Abstract
The etiology and pathophysiological mechanisms of idiopathic pulmonary fibrosis (IPF) are yet to be fully elucidated; however, mining of disease-related microRNAs (miRNAs/miRs) has improved the understanding of the progression of IPF. The aim of the current study was to screen miRNAs associated with IPF using three mathematical algorithms: One-way ANOVA, least absolute shrinkage and selector operation (LASSO) and support vector machine-recursive feature elimination (SVM-RFE). Using ANOVA, three miRNAs and two miRNAs were selected with opposite expression patterns in moderate and severe IPF, respectively. In total, two algorithms, LASSO and SVM-RFE, were used to perform feature selection of miRNAs. miRNAs from patients were also extracted from formalin-fixed paraffin-embedded tissues and detected using reverse transcription-quantitative PCR (RT-qPCR). The intersection of the three algorithms (ANOVA, LASSO and SVM-RFE) was taken as the final result of the miRNA candidates. Three miRNA candidates, including miR-124, hsa-miR-524-5p and hsa-miR-194 were therefore used as biomarkers. The receiver operating characteristic model demonstrated favorable discrimination between IPF and control groups, with an area under the curve of 78.5%. Moreover, RT-qPCR results indicated that miR-124, hsa-miR-524-5p, hsa-miR-194 and hsa-miR-133a were differentially expressed between patients with IPF and age-matched men without fibrotic lung disease. The target genes of these miRNAs were further predicted and Kyoto Encyclopedia of Genes and Genomes enrichment analysis was performed. Collectively, the present results suggested that the identified miRNAs associated with IPF may be useful biomarkers for the diagnosis of this disease.
Keywords: idiopathic pulmonary fibrosis, biomarker, support vector machine-recursive feature elimination, microRNA, bioinformatics
Introduction
Idiopathic pulmonary fibrosis (IPF) is a lung disease associated with a lower average survival time of ~2-5 years after initial diagnosis (1). For instance, Wolters et al (2) reported that 50% of patients have a survival time of 3-5 years. The characteristics of IPF include the proliferation of myofibroblasts, excessive accumulation of extracellular matrix and abnormal proliferation of alveolar epithelial cells (2). Moreover, fibroblasts in patients with IPF may originate from the following three sources: i) Excessive proliferation of resident fibroblasts in lung tissue; ii) the epithelial-mesenchymal transition (EMT) of alveolar epithelial cells into fibroblasts; and iii) excessive inflammation, which attracts fibroblasts from other tissues and facilitates their migration to the lungs (3). In total, two drugs are currently available to slow the progression of IPF, including prednisone and nintedanib (4). However, as the pathogenesis of IPF remains unknown, these drugs can only alleviate the progression of pulmonary fibrosis; thus, the disease cannot be fully treated (4).
MicroRNAs (miRNAs/miRs) are non-coding RNA molecules that have been revealed to interact with a variety of mRNAs and affect the expression of target genes (5). Furthermore, miRNAs serve important roles in the pathogenesis of IPF. For example, Rubio et al (6) found that epigenetic gene silencing mediated by the ribonucleoprotein complex multicomponent RNA-protein complex results from reduced levels of miRNA lethal 7 d in IPF. Bodempudi et al (7) also reported that miR-210 promoted the proliferation of IPF fibroblasts to resist hypoxia. Additionally, miR-101 attenuates IPF by inhibiting SMAD3, thus reducing transforming growth factor (TGF)-β expression and inhibiting fibroblast proliferation and activation (8). Other previous studies have shown that the downregulation of miR-9 targets anoctamin-1, which results in decreased expression levels of TGF-β and SMAD3, slowed progression of IPF and reduced fibroblast proliferation in bleomycin-induced mice (9,10).
Therefore, the aim of the present study was to comprehensively identify and quantify miRNAs involved in the occurrence and development of IPF using machine learning. The results will improve the understanding of miRNAs in patients with IPF, as well as demonstrate the differential expression of miRNAs and the function of target genes during IPF. In addition, the current results may provide miRNA expression profiles and potential biomarkers for IPF, thus improving diagnosis and treatment strategies.
Materials and methods
Data collection and patient population
In the discovery phase, array data (GSE129126) from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) were used as the finding set. This dataset included 28 samples (eight healthy lung tissues and 20 lung tissues from patients with IPF) from individuals with forced vital capacities of >80, 50-80 or <50%, respectively (8). GSE13316 array data were also included as the validation set with 20 samples (ten healthy lung tissues and ten lung tissues from patients with IPF) (11).
Paraffin-embedded tissue samples were collected from 4 men with IPF (age range, 56-73 years; median age, 63 years) or 3 age-matched men without fibrotic lung disease (controls) at Shenyang Thoracic Hospital and Fushun Central Hospital of Liaoning Province. All patients were enrolled between August 2001 and December 2016 (Table SI). The research was approved by the Ethics Committee of Shenyang Thoracic Hospital and Fushun Central Hospital of Liaoning Province. All selected patients or their families provided oral informed consent for participation in the study.
