Summary
Identifying the earliest somatic changes in prostate cancer can give important insights into tumor evolution and aids in stratifying high- from low-risk disease. We integrated whole-genome, transcriptome and methylome analysis of early-onset prostate cancers (diagnosis ≤ 55 years). Characterization across 292 prostate cancer genomes revealed age-related genomic alterations and a clock-like enzymatic-driven mutational process contributing to the earliest mutations in prostate cancer patients. Our integrative analysis identified four molecular subgroups, including a particularly aggressive subgroup with recurrent duplications associated with increased expression of ESRP1, which we validate in 12,000 tissue microarray tumors. Finally, we combined the patterns of molecular co-occurrence and risk-based subgroup information to deconvolve the molecular and clinical trajectories of prostate cancer from single patient samples.
Introduction
One of the biggest unmet clinical needs in prostate cancer (PC) is to stratify clinically indolent from aggressive types, particularly in patients diagnosed at young age. Molecular markers have shown promise in risk stratification, but the utility is complicated by the heterogeneous natural-history. Primary localized PC develops over decades (Pound et al., 1999), with a typical late age-of-onset (median 66 years of age, seer.cancer.gov). Prior studies have revealed a remarkable inter- and intra-tumor heterogeneity in PC (Boutros et al., 2015; TCGA, 2015) associated with poor outcome in primary localized PC (Espiritu et al., 2018). Prior focus on elderly, late onset patients has hindered the identification of the earliest genomic alterations, which could aid in identifying the evolutionary paths and clinical outcome of PC. One of the earliest molecular alterations in PC are ETS fusions involving the fusion of androgen-receptor (AR) responsive promoters and members of the ETS transcription factor (TF) family genes, most notably the TMPRSS2-ERG fusion (Tomlins et al., 2005) present in 50 % of all PC and exhibiting an elevated occurrence in early-onset PC (EOPC) (Tomlins et al., 2005; Weischenfeldt et al., 2013). PC has relatively few somatic point mutations but has frequent genomic structural variants (SVs), several of which are associated with clinical outcome, including disruption or loss of PTEN, TP53, NKX3-1 and MAP3K7 (Kluth et al., 2013; Taylor et al., 2010; TCGA, 2015).
Identifying the molecular evolution and clinical trajectories of PC requires analysis of the earliest somatic mutation events. A particular relevant subset of PC are early detected cancers associated with EOPC (Pritchard et al., 2016; Weischenfeldt and Korbel, 2017; Weischenfeldt et al., 2013), here defined as patients with an age-at-diagnosis at 55 and below, which are likely to develop a severe disease course and eventually require radical treatment. Studies in EOPC, furthermore, offer insights into early mutational processes and evolutionary trajectories of PC.
Results
Patterns of somatic genomic aberrations in EOPC
We applied uniform and comprehensive genomics-based profiling of 292 PC cases (including 203 EOPCs) (Table S1, Figure 1 and S1A). Profiling included whole-genome sequencing (WGS) of tumors and matched peripheral blood from 184 EOPC patients and 85 late-onset (LOPC) patients, methylomes (450k methylome arrays) in 203 EOPC tumors and 45 LOPC tumors and mRNA-seq of 96 EOPC samples. Established somatic and germline variant calling pipelines were used to identify single-nucleotide variants (SNVs), short insertions and deletions (InDels) and SVs. Genome-wide analysis of somatic SNVs revealed an expected lower average number of SNVs per Mb in EOPC (median 0.47, interquartile range (IQR) = 0.49) as compared to LOPC (median 0.53) (Fraser et al., 2017). TP53 was the most frequently affected gene by nonsynonymous SNVs (nsSNV) in the EOPC cohort (6%).
Figure 1. Somatic alteration landscape and age-at-diagnosis.
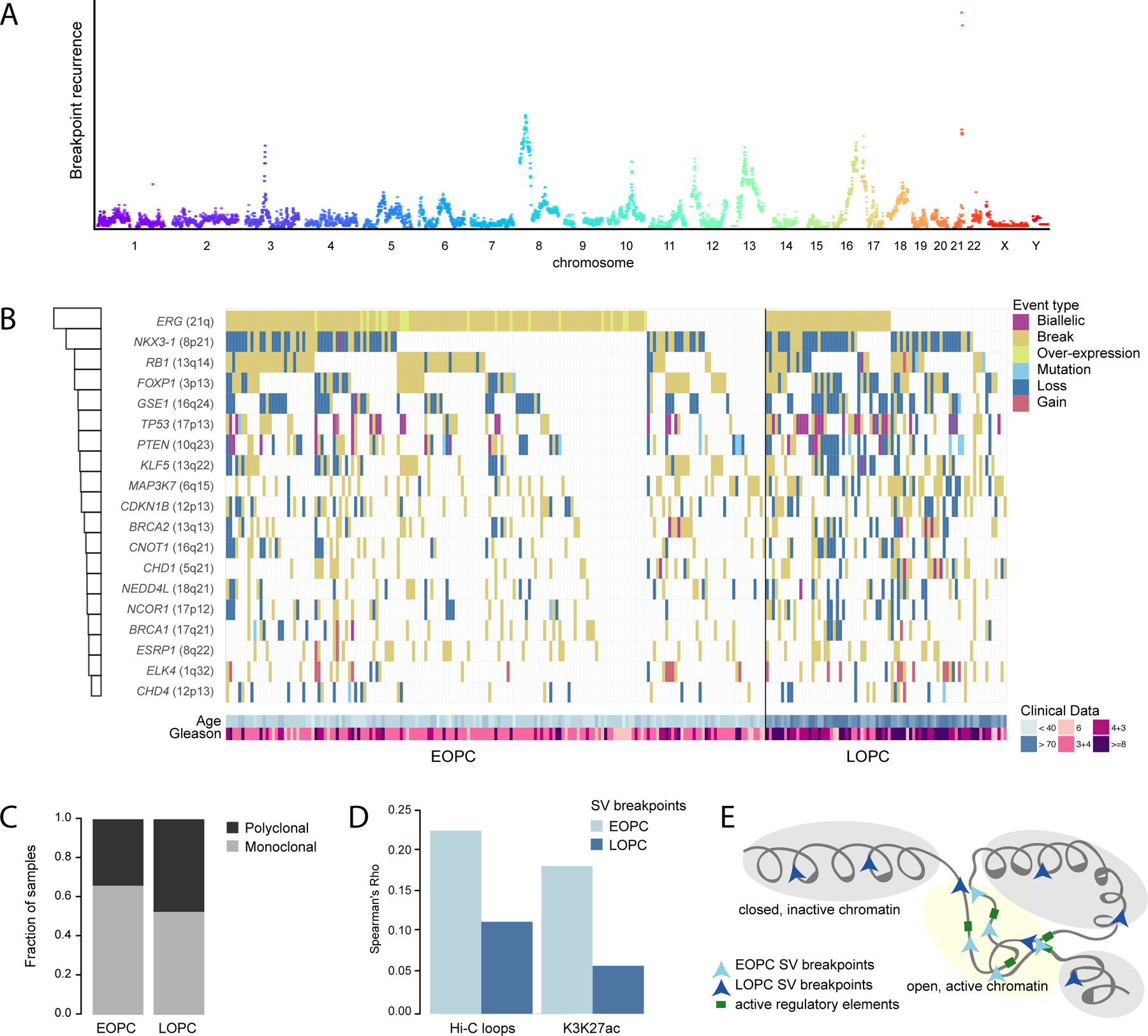
(A) Genome-wide SV breakpoint recurrence pattern across 292 PC samples, color-coded separately for each chromosome.
(B) An Oncoprint summarizing the mutational landscape of RGA regions in PC, color-coded by the mutational event-type and separate into EOPC and LOPC. The barplot at the left quantify the recurrence of each RGA in the PC cohort. The patient age and GS are shown at the bottom.
(C) Fraction of EOPC and LOPC tumors from localized PC associated with either clonal or polyclonal paths (p = 0.18, Chi-square test).
(D) Correlation between breakpoints and Hi-C chromatin loops (combined across eight cell lines) and PC-specific H3K27ac peaks in 1 Mbp bins, separated into localized EOPC and LOPC.
(E) “Chromatin-state”-model of age-associated breakpoint patterns in PC. See also Figure S1 and Table S1.
SVs often involve recurrent fusion gene formation or loss of tumor-suppressor genes in PC (Fraser et al., 2017; Taylor et al., 2010; TCGA, 2015). We confirmed previous findings, namely an increased number of SNVs and SVs with age (p < 0.001) (Figures S1B, S1C). We identified recurrent genomic altered loci (RGA), as breakpoint peak regions at minimum 5% recurrence (Figure 1A, 1B). Our analysis revealed 70% of the EOPC tumor genomes carrying an SV associated with formation of an ETS fusion gene (Figure S1D). The second- and third-most frequently altered loci in EOPC were at chromosome 8p (centered at NKX3-1, 37%) and 3p14 (centered at FOXP1, 30%). We identified PTEN as the gene with the highest rate of biallelic inactivation (12 samples) across the cohort, followed by TP53 (8 samples). Despite being more often affected by SVs, neither NKX3-1 nor FOXP1 underwent recurrent biallelic inactivation, corroborating earlier suggestions of haploinsufficient tumor-suppressive roles of these genes (Locke et al., 2012; Myers et al., 2017).
To identify RGAs associated with age-of-onset, we performed a parallel analysis of LOPC genomes, which revealed similar affected loci but with a more uniform pattern, distinct from that of EOPC (Figure S1D, S1E). LOPC displayed an overall higher proportion of RGAs affected by genomic losses compared to a higher rate of balanced breaks in EOPC (p < 1x10−7 and p < 1x10−4, Fisher’s exact test). Moreover, EOPC exhibited a more monoclonal architecture compared to LOPC (66% and 53%, respectively, Figure 1C and S1F), suggesting that EOPC tend to be primarily associated with a clonal origin, potentially due to the shorter life-span compared to LOPC.
The epigenetic landscape is often altered during cancer progression and impacts on where DNA double-strand breaks occur (Aryee et al., 2013; Urbanucci et al., 2017). We previously showed that breakpoints in EOPC genomes occur more often in the vicinity of AR-binding sites (Weischenfeldt et al., 2013). This raises the possibility that age-associated altered chromatin states impact on breakpoint occurrence. We therefore examined genomic breakpoints from EOPC tumors in relation to specific chromatin regions (Taberlay et al., 2014). This revealed a significant enrichment of breakpoints in EOPC near open chromatin, active enhancers, TF binding and actively transcribed regions (Figure S1G, S1H). Active enhancers are associated with long-range promoter-enhancer DNA-DNA chromatin loops, which can increase the likelihood of SV formation between normally distant loci (Chen et al., 2018). We integrated publicly available Hi-C data, which revealed significant correlation between breakpoints and both the number of chromatin loops and H3K27ac peaks (p < 0.0001 both, Spearman rho = 0.23 and 0.18, respectively) in EOPC, but to a lesser extent in LOPC (p < 0.0001 both, Spearman rho = 0.11 and 0.06 for Hi-C and H3K27ac, respectively, Figure 1D), suggesting that the chromatin state and long-range interactions partake in shaping the SV landscape in EOPC (Figure 1E).
DNA rearrangement recurrence analysis identifies a putative oncogene associated with high cell proliferation and poor outcome
We identified two RGAs in EOPC located at 13q22 (27%) and 8q22 (17%) (Figures 2A, 2B). The minimal overlap peak region at 13q22 centered on KLF5, encoding a transcriptional activator involved in repressing cell proliferation (Xing et al., 2014). Loss of 13q22 was associated with decreased KLF5 mRNA level as well as a global increase in SV and SNV burden (Figure 2C and S2A). We additionally identified a subset of tumors that displayed a marked reduction in KLF5 expression and a differentially methylated CpG site (q value = 0.002, t-test) proximal to the KLF5 promoter in a CpG island shore that was inversely correlated with KLF5 mRNA level (spearman rho = −0.523, q-value = 0.0038, CpG #18 in Figure 2D, S2B). A recent study in mouse embryonic stem cells identified a set of KLF5 targets, including the ubiquitin ligase gene Spop, that was significantly downregulated in response to KLF5 knock-down (Parisi et al., 2010). ChIP-seq data showed binding of KLF5 at the SPOP promoter (Yan et al., 2013) and we identified a positive correlation between KLF5 and SPOP mRNA levels in our PC cohort (Figure 2E, S2C) as well as in the the Cancer Genome Atlas (TCGA) cohort (p < 1x10−4, spearman rho = 0.19), but no association with the SPOP mutation status (Fisher’s exact test).
Figure 2. Recurrent alterations target KLF5 and ESRP1.
