Abstract
Introduction
IgG immunoassays have been developed and used widely for clinical samples and serosurveys for SARS-CoV2, with most detecting antibodies against the spike/receptor-binding-domain or nucleocapsid. Limited information is available on comparative evaluation of IgG immunoassays against a clinical reference standard, i.e., RT-PCR positivity with >20 days of illness. This study addresses the need for comparing clinical performance of IgG immunoassays with respect to this alternate reference standard.
Methods
We compared the performance of three immunoassays, an in-house RBD assay, and two commercial assays, the Diasorin LIAISON SARS-CoV-2 S1/S1 IgG CLIA which detects antibodies against S1/S2 domains of the Spike protein and the Zydus Kavach assay based on inactivated virus using a well-characterized panel of sera. 379 sera and plasma samples from RTPCR positive individuals >20 days of illness in symptomatic or RT-PCR positivity in asymptomatic individuals and 184 samples collected prior to 2019 were used for assay evaluation.
Results
The sensitivity of the assays were 84.7 (95 %CI 80.6–88.1), 82.6 (95 %CI 78.3–86.2) and 75.7 (95 %CI 71.0–79.9) respectively for RBD, LIAISON and Kavach. Kavach and the in-house RBD ELISA showed a specificity of 99.5 % and 100 %, respectively. The RBD and LIAISON (S1/S2) assays showed high agreement (94.7 %; 95 %CI: 92.0, 96.6) and were able to correctly identify more positive sera/plasma than Kavach.
Conclusion
Independent comparisons support the evaluation of performance characteristics of immunoassays. All three assays are suitable for serosurveillance studies, but in low prevalence sites, estimation of exposure may require adjustment based on our findings.
Abbreviations: RBD, Receptor Binding Domain; CLIA, Chemi-Luminescence, Immuno Assay; ELISA, Enzyme Linked ImmunoSorbent Assay; MHRA, The UK, Medicines and Healthcare products Regulatory Agency
Keywords: SARS-CoV-2, Immunoassay, Serology, LIAISON SARS-CoV-2 S1/S1 IgG, Zydus Kavach, RBD IgG ELISA
1. Introduction
Nucleic acid-based diagnostic tests like RT-PCR have shown considerable sensitivity and specificity for detection of active SARS-CoV-2 infections and are being used as the primary diagnostic tool. Serological tests, on the other hand, are important tools for estimation of seroprevalence of the disease at a community level. At an individual level, serological evidence may be a correlate of exposure or vaccine response.
Performance characteristics of serological tests determine their utility and interpretation of results. For SARS-CoV2, many tests have been developed but there are limited direct comparisons. Approximately 390 tests for IgG, IgM, IgA and total antibody have been developed across a range of platforms including chemiluminescence, magnetic bead-based assays, microwell ELISA, lateral flow, etc. using different portions of the spike and nucleocapsid proteins as well as whole inactivated virus [1]. Although the nucleocapsid is more abundant and immunogenic, most assays in use or in development have utilized different regions of the spike protein or whole virus as the capture reagent in immunoassays. This is mainly because anti- spike antibodies are believed to be less cross-reactive based on viral spike homology and are expected to correlate better with neutralizing capacity of convalescent sera. Most of these assays have been developed rapidly, many under emergency use authorization, and hence were evaluated by the developers in a limited set of samples. The performance of these assays in larger sample sets in various real-world setting is necessary to interpret the results of the seroepidemiological studies conducted using these assays.
In this study, we evaluate three serological tests, one in-house ELISA, a commercial ELISA, and a commercial chemiluminescence immunoassay and report their sensitivity based on 379 RT-PCR positive convalescent sera and plasma samples collected from a prospective cohort of COVID-19 positive participants and specificity based on 184 pre-pandemic participants. We also perform head-to-head analyses of their ability to correctly identify IgG positive samples.
2. Materials and methods
2.1. Participants for serological assay comparison
The participants for this study were derived from a longitudinal cohort of COVID-19 positive participants known as the Department of Biotechnology India COVID-19 Consortium cohort, with ongoing recruitment from March 2020 at eight clinical sites in the Delhi- National Capital Region, India. The participants in this cohort are derived from two types of enrollment: i) Suspected COVID-19 patients enrolled at the time of RT-PCR testing at the screening center and ii) RT-PCR confirmed COVID-19 positive patients admitted at one of the clinical sites. The testing by RT-PCR was done at an approved laboratory as per the National Testing Strategy of India [2]. The RT-PCR confirmed COVID-19 patients were followed up at 10–28 days and 6–8 weeks of onset of illness. During the enrollment and follow-up detailed clinical information on the exposure history, clinical features and comorbidities were documented. Venous blood samples are collected, transported, processed and stored per protocol [3]. All enrollments were made after an informed consent process and the study protocol was approved by the Institute Ethics Committees of the participating research institutes and hospitals.
