Abstract
The current SARS-CoV-2 pandemic is accompanied by high morbidity and mortality rates, and there is a compelling need for effective vaccines and therapeutic agents to lessen the severity of COVID-19 disease. Appropriate animal models are essential for testing of vaccines and therapeutics and for mechanistic studies of infection and the host response. The Spike (S) protein of SARS-COV-2 has a high affinity for the human ACE2 receptor, which is expressed on multiple cell types including alveolar epithelial and vascular endothelial cells. Wild-type mice are not susceptible to developing coronavirus-mediated diseases. Accordingly, several human (h)ACE2 transgenic mouse models have been developed for coronavirus research. However, these mice have failed to closely mimic important aspects of the human immunopathological responses to SARS-CoV-2. We report herein that DRAGA (HLA-A2.HLA-DR4.Rag1KO.IL-2Rγc KO.NOD) mice infused with human hematopoietic stem cells from cord blood reconstitute a fully functional human immune system, as well as engraft human epithelial and endothelial cells, sustain SARS-CoV-2 infection, and develop severe COVID-19-like symptoms. In pilot experiments, infected mice developed parenchymal and epithelial lung infiltrations with granzyme B+ and perforin+ CD8+ T cells and alveolar CD61+ microthrombi, mimicking human immunopathological responses to SARS-CoV-2. We propose the DRAGA mouse as a novel pre-clinical tool for studying COVID-19 immunopathology and human immune responses to SARS-CoV-2, including events leading to the cytokine storm and coagulopathies, as well as for testing of candidate vaccines and therapeutics.
Keywords: SARS-CoV-2, COVID-19, humanized mouse model, human ACE2 expression
INTRODUCTION
Infection with Severe Acute Respiratory Syndrome-coronavirus-2 (SARS-CoV-2), the highly transmissible pathogen responsible for the ongoing pandemic COVID-19 disease, results in outcomes ranging from asymptomatic or mild disease to severe pneumonia/acute respiratory distress syndrome (ARDS) in the human population1–4. Many severe COVID-19 patients also experience a hyper-inflammatory response (“cytokine storm”)5 combined with a dysregulated coagulation system that subsequently leads to a variety of extrapulmonary manifestations. The thrombotic complications observed in COVID-19 include several unusual features that differ from disseminated intravascular coagulation (DIC), e.g. elevated fibrinogen and fibrin D-dimer, hyperreactive platelets, and formation of microthrombi in multiple organs, but without the consumption of coagulation factors that typically accompany other severe inflammatory conditions such as sepsis6–17.
Appropriate, clinically relevant animal models are required to evaluate mechanisms of pathogenesis and as platforms for rapid testing of vaccines and potential therapeutics. SARS-CoV-2 fuses with and enters host cells following engagement of its surface spike (S) protein with human angiotensin-converting enzyme 2 (hACE2)18,19, which is expressed on multiple tissues, notably including the lung alveolar epithelium and vascular endothelium, and also in the liver, colon, esophagus, small intestine, duodenum, kidney, brain, and tongue20–22. A number of investigators are currently setting up animal models susceptible to SARS-CoV-2 infection such as mice, ferrets, hamsters, rats, and monkeys23–27, but earlier studies have indicated that, due to species differences, these models cannot recapitulate key aspects of the human anti-viral, immune, and pathological responses to coronaviruses28.
The SARS-CoV-1 and SARS-CoV-2 S proteins bind to human ACE2 with higher affinity than to ACE2 receptors in mice and other potential animal models29. Consistent with this, infection with these coronaviruses does not result in severe disease in wild-type mice. Several non-HIS mouse strains transgenic (Tg) for human ACE2 have been generated to date. The human cytokeratin 18 (K18)-hACE2 Tg mouse known as B6.Cg-Tg(K18-ACE2)2Prlmn/J, developed in 2006 by McCay et al., expresses hACE2 in the lungs, liver, kidney, colon, and brain21,30, showing a particular tropism for brain infection by SARS-CoV-1 and fatal disease following infection21,28,31–36.This Tg mouse challenged with SARS-CoV-2 showed high viral titers in the lung and lung pathology but exhibited variable clinical symptoms37. Similar results were reported by exogenous delivery of human ACE2 with a replication-deficient adenovirus (Ad5-hACE2) in BALB/c and C57BL/6 mice38.
The hepatocyte nuclear factor-3/forkhead homologue 4 (HFH4)-ACE2 Tg mice known as B6JCg-Tg (FOXJ1-ACE2) 1Rba/Mmnc were generated in 2016 by Menachery et al. to provide airway-targeted overexpression of hACE2, and they also express hACE2 in the brain, liver kidney, and GI tract39. These mice were generated to assess possible infection and transmission of the Wuhan Institute of Virology (WIV1)-CoV virus, a SARS-like coronavirus found to replicate in horseshoe bats39. However, these Tg mice showed more attenuated SARS-COV-1 infection compared to WIV1-CoV infection. Another Tg mouse expressing hACE2 was generated recently by Sun et al. using CRISPR/Cas9 technology, which can be infected i.n. with SARS-CoV-2, sustain high viral loads, and develop pneumonia40, although fatalities were not observed in initial studies of this model. Overall, these murine models transgenic for hACE2 expression driven under various promoters have shown different tropisms, viral loads, and pathologies following infection with SARS-CoV-1 and other coronaviruses28,36,41,42.
In order to study mechanisms of viral entry and of human immune responses to SARS-CoV-2, and to assess potential vaccines and therapeutics, a humanized animal model permissive for SARS-CoV-2 infection and showing similar pathology to COVID-19 in humans would be of great benefit. In particular, mouse strains reconstituted with a Human Immune System (HIS) that are permissible for SARS-CoV-2 infection and that show human-like pathology would be highly appealing models. Several HIS-mouse strains have been generated, but many have fallen short in mimicking the human immune system with high fidelity due to limitations. These include poor engraftment of hematopoietic stem cells, inefficient human cell expansion and homeostasis, insufficient numbers of reconstituted human T or B cells, sub-optimal B cell development, lack of immunoglobulin class switching, acute/chronic GVHD, and lack of HLA class I and/or II restriction43–47. Furthermore, restrictions on the use of human fetal tissue for research has focused attention on alternative appropriate donor cell sources, such as umbilical cord blood.