Bioinformatics analysis Identification of differentially expressed miRNAs in patients with IPF
The K-nearest neighbor and β-mixture quintile dilation methods (12) were used to perform imputation and normalization in the GSE129126 dataset. Differences among the three groups were tested using single factor ANOVA with post hoc Fisher's LSD. Results with P<0.05 were considered significant. To further analyze the GEO data, glmnet (version 4.0-2; http://cran.r-project.org/web/packages/glmnet/index.html) (13) and e1071 (version 1.7-3; https://cran.r-project.org/web/packages/e1071/index.html) R packages were used to establish a least absolute shrinkage and selector operation (LASSO) model and a support vector machine-recursive feature elimination (SVM-RFE) model, respectively. After primary filtration, a LASSO algorithm, with penalty parameter tuning conducted using 5-fold cross-validation, was constructed to select candidate miRNAs. In R 3.50 software (14), the minimum absolute contraction of glmnet and selection operator (LASSO) Cox regression were used to determine the best candidate, ignoring miRNAs with a regression coefficient of <0.1. Then, miRNAs from the LASSO and SVM-RFE algorithms and ANOVA were combined; the obtained miRNAs were considered potential markers.
Validation of the IPF signature
The validation set was used for internal validation. The R package ‘pROC’ (version 1.16.2; https://CRAN.R-project.org/package=pROC) was used to analyze the receiver operating characteristic curve (ROC) with area under the curve (AUC) analysis. miRNAs with an AUC >0.7 were considered ideal biomarkers (15).
Construction of the miRNA/mRNA network
miRNA target sites were retrieved using miRDB (version 6.0; www.mirdb.org) (16). mRNAs with a target score >90 were selected as target genes and used to construct the miRNA/mRNA network using Cytoscape software (version 3.8.0; https://cytoscape.org/).
Functional enrichment analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/) annotation system and cumulative hypergeometric distribution were used to determine the enrichment pathways of the targeted mRNAs. ClusterProfiler (version 3.11) (17) was used to annotate and analyze KEGG data. The screening conditions were as follows: Enrichment score >2, and false discovery rate <0.05.
Histological analysis
Formalin-fixed (formalin concentration, 10%; 12-24 h at room temperature), paraffin-embedded lung sections (thickness, 2-5 µm) were deparaffinized, stained with hematoxylin (3-5 min) and eosin (2-3 min) at room temperature and diagnosed by a lung pathologist using a blinded method. Tissue sections were dehydrated in xylene I (15 min), xylene II (5 min), 100% ethanol (twice for 5 min), 95% ethanol (2 min), 85% ethanol (2 min) and 75% ethanol (2 min) at room temperature. After soaking for 2 min in distilled water, the slices were stained with hematoxylin (Beijing Solarbio Science & Technology Co., Ltd.; cat. no. H8070) for 20 min and soaked in tap water for 3 min at room temperature. The slices were then re-differentiated in 1% hydrochloric acid-ethanol solution (5 sec) and washed with tap water for 10 min to return to a blue color. Slices were then re-stained with eosin (Beijing Solarbio Science & Technology Co., Ltd.; cat. no. G1100) for 1 min and washed with distilled water for 30 sec to terminate the staining at room temperature. Samples were then sealed with neutral resin glue and observed under an optical microscope (Nikon Corporation; magnification, x200).
Extraction of miRNAs and detection using reverse transcription-quantitative PCR (RT-qPCR)
A miRNeasy FFPE kit (Qiagen GmbH; cat. no. 217504) was used to extract miRNAs from paraffin-embedded samples from the IPF and control groups. The extracted miRNAs were reverse transcribed to cDNA using miScript II RT kits (Qiagen GmbH; cat. no. 218160) at 42˚C for 60 min. cDNA was stored in a -20˚C refrigerator until subsequent analysis using RT-qPCR, which was performed according to a previously described method to amplify miRNAs (18). Reactions were conducted using SYBR Green PCR Master Mix (Thermo Fisher Scientific, Inc.; cat. no. 4309155) following the manufacturer's instructions for RT-qPCR. Triplicate reactions were performed in a QuantStudio 7 system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the following thermocycling conditions: Initial denaturation at 95˚C for 8 min, followed by 40 cycles of 95˚C for 15 sec, 60˚C for 15 sec, 72˚C for 15 sec, and a final extension step at 55˚C for 15 sec. Using the 2-ΔΔCq method (19), the relative expression levels of hsa-miR-221 hsa-miR-524-5p, hsa-miR-194, hsa-miRPlus-E1092, hsa-miR-17 and hsa-miR-133a (Table SII) were normalized to U6 small nuclear RNA.
Statistical analysis
If the data were normally distributed, the measurement data between the two groups were compared using the independent sample t-test, and the measurement data of ≥3 groups were compared using Fisher's and Welch's one-way ANOVA with post hoc Fisher's LSD. If the results demonstrated that there was a significant difference, then the non-parametric Mann-Whitney test was used for comparison of two groups when the data were of skewed distribution. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using R version 3.5(14). In each experiment, data are presented as the mean ± SEM. All experiments were repeated ≥3 times.