The COVID-19 positive reference standard sera panel (n = 379) was formed using the sera/plasma samples collected ≥20 days of illness or following RT-PCR positivity. This criterion is in alignment with the UK Medicines and Healthcare products Regulatory Agency (MHRA) to improve comparability [4,5]. The duration of illness for asymptomatic participants was calculated from their date of diagnosis. For symptomatic participants, it was calculated from the date of testing or date of onset of symptoms whichever was the earlier. The COVID-19 negative standard panel was built from sera samples collected in the pre-pandemic period (184 from pregnant women enrolled in a pregnancy cohort) to ensure a clean set of negative samples [[4], [5], [6]].
2.2. Index test methods
2.2.1. THSTI In-house RBD IgG ELISA
The in-house RBD based IgG ELISA uses RBD coated dry stable wells. The wells were washed 3 times with PBST (PBS with 0.1 % Tween 20) using 96-well plate-washer (Tecan AG). Two hundred microliter of blocking solution (PBST with 3% skim milk) was added to all the wells. The Plates were incubated at 20 °C for 2 h. After 2 h of incubation, plates were removed from the incubator and the blocking solution was thrown off. Hundred microliter of diluted test serum or control samples (1:50 dilution in PBST with 1% skim milk) were added to the appropriate wells and the plates were incubated at 20 °C. After 2 h of incubation, plates were removed from the incubator and washed 3 times with PBST. Horse Radish Peroxidase labelled goat anti-Human IgG Fcγ-sp. Tracer antibody (Jackson Immuno Research, Pennsylvania, USA) was diluted 1:5000 in PBST with 1% skim milk and 50 μL of diluted tracer was added in each well of the plates. Plates were incubated in a 20 °C incubator for 1 h. After the completion of the incubation period, plates were washed 3 times with PBST and in each well 100 μL of TMB substrate (BD Biosciences) was added and plates were incubated at room temperature for 10 min in dark. Fifty microliter of stop solution (1 M H2SO4) was added in each well and the absorbance was measured using a microplate reader (Biorad, California, USA) at 450 nm with 650 nm as reference wavelength. In each assay, 8 known negative samples with variable background were used as control to calculate the cut-off value. The cut-off value was calculated using the formula: Cut-off = Average OD value of negative control samples + 3* SD of OD value of negative control samples. The test result was considered positive when signal to cut-off ratio > = 1.
2.2.2. Covid Kavach IgG ELISA
Covid Kavach IgG ELISA was developed by the Indian Council of Medical Research’s National Institute of Virology, and manufactured by Zydus [7]. The test was performed as per manufacturer’s instructions. The kit suggests interpretation of the results by a two-pronged method, based on OD value and P/N (Positive/Negative Ratio). When both read-outs are in agreement, then the sample is considered positive or negative. The manufacturer’s instruction does not mention interpretation for samples with a read-out not in agreement for the two criteria. We considered such results negative.
2.2.3. DiaSorin CLIA
The LIAISON SARS-CoV-2 S1/S2 IgG chemiluminescence assay manufactured by DiaSorin was also used for comparison. This test uses S1/S2 antigens to capture specific IgG antibodies. The test was performed as per manufacturer’s instructions, with calibration and positive and negative controls run before each batch of antibody testing as per manufacturer's protocol [8]. The tests were considered positive when the IgG concentration was ≥15 AU/mL, negative when the concentration was <12 AU/mL and equivocal when the concentration was >12 and <15 AU/mL. Equivocal samples were considered negative for sensitivity analysis.
