Abstract
The aim of this study was to investigate the role of high mobility group protein-1 (HMGB1) in the proliferation and migration of lung cancer cells. CCK-8 assays and colony formation assays were used to evaluate the effect of HMGB1 regulation on cancer cell viability and colony formation. Trans-well assays and wound healing assays were also performed. Our data showed that HMGB1 is upregulated in clinical lung cancer tissues compared with non-cancer tissues, and it is differentially expressed in lung cancer cell lines. The knockdown of HMGB1 in A549 lung cancer cells significantly reduced cell proliferation, viability and motility. In contrast, overexpression of HMGB1 in lung cancer H1299 cells significantly increased cell viability and motility. Western blotting showed that HMGB1 could promote epithelial-mesenchymal transition. The Wnt/β-catenin pathway was activated after overexpression of HMGB1 in H1299 cells, while it was inactivated by knocking down HMGB1 in A549 cells. These data suggest that HMGB1 promotes the proliferation and migration of lung cancer cells in vitro. The carcinogenic behavior of HMGB1 can be achieved by activating the Wnt/β-catenin pathway.
Keywords: HMGB1, proliferation, migration, lung cancer, Wnt/β-catenin
Introduction
Lung cancer is the most common primary malignant tumor of the lung.1 Lung cancer can often metastasize to the liver, brain, lung, skeletal system, adrenal gland, pancreas and other organs.2 When cancer spreads to the brain, cerebral palsy can occur due to tissue damage, and life can be endangered.3 Lung cancer does not have any unusual symptoms in the early stage. It has symptoms that are common for respiratory diseases, such as cough, blood in the sputum, low fever, chest pain, and nausea.4 Late-stage lung cancer can have facial, neck edema, hoarseness, shortness of breath, etc. Approximately 70% to 80% of lung cancer patients in clinical practice are in the middle and late stages of clinical treatment.5 Despite the advancement in medical technology, surgical techniques and chemotherapy for advanced lung cancer, treatment is still limited due to a range of adverse reactions and toxicity as well as drug resistance.6 Therefore, the prognosis of lung cancer patients is still very poor, and new treatment strategies and effective therapeutic drugs are urgently needed in clinical practice.
High mobility group box 1 (HMGB1) is a highly conserved nucleoprotein that is widely distributed in mammalian cells.7 HMGB1 is expressed in the heart, lung, kidney and other tissues.8 It is involved in gene transcription regulation after binding to DNA and can maintain nucleosome structure stability, participate in tissue regeneration and participate in tumor migration.9 In liver and brain tissues, HMGB1 is localized outside of the cell and is mainly involved in the body’s inflammatory response. Studies have shown that serum HMGB1 is highly expressed in various kinds of leukemia.10 HMGB1 plays a role in the pathogenesis of inflammation, trauma, immune system diseases, malignant tumors, etc. For example, HMGB1 is positively expressed in a variety of malignant tumor tissues, is associated with tumor invasion and migration and is a potential biomarker for identifying tumors.11 These positive reports have drawn our interest and prompted us to study the role of HMGB1 in lung cancer. The aim of this research was to study the role of HMGB1 in the proliferation and migration of lung cancer cells. Since HMGB1 was found in lung cancer tissues, 30 clinical lung cancer tissues were collected to verify the expression of HMGB1. Thereafter, the possible mechanism of HMGB1 in the progression of lung cancer was systematically studied in vitro. This report provides a large amount of data indicating that HMGB1 may be a key mediator of proliferation and migration of lung cancer cells, and the treatment of HMGB1 is promising for the clinical treatment of lung cancer.
Methods
Human Samples
Thirty patients with clinically diagnosed lung cancer underwent traditional surgery at the Third Affiliated Hospital of Suzhou University. Before the operation, the patients did not receive chemotherapy or radiotherapy. Cancerous and adjacent non-cancerous tissues in each case were obtained with the patient’s consent. The protocol was approved by the Ethics Committee of the Third Affiliated Hospital of Suzhou University (No: SU20190231).
