Abstract
Background
Some COVID-19 cases test positive again for SARS-CoV-2 RNA following negative test results and discharge, raising questions about the meaning of virus detection. Better characterization of re-positive cases is urgently needed.
Methods
Clinical data were obtained through Guangdong's COVID-19 surveillance network. Neutralization antibody titre was determined using microneutralization assays. Potential infectivity of clinical samples was evaluated by cell inoculation. SARS-CoV-2 RNA was detected using three different RT-PCR kits and multiplex PCR with nanopore sequencing.
Findings
Among 619 discharged COVID-19 cases, 87 re-tested as SARS-CoV-2 positive in circumstances of social isolation. All re-positive cases had mild or moderate symptoms at initial diagnosis and were younger on average (median, 28). Re-positive cases (n = 59) exhibited similar neutralization antibodies (NAbs) titre distributions to other COVID-19 cases (n = 218) tested here. No infectious strain could be obtained by culture and no full-length viral genomes could be sequenced from re-positive cases.
Interpretation
Re-positive SARS-CoV-2 cases do not appear to be caused by active reinfection and were identified in ~14% of discharged cases. A robust NAb response and potential virus genome degradation were detected in almost all re-positive cases, suggesting a substantially lower transmission risk, especially through respiratory routes.
Keywords: Re-positive, SARS-CoV-2, COVID-19, Neutralizing antibody, Virology
Research in context.
Evidence before this study
Some COVID-19 cases re-tested positive for SARS-CoV-2 RNA a few days after discharge (termed re-positive cases in this study) raising questions about infectivity and the meaning of virus detection. We searched PubMed for articles in English before 16th June 2020, using the search terms “(“SARS-CoV-2″ OR “COVID-19″) AND (“retested positive” or “recurrence” or “reinfection”) in all fields. We found 10 original research articles and 3 related comments. Most of these studies were case reports focusing on virus dynamics in one or a few cases. No studies provided a systematic investigation of the clinical, immunological and virological characteristics of SARS-CoV-2 re-positive cases.
Added value of this study
This study characterized the SARS-CoV-2 re-positive cases in 619 discharged COVID-19 cases between 23 January and 19 February, Guangdong, China. The findings support and are in accordance to other studies that viruse RNA re-positive tests occur commonly in COVID-19 cases (14% in this study). In all re-positive cases a competent neutralizing antibody response was detected and no infectious strain was isolated, indicating the lower infectivity of the re-positive cases. By whole-genome sequencing and RT-PCR, we find virus genome degradation in all re-positive cases, which likely explains the discrepant clinical diagnoses. These data help to inform recommendations for improvements in COVID-19 molecular surveillance and future interventions.
Implications of all the available evidence
Millions of people have recovered from COVID-19 infection. This study shows that a proportion of such cases re-test as SARS-CoV-2 positive, more so in younger individuals who developed mild symptoms at the initial diagnosis. Re-positive cases, likely represent a much lower virus transmission risk, especially through the respiratory route, suggesting that surveillance and intervention strategies based on frequent re-testing and isolation of COVID-19 discharged cases could be optimised.
Alt-text: Unlabelled box
1. Introduction
Coronavirus disease 2019 (COVID-19) is caused by a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1], which shares high genetic similarity with the most closely-related bat origin SARS-like virus (RaTG13) [2]. As a newly emergent virus, the clinical features of SARS-CoV-2 infection were largely unknown at the beginning of the outbreak but are becoming gradually clearer as a result of global clinical studies [3,4]. The design of risk assessments and successful interventions for COVID-19 now depend on how well we understand the course of SARS-CoV-2 infection.
The COVID-19 pandemic is underway [5]. Social measures for monitoring, controlling and treating COVID-19 vary among and within countries. It has been suggested that the detection of antibodies to SARS-CoV-2 could serve as the basis for an “immunity passport”, however it is currently unclear whether recovered COVID-19 cases have neutralizing antibodies that protect them from a second infection. There have been reports that some recovered COVID-19 cases have re-tested positive for SARS-CoV-2 RNA some time after discharge [6,7]. Since a RT-PCR test that targets a short fragment of the virus genome cannot indicate if an individual is infectious or not, we define in this study these observations as “re-positive” cases, not relapse or repeat infection cases.