2.2.4. Statistical analysis
In the absence of a gold standard for SARS−CoV-2 IgG immunoassays, an alternate reference standard was used for this study, which is SARS−CoV-2 RT-PCR positivity with >20 days duration of illness (symptomatic) or >20 days beyond RT-PCR positivity (asymptomatic). While comparing the three assays, we report sensitivity and specificity of each test with the positive and negative reference standards as defined. In addition, we performed a head-to-head analysis for agreement between these methods. We estimated the global agreement calculated as the sum of the number of positives by both methods and number negative by both divided by the total number of samples. We estimated the bias index defined as the difference in the proportion of positivity for any bias between the methods to check whether one method was superior to the other in correctly identifying positive samples. As the prevalence of positives and negatives may play a role in the interpretation of the kappa statistic, we report the prevalence index defined as difference between probability of positives and probability of negatives. We finally report the kappa statistic adjusted for prevalence and bias known as prevalence and bias-adjusted kappa [9]. We then compared the sensitivity of the three candidate tests across different periods of illness. All analyses were done using the STATA-SE-15 software (Texas, USA) and the Kappa coefficient and related indices were estimated using Cohenkap package for STATA [10].
3. Results
3.1. Samples defined by an Alternate Reference Standard used for comparative evaluation
The reference true positive sample panel consisted of 379 samples from 368 participants; 11 of whom provided two samples at different time-points. The distribution of the duration of illness was bimodal owing to the design of the cohort from which the samples were derived. The means of the sampling window distributions are 23.5 and 49.3 days respectively (Supplementary Fig. 1). Most samples (83.7 %) were obtained from symptomatic individuals. Nearly half of the samples were sera, rest were plasma. The reference negative panel consisted of 184 pre-pandemic samples collected before September 2019.
Comparison of the sensitivity and specificity of candidate assays
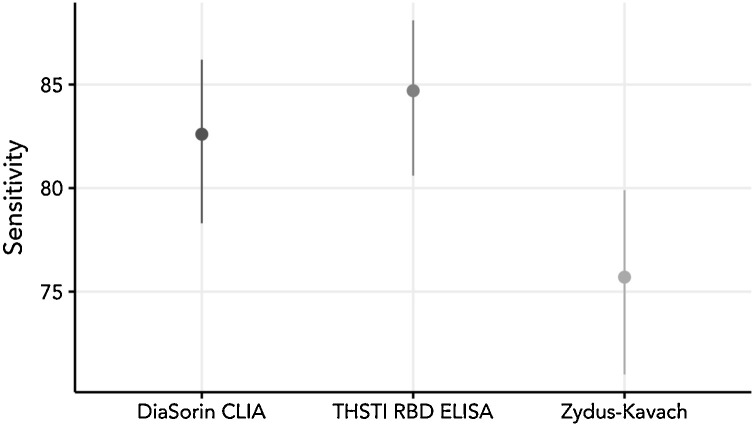
Among the three candidate tests, RBD IgG ELISA demonstrated the highest sensitivity (84.7; 95 %CI: 80.6–88.1) and Zydus Kavach the least (75.7; 95 %CI: 71.0–79.9). Zydus Kavach is interpreted as positive when both test parameters were positive based on cut-0ff and P/N ratio. Six samples were indeterminate in Zydus Kavach test; and 25 samples were positive only by one condition (Cut-off, P/N ratio) by Zydus Kavach. Seven samples were reported as indeterminate by DiaSorin CLIA. When Zydus Kavach was interpreted as positive with one of the two criteria, 25 additional samples were identified as positive, improving the sensitivity to 81.8 %. The specificities of Zydus Kavach and RBD ELISA were 99.5 and 100 % respectively (Table 1 , Fig. 1 ). The specificity of DiaSorin could not be evaluated due to limited availability of pre-pandemic negative sera.
Table 1.
Sensitivity (%) and specificity (%) of Zydus-Kavach, DiaSorin CLIA & THSTI-RBD ELISA.
| Tests | Sensitivity (95 %CI) | Specificity (95 %CI) |
|---|---|---|
| Zydus-Kavach | 75.7 (71.0–79.9) | 99.5 (96.5–99.9) |
| DiaSorin CLIA | 82.6 (78.3–86.2) | --- |
| RBD ELISA | 84.7 (80.6–88.1) | 100 (97.4–100) |
Fig. 1.
Plot of sensitivity (95 % confidence intervals) for the candidate assays.
The dots represent the sensitivity (%) and the bars, 95 % CI.
Total true positive samples evaluated: 379; true negative samples: 184
The specificity of DiaSorin could not be evaluated due to limited availability of pre-pandemic negative sera.
