Abstract
The novel severe acute respiratory syndrome (SARS) coronavirus SARS-CoV-2 walks the planet causing the rapid spread of the CoV disease 2019 (COVID-19) that has especially deleterious consequences for the patients with underlying cardiovascular diseases (CVDs). Entry of the SARS-CoV-2 into the host cell involves interaction of the virus (via the receptor-binding domain (RBD) of its spike glycoprotein) with the membrane-bound form of angiotensin-converting enzyme 2 (ACE2) followed by the virus-ACE2 complex internalization by the cell. Since ACE2 is expressed in various tissues, such as brain, gut, heart, kidney, and lung, and since these organs represent obvious targets for the SARS-CoV-2 infection, therapeutic approaches were developed to either inhibit ACE2 or reduce its expression as a means of prevention of the virus entry into the corresponding host cells. The problem here is that in addition to be a receptor for the SARS-CoV-2 entry into the host cells, ACE2 acts as a key component of the renin-angiotensin-aldosterone system (RAAS) aimed at the generation of a cascade of vasoactive peptides coordinating several physiological processes. In RAAS, ACE2 degrades angiotensin II, which is a multifunctional CVD-promoting peptide hormone and converts it to a heptapeptide angiotensin-(1–7) acting as the angiotensin II antagonist. As protein multifunctionality is commonly associated with the presence of flexible or disordered regions, we analyze here the intrinsic disorder predisposition of major players related to the SARS-CoV-2 – RAAS axis. We show that all considered proteins contain intrinsically disordered regions that might have specific functions. Since intrinsic disorder might play a role in the functionality of query proteins and be related to the COVID-19 pathogenesis, this work represents an important disorder-based outlook of an interplay between the renin-angiotensin-aldosterone system and SARS-CoV-2. It also suggests that consideration of the intrinsic disorder phenomenon should be added to the modern arsenal of means for drug development.
1. Introduction
Coronavirus disease 2019 (COVID-19) caused by the infection with a novel coronavirus (NCoV-19), also known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is rapidly spreading through the globe. This emerging world-wide pandemic is taking its toll in a form of heavy morbidity and mortality (SARS-CoV-2 is more transmissible and lethal than influenza) and has large scale socio-economic impact (Yang et al., 2020). It seems that COVID-19, which was originally reported at the end of December 2019 as a “pneumonia of unknown etiology” in four patients in Wuhan, Hubei Province, China, is now present at each and any corner of the world. Although COVID-19 is characterized by rather mild symptoms of a typical respiratory illness that include fever, cough, sore throat, shortness of breath, as well as mild gastrointestinal (GI) symptoms in some patients, there are also multiple more serious cases, where the infection is causing severe pneumonia and even death, with an increase in risk of mortality of COVID-19 pneumonia being associated with age ≥ 65 years and preexisting concurrent cardiovascular or cerebrovascular diseases (Du et al., 2020). In light of the current COVID-19 outbreak, everyone is guessing what makes SARS-CoV-2 so special.
Although coronaviruses (CoVs) are widespread among vertebrates, they often cause only mild respiratory or enteric infections (Payne, 2017). CoVs belong to the subfamily Coronavirinae of the Coronaviridae family (which also includes the Torovirinae subfamily) in the order Nidovirales (http://ictvonline.org/virusTaxonomy.asp?version=2012). They are divided into four genera, namely α-, β-, γ-, and δ-CoVs, with β-CoVs being further separated into clades a–d (de Groot et al., 2012). α- and β-CoV are able to infect mammals (including humans and domestic animals), while γ- and δ-CoV tend to infect birds. Six CoVs have previously been identified as human-infecting viruses, among which are two α-CoVs, HCoV-229E and HCoV-NL63, and two β-CoVs, HCoV-HKU1 and HCoV-OC43 characterized by low pathogenicity and typically causing mild respiratory symptoms similar to a common cold. The other two known human β-CoVs, Severe Acute Respiratory Syndrome CoV (SARS-CoV) and Middle East Respiratory Syndrome CoV (MERS-CoV), lead to severe and potentially fatal respiratory tract infections and because of that have received special attention as emergent pathogens in humans, with the potential to create global epidemics (Yin and Wunderink, 2018). The emergence of human-infecting CoVs is likely associated with the cross-species transmission events (Drexler et al., 2014).
There is now a new player on the block, a novel coronavirus (NCoV-19), also known as SARS-CoV-2, which is another emerging pathogen currently representing a global threat. The genetic sequence analysis demonstrated that SARS-CoV-2 belongs to the β-coronavirus genera. This enveloped non-segmented positive-sense RNA virus (subgenus Sarbecovirus, Orthocoronavirinae subfamily) shows 79.5% nucleotide identity to SARS-CoV and 52% identity to MERS-CoV (Guo et al., 2020b; Zhu et al., 2020). Despite this similarity, SARS-CoV-2 shows higher levels of infectivity than the SARS-CoV and MERS-CoV did. At the moment, SARS-CoV-2 is showing a rapid worldwide spread, ever increasing morbidity and mortality rates. As a result of this rapid spread of SARS-CoV-2 infection, the WHO designated the SARS-CoV-2 infection disease (COVID-19) as a Public Health Emergency of International Concern on 30 January 2020, and subsequently, on March 11, 2020, declared it a Global Pandemic. It is impossible to provide here an accurate evaluation of the COVID-19-associated morbidity and mortality, since the corresponding numbers are changing on the hourly basis. Although COVID-19 is typically characterized by the symptoms of viral pneumonia, many COVID-19 patients are dying because of a complex organ failure (Du et al., 2020; Zhang, 2020). Older patients, especially those who have underlying illness, such as cardiovascular disease, liver disease, kidney disease or malignant tumors can become severely ill (Chen et al., 2020a; Huang et al., 2020; Wang et al., 2020; Wang et al., 2020c).
Clinical course of COVID-19 is characterized according to the Chinese Clinical Guidance for the COVID-19 Pneumonia Diagnosis and Treatment (7th edition) (Commission, 2020). Incubation period for COVID-19 ranges from 1 to 14 days. The first and main symptoms are nonspecific fever that can be mild to moderate or even absent in some patients, fatigue, and dry cough, which may become associated with respiratory symptoms, such as nasal obstruction, runny nose, and sore throat, myalgia or diarrhea. The condition may progress to become severe within one week of onset to present by dyspnea and/or hypoxia. Some patients deteriorate to ARDS, septic shock, refractory metabolic acidosis, coagulation dysfunction, and multiple organ system failure leading to death (see Supplementary Materials, Table S1).
The renin-angiotensin-aldosterone system (RAAS, also known as renin–angiotensin system, RAS) is a hormone system responsible for the regulation of blood pressure and fluid and electrolyte balance, and for the control of the systemic vascular resistance. RAAS includes a cascade of vasoactive peptides that coordinates key processes in human physiology. Angiotensin-converting enzyme 2 (ACE2), an enzyme that physiologically inhibits RAAS activation, functions as a receptor for both SARS-CoV and SARS-CoV-2, which have been responsible for the SARS epidemic in 2002 to 2004 and for the more recent COVID-19 pandemic, respectively (Gheblawi et al., 2020; Hoffmann et al., 2020; Li et al., 2003b; Wang et al., 2020; Yan et al., 2020b).
2. The renin-angiotensin-aldosterone system and its inhibitors
Juxtaglomerular (JG) cells within the afferent arterioles of the kidney secrete prorenin in its inactive form. Activation of JG cells by decreased blood pressure, or decreased sodium load in the distal convoluted tubule or via β-activation causes the cleavage of prorenin to produce mature renin (Nehme et al., 2019; Ren et al., 2019). Once released, renin acts on its target, angiotensinogen, produced by the liver, converting it into angiotensin I. Angiotensin I (a decapeptide) is physiologically inactive, but serves as a precursor for angiotensin II (an octapeptide). Angiotensin converting enzyme-1 (ACE1) converts angiotensin I to angiotensin II that acts as an agonist for both angiotensin II receptors type 1 and type 2 (AT1R and AT2R, respectively). Angiotensin II is converted, by ACE2, to the heptapeptide angiotensin-(1–7). ACE2 also converts angiotensin I to the nonapeptide angiotensin-(1–9), which is further processed by ACE1 to generate angiotensin-(1–7) that serves as an antagonist for the AT1R receptors and an agonist for the MAS1 receptor (MasR, also known as proto-oncogene Mas).
SARS-CoV-2 infection down-regulates ACE2 expression with subsequent elevation of plasma angiotensin II levels, which are in turn correlate with the total viral load and the degree of lung injury (Gurwitz, 2020). Curiously, a recent study performed by Ziegler et al. demonstrated that human ACE2 is an interferon-stimulated gene (ISG) and that interferon is capable of induction of broader expression of ACE2 in upper airway epithelial cells, suggesting that SARS-CoV-2 can utilize such interferon-driven upregulation of ACE2 to further enhance infection (Ziegler et al., 2020).
ACE1 inhibitors inhibit the conversion of angiotensin I to angiotensin II and angiotensin-(1–9) transformation to angiotensin-(1–7), thereby attenuating angiotensin II vasoconstriction, sodium retention, proinflammatory, and profibrogenic effects. Inhibitors of ACE2 have been developed, but none has been marketed as of yet. Angiotensin receptor blockers (ARBs) inhibit the actions of angiotensin II and angiotensin-(1–7) on the angiotensin AT1Rs. Therefore, ACE2 serves as a key enzyme for the balance between the two main arms of the RAAS: the classic RAAS, which is the ACE1/angiotensin II/ angiotensin II type 1 receptor axis the anti-RAS, which is the ACE2/angiotensin-(1–7)/Mas receptor (MasR) axis (Sarzani et al., 2020).
Finally, the alamandine pathway should be mentioned here (Sarzani et al., 2020). This pathway is based on the ability of ACE2 to interacts with the other group of the angiotensin peptides, alatensins, in which the N-terminal aspartate is replaced by alanine, leading to Ala-angiotensin-(1–7) (known as alamandine) capable of binding to the Mas-related G protein-coupled receptor D (MrgD) (Santos et al., 2019). The effects of the alamandine interaction with MrgD are similar to the effects of the angiotensin-(1–7) stimulating MasR (Sarzani et al., 2020). Therefore, the SARS-CoV-2-induced downregulation of ACE2 leads to a consequent changes in the levels of both angiotensin-(1–7) and alamandine (Sarzani et al., 2020).