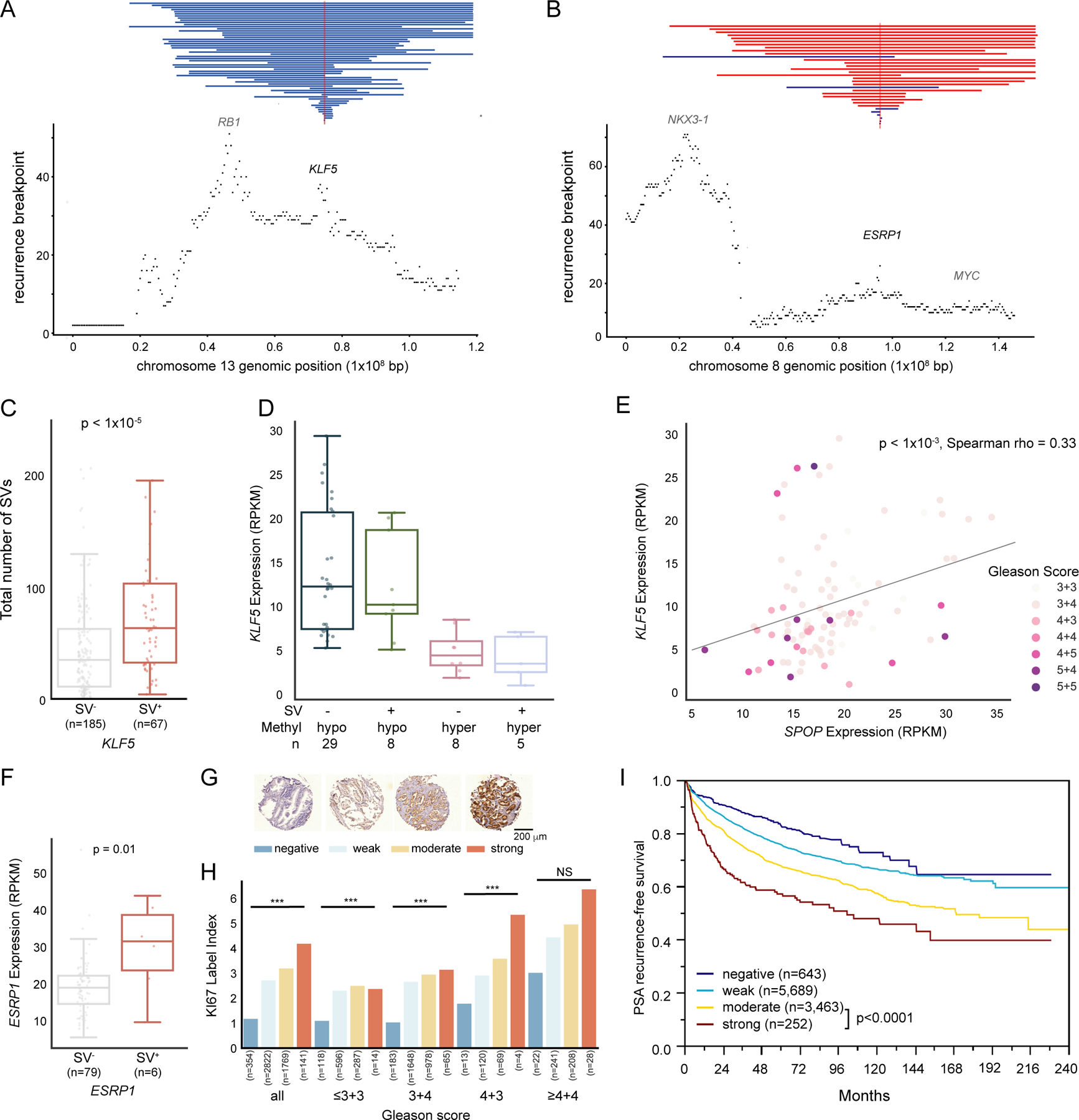
(A, B) SV recurrence plot at 13q22 (A) and 8q22 (B) with vertical red and blue lines represents genomic gain and loss, respectively (n tumor samples = 292). The smallest overlapping SV is shown in case of multiple SVs per tumor sample.
(C) Number of somatic SVs from our total cohort of tumors according to presence (SV+) or absence (SV−) of SVs affecting the KLF5 locus. MWU-based p value.
(D) KLF5 gene expression of different methylation and somatic SV states with x-axis representing KLF5 promoter-proximal methylation status and somatic SV states.
(E) Correlation between KLF5 and SPOP expression, with each dot representing a tumor, color-labeled with GS.
(F) Boxplot of ESRP1 mRNA expression separated by tumors with an SV gain of ESRP1 (SV+) and without (SV−). MWU-based p value.
(G) ESRP1 protein expression stained in 11,954 TMA samples and scored as “negative” (dark blue), “weak” (light blue), “moderate” (yellow) or “strong” (red).
(H) Barplot showing Ki67 labelling index separated by GS and ESRP1 staining. Number of tumors for each category is labelled below each bar. ***: p < 0.001, NS = not significant (a = 0.05). The colors of bars correspond to those in (G).
(I) Kaplan-Meier plot, showing PSA-recurrence-free survival for patients stratified by ESRP1 staining intensity.
Boxplots show median (line), upper and lower quartiles (boxes), and lines extending to 1.5 x IQR (whiskers).
A region at 8q22 displayed recurrent genomic duplications centered on ESRP1 (Figure 2B), with the minimal overlapping region residing 33 Mbp away from MYC. ESRP1 encodes an RNA-binding protein involved in epithelial-to-mesenchymal transition (EMT) and RNA splicing (Jeong et al., 2017). Tumors harboring duplications intersecting ESRP1 displayed significantly increased ESRP1 mRNA expression (> 1.5 fold, Figure 2F). While several duplications overlapped both ESRP1 and MYC, only ESRP1 displayed a significant increase in mRNA level across the affected samples (Figure 2F, S2D). ESRP1 duplications were significantly associated with elevated Gleason Score (GS) (p < 1x10−11, Chi-squared test), in fact, more than any other RGA in the cohort. We therefore pursued immunohistochemistry (IHC)-based validation in 11,954 tumor specimens on tissue-microarrays (TMA) (Figure 2G), which confirmed a significant correlation between increased GS and pT and ESRP1 staining (Figure S2E). High ESRP1 protein level particularly showed association with high GS (>4+4), tumor stage (pT3b-pT4), number of lymph node metastases and preoperative prostate-specific antigen (PSA) levels. Increased ESRP1 protein levels correlated with higher proliferation rate irrespective of GS, as measured by Ki67 index labelling (Figure 2H). Additionally, ESRP1 protein intensity was associated with adverse outcome, with strong ESRP1 staining correlating with significantly shorter time to biochemical recurrence (BCR) (Figure 2I, Figure S2F).
A multivariate analysis revealed ESRP1 to be an independent prognostic marker in four established clinico-pathological parameters and that high ESRP1 expression was associated with shorter BCR irrespective of ERG status (Table S2, Figure S2G, S2H). ESRP1 was particularly discriminative in the biopsy setting, where GS is often underestimated and additional prognostic markers are needed. In summary, we identified recurrent genomic duplications of ESRP1 associated with increased ESRP1 protein expression, higher levels of cell proliferation and elevated GS and tumor stage, and demonstrated that ESRP1 expression is an independent prognostic biomarker in PC.
Enzymatic activity is associated with the earliest detectable mutational processes in prostate genomes
Mutational signatures can be employed to describe intrinsic and exogenous-mediated mutational processes acting on tumor cells (Alexandrov et al., 2013, 2015; Nik-Zainal et al., 2016) (Figure 3A). We observed six mutational signatures: two clock-like signatures (1 and 5), two related to DNA repair defects (3 and 6) and two related to APOBEC cytidine deaminase-attributable mutagenesis (2 and 13). Mutational processes were associated to GS, in particular the APOBEC signatures (2 and 13) and the homologous recombination repair-associated signature 3 (Figure 3B). The clock-like mutational signatures 1 and 5 were the predominant signatures across all tumors and both showed significant association with patient age (Figure 3C).
Figure 3. Age-related mutational signatures in prostate cancer.
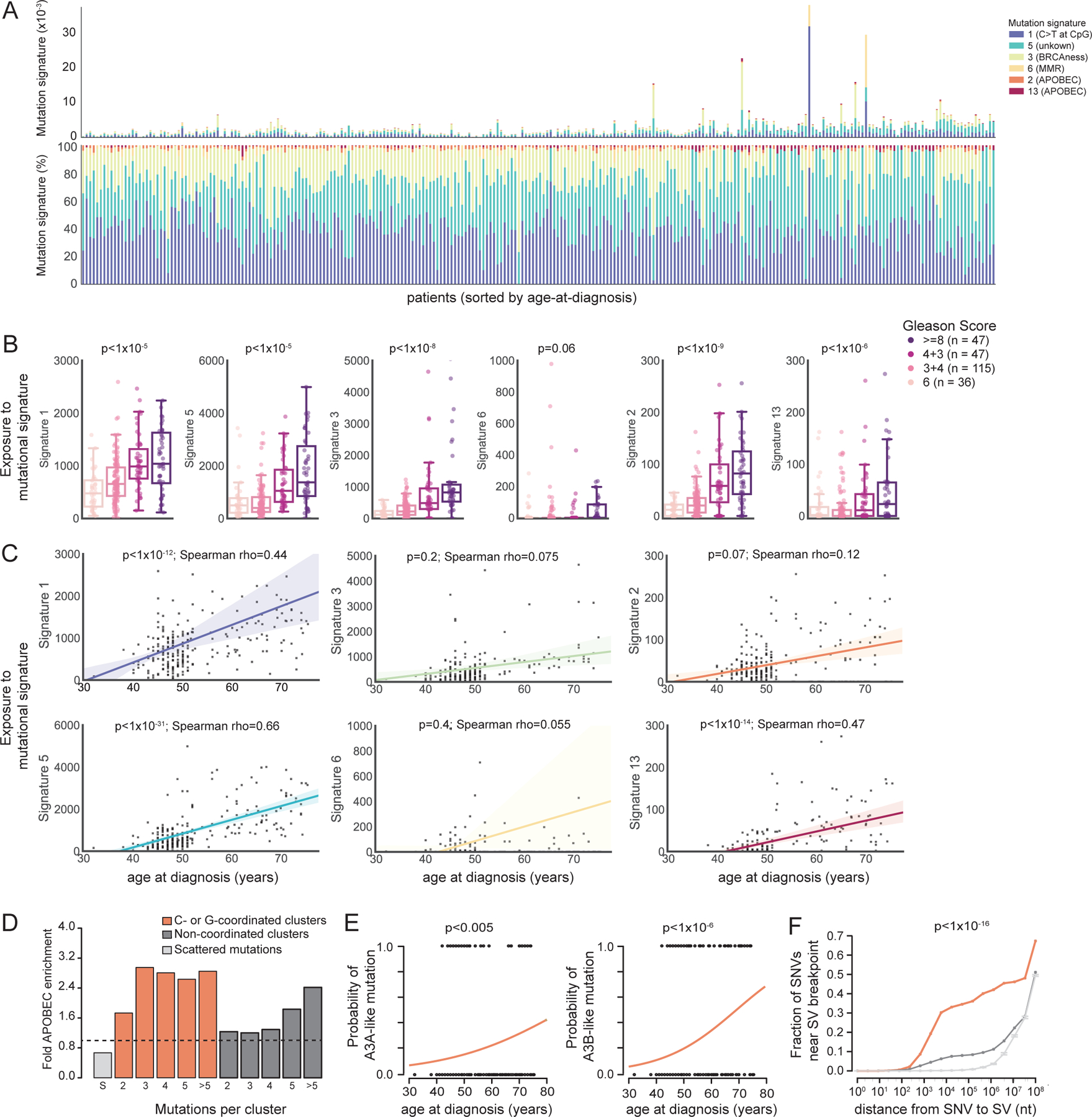
(A) Barplot of the absolute (top) and relative (bottom) proportion of exposure of six mutational signatures (1, 5, 3, 6, 2 and 13, colored bars) per individual tumor of patients with localized PC, sorted by age-at-diagnosis (x-axis, range from 32 to 75 years).
(B) Association between mutation signature burden (y-axis) and GS. POLR p values. Boxplots show median (line), upper and lower quartiles (boxes), and lines extending to 1.5 x IQR (whiskers).
(C) Correlation between the mutation signature burden and age-at-diagnosis.
(D) Fold-enrichment of APOBEC signature in scattered mutations (light grey), C/G clusters (orange) or non-coordinated clusters (dark grey).
(E) Age-association between A3A (“ytCa” signature, left) or A3B (“rtCa” signature, right) in C/G clusters of mutations as a function of age (binomial logistic regression). Generalized linear model (GLM) logit p values.
(F) Fraction of mutations close to SV breakpoint for C/G cluster mutations (orange, n = 1,694), non-coordinated cluster mutations (dark grey, n = 8,408) and non-clustered mutations (light-grey, 100 bootstraps of 456,406 SNVs, 95% confidence interval shown). X-axis displays log10 distance between SNV and breakpoint.
See also Figure S3.
Curiously, we also observed clear signs of a clock-like accumulation of APOBEC-associated signature 2 and 13 mutations in PC (Figure 3C) and could further corroborate this finding using a knowledge-based approach that estimates APOBEC mutagenesis in cancer genomes (p = 5.2x10−3, Spearman’s rho = 0.17). APOBEC proteins are cytidine deaminases that can act to restrict retroelements during the single strand DNA (ssDNA) replication cycle, but can also induce mutations in cancer genomes (Roberts et al., 2012, 2013). These lesions were previously suggested to be driven by APOBEC3A (A3A) and/or APOBEC3B (A3B) (Swanton et al., 2015). APOBEC associated mutations occasionally arise as clusters of C- (or G) strand-coordinated mutational events (C/G clusters) – also termed kataegis events – a mutational phenomenon resulting in localized hypermutation (Nik-Zainal et al., 2012; Roberts et al., 2012). Indeed, we observed a strong enrichment of APOBEC mutagenesis at C/G clusters in PC (Figure 3D).