We have generated a HIS-humanized mouse strain devoid of many of the above limitations: the DRAGA mouse (HLA-DR4. HLA-A2.1. IL-2Rγc KO. RAG KO. NOD)48,49. These mice are HIS-reconstituted after irradiation and infusion with hematopoietic stem cells (HSC) from HLA-matched umbilical cord blood. These mice lack the murine immune system while expressing a long-lived functional HIS. We have previously demonstrated that they respond vigorously by specific T and B cell responses to infection or immunization with various pathogens including malaria protozoans, HIV, ZIKA, Scrub Typhus, and Influenza type A heterosubtypes50–55. We also reported recently that they engraft human endothelial cells (EDs) in the liver50 and both epithelial and endothelial cells in their lungs55.
Herein, we demonstrate that the HIS-DRAGA mouse naturally reconstitutes human ACE2-expressing epithelial cells (ECs) and EDs in the lungs. Importantly, these mice sustained i.n. infection with SARS-COV-2 virus, developing mild to severe COVID-19 disease, including fatal outcomes. We propose that DRAGA mice are an excellent and convenient model for studies of SARS-CoV-2 infection, SARS COVID-19 immunopathology and human immune responses to the virus, including early events leading to cytokine storm and coagulopathies, and for testing of potential vaccines and therapeutics.
RESULTS
HIS-reconstituted DRAGA mice express human ACE2 in lung epithelial and endothelial cells
Both murine ACE2 (mACE2) and human ACE2 (hACE2) mRNA expression were detected in the lungs of HIS-DRAGA mice by RT-PCR (Fig. 1). To detect hACE2 protein, immunoprecipitation (IP) experiments were carried out using a recombinant SARS-CoV-2 S1 protein Receptor Binding Domain(RBD)-mouse Fcγ2a chimeric protein (S1-RBD-mFcγ2a), which is expected to bind hACE2 much more effectively than mACE256, and rat anti-mouse IgG2a-magnetic beads. Immunoprecipitation (IP) experiments were carried out using homogenates from (a) lungs from HIS-reconstituted DRAGA mice; (b) non-reconstituted DRAGA mice (negative control); and (c) commercially obtained human hung tissue (positive control). ELISA assays measuring binding of the immunoprecipitates to S1-RBD-mFcγ2a indicated that hACE2 was expressed in HIS-DRAGA lungs, albeit at average levels that were ~18X less than in human lungs. No hACE2 was detected by ELISA in immunoprecipitates from non-HIS-reconstituted DRAGA lungs. These results confirmed that hACE2 mRNA, as well as hACE2 protein, are expressed in the lungs of HIS-reconstituted DRAGA mice.
Figure 1. Detection of human ACE2 mRNA expression in the lungs of HIS-humanized DRAGA mice.
Upper row, PCR amplicons of hACE2 from the lungs of four HIS-humanized DRAGA mice and three non HIS-humanized DRAGA mice as controls (all non-infected). Positive control was human lung mRNA (Sigma-Aldrich). Lower row (quality control RNA), the PCR amplicons of mACE2 from the lungs of mice in the upper row, as amplified with murine specific primers. NC, negative control (primers alone). As expected, the HIS-DRAGA mice express both murine ACE2 and human ACE2 derived from engrafted human cells.
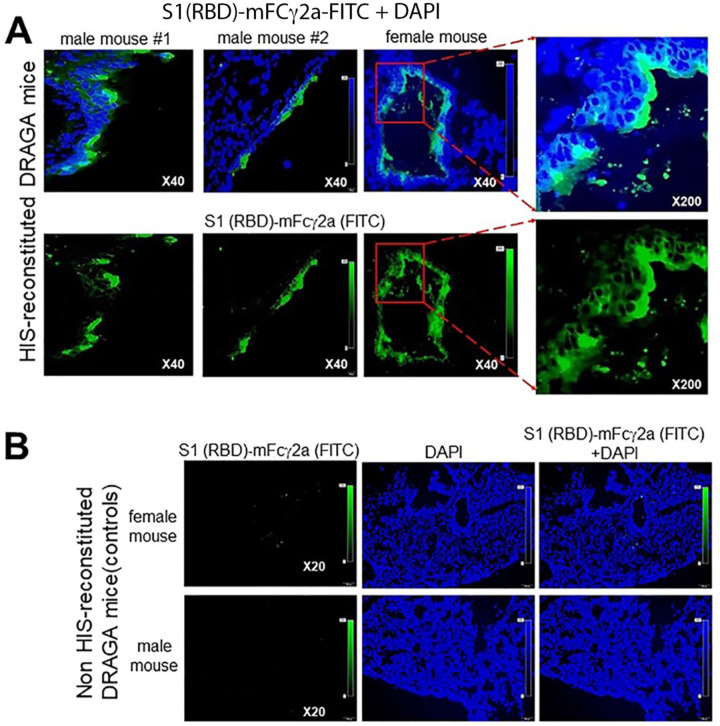
Lung sections from HIS-reconstituted and non-reconstituted DRAGA mice were probed by immunofluorescence microscopy with S1-RBD-mFcγ2a followed by a goat anti-mouse IgG-FITC conjugate to detect hACE2. The images revealed an uneven distribution of fluorescent-labeled cells on the alveolar and bronchiolar outer layers (Fig. 2A), while no staining was detected in lungs of non-reconstituted (control) mice (Fig. 2B). Co-staining with an anti-hCD326-PE conjugate (as a marker for human lung ECs) revealed co-localization of the S1-RBD-mFcγ2 bound hACE2 receptor with CD326+ lung epithelial cells, indicating that hACE2 is expressed on human epithelial cells in the lungs of HIS-reconstituted mice, but not in lungs from non-HIS (control) mice (Fig. 3). In aggregate, these results demonstrated that lungs from HIS-DRAGA mice are reconstituted with human epithelial cells expressing hACE2.