Cell Culture and Transfection
Human lung cancer cell lines, A549, H1650, H358, H1299 and HCC827 (ATCC, MA), were incubated in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). shHMGB1 and shControl were designed and synthesized by Kehao (Xi’an, China). Cells were inoculated into 24-well plates with a density of 1 x 105/well, and the cell density was approximately 50% at the time of transfection. For transfection, a mixture of 1 ml Lipofectamine 2000 (USA) and 50 ml Opti-MEM I was added to serum-free medium (USA) and placed at room temperature for 10 minutes. Serum-free medium was added to a mixture of 2 ml FAM-siRNA and 50 ml Opti-MEM I. After incubating at room temperature for 5 minutes, diluted FAM-siRNA and Lipofectamine 2000 were mixed and incubated for 20 minutes. Then, the cells were incubated with the mix at 37°C and 5% CO2 for 6 hours and replaced with conventional medium.
qRT-PCR
Trizol reagent (1 ml) was added to the cells to extract total RNA. Reverse transcription of RNA into DNA was performed in accordance with the instructions of the PrimeScriptTM RT kit (TaKaRa, Dalian, China). Quantitative RT-PCR was performed using a CFX96-Real-Time System. The PCR procedure was as follows: 40 cycles for 5 seconds at 95°C, denaturation for 10 seconds at 95°C, annealing for 20 seconds at 60°C and elongation for 15 seconds at 72°C. Three replicates were tested for each sample. The relative expression levels of mRNAs were calculated using 2-ΔΔCt with GAPDH as a reference standard (Table 1).
Table 1.
The Sequences of Primers Used in RT-PCR.
| Gene | Primers sequences (5’-3’) | |
|---|---|---|
| HMGB1 | Forward: | TGTCGGGAGGAGCATAAGAACG |
| Reverse: | GGGCGATACTCAGAGCAGAAGC | |
| c-myc | Forward: | TGTCCGTAGAGGAGCATAAGTAC |
| Reverse: | GGGCGATATGCAGAGCAGAAGA | |
| cyclinD1 | Forward: | TGTGACTGAACCCGGAGCATAAG |
| Reverse: | GGGCGATACTCAGAGCAACCGTA | |
| axin | Forward: | TGTCGGGAGGAGCATAAGGACTGA |
| Reverse: | GGGCGATTAGCAACGCAGAGCAGAA | |
| GSK3β | Forward: | TGTCGGGTGACCAGTGCATAAGA |
| Reverse: | GGGCGAAGCTGACGTAGCAGAA | |
| APC | Forward: | TGTCGGATGCATGACAGCATAAG |
| Reverse: | AGGGCGATACTGCAAGCAGAA | |
| GAPDH | Forward: | ACCCAGAAGACTGTGGATGG |
| Reverse: | CCACCCTGTTGCTGTAGCCTA | |
Immunohistochemistry Analysis
Formalin-fixed, paraffin-embedded clinical tumor tissues underwent immunohistochemical analysis. In short, each 4-micron thick slide was deparaffinized, hydrated, treated for antigen recovery, washed with phosphate buffer saline (PBS), and incubated overnight with the primary antibody at 4°C. Then, at room temperature, the slides were washed in PBS and incubated with the secondary antibody (Santa Cruz Biotechnology, Dallas, TX) containing horseradish peroxide for 1 hour. The slide was then stained with 3,3-diaminobenzidine (FASTDAB tablet; Sigma-Aldrich, St. Louis, MO).
Western Blotting
The cells were washed 3 times with cold PBS. Cells were lysed on ice for 30 minutes, and the lysate was centrifuged at 4°C for 3500 rpm for 3 hours; the supernatant contained the total protein. Protein concentration was measured by a bicinchoninic acid (BCA) kit (Sigma). The sample was then mixed with loading buffer and boiled at 100°C for 5 minutes, and then it was run on an SDS gel for separation. The gel separated protein was transferred to a PVDF membrane, and a primary antibody was incubated with the membrane overnight at 4°C. The next day, the skimmed milk buffer was washed away for 1 hour, and the labeled secondary antibody was then incubated with the membrane for 1 hour. Then, the secondary antibody was washed off to reduce anti-luminescence. Electroluminescence (ECL) was used for detection, and an associated computerized image analysis program (USA) was used to quantify the relevant signals by optical density measurement.
Cell Proliferation Assay (CCK-8)
Cells in logarithmic growth phase were inoculated into 12-well plates with a cell density of 5 x 104 cells/mL. After 48 hours of successful transfection, the cells were cultured for 1-5 days, and then 20 ml CCK-8 reagent was added into each well. A Synergy H4 Hybrid Enzyme Marker (Bio Tek, Winooski, VT) was used to measure the signal at 450 nm.