We also identified a significant association between patient age and C/G clusters attributable to APOBEC enzymes (Figure S3A), which was primarily attributable to A3B-like mutagenesis at C/G clusters (Figure 3E). To further substantiate the relevance of A3B-like mutagenesis in PC, we genotyped a known ~30 kb germline APOBEC3B deletion, which results in complete removal of its protein-coding sequence (Middlebrooks et al., 2016). We observed in germline APOBEC3B deletion carriers i) significantly fewer APOBEC-associated signature 2 and 13 mutation, ii) reduced expression levels of A3B in PC and iii) a significant shift from A3B-like to A3A-like mutagenesis (Figure S3B). These findings thus suggest that A3B-like mutagenesis is active at a basal level in prostate cells, and that this endogenous mutagenic process is responsible for the clock-like accumulation of somatic mutations – including the occurrence of localized hypermutation events – in PC. APOBEC-associated mutations have previously been observed to frequently co-localize with SV breakpoints in cancer (Chan and Gordenin, 2015; Roberts et al., 2012). We found a strong enrichment of C/G clusters to co-localize with SV breakpoints compared to both non-coordinated mutation clusters and scattered mutations (Figure 3F), with an increase in co-localization frequency between 1 kb and 10 kb. Several of these APOBEC-associated SV breakpoints resulted in alteration of driver genes in PC, including formation of TMPRSS2-ERG fusion and PTEN, FOXP1 and BRCA2 disruption (Table S2). Our findings demonstrate an age-associated mutational process that involves an endogenous mutagenic enzyme, and suggest that mutations attributable to APOBEC enzymes are likely to contribute to the earliest mutations seen in PC patients.
Germline mutations also are likely to contribute to early lesions in PC patients, for example by modulating somatic mutational processes. Germline protein-truncating variants (PTVs) in DNA damage response (DDR) genes including BRCA1, BRCA2, PALB2, ATM, and CHEK2 have previously been associated with poor outcome and increased frequency of PC metastasis (Na et al., 2017; Pritchard et al., 2016). We detected significant associations between germline PTVs in these DDR genes and somatic SVs and SNVs as well as APOBEC-like signature 2 and the ‘BRCAness’ mutational signature 3 (Figure 4). In summary, we identify three age-associated mutational processes in PC, namely, CpG mutagenesis, signature 5 with unknown etiology and A3B-associated mutagenesis. Tumor genomes harboring pathogenic germline mutations in genes involved in homologous recombination repair exhibited increased genomic instability.
Figure 4. Predisposing germline mutations associate with specific somatic alteration landscapes.
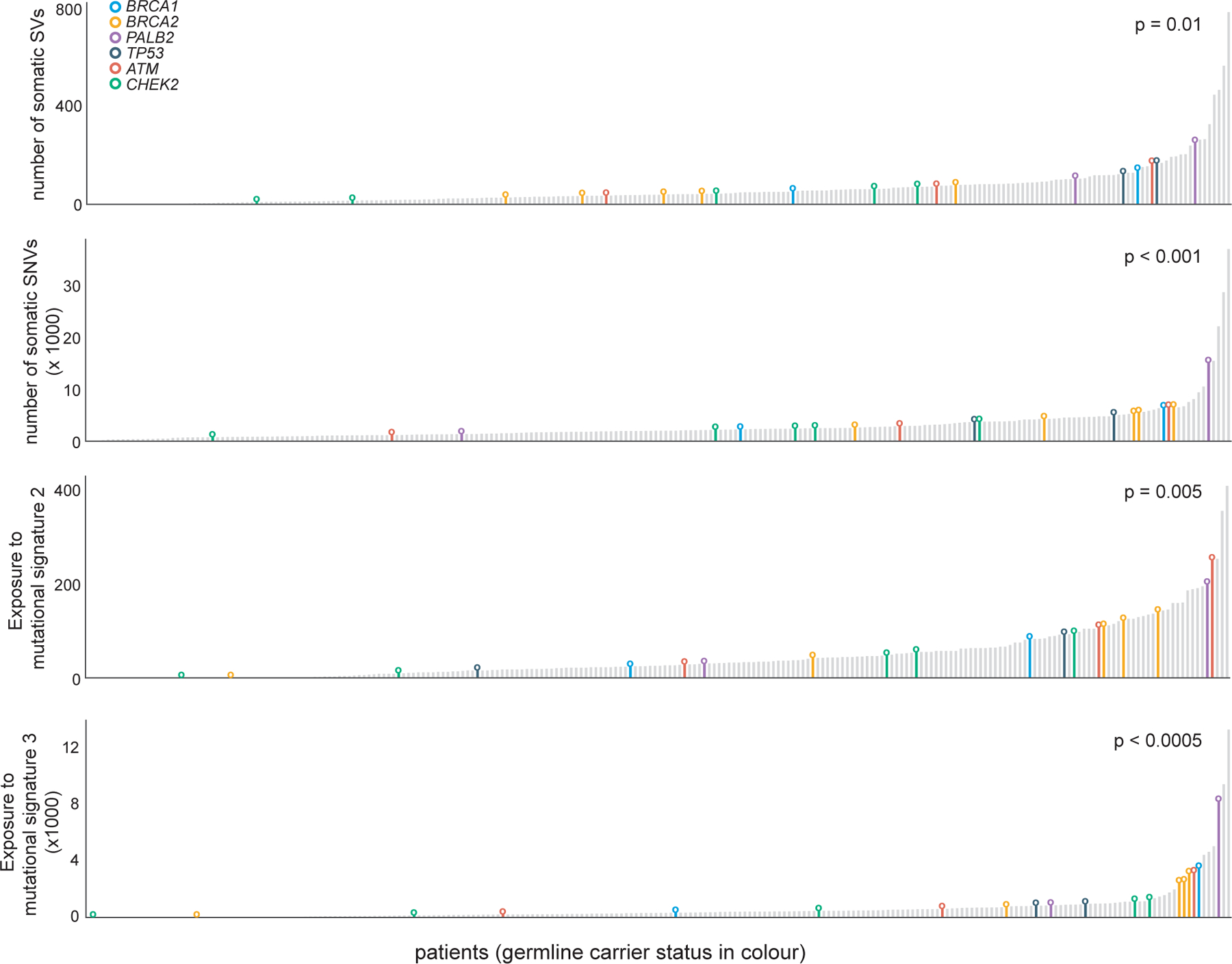
Association between individuals carrying germline PTV in the indicated cancer predisposition gene and total number of somatic SVs, total number of somatic SNVs, exposure to mutational signature 2 and mutational signature 3. X-axis represent patients, sorted in ascending order of the phenotype.
PEPCI, a methylation-based risk score
Normal human prostate tissue is composed of basal, luminal and stromal cells, whereas PC loses basal cells and gains tumor-specific luminal (T-luminal) cells as well as infiltrating immune cells (Bhasin et al., 2015). Given that DNA methylation profiles are cell type (ct) specific, we sought to account for differences in ct composition in methylation analyses by using available reference methylomes (Teschendorff and Zheng, 2017). To this end, we acquired additional resected samples from benign prostate hyperplasia (BPH) cases and PC and performed FACS-sorting to identify the main cts present in PC (STAR Methods) (Figure 5A and S4A–C), which enabled us to identify the ct-identity of every methylation site in the PC genome.
Figure 5. PEPCI, a methylation-based risk group score.
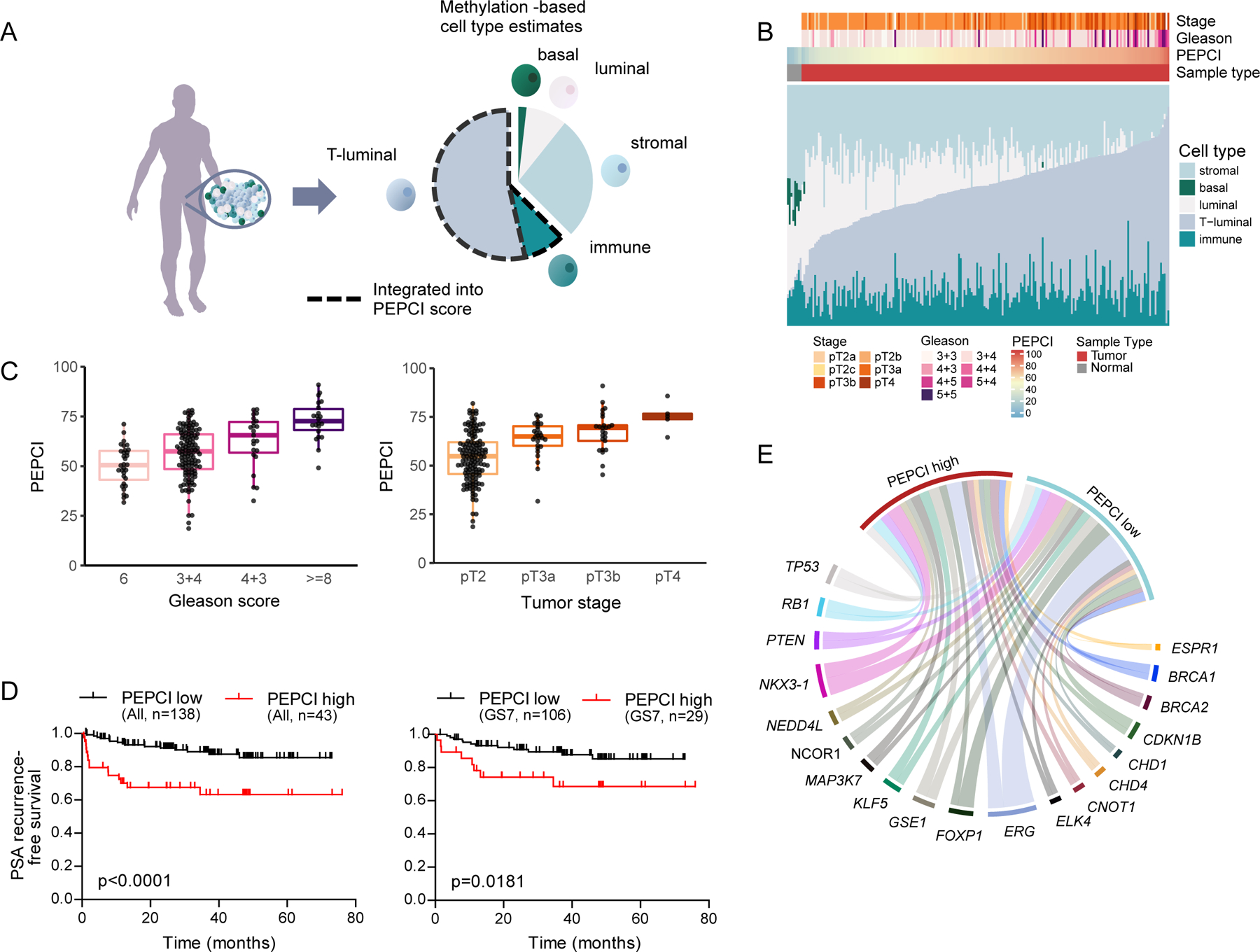
(A) A schematic representation of methylation-based estimation of ct composition of each bulk tumor sample.
(B) Stacked barplots of ct composition, tumor stage, GS and PEPCI per PC.
(C) Association between PEPCI and GS (left) and pT (right). Boxplots show median (line), upper and lower quartiles (boxes), and lines extending to 1.5 x IQR (whiskers).
(D) Kaplan-Meier curves of localized EOPC patients stratified according to PEPCI-high and PEPCI-low, for all cases (left) and for GS7 only (right).
(E) Chord-diagram showing proportions of tumors with a specific RGA and the associated PEPCI-high and -low risk group, colored by each RGA.
We found a recurrent shift from basal and luminal cells to T-luminal cells and infiltrating immune cells in high GS tumors (Figure 5B, S4C, S4D). Given this relevance of T-luminal and immune cell content in identifying high-grade tumors, we combined this information as a Purity-adjusted Epigenetic Prostate Cancer Index (PEPCI) of tumor aggressiveness (Figures 5A, 5B). We found that high PEPCI was strongly associated with high pT (p < 1x10−7, Kruskal Wallis), high GS (p < 1x10−17, Wilcoxon) (Figure 5C) and elevated risk of BCR (log-rank p < 0.0001). Moreover, PEPCI was able to stratify intermediate-risk (GS7, especially GS4+3) cases (Figure 5D, Figures S4E, S4F, Table S3), which we validated in the TCGA cohort of primarily LOPC samples (TCGA, 2015) (Figure S4E, S4F; Table S3). Finally, our PEPCI score was also able to independently predict GS and BCR (Area under the curve (AUC) = 0.831 and 0.702, respectively) (Figure S4G). We examined whether particular RGAs were associated with PEPCI-based risk groups (Figure 5E), which revealed a striking association between PEPCI-high and gain of ESRP1 (odds-ratio = 15.7, FDR-corrected p < 1x10−5, Fisher’s exact test, Table S3).
Integrative analysis identifies molecular subgroups associated with disease progression
We sought to identify pathways and processes that underwent transcriptional deregulation in EOPC. Using the graph theory-based CLICK algorithm (Sharan et al., 2003) on 96 patients with available mRNA-seq data, we identified seven distinct CLICK clusters (abbreviated CC1-7) of co-expressed genes, splitting the patients into CC-high and CC-low expression groups per CC (Figure 6A, S5A; Table S4). We next integrated CC expression profiles, ct-content and PEPCI information to further refine the PEPCI-based risk stratification (Figure 6A–C, see STAR Methods). This led us to identify a prominent PC subgroup 1 of mainly PEPCI-high tumors (19 samples) with high content of T-luminal cells and expression of CC7 (Figure 6B, 6C). CC7 is associated with reactive stroma, which is indicated by an enriched myofibroblast signature and the reactive stroma marker ASPN in CC7 (Barron and Rowley, 2012; Rochette et al., 2017). Subgroup 1 was also associated with prominent loss of CC2 and CC4 gene expression representing normal basal and luminal prostate epithelium (Strand and Goldstein, 2015). CC2 loss and CC7 gain were strongly and independently linked to GS (Figure S5B) and BCR (Figure 6D). Multivariable statistics showed that CC2 adds significant information on top of GS in predicting BCR in both our cohort and in the TCGA cohort (p = 0.003 and p = 0.01, respectively; Table S4). Additionally, CC2-low and CC7-high tumors were associated with specific RGAs, in particular PTEN loss (p < 0.0001, both) and ESRP1 gain (p < 0.0001 and p < 0.0005, respectively, Pearson’s Chi-squared test).