Figure 2. Expression of hACE2 receptor in the lungs of HIS-DRAGA mice.
Panel A, hACE2 protein expression on the outer layer of alveoli, as indicated by the binding of the S1 (RBD)-mFcγ2a chimera to OCT-frozen lung sections from two HIS-reconstituted DRAGA male mice and a female mouse. S1 (RBD)-mFcγ2a binding was revealed with a goat anti-mouse IgG-FITC conjugate (green color). Shown are overlapped images of S1 (RBD)-mFcγ2a binding and DAPI (cell nuclei, blue color), and enlargements of the outer layer of alveolar areas expressing the hACE2 protein (red squares). Panel B, The same staining protocol for OCT-frozen lung sections from two non-HIS-reconstituted DRAGA mice (one female and one male) showed no binding of S1(RBD)-mFcγ2a, reflecting the expected lack of hACE2 expression in these mice.
Figure 3. Co-localization of hACE2 receptor with alveolar epithelial cells of HIS-DRAGA mice.
Panels A&B, co-localization of hACE2 receptors with the hCD326+ epithelial layer of alveoli was revealed by staining with the S1(RBD)-mFcγ2a chimera followed by a goat anti-mouse IgG-FITC conjugate (green) and co-stained with recombinant human anti-hCD326-PE to OCT-frozen lung sections from two HIS-reconstituted DRAGA mice. Also shown are enlargements of the outer layer of alveoli expressing the hACE2 protein on hCD326+ ECs (yellow squares). Panel C, lack of binding of S1 (RBD)-mFcγ2a and the anti-hCD326 antibody to a lung section from a non-HIS-reconstituted DRAGA mouse indicates its expected lack of human ACE2 expression and the lack of human epithelial cells.
DRAGA mice can sustain SARS-CoV-2 infection
Having found that hACE2 is expressed in the lungs of HIS-reconstituted DRAGA mice, we questioned whether these mice are permissive for SARS COV-2 infection. In a pilot experiment, three HIS-DRAGA mice were infected by the intranasal (i.n.) route with 2.8 × 103 (1 female mouse) or 2.8 × 104 pfu (1 male + 1 female mouse) of SARS-CoV-2 live virions in 50 μl saline, 25 μl in each nostril. The male mouse infected with the low dose succumbed to the infection after 24 hours, while both female mice sustained the infection, showing an abrupt loss in body weight, ruff fur, hunched back, and reduced mobility starting 1 day post-infection (dpi). The surviving mouse infected with the low dose of virus regained its original weight and mobility at 10 dpi, while the surviving mouse infected with the high dose was still 10% below its original weight at 14 dpi (Fig. 4). ELISA assays carried out using serum samples at day 0 and at 14 dpi from the two surviving mice indicated that only the mouse infected with the high dose elicited S1 (RBD)-specific human IgM antibodies in sera (titer = 100) by 14 dpi (Supplemental data).
Figure 4. Dynamics of body mass changes in HIS-DRAGA mice infected with SARS-CoV-2 virus.

Shown are daily monitored body weights of a HIS-DRAGA female (F#1) mouse infected i.n. with a low dose of SARS-CoV-2 virus (2.8×103 pfu), a female mouse (F#2) infected i.n. with a high dose of virus (2.8×104 pfu), and a male mouse infected i.n. with a low dose of virus (2.8×103 pfu). The male mouse succumbed 24 hours post-infection and the two surviving female mice were sacrificed at 14 dpi.
SARS-CoV-2 infected HIS-DRAGA mice display human-like lung immunopathology
Lung sections from the two HIS-DRAGA survivors of SARS-CoV-2 infection were further analyzed to characterize pathological changes. As in recent reports based on human autopsies57–59, the H&E and Mason-trichrome staining of lung sections showed heavy parenchymal infiltrates around the large bronchioles and at peripheral areas of the lungs, as well as hCD45+ cells and human (CD3+) T cells dispersed throughout the lung parenchyma, which were more pronounced in the mouse infected with the high dose of virus (Fig. 5). Interestingly, some of the human CD3+ T-cell infiltrates appeared to be fairly organized, such those described in the epithelial lung niches of severely-infected mice and humans with influenza viruses60–64 (Fig. 5E). Further analysis revealed that hCD8+ T cells, and to a lesser extent the hCD4+ T cells, were clustered into the alveolar EC niches and able to egress into the alveolar air space (Fig. 6). Indeed, the alveolar epithelial layer containing large clusters of CD8+ T cells was identified by immunofluorescence using an anti-human CD326 antibody (a human lung epithelial cell marker)65 (Fig. 7). The CD8+ T cells sequestered in CD326+ lung epithelial niches also stained positive for CD103, indicating they were T resident cells (Fig. 8), as well as for perforin and granzyme B, indicating their potential cytotoxic activity (Fig. 9).
Figure 5. Human immune cell infiltrates in the lungs of SARS-CoV-2 infected HIS-DRAGA mice.
Panel A, paraffin-embedded lung sections from HIS-DRAGA mouse F#2 infected i.n. with high dose of virus (2.8×104 pfu) stained with H&E and Mason-Trichrome. Shown are healthy areas and heavily infiltrated areas of the lung parenchyma and around a terminal bronchiole (H&E staining, black arrow). Shown are the epithelial outer layer of the bronchiole (red color), and deposition of collagen fibers (blue color). Panels B & C, Paraffin-embedded lung sections from HIS-DRAGA mouse F#2 infected with high dose of virus (2.8×104 pfu) showing heavily infiltrated lung tissue with hCD45+ cells and human T cells, respectively. Panels D & E, Paraffin-embedded lung sections from HIS-DRAGA mouse F#1 infected with a low dose of virus (2.8×103 pfu) showing lightly infiltrated lung tissue with hCD45+ cells and human T cells, respectively. Of note, the human T cells of mouse F#1 which regained its body weight at 10 dpi, appear to be sequestered within a lung epithelial niche (panel E).