There are four likely explanations for these “re-positive” cases: (i) relapse or recrudescent infection with the “first” SARS-CoV-2 inoculum, (ii) re-infection with a “second” SARS-CoV-2 inoculum, (iii) remnant RNA fragments of the “first” SARS-CoV-2 infection, resulting from intermittent shedding of cells containing viral fragments (iv) laboratory errors, or technical limits of RT-PCR assays, such as inter-run assay variance, kit-to-kit differences in sensitivity, etc. Further epidemiological and laboratory evidence are required to clarify the cause(s) of cases that test re-positive for COVID-19 RNA. Importantly, re-positive detection of SARS-CoV-2 RNA raises questions about the relationship between molecular testing and potential infectivity, for example (i) what percentage of discharged cases test re-positive for COVID-19, (ii) are re-positive cases associated with clinical characters, (iii) what is the immune status of re-positive cases, and (iv) to what extent are re-positive cases infectious.
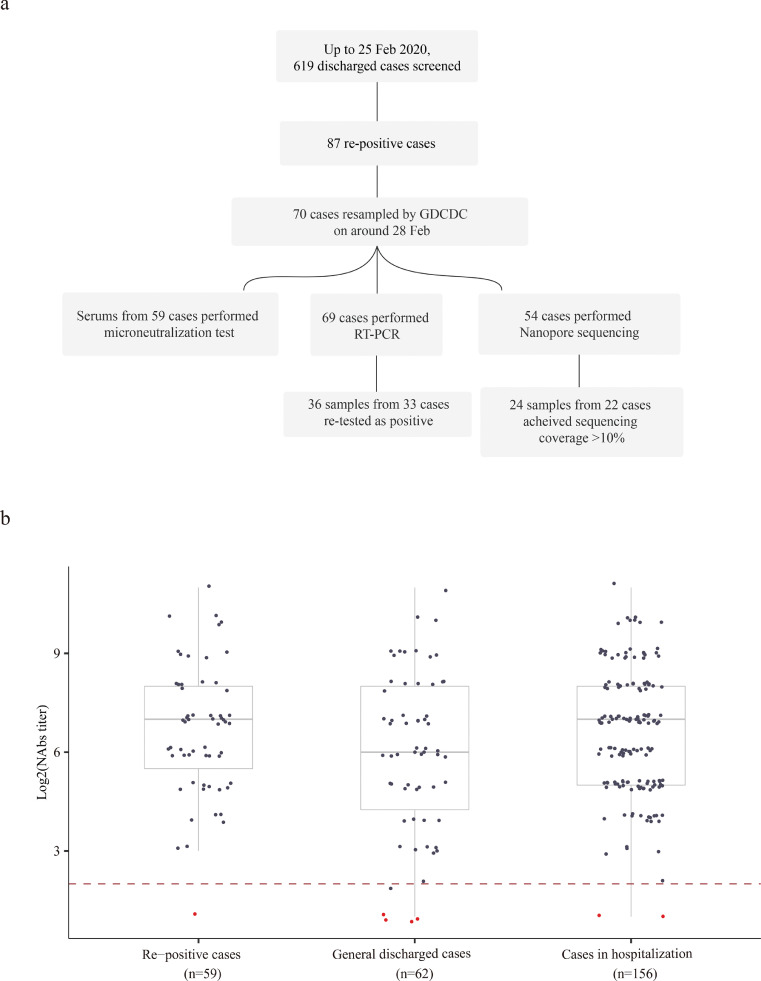
Guangdong Province reported the highest number of COVID-19 cases in China except Hubei. Guangdong launched an enhanced surveillance network and a series of intervention measures in response to the outbreak soon after the first COVID-19 case was reported in December 2019 [8]. Since 23 January, all discharged COVID-19 cases were isolated in designated hotels under medical observation for another 14 days. In this study, we screened 619 recovered COVID-19 cases in Guangdong discharged between 23 January and 19 February. One hundred thirty-seven swabs and 59 serum samples from 70 re-positive cases were collected in order to reveal the immunological and virologic characteristics of the SARS-CoV-2 re-positive cases.
2. Materials and methods
2.1. Discharge criteria and after discharge measures for COVID-19 cases in Guangdong
Guangdong follows the Diagnosis and Treatment Scheme for SARS-CoV-2 released by the National Health Commission of China, with a little modification. The Discharge criteria for COVID-19 cases in Guangdong include: 1) body temperature is normal for more than three days, 2) respiratory symptoms including fever, dry cough and tiredness noticeably improve, 3) pulmonary imaging shows obvious absorption of inflammation, and 4) nucleic acid tests are negative twice consecutively, on both respiratory tract samples (such as sputum and nasopharyngeal swabs) and digestive tract samples (such as stool and anal swabs), with a sampling interval of at least 24 h. The intervention measures for discharged COVID-19 cases are as follows: 1) all discharged COVID-19 cases are isolated in designated hotels for another 14 days; 2) during isolation, discharged cases live in a well-ventilated single room, dine separately, practice hand hygiene, and minimize close contact with others, 3) health status is monitored during isolation and SARS-CoV-2 RT-PCR tests are performed on 7th and 14th days after discharge or more frequently, 4) cases return home only when nucleic acid tests are negative on both respiratory tract samples and digestive tract samples during isolation. Clinical outcome was categorized as mild, moderate, severe, and critical, as previously described [8].