Figure 6. Integrative expression and methylation analysis.
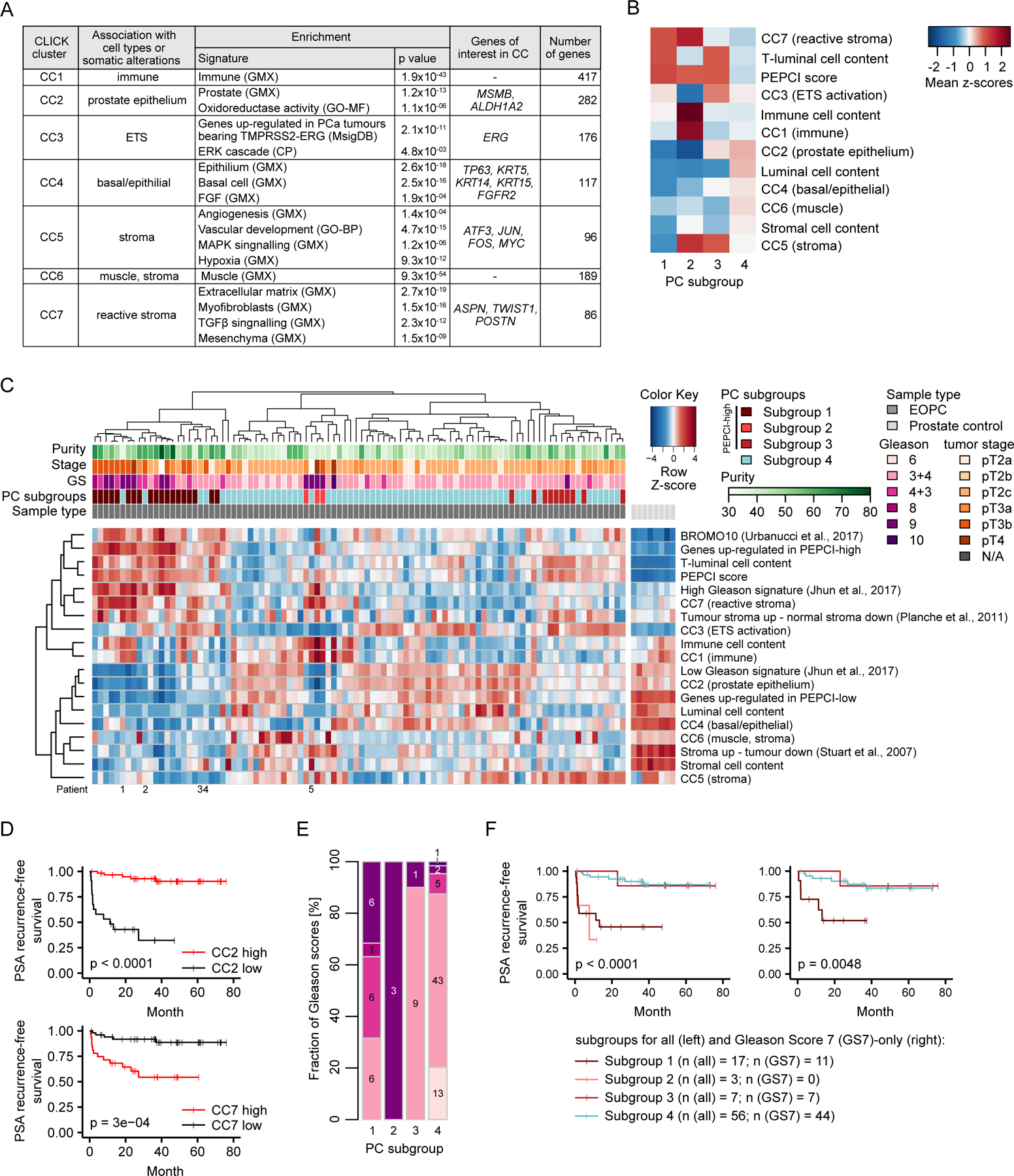
(A) Summary of the most prominent characteristics of CCs. Sources of gene sets are indicated in the brackets: GMX, Genomatix curated gene sets; GO-MF and GO-BF, molecular and biological functions in gene ontology terms, respectively; CP, Genomatix canonical pathways. FDR-corrected p values < 0.05.
(B) Heatmap of the four PC subgroups and their average ct compositions and CC mean pattern values.
(C) Hierarchical clustering heatmap of ct content, CCs, three external gene signatures and indicated PC subgroups across 96 EOPC samples and eight normal prostate controls. CCs and external gene signatures are represented as mean pattern values. Clustering was based on PEPCI-related features and CC information (excluding CC6 due to low information content). Patient number 1 and 3: PEPCI score just below the Inflection point, #2 multi-area sample with varying PEPCI score, #4 and #5: high stromal content.
(D) Kaplan-Meier curves between subgroups in CC2 and CC7 and event-free survival (log-rank test).
(E) Stacked barplot of fraction of GS in the four PC subgroups.
(F) Kaplan-Meier curves of the four PC subgroups using ICGC EOPC samples with available methylation and RNA-seq data (left, n= 83) and a subset of GS7 cases (right, n=62).
See also Table S4 and Figures S5 and S6.
A small group of PEPCI-high tumors, termed PC subgroup 2, was associated with high CC1 (immune), CC5 (stroma) and CC7 (reactive stroma) expression and very high immune cell content, but low T-luminal cell content (Figure 6B, 6C). Consistent with the CC2-low and/or CC7-high expression profiles, subgroups 1 and 2 were strongly associated with high GS (Figure 6E) and shorter time to BCR (Figure 6F).
PC subgroup 3 was associated with high CC5 (stroma) and represented an intermediate risk group. The last subgroup, termed PC subgroup 4, was PEPCI-low and associated with a high fraction of normal-like luminal cells, CC2- and CC4 (basal/epithelial) expression and a known gene signature associated with less aggressive PC (Jhun et al., 2017) (Figure 6F). We observed an enrichment for a TMPRSS2-ERG related gene signature in CC3 (Figure 6A) and a significant enrichment for ETS fusions in CC3-high tumors (p < 1x10−13, Mann–Whitney U test (MWU); Figure S5A). CC3 did not associate strongly with any of the four subgroups.
We validated the CCs and clinical relevance of the subgroups in the TCGA cohort of 462 PC samples with available RNA-seq data (Figure S6A–E). The subgroups showed an improved prediction of BCR compared to GS alone in GS7 cases of the TCGA cohort (p = 0.015; Table S4). Importantly, most PEPCI-high GS7 cases in the TCGA cohort belonged to subgroup 3 (Figure S6F–H), supporting our hypothesis of an intermediate risk group. Comparing subgroups between the two age-of-onset groups identified a higher occurrence of subgroup 4 in EOPC (associated with better prognosis) (p = 0.008, adjusted for GS, Table S4), suggesting age-associated differences in the subgroups. In summary, our integrated analysis of CC signatures and PEPCI score stratified the patients into four prognostic relevant subgroups with distinct differences in the expression of biological pathways.
Tracing the temporal order and clinical trajectories of prostate cancer
Defining the temporal order of somatic events during tumorigenesis can give fundamental insights into the mutational process, clinical trajectories and ultimately guide therapeutic decision making. Prior work have utilized various methods including linear models, tree-based models, clustering or Bayesian approaches to delineate the most likely sequences of somatic events (Lecca et al., 2015; Ramazzotti et al., 2015). A particular relevant question that was not previously addressed is to identify both the most likely next molecular event at any given point and the associated clinical outcome, conditioned on the occurrence of all preceding mutations in that tumor. Our EOPC cohort provides an attractive sample set to address this, due to enrichment of the earliest somatic events and higher clonality (Figure S7A). To identify the temporal order of events in our cohort, we developed PRESCIENT (PREdiction of Sequential Changes In the Evolution of Nascent tumors), a conditional probability-based network model to predict the temporal sequence of somatic events in PC and associated clinical outcome. PRESCIENT uses the probability of observing two events as the exclusive events in the tumor (formulated as Exclusion score (E)), with high Exclusion score as a proxy for early, clonal events. ERG had the single highest exclusion score (EERG,ERG, diagonal on Figure 7A), followed by ERG together with FOXP1 (EERG,FOXP1), with both RGAs having a high level of connections to other RGAs (Figure S7B), suggesting that ERG is frequently occurring as the initial event, followed by FOXP1. A pathway-level analysis showed paths including an initiating ETS fusion event followed by events involved in AR-signaling or cell cycle and subsequent pathway-level events (Figure 7B).
Figure 7. Molecular evolution of prostate cancer.
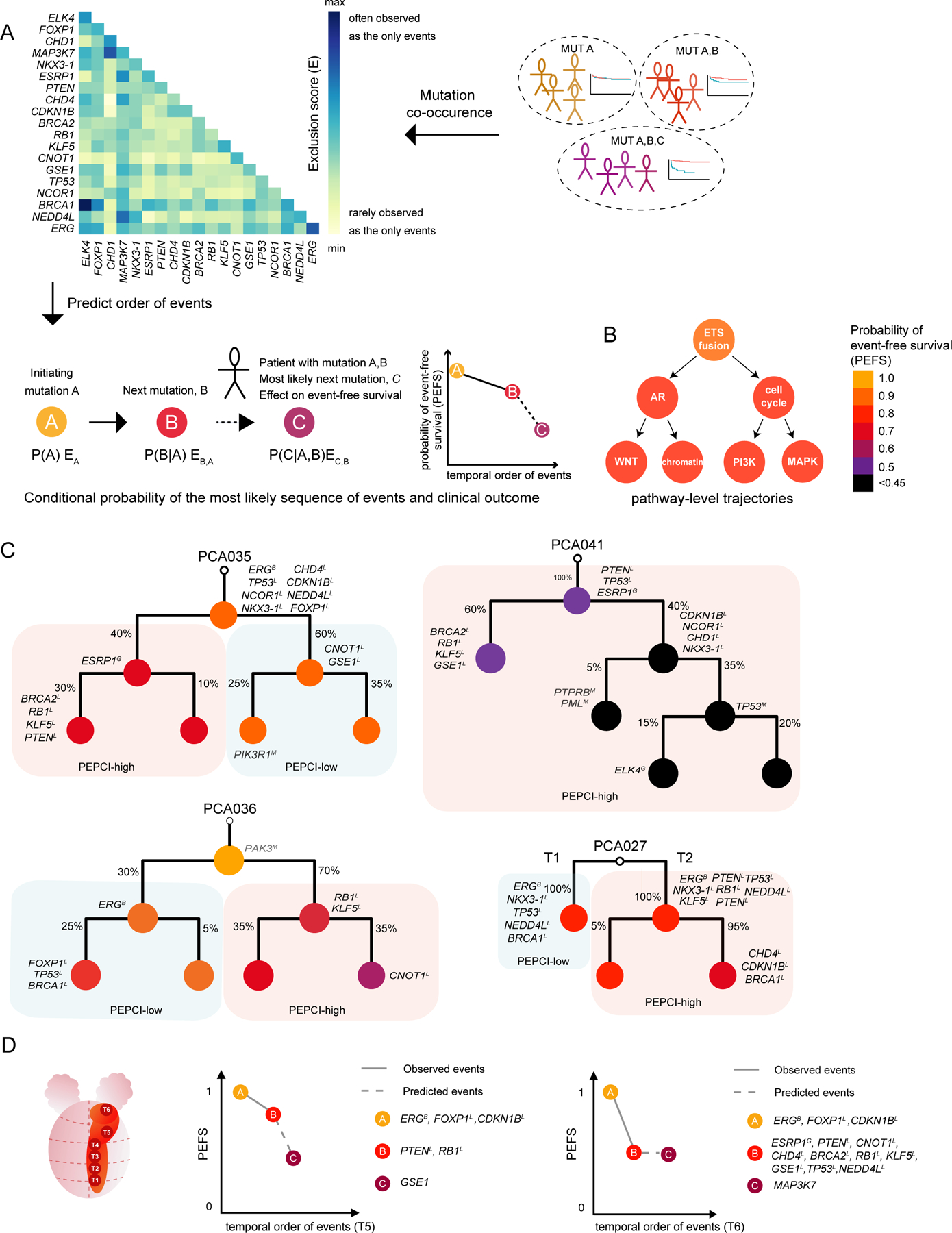
(A) Outline of the PRESCIENT method.
(B) RGAs were labelled with a molecular pathway, which was used as an event in PRESCIENT.
(C) Molecular reconstruction of individual EOPC tumors, using a Bayesian mixture model with each node annotated with RGAs and event-free survival prediction (color-range). The clonal status is annotated with percentages at each branch. Mutations in COSMIC genes are annotated in grey for each node. Branches are labelled by background color based on PEPCI score for PEPCI-high and PEPCI-low.
(D) A schematic representation of PCA035 (left) and application of PRESCIENT prediction to T5 (middle) and T6 (right) regions of the tumor.
See also Figure S7.
For every node in the network, PRESCIENT uses molecular markers to predict the associated Probability of Event-Free Survival (PEFS) (Figure 7A, S7B, S7C). We verified the ability of PRESCIENT to infer the order of mutational events by performing random subsampling and by cross-validation through WGS and reconstruction of the molecular evolution from 40 PC patients developing local metastases. Cross-validation showed robust sensitivity and specificity for PRESCIENT (Figure S7D), providing support that with a given patient’s tumor being molecularly profiled, our probabilistic model is able to predict a patient’s next mutational event more accurately than frequency-based estimates.