Figure 6. Human T-cell infiltrates in the lungs of a SARS-CoV-2 infected HIS-DRAGA mice.
Panel A, paraffin-embedded lung section from the HIS-DRAGA mouse F#1 infected with a low virus dose (2.8×103 pfu, two right panels) which regained its body weight 10 days post-infection. Shown is a large number of hCD8+ T cells sequestered within the alveolar epithelial niches, and enlargement of the epithelial niche (yellow square) containing hCD8+ T cells that egress into the alveolar air space. Panel B, lung section from the same mouse showing CD4+ T cells sequestered in a lung epithelial niche.
Figure 7. Sequestration of CD8+ T cells in the lung epithelial niches of a HIS-DRAGA mouse infected with a low dose of SARS-CoV-2 virus.
Paraffin-embedded lung section from the HIS-DRAGA female mice infected with a high and low dose, respectively, of SARS-CoV-2 virus were co-stained for CD326 as a marker of lung epithelial cells and/or for hCD8+ T cells using specific fluorescent-labeled antibody conjugates. Panel A, A large cluster of hCD3+ T cells sequestered in the CD326+ lung epithelial niche of the female mouse infected with a low viral dose (2.8×103 pfu), which regained the body mass by 10 dpi. Panel B, A few scattered hCD3+ T cells co-localized with the CD326+ lung epithelia in the HIS-DRAGA female mouse infected with a high viral dose (2.8×104 pfu), which did not regain the body mass by 14 dpi. Also shown are enlargements of the CD3+ T-cell clusters in the alveolar epithelia.
Figure 8. CD8+ T cells sequestered in lung epithelial niches of SARS-CoV-2 infected HIS-DRAGA mice express the CD103 marker of T-cell residency.
Shown are representative paraffin-embedded lung sections from HIS-DRAGA mouse F#1 infected with a low virus dose (2.8×103 pfu) that have been double stained for hCD8 and hCD103. Yellow arrowheads in the enlargements show CD103+ CD8+ T resident cells egressing from epithelial niches into the alveolar air space.
Figure 9. Cytotoxic hCD8+ T cells in the lungs of a SARS-CoV-2 infected HIS-DRAGA mouse.
Panel A, paraffin-embedded lung section from the HIS-DRAGA mouse F#1 infected with a low virus dose (2.8×103 pfu, two right panels), which regained its body weight at 10 dpi. Shown are hCD8+ perforin-secreting T cells sequestered in lung epithelial niches, and two enlarged areas of the lung epithelial niches (Yellow Square). Panel B, A lung section from the same mouse showing a large number of hCD8+ granzyme B-secreting T cells that egressed from the lung epithelial niches into the alveolar air space.
Coagulopathies including microthrombi in the lung, sometimes termed “immunothrombi” due to their association with the hyperinflammatory response, are a feature of severe COVID-19 disease in humans6,66. The lungs of the mouse that survived infection with the high viral dose showed clusters of intra-alveolar mouse CD61+ (platelet glycoprotein IIIa) cells (Fig. 10A), and Mason-Trichrome staining revealed intra-alveolar microthrombi and parenchymal hemorrhagic areas (Fig. 10B). Together, these data show immunopathological events in lungs of HIS-DRAGA mice infected with SARS-Cov-2 similar to those seen in autopsy samples from patients with severe COVID-19 disease.
Figure 10. Intra-alveolar hemorrhagic events in the lungs of a SARS-CoV-2 infected HIS-DRAGA mouse.
Panel A, paraffin-embedded lung section from the HIS-DRAGA mouse F#2 infected with high dose of virus (2.8×104 pfu) showing intra-alveolar murine CD61+ cells (enlargement, yellow square). Panel B, lung section from the same mouse showing multiple intra-alveolar micro thrombi (left two panels, black arrows) and a large intra-alveolar hemorrhagic area (enlargement, Yellow square).
DISCUSSION
There is a critical need for appropriate animal models to study SARS-CoV-2 infection and immunopathological responses23. The HIS-DRAGA mouse offers compelling advantages as a pre-clinical tool for SARS-COVID-19 research and for efficient and informative testing of vaccines and potential therapeutics. This mouse is reconstituted from human umbilical cord blood, expresses a long-lived, fully functional human immune system, and importantly also reconstitutes human endothelial cells, epithelial cells and hepatocytes44,46,50,55. In this pilot study, we have demonstrated that HIS-DRAGA mice express hACE2, the primary receptor for SARS-CoV-2, on engrafted human epithelial cells in the lungs (Fig. 3) and on human endothelial cells in the liver and kidneys (Fig. 11). Notably, we have also demonstrated that they are readily infected via the i.n. route with non-mouse-adapted SARS-CoV-2, sustaining the infection for at least 14 days, and that they can develop severe human-like lung immunopathology, including T-cell infiltrates, alveolar damage and microthrombi. As in recent analyses of human autopsy samples from COVID-19 patients57–59, the lung infiltrates of infected DRAGA mice were intra-parenchymal, relatively diffuse, and grouped around terminal bronchioles and at the periphery.
Figure 11. Expression of hACE2 on liver and kidney endothelial cells of HIS-DRAGA mice.
Panel A, hACE2 expression in the livers of a HIS-DRAGA male (left panels) and female (right panels) mouse, as revealed by S1(RBD)-Fcγ2a + goat anti-mouse IgG-FITC (green color). DAPI co-stain (blue color) shows the cell nuclei. Lower left and right panels: same mice as in the upper panels, showing the lack of hACE2 expression (in adjacent liver sections) when stained with DAPI + secondary antibody (goat anti-mouse IgG-FITC) alone. Panel B, a representative kidney section from a HIS-DRAGA mouse stained with S1(RBD)-Fcγ2a + goat anti-mouse IgG-FITC (left panel) + anti-human CD31-Alexa Fluor 594 (endothelial cell marker, center panel). These images are superimposed in the right-most panel, showing co-localization of hACE2 with endothelial cells.
