrometer Featuring Interchangeable, Ambient Ionization Sources for High Throughput Forensic Evidence Screening, pp. 1048–1059 (ref 94). Copyright 2017, with permission from Elsevier.
APPLICATIONS OF AMBIENT IONIZATION MS
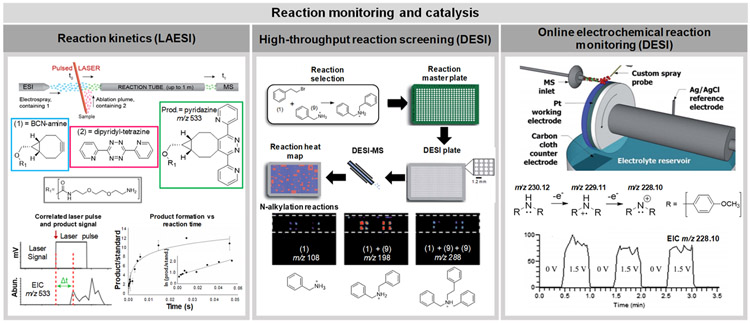
The appealing advantages provided by ambient ionization MS technologies for direct sample analysis including speed and ease of use have motivated researchers to implement ambient ionization MS techniques in many scientific fields, including biomedical and bioanalytical analysis, forensics, environmental science, food and agriculture, reaction monitoring and catalysis, and elemental and isotope analyses. The following section emphasizes recent innovative uses of ambient ionization MS methods within these research areas.
Biomedical Analysis.
Ambient ionization MS techniques have been largely explored in biomedical and clinical applications as a tool for rapid analysis of biological specimens. Next, we describe key developments and applications related to biomedical analysis, including advances in MS imaging of biological tissues, therapeutic drug monitoring, and disease detection and diagnosis.
Advances in Tissue Imaging.
Liquid extraction based ambient ionization MS techniques have been widely used in the imaging mode to map the distribution of biomolecules in thin tissue sections,7,95 in particular metabolites and lipids that are highly abundant in tissues and/or are easily ionized by ESI mechanisms. In the past 2 years, many advances have been made to further expand the breadth and better characterize the molecules detected using ambient ionization MS imaging methods. Research on solvent additives, for example, has been pursued to improve sensitivity in the detection of molecules that are not readily ionized using blends of common solvent systems. Additionally, better chemical characterization of double bonds within lipid species has been explored using chemical reactions and advanced fragmentation techniques. Advances to allow for semitargeted imaging of specific classes of molecules have also been pursued by integrating techniques such as ion mobility (IM) MS and polarimetry into ambient ionization MS workflows.
The use of silver ions as solvent additives in nano-DESI experiments has been recently explored as an approach to enhance sensitivity for detection of prostaglandins (PG). PG are lipid species that are widely present in various tissues and play key roles in biological processes such as pregnancy.96 Yet, PG are found at lower abundance within biological samples when compared to free fatty acids (FA) and glycerophospholipid species, which often results in poor detection by ambient ionization MS analysis. In a study by Duncan et al., the addition of silver ions in the acetonitrile/methanol (9:1) 0.1% formic acid nano-DESI solvent system allowed complexation of silver cations with the PG alkene functional groups, improving the relative abundance of PG species by ~30 times when compared to the relative abundance of the deprotonated ion. This increase in sensitivity allowed detection and imaging of five PG species in mouse uterine tissue that were not detected using traditional nano-DESI, including three PG molecules that had not been previously reported in this tissue type. Quantitative imaging of PG species within mouse uterine tissue was also performed by incorporating internal standards into the nano-DESI solvent. The prostaglandin PGE2 was detected at levels approximately 100 nM across the entire uterine tissue section, although slightly more abundant (up to 310 nM) within the luminal and glandular epithelium. PGF2α was also localized to the luminal and glandular epithelium but at about two-thirds of the concentration of PGE2. Note that addition of dopants to the extraction solvent has been explored for many other liquid extraction techniques, including DESI97 and, more recently, the single-probe. The single-probe utilizes a pulled dual-bore quartz capillary to perform a continuous surface microextraction with a spatial resolution less than 10 μm, followed by ESI of the extract.98 Rao et al. added dicationic compounds to the methanol/water (9:1) solvent system of the single-probe to form cationic complexes with negatively charged FA and glycerophospholipids, which are commonly observed in the negative ion mode.99,100 Using this approach, a larger range of lipid species was detected using positive ion mode MS imaging only, thus improving the MS imaging workflow by precluding the need for repeat MS imaging analysis in the negative ion mode.
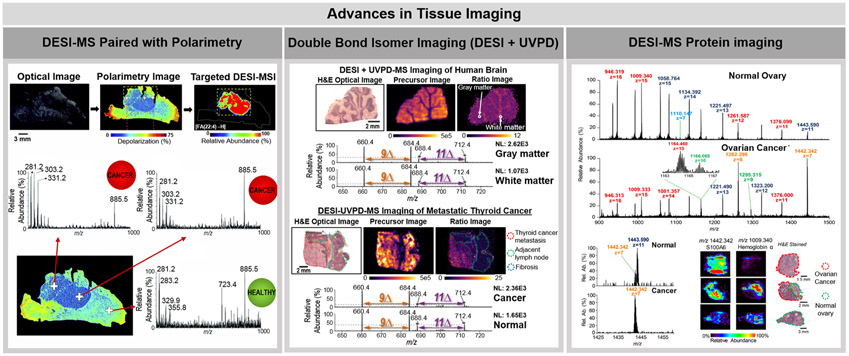
Precise structural characterization of lipid double bond and FA chain position has represented a major analytical challenge in direct lipid analysis by ambient ionization MS.101 Lipids are a diverse class of molecules that present varied chemical structures and play key biological roles in cellular function and disease development.102 While detailed structural characterization of lipids is important to better decipher their biological roles, many lipid species present isomerism in their C═C double bonds and are thus detected at identical m/z using ambient ionization MS imaging alone.101 New developments in the field have focused on improving characterization of lipid species in ambient ionization MS, especially for rapid discrimination between C═C isomers directly from tissue sections.103,104 Tang et al., for example, were able to discriminate C═C isomers of glycerophosphocholines (PC) and FA species using LMJ-SSP by performing Paterno-Buchi reactions in the fused silica capillary post extraction but prior to ionization.103 The Paterno-Buchi reaction produces an oxetane ring product between the C═C double bond of a FA chain and the ketone or aldehyde reagent, which was acetone in this instance. Collision-induced dissociation (CID) fragmentation of the products cleaves the C–C at the position of the initial double bond and the C–O bond of the FA carbonyl, leading to characteristic fragment ions that can unambiguously identify double bond position.105 The signal intensity of the fragment ions diagnostic of double bond location was used for relative and absolute quantitation of PC 16:0_18:1 C═C location isomers in rat brain, lung, liver, spleen, kidney, breast, and spinal cord tissue. A study by Klein et al. also explored C═C lipid isomer profiling by integrating ultraviolet photodissociation (UVPD) with DESI-MS imaging. While other fragmentation methods such as CID and higher-energy collision-induced dissociation (HCD) do not provide information on double bond positioning, UVPD is a high energy activation method that produces diagnostic fragmentation ions by cleaving near the C═C double bond without the need for chemical derivatization.104 Upon UVPD irradiation, fragmentation of unsaturated FA within glycerophospholipids results in diagnostic ion pairs spaced 24 Da apart which shift in mass depending on the double bond placement and can thus be used to identify unsaturation location.106 Using DESI-UVPD, 9Δ and 11Δ lipid isomers of PC 16:0_18:1 were identified and imaged within human brain and metastatic thyroid carcinoma tissue sections. The ion abundance ratio of 9Δ to 11Δ diagnostic ions differed in the gray and white matter regions, with a greater abundance of the 11Δ peaks localized to the gray matter of the brain. Additionally, an increased abundance of the 9Δ peaks was observed in the metastatic thyroid cancer tissue when compared to the adjacent normal lymph node (center panel, Figure 3). Both of these studies have interesting implications in disease as better characterization of double bond positional isomers may improve disease diagnosis and provide new insights into their biological mechanisms.102,107
Figure 3.
Highlights of advances pursued in tissue imaging using ambient ionization MS. The left panel shows the coupling of DESI-MS imaging with polarimetry. Tissue sections were first analyzed using polarimetry to identify regions of polarimetric heterogeneity, which are indicative of cancer. Only cancer regions were then analyzed by DESI-MS imaging, increasing throughput of analysis. Reproduced from Tata, A.; Gribble, A.; Vantura, M.; Ganguly, M.; Bluemke, E.; Ginsberg, H. J.; Jaffray, D. A.; Ifa, D. R.; Vitkin, A.; Zarrine-Afsar, A. Chem. Sci. 2016, 7, 2162–2169 (ref 109), with permission from The Royal Society of Chemistry. The center panel shows the use of DESI-MS paired with UVPD fragmentation to image the spatial distribution of double bond lipid isomers in human brain and metastatic thyroid cancer in lymph node. The precursor images are of m/z 798, which correspond to the lipid PC 16:0_18:1. The ratio image is the of the sum of the double bond diagnostic DESI-UVPD fragments of the 9Δ double bond position over the sum of the double bond diagnostic 11Δ positional isomer fragments (Im/z 660+684)/(Im/z 688+712). The mass spectra shown correspond to the DESI-UVPD-MS/MS of m/z 798. Reprinted from Klein, D. R.; Feider, C. L.; Garza, K. Y.; Lin, J. Q.; Eberlin, L. S.; Brodbelt, J. S. Anal. Chem. 2018, 90, 10100–10104 (ref 106). Copyright 2018 American Chemical Society. The right panel shows the use of DESI-FAIMS-MS for the analysis and imaging of proteins from biological tissue sections. The mass spectra and ion images shown are from normal ovary and ovarian high-grade serous carcinoma. Reprinted from Garza, K. Y.; Feider, C. L.; Klein, D. R.; Rosenberg, J. A.; Brodbelt, J. S.; Eberlin, L. S. Anal. Chem. 2018, 90, 7785–7789 (ref 115). Copyright 2018 American Chemical Society.
Recent improvements in tissue imaging workflows and performance using ambient ionization MS methods have also been achieved by integrating ambient ionization MS sources with other physical and analytical techniques.108,109 For instance, nano-DESI was coupled to shear force microscopy to control the distance between the sample and the probe to account for variations in tissue section thickness.108 Nano-DESI typically requires fine control of the sample-to-probe distance as small alterations in this distance can result in probe clogging and inconsistent results. A sample-to-probe distance of ~1 μm has been suggested as optimal to achieve high spatial resolution data with nano-DESI. Yet, variations of tissue section thickness up to 6 μm were observed in a mouse lung tissue section using shear force microscopy, further justifying the need for this type of system. Integration of nano-DESI with shear force microscopy allowed for real-time feedback of the z-position of the sample that could then be adjusted to account for topographic alterations.
In a different approach, DESI was coupled to polarimetry to identify regions of polarimetric heterogeneity in tissue sections, which are indicative of cancer, prior to DESI imaging.109 DESI-MS imaging was then performed only on the targeted cancer or cancer margin region identified by polarimetry (left panel, Figure 3), decreasing imaging time from 30 to 90 min (entire tissue section) to 1–2 min (targeted cancer region). Expediting DESI imaging could facilitate its use as an intraoperative tool as current methods of cancer margin evaluation typically require 30 minutes or more for sample preparation and analysis.
While ambient ionization MS imaging of lipids and small metabolites has been broadly reported, the use of liquid extraction based ambient ionization MS methods for analysis and imaging of proteins directly from thin tissue sections has been more recently achieved.110-116 Protein detection from biological tissues with ambient ionization MS methods has traditionally been challenging, owing to their large size, the complex sample matrix environment, and poor desorption efficiency.117 In 2015, imaging of proteins directly from biological tissues sections using nano-DESI was demonstrated by Hsu et al.110 Nano-DESI allowed analysis and top-down identification of multiple proteins within MYC-induced lymphoma tissue sections from a mouse model. Interestingly, intact proteins, such as ubiquitin and β-thymosins, were observed at higher relative abundance within healthy tissue while their truncated counterparts were more abundant within the lymphoma regions, a result corroborated with other MS studies of cancer tissue. Nano-DESI was also used in conjunction with light microscopy to perform guided profiling of proteins including hemoglobin, neurohemrythrin, and multiple peptides directly from ganglia of leech.114 DESI-MS has been more recently improved to allow analysis of intact proteins from tissue sections. In a study by Garza et al.,115 optimization of DESI-MS experimental parameters and integration with high-field asymmetric waveform ion mobility spectrometry (FAIMS) were pursued to enable direct protein analysis from thin tissue sections (right panel, Figure 3). Optimization of DESI parameters including spray voltage, spray angle, and solvent system allowed a 10-fold increase in protein signal-to-noise as well as the detection of 10 unique protein species. The addition of FAIMS to the optimized DESI experiment allowed for the selective transmission of protein ions and an improved signal-to-noise ratio, leading to higher quality MS images that more clearly displayed the molecular distributions of proteins in biological tissue sections. Integration of FAIMS to other ambient ionization MS techniques such as LMJ-SSP113 and LESA111 has been previously described to enhance detection and imaging of proteins from biological tissue sections. A subsequent study by Towers et al. also reported the detection of proteins from tissue sections by optimizing DESI-MS experimental parameters including transfer tube temperature and further integrating DESI imaging to a traveling wave IM mass spectrometer.116 While the DESI-MS imaging studies were successful in detecting proteins smaller than 17 kDa, Griffiths et al. described the use of LESA to detect a large native protein complex, specifically the hemoglobin tetramer (~63 kDa) directly from a vascular feature in mouse liver tissue sections.112 Collectively, these studies demonstrate the potential of ambient ionization MS methods for the detection of smaller proteins as well as larger, native protein complexes directly from biological tissue sections.
Therapeutic Drug Monitoring.