We next sought to test our ability to predict aggressiveness of clones from a tumor phylogeny by performing multi-regional WGS of seven EOPC genomes (Figure 7C), followed by clonal reconstruction. For every tumor clone in the tree, we applied our conditional probability model to predict the PEFS (color-scale in Figure 7B). For tumors with divergence in aggressiveness of subclones, i.e. for branches with differences in PEFS, the branch with the shorter PEFS was also the more dominant clone, that is, the tumor clone with the highest relative contribution to the tumor mass from the sampled areas (see e.g. PCA036 in Figure 7C and PCA037 in Figure S7E). This suggested that genomic profiling of several areas of a tumor can be used to identify the more dominant, aggressive clone and pointed towards the utility of applying a probabilistic modelling in order to predict aggressiveness.
PCA041 displayed a particular aggressive molecular evolution involving an early, clonal ESRP1 gain. Both branches maintained an aggressive phenotype, with one branch acquiring KLF5, BRCA1 and RB1 loss and another branch acquiring two parallel biallelic inactivation events of PTEN and TP53 (Figure 7C).
We tested the ability to predict the disease-course based on a single biopsy using PRESCIENT. As a first approximation, we used single-sampled areas from PCA035 to predict both PEFS and the next molecular alteration (Figure 7D). We note that the phylogenetic trees were based on a Bayesian mixture model that uses information from all individual sequenced areas of one tumor and a single sample could therefore in most cases not uniquely be assigned to one branch. We found that the PEFS predicted from a single area of the tumor tended to initiate differently but eventually converged, as exemplified for tumor area T5 (Figure 7D), suggesting that the initiating RGAs in T5 were sufficient to predict the clinical trajectory of the tumor.
Finally, we assessed the potential to identify targeted therapies (Tamborero et al., 2018) based on germline and somatic genomic data. More than 40% of the EOPC patients could be matched with at least one targeted therapy in either pre-clinical test or clinical trial for PC. The targeted agents included PI3K, mTOR and PARP inhibitors (Figure S7F), with the majority of BRCA2 mutated patients showing high levels of somatic BRCAness mutation signature 3 and a high mutational burden (see also Figure 3 and 4), suggesting that these patients could benefit from PARP inhibitors.
In summary, taking advantage of our EOPC cohort, we developed a statistical framework that uses molecular markers to predict the most likely next somatic alteration and associated change in event-free survival.
Discussion
Deciphering the molecular evolution and clinical trajectories of PC require a comprehensive and integrative analysis of the early molecular alterations and mutational mechanisms. A previous survey identified age-associated mutations as the most common mutational process in cancer followed by APOBEC mutagenesis (Alexandrov et al., 2013; Roberts and Gordenin, 2014). Our analysis revealed that APOBEC-associated mutations and kataegis clusters show a clock-like behavior in PC. This mutational process, in PC, is primarily attributable to APOBEC3B activity, likely due to a residual albeit constant enzymatic activity. Our data suggest that APOBEC-attributable mutations occur throughout the development of PC as well as in the normal prostate tissue prior to transformation. Thus, APOBEC mutagenesis and the resulting kataegis events, some of which occur in conjunction with SV formation, are likely to contribute to the earliest mutations seen in PC.
Whereas mutational signatures bear trace of the age and exposure of the tissue, somatic SVs frequently cause tumor promoting gene-dysregulation. We identified recurrent breakpoint hotspots including a potentially clinical relevant biomarker at 8q22, associated with genomic duplication of ESRP1. ESRP1 has been demonstrated to take part in RAF-fusion formation in PC (Palanisamy et al., 2010), although we did not find evidence for ESRP1 fusion events in our cohort. ESRP1 has also been implicated in EMT transition (Ishii et al., 2014) and overexpression has been demonstrated to cause anchorage-independent growth and metastases in colorectal cancer (Fagoonee et al., 2017). ESRP1 was previously shown to be strongly co-expressed with E-cadherin, and increased expression of ESRP1 may therefore lead to accelerated proliferation of an epithelial cell state.
PC is a highly heterogeneous disease, but there is a limited understanding of the ct composition and how this impacts disease progression. Recent genomic surveys have characterized seven subtypes based on somatic alterations (TCGA, 2015), primarily involving ETS family genes, but the ct composition and associated clinical relevance remained unexplored. Analysis of basal and luminal ct composition has shown biological and clinical relevance in another hormone-associated adenocarcinoma - breast cancer (Sotiriou et al., 2003). We pursued a complementary approach by integrating methylation array and RNA-seq data, which led us to identify four molecular subgroups based on ct composition and gene expression patterns in EOPC tumors that showed association with clinical outcome independent of GS. The subgroups were able to stratify intermediate-risk GS7 PC cases, suggesting that the subgroup information can serve as an independent molecular risk score. Tumors that differ in ct composition would likely respond differentially to therapies, and further work will be needed to investigate whether the different subgroups predict response to therapy.
There is an urgent need for biomarkers that can stratify patients who need definitive treatment from those who can follow active surveillance or watchful waiting. Single biomarkers are unlikely to be useful in a heterogeneous and complex disease such as PC, and multiple biomarkers will thus be required to guide clinical decision making. We utilized our comprehensive molecular catalogue of the earliest alterations in PC to develop PRESCIENT. PRESCIENT establishes a knowledge-based framework for future genomics-informed patient stratification and drug-targeting. The current implementation is limited in the sample cohort size, which we expect to improve in both sensitivity and specificity with inclusion of more samples. This will involve highly aggressive tumors, including LOPC and metastatic cancers, to increase the ability to predict event-free survival and therapy response, as well as enable the prediction of secondary alterations associated with metastasis.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joachim Weischenfeldt (joachim.weischenfeldt@bric.ku.dk)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
Tumor samples were collected form 251 PC patients. Data was supplemented with bam files from 41 published tumor and normal WGS samples (Fraser et al., 2017). Informed consent and an ethical vote (institutional reviewing board) were obtained according to the current International Cancer Genome Consortium (ICGC) guidelines (see http://www.icgc.org). Manufacturing of TMAs and their analysis for research purposes as well as patient data analysis has been approved by local laws (HmbKHG, §12,1) and by the local ethics committee (Ethics commission Hamburg, WF-049/09 and PV3652). All work has been carried out in compliance with the Helsinki Declaration. Radical prostatectomy specimens were available from 17,747 patients, undergoing surgery between 1992 and 2014 at the Department of Urology and the Martini Clinics at the University Medical Center Hamburg-Eppendorf. Follow-up data were available for a total of 14,464 patients with a median follow-up of 48 months (range: 1 to 275 months).
Tissue-microarray processing
Archived formalin fixed tissues were used for the TMA analysis, as previously described (Kononen et al., 1998), which involved taking a 0.6 mm core from a representative tissue block from each patient. The tissues were distributed among 39 TMA blocks, each containing 144 to 522 tumor samples. For internal controls, each TMA block also contained various control tissues, including normal prostate tissue.
The usage of archived diagnostic left-over tissues for manufacturing of tissue microarrays and their analysis for research purposes as well as patient data analysis has been approved by local laws (HmbKHG, §12,1) and by the local ethics committee (Ethics commission Hamburg, WF-049/09). Informed consent was exempt based on the „Hamburgisches Krankenhausgesetz“ HmbKHG 312,1.
METHOD DETAILS
Biospecimens and Quality Control
Sample inclusion criteria
Biospecimens were collected from patients diagnosed with PC. Specialized pathologists dissected each prostate immediately after surgery. Dissection followed a predefined scheme to represent the position of each block relative to the entire prostate. This procedure resulted in 60 – 150 pieces of tissues depending on the size of the prostate. An image was taken from each dissected prostate specimen for later reference. After dissection, each tissue block was placed on a separately labeled cork plate covered with a special compound for cryopreservation (OCT) before the tissue was frozen to −20°C. Cryo-sections were taken from each block and presence as well as content of tumor was determined by the pathologist. The tumor cell content is given as the percentage of cancer cells relative to the entire tissue block. If necessary, IHC tumor validation (e.g. AMACR, 34BE12) was performed of the frozen tissue or after secondary paraffin embedding of selected frozen blocks. Uni- and multifocal cancers were distinguished according to the criteria of Wise et al. (Wise et al., 2002). Tumor areas were defined as part of a single focus if they were within 3 mm of each other in any section or within 4mm on adjacent sections.
Except for two patients (PCA125 and PCA176) who received pre-operation hormone therapy with LH-RH analogon, the patients did not receive any neo-adjuvant radiotherapy, androgen deprivation therapy, or chemotherapy prior to the surgical removal of tumor tissue. Tumor samples and one normal prostate control were frozen at −20 °C and subsequently stored at −80 °C. Eight additional normal prostate samples were obtained from a previous project (Börno et al., 2012). DNA and RNA were extracted as described previously (Weischenfeldt et al., 2013).
ESRP1 immunohistochemistry and FISH
TMA sections were freshly cut and used for an IHC staining performed on one day and in one experiment. The slides were deparaffinized with xylene and a descending alcohol series. Antigens were retrieved by heating for 5 minutes in an autoclave at 121°C in Tris-EDTA-Citrate buffer, pH 7.8. To prevent non-specific binding, a hydrogen peroxide blocking solution was applied for 10 minutes. The primary antibody specific for ESRP1 (rabbit polyclonal antibody, Sigma Aldrich Germany, cat#HPA023720; dilution 1:450) was incubated at 37°C for 60 minutes. The antibody (HPA023720) has been validated by the Human Protein Atlas project, which shows i) overlapping staining patterns with other anti-ESRP1 antibodies, ii) a band of appropriate size in western blots, iii) specific binding to ESRP1 on a protein array. The FISH probe mix consisted of a spectrum-orange labeled ESPR1 (8q22.1) probe (made from bacterial artificial chromosomes (BACs) RP11-267M23 and BAC RP11-22C11), and a spectrum-green labeled, commercial centromere 8 probe (#6J37-08; Abbott, Wiesbaden, Germany). To visualize the bound antibody, the EnVision Kit (Dako, Glostrup, Denmark) was used according to the manufactureŕs directions. ESRP1 staining was found in the nucleus and cytoplasm of positive cells. In ESRP1 positive cancers, staining was usually seen in all tumor cells (100%). Hence, the staining intensity in prostate epithelial cells was estimated in four categories for each cancer, i.e. negative (not detectable), weak, moderate and strong staining.
Pathology review
All prostate specimens were analyzed according to a standard procedure, including a complete embedding of the entire prostate for histological analysis (Erbersdobler et al., 1997). Histopathological data were retrieved from the patient’s records, including tumor stage, GS, nodal stage and stage of the resection margin. PSA values were measured following surgery and PSA recurrence was defined as a postoperative PSA of 0.2 ng/ml and increasing in subsequent measurements.
Copy Number Analysis
Copy number and SV profiles for each patient were binned with a 500 kb sliding bin size. Each bin containing a boolean information if an aberration occurred within the bin in the given patient. RGAs were computed by overlapping the binned data of each patient computing a cohort frequency of aberrations for each bin, and selecting the peak-frequency bins within regions with frequency higher than 5%.
DNA Sequencing and Analysis
Whole genome sequencing
DNA library preparation and WGS was performed on Illumina sequencers as described earlier (Weischenfeldt et al., 2013) with a median insert size of 310 bp (sd 57 bp) and a median WGS coverage of 61-fold for tumor and 38-fold for germline control samples.
Read alignment
WGS data was aligned to the human genome Build GRCh37 using BWA-MEM (Li, 2013) according to Pan Cancer Analysis of Whole Genomes (PCAWG) protocol (https://doi.org/10.1101/161638).
Median purity
Tumor purity was calculated as the median of three purity measures, a methylation-based score defined by methylation of selected sites in the promoter of the GSTP1 (Brocks et al., 2014), a score based on allele-specific copy number profiles (Favero et al., 2015), and a score based on the absolute quantification of somatic DNA alterations (Carter et al., 2012).
DNA variant calling
Single-nucleotide variant and SV calling
Somatic SNVs were identified by the PCAWG implementation of the DKFZ SNV pipeline (https://doi.org/10.1101/161638). Subsequently, SNVs overlapping with tandem repeats and strand bias were marked as low-confidence and removed from the consecutive analysis.
Somatic SV discovery was pursued across all samples (matched tumor/normal genome) using the DELLY2 (Rausch et al., 2012) PCAWG analysis workflow (https://github.com/ICGC-TCGA-PanCancer/pcawg_delly_workflow). We used a high-stringency SV set by additionally filtering somatic SVs detected in ≥ 1% of a set of 1105 germline samples from healthy individuals belonging to phase I of the 1000 Genomes Project (1000 Genomes Project Consortium et al., 2015), and by removing somatic SVs present in any of the PCAWG germline samples. For inference of high-stringency SVs we further required at least four supporting read pairs with a minimum mapping quality of 20 and restricted valid somatic SV sizes from 300 bp to 500 Mb. Somatic copy number alterations (SCNAs) were identified using sequenza (Favero et al., 2015), which was applied to tumor and normal bam files. SCNAs near low mappability regions and with logR below 0.2 were removed. Purity parameter was selected to match the median purity (see Median purity section), and ploidy parameter was selected to fit a diploid state.