Results
Identification of differentially expressed miRNAs in patients with IPF using ANOVA
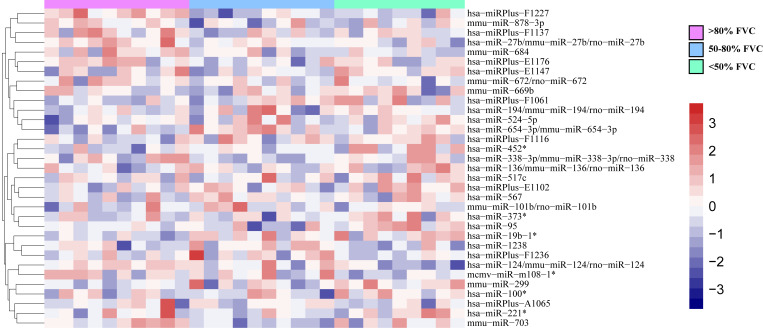
To identify IPF-specific genes and key biomarkers, ANOVA was used to evaluate gene expression at specific disease stages. Using the significance criteria (|Log2FC|≥1; P<0.05) to filter the miRNAs, miRNAs as markers of IPF at different stages determined according to the pre-bronchodilator forced vital capacity % values, such as hsa-miR-124, hsa-miR-133a and hsa-miR-524-5p, were obtained using one-way ANOVA. (Fig. 1).
Figure 1.
Heatmap of differentially expressed miRNAs. Differentially expressed genes screened by ANOVA. Differentially expressed genes are represented by heatmap. Red indicates upregulation, blue indicates downregulation. miR, microRNA; FVC, forced vital capacities.
Feature selection of IPF using LASSO and SVM-RFE
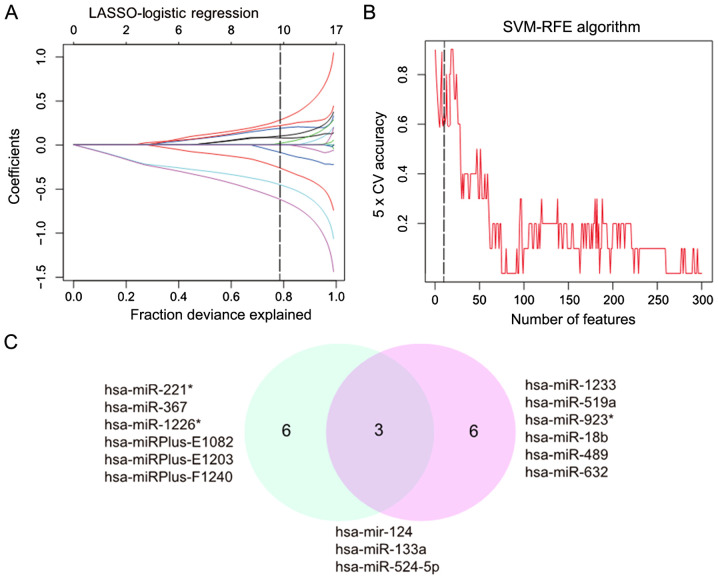
The LASSO algorithm was used to distinguish between healthy lung tissues and mild IPF. In total, nine eigenvalues were obtained (Fig. 2A). Of the 300 differentially expressed miRNAs, nine demonstrated features with non-zero coefficients in the LASSO logistic regression model, including four upregulated and five downregulated miRNAs, and were selected on the basis of the training set for mild IPF (Fig. 2B). From the two algorithms, hsa-miR-124, hsa-miR-133a and hsa-miR-524-5p were identified (Fig. 2C). Among these miRNAs, hsa-miR-124 appeared in the three algorithms, demonstrating the reliability of the association between mild IPF and miR-124.
Figure 2.
Using LASSO and SVM-RFE algorithm to identify the key eigenvalues of mild IPF. (A) Eigenvalues between mild IPF and healthy lungs calculated using LASSO. (B) Eigenvalues between moderate IPF and healthy lungs calculated using SVM-RFE. (C) Venn graph of eigenvalues filtered using SVM-RFE (blue) and LASSO (red). IPF, Idiopathic Pulmonary Fibrosis; LASSO, least absolute shrinkage and selector operation; SVM-RFE, support vector machine-recursive feature elimination; miR, microRNA.
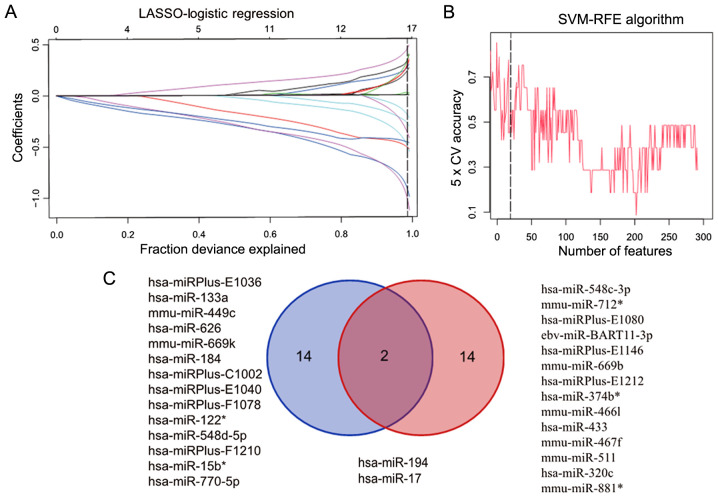
When comparing advanced fibrosis with healthy lung tissue, the LASSO algorithm calculated a total of 16 features (Fig. 3A), and SVM-RFE selected the first 16 features (Fig. 3B). Further analysis identified that hsa-miR-194 and hsa-miR-17 were found in both algorithms (Fig. 3C).