Colony Formation Assay
After successful transfection, 200 cells were inoculated per well of 6-well plates and were grown for 15 days. Cells were fixed in 10% paraformaldehyde for 10 minutes and stained with 0.1% crystal violet for 15 minutes. Finally, the number of clones was observed under a microscope.
Cell Migration and Invasion Assay
After transfection, 600 ml DMEM containing 10% FBS was added to the lower chamber of the transwell, and 300 ml serum-free cell mixture was added into the upper chamber. Invasion requires pre-coating with Matrigel. The cells were incubated for 48 hours at 37°C and 5% CO2. The cells were immobilized in 75% ethanol and counted by crystal violet staining.
Statistical Analysis
The data were analyzed using GraphPad Prism 8 software. Unmatched Student’s t-tests were used to compare the data between 2 groups. Multiple comparisons were made using one-way ANOVA with Bonferroni correction. In vitro experiments were repeated at least 3 times, and the data were expressed as the mean +standard deviation (SD) from at least 3 independent experiments. For all analyses, P < 0.05 is considered significant.
Results
HMGB1 Is Upregulated in Clinical Lung Cancer Tissues and Is Differentially Expressed in Lung Cancer Cell Lines
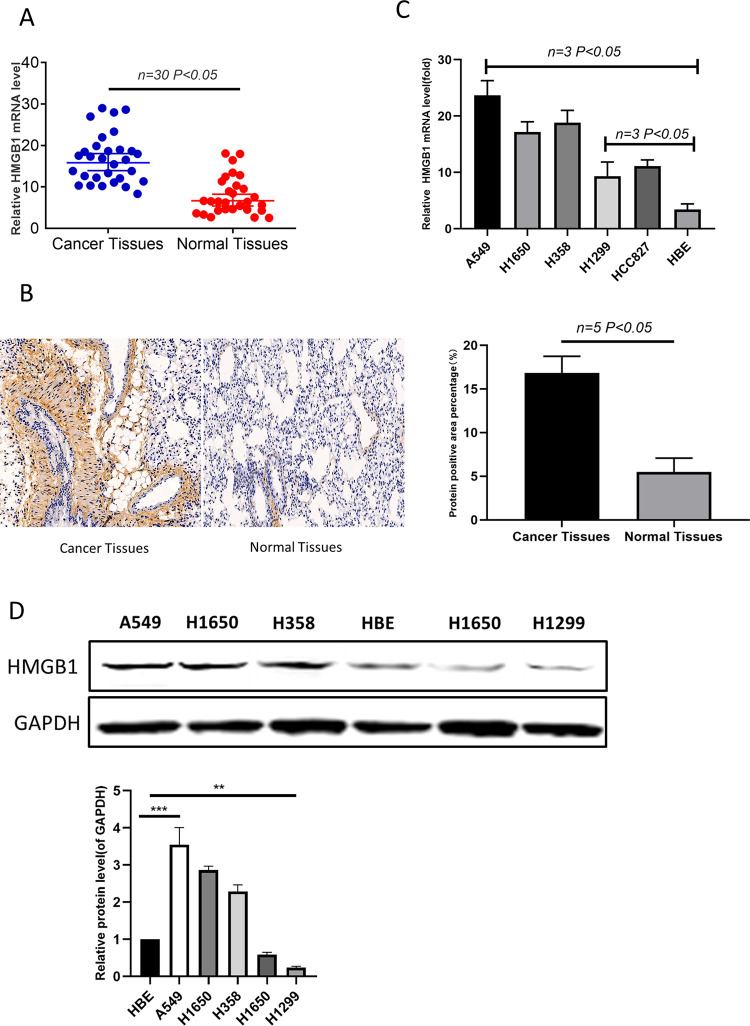
Initially, HMGB1 transcription levels were examined in 64 cases of lung cancer. The results showed that the average level of HMGB1 in cancer tissues was higher than it was in adjacent non-cancerous normal tissues (Figure 1A). Immunohistochemical staining of HMGB1 in cancer tissues was significantly stronger than that in adjacent normal tissues (Figure 1B). These results suggest that HMGB1 is upregulated in lung cancer tissues. There were statistically significant differences in the expression of HMGB1 among patients with different TNM stages and tumor size. There were statistically significant differences in the expression of HMGB1 between patients with different TNM stages and tumor size (Table 2). In addition, HMGB1 was more highly expressed in the A549, H358 and H1650 cell lines compared to the H1299 and HCC827 cell lines (Figure 1C and D). The results showed that there was a difference in the expression of HMGB1 in lung cancer cells. We selected A549 and H1299 as subjects for subsequent studies.