Several ambient ionization MS techniques are well suited for high-throughput analysis of clinical samples including human biofluids and as such provide an effective platform for rapid therapeutic drug monitoring. PSI provides an appealing platform to analyze serum, urine, and blood deposited on paper substrates. PSI using ZrO2 coated paper, for example, was used to quantify several therapeutic drugs including clozapine and amitriptyline present at therapeutic concentrations (10−–102 ng/mL) directly from dried blood spots.118 The use of ZrO2 coated paper reduced the limit of quantitation (LOQ) of amitriptyline to 1.38 ng/mL compared to 241.65 ng/mL achieved with uncoated paper, allowing quantitation within the therapeutic range of this drug (50–200 ng/mL). Quantification of the antibiotic penicillin G in plasma and urine was reported by Hecht et al. using sponge spray ionization (SSI), an ambient ionization MS technique similar to PSI.119 In SSI, a volumetric absorptive microsampling material, termed sponge, is used to absorb a fixed volume of blood, plasma, or urine which are then sprayed directly from the sponge into the mass spectrometer upon application of a spray voltage.120 SSI ensures a fixed volume of matrix is consistently absorbed by the sponge, offering an advantage over other substrate spray techniques that utilize sample deposition methods in their analysis which could arguably introduce experimental error for quantitative analysis. Another technique, coated blade spray (CBS), was also used to quantify pain management drugs, controlled substances, and therapeutic medications spiked in human biofluids.121 CBS uses a stainless steel sheet coated in adsorbent particles to extract analytes of interest from a biological sample deposited on the blade. The blade is then rinsed to remove the excess matrix not adhered to the surface, followed by the application of spray solvent and MS analysis from the blade.122 Using a CBS blade coated in hydrophilic–lipophilic balanced particles, LOQs for 17 common drugs, including fentanyl and methadone, were obtained in solvent, plasma, and whole blood matrixes. Notably, the LOQ obtained for 16 of the 17 drugs analyzed were well below the minimal required levels for all three matrixes.
In addition to biofluid analysis, ambient ionization MS techniques have also been used to quantify drugs in cell cultures and tissue sections, although less extensively. For example, PSI was used to quantify nicotine and cotinine in cell cultures after exposure to tobacco smoke to test the applicability of PSI as an in vitro cytotoxicity assay.123 As tobacco products cannot be tested in vivo practically, in vitro assays are essential for evaluating tobacco exposure over a range of conditions. Using PSI, LOQs of 79.17 ng/mL and 10.79 ng/mL for nicotine and cotinine, respectively, in PBS buffered human tracheobronchial epithelial cells with this method were determined, thus exemplifying the amenability of PSI for use in cytotoxicity assays. In a different study, Luo et al. used air-flow assisted desorption electrospray ionization (AFADESI) to perform quantitative MS imaging of S-(+)-deoxytylophorinidine within mouse brain, heart, kidney, lung, and liver tissue sections.124 In this approach, an inkjet printer was used to print the target molecule directly on a glass slide in order to create a calibration curve for each tissue type. This pseudo-internal standard method combined with normalization by the signal extinction coefficient, a parameter used to determine the tissue-specific ion suppression, allowed on-tissue quantitation of S-(+)-deoxytylophorinidine. The concentrations obtained with quantitative AFADESI were similar to those obtained by LC–MS/MS analysis of the tissues, with a percent difference in concentration of 8.20%, −12.40%, and 8.43% for brain, heart, and kidney, respectively. Note that although the results reported by Luo et al. are promising, recent work has also shown that variations in ion suppression susceptibility across different tissue types as well as unintentional binding of the internal standard to the matrix can prevent accurate quantitation in heterogeneous samples and should thus be considered factors impacting quantitative analyses by ambient ionization MS.125,126
Disease Detection and Diagnosis.
Ambient ionization MS techniques have been widely applied for real time disease detection and diagnosis. Although limited in their quantitative capabilities, the mass spectra obtained using ambient ionization MS techniques have been shown to be highly reproducible and thus can serve as diagnostic fingerprints for discriminating tissue type and disease status. In addition, the molecular information acquired using ambient ionization MS techniques offers new and intriguing insights into the biochemical processes occurring in disease. Ambient ionization MS methods developed for ex vivo and/or in vivo tissue analysis are described in the next section.
Ex Vivo Analysis.
Ambient ionization MS methods have been widely used for ex vivo analysis of cancerous and healthy biological tissues.127-129 DESI-MS imaging, in particular, has been extensively used in cancer research to investigate lipid and metabolite profiles of a variety of cancer and normal tissues including prostate, brain, skin, ovarian, thyroid, and breast130-135 and is strongly suggested as a tool for intraoperative diagnosis and surgical margin evaluation. In a recent study by Margulis et al., DESI-MS imaging was used to detect micrometer-sized basal cell carcinoma (<200 μm) lesions within healthy skin structures from surgically excised tissues.132 Using statistical models to distinguish normal skin and basal cell carcinomas, molecular information acquired with DESI-MS resulted in correct tissue classification with 88.3% and 94.1% overall accuracy on a training and test set, respectively. Another work by Zhang et al. applied DESI-MS imaging to investigate the lipid profiles of oncocytic thyroid tumors, allowing identification of various previously unreported cardiolipin species as potential biomarkers of this aggressive thyroid tumor subtype.134 While the majority of DESI-MS imaging studies have been performed using thin tissue sections, DESI-MS has also been used to directly analyze tissue smears prepared from ex vivo tissue pieces,131,136,137 thus precluding the need and time required for histologic tissue sectioning. Pirro et al., for example, applied DESI-MS to analyze tissue smears of brain tumor surgical margins prepared and analyzed in the operating room within 3 minutes of tissue collection.138 Using this approach, 93% sensitivity and 83% specificity were achieved for glioma diagnosis based on the detection of diagnostic metabolite and lipid profiles, showcasing the potential of DESI-MS to aid surgical resection. In a related study, Pirro et al. showed feasibility of using medical swabs to analyze ex vivo glioma tissues through a fast (<3 min) touch spray ionization workflow in the operating room.139 DESI-MS and touch spray ionization enabled the detection of 2-hydroxyglutarate, an oncometabolite diagnostic of the isocitrate dehydrogenase 1 mutation that is associated with better prognosis for patients,131,138,139 as well as n-acetylaspartic acid, a biomarker of normal brain tissue.
DESI-MS has also been combined with other techniques such as Raman spectroscopy or gene expression analysis to investigate biochemical processes associated with diseases including multiple sclerosis and lung adenocarcinoma.140,141 Bergholt et al. integrated DESI-MS, Raman spectroscopy, and immunofluorescence imaging to investigate the structural and compositional alterations within myelin tissue of multiple sclerosis afflicted mouse models and humans.140 Raman spectroscopy allowed differentiation of normal, demyelinated, and remyelinated myelin based on the vibrational properties of the lipids within myelin sheaths, which was spatially coregistered with the molecular information obtained by DESI-MS imaging of the same tissue. This integrated method facilitated the discovery that reconstructed myelin had a different lipid composition characterized by alterations in the abundance of PC and glycerophosphoethanolamine (PE) lipids when compared to normal myelin in human multiple sclerosis lesions. This study strongly illustrates how multimodal imaging workflows could aid in understanding the biological processes involved in multiple sclerosis and potentially other diseases.
Besides DESI, other ambient ionization MS techniques have also been used to analyze ex vivo biological samples to investigate molecular changes related to human diseases.127-129,142,143 PESI, for example, was used investigate the molecular profiles of head and neck squamous cell carcinomas.127 The PESI probe was dipped in an ethanol/water extract from the tissue biopsy or directly touched to the biopsy to collect analytes, followed by positive and negative ion mode MS analysis. Statistical analysis of the data collected in both polarities yielded predictive accuracies over 90% for cancerous versus noncancerous tissues. In a different study, SESI was applied for exhaled breath analysis to investigate differences in metabolic profiles of breath from patients with cystic fibrosis compared to healthy controls.142 A total of 49 features showed significant differences in their abundance between the breath of cystic fibrosis patients and healthy controls. Interestingly, breath signals from 28 patients could be correlated with six bacterial strains associated with inflammation, and 11 significant features correlated to Stenotrophomonas maltophilia colonization were found in the breath of 12 patients. In another exhaled breath analysis study, SESI was used to investigate the ω-oxidation degradation pathway of FA, as abnormal FA degradation has been associated with a variety of diseases.143 The SESI method allowed for the detection of 33 ω-oxidation metabolites, indicating that these species are abundant within exhaled breath despite ω-oxidation being considered the minor degradation pathway of FA.
In Vivo Analysis.
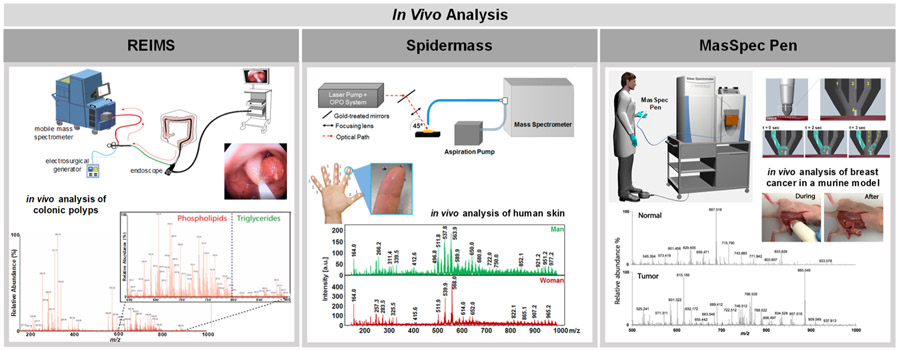
Development of ambient ionization MS techniques as methods and/or devices that allow in vivo tissue analysis has been increasingly explored in an effort to bring MS into the operating room and into the hands of medical professionals for real time diagnosis. This section describes ambient ionization MS platforms developed as suitable platforms for in vivo use, including the iKnife, SpiderMass, the PIRL desorption probe, and the MasSpec Pen.
REIMS was the first hand-held system developed for ex vivo and in vivo analysis and intraoperative applications.144 In a recent study by Alexander et al., REIMS was utilized to compare the molecular profiles obtained from colorectal cancer (n = 75) and colonic adenoma (n = 14) ex vivo tissue samples.86 Using partial least-squares-discriminate analysis (PLS-DA) for statistical analysis, the authors reported 94.4% accuracy in the identification of colorectal cancer compared to adenomas as well as a 90.5% overall accuracy in distinguishing cancer from normal adjacent mucosa (n = 145). In the same study, the authors developed an endoscopic snare REIMS (left panel, Figure 4) and demonstrated feasibility of the device for in vivo use during surgeries performed in five patients. REIMS was also used to discriminate ovarian cancer from normal gynecological tissues both ex vivo and in vivo.85 Frozen ovarian tissues were used to build predictive models that were then validated using fresh tissues, yielding sensitivities of 87.0%, 71.4%, and 87.2% for benign ovary, borderline tumor, and cancer tissue diagnosis, respectively. Moreover, high quality in vivo mass spectra collected from six patients provided evidence on the value of REIMS as an intraoperative ovarian cancer diagnosis tool, although limitations in validation with pathology prevents confirmation of in vivo diagnosis.
Figure 4.
Highlights of in vivo analysis using ambient ionization MS techniques. The left panel shows the use of an endoscopic version of REIMS termed iEndoscope for the analysis of colonic polyps. REIMS desorbs and ionizes molecules using an electrocautery device. A scheme of the endoscopic version of REIMS is shown at the top of the panel while a mass spectrum collected during the analysis of colonic polyps is shown below. Reprinted from Alexander, J.; Gildea, L.; Balog, J.; Speller, A.; McKenzie, J.; Muirhead, L.; Scott, A.; Kontovounisios, C.; Rasheed, S.; Teare, J.; Hoare, J.; Veselkov, K.; Goldin, R.; Tekkis, P.; Darzi, A.; Nicolson, J.; Kinross, J.; Takats, K. Surg. Endosc. 2017, 31, 1361–1370 (ref 86), with permissions from Springer. The center panel shows the use of SpiderMass, a device that utilizes resonant IR laser ablation to generate and desorb ions, in the analysis of human skin. A scheme of the device is shown at the top while mass spectra corresponding to the analysis of human skin of men and women are shown below. Adapted with permission from Scientific Reports, Fatou, B.; Saudemont, P.; Leblanc, E.; Vinatier, D.; Mesdag, V.; Wisztorski, M.; Focsa, C.; Salzet, M.; Fournier, I. Sci. Rep. 6, 2016 (ref 75). The right panel shows the use of the MasSpec Pen, a liquid extraction based method that uses a water droplet to nondestructively desorb diagnostic molecules from a tissue’s surface. A scheme of the MasSpec Pen is shown at the top of the panel, while mass spectra collected in vivo during tumor resection of breast cancer in a mouse model are shown below. Reproduced from Zhang, J.; Rector, J.; Lin, J. Q.; Young, J. H.; Sans, M.; Katta, N.; Giese, N.; Yu, W.; Nagi, C.; Suliburk, J.; Liu, J.; Bensussan, A.; DeHoog, R. J.; Garza, K. Y.; Ludolph, B.; Sorace, A. G.; Syed, A.; Zahedivash, A.; Milner, T. E.; Eberlin, L. S., Sci. Transl. Med., 9, 2017 (ref 40), with permission from the American Association for the Advancement of Science.
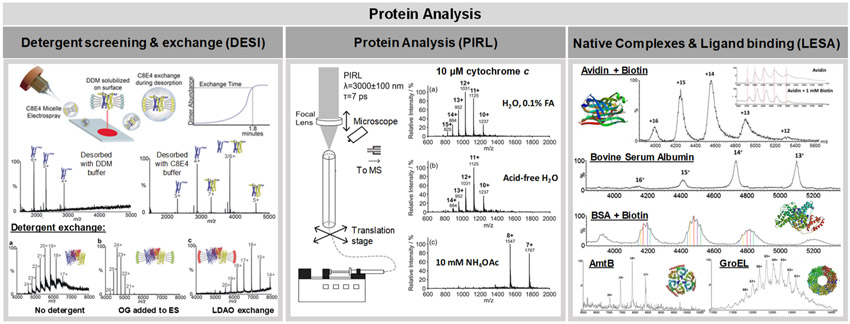
The PIRL and SpiderMass probes are alternative approaches based on IR laser desorption and ionization for direct and in vivo tissue analysis.75,145 The SpiderMass system was applied for ex vivo analysis of an ovarian cancer tissue and an adjacent normal ovarian tissue sample as well as in vivo analysis of human finger skin with little damage to the tissue surface (center panel, Figure 4).75 The positive ion mode mass spectra profiles obtained from the ovarian cancer tissue sample presented high relative abundances of PC lipids that appeared to be characteristic of tissue type. PIRL was used for the ex vivo analysis and typing of xenograft models of medulloblastomas, yielding negative ion mode mass spectral profiles with high relative abundances free FA, diacylglycerols, and various glycerophospholipids and phospholipid fragments. Statistical analysis of the PIRL data provided 98% accuracy in distinguishing medulloblastoma subtypes using PLS-DA with 5% leave-out cross validation.145 While the PIRL-MS probe has not yet been reported for in vivo analysis, its design has strong potential for in vivo use in future applications.