Figure 3.
Using LASSO and SVM-RFE algorithm to identify the key eigenvalues of severe IPF. (A) Eigenvalues between severe IPF and healthy lungs calculated using LASSO. (B) Eigenvalues between severe IPF and healthy lungs calculated using SVM-RFE. (C) Venn graph of eigenvalues filtered using SVM-RFE (blue) and LASSO (red). IPF, Idiopathic Pulmonary Fibrosis; LASSO, least absolute shrinkage and selector operation; SVM-RFE, support vector machine-recursive feature elimination; miR, microRNA.
Verification of biomarkers using ROC curves
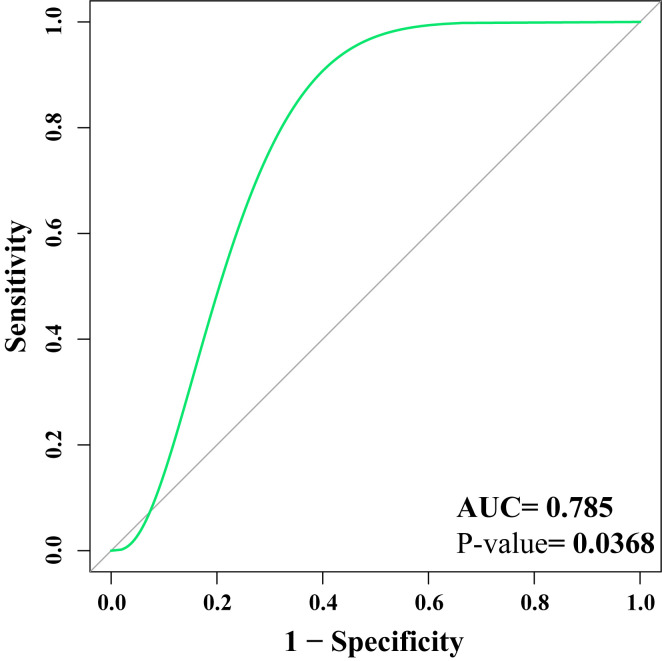
GSE13316 was used as the verification set to assess the potential biomarkers. The results demonstrated that hsa-miR-124, hsa-miR-194 and hsa-miR-524-5p could distinguish between healthy tissue and pulmonary fibrosis tissue (AUC=0.785; P=0.0368), indicating that the biomarkers may be clinically useful (Fig. 4). The ROC curve was above the diagonal, indicating a good sensitivity (96.52%) and specificity (96.32%).
Figure 4.
ROC curves for hsa-miR-124, hsa-miR-194 and hsa-miR-524-5p. ROC curves for hsa-mir-124, hsa-miR-194 and hsa-miR-524-5p. ROC, receiver operating characteristic curve; miR, microRNA; AUC, area under the curve.
Identification of differential miRNA expression using RT-qPCR
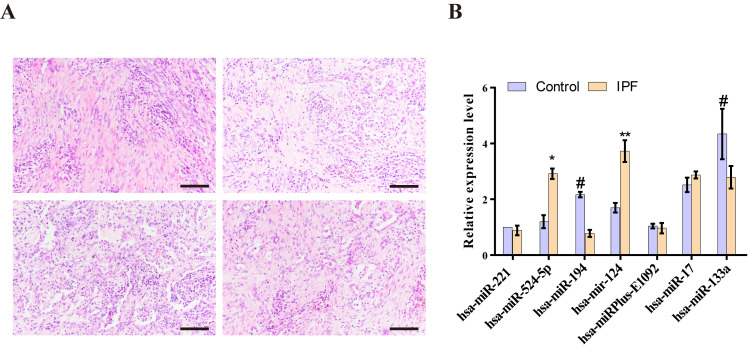
The number of interstitial cells increased and interstitial sclerosis appeared in the lungs of the patients with IPF. Furthermore, inflammatory cell infiltration was found around the interstitial lesion, and an inflammatory reaction was identified in the lung tissue around the lesion area in the patients with IPF (Fig. 5A).
Figure 5.
Biomarkers are significantly expressed in the lung tissues of patients with IPF. (A) Representative images of hematoxylin and eosin stained lung tissues from patients with IPF; magnification, x200; scale bar, 100 µm. (B) Differentially expressed miRNAs in IPF lung tissues and control tissue were measured using reverse transcription-quantitative PCR. Data are presented as the mean ± SEM. *P<0.05, **P<0.01 vs. Control, #P<0.05 vs. IPF.
It was demonstrated that miR-124 (P=0.0086) and hsa-miR-524-5p (P=0.0362) were highly expressed in the IPF group compared with the control (Fig. 5B). These results suggested that these miRNAs may contribute to IPF by reducing the expression of target genes. Moreover, hsa-miR-194 (P=0.0399) and hsa-miR-133a (P=0.0489) were expressed at lower levels in patients with IPF compared with the control group. Therefore, the results indicated that these miRNAs may also contribute to IPF by increasing the expression of target genes. Other differentially expressed miRNAs were detected in the lungs of patients with IPF and age-matched men without fibrotic lung disease; however, the differences were not statistically significant (Fig. 5B).