We employed freebayes (v1.1.0) in single sample- and paired-sample calling mode for discovery of SNVs, multi nucleotide variants, and InDels < 50 bp (used parameters: --min-repeat-entropy 1, --report-genotype-likelihood-max, --alternate-fraction 0.2, and --no-partial-observations), as previously described (http://dx.doi.org/10.1101/208330). Raw variant predictions were further filtered for quality (QUAL > 20, QUAL/AO > 2), strand bias artifacts (SAF > 1, SAR > 1), read position artifacts (RPR > 1, RPL > 1), and normalized for consistent representation across patients with vt (v0.5). Germline variants were annotated with the Ensembl Variant Effect Predictor (VEP) (r81). High impact (i.e. damaging) germline mutations were defined as frameshift, stop gain, start lost, canonical splice site, exon/gene deletions, known (ClinVar; accessed 2017-02-16) damaging non-canonical splice site variants, and somatic mosaic mutations (the latter of which are defined as mutations present in a subset of normal cells). Putative damaging germline mutations were removed if the estimated minor allele frequency (MAF) in at least one continental population was above 1%, which we judged based on 53,105 sequenced individuals that were assigned to known (control) populations and without cancer diagnosis from the ExAC resource (http://exac.broadinstitute.org), the 1000 Genomes Project (http://www.internationalgenome.org), and the NHLBI GO Exome Sequencing Project. Putative gain-of-function (GoF) missense variants in TP53 were further evaluated based on information in the IARC TP53 database (http://p53.iarc.fr/) and annotated as pathogenic if TP53 mutations were classified as “non-functional” based on experimental transcriptional activity assays. Finally, all germline mutations were excluded from the analysis if annotated as benign in ClinVar. We estimated the primary population ancestry (European, African, East Asian, South Asian, and Native American) for all patients using a supervised decomposition approach (https://doi.org/10.1101/208330) and ancestry-informative markers.
Subclonal copy-number analysis
Subclonal copy number were identified by computing the cancer cell fractions based on the B-allele frequency and the depth ratio for each segment. To compute and we postprocessed the DNA copy number segments following the sample-wide analysis with sequenza. A sample-wide analysis provides the copy number state estimate for each segment based on the total clonal contribution and the global cellularity (ρ) and ploidy (ψ) values.
We assume that subclones share the same ploidy but differ in cellularity.
Using the B-allele frequency and depth ratio models previously described (Favero et al., 2015), we used a grid search approach for each segment i to find the optimal value of the local-cellularities and given respectively, the observed values of B-allele frequency and depth ratio for the segment and the copy number and ploidy estimates from the sample-wide analysis.
The and are calculated by dividing the estimated cellularity derived by the depth-ratio model and the B-allele frequency model with the sample-wide cellularity ρ.
We then applied a bivariate Dirichlet process to generate 2D clusters versus . Clusters with both and values between 0.1 and 0.9 were identified as subclonal clusters.
Samples in which the sum of the subclonal segments represent more than 0.1% of the genome are classified as polyclonal, otherwise are classified as monoclonal.
DNA methylation analysis
Sample preparation and Data analysis
Normal and tumor basal, luminal and stromal cell fractions were generated from fluorescence-activated cell sorted (FACS) cell fractions of seven BPH samples (age range 68–90) and seven PC cases (age range 55–79, GS: 3+3 (n = 3), 7 (n = 1), 4+5 (n = 1), 5+4 (n = 2)) obtained from UT Southwestern Medical Center and prepared according to (Henry et al., 2017). DNA was extracted using Qiagen AllPrep DNA/RNA/Protein Mini Kit. For the ICGC EOPC and LOPC cohorts, genomic DNA was extracted from bulk fresh frozen tumor specimen. DNA was submitted to HumanMethylation450 analyses at the Genomics and Proteomics Core Facility of the German Cancer Research Center (Heidelberg). Data quality control, preprocessing and beta-mixture inter-quantile (BMIQ) normalization was done using RnBeads (Assenov et al., 2014) Further data processing included removal of 27598 cross-reactive probes (Chen et al., 2013b) and 39752 sites overlapping with SNPs (dbSNP Build 150, Feb. 2017). Array-based methylation beta values were independently validated using Agena MassArray EpiTyper technology (BLUEPRINT consortium, 2016). For reference-based ct estimation, we used the Houseman algorithm (Houseman et al., 2012) with quality-controlled sorted basal, stromal, normal luminal and T-luminal cell fractions as reference cts. The fraction of infiltrating immune cells for every sample was estimated using the Leukocytes unmethylation for purity (LUMP) algorithm (Aran et al., 2015). We selected the 500 most discriminatory CpG sites between the different cts to compute the ct composition of our EOPC samples (Figure S4B). PEPCI was calculated as the combined fraction of T-luminal cells and immune cells for every sample (Figure 5A). The computation of PEPCI was implemented in a dedicated R package (KEY RESOURCES TABLE).
KEY RESOURCES TABLE
The PEPCI R package provides quantitative information on four cts (basal, stromal, normal luminal and T-luminal cells) and infiltrating immune cells, inferred from methylation data. The PEPCI score represents the combined percentage of T-luminal and immune cells as a measure of tumor aggressiveness.
Ct-specific reference methylomes are used to estimate ct composition in bulk tumor samples, employing the Houseman algorithm, which is a common tool to deconvolute the composition of blood samples. The algorithm selects a specified number of CpG sites with most variable methylation between provided reference cts. This process is predefined in the PEPCI R package, which processes Illumina 450k or EPIC array data and interrogates 500 preselected CpG sites. The location of these sites relative to ChromHMM states of prostate epithelial cells indicates significant enrichment of T-luminal cell hypermethylation in promoter CpG islands, and stromal-specific hypermethylation in enhancer regions (Figure S4C, heatmap legend on the right).
Methylation based principal component analysis was done using RnBeads (Assenov et al., 2014). Enrichment of CpG sites at chromatin states was performed with EpiAnnotator (Pageaud et al., 2018), using ChromHMM states for prostate epithelial cells (Taberlay et al., 2014). Trees representing sample similarities were constructed from methylation-based Euclidean pairwise distances using the algorithm for phylogenetic tree reconstruction of Desper and Cascuel (Desper and Gascuel, 2002). Logistic regression models based on methylation data and their evaluations using receiver operating characteristic (ROC) curves were performed using the R programming language.
Heterogeneity estimates of heterogeneity and multifocal cases were calculated by averaging all pairwise dissimilarities between methylation-based ct fractions of the corresponding multi-area samples using cosine dissimilarity.
For the TCGA PRAD cohort, HumanMethylation450 raw signal intensities of probes for each participant’s tumor sample (n = 498) were downloaded as idat files from the TCGA data portal (https://tcga-data.nci.nih.gov) and processed as described for the ICGC cohort.
Tri-nucleotide mutational signature
To identify mutational signatures, we applied YAPSA (Yet Another Package for Signature Analysis) (Huebschmann et al., 2016), a linear combination decomposition of the mutational catalog with predefined signatures from the COSMIC database (http://cancer.sanger.ac.uk/cosmic/signatures, downloaded June 2016) computed by non-negative least squares (NNLS). To increase specificity, the NNLS algorithm was applied twice; after the first execution, only those signatures whose exposures, i.e. contributions in the linear combination, were higher than a certain cut-off were kept, and the NNLS was run again with the reduced set of signatures. As the detectability of different signatures may vary, signature-specific cut-offs were determined in a random operator characteristic analysis using publicly available data on mutational catalogs of 7,042 cancers (507 samples with WGS; 6,535 samples with whole exome sequencing) (Alexandrov et al., 2013) and mutational signatures from COSMIC. This yielded the following signature-specific cutoffs: AC1: 0; AC2: 0.01045942; AC3: 0.08194056; AC4: 0.01753969; AC5: 0; AC6: 0.001548535; AC7: 0.04013304; AC8: 0.242755; AC9: 0.1151714; AC10: 0.01008376; AC11: 0.09924884; AC12: 0.2106201; AC13: 0.007876626; AC14: 0.1443059; AC15: 0.03796027; AC16: 0.3674349; AC17: 0.002647962; AC18: 0.3325386; AC19: 0.1167454; AC20: 0.1235028; AC21: 0.1640255; AC22: 0.03102216; AC23: 0.03338659; AC24: 0.03240176; AC25: 0.01611908; AC26: 0.09335221; AC27: 0.009320062; AC28: 0.05616434; AC29: 0.05936213; AC30: 0.05915355. We removed mutation signature present in fewer than 10% of the samples.
Knowledge-based analysis of APOBEC-associated mutational processes
Enrichment and mutation load associated with APOBEC mutagenesis were calculated based on prior mechanistic knowledge about mutation motifs associated with certain mutagenic factors and pathways (Chan et al., 2015; Roberts and Gordenin, 2014). Calculations were done for genome-wide mutation calls and for mutation calls only in C/G clusters identified as described in (Chan et al., 2015; Roberts et al., 2012). Briefly, the enrichment with a tri- or tetra-nucleotide motif pXq → pZq were calculated, where X is the mutated nucleotide, Z is the nucleotide after base substitution, p is the −1 nucleotide (or −1 and −2 nucleotides), and q is the +1 nucleotide (within the context of the given mutation type/trinucleotide). For each motif, we also included the reverse complement sequence that would represent the mutagenic process occurring on the opposite DNA strand. To statistically evaluate whether a certain mutation type is enriched in a sample as compared to mutations generated by random mutagenesis, a one-sided Fisher’s exact test was performed. To account for multiple testing, p values obtained were corrected using the Benjamini-Hochberg method.
The MAF of the germline APOBEC3B deletion in our PC cohort was 7.0%, in line with the expected frequency in individuals with a European germline genetic background.
RNA-seq expression analysis
Sample preparation and Data analysis
RNA extraction and sequencing for the samples ICGC_PCA1-12 was performed as described in a previously publication (Weischenfeldt et al., 2013). DNase digested total RNA from additional 109 EOPC samples and 9 control samples was analyzed using RNA6000 nano assays (Agilent 2100 Bioanalyzer) and Qubit 2.0 Fluorometer. Only samples with an RNA Integrity Number (RIN) > 7.0 were included in this study. We used 1–4 micrograms of total RNA from each sample to prepare Truseq stranded sequencing libraries (Illumina). In brief, poly-A enrichment, fragmentation, first and second strand synthesis, A-tailing, and adapter ligation were performed following the manufacturer’s instructions. Libraries were PCR-amplified for 7–20 cycles and qualitatively validated on an Agilent 2100 Bioanalyzer of product size and concentration and on Qubit. Libraries were sequenced 50 bp paired-end on a HiSeq 2000 flowcell according to Illumina’s protocol.
RNA reads were aligned to hg1000 using BWA (v. 0.5.9-r16 for reads up to 51 bases and v. 0.7.7-r441 for reads with 100 bases) and SAMtools. Uniquely mapped reads were annotated using Ensembl v62. Gene expression levels were quantified in reads per kilobase of exon per million mapped reads (RPKM) and corrected for RNA composition effects applying TMM implemented in the R package edgeR (Mortazavi et al., 2008; Robinson and Oshlack, 2010). Multi-area samples of the same patient were merged in to an artificial sample by taking the sum of read counts per gene before RPKM calculation resulting in a cohort of 96 RNA-seq samples. Hierarchical clustering on the most variable genes revealed batch effects across the RNA-seq cohort. Using hierarchical clustering and sample preparation information, a set of 42 tumor RNA-seq samples without batch effects was manually selected and used to identify co-expression clusters applying the algorithm CLICK, part of the software tool EXPANDER (v7.11) (Sharan et al., 2003). CLICK was run with default parameters on log2 transformed and z-score normalized RPKM values of the 1231 most variable genes. Genes were expressed in minimum 3 samples ≥ 0.5 RPKM. 14 CCs were identified showing an overall separation of −0.039 and overall homogeneity of 0.632. Seven out of 14 CCs showed a homogeneity value ≥ 0.6 and were selected for further analyses (named CC1-CC7). An 8th CC also showed a high homogeneity value, but consisted mainly of one protocadherin gene cluster and was discarded. We calculated a mean pattern value for each CC in each of the EOPC samples. A mean pattern value for one sample is defined by taking the trimmed mean across log2 and z-score transformed RPKM values of genes in a particular CC. Based on the mean pattern values obtained for each CC, tumor samples were divided into two subgroups (called CC-high and CC-low) using partitioning around medoids (pam). We compared the subgroups in an CC using edgeR and selected differentially expressed genes following the expression pattern in a CC with |log2(FC)| ≥ log2(2), FDR ≤ 0.01 and difference of median expression > 1, resulting in overall 417, 282, 189, 176, 117, 96, 86 and 21 genes in CC1-CC7, respectively (Table S4). After the establishment of the CCs, the remaining 54 EOPC RNA-seq samples were reintegrated. Mean pattern and CC subgroups were recalculated as described above taking the whole RNA-seq cohort of 96 samples (Table S4). CCs were functionally annotated applying gene set overrepresentation analysis of the GePS Genomatix software (v3.80116). For additional annotation and comparison of the CCs to the literature, we integrated external signatures that are associated with high risk PC (BROMO10), Gleason score, stroma or reactive stroma into the mean pattern matrix (Jhun et al., 2017; Planche et al., 2011; Stuart et al., 2004; Urbanucci et al., 2017).
RPKM values for the TCGA cohort (495 samples) were calculated as described above. Batches with the ID 312 and 320 were excluded due to batch effects resulting in a cohort of 462 samples. To classify TCGA samples into CC subgroups, RPKM values were log transformed and z-score normalized, and mean pattern values for each sample and CC were calculated. Based on the mean pattern values, samples were assigned to the CC subgroup with nearest medoid. Here, the medoids originate from the calculations on the ICGC EOPC cohort.
In TCGA, subgroup 1 (97 samples) and subgroup 3 (143 samples) could be derived from the hierarchical clustering using the same features as in our ICGC EOPC cohort. To annotate subgroup 2 in the TCGA data, we applied criteria defined from our ICGC EOPC cohort. As described above, the samples belonging to the “Immune” subgroup showed a high immune cell content and high expression of CC5 and CC7. Here, the lowest immune cell content in subgroup 2 ranked at the 94th percentile considering a normal distribution, and the mean and standard deviation of the estimated immune cell content across the 96 EOPC samples with RNA-seq. In total nine TCGA samples were assigned to subgroup 2 based on high PEPCI score, CC5-high and CC7-high, and a high immune cell content defined by the 94th percentile of immune cell content values.