2.2. Case definition and specimen collection
The term “re-positive case” in this study refers to discharged cases who retested as SARS-CoV-2 positive using real-time Reverse-Transcription Polymerase Chain Reaction (RT-PCR; see below). In Guangdong, all discharged COVID-19 cases were isolated continuously in designated hotels. Samples including nasopharyngeal swabs, throat swabs and anal swabs, were collected for RT-PCR diagnosis at 7 days and 14 days after discharge, or more frequently. The demographic, clinical and laboratory information of all confirmed COVID-19 cases were retrieved from the Guangdong Provincial COVID-19 surveillance network.
2.3. Viral isolation and RT-PCR
The clinical samples including nasopharyngeal swabs, throat swabs and anal swabs were vortexed and centrifuged at 250×g for 10 min. The suspension was collected and filtered through a 0.22-μm filter for following virus culture. Next, vero E6 cells were inoculated with 100 µl processed patient sample. Cytopathic effect (CPE) were observed daily. If there no CPE was observed at 7 days, cell lysis was collected by centrifugation after three repeated freeze-thaw and 100 µl supernatant was used for the second round of passage. For RT-PCR diagnosis, total RNA was extracted from clinical specimens using the QIAamp Viral RNA mini kit (QIAGEN, Germany) according to the manufacturer's instructions. In this study, three RT-PCR kits were used to conduct nucleic acid testing, in an attempt to avoid false negatives. Kit A (DAAN GENE, Guangzhou, China)and Kit B (BioGerm, Shanghai, China) [9] have primers and probes targeting the open reading frame (ORF1ab) and nucleocapsid protein (N), respectively. Kit C (Liferiver, Shanghai, China) is designed to detect RNA-dependent RNA polymerase (RdRp), envelope protein (E) and N. Kit A and Kit C were included into WHO Emergency Use Listing for detecting SARS-CoV-2 nucleic acid (https://www.who.int/diagnostics_laboratory/200710_eul_sars_cov2_product_list.pdf?ua=1).
2.4. Microneutralization assay
Serum samples were collected from re-positive cases, cases in hospital, and discharged COVID-19 cases more than 21 days post illness onset. Microneutralization antibody assays for SARS-CoV-2 were performed in a BSL-3 laboratory according to standard neutralization test protocols. A local SARS-CoV-2 strain isolated from the first COVID-19 patient in Guangdong (GISAID accession ID: EPI_ISL_403,934) was used in the microneutralization assays. All neutralizing antibody assays were run in 96-well microplates. Serum samples were inactivated at 56 °C for 30 mins before use, diluted two-fold from 1:4 to 1:1024, and then incubated at 37 °C for 2 h with equal volumes of 100 half tissue culture infective doses (100 TCID50). Thereafter, the mixture was added to a 96-well Vero-E6 cell culture plate. Viral-induced CPE was monitored daily for 7 days. All diluted samples were tested in duplicate. Cell, serum and virus controls were included in each plate. Virus back titration was conducted in each test. The antibody titre of the sample was defined as the highest dilution that could inhibit CPE development in 50% of the virus-infected wells.
2.5. High-throughput sequencing
For the multiplex PCR approach, we followed the general multiplex PCR method described in (https://artic.network/ncov-2019) [10]. Briefly, multiplex PCR was performed with two pooled primer mixtures and cDNA reverse-transcribed with random primers was used as a template. After 35 rounds of amplification, PCR products were collected and quantified, followed by end-repairing and barcoding ligation. Around 50 fmol of final library DNA was loaded onto the MinION sequencing device. The ARTIC bioinformatics pipeline for COVID (https://artic.network/ncov-2019) was used to generate consensus sequences and call single nucleotide changes relative to the reference sequence (MN908947). Assembly of the nanopore raw data was performed using the ARTIC bioinformatic pipeline for COVID-19 with minimap2 [11] and medaka (https://github.com/nanoporetech/medaka) for consensus sequence generation. Sequencing data after mapping to SARS-COV-2 reference genome (MN908947.3) have been deposited in the Genome Sequence Archive [12] in BIG Data Center [13], Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under project accession numbers CRA002500, publicly accessible at https://bigd.big.ac.cn/gsa.
