Abstract
The Simons Foundation Autism Research Initiative (SFARI) has launched SPARKForAutism.org, a dynamic platform that is engaging thousands of individuals with autism spectrum disorder (ASD) and connecting them to researchers. By making all data accessible, SPARK seeks to increase our understanding of ASD and accelerate new supports and treatments for ASD.
Rationale
Autism spectrum disorder (ASD) is a common condition affecting 1 in 68 individuals and is behaviorally defined by difficulties with communication and social interaction and by restrictive interests and repetitive behaviors. Nonetheless, there is significant heterogeneity among individuals in severity, symptoms, and associated co-morbidities. The costs of supporting an individual with ASD are between $1.4 million and $2.4 million per person over a lifetime. The causes of ASD and the cellular mechanisms leading to ASD are incompletely understood. Findings from clinical studies that have attempted to understand the brain and behavior in ASD are hampered by a lack of reproducibility. Major challenges for replication are the heterogeneity of ASD and the difficulty in recruiting large numbers of participants for these studies. These challenges have limited the development of effective diagnostic methods and treatments for this condition, and there are currently no approved medications that treat the core symptoms of ASD. Many of the research challenges in ASD are shared with other neurodevelopmental or neuropsychiatric disorders.
Recent studies suggest an important role for genetic factors in ASD risk, estimating ASD heritability to be as high as 0.83 (Sandin et al., 2017). The remaining variance is likely explained by environmental factors or gene-environment interactions. However, our understanding of environmental risk factors and how they interact with genetic risk factors is limited.
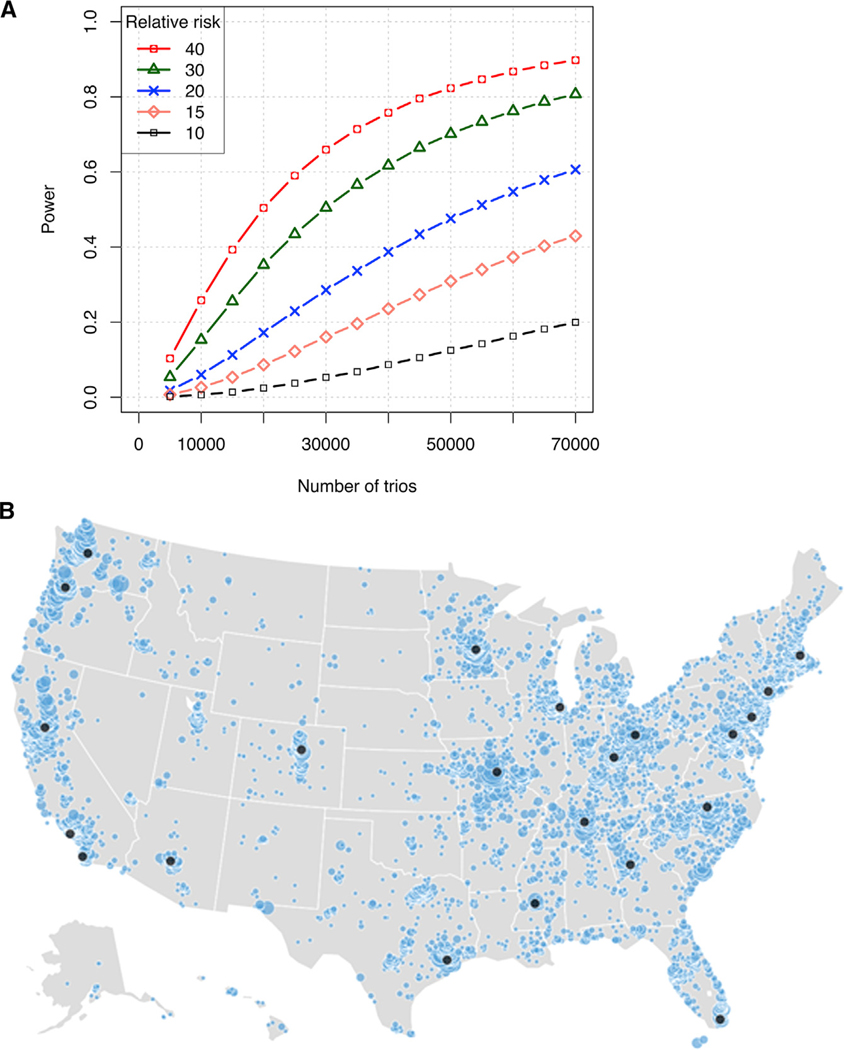
Until recently, the genetic architecture of ASD was largely unknown; however, there are now ~90 genes with rare and mostly de novo (i.e., new in child and not present in either parent) variants of large effect (Sanders et al., 2015). These genetic discoveries were made possible by whole-exome sequencing (WES) of several ASD cohorts, including the Simons Simplex Collection (SSC), a cohort that was also funded by the Simons Foundation Autism Research Initiative (SFARI). These studies also suggested that there are approximately 400–1,000 genes and loci that contribute to ASD risk through de novo variants with large effect size, leaving many ASD risk genes yet to be identified. Inherited, rare, moderate risk variants and common variants of individually small effect also contribute risk to ASD (Gaugler et al., 2014). Large sample sizes are needed to identify common variants of small effect, and to date, only a single genome-wide significant locus of small effect at 10q24.32 has been identified in ASD (Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium, 2017). Based on the analytical framework previously described (Samocha et al., 2014), we estimate that the identification of most of the additional genetic risk factors for ASD (with relatively large effect) will require genomic analysis of at least 50,000 additional affected individuals (Figure 1A). By understanding the underlying mechanisms leading to ASD, we are more likely to develop effective treatments by selecting genetically validated, meaningful therapeutic targets.