Our group has recently developed the MasSpec Pen as a hand-held device for nondestructive analysis of tissues using liquid extraction.40 The MasSpec Pen was applied for ex vivo analysis of 253 tissues, including normal and cancerous lung, ovarian, thyroid, and breast tissues. Using the lasso method to build statistical classifiers using cross-validation, 96.3% accuracy was achieved for discriminating cancer from normal tissues. Furthermore, the MasSpec Pen was used for the analysis of a breast tumors in living mouse models, thus demonstrating feasibility for in vivo tissue analysis (right panel, Figure 4). Collectively, these studies show that the development of ambient ionization MS platforms combined with statistical models and machine learning methods brings the ambient ionization MS field one step closer to enabling online, real-time, in vivo tissue diagnosis to aid in surgical resection.
Bioanalytical Applications.
Applications of ambient ionization MS techniques for the detection of proteins, single cell analysis, the study of bacteria, and explorations of plants and animals and related byproducts have been thoroughly explored in the past 2 years. The following section highlights some of the advances made using ambient ionization MS for bioanalytical studies. To avoid overlap with the Biomedical Analysis section, this section presents topics not directly relatable to disease, pharmaceuticals and drug monitoring, or any molecular imaging directly from tissue sections.
Protein Analysis.
Native analysis of intact proteins and large protein complexes has been recently explored using ambient ionization MS techniques. Ambrose et al., for example, used DESI-MS to investigate mechanisms of protein complex formation (left panel, Figure 5).146 Native DESI mass spectra were obtained from large proteins with molecular weights ranging from 66 to 800 kDa that had been gradually deposited onto glass substrates and subjected to DESI analysis. Upon addition of different additives such as small molecules, lipids, and peptides to the DESI spray solvent, native protein complexes were detected in the mass spectra. For example, outer membrane protein F (OmpF) was detected in its trimeric state (110 kDa, average charge state detected = 24+) when octyl glucoside micelles were added to the DESI spray solvent. Upon substitution of the octyl glucoside with a different detergent, lauryldimethylamine N-oxide, a shift to higher charge states was observed suggesting detergent exchange. These results suggest that DESI could be used as a tool for rapid screening of detergents in membrane protein studies. As current methods for detergent screening are time-consuming and costly, a rapid and relatively inexpensive method such as native DESI could prove beneficial in the purification of membrane proteins, a process where optimal detergent is necessary. In addition, DESI-MS showed potential for the determination of protein–ligand dissociation constants (Kd). OmpF was deposited and incubated with various concentrations of a known ligand, OBS1, on the native DESI stage and the relative intensities of the bound and unbound proteins were plotted versus the OBS1 concentration. Remarkably, this method yielded a Kd value of 0.7 ± 0.34 μM for this protein–ligand interaction, which is in agreement with the reported literature using established techniques.
Figure 5.
Recent applications of ambient ionization MS for protein analysis. The left panel demonstrates the use of detergent additives in DESI-MS solvent to obtain native mass spectra of lipophilic proteins. Reproduced from Native Desorption Electrospray Ionization Liberates Soluble and Membrane Protein Complexes from Surfaces, Ambrose, S.; Housden, N. G.; Gupta, K.; Fan, J.; White, P.; Yen, H.; Marcoux, J.; Kleanthous, C.; Hopper, J. T. S.; Robinson, C.V. Angew. Che. Int. Ed. Engl., Vol. 56 (ref 146). Copyright 2017 Wiley. The center panel shows a schematic of PIRL for protein analysis. A variety of different protein charge states were observed for cytochrome c, including potentially native species. Modified from Lu, Y.; Pieterse, C. L.; Robertson, W. D.; Miller, R. J. D. Anal. Chem. 2018, 90, 4422–4428 (ref 149). Copyright 2018 American Chemical Society. The right panel shows LESA-MS analysis of a range of native protein complexes as well as the analysis of protein–ligand interactions. Reprinted from Int. J. Mass Spectrom., Vol. 420, Mikhailov, V. A.; Griffiths, R. L.; Cooper, H. J. Liquid extraction surface analysis for native mass spectrometry: Protein complexes and ligand binding, pp. 43—50 (ref 150). Copyright 2017, with permission from Elsevier.
To investigate post-translational modifications (PTMs), Shin et al. also applied reactive DESI for in situ, selective derivitization of post-translationally modified peptides, including citrullinated peptides.147 Arginine residues in peptides can be converted to citrulline by the enzyme peptidyl arginine deiminase. However, citrullination is a difficult PTM to resolve and thus identify using MS analysis as it results in a + 1 Da mass shift from the original peptide mass. To address this challenge, a citrulline derivatization agent, phenylglyoxal, was added to the DESI solvent system, allowing detection of a citrullinated peptide at m/z 488 (z = 3+) and m/z 732 (z = 2+) as the derivatized product as well as the underivatized peptide at m/z 450 (z = 3+) and m/z 674 (z = 2+). As this reaction is selective to citrulline residues, this method could be used to resolve various citrullinated peptides from their arginine containing counterparts and be further refined to characterize other PTMs.
Ambient ionization MS techniques have also been used as a tool in native MS to investigate protein complexes in their natural conformations.148 Lu et al. used low energy PIRL pulses (40 μJ focused to 0.3 J/cm2) to vibrationally excite and ionize proteins and peptides from bulk solution (center panel, Figure 5).149 By using a native-like solvent (10 mM ammounium acetate buffer) to dissolve the proteins compared to a traditional ESI solvent (0.1% formic acid in water), the charge-state envelope of the proteins studied was shifted toward lower values. For example, the mass spectrum obtained from cytochrome c presented a narrow distribution of the 7+ and 8+ charge state species, compared to a wider charge-state envelope centered at 12+ for the traditional solvent, which was comparable to the mass spectrum obtained using standard native ESI. These results suggest that proteins were detected in their natively folded conformation and further indicates the soft nature of the ablation method. LESA has also been used as a tool for native protein complex analysis. Mikhailov et al. applied LESA to analyze native, noncovalent protein complexes (right panel, Figure 5), including soluble tetradecameric GroEL (~800 kDa) and trimeric membrane protein AmtB (~140 kDa).150 Formation of ligand-bound complexes were also investigated, including tetrameric avidin (~64 kDa) and bovine serum albumin bound with biotin.
Improvements in PSI have also been pursued to enable high-throughput protein analysis. Paper substrates are commonly less amenable to protein analysis due to significant hydrogen bonding and van der Waals forces that can be formed between the paper substrate and the protein amino acid side chains. Li et al. addressed this limitation by baking polystyrene microspheres into the paper substrate, improving sensitivity by 12–1 348-fold for a range of proteins, including myoglobin and lysozyme.151 In a different study, Zhang et al. integrated solid phase extraction (SPE) into the PSI workflow to allow quick preconcentration of proteins within an antibody-enriched SPE column, which were then eluted onto a carbon nanotube coated porous polyethylene substrate for analysis.152 This platform was applied to detect clinically relevant protein targets from plasma, including the apolipoprotein CL T45S variant, which has been associated with high body mass index and diabetes.
Single-Cell and Cell Culture Analysis.
Research in single-cell analysis is an area of strong scientific interest as it enables new discoveries on cellular and molecular heterogeneity found within cell populations.153 The application of ambient ionization MS techniques to single-cell analysis has been increasingly explored using different strategies, including the extraction of cellular components prior to MS analysis, extraction of analytes from single cells, as well as technical improvements in spatial resolution of desorption/ionization techniques for direct single-cell analysis.
The pressure probe electrospray ionization MS with internal electrode capillary (IEC-PPESI-MS), for example, integrates high spatial resolution sampling and precise post sampling manipulation for single-cell analysis.154 IEC-PPESI-MS utilizes a quartz capillary to extract cell sap from individual cells by directly inserting the capillary into the cell and applying a backpressure to the capillary with a pressure transducer. Once loaded, the capillary is directed toward the mass spectrometer inlet where a high voltage is applied to induce the formation of an electrospray. Using this technique, the metabolic profiles of two adjacent cell types comprising a trichome unit on a tomato plant were compared in situ, allowing characterization of their distinct metabolic composition. Remarkably, less than a picoliter of cell sap is removed for each analysis using IEC-PPESI-MS, which allows subsequent sampling of the same trichome and extensive comparative analyses. Another approach to cell content extraction is the t-probe, in which a t-shaped probe composed of three silica capillaries is inserted into single cells for extraction of cellular contents.155 Solvent flow through the t-probe generates a suction force at the tip of the orthogonal probe, allowing extraction of cellular contents, which are then ionized using nano-ESI. The t-probe allowed online, in situ screening of metabolic changes in individual HeLa cells exposed to the chemotherapy agent irinotecan. A total of 17 metabolites, including pyridoxamine-5′-phosphate and sodiated nicotinaminde adenine dinucleotide, showed significant differences in abundance when comparing control (n = 9 cells) and treatment (n =11 cells) groups, suggesting that metabolic changes associated with drug treatment can be monitored on a single-cell level using the t-probe.
Improvement in the spatial resolution of nano-DESI has also been pursued to enable high-throughput single-cell analysis.156 While typically performed at 150 μm resolution, a silica capillary with an outer diameter of 90 μm was employed in the nano-DESI set up to create a smaller liquid sampling bridge. At the resulting spatial resolution of ~90 μm, human cheek cells (50–100 μm in diameter) were analyzed and expected to be entirely immersed within the solvent bridge. Internal standards added to the nano-DESI solvent system allowed quantification of the combined concentration of the six most abundant PC species in a single human cheek cell as 1.2 pmol. In a different study, Lee et al. developed an ambient ionization MS technique named laser desorption/ionization droplet delivery (LD-IDDMS) for high-resolution (3 μm for mouse brain) and sensitive subcellular imaging of tissue samples and single live-cell secretions.157 In this approach, a pulsed UV laser beam was directed at a surface for desorption and photoionization of molecules while a spray of liquid droplets was directed at the surface for ion capture and delivery to the MS. Single-cell measurements allowed identification of heterogeneity in metabolic profiles of distinct cells during exocytosis and apoptosis processes. For example, upregulation of PC 32:0, 33:1, and 34:1 as well as downregulation of PC 36:1 were observed in single apoptitic cells when compared to individual control cells. The authors suggest that these unprecedented results could pinpoint biological mechanisms driving cell body shrinkage that occurs during apoptosis.
Extraction of analytes from single cells was also demonstrated by Chen et al. using PESI-MS.158 In this platform, an inkjet printer samples a cell suspension, which is continuously agitated to ensure homogeneous cell distribution. Droplets containing single cells are then precisely deposited by the inkjet printer onto the tip of a tungsten ESI needle for PESI-MS analysis. Under optimized cell suspension and inkjet parameters, the number of deposited droplets found to contain exactly one cell was reported to be as high as 43.8%. Using this method, eight human cell lines were analyzed in the positive ion mode allowing detection and identification of 11 different PC species. These results showed that PESI-MS can be used to profile the lipid information of individual cells in different cell lines, which was further illustrated by the excellent separation achieved between cells lines using PCA.
In addition to single-cell analysis, ambient ionization MS techniques including LESA and REIMS have also been recently applied for the direct analysis of populations of cells from distinct cancer cell lines.82,159 Strittmatter et al., for example, used REIMS to analyze the 60 human cancer cell lines from nine different organs that comprise the NCI60 cell line panel.82 Metabolic profiles detected from cells lines were grouped using PCA and hierarchical cluster analysis (HCA). Unsupervised statistical methods were used to evaluate if REIMS profiles were capable of grouping cell lines based on their identity upon replicate analysis. Biological replicates were performed for eight of the cell lines which presented close clustering within the HCA dendrogram due to the similarity in molecular profiles. Only two cell lines, SF-268 and IGROV-1, showed incorrect clustering upon replicate analysis, potentially due to biological variance inherent to cell lines. These results suggest that REIMS could be used as a tool to characterize cell lines based on reproducible metabolic profiles in a simple and rapid (<5 s) manner. Basu et al. have also applied LESA for in vitro analysis of four breast cancer cell lines, which represent distinct molecular subtypes of breast cancer directly from culture wells. Distinct metabolic signatures were detected for each cell line, characterized by variations in the relatives abundances of various lipids including ceramides, PE, and glycerophosphoinisitols (PI).159 As the molecular profile of cell lines can change in as little as 30 minutes after culture media aspiration, in vitro LESA provides an appealing ambient ionization MS platform to perform cell culture analysis directly from their native state.
Bacteria Analysis.
MS analysis of bacteria has been largely accomplished using MALDI-MS, a powerful tool in clinical microbiology.160,161 Ambient ionization MS methodologies have also been increasingly applied to investigate the molecular profiles of bacterial microorganisms, with recent studies focused on rapid speciation, screening, and imaging of live colonies.
Ambient ionization MS techniques have enabled bacterial speciation based on metabolic and lipid profiles. For example, Bardin et al. utilized REIMS to analyze the metabolic profiles of Pseudomonas aeruginosa isolated from cystic fibrosis patients and from noncystic fibrosis related P. aeruginosa, in an effort to improve understanding of these bacteria on a subspecies level.162 PLS-DA of the REIMS data revealed a lower diversity of metabolites detected from the P. aeruginosa bacteria isolated from nonrespiratory related sources than from cystic fibrosis patients as well as discrimination between isolates collected from cystic fibrosis patients and patients with bronchiectasis, another chronic lung infection. This result suggests that the molecular profiles collected by REIMS can differ in isolates exposed to different environmental conditions that can potentially be linked to disease progression and patient prognosis as chronic P. aeruginosa infection is often associated with poorer clinical outcomes. Additionally, the metabolic profiles obtained by REIMS allowed classification of P. aeruginosa strains with 83% accuracy when compared to the bacterial type determined by genome sequencing. In a similar approach, zero-volt PSI was used by Wei et al. as a tool to distinguish bacterial strains based on the detection of bacterial membrane lipids.163 Membrane lipid profiles obtained from eight gram-negative and gram-positive strains allowed strain speciation using PCA. Surface acoustic wave nebulization (SAWN) was also used by Liang et al. to characterize the lipid A profiles of six species of gram-negative bacteria.164 In SAWN, high-frequency acoustic waves are used to nebulize a sample for direct introduction of the gas phase material into the mass spectrometer.165 SAWN allowed the direct detection of lipid profiles of lipid A extracts that were characteristic of each species, suggesting its amenability for bacterial phenotyping.