3. Entry of SARS-CoV-2 into cells
SARS-CoV-2 uses the SARS-CoV receptor ACE2 (expressed in the principal target cells for SARS-CoV-2, such as the heart, kidneys, intestine, and lung alveolar epithelial cells,) for entry and the transmembrane protease, serine 2 (TMPRSS2, also known as serine protease 10) for the spike (S) glycoprotein priming (Hoffmann et al., 2020). The S glycoproteins of coronaviruses have two subunits, S1, and S2. The S1 subunit binds to the ACE2 enzyme, via its RBD, on the cell membrane and S2, fuses with the cell membrane (Kuba et al., 2005). In addition, S protein priming by cellular proteases is needed for viral entry, which can proceed in two ways. (a) A host TMPRSS2 activates the S and cleaves ACE2; (b) TMPRSS2 causes irreversible conformational changes in the S2 subunits, activating them, and facilitating fusion of the virus to the cell membrane. The virus then enters the cell (Glowacka et al., 2011; Heurich et al., 2014). According to Hoffmann et al., ACE2 and TMPRSS2 are essential in airway cells for SARS-CoV-2 infection. A serine protease inhibitor, Camostat mesylate, used in Japan to treat chronic pancreatitis, inhibits the TMPRSS2 and partially blocks the entry of SARS-CoV-2 into bronchial epithelial cells in vitro (Hoffmann et al., 2020).
The efficient COVID-19 transmission from human to human within the population may be explained by the ~10–20 fold higher affinity of the SARS-CoV-2 spike protein to the human angiotensin-converting enzyme-2 (ACE2) than the corresponding affinity of the SARS-CoV spike protein (Wrapp et al., 2020). It is also speculated that the presence of a unique furin cleavage site within the SARS-CoV-2 spike protein, which is a novel feature setting this virus apart from SARS-CoV, and the almost ubiquitous expression of furin-like proteases could participate in expanding SARS-CoV-2 cell and tissue tropism, relative to SARS-CoV, as well as increasing its transmissibility and/or altering its pathogenicity (Chen et al., 2020; Walls et al., 2020; Wrapp et al., 2020; Yan et al., 2020a). In the same line, the distinctive structural differences between the receptor-binding domains (RBDs) of the spike proteins from SARS-CoV and SARS-CoV-2 can represent energetically favorable changes for the more efficient interaction of the SARS-CoV-2 spike protein with the ACE2 receptor. In fact, the local environment present in the ACE2 receptor allows these mutations to produce a significant number of electrostatic stabilizing interactions. Furthermore, the presence of the two capping loops in the RBD of the SARS-CoV-2 spike protein is likely to produce a more stabilization effect over the interaction with the cellular receptor. These two loops around the RBD of SARS-CoV-2 might be promoting the interaction with the ACE2 receptor, improving the binding to the ACE2 by increasing the number of atoms involved. Therefore, the amino acid substitutions and the longer capping loops could explain the increase in the binding affinities in SARS-CoV-2 compared to SARS-CoV. Higher affinity values might be related to the dynamics of the infection and the rapid spread observed for this virus (Ortega et al., 2020). Wan et al., based on their computational analysis, suggested that when all the human-ACE2-favoring residues were combined into one RBD, this RBD would bind to human ACE2 with super affinity and the corresponding spike protein would mediate viral entry into human cells with super efficiency, which may improve the epidemic surveillance (Wan et al., 2020) and the virus virulence.
Viral entry and lung type II pneumocytes infection triggers an inflammatory cascade in the lower respiratory tract. This inflammatory cascade is initiated by the antigen-presenting cells (APC) performing two functions, (i) presenting the foreign antigen to CD4 +-T-helper (Th1) cells, and (ii) releasing interleukin-12 to further stimulate the Th1 cell. The Th1 cells stimulate CD8 +-T-killer cells that will target any cells containing the foreign antigen. In addition, activated Th1 cells stimulate B-cells to produce antigen-specific antibodies (Lake, 2020; Rabi et al., 2020).
The antibody profile against SARS-CoV virus has a typical pattern of IgM and IgG production. The SARS-specific IgM antibodies disappear at the end of week 12, while the IgG antibody can last for a long time, indicating that IgG antibody may mainly play a protective role (Li et al., 2003a). The latest report shows the number of CD4 + and CD8 + T cells in the peripheral blood of SARS-CoV-2-infected patients is significantly reduced. However, these cells are excessively activated, as evidenced by high proportions of HLA-DR (CD4 3.47%) and CD38 (CD8 39.4%) double-positive fractions (Xu et al., 2020). These T-cell responses can inhibit the over activation of innate immunity and clear SARS-CoV-2.
However, uncontrolled systemic inflammatory response resulting from the release of large amounts of pro-inflammatory cytokines (such as IFN-α, IFN-γ, IL-1β, IL-6, IL-12, IL-18, IL-33, TNF-α, TGFβ, etc.) and chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10, etc.) by immune effector cells results in the cytokine storm (Mehta et al., 2020), which, with a suboptimal T-cell response, triggers diffuse alveolar damage with marked pulmonary edema and hyaline membrane formation, causing acute respiratory distress syndrome (ARDS). ARDS is the main cause of death in severe cases of SARS-CoV-2 infection, just like what occurred in SARS-CoV and MERS-CoV infection (Xu et al., 2020).
SARS-CoV-2 virus as one of respiratory viruses normally attach to receptors on the cellular airways with a low level of viraemia detected in some symptomatic patients. Viraemia was detected in six of 41 patients (15%) in China (Huang et al., 2020), but only in one of 12 patients (8%) in the study from Singapore (Young et al., 2020). Another study from China, reported only three PCR-positive samples (1%) out of 307 blood samples (Wang et al., 2020). The results of PCR testing of serum samples in six viraemic patients suggested a very low viral load in specimens (Huang et al., 2020). Yet, multiple organ involvement including the heart, kidney, liver, and gastrointestinal tract have been recently reported in patients with COVID-19 (Chen et al., 2020a).
The action mechanism a COVID-19 infection on the kidney is likely to be multifactorial; (a) SARS-CoV-2 may exert direct cytopathic effects on kidney tissue by entering through an ACE2- related signaling pathways. This is supported by the detection of SARS-CoV-2 genomic fragments in blood and urine of patients with COVID-19 (Huang et al., 2020); (b) Virus-induced cytokines or mediators might exert indirect effects on renal tissue, such as hypoxia, shock, and rhabdomyolysis (Cheng et al., 2020); (c) Deposition of immune complexes of viral antigen may damage the kidney. However, kidney microscopy specimens from patients with SARS showed a normal glomerular aspect and absence of electron-dense deposits (Chu et al., 2005). The etiology of acute myocardial injury caused by SARS-CoV-2 infection might be related to the ACE2-dependent pathway or the cytokine storm, respiratory dysfunction, and hypoxaemia (Danser et al., 2020; South et al., 2020; Zheng et al., 2020).
4. Effects of angiotensin converting enzyme-1 inhibitors
Treatment with agents, which have an impact on the RAAS as ACE inhibitors (ACEi) or ARBs is common in patients with hypertension, cardiac diseases and diabetes. In humans, the effects of RAAS inhibition on ACE2 expression were studied a little. Administration of ACE inhibitors in patients with coronary artery disease and hypertension did not affect angiotensin-(1–7) production, questioning its effect on ACE2 (Campbell et al., 2004; Luque et al., 1996). In other studies, it was found to up-regulate ACE2 receptor expression (Gurwitz, 2020; Vuille-dit-Bille et al., 2015). Data showing the effects of ACE inhibitors, ARBs, and other RAAS inhibitors on lung-specific expression of ACE2 in experimental animal models and in humans are lacking (Vaduganathan et al., 2020). It is not clear whether such up regulation of ACE2 would be desirable or not. Theoretically, increased ACE2 activity could increase ability of SARS-CoV-2 to penetrate cells being its receptor. In contrast, increased production of angiotensin-(1–7) from angiotensin II under the action of ACE2 could lead to increased anti-inflammatory activity and some protection from the lung damage caused by the virus (Danser et al., 2020).
There is no sufficient experimental or clinical data demonstrating the beneficial or adverse outcomes of the background use of ACE1, ARBs or other RAAS antagonists in COVID-19 or among COVID-19 patients with a history of cardiovascular disease treated with such agents. However, severe COVID-19 forms were described in patients with hypertension and diabetes mellitus; i.e., conditions known to be associated with RAAS blockade therapy (Guan et al., 2020), suggesting that patients on ACE inhibitors or ARB could be at a greater risk due to the mechanism by which SARS-CoV-2 enters the cell (Busse et al., 2020). One should keep in mind though that the generality of this hypothesis could be questionable. In fact, although no randomized controlled trials are still available, several observational studies of good quality have been recently published, all denying the hypothesis that patients taking ACE inhibitors or ARB could be at a greater risk of SARS-CoV-2 infection or development of the severe COVID-19 forms (Guo et al., 2020a; Iaccarino et al., 2020; Mancia et al., 2020). Accordingly, the Council on Hypertension of the European Society of Cardiology, the American College of Cardiology, American Heart Association, and Heart Failure Society of America strongly recommend that physicians and patients should continue treatment with their usual anti-hypertensive therapy and not to add or remove any RAAS-related treatments, beyond actions based on standard clinical practice (Bozkurt et al., 2020; de Simone, 2020).
5. Roles of intrinsic disorder in the SARS-CoV-2 – RAAS interplay
Since it is known that viral proteomes contain noticeable levels of intrinsically disordered proteins and viral proteins utilize intrinsic disorder during host cell invasion, and since in its turn host is utilizing intrinsic disorder in fighting the viral infection, we looked at the intrinsic disorder status of viral and host proteins associated with the SARS-CoV-2 – RAAS interplay.