3.2. Comparison of Zydus-Kavach, DiaSorin CLIA & RBD ELISA
When evaluated for agreement between the tests, DiaSorin CLIA and RBD ELISA had highest concordance with a global agreement 0f 94.7 % (95 %CI: 92.0, 96.6). There was minimal bias between the two tests; with just 6 samples positive with DiaSorin labelled negative by RBD ELISA. Among the other 14 discordant samples that were positive by RBD ELISA, five were equivocal by DiaSorin and the rest were negative. The agreement estimated by prevalence and bias-adjusted kappa statistic (0.89) was near perfect between the two tests (Table 2 & Supplementary table-1).
Table 2.
Head-to-head comparison of Zydus-Kavach, DiaSorin CLIA & RBD ELISA.
| Kit | Zydus-Kavach | DiaSorin CLIA | RBD ELISA |
|---|---|---|---|
| Zydus-Kavach | 88.7 % (95 %CI: 85.1, 91.5) | 87.3 % (95 %CI: 83.6, 90.3) | |
| DiaSorin CLIA | 0.77 | 94.7 % (95 %CI: 92.0, 96.6) | |
| RBD ELISA | 0.75 | 0.89 |
On the other hand, head-to-head comparison of DiaSorin CLIA and RBD ELISA against Zydus Kavach demonstrated that the degrees of agreement were modest. The global agreement between the pairs of DiaSorin CLIA and Zydus Kavach, and RBD ELISA and Zydus Kavach were 88.7 % (95 %CI: 85.1, 91.5) and 87.3 % (95 %CI: 83.6, 90.3) respectively (Table 2). DiaSorin CLIA and RBD ELISA were superior to Zydus Kavach ELISA and were able to correctly identify 7% and 9% more IgG positive sera than the latter (Supplementary tables 2 & 3).
Numbers in the cells below the diagonal in the table denote Prevalence and Bias adjusted Kappa statistic. Numbers in the cells above the diagonal in the table denote agreement and 95 %CI calculated as (positive by both methods + negative by both)/ total samples
4. Discussion
Comparison of three assays on a well-characterized sample set showed that the DiaSorin CLIA and the in-house RBD ELISA performed slightly better than the Zydus Kavach ELISA with higher sensitivity and ability to identify more IgG positive samples. These results are important as they help interpretation of the serosurveillance studies that are being conducted in India using these tests.
The sensitivities reported in our study are less than reported by the companies or elsewhere [5,7]. An independent evaluation of DiaSorin CLIA, the sensitivity was reported to be 95.0 % (95 %CI: 92.8, 96.7), which is at least 13 percentage points higher than this report [3]. While unlikely, this difference could be attributed to false positives in our RT-PCR assay since we used RT-PCR positive convalescent samples as reference standards. Similarly, the article reporting internal validation of Zydus Kavach ELISA reported a sensitivity of 92.37 % based on samples that were positive in a micro-neutralization assay as against 75.7 % that we see in our study using sera collected >20 days post-RT-PCR positivity [7]. While we harmonised our definition of positive reference standard with that of the UK MHRA, about 14 % of our participants were asymptomatic, and may have had a lower antibody response as reported in a longitudinal study, albeit with small numbers, which showed that 2/24 of their participants did not seroconvert when followed up 20–28 days into their illness [11].
To overcome the limitation of an imperfect positive reference standard and to improve inferences, the relative performance of these tests was evaluated by head-to-head comparison. RBD ELISA and DiaSorin CLIA were able to identify more positive IgG sera/plasma than Zydus Kavach. However, RBD ELISA is an in-house ELISA developed using similar sample collections. While the sample panel used for evaluation was independent of that used in the development of the RBD ELISA, its true test would be when it is evaluated externally.
Highly sensitive serological assays can assess immune response to SARS-CoV-2 and are needed to determine the extent of spread of the virus, which in turn is critical for assessing case fatality rates and herd immunity. Serological assays also help in assessing development of herd immunity to devise community management strategies that are of crucial importance at this time, and will continue to be relevant in the coming years. Other uses of serological assays can be to assess exposure in high-risk populations such as healthcare workers and assess vaccination strategy at state or national level. Cross-reactivity with SARS-CoV and seasonal coronaviruses in different population and timing of IgM and IgG responses need to continue to be considered. Till date, studies on comparative performance serological assays for SARS-CoV-2 show a range of sensitivity of 84–98 % and specificity of 96–99 % [[12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29]]. Two of the three IgG assays in this study used the Spike protein (Receptor binding domain or RBD in THSTI-In-house ELISA and S1/S2 in DiaSorin CLIA), while the specific antigenic epitope(s) of the inactivated virus in the Kavach assay are not defined. Since RBD in the spike protein is the major site of ACE-2 binding, assays with this target may have more concordance with neutralizing antibodies. The spike protein has more CD4 and CD8 T cell immunodominant epitopes as experimentally shown in SARS-CoV, and since these epitopes are mostly conserved even in SARS-CoV-2 isolates, serological assays targeting RBD or full length S1/S2 are putatively more appropriate for assessing long-term immune status [30].