Before the comparison of subtype fractions in ICGC EOPC, TCGA EOPC (100 cases) and TCGA LOPC (360 cases), GS composition differences were adjusted to the ICGC EOPC cohort (fractions of GS in ICGC: GS6 = 0.135, GS7 = 0.72, GS8 = 0.01, GS > 8 = 0.135). The GS-corrected subtype fractions in the TCGA EOPC and TCGA LOPC cohorts were calculated using 1,000 bootstrap samples. A single resampling was performed by extracting TCGA EOPC/LOPC cases according to the GS, and by drawing a bootstrap sample of these cases for each GS (GS6, GS7, GS8, GS >8) independently. Here, the samples size of a GS bootstrap in TCGA related to the GS fraction in ICGC EOPC (e.g. for GS6 and TCGA LOPC the sample size is equal to 360*0.135). From the GS-corrected bootstrap samples the fraction of subgroups was calculated. The p value of the differences in subgroup fraction was based on a permutation test.
Based on gene expression, the heterogeneity of multi-area samples belonging to the same patient was estimated by taking the average of the pairwise dissimilarity values between multi-area samples. To calculate the pairwise dissimilarity values we took the mean of the cosine dissimilarity between multi-area samples in the different CCs. Here, the cosine dissimilarity was measured across the gene expression values of a particular CC.
Breakpoint-association with chromatin data
Chromatin-related data from PC tissue and cell lines were downloaded from different studies to generate chromatin states (ChromHMM). ChromHMM from normal prostate epithelial cells PrEC and PC3 cells were downloaded from GSE57498. Similar 9 ChromHMM states were produced for LNCaP and VCaP cells using ChromHMM software (Ernst and Kellis, 2012). To learn ChromHMM states in LNCaP cells we computed H3K27me3, H3K27Ac, H3K4me1, and H3K4me3 profiles (Barfeld et al., 2017), CTCF binding profile (ENCODE), and Phospho S5 RNA Pol II binding profile (Massie et al., 2011). For VCaP cells we computed H3K27me3, H3K4me1, H3K27me3 (Yu et al., 2010), RNA pol II profiles (Asangani et al., 2014), and CTCF and H3K27Ac profiles (ENCODE). The resulting 9 chromatin states for PrEC, PC3, LNCaP, and VCaP cells were reduced to 7 by combining “enhancer+CTCF” and “promoter+CTCF” states together with “enhancer” and “promoter”, respectively. The derived 7 ChromHMM states were then used to annotate DNA methylation data. For enrichment analysis of DNA breaks we excluded the states “transcribed” and “bound by CTCF”.
SV breakpoints from EOPC and LOPC samples were assessed by performing intersection between chromatin states and SV breakpoints using bedtools v2.25.0 (Quinlan and Hall, 2010), adding 5 kbp to both sides of the peaks. To compute an expected overlap frequency, breakpoints were randomly shuffled 100 times on the genome (excluding telomere and centromere regions, downloaded from UCSC, hg19) and keeping the SV size and chromosome fixed before performing overlap.
Association of SVs with DNA-DNA interaction and open chromatin marks was measured in a correlation-based analysis between breakpoint density and marks of physically interacting and open chromatin. We divided the genome in 1 Mb sliding windows using a 100 kb step and overlapped each bin with the SV breakpoints, retaining maximum 1 overlap per patient per bin, the number of chromatin loops and PC-specific H3K27ac peaks. Spearman’s Rho was estimated by correlating the EOPC and LOPC breakpoints with the number of Hi-C loops and H3K27ac peaks, respectively.
For Hi-C, we combined the loop annotations from 8 human cell-lines (GM12878, Hela, HMEC, HUVEC, IMR90, K562, KBM7, NHEK), obtained from GSE63525 (Durand et al., 2016; Rao et al., 2014), and removed duplicate chromatin loops. H3K27ac PC-specific peaks were obtained from (Kron et al., 2017).
Exclusion score estimate
Here we define Exclusion score (E) as a metric to evaluate the preference of an aberration to occur as a sole event, or in tandem with many aberrations.
For every patient x, we construct an aberration index Ax that contains a list of all the Ii(x) deleted genes/regions for that patient, e.g. Ax = {BRCA1, ERG, ELK4}. The exclusion score is defined as the fraction of the observed aberration, in this case 1, and the total number of aberrations for that patient:
The exclusion score for a given aberration i, is defined as the mean exclusion score across all patients:
The exclusion score is confined between 0 and 1 and the interpretation is such that, the higher the exclusion score of an aberration, the more frequently it tends to occur as a single event or in combination with very few events and vice versa.
Pairwise exclusion score
In a similar manner we can compute the exclusion score for a pair of aberrations i and j which is the “preference” of that pair to occur with few or many over events,
and compute the mean pairwise exclusion score respectively,
Survival analysis
We employed Random Survival Forests (RSF) to predict the PEFS, using the time (in months) from diagnosis to BCR as the response variable. The model was implemented in R using the randomForestSRC package (Ishwaran et al., 2008), with n.tree = 5,000. Missing data were imputed each time a tree node performed a split on samples with missing values. Split statistics were aggregated over all trees to determine the median split value for each variable (PEPCI 69.1). We chose a random forest approach to binarize the PEPCI-score since it is able to treat continuous right-censored survival data and identify the optimal split that separates the samples into high and low risk groups.
For BCR analyses incorporating GS, samples were grouped into the categories GS6, GS3+4, GS4+3 and GS8-10 for ICGC, and GS6, GS3+4, GS4+3 and GS8, GS9-10 for TCGA. In ICGC, only one case showed GS8 and therefore included into the GS9-10 group. Time to BCR was right-censored at 100 months for TCGA PRAD. Prediction of BCR was analyzed using Kaplan-Meier curves and log-rank test, and cox proportional hazards regression model (CPHM). Fitted CPH models were compared using log-likelihood ratio test. Here, we tested a full model incorporating GS, and the introduced PEPCI groups, CC2/CC7 groups or subgroups against a model incorporating GS only. All analyses were done using R (R-3.3.3) and the R packages survminer (v0.3.1) and survival (v2.41-3) (Therneau and Grambsch, 2000). CPHM and LRT related results were provided in Table S4.
We assessed the predictive advantage of ESRP1 gain over MYC gain by comparing the variable importance (VIMP) values of the two features in a RSF model, using PSA, pT, GS, Age, CC2, and PEPCI, to predict BCR. We trained 1,000 models to get the VIMP distribution of ESRP1 and MYC. A one-sided Wilcoxon test shows that VIMP associated with ESRP1 gain is significantly higher than MYC gain (p value < 2.2e-16)
PRESCIENT tumor evolution model
PRESCIENT (https://bitbucket.org/weischenfeldt/prescient) uses conditional probability to predict the order of molecular events, given a known or observed co-occurrence matrix. It differs from similar algorithms previously published including CAPRI (Ramazzotti et al., 2015) and TO-DAG (Lecca et al., 2015) by the ability of predicting the next event in a progressive manner rather than compute an overall general consensus evolution tree, and use the estimate of event free survival to predict patient outcome based on genomic evolution. Additionally it includes a novel metric, Exclusivity Score, which measure the tendency of an aberration to occurs with other events.
PRESCIENT is based on the following assumptions i) molecular events that are often observed in the same tumor are more likely to be phylogenetically closely related ii) molecular event(s) that are often observed as the exclusive events, are more likely to occur early in the tumor evolution. As molecular events, we used the RGAs and used the presence or absence in each patient to generate a co-occurrence matrix. PRESCIENT constructs an expected frequency distribution F(f) to identify the most probably initiating RGA event. The RGA co-occurrence matrix is bootstrap sampled 10,000 times, and at each iteration, a new exclusion score matrix and RGA co-occurrence frequency matrix is computed. The co-occurrence and exclusion score matrices were used to compute the P(f)i and P(e)i for each RGA respectively, and the probability Pi = P(f)iP(e)i of an RGA i to occur. The first event in the branch corresponds to the event with the highest probability Pi. This RGA is then removed from the RGA co-occurrence matrix, and the resulting subset is used to recompute a new P(f)i and P(e)i for each remaining RGA. The same approach is applied to assign the following RGA in the branch until no events are left in the resulting RGA co-occurrence matrix. Each permutation yields a putative evolution trajectory formed by an ordered series of events.
PRESCIENT associates a probability score for each node in the trajectory. Using these precomputed trajectories, PRESCIENT can take as input a set of RGAs detected in a patient, to predict the most probable next RGAs in the trajectories of the patient and the associated patient disease progression.
Clonal reconstruction
Clonal reconstruction was done for multi-region tumors. To identify the clonal evolution from tumor samples with multiple sequenced tumor regions, we applied the R package Canopy (Jiang et al., 2016), using default parameters. As input for the reconstruction, we used nsSNVs, tumor/normal depth ratio and allele frequency information from sequenza (https://bitbucket.org/sequenza_tools/sequenza_canopy). Reconstructed evolution trees were curated following the guidelines in the R package documentation.
PRESCIENT method validation
From a cohort of 40 patients with 30x WGS primary PC and local lymph node metastasis, we estimated the phylogenetic trees for each patient as described in the heterogeneity cases (with Canopy).
The PRESCIENT method was compared to a naive-frequency approach, in which each RGA have the probability of occurrence given by its frequency in the cohort.
We tested the ability to predict the next RGA in the phylogeny, which served as a true positive set. To compute a true positive prediction, PRESCIENT prediction and the frequency-based prediction were compared to the observed RGA for every node in the tree. A false-positive set was calculated using both methods, by extracting the most abundant RGA in the cohort, which was not present in the patient.
To further assess the robustness of the method, we compared the main evolution trajectory predicted by PRESCIENT with the estimated probability of the same trajectory calculated by random subsampling (10% of the dataset, 10,000 times). We found overall highly similar probabilities, with random-subsampling based probability of the first node within 1.5 standard deviations from PRESCIENT prediction and 1.55 for the second node.
Drug-variant targets
Drug-variants were downloaded from https://www.cancergenomeinterpreter.org (April 15, 2018) (Tamborero et al., 2018). Germline SNP, somatic SNV, SCNA and SV variant types were matched with Biomarker types (e.g. a Biomarker fusion type was matched with SV-mediated fusions and Biomarker deletion matched with SCNA deletion in our dataset). All predicted damaging germline PTVs were considered irrespective of the Biomarker amino acid change. Only Biomarkers associated with a Responsive effect and in pre-clinical or clinical trial or currently in clinical guidelines for PC were considered. BRCAness was scored as tumor genomes with at least 40% contribution of mutation signature 3 and minimum 1,000 somatic SNVs.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical analysis methods are described in the STAR Methods detail subsections.
DATA AND SOFTWARE AVAILABILITY
The raw data for WGS, RNA-seq expression and array-based methylation were submitted to EGA under study identifier EGAS00001002923. Somatic variant calls are available through Mendeley Data doi:10.17632/6gtrrxrn2c.1. Oncological outcome data were collected via the Progether PROMś (patient reported outcome measurement) interface (www.progether.com/proms) and the martini-clinic database.
Softwares used for each analysis are described and referenced in Methods Detail subsection and listed in KEY RESOURCE TABLE.
Supplementary Material
Table S2. ESRP1 IHC and APOBEC-associated SVs, Related to Figure 2
Table S1. Clinico-pathological data, Related to Figure 1
Table S3. Methylation-inferred ct composition, Related to Figure 5
Table S4. CC analysis, Related to Figure 6
Significance.
We used a set of tumors diagnosed early in life and thus harboring the earliest molecular lesions detectable in prostate cancer which led us to identify an APOBEC-driven clock-like mutational process driving the earliest somatic mutations in prostate cancer. We identified somatic alterations of ESRP1, a molecular driver of the disease with a particular value in the pre-operation setting where biomarkers are desperately needed. By integrating DNA methylation and RNA expression data from tumors diagnosed with early-onset, we identified four robust subgroups that readily stratify patients into high and low-risk groups. We combined our cohort of early-onset patients and identified risk-stratification groups to develop a framework to predict the temporal and clinical outcome order of somatic alterations.
Highlights.
Clock-like mutation process attributed to APOBEC3 mediates earliest mutations in PC
Identification of four molecular subgroups that stratifies intermediate-risk disease
Rearrangements at the ESRP1 locus associated with aggressive and proliferative cancer
Development of method to predict clinical trajectories of PC from DNA sequencing data
Gerhauser et al. molecularly characterize prostate cancers diagnosed before 56 years old, which reveals an APOBEC-driving mutational process and identifies an aggressive subgroup with increased expression of ESRP1. They develop a framework to predict the order of somatic alterations and clinical outcome.
Acknowledgements
We thank the patients and families who contributed to this study. The study was funded by the ICGC-EOPC (01KU1001 A) and ICGC-Data Mining (01KU1505A) projects on early-onset prostate cancer by the German Federal Ministry of Education and Research (BMBF). The authors acknowledge the DKFZ, MPI and EMBL Genomics Core Facilities. J.W., F.F. and N.S. were supported by Arvid Nilsson foundation and Rigshospitalets forskningsfond. R.S. and R.T. were supported by Sander Stiftung (#2015.010.1). J.O.K. was, in part, supported by an ERC Starting Grant. C.F. received a stipend by the Helmholtz International Graduate School at the DKFZ. A.U. is supported by the Research Council of Norway (#187615). D.A.G was supported by the US National Institute of Health Intramural Research Program Project Z1AES103266.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Supplemental Information
Supplemental Information include seven figures and four tables (separate).