MS imaging of live bacterial colonies has also been pursued using ambient ionization MS techniques to investigate bacterial antibiotic susceptibility and biocatalytic activity. For example, Li et al. utilized LAESI-IMS-MS to detect lipid and metabolite distribution in Escherichia coli and Bacillus subtilis colonies in response to antibiotic exposure.166 Differential susceptibility was explored in the same agar plate by plating both E. coli and B. subtilis on a paper disk soaked in antibiotic solution (left panel, Figure 6). Distinct spatial distributions of lipid species were observed within the agar plate. For example, PE (16:0/17:1) and (16:0/18:1) were detected at higher abundances within regions of B. subtilis inhabitation while PE (31:1) and (18:1/18:1) were detected at increased abundance near E. coli colonies. The use of LAESI-MS for quicker evaluation of antibiotic susceptibility was explored as well. Molecular imaging of E. coli subjected to 5 h incubation with an antibiotic disk was performed. A one-dimensional diffusion model based on the distributions of a streptomycin fragment at m/z 263.138, indicative of the antibiotic, and PE (16:0/16:1), indicative of colonies, allowed for estimation of a minimum inhibitory concentration value of 285 μg/mL. The typical incubation period required to obtain this result is 16–24 h, illustrating that LAESI can more rapidly determine antibiotic susceptibility than conventional methods. In a related study, Yan et al. utilized a DESI-MS imaging platform coupled to traveling wave IM for direct analysis of living bacterial colonies with the goal of screening biocatalytic activity (right panel, Figure 6).167 Different E. coli colonies were incubated for up to 3 h with different substrates and subjected to DESI-IM-MS imaging at various time points to monitor catalytic activity. Using this approach, in situ detection and imaging of small metabolites within the colonies was achieved after biocatalytic reactions. For example, the authors monitored phenylalanine ammonia lysase (PAL) catalyzed reaction of cinnamic acid to phenylalanine, where PAL was contained within active E. coli cells. While synthetically useful, this reaction presents a challenge for catalytic screening as there is no colorimetric assay available for monitoring. DESI-IM-MS allowed for label-free monitoring of the accumulation of phenylalanine within the E. coli. In another study, simultaneous chemical and topographical imaging of bacterial colonies was accomplished by integrating nano-DESI and shear force microscopy, as previously described.168 The optimized platform was used to image live B. subtilis colonies, which are notable for their complex topographic structures, allowing detection of the cyclic lipopeptide surfactin at high relative abundances at the ridges of the colonies’ aerial structures.
Figure 6.
Highlights of the application of ambient ionization MS for the analysis of live bacterial colonies. The left panel shows LAESI-MS images of live bacterial colonies at different depths, created by imaging, subsequently sputtering the surface, and imaging again. Antibiotic susceptibility of E. coli and B. subtilis were evaluated in the same LAESI-MS image. Reproduced from Molecular Imaging of Growth, Metabolism, and Antibiotic Inhibition in Bacterial Colonies by Laser Ablation Electrospray Ionization Mass Spectrometry, Li, H.; Balan, P.; Vertes, A. Angew. Chem. Int. Ed. Engl., Vol. 55, Issue 48 (ref 166). Copyright 2016 Wiley. The right panel shows the use of DESI-MS imaging to monitor catalytic activity in bacterial colonies. The catalytic reaction monitored was the asymmetric addition of ammonia to cinnamic acid by the PAL enzyme. DESI-MS ion images show the distribution of a membrane lipid (C18:1) to locate colonies, and the distribution of the reaction product (2a). Reprinted from Yan, C.; Parmeggiani, F.; Jones, E. A.; Claude, E.; Hussain, S. A.; Turner, N. J.; Flitsch, S. L.; Barran, P. E. J. Am. Chem. Soc. 2017, 139, 1408–1411 (ref 167). Copyright 2017 American Chemical Society.
Plant and Animal Analysis.
Direct analysis of plants, animals, and related byproducts has been increasingly explored using ambient ionization MS techniques, both in vivo and ex vivo. Recent studies have focused on the metabolic analysis of model organisms in response to a specific stimuli or through their life cycle.
In particular, several ambient ionization MS approaches have been developed for time-resolved analysis of volatile animal byproducts. Li et al., for example, used a confined DART system to study in vivo and real-time release of phermones in butterflies.169 Two Pieris rapae butterflies were placed in connected chambers such that airflow passed from the female butterfly to the male butterfly and then to the contained DART source for ionization and MS analysis at a rate of one mass spectrum per second. Note that confined DART was used to reduce the random diffusion of the gas-phase analytes and improve their ionization efficiency. This experimental approach allowed detection of the pheromone ferrulactone secreted by the male butterfly less than one second after visualization of its partner, with a peak release observed within a minute of visualization. In a related approach, Barrios Collado et al. demonstrated the effectiveness of a low-flow SESI high resolution MS platform for observing volatile organic compounds (VOCs) emitted by plants.170 Humidified air was passed through a glass sample chamber containing a Begonia semperflorens plant for collection of VOCs and was then analyzed every 2 min (250 scans, 1 scan/μs) during three consecutive days by SESI-MS. Monoterpenes, sesquiteroenes, and β-caryophyllene as well as other compounds emitted in association with light cycles were detected and identified. In a separate experiment, molecules associated with herbivore attack or insect attraction, such as methyl jasomonate and hexenyl acetate, were detected after stress was induced on the plant due to mechanical damage to the foliage.
Ambient ionization MS imaging techniques have also been utilized to investigate the metabolism of organisms related to developmental stages. For example, Enomoto et al. applied DESI-MS imaging to determine the spatial distribution of plant hormones involved in seed development, dormancy, and germination in Phaseolus vulgaris seed sections.171 To preserve macroscopic seed structure, whole seeds were briefly treated with carboxymethyl cellulose prior to flash freezing and cryosectioning. Interestingly, abscisic acid, a germination repressor that is crucial to growth regulation, was mainly localized to the seed embryo, a previously unreported finding. In a different study by Stopka et al., LAESI-IM-MS was used for metabolic profiling of the symbiosis between living soybean roots and rhizobia.172 Rhizobia are diazotrophic bacteria that establish themselves within the roots of legumes and lead to nodule development. Using LAESI, the authors found distinct metabolic profiles within the soybean root, the rhizobia, and the root nodule. For example, heme B+ was exclusively detected in the root nodule and free-living rhizobia, while a sodium adducted disaccharide was most abundant within the uninfected root.
Lipid profiles of zebrafish embryos at various stages of development have also been investigated by Pirro et al. using DESI-MS and nano ESI-MS.173 Embryos were analyzed at several time points from 0 to 96 h post fertilization with different solvents in both the positive and negative ion modes. Interestingly, all nutrients necessary for zebrafish development were detected in the embryo yolk at the point of fertilization, including vitellogenins, lipovitellins, and other lipoproteins. 48 hours after fertilization, a large increase in the relative abundance of oleic and eicosapentaenoic acids were observed, which was hypothesized to be related to their need for de novo synthesis of glycerophospholipids. This suggestion is further supported by the increased relative abundance of glycerophos-phoserines (PS) and PI at 72 and 96 h postfertilization time points.
In vivo and real time monitoring of metabolites in living animal models have been accomplished using PESI.174 In a study by Zaitsu et al., in vivo sampling of mouse liver that had undergone hepatic injury was achieved using a semiautomatically manipulated PESI needle with a tip diameter of 700 nm. Hepatic injury, a condition commonly associated with a blunt trauma event, can be induced by radical toxicants such as •CCl3 that can react with the endogenous molecule taurine to reduce its abundance within tissues. Lower relative abundance of taurine was observed in mouse liver that had undergone CCl4-induced hepatic injury compared to the controls, suggesting that PESI can detect metabolic changes associated with hepatic injury. Real-time, in vivo monitoring of metabolites involved in the Krebs cycle was also performed while pyruvic acid was introduced into the heptic portal vein. Notably, a dramatic increase in Krebs cycle activity was observed after pyruvic acid injection, with increased detection of cycle-intermediate fumaric acid occurring within 30 s of administration.
Forensics.
Ambient ionization MS methods have been used in a variety of forensics applications in an effort to provide rapid and robust workflows for field analysis. The key areas of applications within the field of forensics include detection of weapons and explosives, illicit drugs and activities, as well as art conservation and fraud.
DART, PSI, touch spray, and EASI have been applied for the detection of gunshot residue, chemical warfare agents, and explosives.175-180 For instance, Black et al. used DART-MS to analyze gunshot residue from 3D printed firearms composed of a variety of materials. Polymer ions as well as organic compounds such as ethyl centralite and diphenylamine normally associated with gunshot residue were detected in the mass spectra, indicating that DART could provide useful evidence in crimes using 3D printed firearms.175 Swab touch spray was also applied for the analysis of gunshot residue on human hands. A rayon tipped swab was touched to the hands of the shooter, after which solvent and voltage were applied to the swab for direct MS analysis on a portable mass spectrometer.176 Similar to the DART study, compounds associated with gunshot residue including methyl centralite and ethyl centralite were detected in the mass spectra obtained. Paper substrates for PSI have also been applied for the analysis of chemical warfare agents on surfaces and in biological fluids.177,178 McKenna et al. applied PSI for the identification and quantification of chemical warfare agents and their hydrolysis products.177 Compounds commonly used as chemical warfare agents including trimethyl phosphate, diisopropyl methyl-phosphonate, and dimethyl methylphosphonate were detected in blood and urine at limits of detection below those reported in biological fluids found in real chemical warfare agent victims. In a similar approach, Dhummakupt et al. applied PSI using MOF-modified glass substrates to analyze volatile chemical warfare agents called G-agents.178 While PSI is ideal for liquid sample analysis, volatile compounds often evaporate quickly from the paper substrates, thus preventing their detection. Coating the substrate with MOFs increased the absorption of the G-agents to the substrate, which in turn greatly improved their analytical lifetime. For example, the G-agent compounds sarin and soman were detected in the PSI mass spectra for up to 50 min after being spotted in solution on a HKUST-1 coated substrate, which was 10 times longer than the <5 min detection time achieved with paper substrate. This improvement in analytical lifetime could allow PSI-MS to be used as a fieldable method for the detection of chemical warfare agents at a suspected chemical attack location. A DART-MS methodology was also refined for explosive detection by combining Joule heating thermal desorption (JHTD) with DART (left panel, Figure 7).179 JHTD was directly applied to a sample deposited onto a nichrome wire, allowing temperature-controlled analyte desorption and improved monitoring of the thermal degradation and in-source fragmentation. Through this integration, organic and inorganic explosives presenting different desorption temperatures, such as calcium ammonium nitrate and potassium chlorate, were simultaneously detected.
Figure 7.
Highlights of forensics applications of ambient ionization MS techniques. The left panel shows the use of DART combined with JHTD to allow for the detection of various explosives that have different desorption temperatures. Samples were deposited on a nichrome wire loop and rapidly heated by passing dc current through the wire. The desorbed molecules were then ionized by the DART source. Modified from Anal. Methods, Vol. 9, Forbes, T. P.; Sisco, E.; Staymates, M.; Gillen, G. DART-MS analysis of inorganic explosives using high temperature thermal desorption, pp. 4885–5076 (ref 179). Copyright 2017, with permission from Elsevier. The middle panel shows the use of PSI-MS for the analysis of drugs in blood samples. For each drug, two fragment peaks were obtained using selected reaction monitoring (SRM). The graph shown in the bottom of the panel shows the SRM ratio over time of the two fragment peaks for each drug, which can be used to determine the number of scans needed to confidently determine if a drug is present in blood. Reprinted from Anal. Methods, Vol. 9, Jett, R.; Skaggs, C.; Manicke, N. E. Drug screening method development for paper spray coupled to a triple quadrupole mass spectrometer, pp 5037–5043 (ref 183). Copyright 2017, with permission from Elsevier. The right panel shows the use of DESI-MS imaging for the analysis of cross sections of paintings, including a proof-of-concept cross section prepared in the lab with layers of various types of paint and binding media and a historical painting, The Triumph of David. Adapted from Method development for binding media analysis in painting cross sections by desorption electrospray ionization mass spectrometry, Watts, K. E.; Lagalante, A. F. Rapid Commun. Mass Spectrom., Vol. 32 (ref 187). Copyright 2018 Wiley.
PSI-MS workflows have also been developed for rapid screening of drugs of abuse in biological fluids.181-183 For example, Kennedy et al. performed quantitative PSI analysis of fentanyl in urine by utilizing cartridges preloaded with internal standards.182 In a related study, Jett et al. described the development of PSI coupled to a triple quadrupole mass spectrometer for the analysis of 134 drugs and drug metabolites spiked in blood (center panel, Figure 7).183 Using this approach, 30 analytes were detected from dried blood spots at cutoff levels below 100 ng/mL within a 90 s analysis time, illustrating the potential of using PSI for high-throughput drug screening.
DART-MS has also been used to detect nonoxynol, a common spermicide in condom lubricants, in vaginal fluid samples to demonstrate applicability for gathering evidence for sexual assault crimes.184 In the study by Proni et al., vaginal fluid samples were collected using cotton swabs or glass rods before and after contact with a condom and then directly analyzed using DART-MS. In addition to nonoxynol detection, differences in the mass spectral profiles were seen depending on the condom type. This method could thus be used to provide valuable evidence in sexual assault crimes where a condom is used.
Ambient ionization MS techniques have also been applied for direct and gentle analysis of artwork.185-187 Newsome et al., for example, integrated a mechanical shutter to a DART source to allow for discrete sampling of heat sensitive materials, such as photographs, without causing visible damage.185 Aging of paper and documents has also been explored using ambient ionization MS.186 Schedl et al. used reactive PSI to enhance the sensitivity in the detection of chromophores associated with paper aging, which are often present in ppm to ppb concentration ranges. Reactive PSI was accomplished by adding Girard’s reagent T to the spray solvent, which selectively reacted with the chromophore 2,5-dihydroxyaceto-phenone, enhancing sensitivity for analyte detection. The authors demonstrated the value of this approach for studying discoloration and aging of paper through the analysis of both historic paper samples and artificially aged paper.186 DESI has also been applied to analyze binding media in painting cross sections (right panel, Figure 7).187 Painting material has changed over time from lipid containing media to modern acrylic binding media and thus presents distinct chemical composition. To aid in determining painting time period or if unoriginal paint layers have been added to an original piece of artwork, DESI-MS was used to directly analyze a cross section of a painting, without separating the binding media from the paint matrix. Detection of surfactant octyl phenol ethoxylate polymer localized to the acrylic layer and multiple fatty acyl species localized to the oil and egg tempera layers allowed rapid identification of the binding media used in the painting investigated.
Environmental.