5. Conclusion
Overall, this is the first comparative study of SARS-CoV-2 IgG assays in India evaluated independently against a strategically designated reference standard. One limitation of this study is that we were unable to evaluate the specificity of the Diasorin assay due to paucity of negative samples. Nonetheless, the well characterized panels provide useful information for decision-making for choice of serological assays and interpreting serological studies conducted in the country.
Author contribution
SB, GK, SC and RT conceptualized the study; All members of DBT India consortium for COVID-19 Research devised methods, collected samples and clinical data; SC, GB, TS, PK, ND, MD, AD, LC and DD provided reagents and analysed samples; RT, SC and BKD curated and analysed data; SC, RT and GK drafted the manuscript; GK and SB coordinated and supervised the study. All authors contributed in revision and approved the final draft of the manuscript.
Declaration of Competing Interest
None.
Acknowledgement
We deeply thank the Department of Biotechnology, Government of India for supporting the consortium. We are grateful to the leadership and administration of all partner institutions in the consortium for their help and support. We thank all the clinical, laboratory and data management staff for their contributions to this work and the consortium at large.
Footnotes
DBT India Consortium for Covid-19 Research. The members of the consortium are listed at the end of the article.
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104609.
Appendix A.
Members of DBT India consortium for COVID-19 Research
Coordinating Institute: Translational Health Science and Technology Institute (THSTI)
Coordinating Principal Investigator: Dr Shinjini Bhatnagar
Co- Principal Investigator: Dr Gagandeep Kang
Co-Investigators (Clinical): Drs Nitya Wadhwa, Uma Chandramouli Natchu, Ramachandran Thiruvengadam, Shailaja Sopory, Pallavi Kshetrapal, Bapu Koundinya Desiraju, Vandita Bhartia, Mudita Gosain
Co-Investigators (Laboratory): Drs Gaurav Batra, Guruprasad Medigeshi, Susmita Chaudhuri, Niraj Kumar, Tarun Sharma, Chandresh Sharma, Shailendra Mani, Tripti Shrivastava
Clinical Operations Lead: Dr. Monika Bahl
International Centre for Genetic Engineering and Biotechnology (ICGEB): Dr Anmol Chandele
Delhi University, South Campus, New Delhi: Dr Vijay Kumar Chaudhary
National Institute of immunology (NII): Drs. Amulya Panda, Nimesh Gupta
Maulana Azad Medical College and Lok Nayak Jai Prakash Narayan Hospital, New Delhi: Drs. Nandini Sharma, Pragya Sharma, Sonal Saxena, J.C. Passey, Suresh Kumar
ESI Medical College and Hospital, Faridabad, Haryana: Dr Anil K Pandey, Asim Das, Nikhil Verma, Needhi Anand, Sujata Roy Choudhary
Civil Hospital Gurugram (GCH), Haryana: Drs Deepa Sindhu, Jai Singh Malik
Civil Hospital Palwal (PCH), Haryana: Dr Brahmdeep Sindhu
Al-Falah School of Medical Science & Research Centre and Hospital, Dhouj, Haryana: Drs Bhupinder Kaur Anand, Shubham Girdhar
Medanta Hospital, Gurugram, Haryana: Drs Sushila Kataria, Pooja Sharma
Shaheed Hasan Khan Mewati Government Medical College, Nalhar, Nuh, Haryana: Dr Yamini
Lady Hardinge Medical College, New Delhi: Drs. Harish K. Pemde, Tanmaya Talukdar
SGT Medical college, Gurugram, Haryana: Drs. Pankaj Abrol, Mukesh Sharma
Dr. Dang’s Lab, New Delhi: Drs Navin Dang, Manavi Dang, Arjun Dang, Leena Chatterjee, Devjani De
Appendix B. Supplementary data
The following is Supplementary data to this article:
References
- 1.SARS-CoV-2 diagnostic pipeline, FIND. https://www.finddx.org/covid-19/pipeline/ (accessed August 8, 2020).