The female mouse (F#2) that was infected with a high dose of virus (2.8×104 pfu) and did not recover its body mass showed a high titer of S1 (RBD)-specific human IgM antibodies at 14 dpi. We expect that the additional groups of infected HIS-DRAGA mice (e.g., 15 mice currently being infected at Bioqual, Inc.) will develop IgG at later time points (i.e., >21 dpi), similar to HIS-DRAGA mice immunized with an S1-RBD protein construct (manuscript in preparation). In contrast, the female mouse #F1 who recovered the body mass showed no detectable S1 (RBD)-specific human antibodies at 14 dpi, but several granzyme B and perforin positive clusters of human lung-resident CD103+CD8+ T cells in the CD326+ alveolar epithelia at 14 dpi (Figs. 8 & 9), indicating cytotoxic activity. This raises a question that needs to be further addressed, namely, if an early IgM antibody or a lung-resident CD8+ T-cell cytotoxic response to the virus, or a synergistic effect of both IgM and CD8+ T-cell responses can efficiently clear the SARS-CoV-2 infection. The presence of CD4+ and CD8+ T cells reactive to the ORF-1 and NP proteins of SARS-CoV-2 virus in convalescent COVID 19 patients has been reported recently67. The lung infiltrations of SARS-CoV-2 infected HIS-DRAGA mice contained large numbers of T cells, particularly CD8+ T cells, many of which were clustered in the lung epithelial niches and stained positive for granzyme and perforin. Lung-resident CD8+ T cells are developed in mice, monkeys, and humans after a primary viral respiratory infection at the frontline of lung epithelial mucosa60–64, where they provide efficient cross-protection against secondary exposures to the virus68–70. However, exploring the function of lung-resident CD8+ T cells in COVID 19 patients during the infection or those exiting the disease is not feasible, as human studies are restricted to in vitro analyses of lungs collected from cadavers.
The lungs of the surviving infected mice had clusters of cells staining positive for CD61, consistent with the presence of platelets and microthrombi in the alveolar air space, and also mimicking, at least in part, the hemorrhagic areas seen in human lung autopsy samples. Mason-Trichrome staining revealed collagen and fibrin depositions in several areas of the lung. Collagen deposition has been observed in patients with massive destruction of lung tissue following a severe cytokine storm; these are referred to as pulmonary sequels, and they are built during repair of lung tissue damage, late after severe lung infections with upper and lower respiratory viruses71. The infected HIS-DRAGA survivors were sacrificed relatively early after infection (at 14 dpi), in order to evaluate lung pathology. We are now carrying out longer-term studies of 30 infected HIS-DRAGA mice, and is possible that further evidence of collagen sequels and/or fibrin deposition will become apparent at later time points.
This study has several limitations, including analysis of only two mice so far that survived SARS-CoV-2 infection for at least 14 days (although this experiment established lethal and sub-lethal viral doses now being employed in further experiments) and the resulting inability to yet compare tissues from multiple mice. Follow-on studies that are now in progress (analyzing 30 infected mice) will further characterize pathological changes in lungs and other organs of infected HIS-DRAGA mice. Their responses to vaccines and various therapeutics are also of intense interest. The ability to analyze physiological responses as well as tissues harvested from these HIS-mice following initial infections and subsequent challenges with SARS-CoV-2 should allow efficient preclinical testing of vaccines and proposed therapeutics.
MATERIALS AND METHODS
HIS-reconstitution of DRAGA mice:
DRAGA mice express HLA-A2.1 and HLA-DR0401 molecules on a Rag1KO.IL2RγcKO.NOD (NRG) background, and they have been described previously52–55. HLA-A2.1.HLA-DR0401 positive umbilical cord blood samples were obtained from the New York Blood Center (Long Island City, NY, USA). Mice were irradiated (350 rads) and injected intravenously with CD3+ T-cell-depleted cord blood cells (EasySep Human CD3 Positive Selection Kit, Stem Cell Technologies, Vancouver, BC, Canada) containing approximately 105 human HSC (CD34+) as measured by FACS using a mouse anti-human CD34 antibody (BD Biosciences, San Jose, CA, USA) as described49–54. The procedures for assessing human T and B cell reconstitution in peripheral blood by FACS have been described49–52,72. As documented in our previous studies, >90% of DRAGA mice are fully reconstituted. The human reconstitution status, as indicated by analysis of blood from the DRAGA mice used in this study, is shown in Table 1.
Table 1.
Immune parameters of HIS-DRAGA mice
| Mouse* | Gender | Time post-infusion with human stem cells | % human B cells* (CD19+) | % human T cells* (CD3+) | SARS-CoV-2 Challenge dose (pfu) | Cord blood HLA genotype† |
|---|---|---|---|---|---|---|
| M#1 | M | 21 weeks | 4.5 | 13.5 | 2.8×103 | A |
| F#1 | F | 21 weeks | 48.6 | 9.3 | 2.8×103 | A |
| F#2 | F | 21 weeks | 21.9 | 33.4 | 2.8×104 | A |
| a | M | 20 weeks | 18.9 | 28.3 | -- | B |
| b | M | 20 weeks | 0.9 | 41.4 | -- | B |
| c | F | 24 weeks | 52.5 | 17.5 | -- | C |
| d | M | 24 weeks | 7.4 | 16.6 | -- | C |
| e | M | 24 weeks | 1 | 22.1 | -- | C |
| f | M | 24 weeks | 1.2 | 44.7 | -- | C |
| g | M | 24 weeks | 17.6 | 38.3 | -- | C |
| h | F | 20 weeks | 35.2 | 5 | -- | D |
| i | M | 26 weeks | 4.9 | 26.1 | -- | D |
| j | M | 26 weeks | 21.9 | 30.7 | -- | D |
Lungs from uninfected mice a-j were pooled to assess human ACE2 mRNA and protein expression
A = A02:01/A24:02/B13:02/B44:02/DR01:01/DR04:01
B = A02:01/A02:01/B18:01/B44:02/DR04:01/DR11:04
C = A02:01/A02:01/B35:01/B44:02/DR04:01/DR04:04
D = A02:01/A02:01/B08:01/B27:05/DR03:01/DR04:01
RT-PCR detection of hACE2 mRNA in HIS-DRAGA mouse lungs:
RNA was extracted using a Qiagen RNA extraction kit (Qiagen, Hilden, Germany) from lungs of DRAGA and control (non-HSC-infused) mice. Human lung mRNA (Sigma-Aldrich, NJ, USA) served as a positive control. PCR primers specific for hACE2 were: forward, CAGGAAATGTTCAGAAAGCA and reverse, TCTTAGCAGAAAAGGTTGTG). The murine ACE2 specific primers were: forward: AGCAGATGGCCGGAAAGTTG, and reverse: TCTTAGCAGGAAAGGTTGCC. RT-PCR was performed using a One-step RT-PCR kit (Qiagen) for 45 cycles using 1 μg RNA template and 1.6 μM of each primer, following the manufacturer’s instructions. PCR samples were run on 3% agarose gels. PCR bands were purified from the agarose gels and nucleotide sequenced (Eurofins, Coralville, Iowa, USA), which confirmed the identity of human ACE2.