Environmental applications of ambient ionization MS techniques have benefited from their amenability to in situ analyses. The interaction of environmentally relevant products and fouling agents or impurities is a topic of current research, including analysis of oil matrixes,188 petroleum and biodiesel,189-191 and water purification.54,192 For example, Jakka Ravindran et al. utilized DESI-MS to detect molecular signatures of biofouling on water purification membranes.192 Biofilms of P. aeruginosa and B. subtilis were analyzed for biosurfactants common in biofilm generation including rhamnolipids, quorum sensing molecules, as well as heavy metal associated lipids. DESI-MS imaging enabled biofilm detection from a variety of substrates including polymers, membranes, metals, and water purification materials, as surface irregularities promote bacterial adhesion. Another environmentally relevant application of DESI-MS was in the analysis of polar organic compounds on meteorites.193 In the study by Naraoka and Hashiguchi, a flat surface from the meteorite was chipped off and embedded into an indium plate for in situ surface analysis. Many polar organic compounds were detected from the meteorite, including alkylimidazoles and alkylpyridines, without compromising the surface of the chipped meteorite piece due to the gentle nature of DESI-MS analysis.
Food and Agriculture.
Ambient ionization MS techniques have also been extensively used in the fields of food and agricultural analyses, specifically for the detection of food and water contaminants,194-199 food fraud,83,200 and for the discrimination of food products.201,202 PSI, for example, was used by Chen et al. to quantify bisphenol A (BPA) and analogues from food packaging.198 The use of BPA in the production of food packaging has been controversial due to its potential dangers to human health. To analyze BPA directly from packaging materials including water bottles, baby bottles, as well as plastic and paper cups, the materials were cut into small triangles pieces, briefly soaked in organic solvent and dried prior to analysis by PSI. Using this approach, limits of detection (LODs) in the 0.1–0.3 μg/mL range were achieved for BPA and analogues, in a much higher throughput (<2 min per sample) than what achieved with a standard HPLC–MS method. In a different study, microwave plasma torch MS (MPT-MS) was used to investigate the molecular profiles of navel oranges cultivated in three habitats located within close proximity to one another.203 MPT-MS is a plasma-based technique that utilizes a thermal plasma for desorption and ionization of analytes in a process similar to DART and other plasma-based sources. Oranges grown from 12 year old trees within a 3 km radius of each other were directly analyzed by MPT-MS, with the goal of identifying their place of growth in addition to assessing the quality of the produce. The MPT source was angled toward the orange surface and placed coaxially and 10 mm away from the inlet of the MS. Molecules detected from the oranges included vanillin and other volatile and semivolatile sugars, alcohols, and flavonoids. The abundance of protonated 5-hydroxymethyl furfural (m/z 127), a degradation product of hexose, fluctuated with the flavor of the oranges and served as an indicator of fruit quality. Additionally, PCA of the molecular profiles acquired showed separation of the orange rinds and the juice sacs among the three growth locations, thus allowing discrimination among neighboring habitats.
Reaction Monitoring and Catalysis.
Ambient ionization MS techniques have been used to investigate various chemical processes in reaction monitoring, electrochemistry, and catalysis studies. Studies employing ESI have been widely performed and have provided evidence on the accelerated reaction rates occurring in the ESI microdroplets.204,205 In a similar fashion, ambient ionization MS techniques have been used as platforms to study acceleration of reactions in microdroplets as well as to explore reaction mechanisms in bulk solutions.206 Acceleration of many reactions including degradation of the antibiotic Erythromycin A,207 tert-butyloxycarbonyl deprotection of amines,208 and oxidation of oleic209 and pinonic210 acids at air–water interfaces in levitated droplets have been investigated using DART, touch spray, and field-induced droplet ionization, respectively. For example, reactive LAESI was used to monitor the click reaction between various tetrazine compounds and BCN-amine.211 A mid-IR laser was used to desorb a tetrazine compound from a 96 well plate, creating a plume of the compound which was then intercepted by a spray containing BCN-amine (left panel, Figure 8). Metal tubes of varying lengths were placed between the LAESI source and the mass spectrometer to vary the reaction time prior to analysis in order to study reaction kinetics. The speed of the click reaction between dipyridyl-tetrazine and BCN-amine in the microdroplets was at least 2 orders of magnitude faster than what reported in bulk solution.
Figure 8.
Highlights of the application of ambient ionization MS techniques in reaction monitoring and catalysis. The left panel shows the use of LAESI-MS to monitor the formation of click reaction products. The pulsed laser ablates the tetrazine (2) and interacts with the cyclooctyne (1) within the ESI plume, followed by reaction within a varied length reaction tube and sequential MS analysis. The time between the laser pulse that initiated reactant ablation and product detection in the mass spectrometer was monitored and utilized to determine the kinetics of reaction. Adapted from van Greenan, F. A. M. G.; Franssen, M. C. R.; Zuilhof, H.; Nielen, M. W. F. Anal. Chem. 2018, 90, 10409–10416 (ref 211) . Copyright 2018 American Chemical Society. The center panel illustrates the use of DESI-MS for high-throughput reaction screening. The reactants are spotted onto a DESI plate and analyzed, allowing quick identification of successful reactions to then be scaled-up. For example, the technique was used to study 16 alkylation reactions, with the reaction between benzylamine (1) and 2-(bromoethyl)benzene (9) shown. Reproduced from Wleklinski, M.; Loren, B. P.; Ferreira, C. R.; Jaman, Z.; Avramova, L.; Sobreira, T. J. P.; Thompson, D. H.; Cooks, R. G. Chem. Sci. 2018, 9, 1647–1653 (ref 212), with permission of The Royal Society of Chemistry. The right panel shows the use of DESI-MS to detect electrochemically generated nitrenium ions directly from a rotating platinum electrode, confirming that nitrenium is formed through oxidation of 4,4′-dimethoxydiphenylamine, but only when a voltage is applied to the working electrode. Adapted from Brown, T. A.; Hosseini-Nassab, N.; Chen, H.; Zare, R. N. Chem. Sci. 2016, 7, 329–332 (ref 218), with permission of The Royal Society of Chemistry.
As an alternative approach, Wleklinski et al. have suggested the use of droplet accelerated reactions in DESI-MS experiments as a discovery tool in synthetic chemistry for high-throughput screening of reaction conditions.212 To illustrate this concept, 50 nL sized droplets of up to 384 reaction mixtures were deposited onto large (6 144 spots) well plates (center panel, Figure 8). The plate was then sequentially analyzed using DESI-MS to rapidly screen the reaction mixtures in order to determine yield of product formation and thus if any particular reaction should be considered for synthetic optimization and scale-up. For example, screening of n-alkylation and Suzuki cross-coupling reactions were performed at a rate of 10 000 reaction mixtures per hour, allowing quick evaluation and refinement of optimal reaction conditions.
Monitoring electrochemical reactions has also been explored using ambient ionization MS techniques, allowing a unique opportunity to investigate the reaction mechanisms of these processes. Electrochemistry compatible solvent systems typically contain large percentages of aqueous solution and high concentrations of electrolytes, both of which are often detrimental to ESI analysis.213-215 As more amenable approaches to study electrochemical reactions, liquid sample DESI has been applied to the online analysis of reaction products in an electrochemical cell,216 PESI has been used to spatially probe the reactions occurring in bipolar electrode systems,217 and DESI has shown the potential for direct analysis of reactive intermediates generated by electrochemical processes.218 Brown et al., for example, utilized DESI to investigate the oxidation pathways of secondary arylamines.218 The pathway of formation of nitrenium that had been proposed in the literature is through a radical cation intermediate generated by oxidation of the arylamine. In the experiments conducted, a rotating platinum electrode was partially submerged in an electrolyte solution containing the reference and counter electrode and sprayed with a solution of 4,4′-dimethoxydiphenylamine (DMDPA) using DESI (right panel, Figure 8). DMDPA nitrenium ions (m/z 228.102) and a DMDPA radical cation intermediate (m/z 229.109) were formed due to the electrochemical reduction of the DMDPA at the electrode surface and detected in the mass spectrometer. The results obtained suggest that the radical cation intermediate and the nitrenium ion are formed by electrochemical oxidation, which was illustrated by a 10-fold increase in ion signal from the cation when an oxidation potential of 1.5 V was applied to the working electrode.
PSI has also been used to perform online catalytic reactions. Banerjee et al., for example, coated paper substrates with palladium, silver, and gold nanoparticles for Suzuki cross-coupling, reduction of 4-nitrophenol, and oxidation of glucose, respectively.219 In situ catalysis followed by MS analysis allowed for detection of reaction intermediates and products from all three reactions investigated, yielding insights into reaction mechanisms and activity of the catalyst. These results demonstrate that PSI is a simple and quick method to evaluate catalysts for reactions that could potentially be applied more widely for screening of new catalysts in other reactions. Similar methodology was used by other researchers for online catalytic reactions with PSI, including TiO2 coated paper for photo-catalysis of methylene blue degradation220 and platinum nanoparticle coated paper for reduction of the nitro groups in TNT for improved protonation.221
Elemental and Isotope Analysis.
Increasing effort has been directed at improving the amenability of ambient ionization MS for the analysis of elemental222 and isotopic223,224 composition and material characterization.225 Inductively coupled plasma-MS has been traditionally used for elemental analysis, especially analysis of metal atoms for a variety of applications.226 Concurrently, plasma-based ambient ionization MS techniques have also been tested for similar purposes. For example, Evans-Nguyen et al. used MPT-MS for elemental analysis of solid samples, including U.S. pennies, a lead fishing sinker, and brass foil.222 The MPT-MS source allowed for elemental analysis of most of the samples without noticeable damage or significant heating. Additionally, an ion trap mass analyzer was utilized despite being atypical for elemental analysis in order to adapt this technology for on-site analysis as there are currently no fieldable, MS-based elemental analysis tools. To emphasize the use of this technique as an on-site forensic testing method, a mixture of compounds that would typically be found in gun shot residue, including lead, barium, antimony, and the organic molecule methyl centralite were deposited on a substrate. Analysis with the MPT ion source allowed for detection of the methyl centralite, as well as barium and antimony simultaneously, suggesting this technology could allow organic and inorganic composition analysis directly from swabs taken at a crime scene. As an alternate approach, Shiea et al. used electrospray-assisted laser desorption (ELDI) MS to analyze in situ metal atoms from coins.227 Similar to MALDESI and LAESI, ELDI utilizes an IR laser to ablate the sample surface, creating an analyte plume that is intercepted by an ESI generated spray for ionization. In this study, the chelation agent ethylenediaminetetraacetic acid (EDTA) was deposited on the surface of a coin which was then irradiated, desorbing metal–chelator complexes from the sample surface. An ESI plume was then used to ionize these complexes for subsequent mass spectrometric detection. Using this approach, copper, zinc, aluminum, and nickel metal–EDTA complexes were detected from coins from multiple countries, including the United States, Canada, Japan, and Taiwan. The authors propose this method as a technique for the rapid distinguishing of authentic and fake coins as the metal composition of coins is well-regulated and consistent.
CONCLUSIONS
The development of ambient ionization MS has greatly altered the way mass spectrometry is used across many fields by allowing sample analysis to be performed in a simple, quick, and effective manner. Interest in ambient ionization MS has rapidly increased in the past 15 years, greatly expanding the number of techniques and their applications in biomedical, bioanalytical, forensics, environmental, agricultural, and chemical analyses, among other areas. In this review, we highlighted key technological developments and new applications of ambient ionization MS pursued over the past 2 years. Many improvements have focused on expanding ambient MS analyses to a wider variety of analytes, including analysis of nonpolar molecules and large biopolymers from complex samples. Additionally, research has been performed to increase the analytical performance of ambient ionization MS methods including their sensitivity and reliability for qualitative and quantitative analyses, allowing comparable performance to more traditional MS assays for targeted applications such as in Therapeutic Drug Monitoring. New ambient ionization MS platforms that integrate multiple technologies and/or automate sampling have facilitated the use of ambient ionization techniques and expanded their use in research as well as clinical/fieldable applications. Real time MS evaluations of living samples, for example, including in vivo analysis for surgical applications, live screening of bacterial colonies, and real time metabolic monitoring of plants and animals, have demonstrated the power and potential significance of ambient ionization MS techniques in translational and field applications. The use of ambient ionization MS in drug monitoring, detection of explosives, as well as agricultural and environmental contamination have further exemplified the value of these techniques for routine use in regulatory fields.
As ambient ionization MS techniques continue to be refined and further applied across a broad range of scientific fields, increased efforts in improving their analytical performance including lowering LODs and enabling quantitative assays for compounds at trace levels or within complex sample matrixes are needed to further expand their use. More rigorous studies of the extraction and ionization mechanisms employed in ambient ionization MS techniques are still needed to deepen our understanding of the physical and chemical processes driving sampling and chemical analyses. Further, proper validation studies are necessary to consolidate biological and mechanistic discoveries that are enabled by direct analysis of clinical specimens and reaction mixtures, respectively. Lastly, as ambient ionization MS techniques transition from research laboratory tools to devices and tools that can be routinely operated by nonexperts, more work on method automation, hardware integration, and software development will be needed to facilitate their handling, operation, as well as to translate mass spectral data into immediate actionable information. Yet, the ongoing efforts that are already being undertaken to address these remaining challenges combined with the breadth of new applications and developments within the field in recent years strongly suggest that ambient ionization MS techniques have, and will continue to have, an enduring impact on research, development, and our extended communities.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (Grant R00CA190783), the Cancer Prevention Research Institute of Texas (CPRIT, Grant RP170427), and The Welch Foundation (Grant F-1895).
Biography
Clara L. Feider received her B.S. in Chemistry from the University of San Francisco in California in the spring of 2015. Currently, she is a graduate student in Prof. Livia S. Eberlin’s lab at the University of Texas at Austin. Her research focuses on the utilization and development of ambient ionization mass spectrometry techniques towards the improved treatment of endometriosis.
Anna Krieger received her B.A. in Chemistry from Gustavus Adolphus College in Saint Peter, Minnesota in 2017. She is currently a graduate student in Prof. Livia S. Eberlin’s lab at the University of Texas at Austin. Her research is focused on developing the MasSpec Pen for molecular characterization of brain cancers.
Rachel J. DeHoog received her B.S. in Chemistry from Westmont College in 2015 and is currently a graduate student in Prof. Livia S. Eberlin’s lab at the University of Texas at Austin. Her research focuses on developing and applying ambient ionization mass spectrometry techniques to address clinical challenges in the diagnosis and treatment of patients with endocrine diseases.