2.6. Statistical analysis
Statistical analyses were completed using R version 3.5.1 and GraphPad Prism 8.0 (GraphPad Software, Inc., San Diego, CA). Continuous variables that fitted a normal distribution and homogeneity of variance were compared using Student 's t-test and analysis of variance (ANOVA), otherwise they were analysed using a Wilcoxon rank test and Kruskal–Wallis H test. Categorical variables were compared using a Chi-square test or Fisher's Exact Test to assess deviation from the null hypothesis. Spearman's correlation was to assess the correlation between age and neutralization antibody titre. A p-value <0.05 was deemed to be statistically significant.
2.7. Ethics approval
This study was reviewed and approved by the Medical Ethical Committee of Guangdong Provincial Center for Disease Control and Prevention. Data collection and analysis of cases were determined by the Health Commission of Guangdong province to be part of a continuing public health outbreak investigation during the emergency response. Patients were informed about the surveillance before providing written consent, and data were collected and anonymized for analysis.
3. Results
3.1. Clinical characteristics of re-positive cases
A total of 619 COVID-19 cases were discharged from designated hospitals between 23 January and 19 February 2020. These cases were continuously isolated in designated hotels and were all screened for SARS-CoV-2 after discharge (see details in Methods). Up to 25 February 2020, 87 cases (14%) had re-tested as positive for SARS-CoV-2 RNA via RT-PCR and returned to the local designated hospital for isolation and medical observation. The demographic characteristics of the 87 re-positive cases are as follows: (i) gender distribution was equal, with 45 males and 42 females; (ii) re-positive detection of virus RNA was observed in all age groups, ranging from 3 months to 69 years, with a median age of 28 years, which is significantly younger than that of discharged COVID-19 patients in Guangdong in general (median age of 47 years, Table 1). Notably all re-positive cases had only mild (46) or moderate (41) clinical symptoms during initial hospitalization. Based on the provincial surveillance network, 84 of 87 re-positive cases had records on the time of hospitalization, discharge and the first detected as re-positive (Table S1). The median time for discharged COVID-19 cases re-tested as viral RNA positive was 7 days (range 2 to 19 days) post-discharge. Possibly due to their moderate clinical symptoms, re-positive cases had shorter hospital stays (median, 14 days) than general discharged COVID-19 cases (median, 28 days) (Table 1). After discharge, 77 of 87 re-positive cases were asymptomatic, and 10 had a symptom of unproductive cough, mainly at night. Forty-four cases received computerized tomography (CT) examination, and no ground glass opacities were found.
Table 1.
Demographic and clinical data of re-positive and general discharged COVID-19 cases.
| General discharged cases | Re-positive cases | P value | |
|---|---|---|---|
| Demographics | |||
| Age (median, range) | 47(1–90) | 28(0.25–69) | <0.0001 |
| Gender | |||
| Male | 155/303(51.2%) | 45/87(51.7%) | 0.8721 |
| Female | 148/303(48.8%) | 42/87(48.3%) | |
| Clinical classification | |||
| Mild | 28/256(10.9%) | 46/87(52.9%) | <0.0001 |
| Moderate | 167/256(65.2%) | 41/87 (47.1%) | |
| Severe | 61/256(23.8%) | 0/87(0) | |
| Clinical course | |||
| Onset-hospitalization (days) | 3(1–31) | 2(0–12) | 0.00018 |
| Initial hospital stay | 28(7–58) | 14(5–27) | <0.0001 |
| Onset-discharge | 33(8–66) | 17(7–36) | <0.0001 |
| Discharge-re-positive | N/A | 7(2–19) | N/A |
Note: General discharged cases refer to COVID-19 recovered cases detected as SARS-CoV-2 negative in 14 days after discharge. N/A indicates some unavailable data.
3.2. Neutralizing antibody in re-positive cases
An impaired immune response has been associated with fatal COVID-19 infections that exhibit prolonged persistence of viral RNA [14]. One possible explanation for the re-positive detection of SARS-CoV-2 RNA is that some COVID-19 patients may have insufficient immune responses and neutralization antibodies (NAbs) to clear infection completely. To investigate the immunological and virological characteristics of re-positive COVID-19 cases, 70 of 87 re-positive cases were sampled by the Guangdong Provincial Center for Disease Control and Prevention (GDCDC) between 22 February and 1 March 2020, including 59 serum and 137 swab samples (Fig. 1a). Serum samples were collected at a median of 35 days post illness onset (range = 23 to 47 days). In comparison, 218 serum samples from discharged general cases (n = 62) and hospitalized cases (n = 156) were collected with a median duration from illness onset to serum sampling of 31 days (ranging from 21 to 49 days) and 31 days (ranging from 14 to 58 days), respectively. The titre of viral-specific NAbs was estimated by microneutralization assays. As shown in Fig. 1b, 58 of 59 (98.3%) re-positive cases developed NAbs with a titre >4, ranging from 4 to >1024. Our results demonstrated competent immune activation in re-positive cases, which exhibit a distribution of NAbs titres similar to that of cases who detected as negative in 14 days after discharge and COVID-19 cases in hospitalization (Kruskal-Wallis H test, p = 0.2663, Fig. 1b).