Figure 1.
HMGB1 is upregulated in clinical lung cancer tissues, and it is differentially expressed in lung cancer cell lines. (A) The relative mRNA levels of HMGB1 in clinical lung cancer tissues and in the adjacent noncancerous normal tissues; n = 25. (B) HMGB1 protein levels in clinical lung cancer tissues and the adjacent non-cancerous normal tissues; n = 5. (C) The mRNA expression of HMGB1 in a series of lung cancer cell lines was detected by qRT-PCR. (D) Protein levels of HMGB1 in a series of lung cancer cell lines were examined by western blotting.
Table 2.
The Correlation of the Expression of HMGB1and Clinicopathological of the Lung Cancer.
| Characteristics | Number of cases (n = 64) | HMGB1 | x 2 | p |
|---|---|---|---|---|
| Tumor size, cm | ||||
| ≤4 | 37 | 0.124 | 0.301 | 0.032 |
| >4 | 27 | 0.876 | 0.786 | 0.000 |
| pTNM category | ||||
| I/II | 36 | 0.213 | 0.104 | 0.289 |
| III | 28 | 0.764 | 0.643 | 0.000 |
HMGB1 Promotes Lung Cancer Cell Viability In Vitro
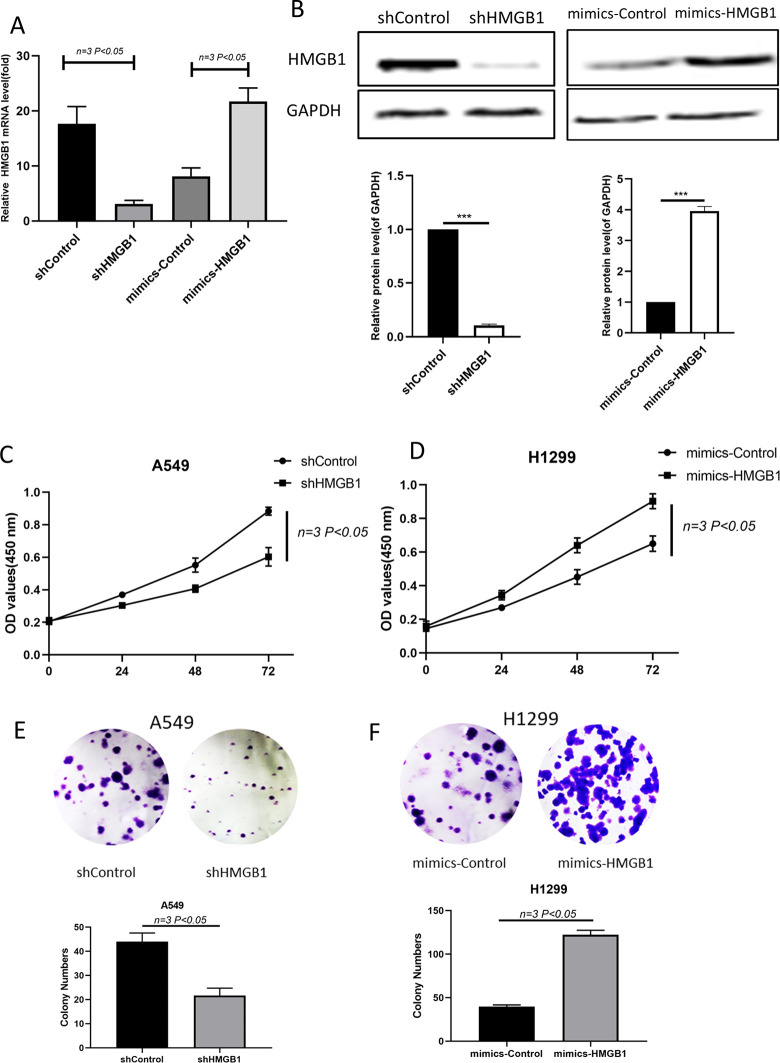
Considering the expression of HMGB1 in lung cancer cell lines, we used a specific shRNA (shHMGB1) targeting HMGB1 to knock down the expression of HMGB1, and we used an expression plasmid with HMGB1 (mimic-HMGB1) to increase the level of HMGB1 in H1299 cells. As shown in Figure 2A and B, shHMGB1 decreased HMGB1 significantly, while the overexpression plasmid increased the expression of HMGB1. Next, we measured cell viability, and it was observed that the knockdown of HMGB1 significantly reduced the rate of cell proliferation in A549 cells (Figure 2C). In contrast, the upregulation of HMGB1 increased cell proliferation in H1299 cells (Figure 2D). In addition, colony formation was measured in 15-day cultures. HMGB1-depleted cells formed only approximately 25 colonies, whereas HMGB1-overexpressing H1299 cells formed more than 121 colonies compared with 52 and 38 colonies in the control A549 and H1299 cells (Figure 2E-F). These observations suggest that HMGB1 promotes cell viability in lung cancer.