Figure 1. Recruitment of the SPARK Cohort.
(A) Statistical power to detect individual ASD risk genes based on excess of de novo variants. For a given sample size (number of trios), we calculate the expected number of de novo likely gene disrupting (LGD) (M1) or D-mis (damaging missense defined by meta-SVM) (Dong et al., 2015) (M2) for each gene by gene-specific background mutation rate calibrated in previous publications (Ware et al., 2015; Homsy et al., 2015). We assume that the observed number of such variants in a risk gene follows a Poisson distribution with mean of M1*R + M2*R/2, in which R is the relative risk of a de novo LGD variant in a risk gene. We then calculate the power of detecting the excess of de novo LGD and D-mis variants at a significance threshold of 2.6 × 3 10−6 to account for ~19,000 protein-coding genes captured by WES and with the available background mutation rate data. The figure shows the average power of all genes given the sample size and R. With WES of 50,000 trios, we achieve greater than 70% power to detect an association based on de novo LGD and D-mis variants if R (relative risk) is 30 or above.
(B) In the first year of recruitment, 18,809 individuals with ASD (higher density indicated by larger circles) and 27,794 of their first-degree relatives consented to participate in SPARK, 81% of whom were recruited through the clinical sites (indicated by black dots). 15,302 of the individuals with ASD are children (<18 years) and 2,787 are adults. 7,610 unaffected full biological siblings were enrolled. The cohort includes 6,309 trios (proband, biological mother, and biological father), 682 sets of multi-gestational pregnancies with individuals with ASD (664 twin, 15 triplet, and three quadruplet sets that included 1,371 individuals with ASD), and 1,944 multiplex families in which at least one first-degree relative of the ASD proband also has ASD.
Clarifying the contributions of environmental exposures is also important, as some of these exposures may be preventable. A recent meta-analysis of 23 studies of airborne pollutants in ASD risk supported an association of ASD with increasing exposure to air pollution in mothers during pregnancy (Lam et al., 2016). Other more common, pregnancy-related factors for ASD include maternal obesity, maternal diabetes, and caesarian section (Modabbernia et al., 2017). This meta-analysis of 80 studies also concluded that birth complications associated with trauma, ischemia, and hypoxia are associated with increased ASD risk (Modabbernia et al., 2017). Significantly larger studies are needed to identify relevant environmental risk factors and accurately estimate their associated risk.
A considerable practical barrier to accelerating clinical research in ASD is the heavy burden of recruiting large numbers of eligible research participants. In ASD and other neuropsychiatric conditions, it is even more challenging to recruit individuals into longitudinal studies, particularly neuroimaging protocols. In order to address these challenges, we saw the need to develop a new paradigm in which an engaged, research-ready cohort could be efficiently and effectively followed longitudinally and informed of new research opportunities.