Livia S. Eberlin received her B.S. in Chemistry from the State University of Campinas in 2008. In 2012, she received her Ph.D. in Analytical Chemistry from Purdue University under the supervision of Prof. R. Graham Cooks. She then pursued her postdoctoral research in the Department of Chemistry at Stanford University under the mentorship of Prof. Richard N. Zare. In 2016, Prof. Eberlin started her role as an assistant professor in the Department of Chemistry at The University of Texas at Austin. Eberlin is the recipient of a NIH/NCI K99/R00 Pathway to Independence Award, a Moore Inventor Fellowship, and a MacArthur Fellowship. Her research program centers around the development and application of ambient ionization mass spectrometry technologies in health related research, with a particular focus on disease detection and diagnosis to improve patient care and clinical outcomes.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Takáts Z; Wiseman JM; Gologan B; Cooks RG Science 2004, 306, 471–473. [DOI] [PubMed] [Google Scholar]
- (2).Cody RB; Laramée JA; Durst HD Anal. Chem 2005, 77, 2297–2302. [DOI] [PubMed] [Google Scholar]
- (3).Gross JH Anal. Bioanal. Chem 2014, 406, 63–80. [DOI] [PubMed] [Google Scholar]
- (4).Ifa DR; Wu CP; Ouyang Z; Cooks RG Analyst 2010, 135, 669–681. [DOI] [PubMed] [Google Scholar]
- (5).Venter A; Nefliu M; Cooks RG TrAC, Trends Anal. Chem 2008, 27, 284–290. [Google Scholar]
- (6).Venter AR; Douglass KA; Shelley JT; Hasman G; Honarvar E Anal. Chem 2014, 86, 233–249. [DOI] [PubMed] [Google Scholar]
- (7).Laskin J; Lanekoff I Anal. Chem 2016, 88, 52–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Harris GA; Galhena AS; Fernández FM Anal. Chem 2011, 83, 4508–4538. [DOI] [PubMed] [Google Scholar]
- (9).Wu C; Dill AL; Eberlin LS; Cooks RG; Ifa DR Mass Spectrom. Rev 2013, 32, 218–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Klampfl CW; Himmelsbach M Anal. Chim. Acta 2015, 890, 44–59. [DOI] [PubMed] [Google Scholar]
- (11).Javanshad R; Venter AR Anal. Methods 2017, 9, 4896–4907. [Google Scholar]
- (12).Alberici RM; Simas RC; Sanvido GB; Romao W; Lalli PM; Benassi M; Cunha IBS; Eberlin MN Anal. Bioanal. Chem 2010, 398, 265–294. [DOI] [PubMed] [Google Scholar]
- (13).Fenn J; Mann M; Meng C; Wong S; Whitehouse C Science 1989, 246, 64–71. [DOI] [PubMed] [Google Scholar]
- (14).Haddad R; Sparrapan R; Eberlin MN Rapid Commun. Mass Spectrom 2006, 20, 2901–2905. [DOI] [PubMed] [Google Scholar]
- (15).Chen H; Venter A; Cooks RG Chem. Commun 2006, 2042–2044. [DOI] [PubMed] [Google Scholar]
- (16).Wu C; Siems WF; Hill HH Anal. Chem 2000, 72, 396–403. [DOI] [PubMed] [Google Scholar]
- (17).Tillner J; Wu V; Jones EA; Pringle SD; Karancsi T; Dannhorn A; Veselkov K; McKenzie JS; Takats ZJ Am. Soc. Mass Spectrom 2017, 28, 2090–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Rejsek J; Vrkoslav V; Pokorny V; Pribyl V; Cvacka J Anal. Chem 2017, 89, 11452–11459. [DOI] [PubMed] [Google Scholar]
- (19).Tillner J; McKenzie JS; Jones EA; Speller AVM; Walsh JL; Veselkov KA; Bunch J; Takats Z; Gilmore IS Anal. Chem 2016, 88, 4808–4816. [DOI] [PubMed] [Google Scholar]
- (20).Van Berkel GJ; Sanchez AD; Quirke JME Anal. Chem 2002, 74, 6216–6223. [DOI] [PubMed] [Google Scholar]
- (21).Roach PJ; Laskin J; Laskin A Analyst 2010, 135, 2233–2236. [DOI] [PubMed] [Google Scholar]
- (22).Kertesz V; Van Berkel GJ J. Mass Spectrom 2010, 45, 252–260. [DOI] [PubMed] [Google Scholar]
- (23).Meurs J; Alexander MR; Levkin PA; Widmaier S; Bunch J; Barrett DA; Kim DH Anal. Chem 2018, 90, 6001–6005. [DOI] [PubMed] [Google Scholar]
- (24).Wang H; Liu J; Cooks RG; Ouyang Z Angew. Chem 2010, 122, 889–892. [DOI] [PubMed] [Google Scholar]
- (25).Dulay MT; Zare RN Rapid Commun. Mass Spectrom 2017, 31, 1651–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Straub RF; Voyksner RD J. Am. Soc. Mass Spectrom 1993, 4, 578–587. [DOI] [PubMed] [Google Scholar]
- (27).Colletes TC; Garcia PT; Campanha RB; Abdelnur PV; Romao W; Coltro WKT; Vaz BG Analyst 2016, 141, 1707–1713. [DOI] [PubMed] [Google Scholar]
- (28).Damon DE; Maher YS; Yin MZ; Jjunju FPM; Young IS; Taylor S; Maher S; Badu-Tawiah AK Analyst 2016, 141, 3866–3873. [DOI] [PubMed] [Google Scholar]
- (29).Liu JJ; He Y; Chen S; Ma M; Yao SZ; Chen B Talanta 2017, 166, 306–314. [DOI] [PubMed] [Google Scholar]
- (30).Salentijn GIJ; Oleschuk RD; Verpoorte E Anal. Chem 2017, 89, 11419–11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kim D; Yim UH; Kim B; Cha S; Kim S Anal. Chem 2017, 89, 9056–9061. [DOI] [PubMed] [Google Scholar]
- (32).Basuri P; Sarkar D; Paramasivam G; Pradeep T Anal. Chem 2018, 90, 4663–4668. [DOI] [PubMed] [Google Scholar]
- (33).Duarte LC; de Carvalho TC; Lobo EO; Abdelnur PV; Vaz BG; Coltro WKT Anal. Methods 2016, 8, 496–503. [Google Scholar]
- (34).Chen J; Tang F; Guo CA; Huo XM; Zhang SC; Wang XH Anal. Bioanal. Chem 2016, 408, 5005–5012. [DOI] [PubMed] [Google Scholar]
- (35).Damon DE; Davis KM; Moreira CR; Capone P; Cruttenden R; Badu-Tawiah AK Anal. Chem 2016, 88, 1878–1884. [DOI] [PubMed] [Google Scholar]
- (36).Wang XT; Zheng YJ; Wang T; Xiong XC; Fang X; Zhang ZP Anal. Methods 2016, 8, 8004–8014. [Google Scholar]
- (37).Hiraoka K; Nishidate K; Mori K; Asakawa D; Suzuki S Rapid Commun. Mass Spectrom. 2007, 21, 3139–3144. [DOI] [PubMed] [Google Scholar]
- (38).Hu B; So P-K; Chen H; Yao Z-P Anal. Chem 2011, 83, 8201–8207. [DOI] [PubMed] [Google Scholar]
- (39).Bernier MC; Li A; Winalski L; Zi Y; Li Y; Caillet C; Newton P; Wang ZL; Fernández FM Rapid Commun. Mass Spectrom 2018, 32, 1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhang JL; Rector J; Lin JQ; Young JH; Sans M; Katta N; Giese N; Yu WD; Nagi C; Suliburk J; Liu JS; Bensussan A; DeHoog RJ; Garza KY; Ludolph B; Sorace AG; Syed A; Zahedivash A; Milner TE; Eberlin LS Sci. Transl. Med 2017, 9, eaan3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Pagnotti VS; Inutan ED; Marshall DD; McEwen CN; Trimpin S Anal. Chem 2011, 83, 7591–7594. [DOI] [PubMed] [Google Scholar]
- (42).Li AY; Paine MRL; Zambrzycki S; Stryffeler RB; Wu JS; Bouza M; Huckaby J; Chang CY; Kumar M; Mukhija P; Fernandez FM Anal. Chem 2018, 90, 3981–3986. [DOI] [PubMed] [Google Scholar]
- (43).Thomson BA J. Am. Soc. Mass Spectrom 1998, 9, 187–193. [Google Scholar]
- (44).Shelley JT; Wiley JS; Chan GCY; Schilling GD; Ray SJ; Hieftje GM J. Am. Soc. Mass Spectrom 2009, 20, 837–844. [DOI] [PubMed] [Google Scholar]
- (45).Takáts Z; Cotte-Rodriguez I; Talaty N; Chen H; Cooks RG Chem. Commun 2005, 1950–1952. [DOI] [PubMed] [Google Scholar]
- (46).Li Z; Zhang JL; Zhang YW; Bai Y; Liu HW J. Anal. Test 2017, 1, 3. [Google Scholar]
- (47).Kogelschatz U; Eliasson B; Egli WJ Phys. IV 1997, 07, C4–47. [Google Scholar]
- (48).Harper JD; Charipar NA; Mulligan CC; Zhang X; Cooks RG; Ouyang Z Anal. Chem 2008, 80, 9097–9104. [DOI] [PubMed] [Google Scholar]
- (49).Na N; Zhang C; Zhao M; Zhang S; Yang C; Fang X; Zhang XJ Mass Spectrom. 2007, 42, 1079–1085. [DOI] [PubMed] [Google Scholar]
- (50).Na N; Zhao M; Zhang S; Yang C; Zhang XJ Am. Soc. Mass Spectrom 2007, 18, 1859–1862. [DOI] [PubMed] [Google Scholar]
- (51).Cody RB Anal. Chem 2009, 81, 1101–1107. [DOI] [PubMed] [Google Scholar]
- (52).Badal SP; Michalak SD; Chan GCY; You Y; Shelley JT Anal. Chem 2016, 88, 3494–3503. [DOI] [PubMed] [Google Scholar]
- (53).Newsome GA; Ackerman LK; Johnson KJ J. Am. Soc. Mass Spectrom 2016, 27, 135–143. [DOI] [PubMed] [Google Scholar]
- (54).Huba AK; Mirabelli MF; Zenobi R Anal. Chim. Acta 2018, 1030, 125–132. [DOI] [PubMed] [Google Scholar]
- (55).Zhou ZP; Lee JK; Kim SC; Zare RN Anal. Chem 2016, 88, 5542–5548. [DOI] [PubMed] [Google Scholar]
- (56).Liu Y; Ma X; Lin Z; He M; Han G; Yang C; Xing Z; Zhang S; Zhang X Angew. Chem 2010, 122, 4537–4539. [DOI] [PubMed] [Google Scholar]
- (57).Bierstedt A; Riedel J Methods 2016, 104, 3–10. [DOI] [PubMed] [Google Scholar]
- (58).Wang H; Fei ZH; Li ZX; Xing R; Liu ZM; Zhang YY; Ding H Talanta 2018, 179, 364–368. [DOI] [PubMed] [Google Scholar]
- (59).Fowble KL; Teramoto K; Cody RB; Edwards D; Guarrera D; Musah RA Anal. Chem 2017, 89, 3421–3429. [DOI] [PubMed] [Google Scholar]
- (60).Van Breemen RB; Snow M; Cotter RJ Int. J. Mass Spectrom. Ion Phys 1983, 49, 35–50. [Google Scholar]
- (61).Shiea J; Huang M-Z; HSu H-J; Lee C-Y; Yuan C-H; Beech I; Sunner J Rapid Commun. Mass Spectrom 2005, 19, 3701–3704. [DOI] [PubMed] [Google Scholar]
- (62).Haarhoff Z; Wagner A; Picard P; Drexler DM; Zvyaga T; Shou WJ Biomol Screening 2016, 21, 165–175. [DOI] [PubMed] [Google Scholar]
- (63).Benham K; Hodyss R; Fernandez FM; Orlando TM J. Am. Soc. Mass Spectrom 2016, 27, 1805–1812. [DOI] [PubMed] [Google Scholar]
- (64).Nemes P; Vertes A Anal. Chem 2007, 79, 8098–8106. [DOI] [PubMed] [Google Scholar]
- (65).Sampson JS; Hawkridge AM; Muddiman DC J. Am. Soc. Mass Spectrom 2006, 17, 1712–1716. [DOI] [PubMed] [Google Scholar]
- (66).Nemes P; Woods AS; Vertes A Anal. Chem 2010, 82, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Fincher JA; Korte AR; Reschke B; Morris NJ; Powell MJ; Vertes A Analyst 2017, 142, 3157–3164. [DOI] [PubMed] [Google Scholar]
- (68).Li H; Vertes A Analyst 2017, 142, 2921–2927. [DOI] [PubMed] [Google Scholar]
- (69).Compton LR; Reschke B; Friend J; Powell M; Vertes A Rapid Commun. Mass Spectrom 2015, 29, 67–73. [DOI] [PubMed] [Google Scholar]
- (70).Robichaud G; Barry JA; Garrard KP; Muddiman DC J. Am. Soc. Mass Spectrom 2013, 24, 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Ekelof M; Muddiman DC Anal. Bioanal. Chem 2018, 410, 963–970. [DOI] [PubMed] [Google Scholar]
- (72).Nazari M; Muddiman DC Analyst 2016, 141, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Bokhart MT; Manni J; Garrard KP; Ekelof M; Nazari M; Muddiman DC J. Am. Soc. Mass Spectrom 2017, 28, 2099–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Woolman M; Gribble A; Bluemke E; Zou J; Ventura M; Bernards N; Wu M; Ginsberg HJ; Das S; Vitkin A; Zarrine-Afsar A Sci Rep. 2017, 7, 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Fatou B; Saudemont P; Leblanc E; Vinatier D; Mesdag V; Wisztorski M; Focsa C; Salzet M; Ziskind M; Fournier I Sci. Rep 2016, 6, 25919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Woolman M; Gribble A; Bluemke E; Zou J; Ventura M; Bernards N; Wu M; Ginsberg HJ; Das S; Vitkin A; Zarrine-Afsar A Sci. Rep. 2017, 7, 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Li Y; Shrestha B; Vertes A Anal. Chem 2007, 79, 523–532. [DOI] [PubMed] [Google Scholar]
- (78).Yung YP; Wickramasinghe R; Vaikkinen A; Kauppila TJ; Veryovkin IV; Hanley L Anal. Chem 2017, 89, 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Berry JL; Day DA; Elseberg T; Palm BB; Hu W; Abdelhamid A; Schroder JC; Karst U; Jimenez JL; Browne EC Anal. Chem 2018, 90, 4046–4053. [DOI] [PubMed] [Google Scholar]