Figure 2.
HMGB1 promotes lung cancer cell viability in vitro. (A) Detection of HMGB1 expression at the mRNA level by qRT-PCR after cells were transfected with shHMGB1 and mimic-HMGB1. (B) Western blots detected HMGB1 expression at the protein level after shHMGB1- and mimic-HMGB1-transfected cells. (C–D) CCK-8 assays were performed to assess how HMGB1 knockdown in A549 cells and HMGB1 overexpression in H1299 cells effected cell viability. (E–F) During the 15-day colony formation assay, colony staining was used to visualize and quantify colony formation, and the results are displayed on the column.
HMGB1 Promotes Cell Migration in Lung Cancer Cell Lines
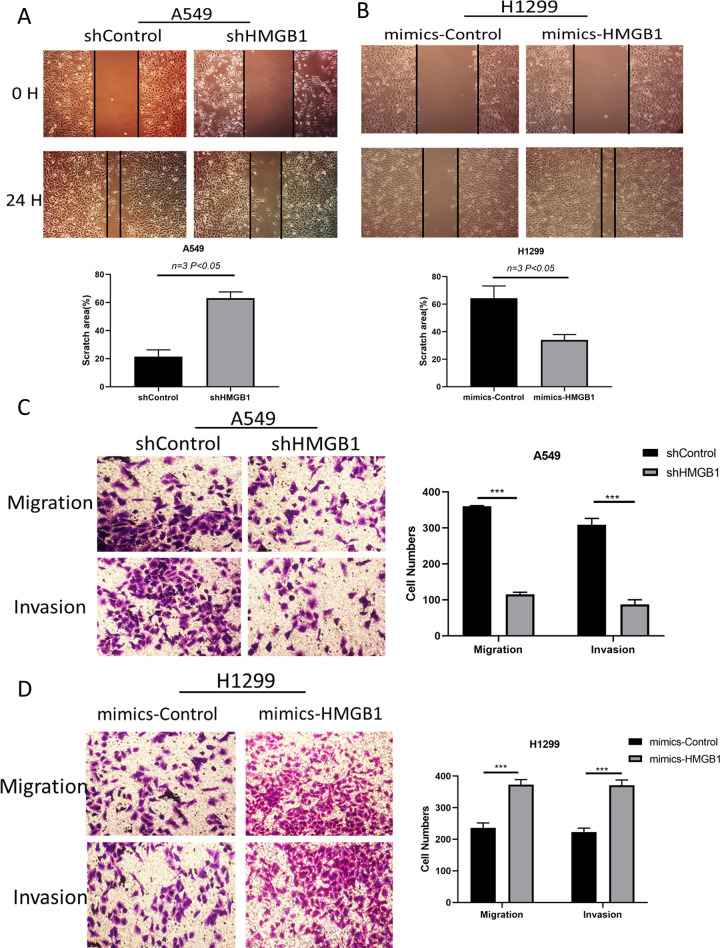
After knockdown of HMGB1, the wound healing rate in A549 cells was inhibited by 65% (Figure 3A). Although HMGB1 overexpression promoted wound healing in H1299 cells, it increased by 45% after HMGB1 was overexpressed in H1299 cells (Figure 3B). Further, trans-well measurements were carried out to verify the above phenomena. In A549 cells, when HMGB1 was depleted, only approximately 120 cells migrated, which was in contrast to more than 360 cells that migrated in the control group. These observations support the fact that knockdown of HMGB1 inhibits cell migration. In addition, the invasion of A549 cells was significantly reduced by up to 70% (Figure 3C). Conversely, when HMGB1 was overexpressed in H1299 cells, both migration and invasion of H1299 cells increased significantly (Figure 3D).