5.1. Functional disorder in the spike glycoprotein from SARS-CoV-2
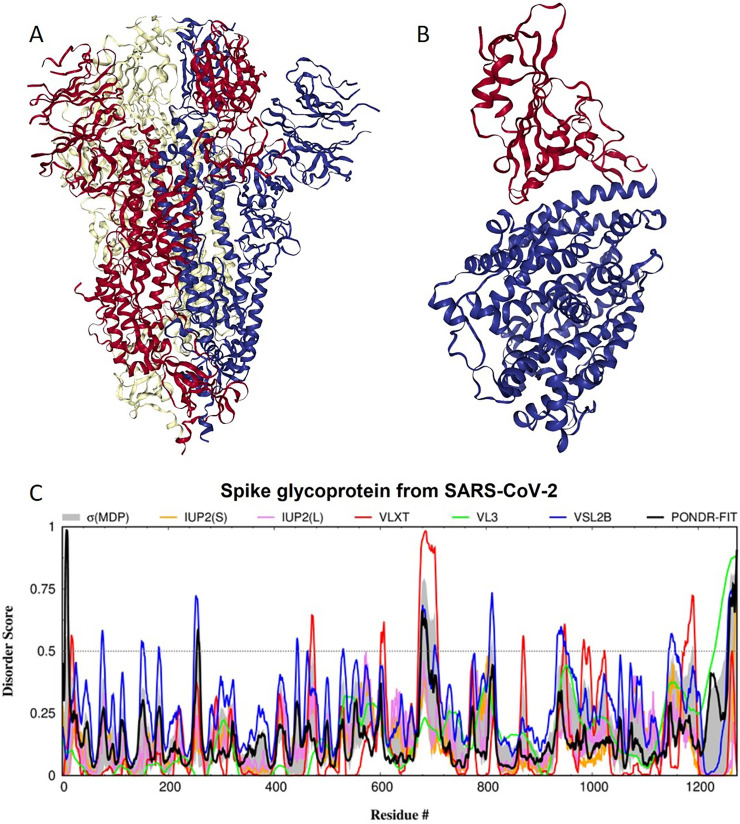
Spike glycoprotein (S) is a large single-pass transmembrane multifunctional protein defining the exterior of the CoV particles, where it forms surface homotrimers (Cavanagh and Davis, 1986; Graham and Baric, 2010) (see Fig. 1A) and interacts with the virion M protein via its C-terminal transmembrane region (Broer et al., 2006; McBride et al., 2007). Importantly, this structure of the S protein resolved to 3.46 Å using cryo-electron microscopy (cryo-EM) (Wrapp et al., 2020) is characterized by the absence of multiple residues (1–26, 67–78, 96–98, 143–155, 177–186, 247–260, 329–334, 444–448, 455–490, 501–502, 621–639, 673–686, 812–814, 829–851, 1147–1288). Therefore, at least 327 residues assembled into the 15 regions ranging in length from 2 to 142 residues are missing from this structure, indicating their high conformational flexibility. The longest missing region is the C-terminal region that includes a single-pass transmembrane region and an intracellular domain.
Fig. 1.
Structure and intrinsic disorder propensity of spike glycoprotein from SARS-CoV-2. A. A 3.46 Å resolution cryo-EM structure of a homotrimeric form of spike glycoprotein from SARS-CoV-2 (PDB ID: 6VSB). B. A X-ray crystal structure (resolution of 2.45 Å) of a complex between the human ACE2 receptor (blue structure) and the RBD of the S protein from SARS-CoV-2 (red structure) (PDB ID: 6M0J). C. Intrinsic disorder profile generated for the S protein from SARS-CoV-2 by DiSpi web-crawler aggregate the results from a set of commonly used predictors of intrinsic disorder, such as PONDR® VLXT (Romero et al., 2001), PONDR® VL3 (Peng et al., 2006), PONDR® VLS2B (Peng et al., 2005), PONDR® FIT (Xue et al., 2010), IUPred2 (Short) and IUPred2 (Long) (Dosztanyi et al., 2005a, Dosztanyi et al., 2005b). This tool enables the rapid generation of disorder profile plots for individual polypeptides as well as arrays of polypeptides.
The S protein consists of an N-terminal signal peptide, a long extracellular domain, a single-pass transmembrane domain, and a short intracellular domain (Broer et al., 2006). Functionally, S protein contains two ectodomains, S1 and S2, with specific biological roles, where binding of the subunit S1 to the host cell receptors (via the receptor-binding domain, RBD) initiates viral infection, whereas the viral entry into the host cells is mediated by the S2 subunit that acts as a class I viral fusion protein moderating the fusion of the virion and cellular membranes (Belouzard et al., 2012; de Haan et al., 2006; Li et al., 2005). Fig. 1B represents a crystal structure of a complex between the human ACE2 receptor and the RBD of the SARS-CoV-2 S protein resolved to 2.45 Å (Lan et al., 2020). In this structure, residues 319–332 and 527–547 (numbering corresponds to the full-length S protein) of the RBD constitute regions of missing electron density; i.e., regions with high conformational flexibility.
Although S-proteins from different CoVs typically show high level of sequence conservation at their C-terminal regions, the N-terminal regions of S proteins display noticeable differences. It was pointed out that SARS-CoV-2 appears to be optimized for binding to the ACE2 receptor. In fact, six amino acids within the RBD (which are L455, F486, Q493, S494, N501, and Y505 in SARS-CoV-2) are crucial for binding to ACE2 receptors (Andersen et al., 2020; Wan et al., 2020). Five of these six residues in SARS-CoV-2 are different from the ACE2 receptor-binding residues in SARS-CoV, suggesting that this sequence variability might be the reason for the observed differences between SARS-CoV-2 and SARS-CoV in their virulence, receptor-mediated binding and entry into the host cell (Andersen et al., 2020; Wan et al., 2020).
S protein is known to undergo maturation, which represents specific posttranslational modification (PTM) that involves two-step proteolytic cleavage. At the first stage, a host cell protease (e.g. furin) nicks the S precursor to generate two separate S1 and S2 subunits that remain to be joined together via a set of the non-covalent interactions. At the next stage that happened after the viral attachment to host cell receptors, S2 subunit undergoes additional cleavage by the host protease TMPRSS2, leading to the release of a fusion peptide and generation of the S2’ subunit that also serves as fusion-mediating subunit (Belouzard et al., 2012; de Haan et al., 2006). In S protein of SARS-CoV, the first and second cleavage sites are located at the residues R667 and R797, respectively. In the SARS-CoV-2 spike, which is a 1273-residue-long protein, the first cleavage site (residue R684; note that here and in the subsequent text, while discussing the functionality of different residues/regions of the SARS-CoV-2 spike protein, numbering of residues in S from SARS-CoV-2 is based on the pairwise alignment of the sequences S proteins from SARS-CoV-2 and SARS-CoV) is located in a close proximity to the C-terminus of a polybasic region introduced to this protein through the insertion of 4 residues and a series of specific mutations. In fact, in S of SARS-CoV, this cleavage site is located within the TVS----LLRSTSQKS region, whereas in spike protein from the SARS-CoV-2, the corresponding region is TQTNSPRRARSVASQS. Altogether, this polybasic region constitutes a novel furin cleavage site of the SARS-CoV-2 spike protein (Andersen et al., 2020), which could represent one of the reasons for the higher binding affinity of the SARS-CoV-2 S protein to ACE2 than S protein from Human SARS (Wrapp et al., 2020). The second cleavage site generating the S2’ subunit in S from SARS-CoV-2 is located at residue R815. Fig. 1C represents an intrinsic disorder profile generated for the SARS-CoV-2 S protein by several commonly used disorder predictors and clearly shows that both cleavage sites associated with the spike maturation are located within the intrinsically disordered protein regions (IDPRs). This in agreement with a well-known fact that the proteolytic digestion is orders of magnitude faster in unstructured as compared to structured protein regions (de Laureto et al., 2006; Fontana et al., 2004; Fontana et al., 1986; Fontana et al., 1997; Iakoucheva et al., 2001; Polverino de Laureto et al., 1995). Therefore, it is extremely important for the protein cleavage process that the sites of cleavage be located in regions that lack structure or possess high structural flexibility.
Furthermore, in the SARS-CoV-2 S protein, fusion peptide (residues 790–808) is located within a flexible region characterized by the mean disorder score of 0.419 ± 0.085. S protein contains two heptad repeat regions that form coiled-coil structure during viral and target cell membrane fusion, assuming a trimer-of-hairpins structure needed for the functional positioning of the fusion peptide. In human SARS-CoV-2 S protein, heptad repeat regions are formed by residues 920–970 and 1163–1202, which have mean disorder scores of 0.441 ± 0.112 and 0.353 ± 0.062, respectively. Another functional region found in S protein is the receptor-binding domain (residues 319–543) containing a receptor-binding motif (residues 440–510) responsible for interaction with human ACE2. Based on the intrinsic disorder predisposition analysis, this motif is characterized by noticeable structural flexibility with the mean disorder score of 0.29 ± 0.12 and contains a short disordered region (residues 442–445). Computational analysis (using specific algorithms, such as MoRFchibi_web (Malhis et al., 2016), ANCHOR (Dosztanyi et al., 2009; Meszaros et al., 2009), MoRFPred (Disfani et al., 2012), and DISOPRED3 (Jones and Cozzetto, 2015)) revealed that the S protein from SARS-CoV-2 contains several molecular recognition features (MoRFs), which are the short interaction-prone disordered regions located within IDPs/IDPRs that undergo a disorder-to-order transition upon binding to their partners (Cheng et al., 2007; Hsu et al., 2013; Mohan et al., 2006; Oldfield et al., 2005; Vacic et al., 2007), which are important for protein-protein interactions, and may initiate an early step in molecular recognition. In fact, SARS-CoV-2 S protein was predicted to contain three MoRFs, residues 1–10, 819–823, and 1265–1272, suggesting that intrinsic disorder could be needed for interaction of this protein with its binding partners. It is likely that the N-terminal MoRF can be engaged in the interaction of this protein with the host receptor, whereas C-terminal MoRF can be related to the S-M interaction and viral assembly. In addition to protein-binding regions, S protein is predicted to contain many RNA- and DNA-binding residues. Therefore, intrinsic disorder in the S protein is crucial for the maturation of this protein and also can be utilized in protein-protein interaction, as well as RNA and DNA binding, interaction with host cell membrane, and further viral infection.