- (80).Robb DB; Covey TR; Bruins AP Anal. Chem 2000, 72, 3653–3659. [DOI] [PubMed] [Google Scholar]
- (81).Lin YN; Wang HX; Rao W; Cui YW; Yu XN; Dai ZY; Shen QJ Agric. Food Chem 2018, 66, 6246–6253 [DOI] [PubMed] [Google Scholar]
- (82).Strittmatter N; Lovrics A; Sessler J; McKenzie JS; Bodai Z; Doria ML; Kucsma N; Szakacs G; Takats Z Anal. Chem 2016, 88, 7507–7514. [DOI] [PubMed] [Google Scholar]
- (83).Black C; Chevallier OP; Haughey SA; Balog J; Stead S; Pringle SD; Riina MV; Martucci F; Acutis PL; Morris M; Nikolopoulos DS; Takats Z; Elliott CT Metabolomics 2017, 13, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Schäfer K-C; Dénes J; Albrecht K; Szaniszló T; Balog J; Skoumal R; Katona M; Tóth M; Balogh L; Takáts Z Angew. Chem., Int. Ed 2009, 48, 8240–8242. [DOI] [PubMed] [Google Scholar]
- (85).Phelps DL; Balog J; Gildea LF; Bodai Z; Savage A; El-Bahrawy MA; Speller AVM; Rosini F; Kudo H; McKenzie JS; Brown R; Takats Z; Ghaem-Maghami S Br.J. Cancer 2018, 118, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Alexander J; Gildea L; Balog J; Speller A; McKenzie J; Muirhead L; Scott A; Kontovounisios C; Rasheed S; Teare J; Hoare J; Veselkov K; Goldin R; Tekkis P; Darzi A; Nicholson J; Kinross J; Takats Z Surg. Endosc 2017, 31, 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Liu CY; Yang JZ; Wang J; Hu YH; Zhao W; Zhou ZY; Qi F; Pan YJ Am. Soc. Mass Spectrom 2016, 27, 1597–1605. [DOI] [PubMed] [Google Scholar]
- (88).Li X; Attanayake K; Valentine SJ; Li P Rapid Commun. Mass Spectrom 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).McEwen CN; Pagnotti VS; Inutan ED; Trimpin S Anal. Chem 2010, 82, 9164–9168. [DOI] [PubMed] [Google Scholar]
- (90).Lu IC; Elia EA; Zhang WJ; Pophristic M; Inutan ED; McEwen CN; Trimpin S Anal. Methods 2017, 9, 4971–4978. [Google Scholar]
- (91).Pagnotti VS; Chubatyi ND; McEwen CN Anal. Chem 2011, 83, 3981–3985. [DOI] [PubMed] [Google Scholar]
- (92).Horan AJ; Apsokardu MJ; Johnston MV Anal. Chem 2017, 89, 1059–1062. [DOI] [PubMed] [Google Scholar]
- (93).Ai WP; Nie HG; Song SY; Liu XY; Bai Y; Liu HW J. Am. Soc. Mass Spectrom 2018, 29, 1408–1415. [DOI] [PubMed] [Google Scholar]
- (94).Lawton ZE; Traub A; Fatigante WL; Mancias J; O’Leary AE; Hall SE; Wieland JR; Oberacher H; Gizzi MC; Mulligan CC J. Am. Soc. Mass Spectrom 2017, 28, 1048–1059. [DOI] [PubMed] [Google Scholar]
- (95).Perez CJ; Bagga AK; Prova SS; Yousefi Taemeh M; Ifa DR Rapid Commun. Mass Spectrom 2018, DOI: 10.1002/rcm.8145. [DOI] [PubMed] [Google Scholar]
- (96).Duncan KD; Fang R; Yuan J; Chu RK; Dey SK; Burnum-Johnson KE; Lanekoff I Anal. Chem 2018, 90, 7246–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Jackson AU; Shum T; Sokol E; Dill A; Cooks RG Anal. Bioanal. Chem 2011, 399, 367–376. [DOI] [PubMed] [Google Scholar]
- (98).Rao W; Pan N; Yang ZJ Am. Soc. Mass Spectrom 2015, 26, 986–993. [DOI] [PubMed] [Google Scholar]
- (99).Rao W; Pan N; Tian X; Yang ZB J. Am. Soc. Mass Spectrom 2016, 27, 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Pan N; Rao W; Kothapalli NR; Liu R; Burgett AW; Yang Z Anal. Chem 2014, 86, 9376–9380. [DOI] [PubMed] [Google Scholar]
- (101).Pulfer M; Murphy RC Mass Spectrom. Rev 2003, 22, 332–364. [DOI] [PubMed] [Google Scholar]
- (102).Spector AA; Yorek MA J. Lipid Res 1985, 26, 1015–1035. [PubMed] [Google Scholar]
- (103).Tang F; Guo CG; Ma XX; Zhang J; Su Y; Tian R; Shi RY; Xia Y; Wang XH; Ouyang Z Anal. Chem 2018, 90, 5612–5619. [DOI] [PubMed] [Google Scholar]
- (104).Klein DR; Feider CL; Garza KY; Lin JQ; Eberlin LS; Brodbelt JS Anal. Chem 2018, 90, 10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Ma X; Xia Y Angew. Chem., Int. Ed 2014, 53, 2592–2596. [DOI] [PubMed] [Google Scholar]
- (106).Klein DR; Brodbelt JS Anal. Chem 2017, 89, 1516–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Ma X; Chong L; Tian R; Shi R; Hu TY; Ouyang Z; Xia Y Proc. Natl. Acad. Sci. U. S. A 2016, 113, 2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Nguyen S; Sontag R; Carson J; Corley R; Ansong C; Laskin JJ Am. Soc. Mass Spectrom 2018, 29, 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Tata A; Gribble A; Ventura M; Ganguly M; Bluemke E; Ginsberg HJ; Jaffray DA; Ifa DR; Vitkin A; Zarrine-Afsar A Chem. Sci 2016, 7, 2162–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Hsu C-C; Chou P-T; Zare RN Anal. Chem 2015, 87, 11171–11175. [DOI] [PubMed] [Google Scholar]
- (111).Griffiths RL; Creese AJ; Race AM; Bunch J; Cooper HJ Anal. Chem. 2016, 88, 6758–6766. [DOI] [PubMed] [Google Scholar]
- (112).Griffiths RL; Cooper HJ Anal. Chem 2016, 88, 606–609. [DOI] [PubMed] [Google Scholar]
- (113).Feider CL; Elizondo N; Eberlin LS Anal. Chem 2016, 88, 11533–11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Hsu CC; Baker MW; Gaasterland T; Meehan MJ; Macagno ER; Dorrestein PC Anal. Chem 2017, 89, 8251–8258. [DOI] [PubMed] [Google Scholar]
- (115).Garza KY; Feider CL; Klein DR; Rosenberg JA; Brodbelt JS; Eberlin LS Anal. Chem 2018, 90, 7785–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Towers MW; Karancsi T; Jones EA; Pringle SD; Claude EJ Am. Soc. Mass Spectrom 2018, 29, 2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Douglass KA; Venter AR J. Mass Spectrom 2013, 48, 553–560. [DOI] [PubMed] [Google Scholar]
- (118).Zheng YJ; Wang Q; Wang XT; Chen Y; Wang X; Zhang XL; Bai ZQ; Han XX; Zhang ZP Anal. Chem 2016, 88, 7005–7013. [DOI] [PubMed] [Google Scholar]
- (119).Hecht M; Evard H; Takkis K; Veigure R; Aro R; Lohmus R; Herodes K; Leito I; Kipper K Anal. Chem. 2017, 89, 11592–11597. [DOI] [PubMed] [Google Scholar]
- (120).Kok MGM; Fillet MJ Pharm. Biomed. Anal 2018, 147, 288–296. [DOI] [PubMed] [Google Scholar]
- (121).Gomez-Rios GA; Tascon M; Reyes-Garces N; Boyaci E; Poole J; Pawliszyn J Sci. Rep 2017, 7, 16104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Gómez-Ríos GA; Pawliszyn J Angew. Chem., Int. Ed 2014, 53, 14503–14507. [DOI] [PubMed] [Google Scholar]
- (123).Keating JE; Minges JT; Randell SH; Glish GL Anal. Methods 2018, 10, 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Luo ZG; He JJ; He JM; Huang L; Song XW; Li X; Abliz Z Talanta 2018, 179, 230–237. [DOI] [PubMed] [Google Scholar]
- (125).Hansen HT; Janfelt C Anal. Chem 2016, 88, 11513–11520. [DOI] [PubMed] [Google Scholar]
- (126).Taylor AJ; Dexter A; Bunch J Anal. Chem 2018, 90, 5637–5645. [DOI] [PubMed] [Google Scholar]
- (127).Ashizawa K; Yoshimura K; Johno H; Inoue T; Katoh R; Funayama S; Sakamoto K; Takeda S; Masuyama K; Matsuoka T; Ishii H Oral Oncol. 2017, 75, 111–119. [DOI] [PubMed] [Google Scholar]
- (128).Sakamoto K; Fujita Y; Chikamatsu K; Tanaka S; Takeda S; Masuyama K; Yoshimura K; Ishii H Transl. Cancer Res. 2018, 7, 758–764. [Google Scholar]
- (129).Zhang JY; Xu JJ; Ouyang YZ; Liu JW; Lu HY; Yu DL; Peng JH; Xiong JW; Chen HW; Wei YP Sci. Rep 2017, 7, 3738.28623324 [Google Scholar]
- (130).Banerjee S; Zare RN; Tibshirani RJ; Kunder CA; Nolley R; Fan R; Brooks JD; Sonn GA Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 3334–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Jarmusch AK; Pirro V; Baird Z; Hattab EM; Cohen-Gadol AA; Cooks RG Proc. Natl. Acad. Sci. U. S. A 2016, 113, 1486–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Margulis K; Chiou AS; Aasi SZ; Tibshirani RJ; Tang JY; Zare RN Proc. Natl. Acad. Sci. U. S. A 2018, 115, 6347–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Sans M; Gharpure K; Tibshirani R; Zhang JL; Liang L; Liu JS; Young JH; Dood RL; Sood AK; Eberlin LS Cancer Res. 2017, 77, 2903–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Zhang JL; Yu WD; Ryu SW; Lin J; Buentello G; Tibshirani R; Suliburk J; Eberlin LS Cancer Res. 2016, 76, 6588–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Porcari AM; Zhang J; Garza KY; Rodrigues-Peres RM; Lin JQ; Young JH; Tibshirani R; Nagi C; Paiva GR; Carter SA; Sarian LO; Eberlin MN; Eberlin LS Anal. Chem 2018, 90, 11324–11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Woolman M; Tata A; Bluemke E; Dara D; Ginsberg HJ; Zarrine-Afsar AJ Am. Soc. Mass Spectrom 2017, 28, 145–153. [DOI] [PubMed] [Google Scholar]
- (137).Pirro V; Jarmusch AK; Alfaro CM; Hattab EM; Cohen-Gadol AA; Cooks RG Analyst 2017, 142, 449–454. [DOI] [PubMed] [Google Scholar]
- (138).Pirro V; Alfaro CM; Jarmusch AK; Hattab EM; Cohen-Gadol AA; Cooks RG Proc. Natl. Acad. Sci. U. S. A 2017, 114, 6700–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Pirro V; Llor RS; Jarmusch AK; Alfaro CM; Cohen-Gadol AA; Hattabd EM; Cooks RG Analyst 2017, 142, 4058–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Bergholt MS; Serio A; McKenzie JS; Boyd A; Soares RF; Tillner J; Chiappini C; Wu V; Dannhorn A; Takats Z; Williams A; Stevens MM ACS Cent. Sci 2018, 4, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Gouw AM; Eberlin LS; Margulis K; Sullivan DK; Toal GG; Tong L; Zare RN; Felsher DW Proc. Natl. Acad. Sci. U. S. A 2017, 114, 4300–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Gaisl T; Bregy L; Stebler N; Gaugg MT; Bruderer T; Garcia-Gomez D; Moeller A; Singer F; Schwarz EI; Benden C; Sinues PML; Zenobi R; Kohler MJ Breath Res. 2018, 12, 036013. [DOI] [PubMed] [Google Scholar]
- (143).Gaugg MT; Bruderer T; Nowak N; Eiffert L; Sinues PML; Kohler M; Zenobi R Anal. Chem 2017, 89, 10329–10334. [DOI] [PubMed] [Google Scholar]
- (144).Schafer KC; Denes J; Albrecht K; Szaniszlo T; Balog J; Skoumal R; Katona M; Toth M; Balogh L; Takats Z Angew. Chem., Int. Ed 2009, 48, 8240–8242. [DOI] [PubMed] [Google Scholar]
- (145).Woolman M; Ferry I; Kuzan-Fischer CM; Wu M; Zou J; Kiyota T; Isik S; Dara D; Aman A; Das S; Taylor MD; Rutka JT; Ginsberg HJ; Zarrine-Afsar A Chem. Sci.2017, 8, 6508–6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Ambrose S; Housden NG; Gupta K; Fan JY; White P; Yen HY; Marcoux J; Kleanthous C; Hopper JTS; Robinson CV Angew. Chem., Int. Ed 2017, 56, 14463–14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Shin E; Cha S Bull. Korean Chem. Soc 2018, 39, 40–44. [Google Scholar]
- (148).Leney AC; Heck AJRJ Am. Soc. Mass Spectrom 2017, 28, 5–13. [DOI] [PubMed] [Google Scholar]
- (149).Lu YF; Pieterse CL; Robertson WD; Miller RJD Anal. Chem 2018, 90, 4422–4428. [DOI] [PubMed] [Google Scholar]
- (150).Mikhailov VA; Griffiths RL; Cooper HJ Int. J. Mass Spectrom 2017, 420, 43–50. [Google Scholar]
- (151).Li J; Zheng YJ; Mi W; Muyizere T; Zhang ZP Anal. Methods 2018, 10, 2803–2811. [Google Scholar]
- (152).Zhang CS; Glaros T; Manicke NE J. Am. Chem. Soc 2017, 139, 10996–10999. [DOI] [PubMed] [Google Scholar]
- (153).Wang DJ; Bodovitz S Trends Biotechnol 2010, 28, 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Nakashima T; Wada H; Morita S; Erra-Balsells R; Hiraoka K; Nonami H Anal. Chem 2016, 88, 3049–3057. [DOI] [PubMed] [Google Scholar]