Fig. 1.
(a) Summary of sampling scheme in this study; (b) Comparison of Nab titres among infections that were re-positive, general discharged COVID-19 cases, and cases in hospitalization. General discharged cases refer to COVID-19 recovered cases detected as SARS-CoV-2 negative in 14 days after discharge. To plot these results into one figure, we recorded 2 for antibody titre less than 4 (highlighted in red), and 2048 for titre larger than 1024. The start point of serum dilution (titre of 4) was highlighted with red dash line. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Viral RNA detection and viral isolation in re-positive cases
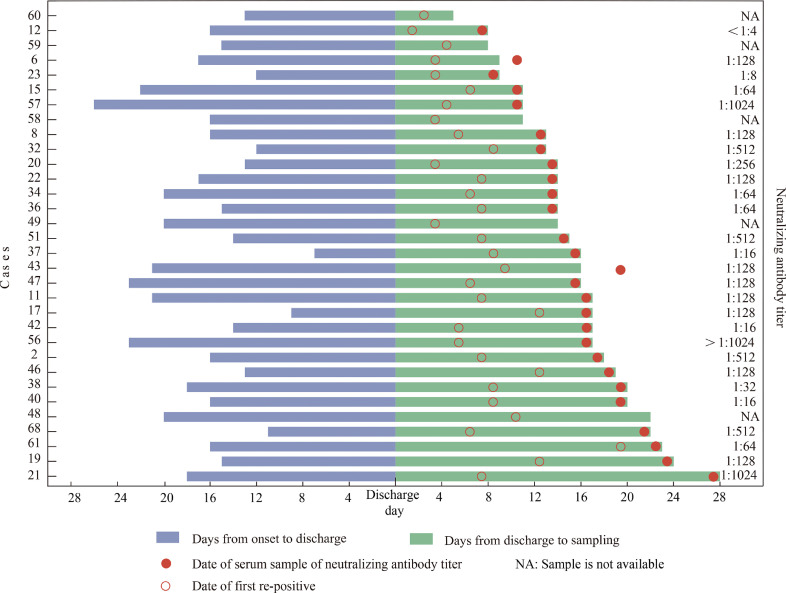
A total of 137 swabs, including 51 nasopharyngeal swabs, 18 throat swabs and 68 anal swabs, were tested using three different RT-PCR kits, in an attempt to reduce the chance of false negatives caused by differences in sensitivity and primer specificity. Fifty re-positive cases had paired nasopharyngeal swabs and anal swabs, and 18 cases had paired throat swabs and anal swabs for viral RNA detection. Thirty-six swabs from 33 cases were detected as positive by at least one RT-PCR kit (Table S2). RT-PCR positive rates were not statistically different for different sample types in 68 paired samples (anal swabs vs nasopharyngeal swabs, and anal swabs vs throat swabs; Chi-square test, p = 0.648; Fisher exact test, p = 0.443). In this cross-sectional analysis, 32 of 33 cases had clear information on the time intervals between illness onset, discharge and sample testing (Fig. 2). We find that the period from discharge to the time tested as re-positive can range in duration from 6 to 28 days (Fig. 2). Importantly, there was no correlation between neutralization antibody titre and the length of time between discharge and the date the case tested as re-positive. For example, for Case 21, SARS-CoV-2 RNA was re-detected at 28 days after discharge and 46 days post symptom onset, yet NAb titre for this case was as high as 1024 (Fig. 2). The 36 RT-PCR positive samples including 14 nasopharyngeal swabs, 3 throat swabs and 19 anal swabs. These RT-PCR positive samples were inoculated into Vero-E6 cell line but no live viruses could be cultured following two rounds of cell passage.
Fig. 2.
Timeline of 32 COVID-19 re-positive cases sampled and tested between 28 February–1 March.