Figure 3.
HMGB1 promotes cell migration in lung cancer cell lines. (A-B) Wound healing was measured with A549 and H1299 cells. ShHMGB1 was transfected into A549 cells, and the HMGB1 plasmid was transfected into H1299 cells before testing. representational images were taken 24 hours after serum-free migration. (C-D) Transwell migration and invasion assays were performed. *P < 0.05; **P < 0.01.
HMGB1 Activates the Wnt/β-Catenin Signaling Pathway in Lung Cancer Cells
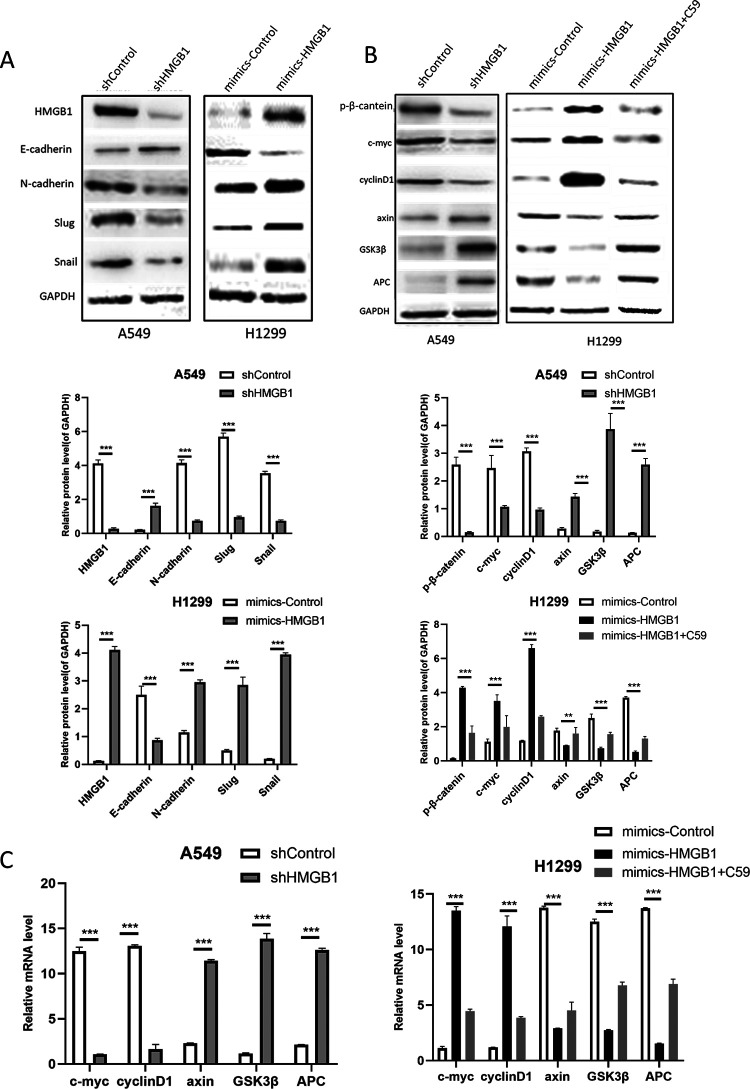
When HMGB1 was depleted in A549 cells, E-cadherin, an epithelial biomarker, was upregulated, while Snail, Slug and N-cadherin, which are mesenchymal biomarkers, were downregulated. In H1299 cells, the opposite expression pattern was observed for the biomarkers (Figure 4A). These data consistently indicate that HMGB1 promotes cell migration and invasion through epithelial-mesenchymal transition (EMT). To explore the possible mechanism of the biological activity of HMGB1 in lung cancer, Western blot analysis was performed on A549 cells and H1299 cells. The Wnt signaling pathway induces EMT conversion by inhibiting glycogen synthase kinase-3β (GSK3β)-mediated phosphorylation and inhibiting β-catenin degradation in the cytoplasm. Consistently, knockdown of HMGB1 in A549 cells downregulated the expression of Wnt/β-catenin factors such as c-myc, cyclinD1, and β-catenin and upregulated axin, GSK3β and APC. After overexpression of HMGB1 in H1299 cells, the expression of these factors was reversed, at the same time, we added the WNT pathway inhibitor C59 to observe the changes of downstream signaling molecules, and found that downstream signaling molecules recovered and decreased accordingly (Figure 4B, C). These data suggest that HMGB1 activates the Wnt/β-catenin signaling pathway in lung cancer cell lines.