References
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, and Stratton MR (2015). Clock-like mutational processes in human somatic cells. Nat. Genet. 47, 1402–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Sirota M, and Butte AJ (2015). Systematic pan-cancer analysis of tumour purity. Nat. Commun 6, 8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, Esopi D, Irizarry RA, Getzenberg RH, Nelson WG, et al. (2013). DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci. Transl. Med 5, 169ra10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. (2014). Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510, 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, and Bock C (2014). Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods 11, 1138–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfeld SJ, Urbanucci A, Itkonen HM, Fazli L, Hicks JL, Thiede B, Rennie PS, Yegnasubramanian S, DeMarzo AM, and Mills IG (2017). c-Myc Antagonises the Transcriptional Activity of the Androgen Receptor in Prostate Cancer Affecting Key Gene Networks. EBioMedicine 18, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron DA, and Rowley DR (2012). The reactive stroma microenvironment and prostate cancer progression. Endocr. Relat. Cancer 19, R187–R204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin JM, Lee BH, Matkin L, Taylor MG, Hu B, Xu Y, Magi-Galluzzi C, Klein EA, and Ting AH (2015). Methylome-wide Sequencing Detects DNA Hypermethylation Distinguishing Indolent from Aggressive Prostate Cancer. Cell Rep 13, 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BLUEPRINT consortium (2016). Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat. Biotechnol 34, 726–737. [DOI] [PubMed] [Google Scholar]
- Börno ST, Fischer A, Kerick M, Fälth M, Laible M, Brase JC, Kuner R, Dahl A, Grimm C, Sayanjali B, et al. (2012). Genome-wide DNA methylation events in TMPRSS2-ERG fusion-negative prostate cancers implicate an EZH2-dependent mechanism with miR-26a hypermethylation. Cancer Discov 2, 1024–1035. [DOI] [PubMed] [Google Scholar]
- Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, et al. (2015). Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat. Genet 47, 736–745. [DOI] [PubMed] [Google Scholar]
- Brocks D, Assenov Y, Minner S, Bogatyrova O, Simon R, Koop C, Oakes C, Zucknick M, Lipka DB, Weischenfeldt J, et al. (2014). Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep 8, 798–806. [DOI] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. (2012). Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol 30, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae M, Danko CG, and Kraus WL (2015). groHMM: a computational tool for identifying unannotated and cell type-specific transcription units from global run-on sequencing data. BMC Bioinformatics 16, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, and Gordenin DA (2015). Clusters of Multiple Mutations: Incidence and Molecular Mechanisms. Annu. Rev. Genet 49, 243–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, Kim J, Kwiatkowski DJ, Fargo DC, Mieczkowski PA, et al. (2015). An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet 47, 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li C, Peng X, Zhou Z, Weinstein JN, Cancer Genome Atlas Research Network, and Liang H (2018). A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell 173, 386–399.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, Gao D, Sirota I, Carver BS, Wongvipat J, et al. (2013a). ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat. Med 19, 1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-A, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, and Weksberg R (2013b). Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desper R, and Gascuel O (2002). Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J. Comput. Biol 9, 687–705. [DOI] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, and Aiden EL (2016). Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Systems 3, 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbersdobler A, Hammerer P, Huland H, and Henke RP (1997). Numerical chromosomal aberrations in transition-zone carcinomas of the prostate. J. Urol 158, 1594–1598. [PubMed] [Google Scholar]
- Ernst J, and Kellis M (2012). ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espiritu SMG, Liu LY, Rubanova Y, Bhandari V, Holgersen EM, Szyca LM, Fox NS, Chua MLK, Yamaguchi TN, Heisler LE, et al. (2018). The Evolutionary Landscape of Localized Prostate Cancers Drives Clinical Aggression. Cell 173, 1003–1013.e15. [DOI] [PubMed] [Google Scholar]
- Fagoonee S, Picco G, Orso F, Arrigoni A, Longo DL, Forni M, Scarfò I, Cassenti A, Piva R, Cassoni P, et al. (2017). The RNA-binding protein ESRP1 promotes human colorectal cancer progression. Oncotarget 8, 10007–10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, Szallasi Z, and Eklund AC (2015). Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol 26, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, Shiah Y-J, Yousif F, Lin X, Masella AP, et al. (2017). Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541, 359–364. [DOI] [PubMed] [Google Scholar]
- Henry GH, Loof N, and Strand DW (2017). OMIP-040: Optimized gating of human prostate cellular subpopulations. Cytometry A 91, 1147–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, and Young RA (2013). Super-enhancers in the control of cell identity and disease. Cell 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, and Kelsey KT (2012). DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebschmann D, Gu Z, and Schlesner M (2016). YAPSA: Yet Another Package for Signature Analysis. R Package Version 0. 99 10. [Google Scholar]
- Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S, Motizuki M, Masuyama K, and Miyazawa K (2014). Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J. Biol. Chem 289, 27386–27399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishwaran H, Kogalur UB, Blackstone EH, and Lauer MS (2008). Random survival forests. Ann. Appl. Stat 2, 841–860. [Google Scholar]
- Jeong HM, Han J, Lee SH, Park H-J, Lee HJ, Choi J-S, Lee YM, Choi Y-L, Shin YK, and Kwon MJ (2017). ESRP1 is overexpressed in ovarian cancer and promotes switching from mesenchymal to epithelial phenotype in ovarian cancer cells. Oncogenesis 6, e389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhun MA, Geybels MS, Wright JL, Kolb S, April C, Bibikova M, Ostrander EA, Fan J-B, Feng Z, and Stanford JL (2017). Gene expression signature of Gleason score is associated with prostate cancer outcomes in a radical prostatectomy cohort. Oncotarget 8, 43035–43047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Qiu Y, Minn AJ, and Zhang NR (2016). Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A 113, E5528–E5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluth M, Hesse J, Heinl A, Krohn A, Steurer S, Sirma H, Simon R, Mayer P-S, Schumacher U, Grupp K, et al. (2013). Genomic deletion of MAP3K7 at 6q12–22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod. Pathol 26, 975–983. [DOI] [PubMed] [Google Scholar]
- Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, and Kallioniemi OP (1998). Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med 4, 844–847. [DOI] [PubMed] [Google Scholar]
- Kron KJ, Murison A, Zhou S, Huang V, Yamaguchi TN, Shiah Y-J, Fraser M, van der Kwast T, Boutros PC, Bristow RG, et al. (2017). TMPRSS2-ERG fusion co-opts master transcription factors and activates NOTCH signaling in primary prostate cancer. Nat. Genet 49, 1336–1345. [DOI] [PubMed] [Google Scholar]
- Lecca P, Casiraghi N, and Demichelis F (2015). Defining order and timing of mutations during cancer progression: the TO-DAG probabilistic graphical model. Front. Genet 6, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM
- Locke JA, Zafarana G, Ishkanian AS, Milosevic M, Thoms J, Have CL, Malloff CA, Lam WL, Squire JA, Pintilie M, et al. (2012). NKX3.1 haploinsufficiency is prognostic for prostate cancer relapse following surgery or image-guided radiotherapy. Clin. Cancer Res 18, 308–316. [DOI] [PubMed] [Google Scholar]
- Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, Warren A, Scott H, Madhu B, Sharma N, et al. (2011). The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J 30, 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middlebrooks CD, Banday AR, Matsuda K, Udquim K-I, Onabajo OO, Paquin A, Figueroa JD, Zhu B, Koutros S, Kubo M, et al. (2016). Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat. Genet 48, 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, and Wold B (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Myers A, du Souich C, Yang CL, Borovik L, Mwenifumbo J, Rupps R, Study C, Lehman A, and Boerkoel CF (2017). FOXP1 haploinsufficiency: Phenotypes beyond behavior and intellectual disability? Am. J. Med. Genet. A 173, 3172–3181. [DOI] [PubMed] [Google Scholar]
- Na R, Zheng SL, Han M, Yu H, Jiang D, Shah S, Ewing CM, Zhang L, Novakovic K, Petkewicz J, et al. (2017). Germline Mutations in ATM and BRCA1/2 Distinguish Risk for Lethal and Indolent Prostate Cancer and are Associated with Early Age at Death. Eur. Urol 71, 740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. (2012). Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, et al. (2016). Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pageaud Y, Plass C, and Assenov Y (2018). Enrichment analysis with EpiAnnotator. Bioinformatics [DOI] [PMC free article] [PubMed]
- Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, Han B, Cao Q, Cao X, Suleman K, et al. (2010). Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat. Med 16, 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi S, Cozzuto L, Tarantino C, Passaro F, Ciriello S, Aloia L, Antonini D, De Simone V, Pastore L, and Russo T (2010). Direct targets of Klf5 transcription factor contribute to the maintenance of mouse embryonic stem cell undifferentiated state. BMC Biol 8, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planche A, Bacac M, Provero P, Fusco C, Delorenzi M, Stehle J-C, and Stamenkovic I (2011). Identification of prognostic molecular features in the reactive stroma of human breast and prostate cancer. PLoS One 6, e18640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, and Walsh PC (1999). Natural history of progression after PSA elevation following radical prostatectomy. JAMA 281, 1591–1597. [DOI] [PubMed] [Google Scholar]
- Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, Garofalo A, Gulati R, Carreira S, Eeles R, et al. (2016). Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl. J. Med 375, 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazzotti D, Caravagna G, Olde Loohuis L, Graudenzi A, Korsunsky I, Mauri G, Antoniotti M, and Mishra B (2015). CAPRI: efficient inference of cancer progression models from cross-sectional data. Bioinformatics 31, 3016–3026. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, and Korbel JO (2012). DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, and Gordenin DA (2014). Hypermutation in human cancer genomes: footprints and mechanisms. Nat. Rev. Cancer 14, 786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al. (2012). Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell 46, 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. (2013). An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet 45, 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, and Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette A, Boufaied N, Scarlata E, Hamel L, Brimo F, Whitaker HC, Ramos-Montoya A, Neal DE, Dragomir A, Aprikian A, et al. (2017). Asporin is a stromally expressed marker associated with prostate cancer progression. Br. J. Cancer 116, 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R, Maron-Katz A, and Shamir R (2003). CLICK and EXPANDER: a system for clustering and visualizing gene expression data. Bioinformatics 19, 1787–1799. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Neo S-Y, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, and Liu ET (2003). Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. U. S. A 100, 10393–10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand DW, and Goldstein AS (2015). The many ways to make a luminal cell and a prostate cancer cell. Endocr. Relat. Cancer 22, T187–T197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart RO, Wachsman W, Berry CC, Wang-Rodriguez J, Wasserman L, Klacansky I, Masys D, Arden K, Goodison S, McClelland M, et al. (2004). In silico dissection of cell-type-associated patterns of gene expression in prostate cancer. Proc. Natl. Acad. Sci. U. S. A 101, 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C, McGranahan N, Starrett GJ, and Harris RS (2015). APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov 5, 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, Statham AL, Kelly TK, Clark SJ, and Jones PA (2014). Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res 24, 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborero D, Rubio-Perez C, Deu-Pons J, Schroeder MP, Vivancos A, Rovira A, Tusquets I, Albanell J, Rodon J, Tabernero J, et al. (2018). Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med 10, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. (2010). Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA (2015). The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, and Zheng SC (2017). Cell-type deconvolution in epigenome-wide association studies: a review and recommendations. Epigenomics 9, 757–768. [DOI] [PubMed] [Google Scholar]
- Therneau TM, and Grambsch PM (2000). Expected Survival. In Modeling Survival Data: Extending the Cox Model, Therneau TM, and Grambsch PM, eds. (New York, NY: Springer New York; ), pp. 261–287. [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, Varambally S, Cao X, Tchinda J, Kuefer R, et al. (2005). Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648. [DOI] [PubMed] [Google Scholar]
- Urbanucci A, Barfeld SJ, Kytölä V, Itkonen HM, Coleman IM, Vodák D, Sjöblom L, Sheng X, Tolonen T, Minner S, et al. (2017). Androgen Receptor Deregulation Drives Bromodomain-Mediated Chromatin Alterations in Prostate Cancer. Cell Rep 19, 2045–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. (2011). Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, and Korbel JO (2017). Genomes of early onset prostate cancer. Curr. Opin. Urol 27, 481–487. [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz HJ, Stehr H, Rausch T, et al. (2013). Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 23, 159–170. [DOI] [PubMed] [Google Scholar]
- Wise AM, Stamey TA, McNeal JE, and Clayton JL (2002). Morphologic and clinical significance of multifocal prostate cancers in radical prostatectomy specimens. Urology 60, 264–269. [DOI] [PubMed] [Google Scholar]
- Xing C, Ci X, Sun X, Fu X, Zhang Z, Dong EN, Hao Z-Z, and Dong J-T (2014). Klf5 Deletion Promotes Pten Deletion–Initiated Luminal-Type Mouse Prostate Tumors through Multiple Oncogenic Signaling Pathways. Neoplasia 16, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, et al. (2013). Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154, 801–813. [DOI] [PubMed] [Google Scholar]
- Yu J, Yu J, Mani R-S, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, et al. (2010). An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 17, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. ESRP1 IHC and APOBEC-associated SVs, Related to Figure 2
Table S1. Clinico-pathological data, Related to Figure 1
Table S3. Methylation-inferred ct composition, Related to Figure 5
Table S4. CC analysis, Related to Figure 6
Data Availability Statement
The raw data for WGS, RNA-seq expression and array-based methylation were submitted to EGA under study identifier EGAS00001002923. Somatic variant calls are available through Mendeley Data doi:10.17632/6gtrrxrn2c.1. Oncological outcome data were collected via the Progether PROMś (patient reported outcome measurement) interface (www.progether.com/proms) and the martini-clinic database.
Softwares used for each analysis are described and referenced in Methods Detail subsection and listed in KEY RESOURCE TABLE.