3.4. Virus whole genome sequencing in re-positive cases
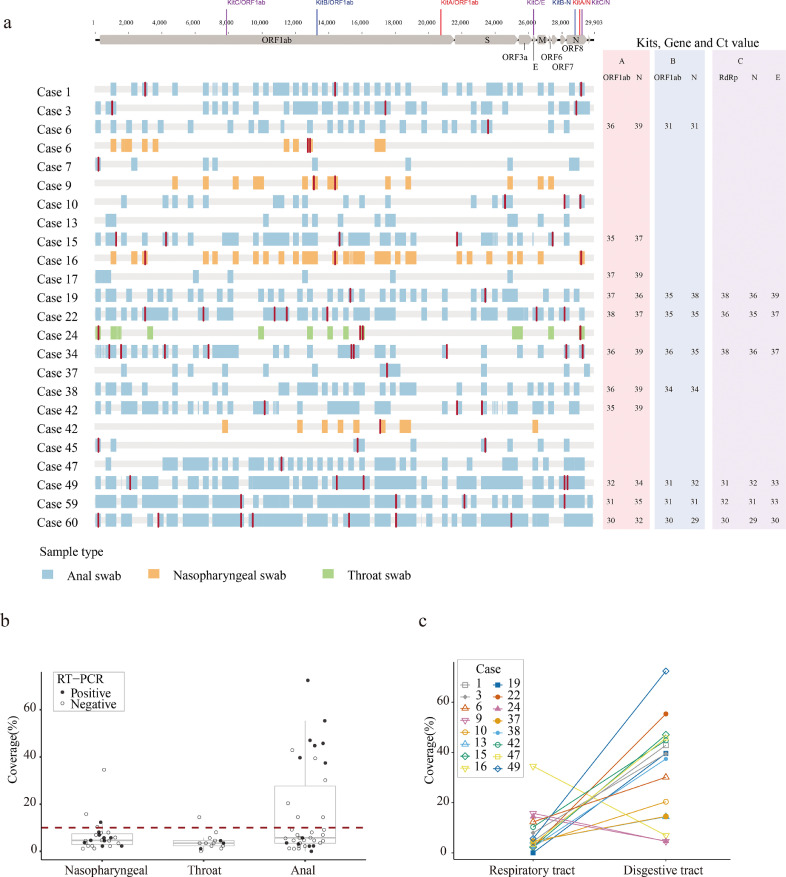
A previously study indicated that virus isolation success was also dependent on viral load, and that samples containing <106 copies/mL (or copies per sample) never yielded an isolate [3]. For acute infection cases, we found that high quality SARS-CoV-2 genomes could be obtained by using a multiplex PCR method, even for samples with a low viral load [8]. However, in this study we obtained zero full-length SARS-CoV-2 genomes by sequencing 94 samples from 54 patients and the corresponding sequencing coverage ranged from 0.00% to 75.48% (Fig. 3a, Table S2).
Fig. 3.
(a) Virus genome sequences (for those samples with genome coverage >10%) obtained from re-positive cases. Single nucleotide polymorphisms (with respect to the reference genome MN908947.3) are coloured in red. Genome sequence fragments are coloured blue, orange to indicate whether they were obtained from anal, nasopharyngeal and throat swabs, respectively. The colored bars at the top indicate the targeting regions for three PCR kits. The corresponding Ct values from three different RT-PCR kits were shown on the right side. (b) Coverage of the consensus sequence among nasopharyngeal swabs, throat swabs and anal swabs. A solid circle refers a RT-PCR positive sample, and a hollow circle refers a RT-PCR negative sample. The red dash line refers to the sequencing coverage of 10%; (c) Coverage of consensus sequence measured from respiratory tract (nasopharyngeal swabs/throat swabs) and matched digestive tract (anal swabs). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Intriguingly, a discrepancy was observed between the results of RT-PCR and multiplex PCR sequencing (Fig. 3a & b). For instance, 21 of 33 samples that were RT-PCR positive did not perform well in sequencing and produced sequences that covered < 10% of the virus genome (Fig. 3b). Conversely, 12 samples detected as negative by three RT-PCR kits gave rise to virus sequences that spanned >10% of the virus genome (Fig. 3a & 3b). This discrepancy might be expected when the viral genome was not intact and the RT-PCR and multiplex PCR primers target different fragments of viral genome (Fig. 3a).
Sequencing results obtained from matched samples of individual cases (anal swabs vs nasopharyngeal swabs, and anal swabs vs throat swabs) showed that the genome coverage of sequences from digestive samples were significantly higher than those of matched respiratory swabs (Fig. 3c). For Case 6 and Case 42, we observed some single nucleotide variants (SNVs) between viral genome sequences achieved from respiratory samples and from digestive samples (Fig. 3a).