Functional enrichment analysis
miRDB was used to predict the mRNAs targeted by miR-124, hsa-miR-194 and hsa-miR-524-5p, and selected genes with a target score >90 as candidate genes. In total, 329 genes targeted by miR-124 were enriched in ‘PI3K/AKT’, ‘cAMP’ and ‘mitogen-activated protein kinase’ (MAPK) signaling pathways. In addition, 63 genes targeted by miR-194 were enriched in the regulation of signaling pathways, such as ‘stem cell pluripotency’, ‘epithelial cell signal transduction’ and ‘lysine degradation’. A total of 401 genes targeted by hsa-miR-524-5p were enriched in the ‘MAPK’, ‘amphetamine addiction’ and ‘cAMP’ signaling pathways. The top six input pathways with P<0.05 enriched by the three miRNAs are presented in Table SIII. Most of these signaling pathways were cascade signaling pathways that regulate ‘growth’, ‘differentiation’ and the ‘EMT’ in fibroblasts.
Discussion
IPF is a progressive disease; however, as current treatments can only delay the disease progression, there is an urgent need for improved methods to treat IPF (20). miRNAs are important regulators of cell function during disease and can serve roles in other cells via exocrine secretion (21,22). The present study analyzed miRNA expression profiles at different stages of IPF to identify miRNAs involved in disease progression. To further reduce the number of differentially expressed miRNAs, LASSO and SVM-RFE were used to calculate the differential gene set. In total, four miRNA were obtained by intersecting the results with ANOVA. At present, to the best of our knowledge, no in-depth studies regarding the expression levels of these three miRNAs in tissue samples of patients with IPF have been published. However, in other diseases, these three miRNAs have been reported to be associated with the extracellular matrix and TGF-β signaling, supporting the relevance of the present analysis in IPF.
Lu et al (23) demonstrated that miR-124 responded to TGF-β1-induced fibrogenic differentiation by regulating Axin-1 expression and activating the Wnt signaling pathway. Panganiban et al (24) revealed significant changes in the serum levels of miR-124-26a, let-7a and let-7d in patients with asthma compared with healthy controls, which suggested that miR-124 may be involved in the development of lung asthma. Moreover, Chen et al (25) demonstrated the inhibitory effects of miR-124 on the DNA repair enzyme poly(ADP) ribose polymerase 1. In addition, overexpression of miR-124 reduces DNA repair ability and leads to a decrease in the drug sensitivity of cells (25). Liang et al (26) also reported that miR-124 may regulate the EMT process by targeting snail family transcriptional repressor 2 to promote breast cancer metastasis, while Cui et al (27) suggested that miR-124 may induce hepatocellular carcinoma metastasis by targeting Slug. Additionally, homeoboxA11 has been shown to induce the formation of type I collagen in scars via the activation of miR-124-3p and the SMAD signaling pathway in the cavernous body (28). Thus, miR-124 contributes to the formation of fibrotic tissue in a variety of diseases by responding to the TGF-β and EMT signaling pathways.
miR-524-5p is a member of the primate-specific chromosome 19 miRNA cluster (C19MC), which is highly homologous to reprogrammed miR-520d-5p. Nguyen et al (29) demonstrated that miR-524-5p regulated stem cell programming by targeting tumor protein p53 and EMT-related genes. Similarly, Liu et al (27) reported that miR-524-5p can positively regulate the expression of distal homeobox 1 and modulate the TGF-β signaling pathway by competing with taurine upregulated-1. It has also been shown that low expression of miR-524-5p in thyroid papillary carcinoma increases the expression levels of Forkhead box protein E1 and integrin subunit α3 in thyroid papillary carcinoma, thus inhibiting cell migration and proliferation and promoting apoptosis (27). Eftekharian et al (30) demonstrated that the expression of miR-524-5p was significantly lower in the peripheral blood of patients with multiple sclerosis compared with healthy controls. Moreover, miR-524-5p can be used as a biomarker of the response to fengomod in patients with multiple sclerosis (30). Zhao et al (31) also revealed that high expression levels of miR-524-3p and miR-524-5p inhibit the TGF-β, Notch and Hippo pathways by targeting SMAD2, hairy and enhancer of split-1 and TEA domain transcription factor 1, respectively. It has also been reported that knockout of H19 imprinted maternally expressed transcript inhibits the activation of the TGF-β/SMAD3 pathway by regulating miR-140. The aforementioned results suggested that miR-524-5p, a member of the C19MC, may serve the same role as miR-140 (32). Furthermore, the current findings indicated that miR-524-5p was involved in signaling pathways and target genes associated with IPF. For instance, miR-524-5p was shown to be downregulated in patients with moderate IPF compared with that in other groups, and was upregulated in healthy lung tissues. Therefore, it was speculated that this miRNA may be involved in the EMT process during the development of IPF.