Figure 4.
HMGB1 activates the Wnt/β-catenin pathway in lung cancer A549 and H1299 cells. (A) Western blotting was used to analyze the major biomarkers of EMT in HMGB1-knockdown A549 cells and HMGB1-overexpressing H1299 cells. (B) Western blot analysis was performed to detect factors involved in Wnt/β-catenin signaling in HMGB1-depleted A549 cells and HMGB1-overexpressing H1299 cells. (C) qRT-PCR was performed to detect factors involved in Wnt/β-catenin signaling in HMGB1-depleted A549 cells and HMGB1-overexpressing H1299 cells.
Discussion
Lung cancer is one of the most common and fatal malignant tumors in the world.12 Improvements in diagnosis and treatment have greatly prolonged the survival of lung cancer patients.2 However, most patients relapse within 5 years, and treatment options are limited.13,14 Accurate molecular characterization of abnormal gene expression involved in the development and progression of lung cancer is essential for identifying novel molecular targets of non-small cell lung cancer (NSCLC), which may improve future clinical outcomes.15
HMGB1 is a highly conserved nuclear protein in cells, and it is generally considered to be a chromatin binding factor that specifically binds to DNA to promote transcription of tumor-associated proteins.16 HMGB1 plays a positive role in a variety of malignant tumor behaviors.10,17 It has been suggested that HMGB1 expression is abnormally high in various tumor tissues.
This research initially confirmed that HMGB1 expression was upregulated in lung cancer tissues. Because HMGB1 is highly expressed in A549 cells, shHMGB1 was used to eliminate the expression of HMGB1. The lowest expression of HMGB1 was found in H1299 cells, so we used expression plasmids to upregulate the expression of HMGB1 in HGB99 cells. Subsequently, in CCK-8 assays, HMGB1 knockdown inhibited proliferation, while HMGB1 overexpression promoted cell proliferation. This effect was further confirmed by anchoring independent colony formation measurements. Cell migration and invasion are also controlled by HMGB1. Reduction of HMGB1 inhibits the wound healing process. The observation of migration and invasion ability shows that the overexpression of HMGB1 reverses the above inhibition and induces EMT. In conclusion, this research identified HMGB1 as an important regulatory protein for the proliferation and migration of lung cancer cells.
An interesting finding was that HMGB1 activates the Wnt/β-catenin pathway. Previous evidence has shown that Wnt/β-catenin plays an important role in many pathophysiological events, including the occurrence and development of tumors. Activated c-myc, cyclin D1 and β-catenin regulate cell growth and survival. Our research found that HMGB1 was a key mediator of the proliferation and migration of lung cancer cells. The malignant behavior of HMGB1 in lung cancer can be achieved by activating the Wnt/β-catenin pathway. In addition, the activation of the Wnt/β-catenin pathway is closely related to tumor angiogenesis in lung cancer. Therefore, the role of HMGB1 in angiogenesis should be further studied in lung cancer to determine whether HMGB1 can induce angiogenesis and promote migration of lung cancer.