While there have been many research studies of individuals with ASD, Simons Foundation Powering Autism Research for Knowledge (SPARK) represents a new era of clinical research that combines online access to participants; ability to recontact and recruit for new research studies; genomic, environmental, and longitudinal behavioral and medical information on all participants; and support for participants through communication of meaningful genetic and other ASD-relevant information. SPARK is founded on principles that emphasize a strong partnership between participants and researchers. SPARK is committed to providing aggregate and individual research results to participants to help keep the goals of research grounded in having a meaningful impact on the lives of individuals and families with ASD. We believe that the challenges and lessons learned from SPARK will be informative to a wide range of researchers working on other conditions.
Approach to Scalable Phenotyping and Genomic Analysis
Currently, approximately 10% of ASD cases have an identifiable genetic etiology of large effect (Tammimies et al., 2015). As more genes are discovered and better methods to predict pathogenicity of genetic variants are developed, it is reasonable to estimate that this percentage will increase to 20% to 30% in the next 5 years. Despite the large number of genes implicated in ASD, several studies have shown that ASD risk genes seem to converge on a limited number of biological pathways, such as targets of the fragile X mental retardation protein (FMRP), synaptic proteins, and chromatin modifiers. Many of these genes are disrupted in 0.1%–0.3% of individuals with ASD. Therefore, recruitment and genomic characterization of at least 50,000 individuals with ASD is needed to yield sufficient numbers of individuals with common genetic etiologies for targeted genetic studies (Figure 1A). Based on previous estimates of relative risk of the most frequent, high-risk ASD genes and background mutation rate of LGD (likely genedisrupting) or damaging missense variants (Ware et al., 2015; Homsy et al., 2015), the SPARK study should identify at least 16–150 individuals with mutations in each of the 100 most common ASD genes.
Building disease- and gene-specific cohorts is labor intensive and time consuming, often requiring inperson clinical visits and phlebotomy to collect phenotypic data and biospecimens. While this approach is feasible for groups of a few thousand individuals, it is not scalable to tens of thousands of individuals and family members. In order to overcome these challenges of scalability, we are collecting phenotypic data and biospecimens remotely so that participants can complete the study protocol online at their convenience. We are intentionally performing less detailed clinical assessments up front because we have the ability to recontact participants and focus future data collection guided by genetic etiology and/or other relevant genomic or phenotypic features. Moreover, this makes participation in ASD research accessible for everyone in the US and not just individuals with access to a major university medical center.
Any individual living in the United States with a professional diagnosis (by a physician, psychologist, or therapist) of ASD plus their parents and an unaffected sibling are eligible to participate in SPARK. Participants consent to share their deidentified data and to be contacted about ASD research studies for which they qualify. Participants can also consent to contribute a saliva sample for genetic analysis and have the option to receive individual genetic results related to ASD should a primary genetic cause of ASD be identified. As part of participation in SPARK, individuals are also asked to complete a battery of online questionnaires (https://www.sfari.org/resource/spark/). Although phenotype information and ASD diagnoses in SPARK are self- or parent-reported, past research on the first web-based registry for ASD, the Interactive Autism Network, suggests that parent-reported diagnosis of ASD is highly valid.
Genomic analyses in SPARK currently include WES and genome-wide genotyping. SPARK is genotyping each participant with a 690,000 SNP array optimized for ethnically diverse cohorts (Infinium Global Screening Array-24 v.1.0), which can capture structural and copy-number variants (CNVs) across the genome and collect genotypes for common variant studies. WES complements genotyping and identifies rare coding variants and, at this time, is more cost effective than whole-genome sequencing.
Return of Individual Genetic Results and Behavioral Information
The return of individual genetic results in genomics studies is currently a topic of national discussion in the research community (Sénécal et al., 2015). Returning individual genetic results in large studies is resource intensive, complex, and often beyond the scope of many research studies. However, many argue that returning individual genetic results is desirable and demonstrates respect and a willingness to partner with research participants. From a scientific standpoint, returning genetic results to participants in SPARK will accelerate research of genetically defined subtypes of ASD since SPARK will increase the numbers of readily recontactable individuals with known genetic findings. Since SPARK is recruiting individuals across the United States, a major challenge is to develop a scalable, centralized process for returning individual genetic results to participants.