5.2. Intrinsic disorder in renin-angiotensin-aldosterone system
The major RAAS components are prorenin/renin, angiotensinogen/angiotensin I/angiotensin II, angiotensin converting enzymes 1 and 2 (ACE1 and ACE2), angiotensin II receptors type 1 and type 2 (AT1R and AT2R), and MAS1 receptor. Structural side of these proteins is studied rather well.
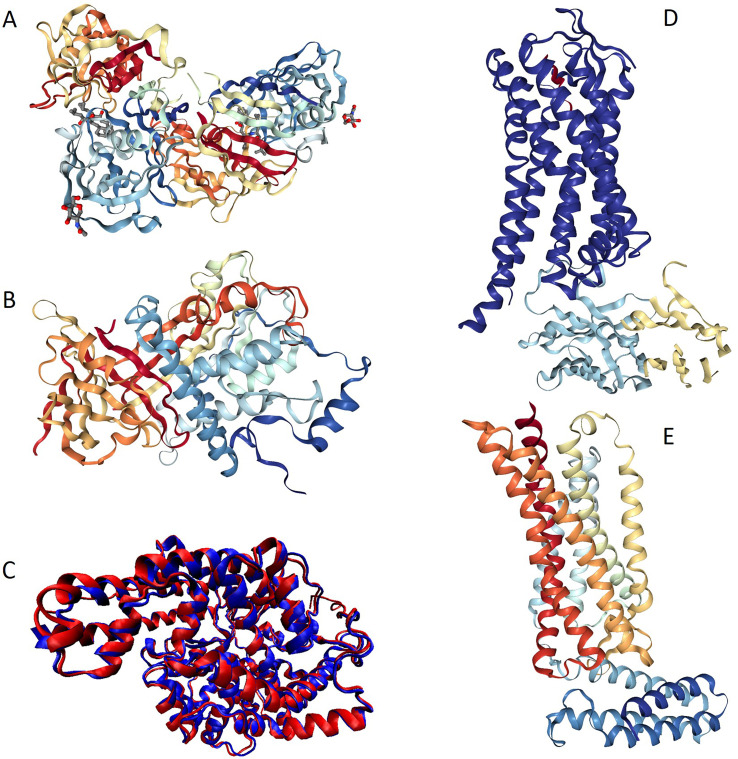
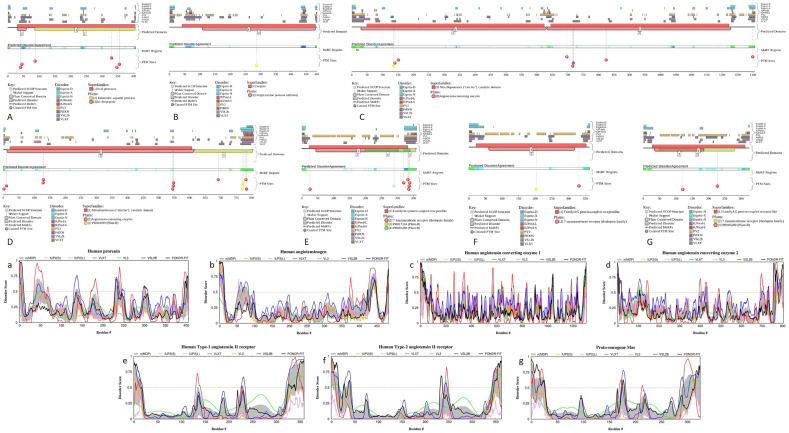
Fig. 2 represents available structures of human renin, angiotensinogen, ACE1, ACE2, as well as AT1R and AT2R. Fig. 2A shows high resolution (1.8 Å) X-ray crystal structure of the homodimeric form of human renin (PDB ID: 1HRN) (Tong et al., 1995). This protein (340 residues) is generated as a result of the maturation of the 406-residue-long prorenin (UniProt ID: P00797) by the removal of a signal peptide (residues 1–23) and propeptide or activation peptide (residues 24–66). Mature renin, which is a highly specific endopeptidase with the only known function to generate angiotensin I from angiotensinogen, is glycosylated at positions N71 and N141 (residue numbering follows that of the prorenin) and has multiple phosphorylation sites (residues S41, S45, Y87, T330, S347, and S355). Fig. 3A and a show that prorenin is predicted to contain several disordered and flexible regions; i.e., regions with the predicted disorder scores exceeding 0.5 and falling within the 0.2–0.5 range, respectively. Proteolytic sites used for the removal of signal peptide and propeptide, as well as glycosylation and phosphorylation sites are located within or in close proximity to the flexible or disordered regions (see Fig. 3A and a), indicating that intrinsic disorder and structural flexibility found in proreinin/renin are functionally important.
Fig. 2.
Structural characterization of the renin-angiotensin-aldosterone system members. A. High resolution (1.8 Å) X-ray crystal structure of human renin (PDB ID: 1HRN) (Tong et al., 1995). B. X-ray crystal structure of human angiotensinogen (PDB ID: 2WXW) (Zhou et al., 2010). C. Overlaid crystal structures of the peptidase M2 1 domain (N-domain, residues 30–658; PDB ID: 3NXQ:A (Anthony et al., 2010)) and the peptidase M2 2 domain (C-domain, residues 642–1232; PDB ID: 2IUL; (Watermeyer et al., 2006)) shown by red and blue colors, respectively. Multiple structure alignment was conducted using the MultiProt algorithm (Shatsky et al., 2004), and VMD platform was used to generate this image (Humphrey et al., 1996). D. Crystal structure of angiotensin II type 1 receptor (AT1R, blue chain) bound to the angiotensin-like peptideS1I8 (red chain) and stabilized by the nanobody (cyan and yellow chains) (PDB ID: (Wingler et al., 2019)). E, crystal structure of the chimera protein of human AT1R and soluble cytochrome b562 from Escherichia coli (PDB ID: 5UNG; (Zhang et al., 2017)).
Fig. 3.
Disorder profiles generated for the members of human RAAS by the D2P2 computational platform (plots A, B, C, D, E, F, and G) and by the DiSpi web-crawler (plots a, b, c, d, e, f, and g). A, a. Prorenin (UniProt ID: P00797); B, b. Angiotensinogen (UniProt ID: P01019); C, c. Angiotensin-converting enzyme 1 (UniProt ID: P12821); D, d. Angiotensin-converting enzyme 2 (UniProt ID: Q9BYF1); E, e. Type-1 angiotensin II receptor (UniProt ID: P30556); F, f. Type-2 angiotensin II receptor (UniProt ID: P50052); G, g. Proto-oncogene Mas (MAS1; UniProt ID: P04201).
Fig. 2B represents X-ray crystal structure of mature human angiotensinogen (PDB ID: 2WXW) (Zhou et al., 2010), which is a 452-residue-long protein generated by the removal of signal peptide (residues 1–33) from a precursor protein (485 residues UniProt ID: P01019). There are two regions of missing electron density in this protein, residues 404–415 and 450–452. Mature angiotensinogen is further processed by cellular proteases to generate a series of peptide hormones with the decreasing length, such as angiotensin I (residues 34–43), angiotensin-(1–9) (residues 34–42), angiotensin II (residues 34–41), angiotensin-(1–7) (residues 34–39), angiotensin-(1–5) (residues 34–38), angiotensin-(1–4) (residues 34–37), angiotensin III (residues 35–41), and angiotensin IV (residues 36–41). Fig. 3B and b represent disorder profiles of human angiotensinogen and show that the aforementioned sites associated with the maturation and processing of this protein and regions of missing electron density are located within the disordered or flexible regions. NMR solution structure of the angiotensin I (PDB ID: 1N9U) is shown in Fig. 4 (Spyroulias et al., 2003), which illustrates that this peptide exists in an extended conformation in solution.
Fig. 4.
NMR solution structure of the angiotensin I (PDB ID: 1N9U; (Spyroulias et al., 2003)).
Metalloprotease ACE1 is a multifunctional enzyme that, in addition to being dipeptidyl carboxypeptidase that hydrolyses peptides by the removal of a dipeptide from the C-terminus, thereby converting the hormone angiotensin I to the active vasoconstrictor angiotensin II (Coates, 2003), also plays a role in degradation of bradykinin and amyloid β-protein (Hemming and Selkoe, 2005) and shows some glycosidase activity cleaving the mannose linkage in the glycosylphosphatidylinositol (GPI)-anchored proteins. In somatic tissues, ACE1 represents a translated tandem duplication that produces a protein with two functional domains, the N-terminal peptidase M2 1 domain (residues 30–630) and the C-terminal peptidase M2 2 domain (residues 631–1232). ACE1 is a type 1 C-terminally membrane anchored ectoenzyme, with the hydrophobic transmembrane anchor included in the C-terminal most part of the protein (Coates, 2003). Human ACE1 undergoes posttranslational processing causing the removal of the N-terminally located signaling peptide (residues 1–29) and, to generate a soluble form, C-terminally located propeptide that serves as the aforementioned hydrophobic transmembrane anchor (residues 1233–1306). Structural information is available for most parts of the soluble form this protein. Fig. 2C represents overlaid crystal structures of the peptidase M2 1 (residues 30–658; PDB ID: 3NXQ:A; (Anthony et al., 2010)) and peptidase M2 2 domains (residues 642–1232; PDB ID: 2IUL; (Watermeyer et al., 2006)) shown by red and blue colors, respectively. It is clearly seen that the N- and C-domains of human AC1 are almost identical structurally (in fact, 577 residues of both domains were included in this structural alignment that is characterized by the RMSD of 0.99 Å). In these structures, residues 159–161 and 640–658 in peptidase M2 1 are missing. These regions, together with signaling peptide and a loop connecting transmembrane anchor with the soluble ACE1 are predicted to be disordered/flexible (see Fig. 3C and c). Furthermore, this protein contains several PTM sites, and its N-terminal region is predicted to possess a MoRF (residues 14–22) (see Fig. 3D).