Xu et al (33) reported that high expression of miR-194-3p inhibits the proliferation and migration of fibroblasts by directly blocking the expression of genes encoding cyclin-dependent kinase 4 and matrix metalloproteinase 2, as well as interacting with RUNX family transcription factor 2 in keloids. Furthermore, Hu et al (34) showed that miR-194 can be used as a biomarker of drug resistance in non-small lung cancer. Downregulation of miR-194 reduces nuclear accumulation of β-catenin and inhibits the Wnt signaling pathway in gastric cancer (35). In addition, the proliferation and infiltration of breast cancer cells are inhibited by knockout of miR-194, which regulates the Wnt/β-catenin signaling pathway (36). Miao et al (37) also revealed that miR-194 targets cadherin 2 (CDH2) to inhibit cell expansion and promote apoptosis in osteosarcoma cells. Thus, miR-194 is involved in the EMT and cell adhesion in a variety of diseases by regulating various genes, such as Wnt and CDH2. Of the three significant miRNAs identified in the present study, including miR-124, hsa-miR-524-5p and hsa-miR-194, two may be used as biomarkers in the diagnosis of IPF. All three miRNAs are related to the EMT process, and the EMT is a key process involved in the pathogenesis of IPF (38-40). Therefore, the present results suggested that the three miRNAs were specific to, and significant for, IPF.
Various changes in miRNA expression have been identified in IPF (41). For example, miR-21 positively regulates IPF by targeting the TGF-β inhibitor SMAD7 and reducing the phosphorylation level of the SMAD2 complex (42). In addition, significant upregulation of serum EVmiR-21-5p is observed in acute and chronic/late fibrosis in a mouse bleomycin-induced lung fibrosis model (43). The upstream region of the miR-154 promoter contains the binding site for the transcription factor SMAD3. In the pathogenesis of IPF, the TGF-β signaling pathway enhances the phosphorylation of SMAD3 and promotes the transcription of miR-154 (44). Subsequently, upregulation of miR-154 protects myofibroblasts from apoptosis by regulating the activity of cyclin (45).
The current results indicated that there were significant differences in the expression levels of miR-124, hsa-miR-524-5p and hsa-miR-194 between patients with IPF and age-matched men without fibrotic lung disease. These findings further support the reliability of the present analysis results. However, due to the small number of samples, additional studies are required to assess the present findings. In addition to verifying the aforementioned three miRNAs, other differences in miRNA expression levels were demonstrated using two machine learning models. For example, hsa-miR-133a expression was found to be significantly different between the experimental and control groups, although the ROC curve AUC was <0.5. Thus, it was suggested that hsa-miR-133a could not be used as a potential biomarker to distinguish pulmonary fibrosis tissue from healthy tissue. Therefore, due to limitations of this study, prospective studies with larger sample sizes are required to confirm the current results. Moreover, additional RT-qPCR experiments should be performed using known IPF biomarkers as reference miRNAs, such as miR-29b and miR-let-7d.
In conclusion, the present study demonstrated the expression patterns of miRNAs in different stages of IPF using ANOVA and identified candidate biomarkers of IPF using two machine learning models. The reliability of these candidate markers was assessed using a validation dataset. The current study successfully identified nine specific miRNAs and obtained a collection of biomarkers including three miRNAs (AUC=0.785). The miRNA expression levels in patients with IPF and age-matched men without fibrotic lung disease were compared using RT-qPCR, and the differences in the expression levels of the four miRNAs obtained were further evaluated using bioinformatics methods.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was supported by the ‘Financial Support for Selected Researchers Back from Abroad (2012)’ from Liaoning Province (grant no. 88030312004).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
QL wrote the article and analyzed the bioinformatics data. ML performed the mathematical modeling and ROC calculations. KZ, HL and HY performed sectioning and analysis of qPCR and IPF data. SM and MZ designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The research was approved by the Ethics Committee of Shenyang Thoracic Hospital and Fushun Central Hospital of Liaoning Province. All selected patients or their families provided informed consent for participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Raghu G, Freudenberger TD, Yang S, Curtis JR, Spada C, Hayes J, Sillery JK, Pope CE II, Pellegrini CA. High prevalence of abnormal acid gastro-oesophageal reflux in idiopathic pulmonary fibrosis. Eur Respir J. 2006;27:136–142. doi: 10.1183/09031936.06.00037005. [DOI] [PubMed] [Google Scholar]