4. Discussion
Millions of people have recovered from COVID-19 infection, and there have been preliminary reports of people testing re-positive for SARS-CoV-2 viral RNA after recovery [7, 8]. Here, we use data from the Guangdong COVID-19 surveillance system to analyze the characteristics of re-positive cases between 23 January and 26 February 2020.
The first question we addressed is the re-positive rate in COVID-19 discharged cases. In this study, the discharge criteria used were those specified by the Diagnosis and Treatment Scheme of SARS-CoV-2 (see Methods for detail). After screening 619 discharged cases, up to 25 February, the percentage of SARS-CoV-2 re-positive cases was around 14%. According to the scheme, all discharged cases are continuously isolated in designated hotels with strict interventions on disease transmission. Thus, the identification of re-positive SARS-CoV-2 during the isolation phase very likely excludes the possibility that re-positive cases are caused by secondary viral infection. Our results also highlight a significant feature of re-positive cases. All re-re-positive cases in our study developed only mild or moderate symptoms in the initial diagnosis, with the median age being significantly lower than that of the general COVID-19 cases (Table 1). The relatively mild symptoms may explain why the median time from onset to discharge in re-positive cases (median 17 days) is slightly lower than that of the other discharged COVID-19 cases (median 33 days). It is unlikely that cases tested as re-positive because they were discharged too early since all re-positive cases tested negative for both nasopharyngeal and anal swabs in two successive tests before discharge. The time from symptom onset to discharge (the time when COVID-19 cases have twice tested negative by PCR) for re-positive cases (median 17, Table 1) is consistent with the time that detectable SARS-CoV-2 RNA is reported on other studies to persist (median 20) in respiratory sites [14,15]. These data indicate that the course in re-positive cases is similar to other COVID-19 cases. The observation of re-positive SARS-CoV-2 RNA cases is not random and mainly observed in young cases without severe clinical symptoms.
Prolonged detection of virus RNA presents a challenge to targeted public health interventions. Therefore, it is important to know if re-positive cases are infectious. One previously proposed reason for prolonged detection of viral RNA in deceased patients is impaired neutralizing ability [14]. Our microneutralization result shows that 58 of 59 (98%) re-positive cases generated specific NAbs to SARS-CoV-2, and their titre distribution is similar to cured COVID-19 cases that were SARS-CoV-2 negative after discharge and to hospitalized COVID-19 cases (Fig. 1b). Recent studies of non-human primates suggests that primary infection enables the animals to develop anamnestic antibody responses following re-challenge by SARS-CoV-2, and that protective efficacy against re-challenge was mediated by rapid immunologic control [16,17]. Therefore, the possibility of active infection in re-positive cases appears to be low. In order to evaluate the possible infectivity we also attempted live virus isolation on clinical samples from re-positive cases. As expected, no viral isolates could be obtained from RT-PCR positive samples, although culture success may be low for samples with higher Ct values [3]. These data provide laboratory evidence that re-positive cases do not represent active infection.
We also performed multiplex PCR combined with high-throughput sequencing on these samples. The discrepancy we observed among different RT-PCR kit results, as well as between RT-PCR and multiplex PCR sequencing results, suggests that the virus genomes detected in re-positive cases may be highly degraded. We recovered only one virus sequence with genome coverage >20% (34.5%) in 23 RT-PCR positive respiratory samples (Fig. 3a). Together with the above results, it is more likely that the re-positive discharged cases are caused by intermittent shedding of cells containing remnant RNA fragments of the initial SARS-CoV-2 infection. The residual risk of infectivity of re-positive cases, especially from the respiratory transmission route, appears to be extremely low.
Several limitations of our study should be noted. First, we did not obtain successively-collected samples from discharged cases, resulting in a potential bias when summarizing the duration from the discharge to firstly re-positive result for viral RNA as well as the time of the re-positive RNA to negative. Secondly, we did not obtain samples during the acute infection for the re-positive cases. Therefore, some virological questions remain, including whether there any genetic differences among SARS-CoV-2 viruses sampled in the acute re-positive phases. Interpretation of the significance of SNVs identified between different samples from re-positive cases is limited by the sample size of our study (Fig. 3a) and should be further clarified in following studies.
The design of appropriate intervention strategies for COVID-19 largely relies on how well we understand the characteristics of SARS-CoV-2 infection. Re-positive detection of viral RNA in some discharged cases poses challenges for some interventions, such as a prolonged isolation phase and created requirements for hospital isolation facilities. Our study shows that NAb response in re-positive cases are comparable to those from other COVID-19 cases. More importantly, no of infective strains could be successfully isolated and no intact viral genomes could be sequenced from all re-positive cases samples. This likely excludes the possibility that re-positive cases represent active infection and indicates a lower risk for disease transmission from such cases. Additional public health messages relating to SARS-CoV-2 re-positive results may to educate the public and improve the allocation of limited medical resources.