Extraction and quantification of hACE2 protein in HIS-DRAGA lungs:
Lungs from 10 HIS-DRAGA and 10 non-reconstituted DRAGA mice were homogenized in the presence of MPER mammalian protein extraction reagent (Fisher Scientific, Waltham, MA, USA) containing complete protease inhibitor cocktail tablets (Roche Diagnostics GmbH, Mannheim, Germany) using tubes loaded with ceramic beads (MP Biologicals, Irvine, CA, USA) in a Fast-prep homogenizer (MP Biologicals). Pooled lung homogenates from the DRAGA and HIS-DRAGA mice, respectively, were sonicated on ice in a Fisher Ultrasonicator for 10 cycles of 10 seconds each, the cellular debris was removed by centrifugation at 5,000 rpm, and the protein in the supernatant was quantified using a BCA reagent (ThermoFisher Scientific, Waltham, MA, USA). 9 mg of clear supernatant was then incubated with gentle shaking (300 strokes per min) in an Eppendorf thermomixer for 1 h at 37°C with 10 μg of the S1 (RBD)-mFcγ2a chimera (ACRO Biosystems, Newark, DE, USA) followed by incubation with gentle shaking for 1 h at 37°C with 50 μl of rat anti-mouse IgG2a-magnetic microbeads (Miltenyi Biotec, Berdisch Gladback, Germany), and the mixture was passed over MACS magnetic columns (Miltenyi Biotec). The hACE2//S1-(RBD)-mFcγ2a//rat anti-mouse IgG2a-bead complexes were eluted according to the manufacturer’s instructions. As a positive control, 2 mg of human lung total protein (Zyagen, San Diego, CA, USA) was analyzed in parallel following the same IP protocol as that used for the clarified supernatants of the mouse lung homogenates.
The amounts of hACE2 extracted from pooled lysates derived from 10 HIS-DRAGA lungs, 10 non HIS-reconstituted DRAGA lungs and human lung total protein, respectively, were measured using a highly sensitive detection kit for human ACE2 protein (human ACE2 ELISA kit PicoKine, Boster Biological Technology, Pleasanton, CA, USA), as per the manufacturer’s instructions. Absorbances were read at 450nm and standard curves were constructed using a four-parameter (4-PL) logistic curve fit formula. Relative amounts of expressed hACE2 in HIS-DRAGA lungs and in total protein from human lung were calculated by comparing OD450 measurements with background subtraction using BioTEK Gen 5 software (BioTek Instruments, Inc., Winooski, VT, USA).
S1 (RBD)-specific antibodies quantified by ELISA:
Semi-quantitative measurements of S1 (RBD)-specific human IgM and IgG antibody titers in sera from infected mice were carried out using SARS-CoV-2-S1-specific human IgM and IgG ELISA kits (ACRO Biosystems, Newark, DE, USA) according to the manufacturer’s instructions.
Infection of mice with SARS-CoV-2 virus:
HIS-DRAGA mice were infected i.n. with SARS-CoV-2 strain USA-WA1/2020 (BEI Resources NR-52281, batch #70033175), courtesy of the Centers for Diseases Control and Prevention (CDC). This virus strain was originally isolated from an oropharyngeal swab of a patient with a respiratory illness who had returned to Washington State, USA, from travel to China and developed COVID-19 in January 2020. Experiments were conducted in a BSL-3 laboratory at Bioqual, Inc. (Rockville, MD, USA) in compliance with local, state, and federal regulations under IACUC protocol #20–019P. The SARS-COV-2 stock was expanded in Vero E6 cells, and the challenging virus was collected at day 5 of culture when the infection reached 90% cytopathic effect. The full viral genome sequence showed 100% identity with the parent virus sequence listed in GenBank (MN985325.1). A plaque forming assay carried out in confluent layers of Vero E6 cells was used to determine the concentration of live virions, reported as plaque-forming units (pfu).
Histopathology of lungs from infected HIS-DRAGA mice:
Lungs harvested from infected mice at 14 dpi were fixed in 10% formalin, embedded in a paraffin block, and 5 μm sections were stained with Hematoxylin/Eosin (HE) or Mason-Trichrome by Histoserv Inc., (Germantown, MD, USA). Microscopic images were captured using an Olympus BX43 microscope (Shinjuku-ku, Tokyo, Japan).