Carboxypeptidase ACE2 converts angiotensin I to angiotensin-(1–9) and angiotensin II to angiotensin-(1–7) (Donoghue et al., 2000; Tipnis et al., 2000; Vickers et al., 2002), hydrolyzes biologically active peptides apelin-13 and dynorphin-13 (Vickers et al., 2002), interacts, in intestine, with the amino acid transporter SL6A19 and regulates its catalytic activity (Camargo et al., 2009; Kowalczuk et al., 2008), and serves as a receptor for SARS-CoV-2, SARS-CoV, and human CoV-NL63, interacting with their spike proteins (Hoffmann et al., 2020; Hofmann et al., 2005; Li et al., 2003b; Wrapp et al., 2020). As it was already pointed out, a crystal structure of a complex between the ACE2 receptor (residues 1–615) and the RBD of the S protein from SARS-CoV-2 was recently solved (Lan et al., 2020) (see Fig. 1B). In this and other structures of ACE2, N-terminal residues 1–18 constitute a region of missing electron density. Furthermore, no structural information is available for the C-terminal region of this protein (residues 616–805). Maturation of human ACE2 involves the removal of a signal peptide (residues 1–17). Also, this protein can be further processed via the removal of C-terminal residues 709–805. Several ACE2 residues are N-glycosylated (N53, N59, N103, N322, N432, N546, and N690). Fig. 3D and d indicate that the N- and C-terminal regions of human ACE2 contain noticeable levels of intrinsically disordered or flexible residues, and glycosylation sites are preferentially located within disordered or flexible regions (see Fig. 3D).
Angiotensin II type 1 receptor (AT1R) is a prototypical G-protein-coupled receptor (GPCR) that serves as a critical regulator of cardiovascular and renal function (Kawai et al., 2017; Wingler et al., 2019). GPCRs are the largest family of transmembrane proteins in humans, acting as cricial regulators of almost every aspect of human physiology. AT1R is one of the key members of the RAAS, promoting a wide range of intracellular signaling pathways resulting in endothelial dysfunction, hypertension, end organ damage, and vascular remodeling (Kawai et al., 2017). AT1R-mediated signaling represents a complex set of signaling pathways that include G protein-dependent signaling, G protein-independent signaling, NADPH oxidase and ROS signaling, transactivation of growth factor receptors, β-arrestin-mediated signaling, signaling pathways initiated by several AT1R interacting proteins, as well as a functional cross-talk between the AT1R signaling pathway and other signaling pathways (Kawai et al., 2017). Fig. 2D represents a crystal structure of AT1R (blue chain) bound to the angiotensin-like peptide S1I8 (red chain) and stabilized by the nanobody (cyan and yellow chains) (PDB ID: 6DO1) and shows that the AT1R structure is typical for seven-transmembrane receptors (Wingler et al., 2019). Fig. 2E illustrates that the angiotensin II type 2 receptor (AT2R) shows very similar structural organization possessing seven transmembrane α-helices characteristic for the GPCR family (PDB ID: 5UNG; (Zhang et al., 2017)). Fig. 3E, e, F, and f show that in line with this high structural similarity, AT1R and AT2R have rather similar disorder profiles, which are characterized by the disordered N- and C-terminal regions and the presence of several flexible regions dispersed within the rather ordered backbone. These receptors also contain several PTM sites embedded within the IDPRs (see Fig. 3E and F). Similar to other members of the GPCR family (Fonin et al., 2019), cytoplasmic C-tails represent the most disordered regions of the AT1R and AT2R. Although no structural information is currently available for the MAS1 receptor, it is likely that this GPCR is structurally similar to AT1R and AT2R. This conclusion is supported by Fig. 3G and g showing remarkable similarity of the disorder profile generated for human MAS1 with the disorder profiles of AT1R and AT2R.
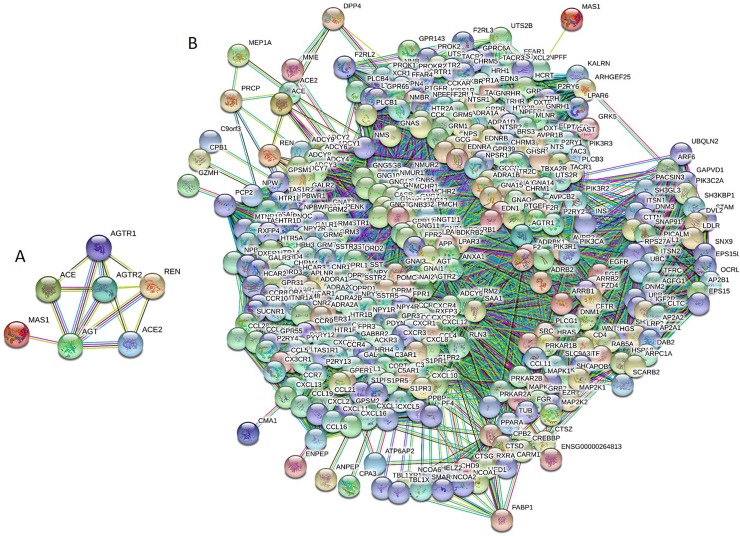
Since binding promiscuity represents one of the important functional features of IDPs/IDPRs (Dunker et al., 2001; Oldfield and Dunker, 2014; Uversky, 2011, Uversky, 2013a, Uversky, 2013b; Uversky and Dunker, 2010), we analyzed interactivity of human RAAS proteins. IDPs and hybrid proteins containing ordered domains and IDRs are considered as binding “professionals”, which are always engaged in interactions with different partners utilizing multiple binding modes (Dunker et al., 2001; Oldfield and Dunker, 2014; Uversky, 2013b; Uversky and Dunker, 2010). IDPs/IDPRs can form static, semi-static, and dynamic complexes (Uversky, 2011, Uversky, 2013a), participate in polyvalent interactions (Mammen et al., 1998; Uversky, 2015), and fold at binding to their partners (Dunker et al., 2002a; Dunker et al., 2002b; Wright and Dyson, 2009). Since degrees of such binding-induced folding can vary in a wide range, the resulting complexes show wide structural and functional heterogeneity (Uversky, 2011, Uversky, 2013a). Some IDPs/IDPRs fold differently at binding to different partners (Dajani et al., 2003; Dunker et al., 2005; Dunker et al., 1998; Dyson and Wright, 2002; Hsu et al., 2013; Kriwacki et al., 1996; Meador et al., 1993; Oldfield and Dunker, 2014; Oldfield et al., 2008; Uversky, 2003; Wright and Dyson, 2009), whereas other IDPs/IDPRs form fuzzy complexes, where significant levels of disorder are preserved at least outside the binding interface (Fuxreiter, 2012; Fuxreiter and Tompa, 2012; Hazy and Tompa, 2009; Permyakov et al., 2003; Sharma et al., 2015; Sigalov et al., 2004; Sigalov et al., 2007; Tompa and Fuxreiter, 2008). Since disorder-based interactions are often characterized by high specificity and low affinity, IDPs/IDPRs are suited well for signaling functions (Schulz, 1979). Finally, many IDPs/IDPRs have highly connected central positions in complex protein-protein interaction (PPI) networks, acting as hubs (Dosztanyi et al., 2006; Dunker et al., 2005; Ekman et al., 2006; Haynes et al., 2006; Patil and Nakamura, 2006; Singh and Dash, 2007; Singh et al., 2007). For evaluation of interactivity of the RAAS proteins, we utilized Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org/) (Szklarczyk et al., 2011). Fig. 5A shows the inter-RAAS PPI network generated by STRING at the high confidence level of 0.7 and demonstrates that these seven proteins form a highly interconnected network with an average node degree of 4.29. Next, we looked at the global interactivity of the whole set of human RAAS proteins by including a first shell interactors (i.e., proteins interacting with RAAS proteins). The resulting network is shown in Fig. 5B. This RAAS-centered PPI network that was built using custom confidence level of 0.901 contains 394 nodes (proteins) connected by 20,014 edges (PPIs). In this network, the average node degree is 102, and its average local clustering coefficient (which defines how close its neighbors are to being a complete clique; the local clustering coefficient is equal to 1 if every neighbor connected to a given node Ni is also connected to every other node within the neighborhood, and it is equal to 0 if no node that is connected to a given node Ni connects to any other node that is connected to Ni) is 0.713. Since the expected number of interactions among proteins in a similarly sized set of proteins randomly selected from human proteome is equal to 3227, this RAAS-centered PPI network has significantly more interactions than expected, being characterized by a PPI enrichment p-value of <10−16. Therefore, these data indicate that human RAAS proteins are promiscuous binders that form a dense PPI network. It is likely that at least in part this high interactivity of the RAAS proteins can be related to their IDPRs.
Fig. 5.
Analysis of the internal (A) and external (B) interactability of the RAAS members conducted by Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org/) that generates a network of predicted associations based on predicted and experimentally-validated information on the interaction partners of a protein of interest (Szklarczyk et al., 2011). In the corresponding network, the nodes correspond to proteins, whereas the edges show predicted or known functional associations. Seven types of evidence are used to build the corresponding network, where they are indicated by the differently colored lines: a green line represents neighborhood evidence; a red line - the presence of fusion evidence; a purple line - experimental evidence; a blue line – co-occurrence evidence; a light blue line - database evidence; a yellow line – text mining evidence; and a black line – co-expression evidence (Szklarczyk et al., 2011).
5.3. Intrinsic disorder in transmembrane protease, serine 2 (TMPRSS2)
Another important player related to the entry of SARS-CoV-2 to host cells is the cellular serine protease TMPRSS2, which is responsible for priming of S protein (Hoffmann et al., 2020). S protein priming by cellular proteases is crucial for viral entry into target cells, since it leads to the cleavage of S protein at the S1/S2 and the S2’ site and allows fusion of viral and cellular membranes via a process driven by the S2 subunit (Belouzard et al., 2012; de Haan et al., 2006). In addition to priming of coronavirus spike glycoprotein that activates it for cathepsin L-independent host cell entry, TMPRSS2 might further promote viral uptake by the ACE2 cleavage (Heurich et al., 2014). It was shown that SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells (Lukassen et al., 2020). TMPRSS2 proteolytically cleaves and activates the fusion glycoproteins F0 of Sendai virus (SeV), human metapneumovirus (HMPV), and human parainfluenza 1, 2, 3, 4a and 4b viruses (HPIV). Furthermore, TMPRSS2 is involved in activation of various influenza viruses, including avian influenza viruses, such as 2013 Asian H7N9 influenza virus, by cleaving the precursor of their surface glycoprotein haemagglutinin (HA) (Shen et al., 2017). Since it also plays a crucial role in the proteolytic activation of SARS-CoV and MERS-CoV (Shen et al., 2017), TMPRSS2 is considered as a suitable target for treatment of influenza virus and coronavirus infections (Laporte and Naesens, 2017; Meyer et al., 2013; Rabaan et al., 2017; Shen et al., 2017; Shin and Seong, 2017; Simmons et al., 2013).