Footnotes
Authors’ Note: Thirty patients with clinically diagnosed lung cancer underwent traditional surgery at the Third Affiliated Hospital of Suzhou University. Before the operation, the patient did not receive chemotherapy or radiotherapy. Cancerous and adjacent non-cancerous tissues in each case were obtained with patient’s consent. The plan was approved by the Ethics Committee of the Third Affiliated Hospital of Suzhou University (No: SU20190231).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Scientific Research Fund of China National Natural Science Foundation (NO.08653125)
ORCID iD: Xiao-peng Xu  https://orcid.org/0000-0002-8902-6332
https://orcid.org/0000-0002-8902-6332
References
- 1. Lin X, Bloom M S, Du Z, Hayo Y. Trends in disability-adjusted life years of lung cancer among women from 2004 to 2030 in Guangzhou, China: a population-based study. Cancer Epidemiol. 2019;63(21):101586. [DOI] [PubMed] [Google Scholar]
- 2. Luu T, Frankel P, Beumer J H, et al. Phase I trial of belinostat in combination with 13-cis-retinoic acid in advanced solid tumor malignancies: a California Cancer Consortium NCI/CTEP sponsored trial. Cancer Chemother Pharmacol. 2019;84(6):1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jurmeister P, Bockmayr M, Seegerer P, et al. Machine learning analysis of DNA methylation profiles distinguishes primary lung squamous cell carcinomas from head and neck metastases. Sci Transl Med. 2019;1 1(509):1453–1464. [DOI] [PubMed] [Google Scholar]
- 4. Kim YJ, Baek DS, Lee S, et al. Dual-targeting of EGFR and neuropilin-1 attenuates resistance to EGFR-targeted antibody therapy in KRAS-mutant non-small cell lung cancer. Cancer Lett. 2019;53(31):535–546. [DOI] [PubMed] [Google Scholar]
- 5. Janssen S, Mehta P, Bartscht T, et al. Prevalence of metastases within the hypothalamic-pituitary area in patients with brain metastases. Radiat Onco l 2019;1 4(1):152–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Levy A, Hendriks L, Le Pechoux C, et al. Current management of limited-stage SCLC and CONVERT trial impact: results of the EORTC Lung Cancer Group survey. Lung Cancer. 2019;136(24):145–147. [DOI] [PubMed] [Google Scholar]
- 7. Das S, Mishra K P, Chanda S, et al. CXCR7: A key neuroprotective molecule against alarmin HMGB1 mediated CNS pathophysiology and subsequent memory impairment. Brain Behav Immun. 2019;72(19):287–298. [DOI] [PubMed] [Google Scholar]
- 8. Xie ZY, Wang FF, Xiao ZH, et al. Long noncoding RNA XIST enhances ethanol-induced hepatic stellate cells autophagy and activation via miR-29b/HMGB1 axis. IUBMB Life. 2019;62(31):1275–1289. [DOI] [PubMed] [Google Scholar]
- 9. Amato J, Cerofolini L, Brancaccio D, et al. Insights into telomeric G-quadruplex DNA recognition by HMGB1 protein. Nucleic Acids Res. 2019;74(41):531–544.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang L, Chai W, Ye F, et al. HMGB1 promotes differentiation syndrome by inducing hyperinflammation via MEK/ERK signaling in acute promyelocytic leukemia cells. Oncotarget. 2017;8(16):27314–27327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Jiang Z, Yan J, et al. HMGB1 as a potential biomarker and therapeutic target for malignant mesothelioma. Dis Markers. 2019;86(64):4183157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohen JT, Lin PJ, Sheinson DM, et al. Are national comprehensive cancer network evidence block affordability ratings representative of real-world costs? An evaluation of advanced non-small-cell lung cancer. J Oncol Pract. 2019;7415(11):e948–e956. [DOI] [PubMed] [Google Scholar]
- 13. Nakada T, Noda Y, Kato D, et al. Risk factors and cancer recurrence associated with postoperative complications after thoracoscopic lobectomy for clinical stage I non-small cell lung cancer. Thorac Cancer. 2019,10(10):1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhong Y, Jiang L, Lin H, et al. Overexpression of KIF18A promotes cell proliferation, inhibits apoptosis, and independently predicts unfavorable prognosis in lung adenocarcinoma. IUBMB Life. 2019, 71(7):942–955. [DOI] [PubMed] [Google Scholar]
- 15. Wang S, Du S, Lv Y, et al. MicroRNA-665 inhibits the oncogenicity of retinoblastoma by directly targeting high-mobility group box 1 and inactivating the Wnt/β-catenin pathway. Cancer Manag Res. 2019. 11(11):3111–3123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Rizk NI, Sallam AM, El-Ansary AR, et al. HMGB1 and SEPP1 as predictors of hepatocellular carcinoma in patients with viral C hepatitis: Effect of DAAs. Clin Biochem. 2019;70(53):8–13. [DOI] [PubMed] [Google Scholar]
- 17. Liu X, Zhou M, Wu J, et al. HMGB1 release from trophoblasts contributes to inflammation during Brucella melitensis infection. Cell Microbiol. 2019;21(10):e13080. [DOI] [PubMed] [Google Scholar]