- 2.Strategy for COVID-19 testing in India. https://www.icmr.gov.in/pdf/covid/strategy/Testing_Strategy_v5_18052020.pdf (accessed August 7, 2020).
- 3.SOP_Specimen_Collection_2019-nCoV.pdf. https://niv.co.in/SOP_Specimen_Collection_2019-nCoV.pdf (accessed August 7, 2020).
- 4.Target Product Profile: enzyme Immunoassay (EIA) Antibody tests to help determine if people have antibodies to SARS-CoV-2, GOV.UK. https://www.gov.uk/government/publications/how-tests-and-testing-kits-for-coronavirus-covid-19-work/target-product-profile-enzyme-immunoassay-eia-antibody-tests-to-help-determine-if-people-have-antibodies-to-sars-cov-2 (accessed August 7, 2020).
- 5.Public Health England Porton Down, Evaluation of sensitivity and specificity of four commercially available SARS-CoV-2 antibody immunoassays, Public Health England, Porton Down Nuffield Department of Medicine, University of Oxford Oxford University Hospitals NHS Foundation Trust, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/898437/Evaluation__of_sensitivity_and_specificity_of_4_commercially_available_SARS-CoV-2_antibody_immunoassays.pdf (accessed August 7, 2020).
- 6.Bhatnagar S., Majumder P.P., Salunke D.M. Interdisciplinary group for advanced research on birth outcomes—DBT india initiative (GARBH-Ini), a pregnancy cohort to study multidimensional correlates of preterm birth in India: study design, implementation, and baseline characteristics of the participants. Am. J. Epidemiol. 2019;188:621–631. doi: 10.1093/aje/kwy284. [DOI] [PubMed] [Google Scholar]
- 7.Sapkal G., Shete-Aich A., Jain R., Yadav P.D., Sarkale P., Lakra R., Baradkar S., Deshpande G.R., Mali D., Tilekar B.N., Majumdar T., Kaushal H., Gurav Y., Gupta N., Mohandas S., Deshpande K., Kaduskar O., Salve M., Patil S., Gaikwad S., Sugunan A.P., Ashok M., Giri S., Shastri J., Abraham P., Gangakhedkar R.R., Team C.S. Development of indigenous IgG ELISA for the detection of anti-SARS-CoV-2 IgG. Indian J. Med. Res. 2020;151:444. doi: 10.4103/ijmr.IJMR_2232_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LIAISON® SARS-CoV-2 S1/S2 IgG https://www.diasorin.com/sites/default/files/allegati_prodotti/liaison_sars-cov-2_s1_s2_igg_0.pdf (accessed August 7, 2020).
- 9.Byrt T., Bishop J., Carlin J.B. Bias, prevalence and kappa. J. Clin. Epidemiol. 1993;46:423–429. doi: 10.1016/0895-4356(93)90018-V. [DOI] [PubMed] [Google Scholar]
- 10.Cohenkap package for STATA software for agreement analysis. http://www.graunt.cat/stata/cohenkap.ado (accessed August 7, 2020).