SPARK returns pathogenic variants in genes definitively associated with ASD to any participant in whom such results are identified and consents to return of genetic information are in place. SPARK utilizes a Medical Genetics Committee that decides annually which genes and CNVs meet stringent criteria for return. Pathogenic variants in genes and CNVs are returned if there are at least three individuals with autism or autistic traits who have confirmed de novo LGD mutations in that gene or locus in at least two different publications. These criteria that variants must meet to be returnable to SPARK participants are intentionally conservative to avoid false positive findings. All genetic results that are returned are confirmed in a CLIA (Clinical Laboratory Improvement Amendments)-certified clinical laboratory that generates a clinical laboratory report that is provided to the SPARK participant. Currently, SPARK participants with returnable genetic results can choose to receive these results through a SPARK-provided genetic counseling service by telephone or through a medical provider the participant designates.
On an annual basis, SPARK will iteratively reanalyze the genomic data of all participants. Therefore, if a participant harbors a genetic variant that is not considered returnable at the initial time of analysis but new knowledge renders that variant returnable in later years, that participant will receive the result when the gene is approved by the Medical Genetics Committee.
Individuals who are aware of their genetic results are contactable for studies focused on their genetic subtype of ASD and may be more motivated to participate in research focused on their genetic condition. Thus far, in a pilot study involving SPARK’s first 500 proband-parent trios, returnable findings have been identified and returned in 5% of participants with ASD using an initial list of 11 CNVs and 78 single genes (https://sparkforautism.org/downloads/SPARK_gene_list.pdf).
An important consideration for SPARK was whether to return incidental findings. SPARK is not actively searching for pathogenic genetic variants unrelated to ASD, so their discovery is not likely. At the time of initial consent, participants set preferences to receive clinically actionable incidental results should they be identified, but the analysis pipelines do not actively interrogate the genes on the American College of Medical Genetics secondary findings list.
In addition to genetic results, SPARK will also share aggregate and individual results of behavioral measures (https://www.sfari.org/resource/spark/). Aggregate reports will provide participants with a lay summary of the SPARK cohort, including demographic and clinical characteristics. When possible, SPARK will provide individual reports for specific questionnaires (e.g., Social Communication Questionnaire–Lifetime) that will display where an individual participant falls in relation to both national norms and other participants in SPARK. Our commitment to the return of individual and aggregate results to SPARK participants is a paradigm that can be more widely applied in other human subject research cohorts and is a crucial component of participant engagement.
Recruitment Strategies and Resources for the Scientific Community
SPARK began nationwide recruitment with 21 clinical sites and a multi-pronged social media strategy. Our rationale for working with clinical sites as a leading recruitment arm for SPARK was their ability to interact with participants, to increase the likelihood that participants had a valid diagnosis of ASD, and to facilitate the contribution of rich clinical data from medical records (Figure 1B). In SPARK’s first 12 months of recruitment, through May 2017, SPARK enrolled 18,089 individuals with ASD and 28,515 of their family members, collected a total of 25,919 saliva samples, and has begun WES and genome-wide genotyping (https://www.sfari.org/resource/spark/). SPARK continues to recruit, on average, 1,100 new participants with ASD every month.
As part of participation in SPARK, all sites were required to delegate institutional oversight of the study to a central institutional review board (IRB). Using a central IRB enabled all sites to follow the same protocol and use the same electronic informed consent forms. Having a central IRB also ensures that SPARK maintains consistency in messaging and outreach strategies and minimizes administrative burden on sites.
SPARK provides the scientific community with rapid access to all genetic and phenotypic data on participants and facilitates research recruitment requests from any qualified researcher. Any academic or industry researcher can apply for access to SPARK data or to invite eligible participants to a research study. Invitations sent to SPARK participants to join research studies are coordinated centrally to prevent research fatigue. SPARK currently does not charge any fees to researchers. If particular subsets of participants (e.g., baby siblings, twins, multiplex families, and genetically defined groups) are frequently requested by multiple investigative teams, policies will be implemented to allow for fair and equitable access among researcher projects.