- 2.Wolters PJ, Blackwell TS, Eickelberg O, Loyd JE, Kaminski N, Jenkins G, Maher TM, Molina-Molina M, Noble PW, Raghu G, et al. Time for a change: Is idiopathic pulmonary fibrosis still idiopathic and only fibrotic? Lancet Respir Med. 2018;6:154–160. doi: 10.1016/S2213-2600(18)30007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selman M, Pardo A. Revealing the pathogenic and aging-related mechanisms of the enigmatic idiopathic pulmonary fibrosis An integral model. Am J Respir Crit Care Med. 2014;189:1161–1172. doi: 10.1164/rccm.201312-2221PP. [DOI] [PubMed] [Google Scholar]
- 4.Antoniou K, Markopoulou K, Tzouvelekis A, Trachalaki A, Vasarmidi E, Organtzis J, Tzilas V, Bouros E, Kounti G, Rampiadou C, et al. Efficacy and safety of nintedanib in a Greek multicentre idiopathic pulmonary fibrosis registry: A retrospective, observational, cohort study. ERJ Open Res. 2020;6:00172–2019. doi: 10.1183/23120541.00172-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kooshapur H, Choudhury NR, Simon B, Mühlbauer M, Jussupow A, Fernandez N, Jones AN, Dallmann A, Gabel F, Camilloni C, et al. Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1. Nat Commun. 2018;9(2479) doi: 10.1038/s41467-018-04871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubio K, Singh I, Dobersch S, Sarvari P, Günther S, Cordero J, Mehta A, Wujak L, Cabrera-Fuentes H, Chao CM, et al. Inactivation of nuclear histone deacetylases by EP300 disrupts the MiCEE complex in idiopathic pulmonary fibrosis. Nat Commun. 2019;10(2229) doi: 10.1038/s41467-019-10066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodempudi V, Hergert P, Smith K, Xia H, Herrera J, Peterson M, Khalil W, Kahm J, Bitterman PB, Henke CA. MiR-210 promotes IPF fibroblast proliferation in response to hypoxia. Am J Physiol Lung Cell Mol Physiol. 2014;307:L283–L294. doi: 10.1152/ajplung.00069.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Xiao X, Yang Y, Mishra A, Liang Y, Zeng X, Yang X, Xu D, Blackburn MR, Henke CA, Liu L. MicroRNA-101 attenuates pulmonary fibrosis by inhibiting fibroblast proliferation and activation. J Biol Chem. 2017;292:16420–16439. doi: 10.1074/jbc.M117.805747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai WJ, Qiu J, Sun J, Ma CL, Huang N, Jiang Y, Zeng J, Ren BC, Li WC, Li YH. Downregulation of microRNA-9 reduces inflammatory response and fibroblast proliferation in mice with idiopathic pulmonary fibrosis through the ANO1-mediated TGF-β-Smad3 pathway. J Cell Physiol. 2019;234:2552–2565. doi: 10.1002/jcp.26961. [DOI] [PubMed] [Google Scholar]
- 10.Kang H. Role of MicroRNAs in TGF-β signaling pathway-mediated pulmonary fibrosis. Int J Mol Sci. 2017;18(2527) doi: 10.3390/ijms18122527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sung J, Loughin C, Marino D, Leyva F, Dewey C, Umbaugh S, Lesser M. Medical infrared thermal imaging of canine appendicular bone neoplasia. BMC Vet Res. 2019;15(430) doi: 10.1186/s12917-019-2180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 14. Team RC: R: A language and environment for statistical computing, 2013. [Google Scholar]
- 15.Xia J, Broadhurst DI, Wilson M, Wishart DS. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics. 2013;9:280–299. doi: 10.1007/s11306-012-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(18) doi: 10.1186/s13059-019-1629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buglyó G, Magyar Z, Romicsné Görbe É, Bánusz R, Csóka M, Micsik T, Berki Z, Varga P, Sápi Z, Nagy B. Quantitative RT-PCR-based miRNA profiling of blastemal Wilms' tumors from formalin-fixed paraffin-embedded samples. J Biotechnol. 2019;298:11–15. doi: 10.1016/j.jbiotec.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Akiyama N, Hozumi H, Isayama T, Okada J, Sugiura K, Yasui H, Suzuki Y, Kono M, Karayama M, Furuhashi K, et al. Clinical significance of serum S100 calcium-binding protein A4 in idiopathic pulmonary fibrosis. Respirology. 2020;25:743–749. doi: 10.1111/resp.13707. [DOI] [PubMed] [Google Scholar]
- 21.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116:751–762. doi: 10.1161/CIRCRESAHA.116.303549. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y, Zhang T, Shan S, Wang S, Bian W, Ren T, Yang D. miR-124 regulates transforming growth factor-β1 induced differentiation of lung resident mesenchymal stem cells to myofibroblast by repressing Wnt/β-catenin signaling. Dev Biol. 2019;449:115–121. doi: 10.1016/j.ydbio.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Panganiban RP, Pinkerton MH, Maru SY, Jefferson SJ, Roff AN, Ishmael FT. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol. 2012;1:154–165. [PMC free article] [PubMed] [Google Scholar]