Immunofluorescence microscopy:
Tissue sections (5μm) from paraffin-embedded cassettes (from infected HIS-DRAGA mice) and from frozen OCT cassettes (from uninfected HIS-DRAGA and from non-reconstituted DRAGA mice) were prepared. Thawed OCT-frozen tissue slides were rehydrated with PBS, and paraffin-embedded sections were de-paraffinized with xylene and rehydrated with graded concentrations of ethanol. Slides were then fixed, permeabilized with fixation/permeabilization buffer (Invitrogen, Waltham, MA, USA), blocked with 3% BSA in PBS for 30 min at 37°C, and stained with fluorochrome-conjugated Abs in PBS containing 0.01% Evans Blue at 37°C for 50 min. To visualize hACE2, slides were probed with the S1 RBD-mFcγ2a chimera (10 μg/ml), washed with PBS, and then incubated with a goat anti-mouse IgG-FITC conjugate (Southern Biotech, Birmingham, AL, USA). Other antibodies to detect antigens of interest were: anti-human CD45-FITC, anti-human CD3-FITC, anti-human CD4-PE, anti-human CD8-PE and anti-human granzyme-B-PE (all from BD Biosciences); anti-human CD326-PE (Miltenyi Biotec), anti-human Perforin-PE (Biolegend, San Diego, CA, USA), anti-human CD103-FITC (BD Pharmingen, Irvine, CA, USA), anti-mouse CD61-PE (Invitrogen), and anti-human CD31- Alexa Fluor 594 (Biolegend). After staining, the slides were washed 3X with PBS, air-dried, and mounted with 10 μl of Vectashield containing DAPI (Vector Laboratories, Burlingame, CA, USA), and images were acquired with a Zeiss Axioscan Confocal microscope equipped with Zeiss Zen software (Jena, Germany).
Supplementary Material
ACKNOWLEDGMENTS:
We acknowledge support from the Department of Medicine and the Collaborative Health Initiative Research Program at Uniformed Services University, Bethesda, MD, the Military Infectious Diseases Research Program (to SC), and NHLBI HL 130448 (to KPP). We thank Dalia Benetiene, Komlan Tevi and Vaneesha Ali for conducting animal experiments at Bioqual, Inc. (Rockville, MD, USA). We thank Prof. David W. Scott for critical reading of the manuscript. The study protocol was reviewed and approved by the Bioqual Institutional Animal Care and Use Committee (#20-019P) and by the Walter Reed Army Institute of Research/Naval Medical Research Center Institutional Animal Care and Use Committee (#19-IDD-24) in compliance with all applicable federal regulations governing the protection of animals and research. This work was supported/funded by work unit number A1210. TDB, KPP and SC are federal employees of the United States government. This work was prepared as part of their official duties. Title 17 U.S.C. 105 provides that `copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. 101 defines a U.S. Government work as work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
REFERENCES
- 1.Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang F, Deng L, Zhang L, Cai Y, Cheung CW, Xia Z. Review of the Clinical Characteristics of Coronavirus Disease 2019 (COVID-19). J Gen Intern Med 2020;35:1545–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye Z, Zhang Y, Wang Y, Huang Z, Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur Radiol 2020;30:4381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev 2020;53:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bikdeli B, Madhavan MV, Jimenez D, et al. COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-Up: JACC State-of-the-Art Review. J Am Coll Cardiol 2020;75:2950–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID-19. Nat Med 2020;26:1017–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desborough MJR, Doyle AJ, Griffiths A, Retter A, Breen KA, Hunt BJ. Image-proven thromboembolism in patients with severe COVID-19 in a tertiary critical care unit in the United Kingdom. Thromb Res 2020;193:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunt BJ, Levi M. Re The source of elevated plasma D-dimer levels in COVID-19 infection. Br J Haematol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020;395:1417–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N Engl J Med 2020;383:120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poissy J, Goutay J, Caplan M, et al. Pulmonary Embolism in Patients With COVID-19: Awareness of an Increased Prevalence. Circulation 2020;142:184–6. [DOI] [PubMed] [Google Scholar]
- 13.Helms J, Severac F, Merdji H, Angles-Cano E, Meziani F. Prothrombotic phenotype in COVID-19 severe patients. Intensive Care Med 2020;46:1502–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helms J, Tacquard C, Severac F, et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: a multicenter prospective cohort study. Intensive Care Med 2020;46:1089–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavoni V, Gianesello L, Pazzi M, Stera C, Meconi T, Frigieri FC. Evaluation of coagulation function by rotation thromboelastometry in critically ill patients with severe COVID-19 pneumonia. J Thromb Thrombolysis 2020;50:281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranucci M, Ballotta A, Di Dedda U, et al. The procoagulant pattern of patients with COVID-19 acute respiratory distress syndrome. J Thromb Haemost 2020;18:1747–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright FL, Vogler TO, Moore EE, et al. Fibrinolysis Shutdown Correlation with Thromboembolic Events in Severe COVID-19 Infection. J Am Coll Surg 2020;231:193–203 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020;181:271–80 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020;367:1444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun 2020;526:135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 2004;203:631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med 2020;14:185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cleary SJ, Pitchford SC, Amison RT, et al. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br J Pharmacol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakdawala SS, Menachery VD. The search for a COVID-19 animal model. Science 2020;368:942–3. [DOI] [PubMed] [Google Scholar]
- 25.Yu J, Tostanoski LH, Peter L, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandrashekar A, Liu J, Martinot AJ, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munster VJ, Feldmann F, Williamson BN, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tseng CT, Huang C, Newman P, et al. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J Virol 2007;81:1162–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luan J, Lu Y, Jin X, Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Commun 2020;526:165–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crackower MA, Sarao R, Oudit GY, et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature 2002;417:822–8. [DOI] [PubMed] [Google Scholar]
- 31.Martina BE, Haagmans BL, Kuiken T, et al. Virology: SARS virus infection of cats and ferrets. Nature 2003;425:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts A, Paddock C, Vogel L, Butler E, Zaki S, Subbarao K. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J Virol 2005;79:5833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts A, Vogel L, Guarner J, et al. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J Virol 2005;79:503–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subbarao K, Roberts A. Is there an ideal animal model for SARS? Trends Microbiol 2006;14:299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang S. Don’t rush to deploy COVID-19 vaccines and drugs without sufficient safety guarantees. Nature 2020;579:321. [DOI] [PubMed] [Google Scholar]