TMPRSS2 is anchored to the plasma membrane and belongs to the family of type II transmembrane serine proteases (TTSPs) (Wu, 2003). TMPRSS2 has a short intracellular N-terminal domain (residues 1–84), a transmembrane α-helical domain acting as signal-anchor (residues 85–105) and a large extracellular domain (residues 106–492) containing low-density lipoprotein (LDL) receptor class A (LDLRA, residues 112–149) that forms a binding site for LDL and calcium, scavenger receptor cysteine-rich (SRCR) domain (residues 150–242) and C-terminal serine protease domain of the chymotrypsin S1 fold (residues 256–489). Activation of TMPRSS2 involves an autocatalytic cleavage between R255 and I256 (Afar et al., 2001), generating non-catalytic and catalytic chains (residues 1–255 and 256–492, respectively). Although after cleavage, the mature TMPRSS2 remains mostly membrane-bound, some number of catalytic chains can be liberated into the extracellular milieu. For example, secreted forms of TMPRSS2 were found in human seminal prostasomes, suggesting the roles of this serine protease in regulation of the sperm function (Chen et al., 2010).
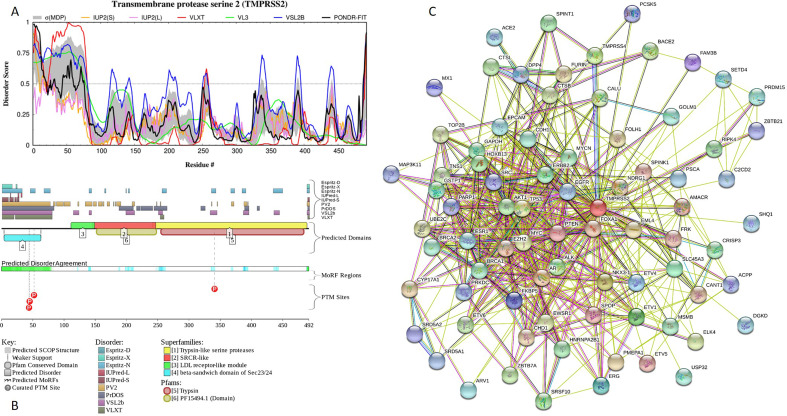
No structural information is available for human TMPRSS2. Fig. 6A and B provide an explanation for this fact showing that TMPRSS2 is expected to contain noticeable levels of intrinsic disorder. In fact, first 70 residues of this protein are predicted to be disordered. Curiously, in addition to the canonical form, TMPRSS2 has an alternatively spliced isoform 2, where M1 is substituted by MPPAPPGGESGCEERGAAGHIEHSRYLSLLDAVDNSKM. This change adds 37 disordered residues and a MoRF (residues 15–38).
Fig. 6.
Intrinsic disorder and interactability of human transmembrane protease TMPRSS2 (UniProt ID: O15393). A. Disorder profile generated by the DiSpi web-crawler. B. Functional disorder profile generated by the D2P2 computational platform. C. TMPRSS2-centered PPI network generated by STRING (http://string-db.org/).
Besides activation of multiple viruses, TMPRSS2 has a number of other biological roles. For example, it activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis (Lucas et al., 2014). In its membrane-anchored form, TMPRSS2 also activates a member of the G-protein-coupled receptors, protease-activated receptor-2 (PAR-2), in prostate cancer cells (Wilson et al., 2005). More generally, perturbations in the regulation of expression of this surface-anchored serine protease and/or distortions in its enzymatic activity serve as contributing factors to the pathogenesis of several cancer types (Martin and List, 2019). To see if this multifunctionality of TMPRSS2 is linked to the binding promiscuity of this protein, we looked at its interactability using STRING platform. Fig. 6C represents TMPRSS2-centered PPI network generated utilizing medium confidence level of 0.4. This network includes 79 proteins connected by 533 interactions, while the expected number of interactions in a random similar size set of proteins is 219. The average node degree of this network is 13.6, the average local clustering coefficient is 0.713, and PPI enrichment p-value is <10−16. It is likely that at least in part this high interactivity of TMPRSS2 can be related to its IDPRs. This information definitely needs to be taken into account while developing drugs targeting TMPRSS2 for treatment of influenza virus and coronavirus infections, since current strategies, which are most frequently used here are primarily focused on finding various serine protease inhibitors; i.e., drugs targeting the catalytic site of TMPRSS2 (Shen et al., 2017). However, since none of the compounds or proteins currently used for inhibition of TMPRSS2 was specifically designed to exclusively target TMPRSS2, all these inhibitors are rather non-specific and may cause unpredictable and deleterious side effects (Shen et al., 2017).
6. Concluding remarks
Assuming that an interplay between the renin-angiotensin-aldosterone system and spike glycoprotein from SARS-CoV-2 contributes to the COVID-19 pathogenesis, one should carefully analyze all the related players. Traditionally, finding potential drug leads involves rational drug design approaches based on the study of the structures and functions of target molecules. Although solving crystal structures of SARS-CoV-2 proteins, RAAS members, and complexes between the viral and human proteins undoubtedly represent important stages for future successes in finding potential anti-COVID-19 drugs, this structural biology side of the potential association of COVID-19 with RAAS should be complemented by careful and diligent investigation of the intrinsic disorder predisposition of corresponding viral and human proteins. This is especially important in light of the common practice of elimination of protein regions that resist crystallization (i.e., segments with high conformational flexibility or IDPRs). If disorder does play a role in the functionality of query proteins and is somehow related to the pathogenesis of COVID-19, then consideration of the intrinsic disorder phenomenon should be added to the modern arsenal of means for drug development. Therefore, this work represents an important step for better understanding the prevalence and functionality of intrinsic disorder in spike glycoprotein from SARS-CoV-2 and in proteins from the human renin-angiotensin-aldosterone system.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Acknowledgment
This work did not receive any financial support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104510.
Appendix A. Supplementary data
Supplementary material
References
- Afar D.E., Vivanco I., Hubert R.S., Kuo J., Chen E., Saffran D.C., Raitano A.B., Jakobovits A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61:1686–1692. [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C.S., Corradi H.R., Schwager S.L., Redelinghuys P., Georgiadis D., Dive V., Acharya K.R., Sturrock E.D. The N domain of human angiotensin-I-converting enzyme: the role of N-glycosylation and the crystal structure in complex with an N domain-specific phosphinic inhibitor, RXP407. J. Biol. Chem. 2010;285:35685–35693. doi: 10.1074/jbc.M110.167866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt B., Kovacs R., Harrington B. 2020. HFSA/ACC/AHA Statement Addresses Concerns Re: Using RAAS Antagonists in COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer R., Boson B., Spaan W., Cosset F.L., Corver J. Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry. J. Virol. 2006;80:1302–1310. doi: 10.1128/JVI.80.3.1302-1310.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse L.W., Chow J.H., McCurdy M.T., Khanna A.K. COVID-19 and the RAAS-a potential role for angiotensin II? Crit. Care. 2020;24:136. doi: 10.1186/s13054-020-02862-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo S.M., Singer D., Makrides V., Huggel K., Pos K.M., Wagner C.A., Kuba K., Danilczyk U., Skovby F., Kleta R., Penninger J.M., Verrey F. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology. 2009;136:872–882. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D.J., Zeitz C.J., Esler M.D., Horowitz J.D. Evidence against a major role for angiotensin converting enzyme-related carboxypeptidase (ACE2) in angiotensin peptide metabolism in the human coronary circulation. J. Hypertens. 2004;22:1971–1976. doi: 10.1097/00004872-200410000-00020. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986;67(Pt 7):1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020;525(1):135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.W., Lee M.S., Lucht A., Chou F.P., Huang W., Havighurst T.C., Kim K., Wang J.K., Antalis T.M., Johnson M.D., Lin C.Y. TMPRSS2, a serine protease expressed in the prostate on the apical surface of luminal epithelial cells and released into semen in prostasomes, is misregulated in prostate cancer cells. Am. J. Pathol. 2010;176:2986–2996. doi: 10.2353/ajpath.2010.090665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (London, England) 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Oldfield C.J., Meng J., Romero P., Uversky V.N., Dunker A.K. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46:13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Luo R., Wang K., Zhang M., Wang Z., Dong L., Li J., Yao Y., Ge S., Xu G. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020;97(5):829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu K.H., Tsang W.K., Tang C.S., Lam M.F., Lai F.M., To K.F., Fung K.S., Tang H.L., Yan W.W., Chan H.W.H., Lai T.S.T., Tong K.L., Lai K.N. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. 2005;67:698–705. doi: 10.1111/j.1523-1755.2005.67130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates D. The angiotensin converting enzyme (ACE) Int. J. Biochem. Cell Biol. 2003;35:769–773. doi: 10.1016/s1357-2725(02)00309-6. [DOI] [PubMed] [Google Scholar]
- Commission C.N.H. 2020. Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment (7th edition) [Google Scholar]
- Dajani R., Fraser E., Roe S.M., Yeo M., Good V.M., Thompson V., Dale T.C., Pearl L.H. Structural basis for recruitment of glycogen synthase kinase 3beta to the axin-APC scaffold complex. EMBO J. 2003;22:494–501. doi: 10.1093/emboj/cdg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danser A.H.J., Epstein M., Batlle D. 2020. Renin-Angiotensin System Blockers and the COVID-19 Pandemic: At Present There Is No Evidence to Abandon Renin-Angiotensin System Blockers. Hypertension (Dallas, Tex. : 1979), Hypertensionaha12015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., Ziebuhr J. In: Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. King A., Adams M., Carstens E.B., Lefkowitz E.J., editors. Elsevier/Academic Press; Amsterdam, Boston: 2012. Family Coronaviridae; pp. 806–820. [Google Scholar]
- de Haan C.A., Te Lintelo E., Li Z., Raaben M., Wurdinger T., Bosch B.J., Rottier P.J. Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range. J. Virol. 2006;80:10909–10918. doi: 10.1128/JVI.00950-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laureto P.P., Tosatto L., Frare E., Marin O., Uversky V.N., Fontana A. Conformational properties of the SDS-bound state of alpha-synuclein probed by limited proteolysis: unexpected rigidity of the acidic C-terminal tail. Biochemistry. 2006;45:11523–11531. doi: 10.1021/bi052614s. [DOI] [PubMed] [Google Scholar]
- de Simone G. 2020. Position Statement of the ESC Council on Hypertension on ACE-Inhibitors and Angiotensin Receptor Blockers. [Google Scholar]