- 25.Chen SM, Chou WC, Hu LY, Hsiung CN, Chu HW, Huang YL, Hsu HM, Yu JC, Shen CY. The effect of microRNA-124 overexpression on anti-tumor drug sensitivity. PLoS One. 2015;10(e0128472) doi: 10.1371/journal.pone.0128472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang YJ, Wang QY, Zhou CX, Yin QQ, He M, Yu XT, Cao DX, Chen GQ, He JR, Zhao Q. MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34:713–722. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang YJ, Wang QY, Zhou CX, Yin QQ, He M, Yu XT, Cao DX, Chen GQ, He JR, Zhao Q. miR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34:713–722. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin J, Zhai HF, Jia ZH, Luo XH. Long non-coding RNA HOXA11-AS induces type I collagen synthesis to stimulate keloid formation via sponging miR-124-3p and activation of Smad5 signaling. Am J Physiol Cell Physiol. 2019;317:C1001–C1010. doi: 10.1152/ajpcell.00319.2018. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen PNN, Choo KB, Huang CJ, Sugii S, Cheong SK, Kamarul T. MiR-524-5p of the primate-specific C19MC miRNA cluster targets TP53IPN1-and EMT-associated genes to regulate cellular reprogramming. Stem Cell Res Ther. 2017;8(214) doi: 10.1186/s13287-017-0666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eftekharian MM, Komaki A, Mazdeh M, Arsang-Jang S, Taheri M, Ghafouri-Fard S. Expression profile of selected microRNAs in the peripheral blood of Multiple Sclerosis patients: A multivariate statistical analysis with ROC curve to find new biomarkers for fingolimod. J Mol Neurosci. 2019;68:153–161. doi: 10.1007/s12031-019-01294-z. [DOI] [PubMed] [Google Scholar]
- 31.Zhao K, Wang Q, Wang Y, Huang K, Yang C, Li Y, Yi K, Kang C. EGFR/c-myc axis regulates TGFβ/Hippo/Notch pathway via epigenetic silencing miR-524 in gliomas. Cancer Lett. 2017;406:12–21. doi: 10.1016/j.canlet.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Cheng Z, Dai L, Jiang T, Jia L, Jing X, An L, Wang H, Liu M. Knockdown of long noncoding RNA H19 represses the progress of pulmonary fibrosis through the transforming growth factor β/Smad3 pathway by regulating microRNA 140. Mol Cell Biol. 2019;39:e00143–00119. doi: 10.1128/MCB.00143-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Z, Guo B, Chang P, Hui Q, Li W, Tao K. The differential expression of miRNAs and a preliminary study on the mechanism of miR-194-3p in Keloids. Biomed Res Int. 2019;2019(8214923) doi: 10.1155/2019/8214923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu S, Yuan Y, Song Z, Yan D, Kong X. Expression profiles of microRNAs in drug-resistant non-small cell lung cancer cell lines using microRNA sequencing. Cell Physiol Biochem. 2018;51:2509–2522. doi: 10.1159/000495921. [DOI] [PubMed] [Google Scholar]
- 35.Peng Y, Zhang X, Lin H, Deng S, Huang Y, Qin Y, Feng X, Yan R, Zhao Y, Cheng Y, et al. Inhibition of miR-194 suppresses the Wnt/β-catenin signalling pathway in gastric cancer. Oncol Rep. 2018;40:3323–3334. doi: 10.3892/or.2018.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang F, Xiao Z, Zhang S. Knockdown of miR-194-5p inhibits cell proliferation, migration and invasion in breast cancer by regulating the Wnt/β-catenin signaling pathway. Int J Mol Med. 2018;42:3355–3363. doi: 10.3892/ijmm.2018.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao J, Wang W, Wu S, Zang X, Li Y, Wang J, Zhan R, Gao M, Hu M, Li J, Chen S. MiR-194 suppresses proliferation and migration and promotes apoptosis of osteosarcoma cells by targeting CDH2. Cell Physiol Biochem. 2018;45:1966–1974. doi: 10.1159/000487973. [DOI] [PubMed] [Google Scholar]
- 38.Hill C, Li J, Liu D, Conforti F, Brereton CJ, Yao L, Zhou Y, Alzetani A, Chee SJ, Marshall BG, et al. Autophagy inhibition-mediated epithelial-mesenchymal transition augments local myofibroblast differentiation in pulmonary fibrosis. Cell Death Dis. 2019;10(591) doi: 10.1038/s41419-019-1820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milara J, Navarro R, Juan G, Peiró T, Serrano A, Ramón M, Morcillo E, Cortijo J. Sphingosine-1-phosphate is increased in patients with idiopathic pulmonary fibrosis and mediates epithelial to mesenchymal transition. Thorax. 2012;67:147–156. doi: 10.1136/thoraxjnl-2011-200026. [DOI] [PubMed] [Google Scholar]
- 40.Senoo T, Hattori N, Tanimoto T, Furonaka M, Ishikawa N, Fujitaka K, Haruta Y, Murai H, Yokoyama A, Kohno N. Suppression of plasminogen activator inhibitor-1 by RNA interference attenuates pulmonary fibrosis. Thorax. 2010;65:334–340. doi: 10.1136/thx.2009.119974. [DOI] [PubMed] [Google Scholar]
- 41.Parker MW, Rossi D, Peterson M, Smith K, Sikström K, White ES, Connett JE, Henke CA, Larsson O, Bitterman PB. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J Clin Invest. 2014;124:1622–1635. doi: 10.1172/JCI71386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. MiR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makiguchi T, Yamada M, Yoshioka Y, Sugiura H, Koarai A, Chiba S, Fujino N, Tojo Y, Ota C, Kubo H, et al. Serum extracellular vesicular miR-21-5p is a predictor of the prognosis in idiopathic pulmonary fibrosis. Respir Res. 2016;17(110) doi: 10.1186/s12931-016-0427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chao CM, Carraro G, Rako Z, Kolck J, Morty RE, Rottier RJ, Bellusci S. MiR-154 controls branching morphogenesis and alveologenesis in lung development involving Tgf-β signaling. Eur Respir J. 2017;50 (Suppl 61)(OA3229) [Google Scholar]
- 45.Milosevic J, Pandit K, Magister M, Rabinovich E, Ellwanger DC, Yu G, Vuga LJ, Weksler B, Benos PV, Gibson KF, et al. Profibrotic role of miR-154 in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2012;47:879–887. doi: 10.1165/rcmb.2011-0377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.