- 11.Chatzidimitriou M., Chatzopoulou F., Gavriilaki E., Chatzivasileiou P., Rousis D., Meletis G., Chatzidimitriou D. Repeated negative serological testing in otherwise healthy patients with COVID-19. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiaa453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastos M.L., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.-P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A., Menzies D., Benedetti A., Khan F.A. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370 doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M.K. Das, A. Chaudhary, A. Bryan, M.H. Wener, S.L. Fink, C. Morishima, Rapid Screening Evaluation of SARS-CoV-2 IgG Assays Using Z-Scores to Standardize Results, (n.d.). https://doi.org/10.3201/eid2610.202632. [DOI] [PMC free article] [PubMed]
- 14.Charlton C.L., Kanji J.N., Johal K., Bailey A., Plitt S.S., MacDonald C., Kunst A., Buss E., Burnes L.E., Fonseca K., Berenger B.M., Schnabl K., Hu J., Stokes W., Zelyas N., Tipples G. Evaluation of six commercial mid to high volume antibody and six point of care lateral flow assays for detection of SARS-CoV-2 antibodies. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.01361-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beavis K.G., Matushek S.M., Abeleda A.P.F., Bethel C., Hunt C., Gillen S., Moran A., Tesic V. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA assay for detection of IgA and IgG antibodies. J. Clin. Virol. 2020;129:104468. doi: 10.1016/j.jcv.2020.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bundschuh C., Egger M., Wiesinger K., Gabriel C., Clodi M., Mueller T., Dieplinger B. Evaluation of the EDI enzyme linked immunosorbent assays for the detection of SARS-CoV-2 IgM and IgG antibodies in human plasma. Clin. Chim. Acta. 2020;509:79–82. doi: 10.1016/j.cca.2020.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egger M., Bundschuh C., Wiesinger K., Gabriel C., Clodi M., Mueller T., Dieplinger B. Comparison of the Elecsys® Anti-SARS-CoV-2 immunoassay with the EDITM enzyme linked immunosorbent assays for the detection of SARS-CoV-2 antibodies in human plasma. Clin. Chim. Acta. 2020;509:18–21. doi: 10.1016/j.cca.2020.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haselmann V., Kittel M., Gerhards C., Thiaucourt M., Eichner R., Costina V., Neumaier M. Comparison of test performance of commercial anti-SARS-CoV-2 immunoassays in serum and plasma samples. Clin. Chim. Acta. 2020;510:73–78. doi: 10.1016/j.cca.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Hörber, J. Soldo, L. Relker, S. Jürgens, J. Guther, S. Peter, R. Lehmann, A. Peter, Evaluation of three fully-automated SARS-CoV-2 antibody assays, (n.d.) 8. [DOI] [PubMed]
- 20.Jääskeläinen A.J., Kuivanen S., Kekäläinen E., Ahava M.J., Loginov R., Kallio-Kokko H., Vapalahti O., Jarva H., Kurkela S., Lappalainen M. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J. Clin. Virol. 2020;129:104512. doi: 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jääskeläinen A.J., Kekäläinen E., Kallio-Kokko H., Mannonen L., Kortela E., Vapalahti O., Kurkela S., Lappalainen M. Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.18.2000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko J.-H., Joo E.-J., Park S.-J., Baek J.Y., Kim W.D., Jee J., Kim C.J., Jeong C., Kim Y.-J., Shon H.J., Kang E.-S., Choi Y.K., Peck K.R. Neutralizing antibody production in asymptomatic and mild COVID-19 patients, in comparison with pneumonic COVID-19 patients. J. Clin. Med. 2020;9 doi: 10.3390/jcm9072268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krüttgen A., Cornelissen C.G., Dreher M., Hornef M., Imöhl M., Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J. Clin. Virol. 2020;128:104394. doi: 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merrill A.E., Jackson J.B., Ehlers A., Voss D., Krasowski M.D. Head-to-Head comparison of two SARS-CoV-2 serology assays. J. Appl. Lab. Med. 2020 doi: 10.1093/jalm/jfaa125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer B., Torriani G., Yerly S., Mazza L., Calame A., Arm-Vernez I., Zimmer G., Agoritsas T., Stirnemann J., Spechbach H., Guessous I., Stringhini S., Pugin J., Roux-Lombard P., Fontao L., Siegrist C.-A., Eckerle I., Vuilleumier N., Kaiser L. Validation of a commercially available SARS-CoV-2 serological immunoassay. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang M.S., Hock K.G., Logsdon N.M., Hayes J.E., Gronowski A.M., Anderson N.W., Farnsworth C.W. Clinical performance of two SARS-CoV-2 serologic assays. Clin. Chem. 2020;66:1055–1062. doi: 10.1093/clinchem/hvaa120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Theel E.S., Harring J., Hilgart H., Granger D. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weidner L., Gänsdorfer S., Unterweger S., Weseslindtner L., Drexler C., Farcet M., Witt V., Schistal E., Schlenke P., Kreil T.R., Jungbauer C. Quantification of SARS-CoV-2 antibodies with eight commercially available immunoassays. J. Clin. Virol. 2020;129:104540. doi: 10.1016/j.jcv.2020.104540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.-P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A., Menzies D., Benedetti A., Ahmad Khan F. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nasab M.G., Saghazadeh A., Rezaei N. SARS-CoV-2-A tough opponent for the immune system. Arch. Med. Res. 2020 doi: 10.1016/j.arcmed.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.