SPARK requires that all data collected by other researchers be shared back with SPARK within 18 months of a study’s completion. This data sharing policy helps ensure that redundant data elements are not unnecessarily collected from participants and that the cohort’s dataset increases over time. By emphasizing data sharing and participant engagement, we hope to encourage and maintain engagement by a cohort that is motivated to participate in research not just by financial incentives or personal gain, but by having a broader understanding of what their participation means for research and by seeing how their participation leads to improvements for the ASD community.
Participant Retention and Partnership
SPARK’s success depends in part on the retention and long-term engagement of participants who feel they are active partners in the research mission. Therefore, we are building a research community founded on meaningful reciprocity. SPARK participants actively guide the initiative through multiple feedback channels. During the planning stage, we surveyed hundreds of individuals with ASD and their family members for input on each aspect of SPARK. We found that return of genetic results, access to ASD-specific educational mobile applications and websites, and access to information about ASD research were the top three incentives for research participation. SPARK participants continue to directly inform the design of SPARK’s online platform. The website underwent extensive user experience testing by participants to improve usability. SPARK also engages a Community Advisory Council that provides input into research priorities and helps guide the development of study materials.
To keep participants engaged, SPARK strives to create positive and meaningful research experiences by providing useful resources, sharing updates about research conducted through SPARK, returning individual behavioral and genetic results when possible, and asking our community to help guide the initiative’s development. SPARK provides families with useful information on a variety of ASD topics through monthly webinars with ASD experts and articles on our website, including ASD research summaries, evidence-based guidance, and personal stories. We provide updates on the study’s progress through monthly newsletters and easy-to-understand data reports. Finally, we require investigators who use SPARK’s recruitment services to summarize their findings and results for research participants, which is another participant engagement tactic that is widely applicable.
SPARK has conducted several pilot studies of the research-matching program. In one study that included a 15-min online questionnaire about environmental exposures during pregnancy, 60% of 2,089 biological mothers queried completed the study within 6 weeks. The high response rate in this study demonstrates the early success of SPARK’s strategies to engage and retain participants.
Challenges and Lessons Learned
The ability to reach thousands of individuals and families affected with ASD through the internet and the ease with which participants can register and consent have resulted in rapid recruitment and biospecimen collection that far exceeds typical clinic-based research. Nevertheless, there are many challenges that we will need to address to reach our goal of recruiting 50,000 families and collecting their biospecimens.
A significant challenge for SPARK is recruitment of complete biological families for genetic analysis of de novo variants. Over the course of the study, in families in which both biological parents are available and have been invited (by the other parent or the individual with ASD) to participate, only 55% complete the registration and consent process. To increase participation from the complete family, SPARK developed social media messaging that used humor to encourage mothers to motivate fathers to complete enrollment in SPARK. In the 2 months prior to the use of this particular message, 60% of second parents accepted the invitation (from the individual with ASD or other parent) to join SPARK. In contrast, in the 2 months after we began using these advertisements, 72% of second parents accepted invitations to join SPARK. The increases we observed support the need for creative and novel social media strategies to reach our recruitment goals.
Another significant issue is the low rate of saliva kit return. After the first 9 months of recruitment, only 39% of saliva kits had been returned. Subsequently, we simplified instructions, added a lottery for an iPad for participants who return their kits within 2 weeks, and sent a physical paper reminder to participants who had not returned their kits. These changes, along with higher levels of engagement with participants through our newsletters, social media channels, and website, improved our return rate to 58%. To further improve the return rate, we will be surveying participants who did not return their kits to better understand their challenges and develop more effective strategies.
Another challenge for the strategy we have adopted for SPARK is the limited depth of the phenotypic information that we can collect directly from participants. Nevertheless, SPARK will acquire much more detailed phenotypes on selected subgroups over time and gather data from electronic medical records and through data-sharing portals, such as Sync for Science (http://syncfor.science/), so we can maximize the number of clinically validated phenotypic data elements. In addition, we require all external investigators to deposit all non-identifiable data collected on SPARK participants into the central SPARK database, which is made broadly available to any qualified researcher. We are committed to developing and supporting SPARK as a longitudinal resource to facilitate transformative advances in autism research, treatment, and support and hope it will serve as a model for other disease groups.