- (155).Liu R; Pan N; Zhu Y; Yang Z Anal. Chem 2018, 90, 11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Bergman HM; Lanekoff I Analyst 2017, 142, 3639–3647. [DOI] [PubMed] [Google Scholar]
- (157).Lee JK; Jansson ET; Nam HG; Zare RN Anal. Chem 2016, 88, 5453–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).Chen FM; Lin LY; Zhang J; He ZY; Uchiyama K; Lin JM Anal. Chem 2016, 88, 4354–4360. [DOI] [PubMed] [Google Scholar]
- (159).Basu SS; Randall EC; Regan MS; Lopez BGC; Clark AR; Schmitt ND; Agar JN; Dillon DA; Agar NYR Anal. Chem 2018, 90, 4987–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (160).Dreisewerd K Anal. Bioanal. Chem 2014, 406, 2261–2278. [DOI] [PubMed] [Google Scholar]
- (161).Singhal N; Kumar M; Kanaujia PK; Virdi JS Front. Microbiol 2015, 6, 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (162).Bardin EE; Cameron SJS; Perdones-Montero A; Hardiman K; Bolt F; Alton E; Bush A; Davies JC; Takats Z Sci. Rep 2018, 8, 10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Wei P; Bag S; Pulliam CJ; Snyder DT; Pielak RM; Cooks RG Anal. Methods 2016, 8, 1770–1773. [Google Scholar]
- (164).Liang T; Schneider T; Yoon SH; Oyler BL; Leung LM; Fondrie WE; Yen G; Huang Y; Ernst RK; Nilsson E; Goodlett DR Int. J. Mass Spectrom 2018, 427, 65–72. [Google Scholar]
- (165).Heron SR; Wilson R; Shaffer SA; Goodlett DR; Cooper JM Anal. Chem 2010, 82, 3985–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Li H; Balan P; Vertes A Angew. Chem., Int. Ed 2016, 55, 15035–15039. [DOI] [PubMed] [Google Scholar]
- (167).Yan CY; Parmeggiani F; Jones EA; Claude E; Hussain SA; Turner NJ; Flitsch SL; Barran PE J. Am. Chem. Soc 2017, 139, 1408–1411. [DOI] [PubMed] [Google Scholar]
- (168).Nguyen SN; Liyu AV; Chu RK; Anderton CR; Laskin J Anal. Chem 2017, 89, 1131–1137. [DOI] [PubMed] [Google Scholar]
- (169).Li Y; Mathews RA J. Insect Physiol 2016, 91–92, 107–112. [DOI] [PubMed] [Google Scholar]
- (170).Barrios-Collado C; Garcia-Gomez D; Zenobi R; Vidal-de-Miguel G; Ibanez AJ; Sinues PML Anal. Chem 2016, 88, 2406–2412. [DOI] [PubMed] [Google Scholar]
- (171).Enomoto H; Sensu T; Sato K; Sato F; Paxton T; Yumoto E; Miyamoto K; Asahina M; Yokota T; Yamane H Sci. Rep 2017, 7, 42977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).Stopka SA; Agtuca BJ; Koppenaal DW; Pasa-Tolic L; Stacey G; Vertes A; Anderton CR Plant J. 2017, 91, 340–354. [DOI] [PubMed] [Google Scholar]
- (173).Pirro V; Guffey SC; Sepulveda MS; Mahapatra CT; Ferreira CR; Jarmusch AK; Cooks RG Mol. BioSyst 2016, 12, 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (174).Zaitsu K; Hayashi Y; Murata T; Yokota K; Ohara T; Kusano M; Tsuchihashi H; Ishikawa T; Ishii A; Ogata K; Tanihata H Anal. Chem 2018, 90, 4695–4701. [DOI] [PubMed] [Google Scholar]
- (175).Black O; Cody R; Edwards D; Cizdziel JV Forensic Chemistry 2017, 5, 26–32. [Google Scholar]
- (176).Fedick PW; Bain RM Forensic Chem. 2017, 5, 53–57. [Google Scholar]
- (177).McKenna J; Dhummakupt ES; Connell T; Demond PS; Miller DB; Nilles JM; Manicke NE; Glaros T Analyst 2017, 142, 1442–1451. [DOI] [PubMed] [Google Scholar]
- (178).Dhummakupt ES; Carmany DO; Mach PM; Tovar TM; Ploskonka AM; Demond PS; DeCoste JB; Glaros T ACS Appl Mater. Interfaces 2018, 10, 8359–8365. [DOI] [PubMed] [Google Scholar]
- (179).Forbes TP; Sisco E; Staymates M; Gillen G Anal. Methods 2017, 9, 4988–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (180).Nascimento Correa D; Melendez-Perez JJ; Zacca JJ; Borges R; Schmidt EM; Eberlin MN; Meurer EC Propellants, Explos., Pyrotech 2017, 42, 370–375. [Google Scholar]
- (181).Michely JA; Meyer MR; Maurer HH Anal. Chem 2017, 89, 11779–11786. [DOI] [PubMed] [Google Scholar]
- (182).Kennedy JH; Palaty J; Gill CG; Wiseman JM Rapid Commun. Mass Spectrom 2018, 32, 1280–1286. [DOI] [PubMed] [Google Scholar]
- (183).Jett R; Skaggs C; Manicke NE Anal. Methods 2017, 9, 5037–5043. [Google Scholar]
- (184).Proni G; Cohen P; Huggins LA; Nesnas N Forensic Sci. Int 2017, 280, 87–94. [DOI] [PubMed] [Google Scholar]
- (185).Newsome GA; Kayama I; Brogdon-Grantham SA Anal. Methods 2018, 10, 1038–1045. [Google Scholar]
- (186).Schedl A; Zweckmair T; Kikul F; Henniges U; Rosenau T; Potthast A Talanta 2017, 167, 672–680. [DOI] [PubMed] [Google Scholar]
- (187).Watts KE; Lagalante AF Rapid Commun. Mass Spectrom 2018, 32, 1324–1330. [DOI] [PubMed] [Google Scholar]
- (188).Da Costa C; Turner M; Reynolds JC; Whitmarsh S; Lynch T; Creaser CS Anal. Chem 2016, 88, 2453–2458. [DOI] [PubMed] [Google Scholar]
- (189).Silva LMA; Alves EG; Simpson AJ; Monteiro MR; Cabral EC; Ifa DR; Venancio T Talanta 2017, 173, 22–27. [DOI] [PubMed] [Google Scholar]
- (190).Ren LM; Han YH; Zhang YH; Zhang YF; Meng XH; Shi Q Energy Fuels 2016, 30, 4486–4493. [Google Scholar]
- (191).Tose L; Murgu M; Vaz B; Romo WJ Am. Soc. Mass Spectrom 2017, 28, 2401–2407. [DOI] [PubMed] [Google Scholar]
- (192).Jakka Ravindran S; Kumar R; Srimany A; Philip L; Pradeep T Anal. Chem 2018, 90, 988–997. [DOI] [PubMed] [Google Scholar]
- (193).Naraoka H; Hashiguchi M Rapid Commun. Mass Spectrom 2018, 32, 959–964. [DOI] [PubMed] [Google Scholar]
- (194).Pu F; Zhang WP; Han C; Ouyang Z Anal. Methods 2017, 9, 5058–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (195).Yong W; Guo TY; Fang PP; Liu JH; Dong YY; Zhang F Int. J. Mass Spectrom 2017, 417, 53–57. [Google Scholar]
- (196).Mirabelli MF; Gionfriddo E; Pawliszyn J; Zenobi R Analyst 2018, 143, 891–899. [DOI] [PubMed] [Google Scholar]
- (197).Gomez-Rios GA; Gionfriddo E; Poole J; Pawliszyn J Anal. Chem 2017, 89, 7240–7248. [DOI] [PubMed] [Google Scholar]
- (198).Chen S; Chang QY; Yin K; He QY; Deng YX; Chen A; Liu CB; Wang Y; Wang LP J. Agric. Food Chem 2017, 65, 4859–4865. [DOI] [PubMed] [Google Scholar]
- (199).Ma Q; Bai H; Li WT; Wang C; Li XS; Cooks RG; Ouyang Z Anal. Chim. Acta 2016, 912, 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Balog J; Perenyi D; Guallar-Hoyas C; Egri A; Pringle SD; Stead S; Chevallier OP; Elliott CT; Takats ZJ Agric. Food Chem 2016, 64, 4793–4800. [DOI] [PubMed] [Google Scholar]
- (201).Martinez-Jarquin S; Moreno-Pedraza A; Cazarez-Garcia D; Winkler R Anal. Methods 2017, 9, 5023–5028. [Google Scholar]
- (202).Gomez-Rios GA; Vasiljevic T; Gionfriddo E; Yu M; Pawliszyn J Analyst 2017, 142, 2928–2935. [DOI] [PubMed] [Google Scholar]
- (203).Wang XC; Yang ML; Wang ZY; Zhang H; Wang GF; Deng M; Chen HW; Luo LP J. Agric. Food Chem 2017, 65, 2488–2494. [DOI] [PubMed] [Google Scholar]
- (204).Bain RM; Pulliam CJ; Cooks RG Chem. Sci 2015, 6, 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (205).Lee JK;Banerjee S; Nam HG; Zare RNQ. Rev. Biophys 2015, 48, 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Cheng S; Wu QH; Xiao H; Chen H Anal. Chem 2017, 89, 2338–2344. [DOI] [PubMed] [Google Scholar]
- (207).Crawford EA; Esen C; Volmer DA Anal. Chem 2016, 88, 8396–8403. [DOI] [PubMed] [Google Scholar]
- (208).Fedick PW; Bain RM; Bain K; Mehari TF; Cooks RF Int. J. Mass Spectrom 2018, 430, 98–103. [Google Scholar]
- (209).Zhang XX; Barraza KM; Upton KT; Beauchamp JL Chem. Phys. Lett 2017, 683, 76–82. [Google Scholar]
- (210).Huang YL; Barraza KM; Kenseth CM; Zhao R; Wang B; Beauchamp JL; Seinfeld JH J. Phys. Chem. A 2018, 122, 6445–6456. [DOI] [PubMed] [Google Scholar]
- (211).van Geenen FAMG; Franssen MCR; Zuilhof H; Nielen MWF Anal. Chem 2018, 90, 10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Wleklinski M; Loren BP; Ferreira CR; Jaman Z; Avramova L; Sobreira TJP; Thompson DH; Cooks RG Chem. Sci 2018, 9, 1647–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (213).Chowdhury SK; Chait BT Anal. Chem 1991, 63, 1660–1664. [DOI] [PubMed] [Google Scholar]
- (214).Tang L; Kebarle P Anal. Chem 1991, 63, 2709–2715. [Google Scholar]
- (215).Lu M; Wolff C; Cui W; Chen H Anal. Bioanal. Chem 2012, 403, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (216).Looi WD; Brown B; Chamand L; Brajter-Toth A Anal. Bioanal. Chem 2016, 408, 2227–2238. [DOI] [PubMed] [Google Scholar]
- (217).Cai Y; Liu PY; Held MA; Dewald HD; Chen H ChemPhysChem 2016, 17, 1104–1108. [DOI] [PubMed] [Google Scholar]
- (218).Brown TA; Hosseini-Nassab N; Chen H; Zare RN Chem. Sci 2016, 7, 329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (219).Banerjee S; Basheer C; Zare RN Angew. Chem., Int. Ed 2016, 55, 12807–12811. [DOI] [PubMed] [Google Scholar]
- (220).Resende SF; Teodoro JAR; Binatti I; Gouveia RL; Oliveira BS; Augusti R Int. J. Mass Spectrom 2017, 418, 107–111. [Google Scholar]
- (221).Sarkar D; Som A; Pradeep T Anal. Chem 2017, 89, 11378–11382. [DOI] [PubMed] [Google Scholar]
- (222).Evans-Nguyen KM; Gerling J; Brown H; Miranda M; Windom A; Speer J Analyst 2016, 141, 3811–3820. [DOI] [PubMed] [Google Scholar]
- (223).Hoegg ED; Barinaga CJ; Hager GJ; Hart GL; Koppenaal DW; Marcus RK J. Anal. At. Spectrom 2016, 31, 2355–2362. [Google Scholar]
- (224).Hoegg ED; Marcus RK; Koppenaal DW; Irvahn J; Hager GJ; Hart GL Rapid Commun. Mass Spectrom 2017, 31, 1534–1540. [DOI] [PubMed] [Google Scholar]
- (225).Liu YM; Nicolau BG; Esbenshade JL; Gewirth AA Anal. Chem 2016, 88, 7171–7177. [DOI] [PubMed] [Google Scholar]
- (226).Becker JS; Dietze HJ Spectrochim. Acta, Part B 1998, 53, 1475–1506. [Google Scholar]
- (227).Shiea C; Huang YL; Cheng SC; Chen YL; Shiea J Anal. Chim. Acta 2017, 968, 50–57. [DOI] [PubMed] [Google Scholar]
- (228).Luo Z; He J; Chen Y; He J; Gong T; Tang F; Wang X; Zhang R; Huang L; Zhang L; Lv H; Ma S; Fu Z; Chen X; Yu S; Abliz Z Anal. Chem 2013, 85, 2977–2982. [DOI] [PubMed] [Google Scholar]
- (229).Ouyang Y; Liu J; Nie B; Dong N; Chen X; Chen L; Wei Y RSC Adv. 2017, 7, 56044–56053. [Google Scholar]
- (230).Peace MR; Stone JW; Poklis JL; Turner JBM; Poklis AJ Anal. Toxicol 2016, 40, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (231).Furter JS; Hauser PC Anal. Methods 2018, 10, 2701–2711. [Google Scholar]
- (232).Jiang J; Chen SH; Li M; Li HM; Chen Y Anal. Lett 2017, 50, 797–805. [Google Scholar]
- (233).Grimm RL; Beauchamp JL J. Phys. Chem. B 2003, 107, 14161–14163. [Google Scholar]
- (234).Zhang H; Zhu L; Luo L; Wang N; Chingin K; Guo X; Chen HJ Agric. Food Chem 2013, 61, 10691–10698. [DOI] [PubMed] [Google Scholar]
- (235).Bartels B; Kulkarni P; Danz N; Bocker S; Saluz HP; Svatos A RSC Adv. 2017, 7, 9045–9050. [Google Scholar]
- (236).Marcus RK; Burdette CQ; Manard BT; Zhang LX Anal. Bioanal. Chem 2013, 405, 8171–8184. [DOI] [PubMed] [Google Scholar]
- (237).Zhan X; Zhao Z; Yuan X; Wang Q; Li D; Xie H; Li X; Zhou M; Duan Y Anal. Chem 2013, 85, 4512–4519. [DOI] [PubMed] [Google Scholar]
- (238).Kim P; Cha S Analyst 2015, 140, 5868–5872. [DOI] [PubMed] [Google Scholar]
- (239).Gholipour Y; Erra-Balsells R; Hiraoka K; Nonami H Anal. Biochem 2013, 433, 70–78. [DOI] [PubMed] [Google Scholar]