- Disfani F.M., Hsu W.L., Mizianty M.J., Oldfield C.J., Xue B., Dunker A.K., Uversky V.N., Kurgan L. MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics. 2012;28:i75–i83. doi: 10.1093/bioinformatics/bts209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z., Csizmok V., Tompa P., Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z., Csizmok V., Tompa P., Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z., Chen J., Dunker A.K., Simon I., Tompa P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J. Proteome Res. 2006;5:2985–2995. doi: 10.1021/pr060171o. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z., Meszaros B., Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antivir. Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R.H., Liang L.R., Yang C.Q., Wang W., Cao T.Z., Li M., Guo G.Y., Du J., Zheng C.L., Zhu Q., Hu M., Li X.Y., Peng P., Shi H.Z. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur. Respir. J. 2020;55(5):2000524. doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K., Garner E., Guilliot S., Romero P., Albrecht K., Hart J., Obradovic Z., Kissinger C., Villafranca J.E. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac. Symp. Biocomput. 1998:473–484. [PubMed] [Google Scholar]
- Dunker A.K., Lawson J.D., Brown C.J., Williams R.M., Romero P., Oh J.S., Oldfield C.J., Campen A.M., Ratliff C.M., Hipps K.W., Ausio J., Nissen M.S., Reeves R., Kang C., Kissinger C.R., Bailey R.W., Griswold M.D., Chiu W., Garner E.C., Obradovic Z. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Du Y., Tu L., Zhu P., Mu M., Wang R., Yang P., Wang X., Hu C., Ping R., Hu P., Li T., Cao F., Chang C., Hu Q., Jin Y., Xu G. Clinical features of 85 Fatal Cases of COVID-19 from Wuhan: a retrospective observational study. Am. J. Respir. Crit. Care Med. 2020;201(11):1372–1379. doi: 10.1164/rccm.202003-0543OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K., Brown C.J., Lawson J.D., Iakoucheva L.M., Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Dunker A.K., Brown C.J., Obradovic Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- Dunker A.K., Cortese M.S., Romero P., Iakoucheva L.M., Uversky V.N. Flexible nets: the roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Dyson H.J., Wright P.E. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Ekman D., Light S., Bjorklund A.K., Elofsson A. What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol. 2006;7:R45. doi: 10.1186/gb-2006-7-6-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonin A.V., Darling A.L., Kuznetsova I.M., Turoverov K.K., Uversky V.N. Multi-functionality of proteins involved in GPCR and G protein signaling: making sense of structure-function continuum with intrinsic disorder-based proteoforms. Cell. Mol. Life Sci. 2019;76:4461–4492. doi: 10.1007/s00018-019-03276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana A., Fassina G., Vita C., Dalzoppo D., Zamai M., Zambonin M. Correlation between sites of limited proteolysis and segmental mobility in thermolysin. Biochemistry. 1986;25:1847–1851. doi: 10.1021/bi00356a001. [DOI] [PubMed] [Google Scholar]
- Fontana A., Polverino de Laureto P., De Filippis V., Scaramella E., Zambonin M. Probing the partly folded states of proteins by limited proteolysis. Fold. Des. 1997;2:R17–R26. doi: 10.1016/S1359-0278(97)00010-2. [DOI] [PubMed] [Google Scholar]
- Fontana A., de Laureto P.P., Spolaore B., Frare E., Picotti P., Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim. Pol. 2004;51:299–321. [PubMed] [Google Scholar]
- Fuxreiter M. Fuzziness: linking regulation to protein dynamics. Mol. BioSyst. 2012;8:168–177. doi: 10.1039/c1mb05234a. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M., Tompa P. Fuzzy complexes: a more stochastic view of protein function. Adv. Exp. Med. Biol. 2012;725:1–14. doi: 10.1007/978-1-4614-0659-4_1. [DOI] [PubMed] [Google Scholar]
- Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S., China Medical Treatment Expert Group for, C Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Zhu Y., Hong Y. Vol. 76. 2020. Decreased Mortality of COVID-19 With Renin-Angiotensin-Aldosterone System Inhibitors Therapy in Patients With Hypertension: A Meta-Analysis. Hypertension (Dallas, Tex. : 1979) pp. e13–e14. [DOI] [PubMed] [Google Scholar]
- Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Military Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. 2020;81(5):537–540. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C., Oldfield C.J., Ji F., Klitgord N., Cusick M.E., Radivojac P., Uversky V.N., Vidal M., Iakoucheva L.M. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2006;2 doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazy E., Tompa P. Limitations of induced folding in molecular recognition by intrinsically disordered proteins. Chemphyschem. 2009;10:1415–1419. doi: 10.1002/cphc.200900205. [DOI] [PubMed] [Google Scholar]
- Hemming M.L., Selkoe D.J. Amyloid beta-protein is degraded by cellular angiotensin-converting enzyme (ACE) and elevated by an ACE inhibitor. J. Biol. Chem. 2005;280:37644–37650. doi: 10.1074/jbc.M508460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu W.L., Oldfield C.J., Xue B., Meng J., Huang F., Romero P., Uversky V.N., Dunker A.K. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 2013;22:258–273. doi: 10.1002/pro.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Iaccarino G., Grassi G., Borghi C., Ferri C., Salvetti M., Volpe M., Investigators S.-R. Vol. 76. 2020. Age and Multimorbidity Predict Death Among COVID-19 Patients: Results of the SARS-RAS Study of the Italian Society of Hypertension. Hypertension (Dallas, Tex. : 1979) pp. 366–372. [DOI] [PubMed] [Google Scholar]
- Iakoucheva L.M., Kimzey A.L., Masselon C.D., Bruce J.E., Garner E.C., Brown C.J., Dunker A.K., Smith R.D., Ackerman E.J. Identification of intrinsic order and disorder in the DNA repair protein XPA. Protein Sci. 2001;10:560–571. doi: 10.1110/ps.29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Forrester S.J., O’Brien S., Baggett A., Rizzo V., Eguchi S. AT1 receptor signaling pathways in the cardiovascular system. Pharmacol. Res. 2017;125:4–13. doi: 10.1016/j.phrs.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczuk S., Broer A., Tietze N., Vanslambrouck J.M., Rasko J.E., Broer S. A protein complex in the brush-border membrane explains a Hartnup disorder allele. FASEB J. 2008;22:2880–2887. doi: 10.1096/fj.08-107300. [DOI] [PubMed] [Google Scholar]
- Kriwacki R.W., Hengst L., Tennant L., Reed S.I., Wright P.E. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake M.A. What we know so far: COVID-19 current clinical knowledge and research. Clin. Med. (London, England) 2020;20:124–127. doi: 10.7861/clinmed.2019-coron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Laporte M., Naesens L. Airway proteases: an emerging drug target for influenza and other respiratory virus infections. Curr. Opin. Virol. 2017;24:16–24. doi: 10.1016/j.coviro.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Chen X., Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (New York, N.Y.) 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Lucas J.M., Heinlein C., Kim T., Hernandez S.A., Malik M.S., True L.D., Morrissey C., Corey E., Montgomery B., Mostaghel E., Clegg N., Coleman I., Brown C.M., Schneider E.L., Craik C., Simon J.A., Bedalov A., Nelson P.S. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4:1310–1325. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S., Lorenz Chua R., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Hennig B.P., Kreuter M., Conrad C., Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39(10):e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque M., Martin P., Martell N., Fernandez C., Brosnihan K.B., Ferrario C.M. Effects of captopril related to increased levels of prostacyclin and angiotensin-(1-7) in essential hypertension. J. Hypertens. 1996;14:799–805. doi: 10.1097/00004872-199606000-00017. [DOI] [PubMed] [Google Scholar]
- Malhis N., Jacobson M., Gsponer J. MoRFchibi SYSTEM: software tools for the identification of MoRFs in protein sequences. Nucleic Acids Res. 2016;44:W488–W493. doi: 10.1093/nar/gkw409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammen M., Choi S.K., Whitesides G.M. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angewandte Chemie-International Edition. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Mancia G., Rea F., Ludergnani M., Apolone G., Corrao G. Renin-Angiotensin-Aldosterone System Blockers and the Risk of Covid-19. N. Engl. J. Med. 2020;382:2431–2440. doi: 10.1056/NEJMoa2006923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C.E., List K. Cell surface-anchored serine proteases in cancer progression and metastasis. Cancer Metastasis Rev. 2019;38:357–387. doi: 10.1007/s10555-019-09811-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C.E., Li J., Machamer C.E. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J. Virol. 2007;81:2418–2428. doi: 10.1128/JVI.02146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador W.E., Means A.R., Quiocho F.A. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science (New York, N.Y.) 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet (London, England) 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszaros B., Simon I., Dosztanyi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D., Sielaff F., Hammami M., Bottcher-Friebertshauser E., Garten W., Steinmetzer T. Identification of the first synthetic inhibitors of the type II transmembrane serine protease TMPRSS2 suitable for inhibition of influenza virus activation. Biochem. J. 2013;452:331–343. doi: 10.1042/BJ20130101. [DOI] [PubMed] [Google Scholar]
- Mohan A., Oldfield C.J., Radivojac P., Vacic V., Cortese M.S., Dunker A.K., Uversky V.N. Analysis of molecular recognition features (MoRFs) J. Mol. Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- Nehme A., Zouein F.A., Zayeri Z.D., Zibara K. An Update on the Tissue Renin Angiotensin System and Its Role in Physiology and Pathology. J. Cardio. Dev. Dis. 2019;6 doi: 10.3390/jcdd6020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield C.J., Dunker A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- Oldfield C.J., Cheng Y., Cortese M.S., Romero P., Uversky V.N., Dunker A.K. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- Oldfield C.J., Meng J., Yang J.Y., Yang M.Q., Uversky V.N., Dunker A.K. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl. 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A., Nakamura H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Payne S. In: Viruses: From Understanding to Investigation. Payne S., editor. Academic Press; London, UK: 2017. Family Coronaviridae; pp. 149–158. [Google Scholar]
- Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K., Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinforma. Comput. Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- Peng K., Radivojac P., Vucetic S., Dunker A.K., Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permyakov S.E., Millett I.S., Doniach S., Permyakov E.A., Uversky V.N. Natively unfolded C-terminal domain of caldesmon remains substantially unstructured after the effective binding to calmodulin. Proteins. 2003;53:855–862. doi: 10.1002/prot.10481. [DOI] [PubMed] [Google Scholar]
- Polverino de Laureto P., De Filippis V., Di Bello M., Zambonin M., Fontana A. Probing the molten globule state of alpha-lactalbumin by limited proteolysis. Biochemistry. 1995;34:12596–12604. doi: 10.1021/bi00039a015. [DOI] [PubMed] [Google Scholar]
- Rabaan A.A., Alahmed S.H., Bazzi A.M., Alhani H.M. A review of candidate therapies for Middle East respiratory syndrome from a molecular perspective. J. Med. Microbiol. 2017;66:1261–1274. doi: 10.1099/jmm.0.000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabi F., Al-Zoubi M., Kasasbeh G., Salameh D., Al-Nasser A. 2020. SARS-CoV-2 and Coronavirus Disease 2019: What We Know So Far. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Lu X., Danser A.H.J. Revisiting the Brain Renin-Angiotensin System-Focus on Novel Therapies. Curr. Hypertens. Rep. 2019;21:28. doi: 10.1007/s11906-019-0937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P., Obradovic Z., Li X., Garner E.C., Brown C.J., Dunker A.K. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Santos R.A.S., Oudit G.Y., Verano-Braga T., Canta G., Steckelings U.M., Bader M. The renin-angiotensin system: going beyond the classical paradigms. Am. J. Physiol. Heart Circ. Physiol. 2019;316:H958–H970. doi: 10.1152/ajpheart.00723.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzani R., Giulietti F., Di Pentima C., Giordano P., Spannella F. Disequilibrium between the classic renin-angiotensin system and its opposing arm in SARS-CoV-2-related lung injury. Am. J. Phys. Lung Cell. Mol. Phys. 2020;319:L325–L336. doi: 10.1152/ajplung.00189.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz G.E. In: Molecular Mechanism of Biological Recognition. Balaban M., editor. Elsevier/North-Holland Biomedical Press; New York: 1979. Nucleotide Binding Proteins; pp. 79–94. [Google Scholar]
- Sharma R., Raduly Z., Miskei M., Fuxreiter M. Fuzzy complexes: Specific binding without complete folding. FEBS Lett. 2015;589(19 Pt A):2533–2542. doi: 10.1016/j.febslet.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Shatsky M., Nussinov R., Wolfson H.J. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- Shen L.W., Mao H.J., Wu Y.L., Tanaka Y., Zhang W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin W.J., Seong B.L. Type II transmembrane serine proteases as potential target for anti-influenza drug discovery. Expert Opin. Drug Discovery. 2017;12:1139–1152. doi: 10.1080/17460441.2017.1372417. [DOI] [PubMed] [Google Scholar]
- Sigalov A., Aivazian D., Stern L. Homooligomerization of the cytoplasmic domain of the T cell receptor zeta chain and of other proteins containing the immunoreceptor tyrosine-based activation motif. Biochemistry. 2004;43:2049–2061. doi: 10.1021/bi035900h. [DOI] [PubMed] [Google Scholar]
- Sigalov A.B., Zhuravleva A.V., Orekhov V.Y. Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie. 2007;89:419–421. doi: 10.1016/j.biochi.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Zmora P., Gierer S., Heurich A., Pohlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G.P., Dash D. Intrinsic disorder in yeast transcriptional regulatory network. Proteins. 2007;68:602–605. doi: 10.1002/prot.21497. [DOI] [PubMed] [Google Scholar]
- Singh G.P., Ganapathi M., Dash D. Role of intrinsic disorder in transient interactions of hub proteins. Proteins. 2007;66:761–765. doi: 10.1002/prot.21281. [DOI] [PubMed] [Google Scholar]
- South A.M., Diz D., Chappell M.C. COVID-19, ACE2 and the Cardiovascular Consequences. Am. J. Physiol. Heart Circ. Physiol. 2020;318(5):H1084–H1090. doi: 10.1152/ajpheart.00217.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyroulias G.A., Nikolakopoulou P., Tzakos A., Gerothanassis I.P., Magafa V., Manessi-Zoupa E., Cordopatis P. Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship. Eur. J. Biochem. 2003;270:2163–2173. doi: 10.1046/j.1432-1033.2003.03573.x. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L.J., von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- Tompa P., Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Tong L., Pav S., Lamarre D., Pilote L., LaPlante S., Anderson P.C., Jung G. High resolution crystal structures of recombinant human renin in complex with polyhydroxymonoamide inhibitors. J. Mol. Biol. 1995;250:211–222. doi: 10.1006/jmbi.1995.0372. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: which way to go? Cell. Mol. Life Sci. 2003;60:1852–1871. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V.N. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem. Soc. Rev. 2011;40:1623–1634. doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. Intrinsic disorder-based protein interactions and their modulators. Curr. Pharm. Des. 2013;19:4191–4213. doi: 10.2174/1381612811319230005. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta. 2013;1834:932–951. doi: 10.1016/j.bbapap.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. The multifaceted roles of intrinsic disorder in protein complexes. FEBS Lett. 2015;589(19 Pt A):2498–2506. doi: 10.1016/j.febslet.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Uversky V.N., Dunker A.K. Understanding protein non-folding. Biochim. Biophys. Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V., Oldfield C.J., Mohan A., Radivojac P., Cortese M.S., Uversky V.N., Dunker A.K. Characterization of molecular recognition features, MoRFs, and their binding partners. J. Proteome Res. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin-Angiotensin-Aldosterone System Inhibitors in Patients with Covid-19. N. Engl. J. Med. 2020;382(17):1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- Vuille-dit-Bille R.N., Camargo S.M., Emmenegger L., Sasse T., Kummer E., Jando J., Hamie Q.M., Meier C.F., Hunziker S., Forras-Kaufmann Z., Kuyumcu S., Fox M., Schwizer W., Fried M., Lindenmeyer M., Gotze O., Verrey F. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids. 2015;47:693–705. doi: 10.1007/s00726-014-1889-6. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020:94. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Du Z., Zhu F., Cao Z., An Y., Gao Y., Jiang B. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet (London, England) 2020;395:e52. doi: 10.1016/S0140-6736(20)30558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Gheblawi M., Oudit G.Y. Angiotensin converting enzyme 2: a double-edged sword. Circulation. 2020 doi: 10.1161/CIRCULATIONAHA.120.047049. In press. [DOI] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients With 2019 novel coronavirus-infected pneumonia in Wuhan, China. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watermeyer J.M., Sewell B.T., Schwager S.L., Natesh R., Corradi H.R., Acharya K.R., Sturrock E.D. Structure of testis ACE glycosylation mutants and evidence for conserved domain movement. Biochemistry. 2006;45:12654–12663. doi: 10.1021/bi061146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S., Greer B., Hooper J., Zijlstra A., Walker B., Quigley J., Hawthorne S. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem. J. 2005;388:967–972. doi: 10.1042/BJ20041066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler L.M., McMahon C., Staus D.P., Lefkowitz R.J., Kruse A.C. Distinctive activation mechanism for angiotensin receptor revealed by a synthetic nanobody. Cell. 2019;176(479–490) doi: 10.1016/j.cell.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.) 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q. Type II transmembrane serine proteases. Curr. Top. Dev. Biol. 2003;54:167–206. doi: 10.1016/s0070-2153(03)54009-1. [DOI] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.) 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T., Xiao R., Lin G. Angiotensin-converting enzyme 2 in severe acute respiratory syndrome coronavirus and SARS-CoV-2: A double-edged sword? FASEB J. 2020;34:6017–6026. doi: 10.1096/fj.202000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology (Carlton, Vic.) 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., Ng O.T., Marimuthu K., Ang L.W., Mak T.M., Lau S.K., Anderson D.E., Chan K.S., Tan T.Y., Ng T.Y., Cui L., Said Z., Kurupatham L., Chen M.I., Chan M., Vasoo S., Wang L.F., Tan B.H., Lin R.T.P., Lee V.J.M., Leo Y.S., Lye D.C., Singapore Novel Coronavirus Outbreak Research, T Epidemiologic features and clinical course of patients infected With SARS-CoV-2 in Singapore. JAMA. 2020;323(15):1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. Imaging changes in severe COVID-19 pneumonia. Intensive Care Med. 2020;46:583–585. doi: 10.1007/s00134-020-05976-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Han G.W., Batyuk A., Ishchenko A., White K.L., Patel N., Sadybekov A., Zamlynny B., Rudd M.T., Hollenstein K., Tolstikova A., White T.A., Hunter M.S., Weierstall U., Liu W., Babaoglu K., Moore E.L., Katz R.D., Shipman J.M., Garcia-Calvo M., Sharma S., Sheth P., Soisson S.M., Stevens R.C., Katritch V., Cherezov V. Structural basis for selectivity and diversity in angiotensin II receptors. Nature. 2017;544:327–332. doi: 10.1038/nature22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17(5):259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A., Carrell R.W., Murphy M.P., Wei Z., Yan Y., Stanley P.L., Stein P.E., Broughton Pipkin F., Read R.J. A redox switch in angiotensinogen modulates angiotensin release. Nature. 2010;468:108–111. doi: 10.1038/nature09505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., 2nd, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.E., Barbry P., Leslie A., Kiem H.P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J., lung-network@humancellatlas.org, H.C.A.L.B.N.E.a., Network, H.C.A.L.B SARS-CoV-2 receptor ACE2 Is an interferon-stimulated gene in human airway epithelial cells and is detected in specific Cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material