Supplementary Material
ACKNOWLEDGMENTS
We are extremely grateful to the thousands of individuals and families who are participating in this study. We are grateful to the many autism advocacy and service organizations that have helped us inform the community about SPARK, including the Autism Society of America and its affiliates, Autism Speaks, Autism Science Foundation, Easter Seals, Arkansas Autism Resource and Outreach Center, Global and Regional Asperger’s Syndrome Partnership, Kentucky Autism Training Center, and Autistic Self Advocacy Network. We thank the members of SPARK’s Community Advisory Council for providing feedback and advice. We thank members of our Scientific and Community Advisory Board and SFARI scientists for advice on our protocol and participant outreach and retention strategies. We thank PreventionGenetics for managing and processing biospecimens, DNA Genotek for handling saliva kit logistics, and Baylor College of Medicine Human Genome Sequencing Center for exome sequencing. The SPARK initiative is funded by the Simons Foundation as part of SFARI.
Footnotes
CONSORTIA
The members of the SPARK Consortium are: Pamela Feliciano, Amy M. Daniels, LeeAnne Green Snyder, Amy Beaumont, Alexies Camba, Amy Esler, Amanda G. Gulsrud, Andrew Mason, Amy Nicholson, Anna Marie Paolicelli, Alexander P. McKenzie, Angela L. Rachubinski, Alexandra N. Stephens, Andrea R. Simon, Anibal Gutierrez, Amy Stedman, Amanda D. Shocklee, Amy Swanson, Brenda Finucane, Brittani A. Hilscher, Brenda Hauf, Brian J. O’Roak, Brooke McKenna, Beverly E. Robertson, Barbara Rodriguez, Brianna M. Vernoia, Bonnie Van Metre, Catherine Bradley, Cheryl Cohen, Craig A. Erickson, Christina Harkins, Caitlin Hayes, Catherine Lord, Christa Lese Martin, Crissy Ortiz, Cesar Ochoa-Lubinoff, Christine Peura, Catherine E. Rice, Cordelia R. Rosenberg, Christopher J. Smith, Carrie Thomas, Cora M. Taylor, Loran Casey White, Corrie H. Walston, David G. Amaral, Daniel Lee Coury, Dustin E. Sarver, Dalia Istephanous, Deana Li, Dzung Cong Nugyen, Emily A. Fox, Eric M. Butter, Elizabeth Berry-Kravis, Eric Courchesne, Eric J. Fombonne, Eugenia Hofammann, Elena Lamarche, Ericka L. Wodka, Emily T. Matthews, Eirene O’Connor, Emily Palen, McLeod F. Gwynette, Fiona Miller, Gabriel S. Dichter, Gabriela Marzano, Gail Stein, Hanna Hutter, Hannah E. Kaplan, Hai Li, Holly Lechniak, Hoa Lam Schneider, Hana Zaydens, Ivette Arriaga, Jennifer A. Gerdts, Joseph F. Cubells, Jeanette M. Cordova, Jaclyn Gunderson, Joseph Lillard, Julie Manoharan, James T. McCracken, Jacob J. Michaelson, Jason Neely, Jessica Orobio, Juhi Pandey, Joseph Piven, Jessica Scherr, James S. Sutcliffe, Jennifer Tjernagel, Jermel Wallace, Kristen Callahan, Katherine Dent, Kathryn A. Schweers, Kira E. Hamer, J. Kiely Law, Kathryn Lowe, Kaela O’Brien, Kaitlin Smith, Katherine G. Pawlowski, Karen L. Pierce, Katherine Roeder, Leonard J. Abbeduto, Leandra N. Berry, Lindsey A. Cartner, Leigh A. Coppola, Laura Carpenter, Lisa Cordeiro, Lindsey DeMarco, Luke P. Grosvenor, Lorrin Higgins, Lark Y. Huang-Storms, Landon Hosmer-Quint, Lynette M. Herbert, Lauren Kasparson, Lisa M. Prock, Lillian D. Pacheco, Laurie Raymond, Laura Simon, Latha V. Soorya, Lucy Wasserburg, Maya Lazar, Michael Alessandri, Melissa Brown, Mary Hannah Currin, Michelle Heyman, Melissa N. Hale, Mark Jones, Michelle Jordy, Michael J. Morrier, Mustafa Sahin, Matthew S. Siegel, Mary Verdi, Meaghan Venezia Parlade, Meredith Yinger, Nicole Bardett, Nathan Hanna, Nina Harris, Natalie Pottschmidt, Nicole Russo-Ponsaran, Nicole Takahashi, Opal Y. Ousley, A. Pablo Juarez, Patricia Manning, Robert D. Annett, Raphael A. Bernier, Renee D. Clark, Rebecca J. Landa, Robin P. Goin-Kochel, Rick Remington, Robert T. Schultz, Stephanie J. Brewster, Stephanie Booker, Sarah Carpenter, Sara Eldred, Sunday Francis, Sandra L. Friedman, Susannah