- 36.McCray PB Jr., Pewe L, Wohlford-Lenane C, et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 2007;81:813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreau GB, Burgess SL, Sturek JM, Donlan AN, Petri WA, Mann BJ. Evaluation of K18-hACE2 Mice as a Model of SARS-CoV-2 Infection. Am J Trop Med Hyg 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J, Zhuang Z, Zheng J, et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell 2020;182:734–43 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menachery VD, Yount BL Jr., Sims AC, et al. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A 2016;113:3048–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun SH, Chen Q, Gu HJ, et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe 2020;28:124–33 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Netland J, Meyerholz DK, Moore S, Cassell M, Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol 2008;82:7264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lutz C, Maher L, Lee C, Kang W. COVID-19 preclinical models: human angiotensin-converting enzyme 2 transgenic mice. Hum Genomics 2020;14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis MM. A prescription for human immunology. Immunity 2008;29:835–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akkina R. New generation humanized mice for virus research: comparative aspects and future prospects. Virology 2013;435:14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akkina R. Human immune responses and potential for vaccine assessment in humanized mice. Curr Opin Immunol 2013;25:403–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brehm MA, Wiles MV, Greiner DL, Shultz LD. Generation of improved humanized mouse models for human infectious diseases. J Immunol Methods 2014;410:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ernst W. Humanized mice in infectious diseases. Comp Immunol Microbiol Infect Dis 2016;49:29–38. [DOI] [PubMed] [Google Scholar]
- 48.Danner R, Chaudhari SN, Rosenberger J, et al. Expression of HLA class II molecules in humanized NOD.Rag1KO.IL2RgcKO mice is critical for development and function of human T and B cells. PLoS One 2011;6:e19826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majji S, Wijayalath W, Shashikumar S, et al. Differential effect of HLA class-I versus class-II transgenes on human T and B cell reconstitution and function in NRG mice. Sci Rep 2016;6:28093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wijayalath W, Majji S, Villasante EF, Brumeanu TD, Richie TL, Casares S. Humanized HLA-DR4.RagKO.IL2RgammacKO.NOD (DRAG) mice sustain the complex vertebrate life cycle of Plasmodium falciparum malaria. Malar J 2014;13:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J, Peachman KK, Jobe O, et al. Tracking Human Immunodeficiency Virus-1 Infection in the Humanized DRAG Mouse Model. Front Immunol 2017;8:1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Majji S, Wijayalath W, Shashikumar S, Brumeanu TD, Casares S. Humanized DRAGA mice immunized with Plasmodium falciparum sporozoites and chloroquine elicit protective pre-erythrocytic immunity. Malar J 2018;17:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang L, Morris EK, Aguilera-Olvera R, et al. Dissemination of Orientia tsutsugamushi, a Causative Agent of Scrub Typhus, and Immunological Responses in the Humanized DRAGA Mouse. Front Immunol 2018;9:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendoza M, Ballesteros A, Qiu Q, et al. Generation and testing anti-influenza human monoclonal antibodies in a new humanized mouse model (DRAGA: HLA-A2. HLA-DR4. Rag1 KO. IL-2Rgammac KO. NOD). Hum Vaccin Immunother 2018;14:345–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mendoza M, Gunasekera D, Pratt KP, Qiu Q, Casares S, Brumeanu TD. The humanized DRAGA mouse (HLA-A2. HLA-DR4. RAG1 KO. IL-2R g c KO. NOD) establishes inducible and transmissible models for influenza type A infections. Hum Vaccin Immunother 2020:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020;181:281–92 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bradley BT, Maioli H, Johnston R, et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou B, Zhao W, Feng R, et al. The pathological autopsy of coronavirus disease 2019 (COVID-2019) in China: a review. Pathog Dis 2020;78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sauter JL, Baine MK, Butnor KJ, et al. Insights into pathogenesis of fatal COVID-19 pneumonia from histopathology with immunohistochemical and viral RNA studies. Histopathology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purwar R, Campbell J, Murphy G, Richards WG, Clark RA, Kupper TS. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One 2011;6:e16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner DL, Bickham KL, Thome JJ, et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol 2014;7:501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Bree GJ, van Leeuwen EM, Out TA, Jansen HM, Jonkers RE, van Lier RA. Selective accumulation of differentiated CD8+ T cells specific for respiratory viruses in the human lung. J Exp Med 2005;202:1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pichyangkul S, Yongvanitchit K, Limsalakpetch A, et al. Tissue Distribution of Memory T and B Cells in Rhesus Monkeys following Influenza A Infection. J Immunol 2015;195:4378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park SL, Zaid A, Hor JL, et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol 2018;19:183–91. [DOI] [PubMed] [Google Scholar]
- 65.Litvinov SV, Velders MP, Bakker HA, Fleuren GJ, Warnaar SO. Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J Cell Biol 1994;125:437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levi M, Hunt BJ. Thrombosis and coagulopathy in COVID-19: An illustrated review. Res Pract Thromb Haemost 2020;4:744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Bert N, Tan AT, Kunasegaran K, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020. [DOI] [PubMed] [Google Scholar]
- 68.Wu T, Hu Y, Lee YT, et al. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J Leukoc Biol 2014;95:215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Piet B, de Bree GJ, Smids-Dierdorp BS, et al. CD8(+) T cells with an intraepithelial phenotype upregulate cytotoxic function upon influenza infection in human lung. J Clin Invest 2011;121:2254–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slutter B, Pewe LL, Kaech SM, Harty JT. Lung airway-surveilling CXCR3(hi) memory CD8(+) T cells are critical for protection against influenza A virus. Immunity 2013;39:939–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z, Wang S, Goplen NP, et al. PD-1(hi) CD8(+) resident memory T cells balance immunity and fibrotic sequelae. Sci Immunol 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wijayalath W, Danner R, Kleschenko Y, et al. HLA class II (DR0401) molecules induce Foxp3+ regulatory T cell suppression of B cells in Plasmodium yoelii strain 17XNL malaria. Infect Immun 2014;82:286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.