Funding sources
This work was supported by grants from Guangdong Provincial Novel Coronavirus Scientific and Technological Project (2020111107001), China Evergrande Group (2020GIRHHMS11), Science and Technology Planning Project of Guangdong(2018B020207006), National Science and Technology Project (2020YFC0846800). The funding sources of this study did not play any role in the study design, the collection, analysis and interpretation of the data, in the writing of the manuscript and in the decision to submit the paper for publication.
Author contributions
JL, JJP, QLX contributed equally to this paper and JL, CWK designed the study. CWK, JFS, MK, XHT and TS had full access to all data in the study, and take responsibility for the integrity and accuracy of the data. JL, JJP, QLX, OP and JFS contributed to data collection, data analysis and data interpretation. JL, JJP, QLX, LdP, LNY, WJM and OP contributed to literature search writing of the manuscript. ZL and HFL worked on laboratory data and virus genome sequencing. CWK, RYY, LLZ, PPZ and CML engaged in sample collection, viral RNA detection and microneutralization assay. All authors contributed to data acquisition, data interpretation, and all reviewed and approved the final version of the manuscript. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. CWK is the guarantor.
Declaration of Competing Interests
All authors: No reported conflicts of interest.
Acknowledgments
We thank the laboratory and administrative personnel at Guangdong Provincial Center for Disease Control for their contribution to the follow-up investigation. We also acknowledge the contributions of other clinical, public health and technical staff from COVID-19 designated hospitals and city-level center for disease control and prevention.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102960.
Contributor Information
Jiufeng Sun, Email: sunjiuf@163.com.
Changwen Ke, Email: kecw1965@aliyun.com.
Appendix. Supplementary materials
References
- 1.Wu F., Zhao S., Yu B., Chen Y.-.M., Wang W., Song Z.-.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020:1–8. doi: 10.1038/s41586-020-2008-3. Feb 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-.L., Wang X.-.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020:1–4. doi: 10.1038/s41586-020-2012-7. Feb 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature [Internet] 2020 doi: 10.1038/s41586-020-2196-x. http://www.nature.com/articles/s41586-020-2196-x Apr 1 [cited 2020 Apr 10]; Available from: [DOI] [PubMed] [Google Scholar]
- 4.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020:1–4. doi: 10.1038/s41591-020-0869-5. Apr 15. [DOI] [PubMed] [Google Scholar]
- 5.WHO . 2020. Coronavirus disease (COVID-2019) situation reports [Internet]https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Available from: [Google Scholar]
- 6.Chen D., Xu W., Lei Z., Huang Z., Liu J., Gao Z. Recurrence of positive SARS-CoV-2 RNA in COVID-19: a case report. Int J Infect Dis. 2020;93:297–299. doi: 10.1016/j.ijid.2020.03.003. Apr 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Hu Y., Yu Y., Zhang X., Li B., Wu J. Positive result of Sars-Cov-2 in faeces and sputum from discharged patient with COVID-19 in Yiwu, China. J Med Virol. 2020 doi: 10.1002/jmv.25905. Apr 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J., du Plessis L., Liu Z., Hill V., Kang M., Lin H. Genomic epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell. 2020 doi: 10.1016/j.cell.2020.04.023. Apr;S0092867420304864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. Mar 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quick J., Grubaugh N.D., Pullan S.T., Claro I.M., Smith A.D., Gangavarapu K. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12(6):1261–1276. doi: 10.1038/nprot.2017.066. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. Sep 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T. GSA: genome sequence archive. Genom Proteom Bioinform. 2017;15(1):14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Genomics Data Center Members and Partners Database resources of the national genomics data center in 2020. Nucl Acids Res. 2020;48(D1):D24–D33. doi: 10.1093/nar/gkz913. Jan 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.To K.K.-.W., Tsang O.T.-.Y., Leung W.-.S., Tam A.R., Wu T.-.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis [Internet] 2020;0(0) doi: 10.1016/S1473-3099(20)30196-1. https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(20)30196-1/abstract Mar 23 [cited 2020 Apr 13]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng W., Bao L., Liu J., Xiao C., Liu J., Xue J. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science [Internet] 2020 doi: 10.1126/science.abc5343. https://science.sciencemag.org/content/early/2020/07/01/science.abc5343 Jul 2 [cited 2020 Jul 9]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020 doi: 10.1126/science.abc4776. May 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.