Homer, Susan Hepburn, Suma Jacob, Stephen Kanne, Soo J. Lee, Sarah A. Mastel, Samantha Plate, Shanping Qiu, Sophia Sandhu, Samantha Thompson, Sabrina White, Vincent J. Myers, Vini Singh, Wha S. Yang, Zachary Warren, Alpha Amatya, Andrea J. Ace, Ahmad S. Chatha, Alex E. Lash, Ben Negron, Chris Rigby, Curtis Ridenour, Colleen M. Stock, Danielle Schmidt, Ian Fisk, John Acampado, Jay L. Nestle, Jay A. Nestle, Kevin Layman, Martin E. Butler, Matt Kent, Malcolm D. Mallardi, Nicholas Carriero, Noah Lawson, Natalia Volfovsky, Ron Edgar, Richard Marini, Rishiraj Rana, Swami Ganesan, Swapnil Shah, Tyler Ramsey, Wubin Chin, William Jensen, Anthony D. Krentz, Angela J. Gruber, Aniko Sabo, Andrei Salomatov, Christine Eng, Donna Muzny, Irina Astrovskaya, Richard A. Gibbs, Xinwei Han, Yufeng Shen, Louis F. Reichardt, and Wendy K. Chung.
DECLARATION OF INTERESTS
Mustafa Sahin has received research funding from Roche, Novartis, Pfizer, Navitor, Rugen, Ibsen, Neuren, and LAM Therapeutics and has served on the Scientific Advisory Board of Sage Therapeutics. Richard A. Gibbs, Donna Muzny, and Aniko Sabo are employees of Baylor College of Medicine, which has part ownership of Baylor Genetics Laboratories and Codified Genomics, each of which derive income from genetic testing. Catherine Lord receives royalties from Western Psychological Services, publisher of the Social Communication Questionnaire. Anthony D. Krentz is an employee of PreventionGenetics and a member of PrevGen Employees, LLC, which owns units in PreventionGenetics. Angela J. Gruber is an employee of PreventionGenetics.
SUPPLEMENTAL INFORMATION
Supplemental Information includes a list of consortium members and can be found with this article online at https://doi.org/10.1016/j.neuron.2018.01.015.
REFERENCES
- Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium (2017). Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, and Liu X. (2015). Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet 24, 2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J, et al. (2014). Most genetic risk for autism resides with common variation. Nat. Genet 46, 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. (2015). De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J, Sutton P, Kalkbrenner A, Windham G, Halladay A, Koustas E, Lawler C, Davidson L, Daniels N, Newschaffer C, and Woodruff T. (2016). A systematic review and meta-analysis of multiple airborne pollutants and autism spectrum disorder. PLoS ONE 11, e0161851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modabbernia A, Velthorst E, and Reichenberg A. (2017). Environmental risk factors for autism: an evidence-based review of systematic reviews and meta-analyses. Mol. Autism 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, et al. (2014). A framework for the interpretation of de novo mutation in human disease. Nat. Genet 46, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, et al. ; Autism Sequencing Consortium (2015). Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, and Reichenberg A. (2017). The heritability of autism spectrum disorder. JAMA 318, 1182–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sénécal K, Rahimzadeh V, Knoppers BM, Fernandez CV, Avard D, and Sinnett D. (2015). Statement of principles on the return of research results and incidental findings in paediatric research: a multi-site consultative process. Genome 58, 541–548. [DOI] [PubMed] [Google Scholar]
- Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, Yuen RK, Uddin M, Roberts W, Weksberg R, et al. (2015). Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA 314, 895–903. [DOI] [PubMed] [Google Scholar]
- Ware JS, Samocha KE, Homsy J, and Daly MJ (2015). Interpreting de novo variation in human disease using denovolyzeR. Curr. Protoc. Hum. Genet 87, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.